Abstract
Although bats are increasingly recognised as potential reservoir hosts of human zoonotic pathogens, bacteria in bats are still poorly studied. To investigate the DNA faecal prevalence of the bacterium Anaplasma phagocytophilum, we sampled 23 lesser horseshoe bat (Rhinolophus hipposideros) maternity colonies located in buildings (churches, barns) in rural villages of eastern France. A total of 552 faecal samples were collected from 278 individuals. Anaplasma phagocytophilum DNA was detected in the faeces of 63 individuals (22.7%). Such high prevalence might suggest persistent infection in bats and/or a frequent consumption of insect preys carrying bacteria. Faecal DNA prevalence varied highly among colonies but was not related to the colony size. Faecal DNA prevalence was the highest in the Jura Department, where the density of ticks is known to be the highest across the study area. Because the sampled bats live in close proximity to humans, we discuss how concerning the presence of A. phagocytophilum DNA in bat guano is for humans frequenting places of worship that shelter bats. We also advocate future research to understand what a high faecal DNA prevalence in bat guano really implicates in terms of bacteria transmission.
Key words: Anaplasma, bacteria, bat faeces, chiroptera
Introduction
The emergence of zoonotic diseases represents a substantial threat to global biodiversity and human health [1]. Due to a unique combination of life-history traits such as a long lifespan (average 10–20 years), high survival, great population densities, roosting behaviour and the ability to fly, bats are increasingly considered to be ideal reservoir hosts for emerging zoonotic pathogens [2–5]. Bats can roost in close contact with both domestic animals and humans, contaminating their local environment with guano and urine. They can indirectly transmit infectious agents to humans through intermediate hosts or arthropod vectors or directly through bat meat consumption, accidental bites and in some cases aerosol transmission [6]. Most documented investigations involving bats and infectious agents are largely limited to viruses responsible for diseases in humans, such as rabies and other lyssaviruses, Ebola, Nipah and Marburg viruses, severe acute respiratory syndrome and Middle East respiratory syndrome coronaviruses (see reviews in 3 and 5).
Unlike viruses, bacteria in bats and their putative threat to human health remain poorly studied [5]. Recently, bat ectoparasites (ticks, mites, fleas, cimicid bugs and bat flies) have been identified as potential vectors for pathogens such as Anaplasma, Bartonella, Borrelia, Rickettsia and Spiroplasma species [7–9]. High prevalence of the haemotrophic vector-borne bacteria Bartonella spp. were detected in the blood and organ tissues of several bat species, suggesting that bat ectoparasites are frequently implicated in transmitting bacteria [10–13]. Veikkolainen et al. [14] also found Bartonella spp. DNA in faeces of vespertilionid bats, which might have originated either from bleeding into the bat intestine or from the insect prey of the bats, which include abundant blood-feeding bat ectoparasites. In the same study, the authors isolated B. mayotimonensis, identified as an aetiologic agent of endocarditis in humans, by culturing of peripheral blood samples of Daubenton's bats (Myotis daubentonii). Because Bartonella spp. are haemotrophic bacteria, it is unclear if viable bacteria can survive in bat faeces and be involved in the transmission to other hosts, including humans. Veikkolainen et al. [14] hypothesised that the faeces of blood-fed bat ectoparasites might transmit Bartonella spp. to human hosts through superficial scratching or trauma to tissue in the skin, which has yet to be confirmed experimentally. This does not exclude the possibility that humans might be exposed to pathogens from ectoparasite bites [15]. The presence of bacteria associated with human pathogens in bat guano has been confirmed in the Leschenault's Rousette (Rousettus leschenaultia) in India and in four insectivorous bat species in South Africa [16, 17]. While the presence of bacteria in bat faeces has become clearer [18], their prevalence in bat faeces is largely unknown, although this is important information to better understand the role of bats in the transmission of potentially pathogenic bacteria.
In the present study, we focused on a pathogenic bacterium commonly spread among wildlife in Europe [19] and causing emerging diseases in humans: Anaplasma phagocytophilum which is transmitted through blood-feeding arthropod vectors. Anaplasma phagocytophilum is an obligate gram-negative intracellular bacterium that infects neutrophils. It can cause influenza-like symptoms in both animal and human hosts and on rare occasions is even a fatal condition in humans [20].
In this context, we investigated the faecal DNA prevalence of A. phagocytophilum in lesser horseshoe bat (Rhinolophus hipposideros) maternity colonies located in the Franche-Comté region (eastern France). The lesser horseshoe bat is a non-migratory species, with winter and maternity roosts located less than 30 kms apart [21]. Females roost at the end of spring when they can form large colonies (generally 10–200 females) to give birth and raise their single offspring throughout the summer [22]. Maternity roosts are generally located in close proximity to humans, in old buildings such as churches or barns.
The aims of the present study were (i) to assess the prevalence of lesser horseshoe bats carrying A. phagocytophilum DNA in their faeces; and (ii) to explore whether bacterial DNA prevalence in bat faeces varied among colonies and was related to colony size. The tendency of bat females to share roosts, crowded together reaching large population densities, creates the potential for the frequent transmission of ectoparasites, which can transmit pathogenic microorganisms. In vespertilionid bats, adult females can host more ectoparasites than males, likely due to the clustering of many female bats in maternity colonies or to the physiological stresses of reproduction [23–25]. We thus hypothesised that roosts with larger lesser horseshoe bat maternity colonies would have higher vector-borne bacterial DNA faecal prevalence. Taking advantage of recent developments in non-invasive approaches in wildlife studies, the bats were studied exclusively through sampling and analysing 552 faecal samples from 23 maternity roosts.
Materials and methods
Ethics statement
Field sampling was carried out with the authorisation of the owners of the buildings hosting bat roosts. This study was based on droppings, no animal was captured or disturbed during the study period. No particular permit was required.
Faecal collection and storage
Faecal samples were collected from 23 lesser horseshoe bat maternity roosts located in three administrative departments of the Franche-Comté region, eastern France: Doubs (13 maternity roosts), Haute-Saône (three maternity roosts) and Jura (seven maternity roosts) (Fig. 1). All roosts were located in buildings (churches, barns) surrounded mainly by woodlands and shrublands, grasslands and arable lands [26]. Sampling was conducted in June–July 2011, when individuals were abundant within the roost and before the birth of offspring. Plastic sheets were placed on the ground beneath the main clusters of bats and were left for 15 days. In each roost, 24 faeces samples were randomly collected directly from the plastic sheets, placed individually in 2 ml microtubes containing 90% alcohol and stored at room temperature.
Fig. 1.
Locations of the 23 lesser horseshoe bat maternity roosts sampled in the Franche-Comté region.
Because samples were collected without identification of the individual that excreted the faeces, each sample was genotyped to assign it to an individual. To check the expected sex structure of the colonies (i.e. essentially pregnant and lactating females and a few isolated males), all individuals were sexed using molecular typing. DNA extraction from bat faeces, genotyping and molecular sexing are detailed in [26] and in the Technical Appendix 1.
Molecular detection of A. phagocytophilum in bat faeces
The faecal DNA prevalence of A. phagocytophilum was measured using molecular diagnosis with the DNA extracted from faeces. Only samples with a reliable genotype were tested for the presence of bacterial DNA. Because bacterial DNA in bat faeces was expected to be fragmented, we used polymerase chain reaction (PCR) primers amplifying short DNA sequences. The amplification of A. phagocytophilum was performed with the PCR primers Msp2 – 3F (5′ – CCAGCGTTTAGCAAGATAAGAG – 3′) and Msp2 – 3R (5′ – GCCCAGTAACAACATCATAAGC – 3′), amplifying a 334 bp fragment of the msp2 gene [27]. The PCR amplifications were conducted in a reaction mixture (25 µl) consisting of 2 µl of DNA extract, 1 × HotStarTaq Master Mix (Qiagen, Courtaboeuf, France), 0.2 µM of each primer and PCR-grade water. A negative control (PCR-grade water) was included for every 12 samples. The amplification cycling programme consisted of an activation step of 15 min at 95 °C followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 60 s and primer extension at 74 °C for 90 s. A final extension was performed at 72 °C for 10 min. The PCR products were separated and visualised using the QIAxcel device and a QIAxcel DNA high-resolution kit (Qiagen).
Samples PCR-positive for A. phagocytophilum were confirmed by sequencing. The amplified products were purified using the QIAquick PCR Purification Kit (Qiagen) according to the manufacturer's instructions. Direct sequencing of the PCR products was performed with an automated sequencer (Applied Biosystems 3130 Genetic Analyzer) using the primers employed for the PCR reactions. A homology search of the sequences generated in this study was then performed in February 2016 by conducting an online search with the NCBI Basic Local Alignment Search Tool for nucleotides (BLASTN) in the GenBank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Faecal DNA prevalence
Because several faecal samples could have been excreted by the same individual, each sample was assigned to one individual with the genotyping analyses. Among the 425 samples with a consensus genotype obtained for the eight microsatellite markers, 199 were unique and 79 had a genotype that was present at least twice in the dataset. Some of the individuals with at least two tests for A. phagocytophilum might have discordant diagnostics during the sampling period (15 days). The faecal DNA prevalence was then estimated by resampling to take into account that individuals could have both positive and negative PCR results. To estimate the prevalence of A. phagocytophilum in each bat colony, the number of DNA-positive individuals was divided by the total number of individuals sampled in the maternity roost. One sample was randomly selected for each individual and the faecal DNA prevalence was calculated. This procedure was repeated 100 times to estimate a mean individual faecal prevalence per maternity roost, i.e. the mean number of DNA-positive individuals among the 100 replicates divided by the number of individuals sampled in the maternity roost. This faecal DNA prevalence represents the mean proportion of individuals with bacterial DNA in their faeces. We estimated 95% binomial confidence intervals of the faecal DNA prevalence using a bootstrap procedure and the R package Hmisc.
Colony size
A statistical approach was used to evaluate whether colony size could be related to faecal bacterial DNA prevalence. Bat colony size is sometimes difficult to readily assess by visual estimation within maternity roosts. The number of bats in the roost varies daily and individuals are difficult to count when they are located in inaccessible places or when they are flying within the roost. The size of each colony was thus estimated via statistical methods, using the sequential Bayesian estimator method developed by Petit and Valière [28] and implemented in the R 2.14.0 software (R Development Core Team. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria, 2011. http://www.R-project.org) using a script provided by Eric Petit (University Rennes 1). A bootstrap-method for correlations using a Spearman rank correlation test in the R software was used to investigate the relationships between the prevalence of A. phagocytophilum DNA and colony size.
Results
A total of 552 samples were collected from 23 maternity colonies. We obtained 425 (76%) consistent genetic profiles from 278 distinct individuals (9–20 individuals per colony, Table 1). Among the 278 individuals sampled, 213 were females and four were males (61 were undetermined due to a failure in PCR-control amplification). With colonies containing from 16 to 153 individuals (according to statistical estimates), a variable portion of the colony was sampled in each roost (the number of individuals sampled and the estimates of colony size are given Table 1).
Table 1.
Samples collected in 23 lesser horseshoe bat maternity roost during June–July 2011
| Department | Roost | Effective size | Genotypes | Individuals | PCR-positive | Faecal DNA prevalence |
|---|---|---|---|---|---|---|
| Doubs | AL | 153 (60; 244) | 22 | 20 | 1 | 0.05 (0.00; 0.24) |
| BA | 24 (14; 44) | 20 | 12 | 1 | 0.08 (0.00; 0.35) | |
| BE | 49 (20; 124) | 19 | 14 | 0 | 0.00 (0.00; 0.22) | |
| CD | 16 (10; 28) | 17 | 9 | 4 | 0.44 (0.19; 0.81) | |
| CL | 30 (17; 56) | 22 | 14 | 11 | 0.79 (0.52; 0.92) | |
| CM | 18 (12; 29) | 21 | 11 | 2 | 0.18 (0.05; 0.48) | |
| EC | 82 (28; 246) | 20 | 16 | 0 | 0.00 (0.00; 0.19) | |
| EP | 27 (15; 50) | 21 | 13 | 2 | 0.15 (0.04; 0.42) | |
| ET | 16 (10; 28) | 17 | 9 | 3 | 0.33 (0.06; 0.64) | |
| FO | 59 (20; 181) | 17 | 13 | 1 | 0.08 (0.00; 0.42) | |
| LP | 37 (19; 76) | 22 | 15 | 0 | 0.00 (0.00; 0.30) | |
| RA | 51 (18; 158) | 16 | 12 | 5 | 0.42 (0.19; 0.68 | |
| SO | 39 (19; 88) | 20 | 14 | 6 | 0.43 (0.16; 0.67) | |
| Haute-Saône | CH | 24 (14; 44) | 20 | 12 | 1 | 0.08 (0.00; 0.45) |
| EM | 31 (15; 68) | 12 | 10 | 1 | 0.10 (0.01; 0.40) | |
| FB | 38 (14; 115) | 14 | 10 | 1 | 0.10 (0.01; 0.40) | |
| Jura | AM | 16 (11; 25) | 23 | 11 | 9 | 0.82 (0.52; 0.95) |
| AR | 28 (12; 70) | 15 | 10 | 4 | 0.40 (0.17; 0.69) | |
| BT | 107 (36; 231) | 11 | 10 | 1 | 0.10 (0.00; 0.40) | |
| CV | 20 (10; 43) | 20 | 10 | 1 | 0.10 (0.01; 0.40) | |
| GI | 18 (12; 29) | 21 | 11 | 0 | 0.00 (0.00; 0.26) | |
| MF | 38 (16; 95) | 17 | 12 | 3 | 0.25 (0.09; 0.53) | |
| OF | 18 (11; 33) | 18 | 10 | 6 | 0.60 (0.31; 0.83) | |
| Total | 469 (417; 530) | 425 | 278 | 63 | 0.23 (0.18; 0.28) |
Department, administrative French department; Roost, code name of the village hosting the maternity roost; Effective size, estimated number of individuals in the colony based on a sequential Bayesian estimator; Genotypes, number of reliable genotypes based on 24 collected faeces samples; PCR-positive, mean number of individuals positive after 100 resampling tests for A. phagocytophilum; Individuals, number of distinct individuals; Faecal DNA prevalence, mean number of individuals positive/number of distinct individuals, numbers in brackets are confidence intervals (95%) based on a bootstrap procedure.
Sequence analysis of DNA from samples PCR-positive for A. phagocytophilum revealed one unique DNA sequence (see Technical Appendix 2). The closest match in Genbank was A. phagocytophilum (GenBank accession number KX591651) with a similarity score of 100% with the sequence 11AL05 (292/292).
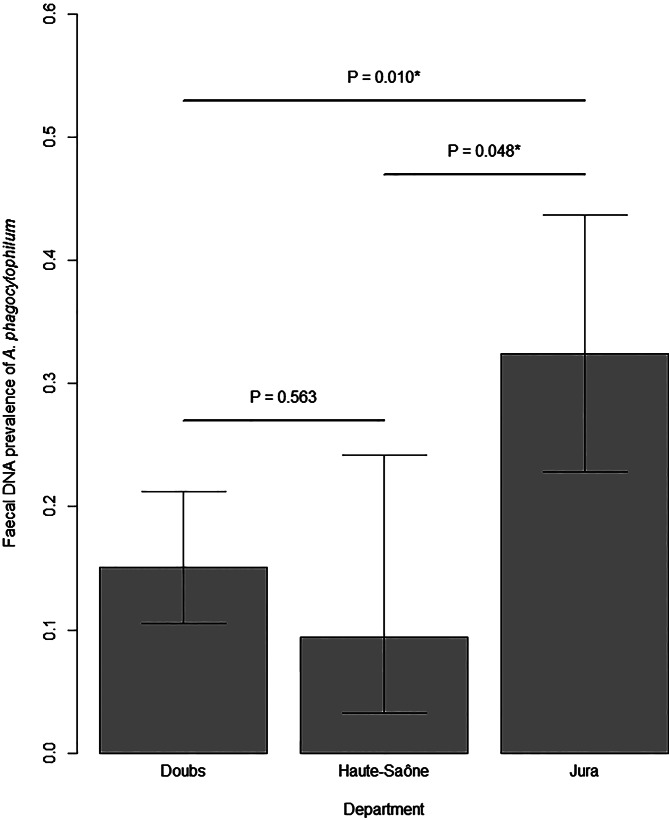
Anaplasma phagocytophilum DNA was detected in 63 of the 278 individuals (0.23, 95% CI 0.18–0.28). The faecal DNA prevalence of A. phagocytophilum was highly variable depending on the colony, with values between 0.00 and 0.82 (Table 1). The faecal DNA prevalence of A. phagocytophilum was significantly the highest in the Jura Department (Fig. 2). The faecal DNA prevalence was not related to the effective size of the colony estimated by a sequential Bayesian estimator and a bootstrap procedure using a Spearman's rank correlation test (P = 0.880).
Fig. 2.
Faecal DNA prevalence of Anaplasma phagocytophilum in lesser horseshoe bats sampled in three departments of the Franche-Comté region. Probabilities resulting from the pairwise comparison of proportional tests are given for each pair of departments. Significant differences in the faecal DNA prevalence of two departments are symbolised by * (P < 0.05).
Discussion
This study is the first investigation of the faecal DNA prevalence of A. phagocytophilum in a rhinolophid bat species and provides evidence that A. phagocytophilum DNA is prevalent in bat populations. DNA from Anaplasma spp. has been previously detected in mites (Spinturnix psi) collected off bats [9]. In the present study, we report the faecal prevalence of A. phagocytophilum DNA in 23 lesser horseshoe bat colonies located in France based on the sampling of 278 individuals. The overall proportion of individuals with bacterial DNA in their faeces was high (22.7%). Such a high prevalence of bacterial DNA in lesser horseshoe bat colonies might suggest persistent infection of long-lived bats, as proposed for Bartonella sp. in other bat species [12]. This high prevalence could also reflect the presence of bacterial DNA in the insect preys of the bats (which can include mosquitoes), which is largely unknown.
Individual faecal DNA prevalence was highly variable among lesser horseshoe bat colonies (0–82%). One can assume that the high bacterial DNA prevalence reported in some roosts can indirectly reflect high exposure of bats to ectoparasites. Interestingly, faecal DNA prevalence was the highest in the Jura Department. According to the French National Institute for Health Survey, Jura Department is the department of the study area with the highest number of reported cases of tick bites in humans and the highest density of ticks (unpublished data).
The variability in prevalence among colonies was not related to the colony size. This result contradicts our hypothesis that roosts with larger lesser horseshoe bat colonies would have a higher prevalence of vector-borne bacteria. We speculated that if large colonies can be associated with high ectoparasite loads, as shown in the big brown bat, Eptesicus fuscus [25], the chance that an arthropod vector carrying bacteria will parasitise a bat would be the highest in large colonies. Our finding could reflect the high complexity in the study of vector-borne microorganism transmission in bats. Furthermore, the faecal DNA prevalence of bacteria is not directly related to the proportion of infested hosts, as it may reflect both bat bleeding into the intestine or ingestion of ectoparasites carrying bacteria [14].
The high prevalence of A. phagocytophilum in faecal material in close proximity to humans (churches and barns in rural villages) raises one question: to what extent is high bacterial DNA prevalence in bat faeces a concern for humans? In the present study, all the maternity colonies were located in buildings, mainly in church bell towers. Sometimes, guano had spread onto the stairs leading to the bell tower or even into the church nave. The churchgoers regularly participate in the cleaning of the church, but the guano in the bell tower is removed more rarely, only when it reaches substantial quantities. Although we can assume that A. phagocytophilum cannot be transmitted by bat bites and likely not via direct contact with faeces, it is not excluded that bat ectoparasites can directly transmit bacteria to humans frequenting bell towers. However, even though anaplasmosis is an emerging disease in France, human cases remain rare [29], contrasting with the high faecal DNA prevalence found in bat guano. However, in Europe, human prevalence studies on tick-borne diseases are mainly conducted on workers exposed to tick bites, such as forestry workers or farmers [30, 31]. Our study shows that surveys should also pay particular attention to churchgoers that frequent places of worship sheltering bat colonies to evaluate the risks of disease transmission by tick bites in such communities.
Even if bat guano is unlikely to directly transmit haemotrophic bacteria to other hosts, bats appear as epidemiologically connected to them. A future perspective of this work would be to investigate if blood-feeding arthropods parasitising bats would be good bioindicators of risk transmission to other hosts, including humans. Moreover, because bat guano can reflect both bat and insect preys A. phagocytophilum contamination, it could be easily used as a bioindicator of the spread of this bacterium in the environment. In the present study, we highlighted a high variability in faecal DNA prevalence of A. phagocytophilum among colonies and we assume that such data could serve as a basis for eco-epidemiological studies, for example by linking prevalence data with landscape components. Bats have been proposed as bioindicator taxa that show measurable responses to climate change, habitat loss and environmental pollution [32, 33] and we assume that they also could serve as a good indicator of bacteria circulation in ecosystems. Bats exhibit several life-history traits favourable to disease transmission: the colonial structure of their populations, close physical contact, heavy ectoparasite infestations and a long lifespan. Moreover, bat guano is an easy-to-collect material which analysis recently benefited from many technical optimisations.
To conclude, further research is needed to clearly understand the high faecal DNA prevalence of A. phagocytophilum we found in this study. The first point to elucidate is probably from which host (bat and/or blood-feeding arthropods) the DNA from A. phagocytophilum comes from. Bats with their guano positive for A. phagocytophilum DNA and showing a positive blood test and clinical signs might provide some answer elements. Because bat captures are difficult to proceed in maternity roosts for ethical reasons, one can hardly imagine collecting their body condition as well as the presence of bacteria in their blood and/or their ectoparasites. However, such a study might be considered on corpses sometimes found in roosts and could be envisaged with a large screening of arthropod-borne bacteria in both bats and their ectoparasites.
Acknowledgements
The authors sincerely thank Sébastien Roué and all the persons who participated in the field work and in genotyping analyses, especially Pierline Tournant and Pierre-Emmanuel Baurand. The authors also thank Eric Petit for providing an R script for the estimation of bat colony size and Kristin Muehldorfer and two anonymous reviewers for their helpful advices on previous versions of the manuscript. The study sites in Franche-Comté belong partly to the Long Term Ecological Research (LTER) Network ‘Zone Atelier Arc Jurassien’ (http://zaaj.univ-fcomte.fr/).
Financial support
This work was supported by the Région Franche-Comté, the Fondation pour la Recherche sur la Biodiversité (FRB) and the French Ministry of Ecology, Energy, Sustainable Development and the Sea (ITTECOP program) as part of the Graphab project managed by the USR 3124 MSHE Ledoux.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268818001279.
click here to view supplementary material
Conflict of interest
None.
References
- 1.Daszak P, Cunningham AA and Hyatt AD (2000) Wildlife ecology – emerging infectious diseases of wildlife – threats to biodiversity and human health. Science 287, 443–449. [DOI] [PubMed] [Google Scholar]
- 2.Calisher CH et al. (2006) Bats: important reservoir hosts of emerging viruses. Clinical Microbiology Reviews 19, 531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayman DTS et al. (2013) Ecology of zoonotic infectious diseases in bats: current knowledge and future directions. Zoonoses and Public Health 60, 2–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brook CE and Dobson AP (2015) Bats as “special” reservoirs for emerging zoonotic pathogens. Trends in Microbiology 23, 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allocati N et al. (2016) Bat–man disease transmission: zoonotic pathogens from wildlife reservoirs to human populations. Cell Death Discovery 2, 16048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han HJ et al. (2015) Bats as reservoirs of severe emerging infectious diseases. Virus Research 205, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loftis AD et al. (2005) Detection of Rickettsia, Borrelia, and Bartonella in Carios kelleyi (Acari: Argasidae). Journal of Medical Entomology 42, 473–480. [DOI] [PubMed] [Google Scholar]
- 8.Reeves W et al. (2005) Molecular evidence for novel bartonella species in Trichobius major (Diptera: Streblidae) and Cimex adjunctus (Hemiptera: Cimicidae) from two southeastern bat caves, U.S.A. Journal of Vector Ecology: Journal of the Society for Vector Ecology 30, 339–341. [PubMed] [Google Scholar]
- 9.Reeves WK, Dowling APG and Dasch GA (2006) Rickettsial agents from parasitic dermanyssoidea (Acari: Mesostigmata). Experimental & Applied Acarology 38, 181–188. [DOI] [PubMed] [Google Scholar]
- 10.Concannon R et al. (2005) Molecular characterization of haemoparasites infecting bats (Microchiroptera) in Cornwall, UK. Parasitology 131, 489–496. [DOI] [PubMed] [Google Scholar]
- 11.Kosoy M et al. (2010) Bartonella spp. in bats, Kenya. Emerging Infectious Diseases 16, 1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai Y et al. (2011) Bartonella spp. in bBats, Guatemala. Emerging Infectious Diseases 17, 1269–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin JW et al. (2012) Identification of novel Bartonella spp. in bats and evidence of Asian gray shrew as a new potential reservoir of Bartonella. Veterinary Microbiology 156, 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veikkolainen et al. (2014) Bats as reservoir hosts of human bacterial pathogen, Bartonella mayotimonensis. Emerging Infectious Diseases 20, 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill JS et al. (2004) Detection of human blood in the bat tick Carios (Ornithodoros) kelleyi (Acari: Argasidae) in Iowa. Journal of Medical Entomology 41, 1179–1181. [DOI] [PubMed] [Google Scholar]
- 16.Banskar S et al. (2016) Microbiome analysis reveals the abundance of bacterial pathogens in Rousettus leschenaultia guano. Scientific Reports 6, 36948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dietrich M et al. (2017) The excreted microbiota of bats: evidence of niche specialisation based on multiple body habitats. FEMS Microbiology Letters 364, fnw284. [DOI] [PubMed] [Google Scholar]
- 18.Mühldorfer K (2013) Bats and bacterial pathogens: a review. Zoonoses and Public Health 60, 93–103. [DOI] [PubMed] [Google Scholar]
- 19.Stuen S (2007) Anaplasma Phagocytophilum – the most widespread tick-borne infection in animals in Europe. Veterinary Research Communications 31, 79–84. [DOI] [PubMed] [Google Scholar]
- 20.Ismail N, Bloch KC and McBride JW (2010) Human ehrlichiosis and anaplasmosis. Clinics in Laboratory Medicine 30, 261–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaisler J and Chytil J (2002) Mark-recapture results and changes in bat abundance at the cave of Na Turoldu, Czech Republic. Folia Zoologica (Czech Republic) 51, 1–10. [Google Scholar]
- 22.Gaisler J (1963) The ecology of lesser horseshoe bat (Rhinolophus hipposideros hipposideros Bechstein, 1800) in Czechoslovakia, part I. Vestnik Ceskoslovenske Spolecnosti Zoologicke 27, 211–233. [Google Scholar]
- 23.Zahn A et al. (2008) Foraging activity of Rhinolophus hipposideros on the Island of Herrenchiemsee, Upper Bavaria. Mammalian Biology – Zeitschrift Für Säugetierkunde 73, 222–229. [Google Scholar]
- 24.Zahn A and Rupp D (2004) Ectoparasite load in European vespertilionid bats. Journal of Zoology 262, 383–391. [Google Scholar]
- 25.Pearce RD and O'Shea TJ (2007) Ectoparasites in an urban population of big brown bats (Eptesicus fuscus) in Colorado. Journal of Parasitology 93, 518–530. [DOI] [PubMed] [Google Scholar]
- 26.Afonso E et al. (2016) Is the lesser horseshoe bat (Rhinolophus hipposideros) exposed to causes that may have contributed to its decline? A non-invasive approach. Global Ecology and Conservation 8, 123–137. [Google Scholar]
- 27.Massung RF and Slater KG (2003) Comparison of PCR assays for detection of the agent of human granulocytic ehrlichiosis, Anaplasma phagocytophilum. Journal of Clinical Microbiology 41, 717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petit E and Valiere N (2006) Estimating population size with noninvasive capture-mark-recapture data. Conservation Biology 20, 1062–1073. [DOI] [PubMed] [Google Scholar]
- 29.Edouard S et al. (2012) Emergence of human granulocytic anaplasmosis in France. Ticks and Tick-Borne Diseases 3, 403–405. [DOI] [PubMed] [Google Scholar]
- 30.Chmielewska-Badora J et al. (2012) Serological survey in persons occupationally exposed to tick-borne pathogens in cases of co-infections with Borrelia burgdorferi, Anaplasma phagocytophilum, Bartonella spp. and Babesia microti. Annals of Agricultural and Environmental Medicine 19, 271–274. [PubMed] [Google Scholar]
- 31.Rigaud E et al. (2016) Seroprevalence of seven pathogens transmitted by the Ixodes ricinus tick in forestry workers in France. Clinical Microbiology and Infection 22, 735.e1–735.e9. [DOI] [PubMed] [Google Scholar]
- 32.Jones G et al. (2009) Carpe noctem: the importance of bats as bioindicators. Endangered Species Research 8, 93–115. [Google Scholar]
- 33.Zukal J, Pikula J and Bandouchova H (2015) Bats as bioindicators of heavy metal pollution: history and prospect. Mammalian Biology-Zeitschrift für Säugetierkunde 80, 220–227. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268818001279.
click here to view supplementary material