Abstract
Measuring individual circadian phase is important to diagnose and treat circadian rhythm sleep-wake disorders and circadian misalignment, inform chronotherapy, and advance circadian science. Initial findings using blood transcriptomics to predict the circadian phase marker dim-light melatonin onset (DLMO) show promise. Alternatively, there are limited attempts using metabolomics to predict DLMO and no known omics-based biomarkers predict dim-light melatonin offset (DLMOff). We analyzed the human plasma metabolome during adequate and insufficient sleep to predict DLMO and DLMOff using one blood sample. Sixteen (8 male/8 female) healthy participants aged 22.4 ± 4.8 years (mean ± SD) completed an in-laboratory study with 3 baseline days (9 h sleep opportunity/night), followed by a randomized cross-over protocol with 9-h adequate sleep and 5-h insufficient sleep conditions, each lasting 5 days. Blood was collected hourly during the final 24 h of each condition to independently determine DLMO and DLMOff. Blood samples collected every 4 h were analyzed by untargeted metabolomics and were randomly split into training (68%) and test (32%) sets for biomarker analyses. DLMO and DLMOff biomarker models were developed using partial least squares regression in the training set followed by performance assessments using the test set. At baseline, the DLMOff model showed the highest performance (0.91 R2 and 1.1 ± 1.1 h median absolute error ± interquartile range [MdAE ± IQR]), with significantly (p < 0.01) lower prediction error versus the DLMO model. When all conditions (baseline, 9 h, and 5 h) were included in performance analyses, the DLMO (0.60 R2; 2.2 ± 2.8 h MdAE; 44% of the samples with an error under 2 h) and DLMOff (0.62 R2; 1.8 ± 2.6 h MdAE; 51% of the samples with an error under 2 h) models were not statistically different. These findings show promise for metabolomics-based biomarkers of circadian phase and highlight the need to test biomarkers that predict multiple circadian phase markers under different physiological conditions.
Keywords: circadian rhythm, circadian misalignment, personalized medicine, sleep restriction, sleep loss, biomarker, metabolomics
Introduction
Shift work, jet lag, and social jet lag are prevalent in modern society and can contribute to circadian rhythm sleep-wake disorders (CRSWDs) and circadian misalignment, defined as misalignment between behavioral (e.g., sleep-wake and fasting-feeding cycles) and circadian (central and peripheral clocks) rhythms (Sack, 2009; Drake and Wright, 2017; Baron and Reid, 2014; Mason et al., 2020; Sletten et al., 2020). CRSWDs and circadian misalignment are associated with adverse health outcomes including risk of cardiometabolic (diabetes, cardiovascular disease, cancer, obesity) and psychiatric (anxiety, depression, schizophrenia) disorders (Scheer et al., 2009; Baron and Reid, 2014). To treat CRSWDs and related circadian misalignment most effectively, it is essential to know the individual circadian phase of a patient (Mullington et al., 2016; Duffy et al., 2021). This knowledge is also critical for chronotherapy where treatments are administered based on individual circadian phase. Findings showing time-of-day variation in responses to vaccinations (Long et al., 2016), surgeries and wound healing (Labrecque and Belanger, 1992; Hoyle et al., 2017; Montaigne et al., 2018), drug metabolism (Labrecque and Belanger, 1992; Ohdo et al., 2001; Ruben et al., 2018), cancer treatment (Hrushesky, 1985; Lévi et al., 1997; Innominato et al., 2012), and heart disease/blood pressure medications (Lemmer et al., 1992; Ruben et al., 2018; Hermida et al., 2020) support the concept of chronotherapy. In primates, ~82% of protein-coding genes that are drug targets show 24-h time-of-day patterns in transcription (Mure et al., 2018), suggesting optimal timing of drug administration will help achieve optimal efficacy. Similarly, in a study of post-mortem human tissues, ~50% of protein-coding genes were found to cycle in at least 1 of the 13 tissues studied, and 12% of those cycling genes encoded proteins that are either drug targets or involved in drug metabolism, again highlighting the need for precisely timed drug administration based on individual circadian phase (Ruben et al., 2018). However, circadian phase can vary by 5 to 6 h between healthy individuals maintaining consistent sleep schedules (Wright et al., 2005; Sletten et al., 2010), and can vary by 10 to 12 h between shift workers (Smith and Eastman, 2008). Therefore, developing methods for assessing individual circadian phase is critical to advancing sleep and circadian-based research and medicine (Mullington et al., 2016; Depner, Cheng, et al., 2020; Duffy et al., 2021).
Multiple methods are used to estimate circadian phase in humans with dim-light melatonin analysis being the gold standard for assessing the master circadian clock (Benloucif et al., 2008; Broussard et al., 2017). For dim-light melatonin analysis, the beginning of the circadian night is defined as the time that endogenous melatonin concentration reaches a predetermined threshold in blood or saliva, known as dim-light melatonin onset (DLMO; Klerman et al., 2002; Vessely and Lewy, 2002; Wright et al., 2013). Alternatively, dim-light melatonin offset (DLMOff), the end of the circadian night (Wehr et al., 2001; Wright et al., 2013; Stothard et al., 2017), is defined as the time that endogenous melatonin concentration decreases below the predetermined threshold. DLMO and DLMOff analyses require frequent samples collected over 24 h in dim-light with controlled timing of food intake and posture, making clinical implementation a challenge (Alfred et al., 1980; Wright et al., 2001; Duffy and Dijk, 2002; Broussard et al., 2017). Therefore, developing and validating methods to assess circadian phase from a single or very few samples will advance the field.
The preferred approach for developing omics-based sleep and circadian biomarkers, as outlined by Mullington et al. (2016) in a white paper report, is to first explore the performance of all omics-based approaches to identify candidate biomarkers using tightly controlled in-lab protocols with highly phenotyped participants. Then, in large-scale follow-up validation studies, it is critical to define biomarker performance in the precise context and population of intended use (Dijk and Duffy, 2020). Several omics-based prediction methods to assess circadian phase have been developed (Ueda et al., 2004; Kasukawa et al., 2012; Hughey, 2017; Laing et al., 2017; Braun et al., 2018; Wittenbrink et al., 2018; Braun et al., 2019; Laing et al., 2019). The performance of these omics-based prediction methods were quantified under varying protocols of circadian alignment and misalignment (Hughey, 2017; Laing et al., 2017; Braun et al., 2018), total sleep deprivation with and without prior sleep debt (Hughey, 2017; Laing et al., 2017; Braun et al., 2018), constant routine (Kasukawa et al., 2012; Braun et al., 2018; Wittenbrink et al., 2018), and free-living conditions (Wittenbrink et al., 2018), with published findings focusing exclusively on DLMO. Recent work demonstrates the physiological relevance of assessing DLMOff in addition to DLMO, as a greater duration of morning wakefulness during the biological night, termed “morning circadian misalignment,” is induced by insufficient sleep and is associated with reduced insulin sensitivity, an established risk factor for diabetes (Eckel et al., 2015; Depner et al., 2018; Simon et al., 2019). Yet, systematic investigations to develop and test omics-based biomarkers predicting DLMOff have not been performed and few of the published findings to date have focused on the potential of metabolomics for developing biomarkers of circadian phase in humans.
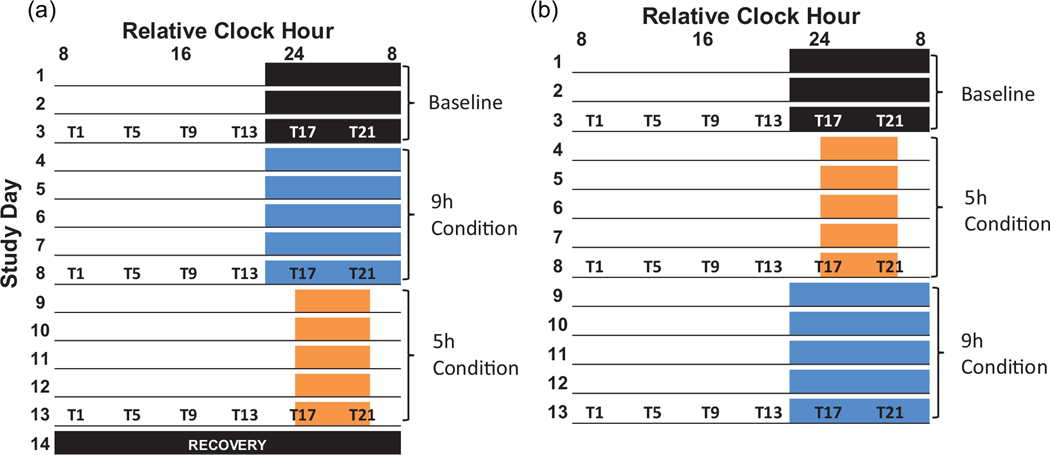
Our primary aim was to use small molecule compounds detected by untargeted metabolomics to identify candidate biomarkers capable of predicting DLMO and DLMOff in individual participants using a single blood sample, regardless of time-of-day, sleep history, or fasting status. Our secondary aim was to compare DLMO versus DLMOff biomarker model performance. We used a crossover, counterbalanced 14- to 15-day protocol with 3-day baseline (BL) assessments followed by 5-day conditions of 9 and 5 h time-in-bed to accomplish our aims (Figure 1), consistent with best practices recommended by the field (Mullington et al., 2016).
Figure 1.
Study Protocol. Colored boxes represent scheduled sleep and underlines represent scheduled wakefulness. The black box on study day 14 indicates a recovery sleep opportunity required for participants in condition order A before discharge from the lab. Blood draws for metabolomics analyses are represented by T1, T5, T9, T13, T17, and T21, where the number refers to hours since scheduled waketime (e.g. T5 = 5 hours since scheduled waketime). All protocol events were scheduled according to each participant’s habitual bedtime. For example, the T1 blood collection for a subject would occur at 10:00am if their wake time was scheduled for 9:00am, while another subject’s T1 blood collection would occur at 8:00am if their wake time was scheduled for 7:00am.
Materials and Methods
Participants
Sixteen participants (8 women) aged 22.4 ± 4.8 years with body mass index (BMI) 22.9 ± 2.4 kg/m2 completed the study (Suppl. Table S1). Of the 16 participants, 13 were Non-Hispanic White (8 male/5 female), 1 female was Non-Hispanic Asian, 1 female was Non-Hispanic White and Native Hawaiian or Other Pacific Islander, and 1 female was Hispanic or Latino. All participants provided written, informed consent, and all procedures were approved by the Scientific and Advisory Review Committee of the Colorado Clinical and Translational Sciences Institute, the Colorado Multiple Institutional Review Board (IRB), and the University of Colorado Boulder IRB. Screening procedures verified participants were free of medical, psychiatric, and sleep disorders. Inclusion, exclusion, and demographic information are fully detailed in Markwald et al. (2013).
Study Protocol
Data were collected in a previously described crossover, counterbalanced 14- to 15-day laboratory protocol with 3-day BL assessments followed by 9 h and 5 h time-in-bed conditions, lasting 5 days each (Figure 1; Markwald et al., 2013). Participants maintained consistent ~9-h sleep schedules for 1 week prior to the in-laboratory study, confirmed by daily sleep diary, time-stamped call-ins, and wrist actigraphy (Markwald et al., 2013). BL assessments consisted of 9-h sleep opportunities timed according to each participant’s habitual bed and wake times. Participants consumed investigator-provided energy balanced diets at home for 3 days prior to the in-laboratory study and during the 3-day in-laboratory BL assessments. Energy intake was ad libitum in the 9-h and 5-h conditions (Markwald et al., 2013). Blood was collected every 4 h over the final 24 h of each condition (as indicated by T1, T5, etc.), with T1 representing 1 h after scheduled waketime (Figure 1). In total, 18 blood sample collections were scheduled for each participant (six samples per each of the three conditions).
Melatonin
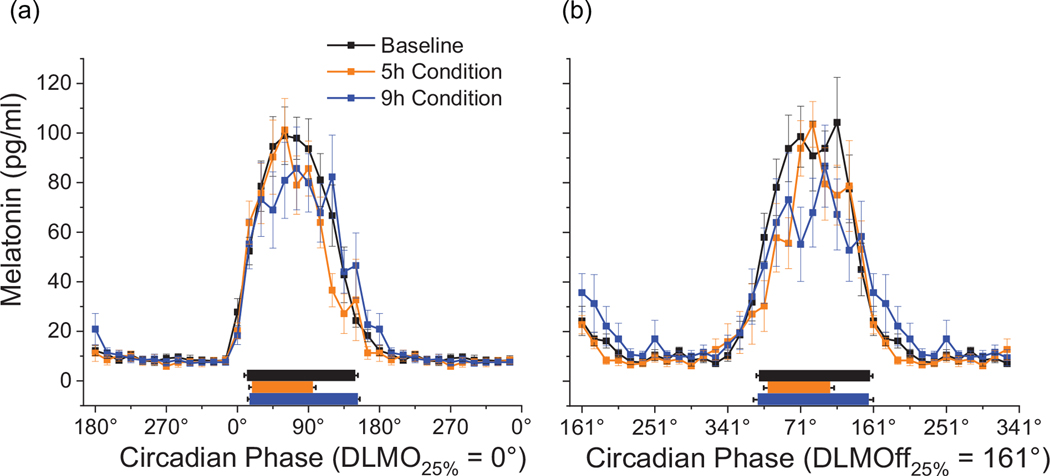
Blood was collected in dim-light (< 8 lux maximum in the angle of gaze at eye level) every hour over the final 24 h of each condition for melatonin analysis during the in-laboratory protocol (Markwald et al., 2013). The threshold for independently calculating DLMO and DLMOff was defined as 25% of the three-harmonic fit of the peak-to-trough amplitude for each participant (Wright et al., 2001, 2013; Markwald et al., 2013). DLMO and DLMOff were calculated independently for each condition for each participant as the linear interpolated point in time when melatonin increased above (DLMO) and decreased below (DLMOff) the defined threshold for that given participant. Each sample was assigned to circadian degrees with DLMO assigned to 0° (Figure 2a). The average duration from DLMO to DLMOff was 10.7 h, or 161°. Therefore, DLMOff was assigned to 161° (Figure 2b). Timing of blood sample collection was referenced individually to DLMO and DLMOff for each participant for biomarker model development, thus DLMO and DLMOff calculations were independent of each other.
Figure 2.
24h Melatonin Profile. Dim-light melatonin data are referenced to DLMO (A) and DLMOff (B). DLMO was assigned to 0º. Sleep opportunities are shown separately for each condition represented by the black (BL), orange (5h) and blue (9h) rectangles. Data are mean ± SEM. Data are double plotted. DLMO, dim-light melatonin onset; DLMOff, dim-light melatonin offset.
Untargeted Metabolomics
Procedures for sample preparation, liquid chromatography, mass spectrometry, and data extraction for untargeted metabolomics analyses were conducted similar to prior studies (Yang et al., 2013; Cruickshank-Quinn et al., 2014; Hughes et al., 2014), as detailed in Depner, Cogswell, et al. (2020) and described in Supplemental Materials. Prior to biomarker model development, metabolomics data were filtered by first removing all compounds with >50% missing values (i.e., missing value is an occurrence when a compound was not detected in a sample) and then sequentially removing compounds with the largest number of missing values until the complete dataset had a total of 10% missing values. After filtering, the 10% missing values were imputed using the Bayesian Principal Components Analysis module in MetaboAnalyst (Chong et al., 2018). Then the hydrophilic dataset was batch corrected using the ComBat method (Johnson et al., 2007) within MetaboAnalyst (Chong et al., 2018) to account for batch effects between mass spectrometry data acquisition days, and finally all data were log2 normalized. The final data set consisted of 4,209 compounds derived from 26,301 metabolomics features. We performed tandem mass spectrometry (MS/MS) on the 320 compounds comprising our DLMO and DLMOff biomarker models to provide additional information for compound annotation as described in Supplemental Materials and detailed in Depner, Cogswell, et al. (2020). Hereafter compounds with annotations are referred to as “annotated compounds” and compounds without annotation matches are referred to as “unknown compounds.” Compound annotations, ID numbers, Metabolomics Standards Initiative (MSI) levels, and biomarker model information are in Supplementary Table S2.
Data Processing
Training and Test Data Sets
The workflow outlining biomarker model development is presented in Supplementary Figure S1. All plasma samples were randomly assigned into either the training set (68%) or test set (32%) using the “caret” package in R (3.5.2) (Kuhn, 2008), stratifying by time points so samples from all time points were evenly represented in the training and test sets. For DLMO, there were 133 plasma samples in the training set and 68 plasma samples in the test set. The DLMO training set consisted of 66 samples from females and 67 samples from males, and the test set consisted of 40 samples from females and 28 samples from males. The training set consisted of 33% from the 5-h condition, 35% from the 9-h condition, and 32% from the BL condition; and the test set was 30% from the 5-h condition, 40% from the 9-h condition, and 30% from the BL condition. For DLMOff, there were 112 plasma samples in the training set and 53 plasma samples in the test set. The training set consisted of 60 samples from females and 52 samples from males, and the test set consisted of 31 samples from females and 22 samples from males. The training set consisted of 39% of samples from the 5-h condition, 31% from the 9-h condition, and 30% from the BL condition and the test set was 32% from the 5-h condition, 36% from the 9-h condition, and 32% from the BL condition. This resulted in 201 total plasma metabolomics samples in our DLMO dataset and 165 total plasma metabolomics samples in our DLMOff dataset. The number of samples is different for DLMO and DLMOff biomarker models because missing samples for melatonin analyses differentially affected DLMO and DLMOff calculations. Missing samples were primarily due to failure of the intravenous catheter used for blood collection. When missed blood draws impeded the calculation of DLMO or DLMOff, the corresponding metabolomics data were removed prior to sample randomization into training and test datasets and therefore such missing samples did not influence biomarker development or performance calculations.
Confounding Features Affected by Energy Balance
During ad libitum energy intake, participants on average consumed excess calories and were in positive energy balance (Markwald et al., 2013). We identified 95 compounds most affected by positive energy balance and excluded these 95 compounds from all biomarker models, resulting in 4,114 compounds to construct the biomarker models. The analysis identifying the 95 compounds most affected by positive energy balance was based on area under the receiver operator characteristic (AUROC) plots when comparing BL with controlled energy balanced food intake versus 9 h with ad libitum energy intake, facilitating comparison between energy-balanced conditions versus positive energy balance without the confounding impact of insufficient sleep. The 95 compounds most affected by positive energy balance are the compounds with an AUROC ≥ 0.70 when comparing BL versus 9-h samples, as fully detailed in (Depner, Cogswell, et al., 2020).
A Priori Feature Selection for 24-H Time-of-day Patterns
Prior findings indicate 2% to 33% of plasma metabolites in humans (out of 132–281 total metabolites analyzed; Dallmann et al., 2012; Chua et al., 2013, 2015; Skene et al., 2018) show circadian regulation regardless of patterns of food intake or sleep. Because circadian regulated metabolites are more likely to be circadian biomarkers, our primary outcomes focused on biomarker models using only compounds (annotated and unknown) that displayed 24-h time-of-day patterns. We used the MetaCycle package (v1.1.0) (Wu et al., 2016) in R (v3.5.2) to identify compounds with 24-h time-of-day patterns in at least one condition as described previously (Depner, Cogswell, et al., 2020). In total, 985 out of 4,114 compounds (~24%) showed ~24-h time-of-day patterns in at least one condition. Biomarker models based on all compounds regardless of 24 h time-of-day patterns (Suppl. Figs. S2 and S3; Table S3) and biomarker models analyzed by timepoint (Suppl. Table S4) are in Supplemental Materials.
Biomarker Model Development
We used partial least squares regression (PLSR; Boulesteix and Strimmer, 2007) to build biomarker models based on strong performance in previous transcriptomics-based work (Laing et al., 2017; Kervezee, Cuesta, et al., 2019). PLSR is a supervised multivariate feature selection technique that uses multiple collinear features (compounds) to predict a response variable (DLMO or DLMOff). PLSR detects features accounting for most of the variability in the response variable by reducing the predictor dimensions, maximizing covariance between the predictors and the response, and extracting latent factors that carry the most absolute “weight” (factors that explain most of the variation), defined as components. PLSR was used twice on the training set, once to select the optimal number of features (compounds) and components to include in a biomarker model and once to generate the biomarker model from the training data using the identified optimal number of features (compounds) and components using leave-one-out cross-validation (LOOCV). We tested combinations ranging from 20 to 500 features (compounds) and 3 to 15 components, resulting in a total of 169 combinations tested for developing each biomarker model. We then applied each biomarker model to the test set to predict circadian phase for final performance assessments. We used published code from Laing et al. (2017) for all PLSR calculations.
Performance Metrics for Biomarker Models
- The value of R2 was calculated during biomarker model development and performance assessments.
- For biomarker development using LOOCV, the highest R2 was used to determine the optimal number of features (compounds) and components for each biomarker model.
- For performance assessments, R2 was used to determine model fit in the test set.
Median absolute error (MdAE ± interquartile range [IQR] in hours) was calculated as the median of the differences between the observed and predicted phase of each sample in the test set. We also report the standard deviation of error (SD) to allow direct comparison with prior findings from circadian omics-based biomarker studies including from Laing et al. (2017) that also used the PLSR approach.
Cumulative error is reported as the proportion of samples in the test set under a given amount of error in hours.
Biomarker Model Comparisons
Wilcoxon t-tests were applied to the test set to detect significant differences in prediction error between and within biomarker models for primary and secondary outcomes, to test for condition order effects within biomarker models (condition order A vs. condition order B), and for exploratory analyses of sex and time-of-day differences within biomarker models. Prediction error was calculated as the absolute value of predicted circadian phase minus actual circadian phase for each sample in degrees. For a priori multiple comparisons between conditions and time points, we applied modified Bonferroni corrections to maintain familywise error rate and thus reduce Type I errors (Keppel, 1991).
Pathway and Functional Analyses
Annotated compounds were converted to Human Metabolome Database IDs (HMDB, v4.0) (Wishart et al., 2018) and searched using the MetaboAnalyst Pathway Analysis tool (Chong et al., 2018) to match compounds with biochemical pathways from the Small Molecule Pathway Database (Jewison et al., 2014) and Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa and Goto, 2000). Within MetaboAnalyst, statistical significance was set at the 10% false discovery rate (FDR) level to correct for multiple comparisons. To further interrogate biochemical functions associated with the top-performing biomarker model, we developed a gene-metabolite interaction network using the Network Explorer module in MetaboAnalyst (Wishart et al., 2018; Depner, Cheng, et al., 2020). Briefly, the gene-metabolite network consists of first-order neighbors that directly interact with the uploaded compound HMDB IDs from annotated compounds. Genes and compounds are represented by the network nodes whereas the interactions among the genes and compounds, based on the MetPriCNet database, are represented by the network edges. Enriched gene ontology biological processes associated with the gene-metabolite interaction network were identified within MetaboAnalyst.
Results
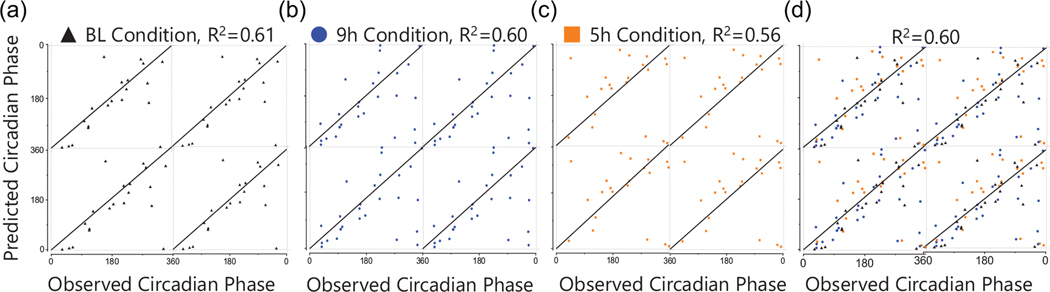
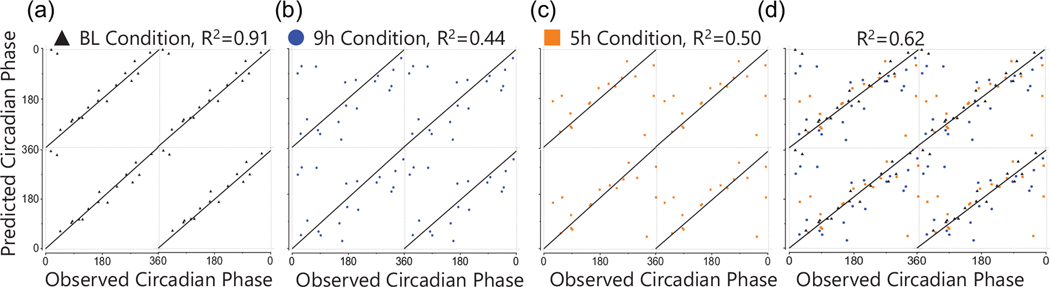
Using annotated and unknown compounds with 24-h time-of-day patterns, we generated one biomarker model predicting DLMO and one predicting DLMOff (Figures 3 and 4). The DLMO biomarker model consisted of 5 components and 100 compounds, the DLMOff biomarker model consisted of 13 components and 300 compounds, and 79 compounds overlapped between the DLMO and DLMOff biomarker models. At BL, the DLMOff biomarker model had a lower (p < 0.01) prediction error by ~1.6 h versus the DLMO biomarker model (Table 1, Figures 3a and 4a). For the 9-h and 5-h conditions and when assessed with all conditions together, the prediction error for the DLMOff biomarker model was not statistically different versus the DLMO biomarker model.
Figure 3.
Performance of the Biomarker Model Predicting DLMO. The X-axis represents the observed circadian degree of the sample and the Y-axis represents the predicted circadian degree of the sample. (A) baseline samples in test set; (B) 9h samples in test set; (C) 5h samples in test set; (D) all samples from all conditions in test set. The line of identity through the center indicates a perfect prediction. BL, baseline.
Figure 4.
Performance of the Biomarker Model Predicting DLMOff. The X-axis represents the observed circadian degree of the sample and the Y-axis represents the predicted circadian degree of the sample. A) baseline samples in test set; (B) 9h samples in test set; (C) 5h samples in test set; (D) all samples from all conditions in test set. The line of identity through the center indicates a perfect prediction. BL, baseline.
Table 1.
DLMO and DLMOff biomarker model performance metrics for overall model and by condition.
| R2 | MDAE (hours) | SD of error (hours) | IQR (hours) | |
|---|---|---|---|---|
| DLMO Prediction (100) | ||||
|
| ||||
| All conditions (overall) | 0.60 | 2.2 | 2.7 | 2.8 |
| Baseline Condition Only | 0.61 | 2.2 | 2.7 | 1.9 |
| 9h Condition Only | 0.60 | 2.1 | 2.9 | 3.0 |
| 5h Condition Only | 0.56 | 2.6 | 2.5 | 4.2 |
|
| ||||
| DLMOff Prediction (300) | ||||
|
| ||||
| All conditions (overall) | 0.62 | 1.8 | 2.8 | 2.6 |
| Baseline Condition Only*† | 0.91 | 1.1 | 0.9 | 1.1 |
| 9h Condition Only | 0.44 | 3.7 | 2.6 | 4.5 |
| 5h Condition Only | 0.50 | 3.3 | 3.4 | 2.8 |
DLMO, dim-light melatonin onset; DLMOff, dim-light melatonin offset; MdAE, Median Absolute Error; SD, Standard Deviation; IQR, Interquartile Range. Numbers in parentheses refer to the number of metabolites included in each model.
P < 0.001 versus 9h condition within the DLMOff biomarker model
P < 0.01 versus baseline condition in the DLMO biomarker model.
Within the DLMO biomarker model, there were no significant differences between conditions (Table 1 and Figure 3). When assessed with all conditions together, R2 was 0.60, MdAE was ~2.2 h, and 44% of the test samples had an error under 2 h (Table 1 and Figure 5). In addition, there were no significant differences between time points (Suppl. Table S4) or sex (Suppl. Table S5), but there was an order effect where participants completing the 5-h condition first had a lower (P < 0.05) prediction error by ~1.2 h versus participants completing the 9-h condition first (Suppl. Table S5).
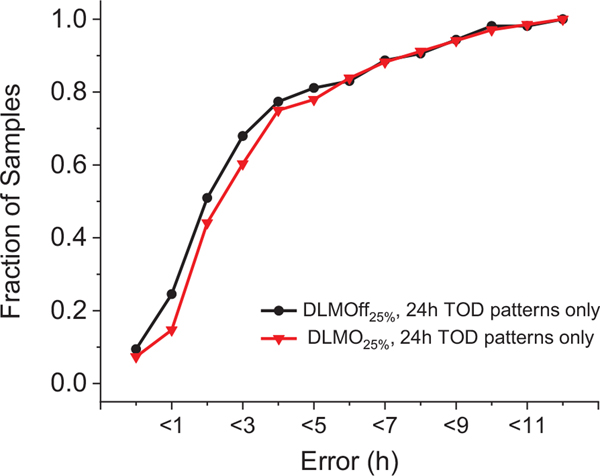
Figure 5.
Cumulative Error for DLMO and DLMOff Biomarker Models. Data are plotted as proportion of samples versus cumulative hours of error. MdAE, median absolute error; TOD, time of day; DLMO, dim-light melatonin onset; DLMOff, dim-light melatonin offset.
Within the DLMOff biomarker model, the BL condition had a lower (p < 0.001) prediction error by ~2.9-h versus the 9-h condition and showed a non-significant trend for a lower (p = 0.10) prediction error by ~2.0 h versus the 5-h condition (Table 1 and Figure 4). When assessed with all conditions together, R2 was 0.62, MdAE was ~1.8 h, and 51% of the test samples had an error under 2 h (Table 1 and Figure 5). In addition, there were no significant differences between time points (Suppl. Table S4), sex, or condition order (Suppl. Table S5).
Functional analyses focused on the DLMOff biomarker model because of the higher overall R2 and significantly lower prediction error in the BL condition of the DLMOff biomarker model versus the DLMO biomarker model. HMDB IDs were available for 65 out of 300 compounds in the DLMOff biomarker model, thus subsequent results are based on these 65 annotated compounds. Pathway analysis identified Glycerophospholipid Metabolism (KEGG pathway entry hsa00564) as the only significant (FDR = 0.018) pathway associated with the annotated compounds in the DLMOff biomarker model. Within Glycerophospholipid Metabolism, there were five KEGG Metabolite hits (Phosphatidylethanolamine [PE], Phosphatidylcholine [PC], Lysophosphatidylethanolamine [LysoPE], Lysophosphatidylcholine [LysoPC], and Choline). These five KEGG Metabolite hits corresponded to 19 annotated HMDB compound IDs from our DLMOff biomarker model consisting of choline, 2 PEs, 9 PCs, 3 LysoPEs, and 4 LysoPCs. We also developed a gene-metabolite interaction network from the annotated compounds in the DLMOff biomarker model. The top 10 gene ontology biological processes associated with this network are listed in Supplementary Table S6. Several gene ontology biological processes associated with the DLMOff biomarker model overlapped with Glycerophospholipid Metabolism from the pathway analysis. Of the top 10 gene ontology biological processes associated with the DLMOff biomarker model, alcohol metabolic process, cellular biogenic amine metabolic process, response to drug, cellular amino acid metabolic process, response to steroid hormone stimulus, and carboxylic acid metabolic process do not directly overlap with Glycerophospholipid Metabolism. No pathway hits associated with the DLMO biomarker model were below the 10% FDR significance level.
Discussion
We applied untargeted metabolomics analyses to human plasma samples collected under varying conditions of sleep duration and food intake to develop and test candidate biomarker models of the circadian phase markers DLMO and DLMOff. At BL, the DLMOff biomarker model had significantly lower prediction error versus the DLMO biomarker model, but when based on all three conditions, performance metrics were similar between the DLMOff and DLMO biomarker models. Within the DLMOff biomarker model, the BL condition had significantly lower prediction error versus the 9-h condition and showed superior performance metrics (R2, MdAE, and IQR) versus the 9-h and 5-h conditions. Within the DLMO biomarker model, prediction errors and performance metrics were similar across conditions. Although preliminary, our findings show promise for predicting DLMOff using metabolomics and highlight the importance of considering DLMO and DLMOff as circadian phase biomarkers in future studies.
To date, findings from just one known study have described a metabolomics-based biomarker predicting DLMO (Kasukawa et al., 2012), with no known findings using any omics-based platform to predict DLMOff. The DLMO biomarker model from Kasukawa et al. consists of 58 metabolomic compounds and predicts circadian phase with ~3 h of error when using two antiphase blood samples during constant routine conditions with total sleep deprivation. These findings from Kasukawa et al. are based on three participants in the development phase and the same three participants were used (plus an additional three participants) in the validation phase. In addition, all participants were male, thus the generalizability of this biomarker model has not been tested. In comparison, our biomarker models use one sample to predict melatonin phase in males and females within ~2 h of error using 100 compounds and 300 compounds for DLMO and DLMOff, respectively.
Compared with these metabolomics approaches, recent transcriptomics approaches have produced better performing circadian phase biomarkers to date. Hughey et al. developed Zeitzeiger that predicts DLMO within 2.1 ± 2.8 h (MdAE ± IQR) of error using 15 genes in 60 participants under controlled conditions of sleep restriction, forced desynchrony, and 1 night of total sleep deprivation. However, these findings used sunrise to estimate circadian phase and are based on average DLMO per condition for one dataset, as opposed to individual DLMO data. Laing et al. used transcriptomics to predict DLMO using PLSR and achieved an R2 of 0.74 using one blood sample and an R2 of 0.90 using two antiphase blood samples and 100 transcripts under controlled conditions of sleep in and out of phase with the circadian clock, and total sleep deprivation with and without prior sleep debt. We achieved a similar R2 of 0.91 when using one blood sample to predict DLMOff in the BL condition (adequate sleep with controlled energy balanced food intake) with 300 compounds; however, our validation dataset had less variability in sleep conditions compared with the data from Laing and colleagues. Braun et al. published a transcriptomics-based prediction algorithm using elastic net regularization and achieved MdAEs ranging from 1 h 12 min to 1 h 51 min under controlled conditions of adequate and insufficient sleep that occurred in and out of phase with the circadian clock. This algorithm works well and is generalizable, but the within-participant normalization step will not work unless there are two blood samples to calculate the mean over a circadian period, and the two blood samples must be taken 10 to 14 h apart. The BodyTime approach published by Wittenbrink et al. (2018) used the Zeitzeiger algorithm followed by a migration of the chosen transcripts to the Nanostring platform. This migration step is unique and improved the accuracy of the biomarker, indicating future research should focus on improving analysis platforms and statistical tools. The final BodyTime method uses two genes to predict DLMO from two blood samples with an error of 0.8 h ± 1.1 (MdAE ± IQR), and 14 genes to predict DLMO from one sample also with an error of 0.8 h ± 1.1 (MdAE ± IQR) following conditions of “free-living” adequate sleep in individuals with early morning or late chronotypes. Considering our success with DLMOff, the accuracy of transcriptomics approaches to predict DLMOff warrant investigation. Because these transcriptomics approaches generally produced superior circadian phase biomarkers compared with metabolomics approaches, it may suggest that circadian regulation of gene expression is stronger than circadian regulation of plasma metabolites, consistent with prior findings showing plasma metabolites are heavily influenced by the behavioral cycle, and especially fasting-food intake patterns (Skene et al., 2018; Kervezee, Cermakian, and Boivin, 2019). Still, all these omic biomarkers published to date require further large-scale validation studies to fully quantify their performance in the precise setting and population of expected use such as shift workers, individuals with CRSWDs, chronotherapy, research settings, older age, and different racial/ethnic backgrounds. Such validation studies are a critical next step in the biomarker development pipeline following these initial studies designed to identify candidate biomarkers (Mullington et al., 2016; Dijk and Duffy, 2020). Although we did not detect sex differences in our biomarker models, it is important for future studies to include men and women and analyze for potential sex differences given the known sex differences in intrinsic period of the human circadian system (Duffy et al., 2011).
Biomarkers using other methods such as actigraphy, temperature, blue light, and heart rate to predict circadian phase have been developed. Cheng et al. (2021) used wrist actigraphy data to develop a model that predicted DLMO in night shift workers with an absolute mean error of 2.88 h. Stone et al. (2019) built artificial neural network models using wrist temperature and blue light sensors to predict urinary aMT6s acrophase. This model predicted phase within 41 ± 32 min (MdAE ± IQR) on a fixed sleep schedule, but accuracy decreased when generalizing to night shift work. Findings from another study (Gil et al., 2013) based on an autoregressive model to predict DLMO using light and heart rate showed a prediction error of 2 h ± 40 min. From a clinical perspective, the most convenient biomarker approach is likely a single blood sample. These other approaches could be useful in a range of other settings, especially where tracking changes over consecutive days is needed, such as research with active duty military personnel or in the transportation industry (Depner, Cheng, et al., 2020).
Functional analyses consistently showed phospholipid/glycerophospholipid metabolism was associated with the DLMOff biomarker model. Phospholipids have a range of functions including stabilizing cell membranes, acting as cofactors, and providing a reservoir for signaling molecules (Lagace and Ridgway, 2013), and some have well characterized 24-h time-of-day variations (Ang et al., 2012; Kervezee, Cermakian, and Boivin, 2019). Many lysophospholipids show strong 24-h time-of-day patterns during circadian alignment, but lose their rhythmicity during circadian misalignment, suggesting some regulation by behavioral cycles such as fasting-food intake or sleep-wake (Kervezee, Cermakian, and Boivin, 2019). Notably, previous findings show phospholipid metabolism in humans is also affected by insufficient sleep, suggesting regulation by the behavioral cycle (Weljie et al., 2015; Depner, Cogswell, et al., 2020). Because there are several thousand species of phospholipids, targeted lipidomics analyses and utilization of circadian protocols (e.g., forced desynchrony, constant routine, simulated shift work; Broussard et al., 2017) are required to dissect behavioral versus circadian regulation of phospholipid metabolism, which could help refine the selection of specific phospholipids most appropriate for circadian phase biomarkers. Furthermore, because altered phospholipid metabolism has been linked to risk of metabolic (Funai et al., 2016; Meikle and Summers, 2017) and neurodegenerative disorders, including Alzheimer’s Disease (Mapstone et al., 2014), a more complete understanding of wake-sleep/food intake-fasting and circadian regulation of phospholipid metabolism could help identify mechanisms underlying increased disease risk associated with sleep and circadian disruption.
Gene ontology biological processes associated with the DLMOff model include amino acid metabolism, biogenic amine metabolism, and carboxylic acid metabolism. Using targeted metabolomics and a simulated night shift-work protocol, Kervezee, Cermakian, and Boivin (2019) identified seven circadian influenced metabolites and class over-representation analysis of these metabolites overlaps with our findings for amino acids and biogenic amines. Although we did not conduct a circadian protocol, the overlap in enriched pathways with a circadian protocol (simulated night shift work) lends support to our findings that metabolites related to these pathways could be used to predict circadian phase and that the circadian clock has a role in regulating these pathways.
Two lipoprotein remodeling pathways—(1) high-density lipoprotein (HDL) remodeling and (2) plasma lipoprotein assembly, remodeling, and clearance—were associated with the DLMOff biomarker model. The circadian system is implicated in regulating various lipoprotein functions, such as absorption, storage, and transport in coordination with feeding and resting cycles (Gooley and Chua, 2014). Circadian oscillations of very low-density lipoprotein (VLDL) triacylglycerol levels in plasma caused by lipoprotein lipase activity and VLDL secretion rates have been reported (Marrino et al., 1987), as well as daily patterns in HDL phospholipids (Mirani-Oostdijk et al., 1985). These prior findings and our data support the concept that lipoprotein metabolism and transport are, in part, under circadian control and related plasma metabolites could play a role in predicting circadian phase.
Beyond these functional analyses, two annotated compounds (MSI 2 level) warrant discussion. First, cortisol was in the DLMO biomarker model. Given the well-established circadian regulation of cortisol (Buijs et al., 1999; Broussard et al., 2017), this supports the concept that circadian regulated metabolites will most likely produce the best performing biomarkers of circadian phase in humans. This finding also suggests there are potential benefits of including cortisol in other omics-based biomarkers of circadian phase, especially biomarkers of DLMO, and warrants further investigation as other omics biomarkers of circadian phase also show enrichment in glucocorticoid signaling pathways (Kasukawa et al., 2012; Laing et al., 2017). Second, glycoursodeoxycholic acid, a secondary bile acid produced by modification of the gut microbiome, was in the DLMO and DLMOff biomarker models. This finding suggests secondary bile acids may, in part, be regulated by the circadian system, consistent with previous data from rodent models (Zhang et al., 2011; Eggink et al., 2017). Because changes in bile acid metabolism have been linked to adverse cardiometabolic risk (Tripathi et al., 2018), targeted profiling of the known secondary bile acids in humans is needed to fully understand the potential circadian regulation of secondary bile acids, their utility in predicting circadian phase, and the effect of circadian misalignment on secondary bile acids (Parkar et al., 2019; Withrow et al., 2021).
Our study has some limitations. Given the protocol, we were unable to determine if metabolites with 24-h time-of-day patterns were regulated by the circadian or behavioral cycles. Prior data indicate the majority of 24-h time-of-day patterns in targeted blood metabolomics analyses are driven by the behavioral cycle (food intake/fasting, wakefulness/sleep; Skene et al., 2018; Kervezee, Cermakian, and Boivin, 2019). An important next step in the biomarker development pipeline is to determine if metabolite biomarkers using exclusively circadian regulated metabolites improves overall biomarker performance. Circadian protocols designed to identify specific circadian regulated metabolites rather than metabolites with 24-h time-of-day patterns regulated by circadian or behavioral cycles are required to achieve this next step. It is evident that positive energy balance under conditions of ad libitum energy intake influences the plasma metabolome and this likely affected the training/development of our biomarker models. This is highlighted by the fact that our DLMOff biomarker model performed best in the BL condition with controlled energy balanced food intake and adequate sleep opportunities. We previously reported a condition order effect for food intake where participants who completed the 9-h condition first had similarly elevated food intake in the 9-h and 5-h conditions, whereas participants who completed the 5-h condition first decreased their food intake when transitioning from the 5-h to 9-h condition (Markwald et al., 2013). This condition order effect potentially contributed to the condition order effect observed in our DLMO biomarker model. Although the impact of increased food intake and positive energy balance is a perceived limitation, our findings highlight the importance of biomarker validation under varying conditions including ad libitum energy intake and sleep duration as these reflect “real-world” conditions. As an additional limitation, we did not have a completely independent validation cohort and the same participants were present in the training and test sets, potentially leading to overfitting of our biomarker models. Ultimate performance assessments will require a completely independent validation cohort. There are also limitations to the metabolomics platform. For example, ~75% and 73% of the compounds were not annotated (unknown compounds) in the DLMO and DLMOff biomarker models, respectively, and MS/MS analyses identified potential redundant annotations for 29 compounds. Validating each of these annotations will require confirmed standards and/or targeted assays. Our best performing biomarker used 300 compounds. To consider a biomarker for clinical use, it would be preferred to use a smaller cluster of compounds. Finally, our findings are based on a relatively homogeneous cohort of young healthy adults, limiting applications to other populations. Prior data show factors such as BMI, age, racial and ethnic background, and chronic disease status can influence the human plasma metabolome (Shen et al., 2013; Isherwood et al., 2017; Sebastiani et al., 2017; van Valkengoed et al., 2017; Vardarajan et al., 2020). Thus, further biomarker development and performance assessments across these various populations could help create a more robust biomarker and are a critical next step for clinical and research implementation of any biomarker of circadian phase in humans.
In summary, our findings show plasma metabolomics can be used to discover biomarker candidates that predict circadian phase with a single blood sample, especially for predicting DLMOff under controlled conditions. Current transcriptomics work is also promising, but none are yet ready for clinical implementation. In addition, circadian biomarker efforts have focused on healthy adults, and no omics-based biomarker to date, ours included, has been tested in children or adults with chronic health conditions, and as biomarker efforts progress, a combination of platforms (transcriptomics, proteomics, metabolomics, wearable light, and actigraphy data) and different circadian phase markers (DLMO, DLMOff, temperature, heart rate) should be tested. Ultimately, developing a validated and easily implemented biomarker of individual circadian phase in humans is a high research priority of the sleep and circadian fields. When successful, such a biomarker will serve to advance sleep and circadian science and medicine.
Supplementary Material
Acknowledgements
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Institute of Health (R01HL085705, R01HL109706, R01HL132150, K01HL145099, F32DK111161, UL1TR000154) and the Sleep Research Society Foundation (011-JP-16).
Footnotes
Conflict of Interest Statement
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: C.M.D. reports travel reimbursement fees from the Sleep Research Society. A.W.M. reports speaker honorarium or travel reimbursement fees from the Utah Sleep Research Society and the California Precast Concrete Association. E.L.M. is supported by resources from the Geriatric Research, Education, and the Clinical Center at the Denver VA Medical Center. The contents do not represent the views of the US Department of Veterans Affairs or the US Government. K.P.W. reports during the conduct of the study being a board member of the Sleep Research Society; chair of the American Academy of Sleep Medicine Clinical Practice Guideline for the Treatment of Adults with Shift Work Disorder and Jet Lag Disorder Workgroup; receiving research support from the NIH, the Office of Naval Research, the PAC-12 conference, and consulting for Circadian Therapeutics, LTD., Circadian Biotherapies, Inc. Philips Respironics, and the US Army Medical Research and Materiel Command-Walter Reed Army Institute of Research outside the submitted work.
References
- Alfred LJ, Wehr TA, Goodwin FK, Newsome DA, and Markey SP (1980) Light Suppresses Melatonin Secretion in Humans. Science 210:1267–1269. [DOI] [PubMed] [Google Scholar]
- Ang JE, Revell V, Mann A, Mantele S, Otway DT, Johnston JD, Thumser AE, Skene DJ, and Raynaud F (2012) Identification of human plasma metabolites exhibiting time-of-day variation using an untargeted liquid chromatography-mass spectrometry metabolomic approach. Chronobiol Int 29:868–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron KG, and Reid KJ (2014) Circadian misalignment and health. Int Rev Psychiatry 26:139–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benloucif S, Burgess HJ, Klerman EB, Lewy AJ, Middleton B, Murphy PJ, Parry BL, and Revell VL (2008) Measuring melatonin in humans. J Clin Sleep Med 4:66–69. [PMC free article] [PubMed] [Google Scholar]
- Boulesteix AL, and Strimmer K (2007) Partial least squares: a versatile tool for the analysis of high-dimensional genomic data. Brief Bioinform 8:32–44. [DOI] [PubMed] [Google Scholar]
- Braun R, Kath WL, Iwanaszko M, Kula-Eversole E, Abbott SM, Reid KJ, Zee PC, and Allada R (2018) Universal method for robust detection of circadian state from gene expression. Proc Natl Acad Sci U S A 115:E9247-E9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun R, Kath WL, Iwanaszko M, Kula-Eversole E, Abbott SM, Reid KJ, Zee PC, and Allada R (2019) Reply to Laing et al.: Accurate prediction of circadian time across platforms. Proc Natl Acad Sci U S A 116:5206–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussard JL, Reynolds AC, Depner CM, Ferguson SA, Dawson D, and Wright KP (2017) Circadian Rhythms Versus Daily Patterns in Human Physiology and Behavior. In Biological Timekeeping: Clocks, Rhythms and Behaviour, Kumar V, ed, pp 279–295, Springer; India, New Delhi. [Google Scholar]
- Buijs RM, Wortel J, Van Heerikhuize JJ, Feenstra MG, Ter Horst GJ, Romijn HJ, and Kalsbeek A (1999) Anatomical and functional demonstration of a multisynaptic suprachiasmatic nucleus adrenal (cortex) pathway. Eur J Neurosci 11:1535–1544. [DOI] [PubMed] [Google Scholar]
- Cheng P, Walch O, Huang Y, Mayer C, Sagong C, Cuamatzi Castelan A, Burgess HJ, Roth T, Forger DB, and Drake CL (2020) Predicting circadian misalignment with wearable technology: Validation of wrist-worn actigraphy and photometry in night shift workers. Sleep. doi: 10.1093/sleep/zsaa180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, and Xia J (2018) MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 46:W486-W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua EC, Shui G, Cazenave-Gassiot A, Wenk MR, and Gooley JJ (2015) Changes in Plasma Lipids during Exposure to Total Sleep Deprivation. Sleep 38:1683–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua EC, Shui G, Lee IT, Lau P, Tan LC, Yeo SC, Lam BD, Bulchand S, Summers SA, Puvanendran K, Rozen SG, Wenk MR, and Gooley JJ (2013) Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc Natl Acad Sci U S A 110:14468–14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank-Quinn C, Quinn KD, Powell R, Yang Y, Armstrong M, Mahaffey S, Reisdorph R, and Reisdorph N (2014) Multi-step preparation technique to recover multiple metabolite compound classes for in-depth and informative metabolomic analysis. J Vis Exp. doi: 10.3791/51670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann R, Viola AU, Tarokh L, Cajochen C, and Brown SA (2012) The human circadian metabolome. Proc Natl Acad Sci U S A 109:2625–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depner C, Cheng PC, Devine JK, Khosla S, de Zambotti M, Robillard R, Vakulin A, and Drummond SPA (2020a) Wearable Technologies for Developing Sleep and Circadian Biomarkers: A Summary of Workshop Discussions. Sleep 43(2):zsz254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depner C, Cogswell DT, Bisesi PJ, Markwald RR, Cruickshank-Quinn C, Quinn KD, Melanson EL, Reisdorph N, and Wright KP Jr., (2020b) Developing Preliminary Blood Metabolomics-based Biomarkers of Insufficient Sleep in Humans. Sleep zsz321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depner C, Melanson EL, McHill AW, and Wright KP Jr., (2018) Mistimed food intake and sleep alters 24-hour time-of-day patterns of the human plasma proteome. Proc Natl Acad Sci U S A 115:E5390-E5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijk DJ, and Duffy JF (2020) Novel Approaches for Assessing Circadian Rhythmicity in Humans: A Review. J Biol Rhythms 35:421–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake CL, and Wright KP (2011) Shift Work, Shift-Work Disorder, and Jet Lag. In Principles and Practice of Sleep Medicine, pp 784–798. [Google Scholar]
- Duffy JF, Abbott SM, Burgess HJ, Crowley SJ, Emens JS, Epstein LJ, Gamble KL, Hasler BP, Kristo DA, Malkani RG, Rahman SA, Thomas SJ, Wyatt JK, Zee PC, and Klerman EB (2021) Workshop report. Circadian rhythm sleep-wake disorders: gaps and opportunities. Sleep. 2021 doi: 10.1093/sleep/zsaa281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JF, Cain SW, Chang AM, Phillips AJ, Münch MY, Gronfier C, Wyatt JK, Dijk DJ, Wright KP Jr., and Czeisler CA(2011) Sex difference in the near-24-hour intrinsic period of the human circadian timing system. Proc Natl Acad Sci U S A 108 Suppl 3:15602–15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JF, and Dijk DJ (2002) Getting through to circadian oscillators: why use constant routines? J Biol Rhythms 17:4–13. [DOI] [PubMed] [Google Scholar]
- Eckel RH, Depner CM, Perreault L, Markwald RR, Smith MR, McHill AW, Higgins J, Melanson EL, and Wright KP Jr., (2015) Morning Circadian Misalignment during Short Sleep Duration Impacts Insulin Sensitivity. Curr Biol 25:3004–3010. [DOI] [PubMed] [Google Scholar]
- Eggink HM, Oosterman JE, de Goede P, de Vries EM, Foppen E, Koehorst M, Groen AK, Boelen A, Romijn JA, la Fleur SE, Soeters MR, and Kalsbeek A (2017) Complex interaction between circadian rhythm and diet on bile acid homeostasis in male rats. Chronobiol Int 34:1339–1353. [DOI] [PubMed] [Google Scholar]
- Funai K, Lodhi IJ, Spears LD, Yin L, Song H, Klein S, and Semenkovich CF (2016) Skeletal Muscle Phospholipid Metabolism Regulates Insulin Sensitivity and Contractile Function. Diabetes 65:358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil EA, Aubert XL, Most EI, and Beersma DG (2013) Human circadian phase estimation from signals collected in ambulatory conditions using an autoregressive model. J Biol Rhythms 28:152–163. [DOI] [PubMed] [Google Scholar]
- Gooley JJ, and Chua EC (2014) Diurnal regulation of lipid metabolism and applications of circadian lipidomics. J Genet Genomics 41:231–250. [DOI] [PubMed] [Google Scholar]
- Hermida RC, Crespo JJ, Domínguez-Sardiña M, Otero A, Moyá A, Ríos MT, Sineiro E, Castiñeira MC, Callejas PA, Pousa L, Salgado JL, Durán C, Sánchez JJ, Fernández JR, Mojón A, and Ayala DE (2019) Bedtime hypertension treatment improves cardiovascular risk reduction: the Hygia Chronotherapy Trial. Eur Heart J. [DOI] [PubMed] [Google Scholar]
- Hoyle NP, Seinkmane E, Putker M, Feeney KA, Krogager TP, Chesham JE, Bray LK, Thomas JM, Dunn K, Blaikley J, and O’Neill JS (2017) Circadian actin dynamics drive rhythmic fibroblast mobilization during wound healing. Sci Transl Med 9(415):eaal2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrushesky WJM (1985) Circadian Timing of Cancer Chemotherapy. Science 228:73–75. [DOI] [PubMed] [Google Scholar]
- Hughes G, Cruickshank-Quinn C, Reisdorph R, Lutz S, Petrache I, Reisdorph N, Bowler R, and Kechris K (2014) MSPrep--summarization, normalization and diagnostics for processing of mass spectrometry-based metabolomic data. Bioinformatics 30:133–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughey JJ (2017) Machine learning identifies a compact gene set for monitoring the circadian clock in human blood. Genome Med 9:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innominato PF, Giacchetti S, Bjarnason GA, Focan C, Garufi C, Coudert B, Iacobelli S, Tampellini M, Durando X, Mormont MC, Waterhouse J, and Levi FA (2012) Prediction of overall survival through circadian rest-activity monitoring during chemotherapy for metastatic colorectal cancer. Int J Cancer 131:2684–2692. [DOI] [PubMed] [Google Scholar]
- Isherwood CM, Van der Veen DR, Johnston JD, and Skene DJ (2017) Twenty-four-hour rhythmicity of circulating metabolites: effect of body mass and type 2 diabetes. Faseb J 31:5557–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewison T, Su Y, Disfany FM, Liang Y, Knox C, Maciejewski A, Poelzer J, Huynh J, Zhou Y, Arndt D, Djoumbou Y, Liu Y, Deng L, Guo AC, Han B, Pon A, Wilson M, Rafatnia S, Liu P, and Wishart DS (2014) SMPDB 2.0: Big Improvements to the SmallMolecule Pathway Database. Nucleic Acids Res 42:D478–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, and Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8:118–127. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, and Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasukawa T, Sugimoto M, Hida A, Minami Y, Mori M, Honma S, Honma K, Mishima K, Soga T, and Ueda HR (2012) Human blood metabolite timetable indicates internal body time. Proc Natl Acad Sci U S A 109:15036–15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppel G (1991) Design and Analysis: A Researcher’s Handbook Prentice Hall Englewood Cliffs, New Jersey. [Google Scholar]
- Kervezee L, Cermakian N, and Boivin DB (2019a) Individual metabolomic signatures of circadian misalignment during simulated night shifts in humans. PLoS Biol 17:e3000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervezee L, Cuesta M, Cermakian N, and Boivin DB (2019b) The Phase-Shifting Effect of Bright Light Exposure on Circadian Rhythmicity in the Human Transcriptome. J Biol Rhythms 34:84–97. [DOI] [PubMed] [Google Scholar]
- Klerman EB, Gershengorn HB, Duffy JF, and Kronauer RE (2002) Comparions of the Variability of Three Markers if the Human Circadian Pacemaker. J Biol Rhythms 17:181–193. [DOI] [PubMed] [Google Scholar]
- Kuhn M (2008) Building Predictive Models in R Using the caret Package. Journal of Statistical Software 28. [Google Scholar]
- Labrecque G, and Belanger PM (1992) Biological Rhythms In the Absorption Distribution, Metabolism, and Excretion of Drugs. Pharmacol Ther 52:95–107. [DOI] [PubMed] [Google Scholar]
- Lagace TA, and Ridgway ND (2013) The role of phospholipids in the biological activity and structure of the endoplasmic reticulum. Biochim Biophys Acta 1833:2499–2510. [DOI] [PubMed] [Google Scholar]
- Laing EE, Moller-Levet CS, Archer SN, and Dijk DJ (2019) Universal and robust assessment of circadian time? Proc Natl Acad Sci U S A 116:5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing EE, Moller-Levet CS, Poh N, Santhi N, Archer SN, and Dijk DJ (2017) Blood transcriptome based biomarkers for human circadian phase. Elife 6 :e20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmer B, Scheidel B, and Behne S (1992) Chronopharmacokinetics and Chronopharmacodynamics of Cardiovascular Active Drugs: Propranolol, Organic Nitrates, Nifedipine. Annals New York Academy of Sciences. [DOI] [PubMed] [Google Scholar]
- Lévi F, Zidani R, and Misset J-L (1997) Randomised multicentre trial of chronotherapy with oxaliplatin, fluorouracil, and folinic acid in metastatic colorectal cancer. The Lancet 350:681–686. [DOI] [PubMed] [Google Scholar]
- Long JE, Drayson MT, Taylor AE, Toellner KM, Lord JM, and Phillips AC (2016) Morning vaccination enhances antibody response over afternoon vaccination: A cluster-randomised trial. Vaccine 34:2679–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapstone M, Cheema AK, Fiandaca MS, Zhong X, Mhyre TR, MacArthur LH, Hall WJ, Fisher SG, Peterson DR, Haley JM, Nazar MD, Rich SA, Berlau DJ, Peltz CB, Tan MT, Kawas CH, and Federoff HJ (2014) Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med 20:415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwald RR, Melanson EL, Smith MR, Higgins J, Perreault L, Eckel RH, and Wright KP Jr., (2013) Impact of insufficient sleep on total daily energy expenditure, food intake, and weight gain. Proc Natl Acad Sci U S A 110:5695–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrino P, Gavish D, Shafrir E, and Eisenberg S (1987) Diurnal variations of plasma lipids, tissue and plasma lipoprotein lipase, and VLDL secretion rates in the rat. A model for studies of VLDL metabolism. Biochim Biophys Acta 920:277–284. [DOI] [PubMed] [Google Scholar]
- Mason IC, Qian J, Adler GK, and Scheer F (2020) Impact of circadian disruption on glucose metabolism: implications for type 2 diabetes. Diabetologia 63:462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikle PJ, and Summers SA (2017) Sphingolipids and phospholipids in insulin resistance and related metabolic disorders. Nat Rev Endocrinol 13:79–91. [DOI] [PubMed] [Google Scholar]
- Mirani-Oostdijk CP, Havekes L, Van Gent CM, Frölich M, Jansen H, and Terpstra J (1985) Diurnal changes in serum triglycerides as related to changes in lipolytic enzymes, lipoproteins and hormones in patients with primary endogenous hypertriglyceridaemia on a carbohydrate-rich diet. Atherosclerosis 57:129–137. [DOI] [PubMed] [Google Scholar]
- Montaigne D, Marechal X, Modine T, Coisne A, Mouton S, Fayad G, Ninni S, Klein C, Ortmans S, Seunes C, Potelle C, Berthier A, Gheeraert C, Piveteau C, Deprez R, Eeckhoute J, Duez H, Lacroix D, Deprez B, Jegou B, Koussa M, Edme J-L, Lefebvre P, and Staels B (2018) Daytime variation of perioperative myocardial injury in cardiac surgery and its prevention by Rev-Erbα antagonism: a single-centre propensity-matched cohort study and a randomised study. The Lancet 391:59–69. [DOI] [PubMed] [Google Scholar]
- Mullington JM, Abbott SM, Carroll JE, Davis CJ, Dijk DJ, Dinges DF, Gehrman PR, Ginsburg GS, Gozal D, Haack M, Lim DC, Macrea M, Pack AI, Plante DT, Teske JA, and Zee PC (2016) Developing Biomarker Arrays Predicting Sleep and Circadian-Coupled Risks to Health. Sleep 39:727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, Ngotho M, Kariuki T, Dkhissi-Benyahya O, Cooper HM, and Panda S (2018) Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 359(6381):eaao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohdo S, Koyanagi S, Suyama H, Higuchi S, and Aramaki H (2001) Changing the Dosing Schedule Minimizes the Disruptive Effects of Interferon on Clock Function. Nat Med 7:356–360. [DOI] [PubMed] [Google Scholar]
- Parkar SG, Kalsbeek A, and Cheeseman JF (2019) Potential Role for the Gut Microbiota in Modulating Host Circadian Rhythms and Metabolic Health. Microorganisms 7(2):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben MD, Wu G, Smith DF, Schmidt RE, Francey LJ, Lee YY, Anafi R, and Hogenesch JB (2018) A database of tissue-specific rhythmically expressed human genes has potential applications in circadian medicine. Sci Transl Med 10(458):eaat8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack RL (2009) The pathophysiology of jet lag. Travel Med Infect Dis 7:102–110. [DOI] [PubMed] [Google Scholar]
- Scheer FA, Hilton MF, Mantzoros CS, and Shea SA (2009) Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A 106:4453–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, Thyagarajan B, Sun F, Schupf N, Newman AB, Montano M, and Perls TT (2017) Biomarker signatures of aging. Aging Cell 16:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Yan L, Liu S, Ambrosone CB, and Zhao H (2013) Plasma metabolomic profiles in breast cancer patients and healthy controls: by race and tumor receptor subtypes. Transl Oncol 6:757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon SL, McWhirter L, Diniz Behn C, Bubar KM, Kaar JL, Pyle L, Rahat H, Garcia-Reyes Y, Carreau AM, Wright KP, Nadeau KJ, and Cree-Green M (2019) Morning Circadian Misalignment Is Associated With Insulin Resistance in Girls With Obesity and Polycystic Ovarian Syndrome. J Clin Endocrinol Metab 104:3525–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene DJ, Skornyakov E, Chowdhury NR, Gajula RP, Middleton B, Satterfield BC, Porter KI, Van Dongen HPA, and Gaddameedhi S (2018) Separation of circadian- and behavior-driven metabolite rhythms in humans provides a window on peripheral oscillators and metabolism. Proc Natl Acad Sci U S A 115:7825–7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten TL, Cappuccio FP, Davidson AJ, Van Cauter E, Rajaratnam SMW, and Scheer F (2020) Health consequences of circadian disruption. Sleep 43(1):zsz194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten TL, Vincenzi S, Redman JR, Lockley SW, and Rajaratnam SM (2010) Timing of sleep and its relationship with the endogenous melatonin rhythm. Front Neurol 1:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MR, and Eastman CI (2008) Night shift performance is improved by a compromise circadian phase position: study 3. Circadian phase after 7 night shifts with an intervening weekend off. Sleep 31:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JE, Phillips AJK, Ftouni S, Magee M, Howard M, Lockley SW, Sletten TL, Anderson C, Rajaratnam SMW, and Postnova S (2019) Generalizability of A Neural Network Model for Circadian Phase Prediction in Real-World Conditions. Sci Rep 9:11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard ER, McHill AW, Depner CM, Birks BR, Moehlman TM, Ritchie HK, Guzzetti JR, Chinoy ED, LeBourgeois MK, Axelsson J, and Wright KP Jr., (2017) Circadian Entrainment to the Natural Light-Dark Cycle across Seasons and the Weekend. Curr Biol 27:508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, and Knight R (2018) The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol 15:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Minami Y, Honma S, Honma K, Iino M, and Hashimoto S (2004) Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles. Proc Natl Acad Sci U S A 101:11227–11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Valkengoed IGM, Argmann C, Ghauharali-van der Vlugt K, Aerts J, Brewster LM, Peters RJG, Vaz FM, and Houtkooper RH (2017) Ethnic differences in metabolite signatures and type 2 diabetes: a nested case-control analysis among people of South Asian, African and European origin. Nutr Diabetes 7:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardarajan B, Kalia V, Manly J, Brickman A, Reyes-Dumeyer D, Lantigua R, Ionita-Laza I, Jones DP, Miller GW, and Mayeux R (2020) Differences in plasma metabolites related to Alzheimer’s disease, APOE epsilon4 status, and ethnicity. Alzheimers Dement (N Y) 6:e12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessely LH, and Lewy AJ (2002) Melatonin as a hormone and as a marker for circadian phase position in humans. In Hormones, Brain and Behavior, pp 121–141, Elsevier. [Google Scholar]
- Wehr TA, Aeschbach D, and Duncan WC Jr., (2001) Evidence for a biological dawn and dusk in the human circadian timing system. J Physiol 535:937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weljie AM, Meerlo P, Goel N, Sengupta A, Kayser MS, Abel T, Birnbaum MJ, Dinges DF, and Sehgal A (2015) Oxalic acid and diacylglycerol 36:3 are cross-species markers of sleep debt. Proc Natl Acad Sci U S A 112:2569–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhai S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, and Scalbert A (2018) HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 46:D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withrow DB SJ; Depner CM; Gonzalez A; Reynolds AC; Wright KP Jr. (2021) Sleep and circadian disruption and the gut microbiome-possible links to dysregulated metabolism. Current Opinion in endocrine and metabolic research 17:26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenbrink N, Ananthasubramaniam B, Munch M, Koller B, Maier B, Weschke C, Bes F, de Zeeuw J, Nowozin C, Wahnschaffe A, Wisniewski S, Zaleska M, Bartok O, Ashwal-Fluss R, Lammert H, Herzel H, Hummel M, Kadener S, Kunz D, and Kramer A (2018a) High-accuracy determination of internal circadian time from a single blood sample. J Clin Invest 128:3826–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenbrink N, Ananthasubramaniam B, Munch M, Koller B, Maier B, Weschke C, Bes F, de Zeeuw J, Nowozin C, Wahnschaffe A, Wisniewski S, Zaleska M, Bartok O, Ashwal-Fluss R, Lammert H, Herzel H, Hummel M, Kadener S, Kunz D, and Kramer A (2018b) High-accuracy determination of internal circadian time from a single blood sample. J Clin Invest 128:3826–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KP Jr., Gronfier C, Duffy JF, and Czeisler CA(2005) Intrinsic period and light intensity determine the phase relationship between melatonin and sleep in humans. J Biol Rhythms 20:168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KP Jr., Hughes RJ, Kronauer RE, Dijk DJ, and Czeisler CA(2001) Intrinsic near-24-h pacemaker period determines limits of circadian entrainment to a weak synchronizer in humans. Proc Natl Acad Sci U S A 98:14027–14032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KP Jr., McHill AW, Birks BR, Griffin BR, Rusterholz T, and Chinoy ED(2013) Entrainment of the human circadian clock to the natural light-dark cycle. Curr Biol 23:1554–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Anafi RC, Hughes ME, Kornacker K, and Hogenesch JB (2016) MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics 32:3351–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Cruickshank C, Armstrong M, Mahaffey S, Reisdorph R, and Reisdorph N (2013) New sample preparation approach for mass spectrometry-based profiling of plasma results in improved coverage of metabolome. J Chromatogr A 1300:217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YK, Guo GL, and Klaassen CD (2011) Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS One 6:e16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.