Abstract
Background:
KRAS mutations are the most common oncogenic driver mutations of non-small cell lung cancer (NSCLC) in the Western world. Mutations of the KRAS gene are most prevalent in the patient population of current and former cigarette smokers. With the recent pivotal approval of a targeted inhibitor therapy for patients with KRAS p.G12C mutated and pretreated NSCLC, analysis of the heterogeneity of KRAS mutations and concomitant molecular alterations in patients with these tumors at all clinical stages is indicated.
Methods:
In this retrospective analysis, patient pathology records were reviewed for all cases receiving a pathologic diagnosis of NSCLC within our hospital system. All data were collected with IRB approval. Cases of indeterminate tumor type favoring a non-lung primary, as well as non-adenocarcinoma NSCLC (eg, squamous) were excluded from the cohort. In this hospital system, molecular testing for KRAS mutations is part of a molecular biomarker panel that is reflex ordered at initial diagnosis by the pathologist and may be performed as a single gene test or as a solid organ cancer hotspot panel by next generation sequencing. For each patient, KRAS mutational status and specific KRAS mutations, if present, were collated. Additional information assessed for this study included patient demographics (age, gender, and smoking history), tumor staging if available, PD-L1 expression levels by immunohistochemistry (IHC), and the presence of other genetic alterations (EGFR, ALK, and STK11).
Results:
Between January 1, 2017 and January 1, 2019, there were 276 patients diagnosed with NSCLC of all stages who had KRAS mutational analysis performed in our hospital system and who met the criteria for inclusion into the study cohort. A KRAS driver mutation was detected in 29% of these patients. The most frequently identified KRAS mutation was p.G12C (38%), followed by p.G12D (21%) and p.G12V (13%). KRAS-mutated lung adenocarcinoma was significantly associated with current or former patient smoking status in this cohort (29/202 (14%) smokers and 1/74 (1%) non-smokers; P = .0006). PD-L1 expression of at least 1% by IHC was present in 43% of KRAS-mutated lung adenocarcinomas and 45% of non-KRAS-mutated adenocarcinomas. In this study, KRAS mutations were not found to co-occur with gene alterations in EGFR, ALK, or STK11. In 48% of cases, at least one genetic alteration (KRAS, ALK, EGFR, or STK11) was identified.
Conclusions:
In this study cohort, KRAS-mutated lung adenocarcinoma demonstrated significant mutational heterogeneity, which is consistent with previously published studies. KRAS mutational status was also significantly associated with a current or former smoking history. Notably, p.G12C was the most frequently identified KRAS mutation in this cohort, with a frequency of 38%. This finding is particularly relevant given the recent approval of a KRAS p.G12C-specific targeted inhibitor therapy and the continued development of additional KRAS targeted therapies that may prove effective in treating NSCLC. These findings also highlight the necessity of considering molecular testing for KRAS mutations in patients with NSCLC and a smoking history, as this population most frequently harbors KRAS mutations and may benefit from these emerging targeted therapies.
Keywords: Lung adenocarcinoma, KRAS mutations, Molecular biomarkers, oncogene proteins, targeted therapy
Introduction
Lung cancer accounts for 11.6% of diagnosed malignancies and is the leading cause of cancer related death worldwide, with a 5-year overall survival of 16.8%.1,2 Lung cancer is divided into 2 main histological subsets; small cell carcinoma (SCLC) and non-small cell lung cancer (NSCLC). NSCLC comprises nearly 85% of all lung cancers, with adenocarcinoma comprising the primary subtype (approximately 60%). 3 While lung cancer is significantly associated with smoking, 15% of NSCLC cases in men and 53% in women occur in patients who have no reported smoking history. 4
The Cancer Genome Atlas (TCGA) performed genomic studies of NSCLC, which reported substantial molecular diversity.5,6 Common gene alterations in lung adenocarcinoma include mutations in EGFR, BRAF, ERBB2 exon 20 insertions, mutations causing MET exon 14 skipping, and translocations in ALK, ROS1, and RET. 7 However, most lung adenocarcinomas either harbor a KRAS mutation or are devoid of a known oncogenic driver mutation. The RAS family of genes encode small enzymes that hydrolyze guanosine triphosphate (GTPases), linking upstream cell surface receptors including EGFR, FGFR, and ERBB2-4 to downstream proliferation and survival pathways including RAF-MEK-ERK, PI3K-AKT-mTOR, and RALGDS-RA. 8 The RAS genes include KRAS, NRAS, and HRAS, and mutations of these genes are frequent in various malignancies, occurring in approximately 30% of all human cancers. 9 KRAS accounts for 86% of RAS-mutated cancer cases, while NRAS is mutated in 11% and HRAS in 3%. 10
Mutated KRAS is the most frequently detected oncogene in NSCLC, occurring in 20% to 40% of patients with lung adenocarcinoma, and is more commonly detected in Western populations and current or former smokers. 11 The most frequently reported KRAS mutations in NSCLC include p.G12C, p.G12V, and p.G12D. Currently, there is no recommendation for laboratory testing of KRAS mutational status in NSCLC, 2 though the presence of a KRAS mutation does convey key prognostic and therapeutic information that may be used in the clinical management of patients with NSCLC. The KRAS p.G12C mutation holds particular relevance in the clinical setting. A recently published phase 1 trial reported the confirmed objective response and disease control of heavily pretreated advanced solid tumors, including NSCLC, with the KRAS p.G12C inhibitor Sotorasib (AMG510).12,13 This drug has very recently received U.S. Food and Drug Administration (FDA) approval in patients with locally advanced or metastatic NSCLC. Additional KRAS p.G12C specific inhibitors and other KRAS targeted inhibitors are currently in earlier stages of development.14 -16
KRAS mutations in NSCLC that co-occur with other known genetic alterations have also been a significant area of interest. Some studies have reported that KRAS-mutated lung adenocarcinoma may have differential sensitivities to treatment with certain chemotherapeutic agents and immune checkpoint inhibitors in the presence of specific concomitant genetic alterations.17(p1),18(p1),19 The efficacy of these novel treatments may be associated with the presence of co-mutations, especially TP53.17(p53),19(p53). The most frequently reported gene alterations co-occurring with KRAS mutations in NSCLC are TP53 (40%), STK11 (32%), and CDKN2A (19.8%). 20
With recent advancements in personalized medicine, including the approval of a KRAS mutation-specific targeted inhibitor therapy and other KRAS targeted therapies in development and clinical trials, the possibilities of precision driven medicine in patients with NSCLC are now a reality. It is therefore necessary to better define the KRAS mutations and co-occurring genetic alterations in NSCLC at all stages within a “real world” standard patient population, and to determine the distribution of these patients that may be candidates for targeted therapies. The aim of this retrospective study was to determine the proportion and characteristics of KRAS-mutated lung adenocarcinoma in a standard patient population with reflex ordered molecular biomarker testing at diagnosis, to specifically examine the frequency of KRAS p.G12C mutations, and to assess for concomitant EGFR, ALK, STK11 gene alterations and PD-L1 expression status by IHC which may have prognostic and therapeutic implications for patients with KRAS-mutated lung adenocarcinoma.
Materials and Methods
The cohort for this retrospective study includes patients with a pathologic diagnosis of lung adenocarcinoma who received molecular testing of their tumor for KRAS mutations within our multi-hospital system over a period of 2 years. All data for this study was collected with IRB approval. A data review using the laboratory information system SoftPathDx (SCC Soft Computer) was performed to query all cases of lung adenocarcinoma from January 1, 2017 to January 1, 2019 diagnosed in our hospital system. Inclusion criteria included all cases (biopsies and surgical specimens, all stages) with the search terms “lung,” “adenocarcinoma,” and “MOLECULAR DIAGNOSTICS” in the interpretation or results free-text fields which were signed out within the above dates. Surgical pathology cases with a diagnosis of “adenocarcinoma” were included, only if a radiographically diagnosed lung mass was identified to the exclusion of any other radiographically diagnosed malignancy. Exclusion criteria included: all cytopathology cases without the mention of lung as part of the diagnostic report and all cases with evidence of a non-lung origin for the tumor. Each of these excluded cases were manually reviewed to confirm that the final surgical pathology report did not describe a primary or metastatic lung adenocarcinoma. Cases of indeterminate tumor type favoring a non-lung primary, as well as non-adenocarcinoma NSCLC (eg, squamous) were excluded from this cohort. Patient demographic information, such as age, gender, and cigarette smoking history, and tumor staging information if available was also collected for each case of lung adenocarcinoma included in the study cohort. Only pathologic staging was considered; clinical staging for patients without pathologic staging (eg, biopsy) was not analyzed in this study.
Within our hospital system, a panel of molecular biomarkers are reflex ordered by the pathologist at the time of initial diagnosis of lung adenocarcinoma at all stages. 21 This reflex ordered panel includes analysis for gene mutations in EGFR and KRAS, gene rearrangements in ALK, and PD-L1 expression by immunohistochemistry, in addition to other molecular biomarkers. Molecular testing for tumor-associated gene mutations in EGFR and KRAS at our institution may be performed clinically on DNA or total nucleic acid (TNA) extracted from formalin fixed, paraffin embedded (FFPE) tumor tissue either as single gene testing using polymerase chain reaction followed by single base extension and mass spectrometry genotyping (iPLEX HS Lung assay panel and Sequenom MassARRAY instrument, Agena Biosciences), or as part of a solid organ tumor hotspot panel by next generation sequencing (Ion AmpliSeq Cancer Hotspot v2 panel and Ion Proton instrument, Life Technologies) for extended mutational analysis if requested by the clinician. The Ion AmpliSeq Cancer Hotspot panel for solid organ tumors targets over 200 regions in 50 cancer related genes (list of genes covered by this panel is included in Supplemental Table 1). Molecular testing for tumor-associated gene rearrangements in ALK was performed on RNA or TNA extracted from FFPE tumor tissue using a targeted next generation sequencing panel for cancer translocations in lung tumors (FusionPlex Comprehensive Thyroid and Lung CTL panel, Archer Dx and NextSeq550 instrument, Illumina). All patient specimens included in this study cohort received molecular testing for the presence of gene alterations in KRAS, EGFR, and ALK as detailed above. For a subset of the cases, an extended gene mutation analysis was performed using the next generation sequencing panel and STK11 mutation status was also available.
For each patient included in the study cohort, KRAS mutational status and specific mutations, if present, were collated. PD-L1 status by immunohistochemistry (IHC) and the concomitant presence of other gene alterations (EGFR, ALK, and STK11) were also collected for each case. Pathologic staging information was only available in those patients that underwent a lobectomy or resection procedure; staging information was not available in cases where the diagnosis of lung adenocarcinoma was made by tissue biopsy. PD-L1 expression by immunohistochemistry was performed using a clinically validated laboratory developed test according to standard procedure at Houston Methodist Hospital (SP142 antibody, Ventana). The percentage of tumor cells expressing PD-L1 was recorded, and PD-L1 immunohistochemical interpretation followed the same criteria as the Atezolizumab trial using the Ventana SP142 antibody 22 and as described previously by our instutition. 23 Less than 1% staining of tumor cells and inflammatory cells was considered as “negative,” greater than 50% staining of tumor cells or 10% staining of inflammatory cells (of any intensity) was considered “high,” and intermediate staining proportion other than the above was considered “low.”
Statistical analysis and final visualization of results were completed in GraphPad Prism 7 and Adobe Illustrator. The RAWGraphs software suite was used to generate beeswarm plots (Figures 2 and 3). 24 In this study, Fisher’s exact test was used to calculate statistical significance for 2 × 2 contingency tables using categorical data. An unpaired student’s t-test was used to calculate statistical significance for continuous variables. Confidence intervals for proportions were calculated using the modified Wald method at 95% confidence level. A P-value less than .05 was considered statistically significant.
Figure 2.
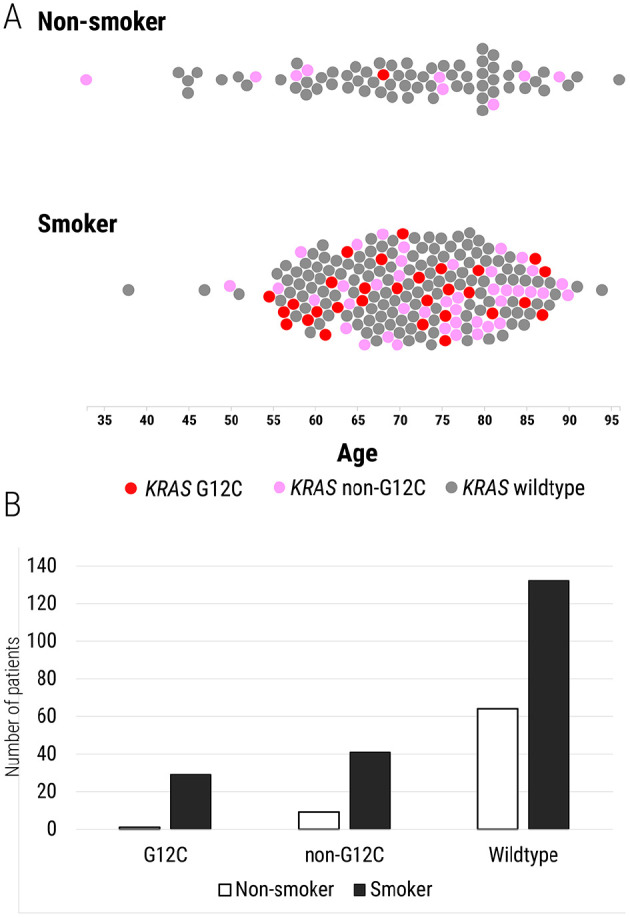
Distribution of KRAS p.G12C mutations in lung adenocarcinoma patients with a history of cigarette smoking (current and former) and those with no smoking history. (A) Each patient in the cohort is denoted by a circle. Those patients with KRAS p.G12C mutations identified are indicated with a red circle; all non-p.G12C KRAS mutations are indicated with a pink circle. Patients with KRAS-wildtype tumors are shown as a gray circle. The X-axis denotes the age of the patient. (B) Bar chart showing the number of current/former smokers (black) and non-smokers (white) with varying KRAS mutation status.
Figure 3.
Distribution of KRAS p.G12C mutations in female and male patients with lung adenocarcinoma. Bar chart shows the number of female (light blue) and male (dark blue) patients with varying KRAS mutation status.
Results
In our hospital system, 276 patients were identified who met the inclusion and exclusion criteria outlined above and were included in the study cohort. All patients in the cohort received molecular biomarker testing at our institution for gene alterations in KRAS, EGFR, and ALK. Gene mutation analysis for KRAS and EGFR was reflex-ordered and tested by the single gene methodology described in 261 of the 276 patient specimens included in the cohort (94.5% of cases). The solid organ tumor hotspot panel (Ion AmpliSeq Cancer Hotspot v2) was clinician-ordered for extended mutational analysis, including STK11 gene mutations, and was performed in 15 of the 276 (5.4%) patients included in this study cohort.
A KRAS mutation was identified in 80/276 (29%) lung adenocarcinomas included in this study. As shown in Figure 1, the specific KRAS mutations identified in this cohort included: p.G12C (30/80, 38%), p.G12D (17/80, 21%), p.G12V (10/80, 13%), p.G12S (7/80, 9%), p.G12A (4/80, 5%), p.G13D (4/80, 5%), p.Q61H (2/80, 3%), p.G12R (1/80, 1%), p.Q61R (1/80, 1%), p.Q61E (1/80, 1%), p.G13C (1/80, 1%), p.G13V (1/80, 1%), and p.Q61L (1/80, 1%) (Figure 1).
Figure 1.
Distribution of KRAS mutations. Left: Proportion of KRAS-wildtype and KRAS-mutated (in red) lung adenocarcinoma cases. Right: All KRAS mutations identified in the cohort, along with the number of cases harboring each specific mutation.
Overall, 86.5% of the patients included in this study cohort were over the age of 60 (Table 1). There was no significant statistical difference noted in KRAS mutational status with the age of the patient; a mean age of 72.0 versus 71.0 years was observed for KRAS mutant and wildtype tumors, respectively (CI = −1.74 to 3.74). The proportion of KRAS p.G12C mutations in this cohort was well-distributed with respect to patient age, though no p.G12C mutations were noted in patients under 50 years of age. Approximately two-thirds of the KRAS p.G12C mutated lung adenocarcinomas in this cohort occurred in patients between 60 and 79 years of age.
Table 1.
KRAS mutation status among different age groups of patients with lung adenocarcinoma.
| Age groups | KRAS G12C | KRAS non-G12C | KRAS wildtype | All patients |
|---|---|---|---|---|
| 30-39 | 1 | 1 | 2 | |
| 40-49 | 6 | 6 | ||
| 50-59 | 4 | 6 | 18 | 28 |
| 60-69 | 10 | 9 | 62 | 81 |
| 70-79 | 11 | 17 | 64 | 92 |
| 80-89 | 5 | 16 | 40 | 61 |
| 90-100 | 1 | 5 | 6 | |
| All patients | 30 | 50 | 196 | 276 |
| Mean age | 70.3 | 73.0 | 71.0 | 71.3 |
Abbreviation: KRAS, Kirsten rat sarcoma virus gene.
The number of patients in each age group for this cohort are shown according to their KRAS mutation status (KRAS wildtype, KRAS p.G12C-mutated, or KRAS non-p.G12C-mutated). The total number of patients in each KRAS mutation status group is shown in bold. The All patients column shows the total number of patients in each age group. The mean patient age for KRAS-wildtype, KRAS p.G12C-mutated, and KRAS non-p.G12C-mutated tumors is also shown.
Among the 80 patients with KRAS-mutated lung adenocarcinomas in this study cohort, 70 (88%) had a current or former history of cigarette smoking. There were 132/207 (64%) KRAS-wildtype cases in which the patient was a current or former smoker (Table 2). Compared to never smokers (1/74, 1% with KRAS p.G12C mutation), current or former smokers were more likely to harbor KRAS p.G12C mutations (OR 12.2; 95% CI 1.6-91.5, P = .0006) (Figure 2). Though not statistically significant, non-p.G12C KRAS mutations were also more frequently identified in current and former cigarette smokers (41/202, 20%) than in never smokers (9/74, 12%) (OR 1.83; 95% CI 0.85-4.0, P = .08).
Table 2.
KRAS mutation status stratified by patient cigarette smoking history.
| Smoking status | KRAS G12C | KRAS non-G12C | KRAS wildtype | All patients |
|---|---|---|---|---|
| Unknown | 1 | 5 | 6 | |
| Non-smoker | 1 | 8 | 59 | 68 |
| Former smoker | 23 | 36 | 97 | 156 |
| Current smoker | 6 | 5 | 35 | 46 |
| All patients | 30 | 50 | 196 | 276 |
Abbreviation: KRAS, Kirsten rat sarcoma virus gene.
The number of patients in each smoking status group for this cohort are shown according to their KRAS mutation status (KRAS wildtype, KRAS p.G12C-mutated, or KRAS non-p.G12C-mutated). The total number of patients in each KRAS mutation status group is shown in bold. The All patients column shows the total number of patients in each smoking status group.
There was no significant gender predilection noted among patients with KRAS-mutated lung adenocarcinoma in this study. As shown in Figure 3, there were a total of 46 female patients with KRAS-mutated lung adenocarcinoma out of 154 (30%) in this study, and there were 34 male patients with KRAS-mutated lung adenocarcinoma out of 122 (28%). Interestingly, KRAS p.G12C mutations were more frequently identified in female patients than male patients, though this finding was not statistically significant (46% females vs 26% males with KRAS p.G12C mutations), (OR 2.33; 95% CI = 0.90-6.08, P = .0636) (Figure 3, Table 3).
Table 3.
KRAS mutation status stratified by patient gender.
| Gender | KRAS G12C | KRAS non-G12C | KRAS wildtype | All patients |
|---|---|---|---|---|
| Female | 21 | 25 | 108 | 154 |
| Male | 9 | 25 | 88 | 122 |
| All patients | 30 | 50 | 196 | 276 |
Abbreviation: KRAS, Kirsten rat sarcoma virus gene.
The number of female and male patients included in this cohort are shown according to their KRAS mutation status (KRAS wildtype, KRAS p.G12C-mutated, or KRAS non-p.G12C-mutated). The total number of patients in each KRAS mutation status group are shown in bold. The All patients column indicates the total numbers of female and male patients in this study cohort.
KRAS mutations were found to be mutually exclusive with other recurrent genomic alterations in lung adenocarcinomas, including EGFR, ALK, and STK11 in this study (Figure 4). There were 44/276 (15%) EGFR-mutated, KRAS-wildtype cases, 8/276 (3%) ALK-rearranged, KRAS-wildtype cases, and 1/15 (7%) STK11-mutated, KRAS-wildtype case. Overall, 133 patients (48%) had at least one gene alteration (in KRAS, ALK, EGFR, or STK11) identified by molecular biomarker testing of their tumor tissue.
Figure 4.
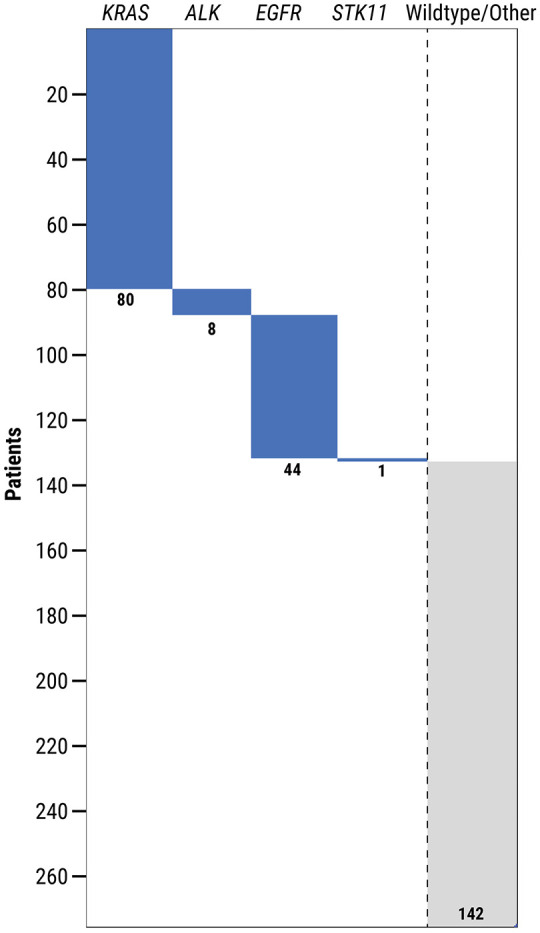
Co-occurrence of KRAS mutations with somatic alterations in ALK, EGFR, and STK11 genes in patients with lung adenocarcinoma. Shown is a submatrix plot with detected gene alterations denoted by a colored bar (blue). Patients without any of these specific genomic alterations are classified as “wild type” (gray).
In this study cohort, PD-L1 expression by IHC using the Ventana SP142 antibody was available for 242/276 (88%) of cases (Table 4). Of these cases, 134/242 (55%) were interpreted using our institutional criteria as negative, 66/242 (27%) were low, and 42/242 (17%) were high. No statistically significant difference in PD-L1 expression levels was noted between females (56/132, 42%) and males (52/110, 47%). PD-L1 expression levels were also not found to be significantly different between KRAS-mutated (29/67, 43%) and KRAS-wildtype tumors (79/175, 45%) in this cohort. In addition, the tumors harboring various KRAS mutations (p.G12C, p.G12V, p.G12D, etc.) also exhibited no statistically significant differences in the levels of PD-L1 expression in tumor cells, as assessed by IHC using the scoring criteria described. Representative histologic images of lung adenocarcinoma cases included in this study cohort showing high and low PD-L1 expression by IHC in KRAS-wildtype and KRAS-mutated tumors are shown in Supplemental Figure 1.
Table 4.
Analysis of PD-L1 expression by immunohistochemistry using the Ventana SP142 antibody.
| TC | IC negative | IC low | IC high | All patients |
|---|---|---|---|---|
| Negative | 134 | 14 | 5 | 153 |
| Low | 39 | 13 | 11 | 63 |
| High | 18 | 5 | 3 | 26 |
| All patients | 191 | 32 | 19 | 242 |
| KRAS mutation | PD-L1 negative | PD-L1 low | PD-L1 high | All patients |
| G12C | 11 | 9 | 5 | 25 |
| G12D | 8 | 2 | 3 | 13 |
| G12V | 5 | 2 | 1 | 8 |
| Other mutation | 14 | 5 | 2 | 21 |
| Wildtype | 96 | 48 | 31 | 175 |
| All patients | 134 | 66 | 42 | 242 |
Abbreviation: KRAS, Kirsten rat sarcoma virus gene; IC, inflammatory cell (expression); PD-L1, programed death-ligand 1; TC, tumor cell (expression).
Top: PD-L1 expression of tumor cells (TC) and inflammatory cells (IC) at negative (<1% staining), low (<50% TC, <10% IC), and high levels, as defined by institutional criteria. Bottom: PD-L1 expression status versus KRAS mutation status. The total numbers of patients in each group are shown in bold.
Pathologic staging was available in only 17 of the 276 lung adenocarcinoma cases included in this study, all 17 were patients who had a lobectomy procedure performed. Of these cases, the following distribution of pathologic stage information was obtained: IA (1), IA1 (1), IA2 (5), IA3 (4), IB (7), IIB (3), IIIA (3), N/A (5). The T stage information was pT1a (1), pT1b (6), pT1c (4), pT2a (8), pT3 (2), pT4 (3), ypT1c (1), ypT2a (2), ypT3 (2). The N stage information was pN0 (24), pN2b (2), ypN1 (1), ypN2 (2). There were no pM1 cases in this dataset. No significant differences were noted in the frequency or distribution of specific KRAS mutations based on the tumor stage in this study cohort.
Discussion
KRAS mutations were identified in 29% of the lung adenocarcinomas included in this study. This finding is comparable to the KRAS mutation frequencies previously described in the literature for NSCLC. The Cancer Genome Atlas and Clinical Lung Cancer Genome Project both report that mutated KRAS is identified in approximately 30% of lung adenocarcinomas.5,25 The most frequently mutated KRAS codon in this cohort was p.G12 (86%), followed by p.G13 (8%) and p.Q61 (6%). In this study, we identified an association between the presence of a KRAS mutation and a current or former patient smoking history, which is also consistent with previous reports.26 -29 In this study, we did not identify any significant differences in PD-L1 expression levels by IHC between KRAS-mutated and KRAS-wildtype lung adenocarcinomas using immunohistochemical analysis with the Ventana SP142 PD-L1 antibody.30,31(p1) We found that 43% of KRAS-mutated tumors had 1% or greater PD-L1 expression, which was comparable to the 45% of KRAS-wildtype tumors with 1% or greater expression. Additionally, no significant association with increased PD-L1 expression and the presence of specific KRAS mutations (such as p.G12C or p.G12V) was identified. Supplemental Figure 1 includes representative images of the morphology and high or low PD-L1 immunohistochemical expression levels in KRAS-wildtype and KRAS-mutated tumors included in this cohort.
It has been reported in some previous studies that KRAS mutations may uncommonly be accompanied by co-mutations in tumor suppressor genes, such as TP53 and STK11.7,32 In the current study, KRAS mutations were not found to co-occur with somatic alterations in EGFR, ALK, or STK11 genes. This may be more reflective of the molecular biomarker ordering practices in our hospital system and the community setting, rather than the actual prevalence of these co-occurring mutations in patients with KRAS-mutated lung adenocarcinoma. One potential limitation of this study is that only 15 patients received extended mutational analysis for molecular biomarkers performed using a targeted NGS panel approach, which includes mutational assessment of additional genes like STK11 or TP53. However, all patients included in this study did have gene mutation and gene rearrangement analysis performed for KRAS, EGFR, and ALK alterations, which are included in the reflex ordered molecular biomarker testing for lung adenocarcinomas approved within our hospital system. Studies examining the prognostic impact of co-occurring TP53 and/or STK11 mutations with KRAS mutations in NSCLC have thus far been inconclusive.33,34 Additional clinical trials are underway to further investigate the prognostic significance of co-occurring mutations, as well as responses to immune checkpoint inhibitor therapies. 35
One objective of this study was to examine the frequency and distribution of KRAS mutations in lung adenocarcinomas of all stages within a single hospital system, particularly in light of the recent approval of the KRAS p.G12C specific inhibitor Sotorasib (AMG510). 12 KRAS p.G12C mutations are identified in 13% of NSCLC cases overall; and the success of a direct KRAS inhibitor in patients with heavily pretreated advanced lung adenocarcinomas will likely alter the future therapeutic landscape for patients with KRAS mutated lung cancer. 36 In our study cohort, 29% of lung adenocarcinomas had a KRAS mutation identified, and 38% of those were KRAS p.G12C mutations. Among all lung adenocarcinomas included in this study, 10.9% had a KRAS p.G12C mutation identified. As expected, we found that a history of cigarette smoking was highly associated with presence of a KRAS p.G12C mutation. In the current study, KRAS p.G12C mutations were found to occur more frequently in female patients than in male patients, though that finding did not reach statistical significance. Interestingly, approximately two-thirds of the KRAS p.G12C mutations identified in this cohort of lung adenocarcinomas occurred in patients between the ages of 60 to 79, while no KRAS p.G12C mutations were identified in patients less than 50 years of age. In a subset of patients with KRAS-mutated NSCLC treated at our institution, we have observed a numerically lower 12-month lower survival rate in the cohort associated with KRAS p.G12C mutations, with over one-quarter of these patients having a history of previous and second primary malignancies. 37
The identification of specific oncogenic driver mutations in patients with lung adenocarcinoma has led to improved and timely targeted therapeutic options. One recently expanding area of inquiry has been the search for effective anti-RAS targeted inhibitors for KRAS-mutated lung adenocarcinomas and potentially other types of KRAS-mutated cancers as well. The most promising of these currently are specific small molecular inhibitors targeting the KRAS p.G12C mutation in NSCLC patients, though others are in various stages of development and clinical trials. Previous studies, including this one, indicate that a KRAS p.G12C mutation is present in approximately 13% of NSCLC and accounts for approximately half of all KRAS mutations occurring in NSCLC patients.33,38 In addition to the KRAS p.G12C inhibitor Sotorasib (AMG510), which has already been granted fast track designation for the treatment of previously treated metastatic NSCLC, clinical trials and further research are underway to interrogate potential targeted therapies for additional KRAS mutations including p.G12D and p.G12V.39 -42 Given the reported responses to this recently approved targeted KRAS inhibitor and the potential clinical benefit for many NSCLC patients, it will become even more important to identify those patients with KRAS p.G12C mutations and to further investigate the effects of concomitant mutations in this population. Additionally, updated testing recommendations for KRAS mutations in patients with NSCLC should be considered for appropriate and timely clinical management.
Conclusion
This study details a single hospital system’s retrospective experience regarding KRAS mutations in lung adenocarcinoma of all stages among a standard patient population. The frequency and distribution of KRAS mutations identified in our cohort was similar to previous reports in the literature. An association between current or former smoking status and the presence of KRAS mutations in NSCLC was also noted in this study, as previously reported. We did not identify any co-occurring genomic alterations of EGFR, ALK, or STK11 in our cohort of KRAS-mutated lung adenocarcinomas, though this assessment was somewhat limited by the infrequent utilization of a targeted NGS panel for extended gene mutational analysis in these patients. PD-L1 expression levels by immunohistochemistry in the context of KRAS mutation status were also examined in this study, and no differences were noted in the overall levels of expression between KRAS-wildtype and KRAS-mutated tumors or among those with varying specific KRAS mutations. Our institution has separately examined clinical outcomes and clinical staging for a subset of the KRAS-mutated adenocarcinomas identified in this study. 37
Overall, this study does describe KRAS mutation frequencies in a relatively large dataset of lung adenocarcinoma patients diagnosed in a large hospital system, which includes 6 community hospitals and an academic medical center. This study also serves to highlight the need for consideration of updated testing recommendations for KRAS mutation analysis in patients with NSCLC. Importantly, we were able to ascertain that over 10% of our lung adenocarcinoma patient population harbors a KRAS p.G12C mutation, which now has significant implications for targeted therapeutic options in NSCLC and clinical management of these patients.
Supplemental Material
Supplemental material, sj-docx-1-pat-10.1177_2632010X221102054 for Molecular Signatures of KRAS-Mutated Lung Adenocarcinoma: Analysis of Concomitant EGFR, ALK, STK11, and PD-L1 Status by Jim Hsu, Joseph F Annunziata, Ethan Burns, Eric H Bernicker, Randall J Olsen and Jessica S Thomas in Clinical Pathology
Supplemental material, sj-jpg-2-pat-10.1177_2632010X221102054 for Molecular Signatures of KRAS-Mutated Lung Adenocarcinoma: Analysis of Concomitant EGFR, ALK, STK11, and PD-L1 Status by Jim Hsu, Joseph F Annunziata, Ethan Burns, Eric H Bernicker, Randall J Olsen and Jessica S Thomas in Clinical Pathology
Footnotes
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of Conflicting Interests: The author declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: JST provided concept and design. JH, JFA performed data collection and analysis. EB provided selected data collection. JH, JFA wrote the manuscript with input from all authors. EHB, RJO, and JST provided multiple rounds of editing, revisions, and feedback. All authors read and approved the final manuscript.
Ethical Statements and Informed Consent: This study was a retrospective observational study with no cost or intervention for patients in the study. Data were collected solely from medical and pathologic records and anonymized. Basic demographic information (sex, age in years) was retained. Identifiers and other protected health information (PHI) remained confidential.
ORCID iDs: Jim Hsu  https://orcid.org/0000-0003-4803-4819
https://orcid.org/0000-0003-4803-4819
Jessica S Thomas  https://orcid.org/0000-0003-0053-3467
https://orcid.org/0000-0003-0053-3467
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [DOI] [PubMed] [Google Scholar]
- 2. Ettinger DS, Wood DE, Aggarwal C, et al. NCCN guidelines insights: non-small cell lung cancer, version 1.2020. J Natl Compr Canc Netw. 2019;17:1464-1472. [DOI] [PubMed] [Google Scholar]
- 3. Howlader N, Noone A, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2016. National Cancer Institute, SEER. Published April 2019. Accessed September 11, 2020. https://seer.cancer.gov/csr/1975_2016/ [Google Scholar]
- 4. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. [DOI] [PubMed] [Google Scholar]
- 5. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hammerman PS, Lawrence MS, Voet D, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. La Fleur L, Falk-Sörqvist E, Smeds P, et al. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer. 2019;130:50-58. [DOI] [PubMed] [Google Scholar]
- 8. Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11-22. [DOI] [PubMed] [Google Scholar]
- 9. Zinatizadeh MR, Momeni SA, Zarandi PK, et al. The role and function of ras-association domain family in cancer: a review. Genes Dis. 2019;6:378-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov. 2014;13:828-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ludovini V, Ricciuti B, Tofanetti F, et al. KRAS mutation and DNA repair and synthesis genes in non-small-cell lung cancer. Mol Clin Oncol. 2018;9:689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong DS, Fakih MG, Strickler JH, et al. KRASG12C inhibition with sotorasib in advanced solid tumors. N Engl J Med. 2020;383:1207-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217-223. [DOI] [PubMed] [Google Scholar]
- 14. Janes MR, Zhang J, Li LS, et al. Targeting KRAS mutant cancers with a covalent g12c-specific inhibitor. Cell. 2018;172:578-589.e17. [DOI] [PubMed] [Google Scholar]
- 15. Christensen JG, Olson P, Briere T, Wiel C, Bergo MO. Targeting krasg12c -mutant cancer with a mutation-specific inhibitor. J Intern Med. 2020;288:183-191. [DOI] [PubMed] [Google Scholar]
- 16. Nagasaka M, Li Y, Sukari A, Ou SI, Al-Hallak MN, Azmi AS. KRAS G12C Game of Thrones, which direct KRAS inhibitor will claim the iron throne? Cancer Treat Rev. 2020;84:101974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Biton J, Mansuet-Lupo A, Pécuchet N, et al. TP53, STK11, and EGFR mutations predict tumor immune profile and the response to anti-PD-1 in lung adenocarcinoma. Clin Cancer Res. 2018;24:5710-5723. [DOI] [PubMed] [Google Scholar]
- 18. Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8:822-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dong ZY, Zhong WZ, Zhang XC, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23:3012-3024. [DOI] [PubMed] [Google Scholar]
- 20. Adderley H, Blackhall FH, Lindsay CR. KRAS-mutant non-small cell lung cancer: converging small molecules and immune checkpoint inhibition. EBioMedicine. 2019;41:711-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anand K, Phung TL, Bernicker EH, Cagle PT, Olsen RJ, Thomas JS. Clinical utility of Reflex ordered testing for molecular biomarkers in lung adenocarcinoma. Clin Lung Cancer. 2020;21:437-442. [DOI] [PubMed] [Google Scholar]
- 22. Herbst RS, Giaccone G, de Marinis F, et al. Atezolizumab for first-line treatment of PD-L1–selected patients with NSCLC. N Engl J Med. 2020;383:1328-1339. [DOI] [PubMed] [Google Scholar]
- 23. Driver BR, Miller RA, Miller T, et al. Programmed Death Ligand-1 (PD-L1) expression in either tumor cells or tumor-infiltrating immune cells correlates with solid and high-grade lung adenocarcinomas. Arch Pathol Lab Med. 2017;141:1529-1532. [DOI] [PubMed] [Google Scholar]
- 24. RAWGraphs. Rawgraphs/Raw. RAWGraphs; 2020. Accessed October 26, 2020. https://github.com/rawgraphs/raw
- 25. Clinical Lung Cancer Genome Project (CLCGP), Network Genomic Medicine (NGM). A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5:209ra153. doi: 10.1126/scitranslmed.3006802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sholl LM, Aisner DL, Varella-Garcia M, et al. Multi-institutional oncogenic driver mutation analysis in lung adenocarcinoma: the Lung Cancer Mutation Consortium experience. J Thorac Oncol. 2015;10:768-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thunnissen FB, Prinsen C, Hol B, et al. Smoking history and lung carcinoma: KRAS mutation is an early hit in lung adenocarcinoma development. Lung Cancer. 2012;75:156-160. [DOI] [PubMed] [Google Scholar]
- 28. Kim HR, Ahn JR, Lee JG, et al. The impact of cigarette smoking on the frequency of and qualitative differences in KRAS mutations in Korean patients with lung adenocarcinoma. Yonsei Med J. 2013;54:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gou LY, Niu FY, Wu YL, Zhong WZ. Differences in driver genes between smoking-related and non-smoking-related lung cancer in the Chinese population. Cancer. 2015;121(Suppl 17):3069-3079. [DOI] [PubMed] [Google Scholar]
- 30. Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018-2028. [DOI] [PubMed] [Google Scholar]
- 31. Liu C, Zheng S, Jin R, et al. The superior efficacy of anti-PD-1/PD-L1 immunotherapy in KRAS-mutant non-small cell lung cancer that correlates with an inflammatory phenotype and increased immunogenicity. Cancer Lett. 2020;470:95-105. [DOI] [PubMed] [Google Scholar]
- 32. Bange E, Marmarelis ME, Hwang WT, et al. Impact of KRAS and TP53 co-mutations on outcomes after first-line systemic therapy among patients with STK11-Mutated advanced non-small-cell lung cancer. JCO Precis Oncol. 2019;3:1-11. doi: 10.1200/PO.18.00326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arbour KC, Jordan E, Kim HR, et al. Effects of Co-occurring genomic alterations on outcomes in patients with KRAS-Mutant Non-Small cell lung cancer. Clin Cancer Res. 2018;24:334-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Facchinetti F, Bluthgen MV, Tergemina-Clain G, et al. LKB1/STK11 mutations in non-small cell lung cancer patients: Descriptive analysis and prognostic value. Lung Cancer. 2017;112:62-68. [DOI] [PubMed] [Google Scholar]
- 35. Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET Registry. Ann Oncol. 2019;30:1321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fakih M, O’Neil B, Price TJ, et al. Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG 510, a novel small molecule KRASG12C inhibitor, in advanced solid tumors. J Clin Oncol. 2019;37:3003-3003. [Google Scholar]
- 37. Burns EA, Ensor JE, Hsu J, Thomas JS, Olsen RJ, Bernicker EH. Outcomes and prognostic contributors in patients with KRAS mutated non-small cell pulmonary adenocarcinomas: a single institution experience. J Thorac Dis. 2021;13:4785-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pakkala S, Ramalingam SS. Personalized therapy for lung cancer: striking a moving target. JCI Insight. 2018;3:120858. doi: 10.1172/jci.insight.120858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Elicio Therapeutics. First in human phase 1/2 trial of ELI-002 immunotherapy as treatment for subjects with Kirsten Rat Sarcoma (KRAS) mutated pancreatic ductal adenocarcinoma and other solid tumors. clinicaltrials.gov; 2021. Accessed August 1, 2021. https://clinicaltrials.gov/ct2/show/NCT04853017
- 40. Verastem, Inc. A phase 2 study of VS-6766 (dual RAF/MEK inhibitor) as a single agent and in combination with defactinib (FAK Inhibitor) in recurrent KRAS-mutant (KRAS-MT) non-small cell lung cancer (NSCLC). clinicaltrials.gov; 2021. Accessed August 1, 2021. https://clinicaltrials.gov/ct2/show/NCT04620330
- 41. Hung PS, Huang MH, Kuo YY, Yang JC. The inhibition of Wnt restrain KRASG12V-Driven metastasis in non-small-cell lung cancer. Cancers. 2020;12:E837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hofmann MH, Gmachl M, Ramharter J, et al. BI-3406, a potent and selective SOS1–KRAS interaction inhibitor, is effective in KRAS-driven cancers through combined MEK inhibition. Cancer Discov. 2021;11:142-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-pat-10.1177_2632010X221102054 for Molecular Signatures of KRAS-Mutated Lung Adenocarcinoma: Analysis of Concomitant EGFR, ALK, STK11, and PD-L1 Status by Jim Hsu, Joseph F Annunziata, Ethan Burns, Eric H Bernicker, Randall J Olsen and Jessica S Thomas in Clinical Pathology
Supplemental material, sj-jpg-2-pat-10.1177_2632010X221102054 for Molecular Signatures of KRAS-Mutated Lung Adenocarcinoma: Analysis of Concomitant EGFR, ALK, STK11, and PD-L1 Status by Jim Hsu, Joseph F Annunziata, Ethan Burns, Eric H Bernicker, Randall J Olsen and Jessica S Thomas in Clinical Pathology




