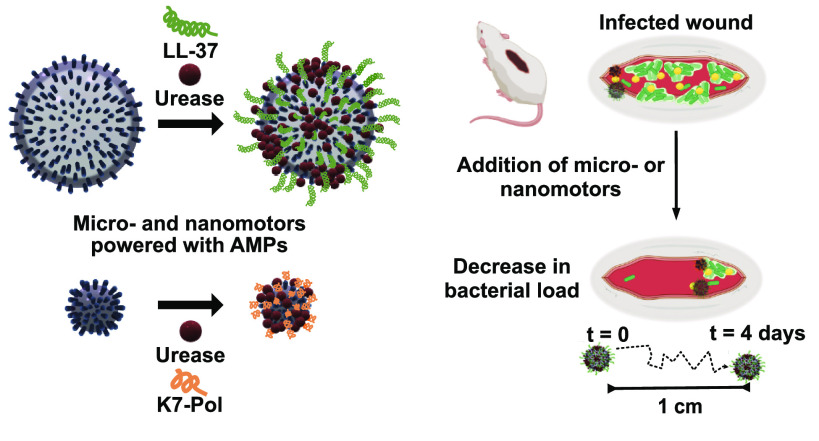
Abstract
The increasing resistance of bacteria to existing antibiotics constitutes a major public health threat globally. Most current antibiotic treatments are hindered by poor delivery to the infection site, leading to undesired off-target effects and drug resistance development and spread. Here, we describe micro- and nanomotors that effectively and autonomously deliver antibiotic payloads to the target area. The active motion and antimicrobial activity of the silica-based robots are driven by catalysis of the enzyme urease and antimicrobial peptides, respectively. These antimicrobial motors show micromolar bactericidal activity in vitro against different Gram-positive and Gram-negative pathogenic bacterial strains and act by rapidly depolarizing their membrane. Finally, they demonstrated autonomous anti-infective efficacy in vivo in a clinically relevant abscess infection mouse model. In summary, our motors combine navigation, catalytic conversion, and bactericidal capacity to deliver antimicrobial payloads to specific infection sites. This technology represents a much-needed tool to direct therapeutics to their target to help combat drug-resistant infections.
Keywords: nanomotors, antimicrobial peptides, nanoparticles, self-propulsion, bacterial infection, autonomous treatment
Introduction
Bacterial infections are predicted to kill 10 million people by 2050, which corresponds to one death every three seconds. Today, they are the fourth leading cause of death in hospitals in the U.S. and cause millions of deaths worldwide, constituting a major public health threat.1 Indeed, bacteria are becoming increasingly resistant to broad-spectrum therapies, such as antibiotics, and approaches for countering recalcitrant infections are urgently needed.1000−6 Currently available antibiotics are limited by their broad and deleterious off-target effects due to the lack of delivery methods that effectively release payloads at the infection site. Advances in nanomedicine have contributed to the development of antibiotic-loaded nanoparticles,7 but these delivery systems only counter bacterial biofilms at the material interface and can become inert over time due to protein adhesion.8 Despite these drastic interventions, infections often remain untreated and untreatable.9,10 Hence, approaches for effectively delivering antimicrobial payloads to the infection site are urgently needed.11
Micro- and nanoparticles can be designed to achieve self-propulsion by converting diverse energy sources into mechanical motion, yielding swimming micro- and nanomotors. Depending on their composition, particles with active motion can be designed for a variety of applications ranging from environmental roles12 to biomedicine.13,14 Biohybrid active motion has been extensively explored in the past decade in the form of micro- and nanosized bioactive motors for minimally invasive interventions, given their potential for active navigation to reach otherwise inaccessible areas.15−17 Recent breakthroughs have been reported utilizing these systems for biomedical applications, including targeted and enhanced drug delivery,18,19 cell manipulation,20,21 microsurgery,22,23 biochemical sensing,16,24,25 and diagnostics.26,27 Bioactive micro- and nanomotors have also been applied to exploit their energy conversion to combat planktonic bacteria21,28−30 and biofilms.31−34 Indeed, the enhanced mixing and towing force of these moving colloids improves the efficiency of their interaction with bacteria, enabling more effective infection eradication than when using passive particles.12 Their navigation increases the probability of contact with the surrounding bacteria and also enhances their penetration capacity.35 However, the antibacterial efficacy of micro- and nanomotors has not yet been extensively reported in clinically relevant mouse models,36−38 an essential step to eventually translate these applications into the clinic.
In this work, we present bioactive micro- and nanomotors that deliver antimicrobial peptide (AMP) payloads to the infection site through the combination of active motion, catalytic conversion, and bactericidal capabilities. AMPs have emerged as promising antimicrobials39,40 whose amphipathic character enables their interaction with, and subsequent disruption of, bacterial membranes.41 Clinical translation of numerous antimicrobials, including AMPs, is currently restricted by their limited bioavailability, susceptibility to enzymatic degradation, and low penetrability toward the target infection.42 Thus, efficient delivery methods are required for these molecules to more readily reach their target area.35 Delivery vehicles such as unbound liposomes43 and nanoparticles44 have been used to encapsulate peptides for the treatment of microbial biofilms43,44 and cancer;45 however these methods rely on passive interactions with the infected area and passive movement through the infection site, e.g., an infected wound.
Here, we used the enzyme urease (a biocompatible source of active motion)46,47 to deliver linear, and thus susceptible to proteolytic degradation, cationic AMPs (LL-37 and K7-Pol) on silica-based micro- and nanoparticles that actively navigate in liquid toward the infection site (Figure 1). LL-37 is an amphipathic α-helical and cationic peptide with a 37-amino acid long sequence (LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES). This natural peptide is widely known for its antimicrobial, wound healing, and immunomodulatory properties,48 making it a promising candidate for testing in animal models. K7-Pol (ILGTILKLLSKL-NH2) is a potent synthetic antimicrobial agent derived from the wasp venom polybia-CP, a mastoparan-like peptide with antimicrobial and immunomodulatory activities49 that presents an amphipathic α-helical and cationic structure with an amidated C-terminus. K7-Pol was shown to present broad-spectrum activity against Gram-positive and Gram-negative bacteria and fungi50 at low nanomolar concentrations and against parasites51 and cancer cells52 at low micromolar doses. Despite the potent antimicrobial activity of these AMPs, their stand-alone broad-spectrum activity and susceptibility to protease-mediated degradation lead to off-target effects and reduced activity in vivo. Our autonomous motors effectively delivered peptide antibiotic payloads to the infected area, displayed bactericidal activity in vitro at low doses (7.8–15.6 μg mL–1), targeted bacteria by rapidly depolarizing their membrane, and demonstrated in vivo anti-infective efficacy in a clinically relevant mouse model.
Figure 1.
Bioactive micro- and nanomotors coated with antimicrobial peptides for the autonomous treatment of infections. Schematic of the AMP-coating process of the urease micro- and nanomotors and their autonomous propulsion to target pathogenic infections both in vitro and in vivo. Briefly, AMP–urease motors will encounter and hydrolyze urea (yellow spheres) in solution. The hydrolysis reaction will propel the motors, and when exposed to bacterial membranes, the AMPs onto their surface will act as antimicrobials and lyse bacterial cells in controlled (e.g., in vitro assays) and complex (e.g., infected wound) biological environments.
Results and Discussion
Synthesis and Characterization of Bioactive Micromotors
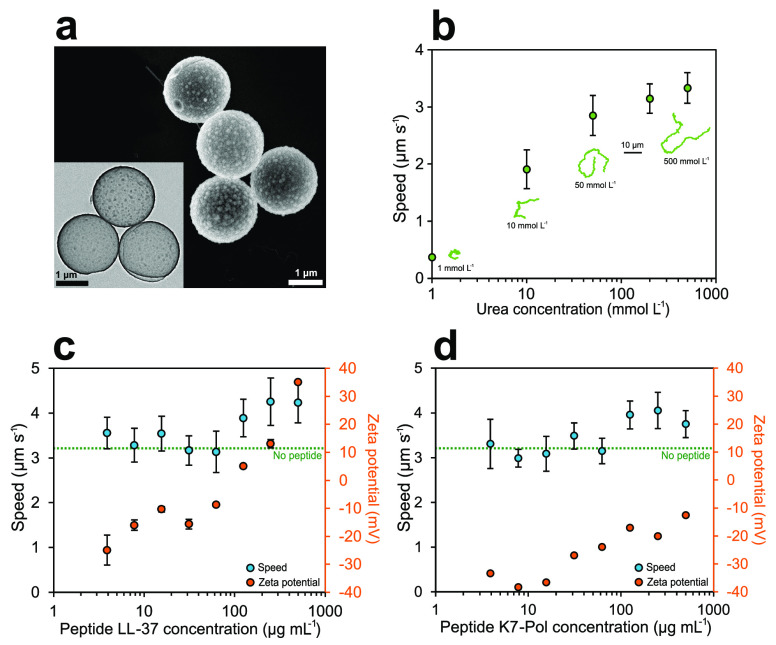
Bioactive micromotors were fabricated as previously reported53,54 through a modified Stöber method55 (Figure S1) (see Methods section for details). Briefly, commercial spherical beads made of polystyrene (PS) with a diameter of 2 μm were used to grow silicon dioxide (SiO2) on their surface through a combination of tetraethylorthosilicate (TEOS) and 3-aminopropyltriethoxysilane (APTES) as silica precursors. Later, N,N-dimethylformamide (DMF) was used to dissolve the PS core and obtain hollow silica microparticles (HSMPs). We characterized the shape, size, and morphology of the microparticles by scanning electron microscopy (SEM) (Figure 2a) and transmission electron microscopy (TEM) (inset Figure 2a), revealing an average microparticle diameter of 1.87 ± 0.01 μm (n = 91, mean ± standard error of the mean). We used TEM to measure the silica particle shell as 48 ± 1 nm (n = 120, mean ± standard error of the mean).
Figure 2.
Effect of antimicrobial peptides on the active motion of bioactive micromotors. (a) SEM micrograph of the hollow silica microcapsules. Inset: TEM micrograph of the hollow silica microparticles. (b) Average speed of urease micromotors for different concentrations of urea. Inset: Representative 15 s trajectories for different concentrations of urea. (c) Average speed and zeta potential of urease micromotors for different concentrations of LL-37 peptide used to functionalize the silica surface. (d) Average speed and zeta potential of urease micromotors for different concentrations of K7-Pol peptide used to functionalize the silica surface. All results are shown as the mean ± standard error of the mean.
To obtain bioactive micromotors, urease was attached to silica with a glutaraldehyde (GA) linker (Figure 1) on the amino groups of the bare silica particle, which were confirmed by dynamic light scattering (DLS), revealing a zeta potential of 2.64 ± 0.40 mV. For motion experiments, urease micromotors were recorded at 25 FPS for 20–25 s. The videos were analyzed using a custom-designed Python-based software in order to extract the trajectories of the micromotors, the mean squared displacement (MSD), and the speed (see Methods section for details).56,57 The active motion of urease microparticles was studied as a function of urea (substrate of urease) in water. When the urea concentration increased, the micromotors showed higher MSD (Figure S2) and speed (Figure 2b), which increased in a similar fashion to the Michaelis–Menten saturation curve, as observed in previous publications of urease-powered motion.54,58 The self-propulsion capacity reached a plateau with a maximum speed of 3.3 ± 0.3 μm s–1 when adding 500 mmol L–1 urea. The increment in self-propulsion was clearly distinguished by tracking the trajectories of the individual urease micromotors (Figure 2b, inset, and Video 1).
Preparation of AMP-Coated Bioactive Micromotors
Once the motion behavior of urease micromotors was thoroughly studied and characterized, we incorporated human cathelicidin LL-37 and peptide K7-Pol onto the surface of the silica microparticles (Figure 1). In the last functionalization step to yield the peptide-modified bioactive micromotors, we added both urease and AMP to be anchored to the silica surface by the GA linker (Figure S1) (see Methods section for details). Different concentrations of each peptide were attached to the urease micromachine. By analyzing their electrophoretic mobility through DLS, we observed a considerable increase in the zeta potential (represented by the electrical charge surrounding the particle) when more AMP was added (Figure 2c and d), indicating that the positively charged AMPs were properly coated onto the micro- and nanomotors. For concentrations of LL-37 higher than 125 μg mL–1, the microparticle zeta potential changed to positive, reaching a maximum of 35.1 ± 1.4 mV when adding 500 μg mL–1 of LL-37 (Figure 2c). In the case of K7-Pol, the zeta potential also increased with higher peptide loads, up to −12.6 ± 0.2 when adding 500 μg mL–1 of K7-Pol, but never reached a net positive charge (Figure 2d). The presence of multiple positively charged amino acids in LL-37’s amphipathic sequence (pI = 11.13) (i.e., five lysine (K) and five arginine (R) residues) explains the drastic change toward a positive zeta potential. In the case of the K7-Pol sequence (pI = 10.6), the presence of two lysine (K) residues and the amidated C-terminus also caused an increase in zeta potential but are insufficient to reach a net positive value.
After ensuring the attachment of peptides onto the urease micromotors, we tested their motion using 200 mmol L–1 of urea in water, as this constituted the lowest urea concentration needed for maximum speed (Figure 2b). For LL-37-modified micromotors, no significant differences were detected in the urease micromotors’ speed (3.1 ± 0.3 μm s–1) when the AMP concentration used was below 62.5 μg mL–1 (Figure 2c). Nonetheless, higher doses of AMP (250 μg mL–1) caused a significant boost in self-propulsion, increasing the speed by 38.7% and reaching 4.3 ± 0.5 μm s–1. This increase in directional motion was directly observable by looking at the tracking trajectories extracted from the videos (Video 2). In the case of K7-Pol-modified micromotors, self-propulsion was also observed when adding K7-Pol, which increased the speed by 32.2% for higher concentrations of peptide, as indicated by the trajectory tracking experiments (Video 3). In this case, the maximum speed reached was 4.1 ± 0.4 μm s–1 at 250 μg mL–1 of K7-Pol (Figure 2d).
While the underlying motion mechanism powered by urease has still not been resolved in the literature, it has recently been pointed out that the electric field generation by the release of ionic products could play a significant role.53,59 In this regard, the fact that higher concentrations of peptide led to higher propulsion capabilities could be explained by an increase in the surface net charge of the particle, leading to higher conductivity and enhanced electric field generation.
Synthesis and Characterization of Bioactive Nanomotors
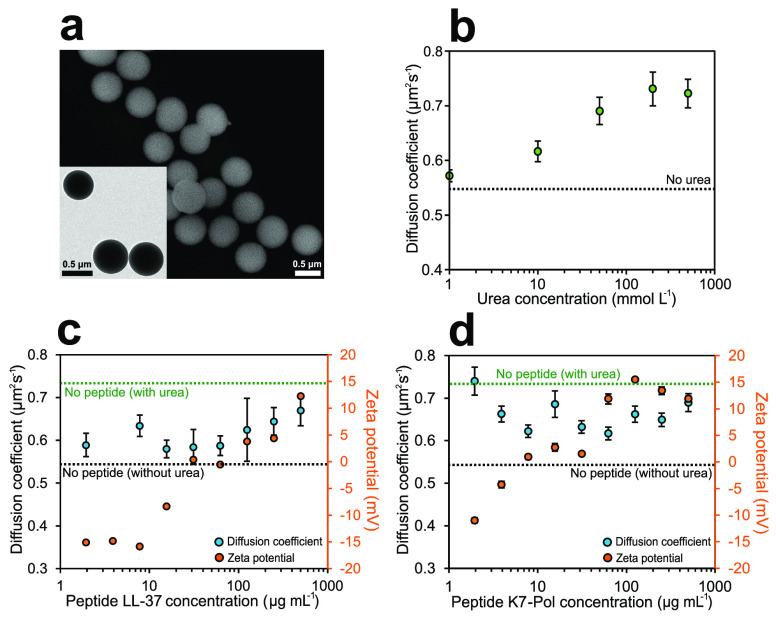
Mobil Composition of Matter No. 41 (MCM-41) mesoporous silica nanoparticles (MSNPs) were chosen as a base material for the bioactive nanomotors because of their biocompatibility and ease of surface modification,60,61 as well as their extensive use for biomedical applications.62 Urease nanomotors were fabricated as recently reported35,63 using a modified Stöber method64 (Figure S3) (see Methods section for details). The hydrolysis and condensation of TEOS were used to synthesize silica nanoparticles in aqueous media using triethanolamine (TEOA) as a basifier and hexadecyltrimethylammonium bromide (CTAB) as the structure-directing agent. Next, to obtain the mesoporous silica nanoparticles, the CTAB surfactant was removed by acidifying the solution in methanol. The generated MSNP were then treated with APTES to modify the silica surface with amine groups. The morphology of the nanoparticles was characterized by both SEM and TEM (Figure 3a), displaying an average diameter of 694.698 ± 0.003 nm (n = 63, average size ± standard error of the mean). A clear radial mesoporosity was observed when increasing the magnification of TEM micrographs (Figure S4). The amino groups on the surface of the bare MSNP were later used to attach the urease enzyme through the GA linker to yield urease nanomotors, which were first confirmed through DLS, revealing a zeta potential of 16.22 ± 3.94 mV.
Figure 3.
Effect of antimicrobial peptides on the active motion of bioactive nanomotors. (a) SEM micrograph of the mesoporous silica nanoparticles. Inset: TEM micrograph of the mesoporous silica nanoparticles. (b) Diffusion coefficient of urease nanomotors for different concentrations of urea. (c) Diffusion coefficient and zeta potential of urease nanomotors for different concentrations of LL-37 peptide used to functionalize the silica surface. (d) Diffusion coefficient and zeta potential of urease nanomotors for different concentrations of K7-Pol peptide used to functionalize the silica surface. All results are shown as the mean ± standard error of the mean.
For the motion experiments, we studied the electrophoretic mobility of urease nanomotors through DLS to obtain both their diffusion coefficient and apparent hydrodynamic radii (see Methods section for details). The active motion of urease nanomotors was studied as a function of urea concentration in phosphate-buffered saline (PBS), and for all conditions a sharp population peak was observed (Figure S5), confirming monodispersity of the sample. The diffusion of nanomotors was enhanced when increasing the urea concentration (Figure 3b), from a base Brownian diffusion of 0.54 ± 0.01 μm2 s–1 when no substrate was present to a maximum enhanced diffusion of 0.73 ± 0.03 μm2 s–1 at 200 mmol L–1 urea. This 35.2% increase in diffusion as a function of urea concentration and the further saturation of active motion resemble the dynamics of a Michaelis–Menten kinetics curve, as previously reported for urease-powered enhanced diffusion.18,35,63,65 The apparent hydrodynamic radii decreased from 451.7 nm to a minimum of 329.2 nm at 200 mmol L–1 urea, since when increasing the diffusion coefficient (D) with more urea, the radii values obtained though the Stokes–Einstein equation also decreased accordingly to match the diffusion measured (Figure S5) (see Methods section for details).
Preparation of AMP-Coated Bioactive Nanomotors
Upon determining the motion behavior of urease nanomotors, the fabrication process was modified to incorporate the human cathelicidin peptide LL-37 and the potent synthetic antimicrobial peptide K7-Pol onto the surface of the silica nanoparticles (Figure 1).
For the last functionalization step, we added both urease and AMP to the solution containing nanoparticles, to enable binding of urease and the AMP to the GA linker onto the silica surface (see Methods section for details). A range of AMP concentrations were attached to the urease nanomotors, and analysis of their electrophoretic mobility using DLS revealed a considerable increase in the zeta potential (represented by the electrical charge surrounding the particle) with increasing levels of peptide (Figure 3c and d). A positive zeta potential of the nanomotors was reached by adding 31.25 μg mL–1 of LL-37, and a maximum of 12.3 ± 0.2 mV was measured at 500 μg mL–1 of LL-37 (Figure 3c). In the case of K7-Pol, at least 7.81 μg mL–1 of K7-Pol were required to obtain a positive zeta potential (Figure 3d), reaching a maximum of 15.5 ± 0.2 mV at 125 μg mL–1 of K7-Pol, which was saturated for higher AMP concentrations. These data are consistent with the presence of two lysine residues (K) and the amidated C-terminus of the K7-Pol sequence, yielding a pI of 10.6, and the presence of five lysine (K) and five arginine (R) residues in LL-37’s amphipathic sequence with a pI of 11.13.
Once we confirmed the attachment of the peptides to the urease nanomotors, we tested their motion capabilities at 200 mmol L–1 urea in PBS, as this was the urea concentration that allowed maximum diffusion. The diffusion of LL-37-coated nanomotors decreased when attaching the peptide; however the particles still showed active motion under all conditions tested (Figure 3c and d). The lowest diffusion was 0.58 ± 0.02 μm2 s–1 at 15.62 μg mL–1 LL-37, representing a 78.9% reduced diffusion, and there were no significant differences in diffusion at the different AMP concentrations tested (Figure 3c). Figure 3d shows that attachment of K7-Pol onto the nanoparticle surface also resulted in decreased enhanced diffusion at concentrations higher than 1.95 μg mL–1 (Figure 3d). The maximum decrease in enhanced diffusion was 61.3%, from 0.73 ± 0.03 μm2 s–1 without peptide to 0.62 ± 0,02 μm2 s–1 at 62.5 μg mL–1 K7-Pol. Hence, the enhanced diffusion decrease when attaching AMPs is likely due to direct competition of the peptides with urease for the available functional groups on the silica surface, causing a decrease in both enzyme attachment and active motion. However, for the purpose of fabricating active antimicrobial tools, it is important to note that the peptide–urease nanomotors still presented active motion for all the different peptide concentrations tested.
In Vitro Antimicrobial Activity of Micro- and Nanomotors against Pathogenic Bacteria
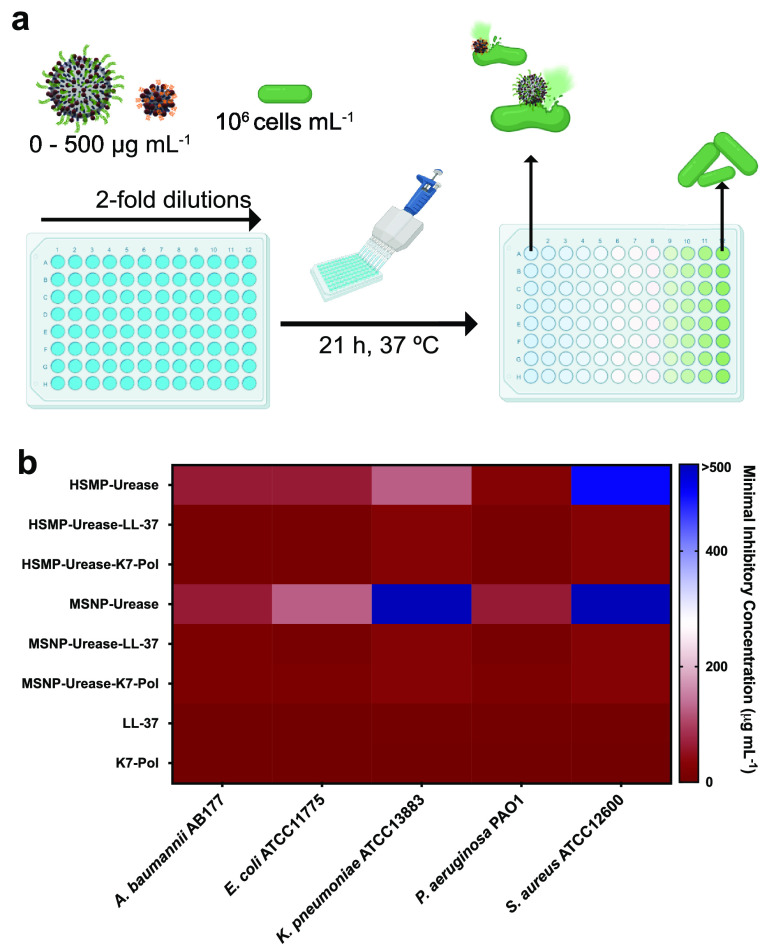
The AMP-modified bioactive micro- and nanomotors were tested against bacteria from the ESKAPE pathogen list declared by the World Health Organization as critical threats for humans: Acinetobacter baumannii AB177, Escherichia coli ATCC11775, Klebsiella pneumoniae ATCC13883, Pseudomonas aeruginosa PAO1, and Staphylococcus aureus ATCC12600. Minimum inhibitory concentration (MIC) values were determined as the lowest concentration of our motors tested in the presence of 200 mmol L–1 urea that inhibited 100% of bacterial growth. The MIC values for each condition were assessed by determining the optical density of the solution at 600 nm and ranged from 7.8 to 125 μg mL–1 depending on the bacterial strain (Figure 3 and Table S1). Both micro- and nanomotors exhibited 4- to 16-fold enhanced antimicrobial activity when the AMPs were incorporated (Figure 3). Interestingly, AMP-modified motors displayed activity against K. pneumoniae and S. aureus, whereas neither the bare micro- nor the nanomotors presented activity toward S. aureus, and the bare nanomotors were inactive against K. pneumoniae.
Mechanism of Action Studies
Mechanism of action (MoA) studies were carried out to explore whether AMP-modified micro- and nanomotors killed bacterial cells by permeabilizing their outer membrane or, instead, acted by depolarizing their cytoplasmic membrane at their MIC. To assess if the AMP-modified bioactive motors were able to permeabilize the outer membrane of A. baumannii and K. pneumoniae cells, we used the fluorescent probe NPN [1-(N-phenylamino)naphthalene]. NPN exhibits weak fluorescence emission in aqueous environments and can permeate the bacterial outer membrane when damaged. The probe interacts with the lipidic environment of permeable outer membranes and displays an increased intensity of fluorescence emission (Figure S6). As a positive control, we used the FDA-approved and widely used peptide antibiotic polymyxin B (PMB), a well-known membrane permeabilizer. When we exposed A. baumannii or K. pneumoniae cells to the AMP-modified micro- and nanomotors and PMB, only PMB was able to permeabilize the outer membrane (Figure S6).
To evaluate whether the AMP–urease micro- and nanomotors were able to depolarize the cytoplasmic membrane of A. baumannii and K. pneumoniae cells, we used the probe DiSC3-5 (3,3′-dipropylthiadicarbocyanine iodide). DiSC3-5, a potentiometric probe, works by accumulating in the cytoplasmic membrane and aggregating at high concentrations, causing fluorescence quenching.66,67 When the cytoplasmic membrane is depolarized, DiSC3-5 migrates to the cytoplasm, leading to increased fluorescence emission intensity (Figure 5a). The micro- and nanomotors with and without AMP had similar ability to depolarize the cytoplasmic membrane of A. baumannii (Figure 5b). However, the AMP–urease motors presented increased depolarization of the K. pneumoniae membrane compared with their bare counterparts (Figure 5c). This result is in accordance with the observed depolarizing effect of the peptides.68 K7-Pol–urease nanomotors showed a slightly increased depolarization effect than LL-37–urease nanomotors. This was not entirely unexpected, since mastoparans, such as K7-Pol and all polybia-CP-derived peptides, depolarize bacterial membranes more efficiently than defensins (e.g., LL-37).66 All experiments were performed in the presence of 200 mmol L–1 urea.
Figure 5.
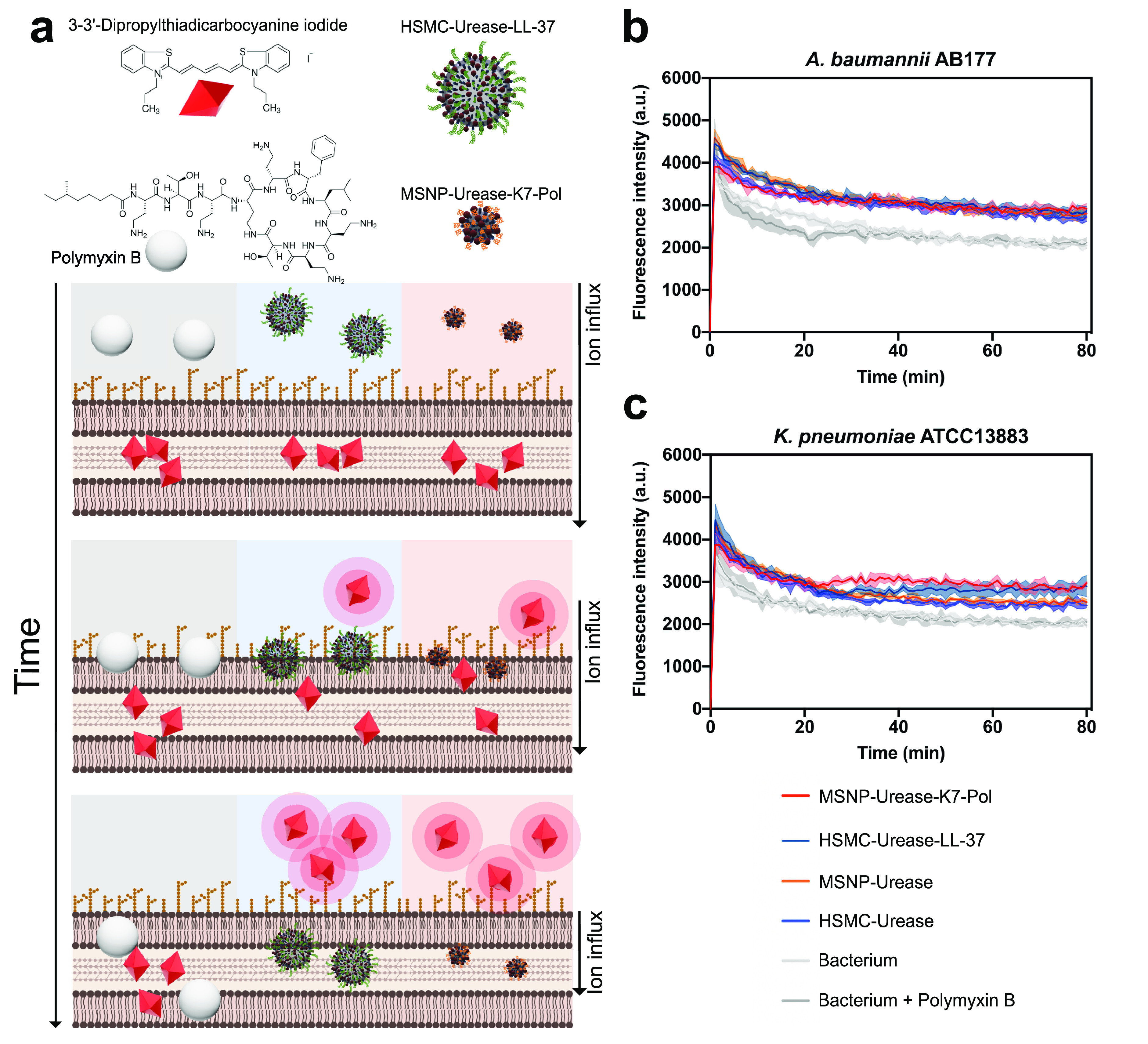
Mechanism of action of the antimicrobial motors. (a) Bioactive micro- and nanomotors cause the depolarization of bacterial membranes at their MIC concentration against (b) A. baumannii AB177 and (c) K. pneumoniae ATCC13883. Briefly, micro- and nanomotors functionalized with LL-37 and K7-Pol, respectively, enabled the higher depolarization of K. pneumoniae cells than the nonfunctionalized motors. When A. baumannii cells were exposed to them both, functionalized and nonfunctionalized motors presented depolarizing effect. The potent permeabilizer antimicrobial polymyxin B was used as a negative control for depolarization. Assays were performed in three independent replicates. This figure was created with BioRender.com.
Anti-infective Activity against A. baumannii in a Skin Abscess Animal Model
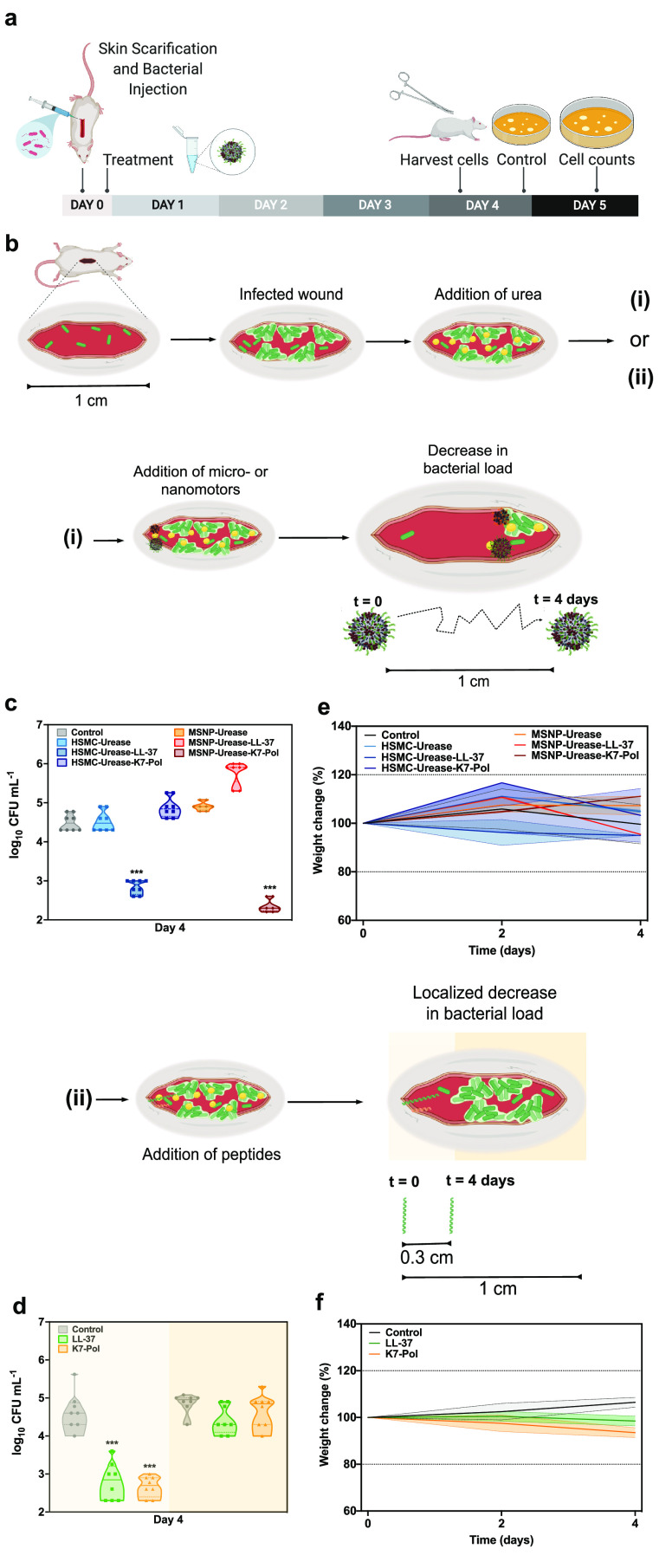
All the combinations of micro- and nanomotors and peptides and their bare urease micro- and nanomotors counterparts were tested in vivo against A. baumannii at 2-fold their MIC, 15.6 and 31.2 μg mL–1, respectively (Figure 6a). Skin infection was induced by administering an A. baumannii solution at 107 CFU mL–1 on the back of mice previously scratched with a needle.66,69,70 The treated groups were administered with a single dose of free AMPs and micro- or nanoparticles (with and without AMP) 2 h postinfection. The samples were added to one extremity of the infected wound, and 100 μL of a 200 mmol L–1 urea solution was spread over the entire length of the wound (Figure 6b). Four days after a single treatment dose, the tissue was homogenized and the CFU bacterial load was quantified, as this quantitative method accurately accounts for the number of bacterial cells in the infected area.71 The most active systems were LL-37–urease micromotors and K7-Pol–urease nanomotors, significantly reducing the bacterial load by 2 and 3 orders of magnitude, respectively (Figure 6c), to levels that can be cleared up by the immune response (<103 CFU mL–1).
Figure 6.
Anti-infective activity of the antimicrobial motors in vivo. (a) Mice had their dorsal region shaved, scratched (1-cm-long wound), and infected with A. baumannii AB177. After 1 h, functionalized and nonfunctionalized micro- and nanomotors or peptides were added to the infection site. Mice were euthanized, the tissue from the infection site was harvested, and the bacterial cells of treated and untreated samples were counted by plating. (b) Schematic representation of the wound site infected and the addition of urea before the treatment with (i) micro- and nanomotors or (ii) peptides free is solution. Antimicrobial micro- and nanomotors self-propelled, driven by urea, through a distance of 1 cm to enable the autonomous treatment of the target infected area. On the other hand, peptides by themselves exhibited antimicrobial activity only within the area they were administered and did not clear the infection at a distance. Briefly, after the infection was established, urea was spread over the entire length of the wound (1 cm). Next, the micro- and nanomotors coated with peptides and the peptides alone were inoculated to one of the extremities of the infected wound. (c) Four days postinfection, 1 cm2 of the infected area was excised and the ability of the micro- and nanomotors to travel throughout the wound alone and when functionalized with peptides was assessed. (d) Treatment with peptides alone decreased bacterial counts only in the extremity where they were administered (light yellow background), as revealed by similar bacterial counts detected in areas at a distance from the administration site (dark yellow background) and those of untreated control groups. (e) Mouse weight was monitored throughout the experiments, serving as a proxy to assess the toxicity of both micro- and nanomotors and (f) peptides in solution. None of the treatment groups led to toxicity in mice. Eight animals were used per group. This figure was created with BioRender.com.
On the contrary, treatment with AMPs free in solution (Figure 6b) that were administered in one of the extremities of the wound displayed anti-infective activity only locally where the peptides were administered (Figure 6d). More specifically, peptide treatment reduced CFU counts by 2–3 orders of magnitude in the administration site (∼1/3 of the whole extent of the wound), whereas infected areas farther away from the administration site remained infected at levels similar to the untreated controls. Interestingly, for LL-37, which presents immunomodulatory and wound-healing properties,72 we also observed partial wound healing in the area where the peptides were added (Figure S7). To test the effect of the motion in the complex in vivo environment, we treated mice with all the systems in the absence of urea and compared them to an untreated control (Figure S8a). All peptide-loaded micro- and nanomotor systems were active against bacteria in the administration site (Figure S8b). Stand-alone treatment with the urease micromotors and urease nanomotors did not decrease bacterial load significantly (Figure S8b). Peptides free in solution and peptide-loaded micro- and nanomotors reduced CFU counts by 3 orders of magnitude. On the other hand, none of the systems tested in the absence of urea were able to significantly reduce CFU counts far from the administration site (Figure S8b). This contrasts with peptide-loaded micro- and nanomotors in the presence of urea, which led to anti-infective activity in vivo at a distance from the administration site (Figure 6). No side effects (e.g., itchiness, redness, swelling) or in vivo toxicity was observed under any of the conditions tested (Figure S8c). To assess whether the peptides free in solution and the AMP-modified bioactive micro- and nanomotors were toxic to the animals, the weight of the mice was monitored throughout the experiment. Variations of up to 20% in weight are a widely used proxy of distress, morbidity, and overall toxicity.50,70 No side effects (e.g., itchiness, redness, swelling) or in vivo toxicity was observed under any of the conditions tested (Figures 6e,f and S8c). The LL-37–urease nanomotors and K7-Pol–urease micromotors and the motors without peptide presented bacteriostatic effects (Figure 4).
Figure 4.
Antimicrobial activity of bioactive micro- and nanomotors functionalized with antimicrobial peptides. (a) Schematic depicting experimental design of in vitro biological activity assays. Briefly, 105 bacterial cells and peptides, the antibiotic polymyxin B, or urease micro- and nanomotors (0–500 μg mL–1) were added to a 96-well plate and incubated at 37 °C. One day after the exposure, the solution in each well was measured in a microplate reader (600 nm) to check inhibition of bacteria compared to the untreated controls. (b) Heat map of the antimicrobial activity of each system against five bacterial strains: A. baumannii AB177, E. coli ATCC11775, K. pneumoniae ATCC13883, P. aeruginosa PAO1, and S. aureus ATCC12600. Assays were performed in three independent replicates, and heat map OD600 values are the arithmetic mean of the replicates in each condition. This figure was created with BioRender.com.
To shed light on the underlying mechanisms associated with the differences in antimicrobial activity observed between our in vitro and in vivo experiments, we investigated the speed of each of the motors. No significant differences were observed when comparing the speed of both AMP-modified micromotors nor when comparing the diffusion coefficient of both AMP-modified urease nanomachines (Figure S9). Thus, we hypothesize that LL-37–urease micromotors and K7-Pol–urease nanomotors presented activity in the animal model because less peptide is needed to obtain more positively charged systems for those combinations. LL-37–urease micromotors presented zeta potential values of −10.3 ± 1.9, while K7-Pol–urease micromotors showed −36.6 ± 0.1 at the concentration used in the animal model (15.6 μg mL–1). On the other hand, K7-Pol–urease nanomotors presented zeta potential values of 1.5 ± 0.2, whereas LL-37–urease nanomotors exhibit a zeta potential value of 0.4 ± 0.2 at the concentration used in vivo (31.2 μg mL–1) (Figures 2 and 3). These values indicate that the initial electrostatic interactions between the negatively charged bacterial membranes and the AMP–urease motors are likely crucial to trigger depolarization and subsequent bacterial death.
Conclusion
We report the fabrication and detailed characterization of urease micro- and nanomotors that actively deliver peptides to combat clinically relevant infections. The motion dynamics of each particle was studied under different concentrations of urea, and active motion was confirmed upon functionalization with the antimicrobial peptides. The antimicrobial micro- and nanomotors displayed broad-spectrum bactericidal activity in vitro against both Gram-negative (A. baumannii, E. coli, K. pneumoniae, P. aeruginosa) and Gram-positive (S. aureus) bacterial pathogens. An in-depth mechanistic study confirmed that both bare urease micro- and nanomotors targeted bacteria by depolarizing their membrane, and coating them with antimicrobial peptides led to enhanced bactericidal activity and bacterial membrane depolarization. Finally, the AMP-modified micro- and nanomotors demonstrated autonomous propulsion to reduce A. baumannii infections by up to 3 orders of magnitude in a preclinical murine infection model, whereas free peptides in solution were not able to exert antimicrobial activity at a distance from the initial administration site. We show that differences in surface net positive charge of the micro- and nanomotors coated with peptides influenced antimicrobial activity in vivo by increasing the activity range along the wound, but not in less complex environments (in vitro). This work constitutes a relevant step toward the implementation of bioactive and autonomous micro- and nanomotors for the treatment of infectious diseases.
Methods
Chemicals
Two micrometer microparticles based on polystyrene (Sigma-Aldrich cat. no. 78452), ethanol 99% (PanReac AppliChem cat. no. 131086-1214), ammonium hydroxide solution 28–30% (Sigma-Aldrich cat. no. 221228), 3-aminopropyltriethoxysilane 99% (Sigma-Aldrich cat. no. 440140), triethanolamine 99% (Sigma-Aldrich cat. no. 90279), hexadecyltrimethylammonium bromide 99% (Sigma-Aldrich cat. no. 52365), methanol (MeOH) 99% (PanReac AppliChem cat. no. 361091-1611), hydrochloric acid (HCl) 37% (PanReac AppliChem cat. no. 211020-1611), tetraethylorthosilicate 99% (Sigma-Aldrich cat. no. 86578), N,N-dimethylformamide 99% (Acros Organics cat. no. 423640010), 1× phosphate-buffered saline (Thermo Fisher Scientific cat. no. 70011-036), glutaraldehyde (25 wt %) (Sigma-Aldrich cat. no. G6257), urease from Canavalia ensiformis (Jack bean) (Sigma-Aldrich cat. no. U4002), urea (Sigma-Aldrich cat. no. U5128) were used.
Instruments
SEM images were captured by a FEI NOVA NanoSEM 230. TEM images were captured by a Zeiss EM 912. The zeta potential (ζ potential) measurements were performed with a Zetasizer Nano S from Malvern Panalytical. The hydrodynamic radius and diffusion coefficient measurements were performed using a Möbius from Wyatt Technology. The optical videos of urease micromotors were recorded using the camera (Hamamatsu digital camera C11440) of an inverted optical microscope (Leica DMi8). The optical density (OD) of the antibacterial assays was measured at 600 nm in a Thermo Scientific Varioskan LUX fluorescence spectrophotometer.
Antimicrobial Peptide Synthesis
The peptides LL-37 and K7-Pol were purchased from Aapptec (Kentucky, USA). They were purified by high-performance liquid chromatography (HPLC). The peptide purity used in all assays was higher than 95%.
Synthesis of Hollow Silica Microparticles
The HSMPs were fabricated by mixing 500 μL of PS beads with a diameter of 2 μm (Sigma-Aldrich cat. no. 78452) with 1 mL of ethanol 99% (Panreac Applichem cat. no. 131086-1214) and 0.8 mL of ultrapure water. Next, 50 μL of ammonium hydroxide 28–30% (Sigma-Aldrich cat. no. 221228) was added, and the solution was magnetically stirred for 5 min. Then, 5 μL of APTES 99% (Sigma-Aldrich cat. no. 440140) was added, and the reaction was stirred for 6 h. After, 15 μL of TEOS ≥99% (Sigma-Aldrich cat. no. 86578) was added, and the solution was allowed to react overnight (17 h) under continuous mixing. Next, the PS particles covered with silica were washed three times with ethanol (centrifugation of microparticles at 1503 rcf during 4 min). The PS was then dissolved from inside the silica shell with four washes of DMF ≥99.8% (Acros Organics cat. no. 423640010), with a 15 min mixing step in each wash. Afterward, three washes of ethanol 99% were performed to the HSMP solution, and the final samples were stored at room temperature.
Functionalization of HSMPs with Urease and AMPs
The HSMPs were washed three times with ultrapure water and one time with 1× PBS (pH = 7.4) (Thermo Fischer Scientific cat. no. 70011-036). Then, the particles were suspended in a 2.5 wt % GA solution (Sigma-Aldrich cat. no. G6257) in 1× PBS and kept mixing at room temperature for 3 h. Next, the silica microparticles with GA were washed three times with 1× PBS (pH = 7.4) and resuspended again in 1× PBS (pH = 7.4) with 3 mg mL–1 of urease powder from Canavalia ensiformis (Jack bean) (Sigma-Aldrich cat. no. U4002). When fabricating micromotors with peptides attached, different concentrations of AMP (500, 250, 125, 62.5, 31.25, 15.62, 7.81, 3.9, and 0 μg mL–1) were added simultaneously with urease. The solution was kept mixing overnight (16 h) and then washed three times with 1× PBS (pH = 7.4). Then, the urease micromotors in a 1× PBS (pH = 7.4) solution was divided in aliquots and stored at 4 °C for further use.
Optical Video Recording of Bioactive Micromotor Motion
To study the active motion of urease micromotors, optical videos were recorded using a digital camera (Hamamatsu digital camera C11440) coupled with an inverted optical microscope (Leica DMi8). The 63× water immersion objective was used to record the micromotors placed on a glass slide, which were thoroughly mixed with the urea aqueous solutions (enzymatic substrate) at different concentrations (500, 200, 50, 10, 1, and 0 mmol L–1), specifically selected for the urease Michaelis–Menten kinetics saturation, as reported in BRENDA, the Comprehensive Enzyme Information System (https://www.brenda-enzymes.org/). For the motion experiments as a function of different AMP concentrations (500, 250, 125, 62.5, 31.25, 15.62, 7.81, 3.9, and 0 μg mL–1), 200 mM urea was used to ensure optimal catalytic rate and active motion. The micromotors solution was put on a glass slide covered with a coverslip, and videos of 25 FPS and 20–25 s were recorded for 3 min. For each condition studied of urea and AMP concentration, 10–14 individual urease micromotors were recorded.
Data Analysis of Bioactive Micromotor Motion
The videos were analyzed using custom-designed tracking Python software to obtain the tracking trajectories of the microparticle displacement. From the x and y axes values over time, the MSD was calculated using the following equation:
| 1 |
where t is the time, ri(t) is the position of the particle in the coordinate i at time t, n = 2 are the dimensions of 2D analysis, and ⟨·⟩ denotes the ensemble and time average. The velocity (v) was then extracted from fitting the MSD to
| 2 |
where Dt is the diffusion coefficient and v is the speed, since we analyze the propulsive regime when t ≪ τr, with τr being the rotational diffusion time and t the time of MSD represented.56,57 The results are presented as mean ± standard error of the mean.
The τr was calculated to be 4.575 ± 0.044 s, which is
| 3 |
where Dr is the rotational diffusion coefficient (Dr = 0.219 ± 0.002 s–1), which depends on the radius of the particle, as it can be observed in the Stokes–Einstein equation,
| 4 |
where kB is the Boltzmann constant, T is the absolute temperature, η is the solvent viscosity, and r is the radius of the diffusing particle. Hence, τr depends on the temperature (T = 24 ± 1 °C), the solvent viscosity (η = 0.9107 × 10–3 kg m–1 s–1), and the radius of the particle (r = 0.936 ± 0.003 μm). A nano/micromotor analysis tool was used to compile and visualize the resulting files (https://github.com/rafamestre/NMAT-nanomicromotor-analysis-tool).
Synthesis of Mesoporous Silica Nanoparticles
The MSNPs were synthesized through the sol–gel methodology.64 Briefly, 570 mg of CTAB (Sigma-Aldrich cat. no. 52365) and 35 g of TEOA (Sigma-Aldrich cat. no. 90279) were dissolved in 20 mL of ultrapure water and heated at 95 °C in a silicon oil bath. This solution was stirred for 30 min, after which 1.5 mL of TEOS (Sigma-Aldrich cat. no. 86578) was added dropwise. The solution was then stirred at 95 °C for 2 h stirring in a silicon oil bath. Then, the resulting particles were collected by centrifugation and washed three times with ethanol (centrifugation of nanoparticles at 1503 rcf for 5 min). To remove the CTAB from the MSNP pores, the particles were suspended in a solution composed by 30 mL of methanol (PanReac AppliChem cat. no. 361091-1611) and 1.8 mL of HCl (PanReac AppliChem cat. no. 211020-1611) and left in the reflux at 80 °C for 24 h. After collecting the particles by centrifugation, three washes of ethanol were applied with 10 min of sonication for each wash. Finally, three aliquots of 0.5 mL were taken from the resulting solution to centrifuge them and determine the concentration by measuring the weight before and after air-drying the solvent.
Amino Modification of the MSNP Surface
The silica surface of the MSNPs was modified with APTES to incorporate amino groups, by adjusting an already reported method.73 First, a solution of 2 mg mL–1 of MSNP was prepared in a round-bottom flask under magnetic stirring and with a stable temperature of 50 °C. Next, APTES was added to a stirring solution to obtain a final concentration of 5 mM, and it was left under reflux at 50 °C for 24 h. After this process, the resulting MSNPs-NH2 were washed three times with ultrapure water, and three aliquots of 0.5 mL were taken to determine the concentration by measuring the weight before and after air-drying the solvent inside.
Functionalization of MSNP-NH2 with Urease and AMPs
The resulting MSNP-NH2 were diluted in ultrapure water to obtain a 1 mL solution of 1 mg mL–1 of nanoparticles, then centrifuged, resuspended in 900 μL of PBS (pH = 7.4), and sonicated for 10 min. After this, 100 μL of GA at 25 wt % was added and left to mix for 2.5 h at room temperature. The MSNP-NH2 were then washed three times with PBS, applying a 10 min sonication for each wash. Next, the silica nanoparticles functionalized with GA were washed three times with 1× PBS (pH = 7.4) and resuspended again in 1× PBS (pH = 7.4) with 3 mg mL–1 of urease powder from Canavalia ensiformis (Jack bean). When fabricating silica nanomotors with attached peptides, different concentrations of AMP (500, 250, 125, 62.5, 31.25, 15.62, 7.81, 3.9, 1.95, and 0 μg mL–1) were added concomitantly with urease. The solution was kept mixing overnight (16 h) and then washed three times with 1× PBS (pH = 7.4). Afterward, the solution of urease nanomotors in 1× PBS (pH = 7.4) was divided in aliquots and stored at 4 °C to be used for further experiments.
Analysis of Bioactive Nanomotor Motion
The Möbius from Wyatt Technology was used to analyze the electrophoretic mobility through DLS and extract both the apparent hydrodynamic radius and the diffusion coefficient of the urease nanomotors. Active motion of urease nanomotors was studied as a function of different concentrations of urea (500, 200, 50, 10, 1, and 0 mmol L–1) and AMPs (500, 250, 125, 62.5, 31.25, 15.62, 7.81, 3.9, 1.95, and 0 μg mL–1). The hydrodynamic radius is correlated with the diffusion coefficient according to the following Einstein–Stokes equation:
| 5 |
where kB is the Boltzmann constant, T is the absolute temperature, η is the solvent viscosity, and rh is the hydrodynamic radius of the diffusing particle. The electrophoretic mobility was studied using an acquisition time of 5 s, with a laser of 532 nm wavelength and a detector angle of 163.5°. For each condition, the diffusion coefficient was calculated for the average of 17–26 acquisitions obtained directly from the analysis of the scattering data on the Dynamics software. All the results are presented as mean ± standard error of the mean.
Zeta Potential Analysis of Bioactive Micro- and Nanomotors
The Zetasizer Nano S from Malvern Panalytical was used to study the electrophoretic mobility through DLS and obtain the zeta potential of the micro- and nanomotors. The zeta potential of AMP-modified urease micromotors was analyzed as a function of peptide concentration (500, 250, 125, 62.5, 31.25, 15.62, 7.81, 3.9, and 0 μg mL–1), and the zeta potential of AMP-modified urease nanomotors as a function of peptide concentration (500, 250, 125, 62.5, 31.25, 15.62, 7.81, 3.9, 1.95, and 0 μg mL–1). For each condition, the resulting value was the mean of three measurements (which in its turn were an average of a minimum of 10 acquisitions), detected using a scattering angle of 173° and calculated using the Henry equation. The results are presented as mean ± standard error of the mean. The isoelectric points (pI) of the AMP were calculated through the Web service Isoelectric Point Calculator (IPC) for peptides (http://isoelectric.org/).
Bacterial Strains and Media
In this study, we used the following pathogenic strains: Escherichia coli ATCC11775, Acinetobacter baumannii AB177, Pseudomonas aeruginosa PAO1, Staphylococcus aureus ATCC12600, and Klebsiella pneumoniae ATCC13883. All bacterial strains were grown and plated on Luria–Bertani (LB) plates, except for P. aeruginosa, which was grown in Pseudomonas Isolation agar plates. All the agar plates were incubated overnight at 37 °C. After the incubation period, we transferred one colony to 5 mL of LB broth and incubated it overnight at 37 °C (250 rpm). On the next day, we prepared inocula by diluting the bacterial overnight solutions 1:100 in 5 mL of LB broth and incubating at 37 °C until logarithmic phase (OD600 = 0.3–0.5) was reached.
Antibacterial Assays
MICs of micro- and nanomotors were determined using the broth microdilution technique in LB with an initial inoculum of 5 × 106 cells mL–1 in untreated polystyrene microtiter plates (Corning, USA). The detailed methodology is described by Cesaro etal.74 Briefly, capsules were added to the plate as solutions in LB broth in concentrations ranging from 0 to 500 μg mL–1. The MIC value was considered as the lowest concentration of the antimicrobial system that inhibited the visible growth of bacteria. After 24 h of incubation at 37 °C, the plates were read in a spectrophotometer at 600 nm. All assays were done in three independent replicates.
Membrane Depolarization Assays
The cytoplasmic membrane depolarization activity of the AMP–urease motors was determined by measurements of fluorescence of the membrane-potential-sensitive dye DiSC3(5). Briefly, A. baumannii AB177 and K. pneumoniae ATCC13883 were grown at 37 °C with agitation until they reached mid log phase (OD600 = 0.5). The cells were then centrifuged and washed twice with HEPES buffer (5 mmol L–1) with 20 mmol L–1 glucose at pH 7.2 and diluted 1:10 in the same buffer with KCl (0.1 mol L–1). The cells (100 μL) were then incubated for 15 min with 20 nmol L–1 of DiSC3(5) until fluorescence emission values were stable, indicating the incorporation of the dye into the bacterial cytoplasmic membrane. Membrane depolarization was tracked over 60 min by the change in the fluorescence emission intensity of DiSC3(5) (λex = 622 nm, λem = 670 nm), after the addition of the micro- and nanomotors (100 mL solution at MIC values).
Membrane Permeabilization Assay
The membrane permeability of the AMP–urease motors was determined by using the NPN uptake assay. A. baumannii AB177 and K. pneumoniae ATCC13883 were grown to an OD600 of 0.4, centrifuged (10 000 rpm at 4 °C for 10 min), and washed and resuspended in HEPES (5 mmol L–1) buffer with 5 mmol L–1 glucose at pH 7.4. NPN solution (4 μL at 0.5 mmol L–1) was added to the bacterial solution (100 μL) in a white 96-well plate. The background fluorescence was recorded at λex = 350 nm and λem = 420 nm. Micro- and nanomotors functionalized with urease and AMPs in water (100 μL solution at their MIC values) were added to the 96-well plate, and fluorescence was recorded for 45 min.
Skin Abscess Infection Mouse Model
A. baumannii AB177 cells were grown in tryptic soy broth (TSB) medium. Subsequently, cells were washed twice with sterile PBS (pH 7.4, 13 000 rpm for 1 min) and resuspended to a final concentration of 5 × 106 CFU/20 μL. Female 6-week-old CD-1 mice, anesthetized with isoflurane, had the fur on their backs removed followed by a 1-cm-long superficial linear skin abrasion with a needle. The wound was made to damage only the stratum corneum and upper layer of the epidermis. A single aliquot of 20 μL containing the bacterial load was inoculated over the wound area. Four hours after the infection, free peptides and micro- and nanomotors functionalized with urease and AMPs at their MIC value were administered to the infected area in the presence (100 μL of a 200 mmol L–1 solution) and absence of urea. Animals were euthanized, and the wound area was excised 4 days postinfection. The tissue was then homogenized for 20 min (25 Hz), and the resulting solution with suspended bacteria in PBS was 10-fold serially diluted for CFU quantification. Mannitol salt agar plates were used for easy differentiation of A. baumannii cells present in the homogenized tissue. Two independent experiments were performed with 8 mice per group in each condition.
Acknowledgments
This work was supported by the Spanish MINECO (project RTI2018-095622-B-I00), the Catalan AGAUR (project 2017 SGR 238), and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 866348: iNanoSwarms). It was also funded by the grant RTI2018-098164-B-I00 from the MICIN/AEI/10.13039/5011000110333 and the “FEDER Una manera de hacer Europa” (BOTSinFluids project), the CERCA program by the Generalitat de Catalunya, and the “Centro de Excelencia Severo Ochoa”, funded by Agencia Estatal de Investigación (CEX2018-000789-S). X.A. thanks the Spanish MINECO for the Severo Ochoa program (SEV-2014-0425) for the Ph.D. fellowship (PRE2018-083712). C.F.-N. holds a Presidential Professorship at the University of Pennsylvania, is a recipient of the Langer Prize by the AIChE Foundation, and acknowledges funding from the Institute for Diabetes, Obesity, and Metabolism, the Penn Mental Health AIDS Research Center of the University of Pennsylvania, the Nemirovsky Prize, the Dean’s Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania, the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM138201, and the Defense Threat Reduction Agency (DTRA; HDTRA11810041 and HDTRA1-21-1-0014). A.B. acknowledges funding from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, 2016/10585-4 and 2019/15871-3). The authors acknowledge the support of Ana Hortelão with the mesoporous silica particle synthesis and the TEM/SEM investigations. Some of the figures were created with BioRender.com.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.1c11013.
Schematic representation of the synthesis of AMP-coated micro- and nanomotors; mean square displacement of urea micromotors; TEM micrographies of the silica nanoparticles; hydrodynamic radius of urease nanomotors in the presence of different urea concentrations; membrane permeabilization experiments with urease micro- and nanomotors functionalized with peptides; images of the infection site on the back of mice used for the animal model; comparison of the active motion of micro- and nanomotors with different antimicrobial peptide coating (PDF)
Video 1: Tracking trajectory of urease micromotors for different urea concentrations (0, 1, 10, 50, 200, 500 mM) over 20 seconds (AVI)
Video 2: Tracking trajectory of urease micromotors functionalized with different LL-37 peptide concentrations (0, 7.81, 31.25, 125, 500 μg mL–1) with 200 mM urea and the control without urea over 20 seconds (AVI)
Video 3: Tracking trajectory of urease micromotors functionalized with different K7-Pol peptide concentrations (0, 7.81, 31.25, 125, 500 μg mL–1) with 200 mM urea and the control without urea over 20 seconds (AVI)
Author Contributions
# X.A. and M.D.T.T. contributed equally.
Author Contributions
S.S. and C.F.-N. conceived the idea and project. X.A., T.P., and S.S. designed the micro- and nanomotors and conducted their fabrication, characterization, and motion experiments. X.A. performed the micro- and nanomotors fabrication, characterization, and motion experiments. M.D.T.T. and C.F.-N. designed all biological experiments. M.D.T.T. and A.B. performed all biological activity experiments, mechanism of action, and mouse work. X.A., M.D.T.T., and C.F.-N. wrote the first draft of the manuscript. A.B., T.P., and S.S. revised the manuscript. S.S. and C.F.-N. supervised and administered the work.
The authors declare no competing financial interest.
Notes
The preprint version of this work is available at Xavier Arqué, Marcelo D. T. Torres, Tania Patiño, Andreia Boaro, Samuel Sánchez, Cesar de la Fuente-Nunez. Autonomous treatment of bacterial infections in vivo using antimicrobial micro- and nanomachines. BiorXiv 2021, 2021.11.11.468250 (10.1101/2021.11.11.468250, accessed 04/05/2022).
Supplementary Material
References
- Bryers J. D. Medical Biofilms. Biotechnol. Bioeng. 2008, 100 (1), 1–18. 10.1002/bit.21838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente-Nunez C.; Reffuveille F.; Fernandez L.; Hancock R. E. Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 2013, 16, 580–589. 10.1016/j.mib.2013.06.013. [DOI] [PubMed] [Google Scholar]
- Chambers H. F.; DeLeo F. R. Waves of Resistance: Staphylococcus Aureus in the Antibiotic Era. Nat. Rev. Microbiol. 2009, 7 (9), 629–641. 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxminarayan R.; Duse A.; Wattal C.; Zaidi A. K. M.; Wertheim H. F. L.; Sumpradit N.; Vlieghe E.; Hara G. L.; Gould I. M.; Goossens H.; et al. Antibiotic Resistance-the Need for Global Solutions. Lancet Infect. Dis. 2013, 13 (12), 1057–1098. 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- Koo H.; Allan R. N.; Howlin R. P.; Stoodley P.; Hall-Stoodley L. Targeting Microbial Biofilms: Current and Prospective Therapeutic Strategies. Nat. Rev. Microbiol. 2017, 15 (12), 740–755. 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Reisman I.; Ronin I.; Gefen O.; Braniss I.; Shoresh N.; Balaban N. Q. Antibiotic Tolerance Facilitates the Evolution of Resistance. Science 2017, 355 (6327), 826–830. 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- Magana M.; Pushpanathan M.; Santos A. L.; Leanse L.; Fernandez M.; Ioannidis A.; Giulianotti M. A.; Apidianakis Y.; Bradfute S.; Ferguson A. L.; et al. The Value of Antimicrobial Peptides in the Age of Resistance. Lancet Infect. Dis. 2020, 20, e216. 10.1016/S1473-3099(20)30327-3. [DOI] [PubMed] [Google Scholar]
- Horev B.; Klein M. I.; Hwang G.; Li Y.; Kim D.; Koo H.; Benoit D. S. W. PH-Activated Nanoparticles for Controlled Topical Delivery of Farnesol to Disrupt Oral Biofilm Virulence. ACS Nano 2015, 9 (3), 2390–2404. 10.1021/nn507170s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuson H. H.; Weibel D. B. Bacteria-Surface Interactions. Soft Matter 2013, 9 (17), 4368–4380. 10.1039/c3sm27705d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pozo J. L.; Patel R. The Challenge of Treating Biofilm-Associated Bacterial Infections. Clin. Pharmacol. Ther. 2007, 82 (2), 204–209. 10.1038/sj.clpt.6100247. [DOI] [PubMed] [Google Scholar]
- Tse B. N.; Adalja A. A.; Houchens C.; Larsen J.; Inglesby T. V.; Hatchett R. Challenges and Opportunities of Nontraditional Approaches to Treating Bacterial Infections. Clin. Infect. Dis. 2017, 65 (3), 495–500. 10.1093/cid/cix320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalelkar P. P.; Riddick M.; García A. J. Biomaterial-Based Antimicrobial Therapies for the Treatment of Bacterial Infections. Nat. Rev. Mater. 2022, 7, 39. 10.1038/s41578-021-00362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar J.; Vilela D.; Villa K.; Wang J.; Sánchez S. Micro- and Nanomotors as Active Environmental Microcleaners and Sensors. J. Am. Chem. Soc. 2018, 140 (30), 9317–9331. 10.1021/jacs.8b05762. [DOI] [PubMed] [Google Scholar]
- Wang H.; Pumera M. Fabrication of Micro/Nanoscale Motors. Chem. Rev. 2015, 115 (16), 8704–8735. 10.1021/acs.chemrev.5b00047. [DOI] [PubMed] [Google Scholar]
- Katuri J.; Ma X.; Stanton M. M.; Sánchez S. Designing Micro-and Nanoswimmers for Specific Applications. Acc. Chem. Res. 2017, 50 (1), 2–11. 10.1021/acs.accounts.6b00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng F.; Tu Y.; Wilson D. A. Micro/Nanomotors towards: In Vivo Application: Cell, Tissue and Biofluid. Chem. Soc. Rev. 2017, 46 (17), 5289–5310. 10.1039/C6CS00885B. [DOI] [PubMed] [Google Scholar]
- Li J.; Esteban-Fernández de Ávila B.; Gao W.; Zhang L.; Wang J. Micro/Nanorobots for Biomedicine: Delivery, Surgery, Sensing, and Detoxification. Sci. Robot. 2017, 2 (4), eaam6431. 10.1126/scirobotics.aam6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Moolenbroek G. T.; Patiño T.; Llop J.; Sánchez S. Engineering Intelligent Nanosystems for Enhanced Medical Imaging. Adv. Intell. Syst. 2020, 2 (10), 2000087. 10.1002/aisy.202000087. [DOI] [Google Scholar]
- Hortelão A. C.; Patiño T.; Perez-Jiménez A.; Blanco À.; Sánchez S. Enzyme-Powered Nanobots Enhance Anticancer Drug Delivery. Adv. Funct. Mater. 2018, 28 (25), 1–10. 10.1002/adfm.201705086. [DOI] [Google Scholar]
- Medina-Sánchez M.; Xu H.; Schmidt O. G. Micro- and Nano-Motors: The New Generation of Drug Carriers. Ther. Delivery 2018, 9 (4), 303–316. 10.4155/tde-2017-0113. [DOI] [PubMed] [Google Scholar]
- Villa K.; Krejčová L.; Novotný F.; Heger Z.; Sofer Z.; Pumera M. Cooperative Multifunctional Self-Propelled Paramagnetic Microrobots with Chemical Handles for Cell Manipulation and Drug Delivery. Adv. Funct. Mater. 2018, 28 (43), 1–8. 10.1002/adfm.201804343. [DOI] [Google Scholar]
- Villa K.; Novotný F.; Zelenka J.; Browne M. P.; Ruml T.; Pumera M. Visible-Light-Driven Single-Component BiVO4Micromotors with the Autonomous Ability for Capturing Microorganisms. ACS Nano 2019, 13 (7), 8135–8145. 10.1021/acsnano.9b03184. [DOI] [PubMed] [Google Scholar]
- Lee S.; Kim J. Y.; Kim J.; Hoshiar A. K.; Park J.; Lee S.; Kim J.; Pané S.; Nelson B. J.; Choi H. A Needle-Type Microrobot for Targeted Drug Delivery by Affixing to a Microtissue. Adv. Healthc. Mater. 2020, 9 (7), 1–6. 10.1002/adhm.201901697. [DOI] [PubMed] [Google Scholar]
- Kagan D.; Benchimol M. J.; Claussen J. C.; Chuluun-Erdene E.; Esener S.; Wang J. Acoustic Droplet Vaporization and Propulsion of Perfluorocarbon-Loaded Microbullets for Targeted Tissue Penetration and Deformation. Angew. Chemie - Int. Ed. 2012, 51 (30), 7519–7522. 10.1002/anie.201201902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano S.; Esteban-Fernández De Ávila B.; Yáñez-Sedeño P.; Pingarrón J. M.; Wang J. Nano/Microvehicles for Efficient Delivery and (Bio)Sensing at the Cellular Level. Chem. Sci. 2017, 8 (10), 6750–6763. 10.1039/C7SC02434G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaboldi S.; Salinas G.; Karajić A.; Garrigue P.; Benincori T.; Bonetti G.; Cirilli R.; Bichon S.; Gounel S.; Mano N.; et al. Direct Dynamic Read-out of Molecular Chirality with Autonomous Enzyme-Driven Swimmers. Nat. Chem. 2021, 13 (12), 1241–1247. 10.1038/s41557-021-00798-9. [DOI] [PubMed] [Google Scholar]
- Chałupniak A.; Morales-Narváez E.; Merkoçi A. Micro and Nanomotors in Diagnostics. Adv. Drug Delivery Rev. 2015, 95, 104–116. 10.1016/j.addr.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Yuan K.; Jiang Z.; Jurado-Sánchez B.; Escarpa A. Nano/Micromotors for Diagnosis and Therapy of Cancer and Infectious Diseases. Chem. - A Eur. J. 2020, 26 (11), 2309–2326. 10.1002/chem.201903475. [DOI] [PubMed] [Google Scholar]
- Soto F.; Kupor D.; Lopez-Ramirez M. A.; Wei F.; Karshalev E.; Tang S.; Tehrani F.; Wang J. Onion-like Multifunctional Microtrap Vehicles for Attraction–Trapping–Destruction of Biological Threats. Angew. Chemie - Int. Ed. 2020, 59 (9), 3480–3485. 10.1002/anie.201913872. [DOI] [PubMed] [Google Scholar]
- Vilela D.; Stanton M. M.; Parmar J.; Sánchez S. Microbots Decorated with Silver Nanoparticles Kill Bacteria in Aqueous Media. ACS Appl. Mater. Interfaces 2017, 9 (27), 22093–22100. 10.1021/acsami.7b03006. [DOI] [PubMed] [Google Scholar]
- Xu D.; Zhou C.; Zhan C.; Wang Y.; You Y.; Pan X.; Jiao J.; Zhang R.; Dong Z.; Wang W.; et al. Enzymatic Micromotors as a Mobile Photosensitizer Platform for Highly Efficient On-Chip Targeted Antibacteria Photodynamic Therapy. Adv. Funct. Mater. 2019, 29 (17), 1–11. 10.1002/adfm.201807727. [DOI] [Google Scholar]
- Stanton M. M.; Park B. W.; Vilela D.; Bente K.; Faivre D.; Sitti M.; Sánchez S. Magnetotactic Bacteria Powered Biohybrids Target E. Coli Biofilms. ACS Nano 2017, 11 (10), 9968–9978. 10.1021/acsnano.7b04128. [DOI] [PubMed] [Google Scholar]
- Hwang G.; Paula A. J.; Hunter E. E.; Liu Y.; Babeer A.; Karabucak B.; Stebe K.; Kumar V.; Steager E.; Koo H. Catalytic Antimicrobial Robots for Biofilm Eradication. Sci. Robot. 2019, 4 (29), 1–14. 10.1126/scirobotics.aaw2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K.; Jurado-Sánchez B.; Escarpa A. Dual-Propelled Lanbiotic Based Janus Micromotors for Selective Inactivation of Bacterial Biofilms. Angew. Chem. 2021, 133, 2–12. 10.1002/ange.202011617. [DOI] [PubMed] [Google Scholar]
- Vilela D.; Blanco-Cabra N.; Eguskiza A.; Hortelao A. C.; Torrents E.; Sanchez S. Drug-Free Enzyme-Based Bactericidal Nanomotors Against Pathogenic Bacteria. ACS Appl. Mater. Interfaces 2021, 13 (13), 14964–14973. 10.1021/acsami.1c00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortelão A. C.; Carrascosa R.; Murillo-Cremaes N.; Patiño T.; Sánchez S. Targeting 3D Bladder Cancer Spheroids with Urease-Powered Nanomotors. ACS Nano 2019, 13 (1), 429–439. 10.1021/acsnano.8b06610. [DOI] [PubMed] [Google Scholar]
- De Ávila B. E. F.; Angsantikul P.; Li J.; Angel Lopez-Ramirez M.; Ramírez-Herrera D. E.; Thamphiwatana S.; Chen C.; Delezuk J.; Samakapiruk R.; Ramez V.; et al. Micromotor-Enabled Active Drug Delivery for in Vivo Treatment of Stomach Infection. Nat. Commun. 2017, 8 (1), 1–8. 10.1038/s41467-017-00309-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Song Z.; Deng G.; Jiang K.; Wang H.; Zhang X.; Han H. Gastric Acid Powered Nanomotors Release Antibiotics for In Vivo Treatment of Helicobacter Pylori Infection. Small 2021, 17, 2006877. 10.1002/smll.202006877. [DOI] [PubMed] [Google Scholar]
- Xie L.; Pang X.; Yan X.; Dai Q.; Lin H.; Ye J.; Cheng Y.; Zhao Q.; Ma X.; Zhang X.; et al. Photoacoustic Imaging-Trackable Magnetic Microswimmers for Pathogenic Bacterial Infection Treatment. ACS Nano 2020, 14 (3), 2880–2893. 10.1021/acsnano.9b06731. [DOI] [PubMed] [Google Scholar]
- Torres M. D. T.; Sothiselvam S.; Lu T. K.; de la Fuente-Nunez C. Peptide Design Principles for Antimicrobial Applications. J. Mol. Biol. 2019, 431 (18), 3547–3567. 10.1016/j.jmb.2018.12.015. [DOI] [PubMed] [Google Scholar]
- Der Torossian Torres M.; De La Fuente-Nunez C. Reprogramming Biological Peptides to Combat Infectious Diseases. Chem. Commun. 2019, 55 (100), 15020–15032. 10.1039/C9CC07898C. [DOI] [PubMed] [Google Scholar]
- Mercer D. K.; Torres M. D. T.; Duay S. S.; Lovie E.; Simpson L.; von Köckritz-Blickwede M.; de la Fuente-Nunez C.; O’Neil D. A.; Angeles-Boza A. M. Antimicrobial Susceptibility Testing of Antimicrobial Peptides to Better Predict Efficacy. Front. Cell. Infect. Microbiol. 2020, 10, 326. 10.3389/fcimb.2020.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M. D. T.; Cao J.; Franco O. L.; Lu T. K.; de la Fuente-Nunez C. Synthetic Biology and Computer-Based Frameworks for Antimicrobial Peptide Discovery. ACS Nano 2021, 15 (2), 2143–2164. 10.1021/acsnano.0c09509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forier K.; Raemdonck K.; De Smedt S. C.; Demeester J.; Coenye T.; Braeckmans K. Lipid and Polymer Nanoparticles for Drug Delivery to Bacterial Biofilms. J. Controlled Release 2014, 190, 607–623. 10.1016/j.jconrel.2014.03.055. [DOI] [PubMed] [Google Scholar]
- Baelo A.; Levato R.; Julián E.; Crespo A.; Astola J.; Gavaldà J.; Engel E.; Mateos-Timoneda M. A.; Torrents E. Disassembling Bacterial Extracellular Matrix with DNase-Coated Nanoparticles to Enhance Antibiotic Delivery in Biofilm Infections. J. Controlled Release 2015, 209, 150–158. 10.1016/j.jconrel.2015.04.028. [DOI] [PubMed] [Google Scholar]
- Dai T.; Li N.; Han F.; Zhang H.; Zhang Y.; Liu Q. AMP-Guided Tumour-Specific Nanoparticle Delivery via Adenosine A1 Receptor. Biomaterials 2016, 83, 37–50. 10.1016/j.biomaterials.2016.01.011. [DOI] [PubMed] [Google Scholar]
- Ma X.; Hortelão A. C.; Patiño T.; Sánchez S. Enzyme Catalysis to Power Micro/Nanomachines. ACS Nano 2016, 10 (10), 9111–9122. 10.1021/acsnano.6b04108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patiño T.; Arque X.; Mestre R.; Palacios L.; Sánchez S. Fundamental Aspects of Enzyme-Powered Micro- and Nanoswimmers. Acc. Chem. Res. 2018, 51 (11), 2662–2671. 10.1021/acs.accounts.8b00288. [DOI] [PubMed] [Google Scholar]
- Lima S. M. F.; Freire M. S.; Gomes A. L. O.; Cantuária A. P. C.; Dutra F. R. P.; Magalhães B. S.; Sousa M. G. C.; Migliolo L.; Almeida J. A.; Franco O. L.; et al. Antimicrobial and Immunomodulatory Activity of Host Defense Peptides, Clavanins and LL-37, In Vitro : An Endodontic Perspective. Peptides 2017, 95, 16–24. 10.1016/j.peptides.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Silva O. N.; Torres M. D. T.; Cao J.; Alves E. S. F.; Rodrigues L. V.; Resende J. M.; Lião L. M.; Porto W. F.; Fensterseifer I. C. M.; Lu T. K.; et al. Repurposing a Peptide Toxin from Wasp Venom into Antiinfectives with Dual Antimicrobial and Immunomodulatory Properties. Proc. Natl. Acad. Sci. U. S. A. 2020, 118 (3), e2025351118. 10.1073/pnas.2012379117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M. D. T.; Pedron C. N.; Higashikuni Y.; Kramer R. M.; Cardoso M. H.; Oshiro K. G. N.; Franco O. L.; Silva Junior P. I.; Silva F. D.; Oliveira Junior V. X.; et al. Structure-Function-Guided Exploration of the Antimicrobial Peptide Polybia-CP Identifies Activity Determinants and Generates Synthetic Therapeutic Candidates. Commun. Biol. 2018, 1 (1), 221. 10.1038/s42003-018-0224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire K. A.; Torres M. D. T.; Lima D. B.; Monteiro M. L.; Bezerra de Menezes R. R. P. P.; Martins A. M. C.; Oliveira V. X. Jr Wasp Venom Peptide as a New Antichagasic Agent. Toxicon 2020, 181, 71–78. 10.1016/j.toxicon.2020.04.099. [DOI] [PubMed] [Google Scholar]
- Torres M. D. T.; Silva A. F.; Andrade G. P.; Pedron C. N.; Cerchiaro G.; Ribeiro A. O.; Oliveira V. X.; Fuente-Nunez C. The Wasp Venom Antimicrobial Peptide Polybia-CP and Its Synthetic Derivatives Display Antiplasmodial and Anticancer Properties. Bioeng. Transl. Med. 2020, 5 (3), 1–7. 10.1002/btm2.10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arqué X.; Andrés X.; Mestre R.; Ciraulo B.; Ortega Arroyo J.; Quidant R.; Patiño T.; Sánchez S. Ionic Species Affect the Self-Propulsion of Urease-Powered Micromotors. Research 2020, 2020, 1–14. 10.34133/2020/2424972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arqué X.; Romero-Rivera A.; Feixas F.; Patiño T.; Osuna S.; Sánchez S. Intrinsic Enzymatic Properties Modulate the Self-Propulsion of Micromotors. Nat. Commun. 2019, 10, 2826. 10.1038/s41467-019-10726-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi G.; Wang Y.; Estevez L.; Switzer A. K.; Duan X.; Yang X.; Giannelis E. P. Facile and Scalable Synthesis of Monodispersed Spherical Capsules with a Mesoporous Shell. Chem. Mater. 2010, 22 (9), 2693–2695. 10.1021/cm100174e. [DOI] [Google Scholar]
- Howse J. R.; Jones R. A. L.; Ryan A. J.; Gough T.; Vafabakhsh R.; Golestanian R. Self-Motile Colloidal Particles: From Directed Propulsion to Random Walk. Phys. Rev. Lett. 2007, 99, 048102. 10.1103/PhysRevLett.99.048102. [DOI] [PubMed] [Google Scholar]
- Dunderdale G.; Ebbens S.; Fairclough P.; Howse J. Importance of Particle Tracking and Calculating the Mean-Squared Displacement in Distinguishing Nanopropulsion from Other Processes. Langmuir 2012, 28 (30), 10997–11006. 10.1021/la301370y. [DOI] [PubMed] [Google Scholar]
- Ma X.; Wang X.; Hahn K.; Sánchez S. Motion Control of Urea-Powered Biocompatible Hollow Microcapsules. ACS Nano 2016, 10 (3), 3597–3605. 10.1021/acsnano.5b08067. [DOI] [PubMed] [Google Scholar]
- Zhan X.; Wang J.; Xiong Z.; Zhang X.; Zhou Y.; Zheng J.; Chen J.; Feng S. P.; Tang J. Enhanced Ion Tolerance of Electrokinetic Locomotion in Polyelectrolyte-Coated Microswimmer. Nat. Commun. 2019, 10 (1), 3921. 10.1038/s41467-019-11907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hincapié-Rojas D. F.; Rojas-Hernández S. P.; Castaño-González F.; Parra-Castaño K. N.; Giraldo-Torres L. R. Mesoporous Silica Nanoparticles Obtention, Functionalization and Biomedical Applications: A Review. Dyna 2020, 87 (215), 239–253. 10.15446/dyna.v87n215.88586. [DOI] [Google Scholar]
- Tang F.; Li L.; Chen D. Mesoporous Silica Nanoparticles: Synthesis, Biocompatibility and Drug Delivery. Adv. Mater. 2012, 24 (12), 1504–1534. 10.1002/adma.201104763. [DOI] [PubMed] [Google Scholar]
- Li Z.; Barnes J. C.; Bosoy A.; Stoddart J. F.; Zink J. I. Mesoporous Silica Nanoparticles in Biomedical Applications. Chem. Soc. Rev. 2012, 41 (7), 2590. 10.1039/c1cs15246g. [DOI] [PubMed] [Google Scholar]
- Llopis-Lorente A.; Garciá-Fernández A.; Murillo-Cremaes N.; Hortelaõ A. C.; Patinõ T.; Villalonga R.; Sancenón F.; Martínez-Máñez R.; Sánchez S. Enzyme-Powered Gated Mesoporous Silica Nanomotors for on-Command Intracellular Payload Delivery. ACS Nano 2019, 13 (10), 12171–12183. 10.1021/acsnano.9b06706. [DOI] [PubMed] [Google Scholar]
- He Q.; Cui X.; Cui F.; Guo L.; Shi J. Size-Controlled Synthesis of Monodispersed Mesoporous Silica Nano-Spheres under a Neutral Condition. Microporous Mesoporous Mater. 2009, 117 (3), 609–616. 10.1016/j.micromeso.2008.08.004. [DOI] [Google Scholar]
- Ma X.; Jannasch A.; Albrecht U. R.; Hahn K.; Miguel-López A.; Schäffer E.; Sánchez S. Enzyme-Powered Hollow Mesoporous Janus Nanomotors. Nano Lett. 2015, 15 (10), 7043–7050. 10.1021/acs.nanolett.5b03100. [DOI] [PubMed] [Google Scholar]
- Silva O. N.; Torres M. D. T.; Cao J.; Alves E. S. F.; Rodrigues L. V.; Resende J. M.; Lião L. M.; Porto W. F.; Fensterseifer I. C. M.; Lu T. K.; et al. Repurposing a Peptide Toxin from Wasp Venom into Antiinfectives with Dual Antimicrobial and Immunomodulatory Properties. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (43), 26936–26945. 10.1073/pnas.2012379117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M. D. T.; Voskian S.; Brown P.; Liu A.; Lu T. K.; Hatton T. A. A.; de la Fuente-Nunez C. Coatable and Resistance-Proof Ionic Liquid for Pathogen Eradication. ACS Nano 2021, 15 (1), 966–978. 10.1021/acsnano.0c07642. [DOI] [PubMed] [Google Scholar]
- Säll J.; Carlsson M.; Gidlöf O.; Holm A.; Humlén J.; Öhman J.; Svensson D.; Nilsson B.-O.; Jönsson D. The Antimicrobial Peptide LL-37 Alters Human Osteoblast Ca 2+ Handling and Induces Ca2+-Independent Apoptosis. J. Innate Immun. 2013, 5 (3), 290–300. 10.1159/000346587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M. D. T.; Pedron C. N.; Higashikuni Y.; Kramer R. M.; Cardoso M. H.; Oshiro K. G. N.; Franco O. L.; Silva Junior P. I.; Silva F. D.; Oliveira Junior V. X.; et al. Structure-Function-Guided Exploration of the Antimicrobial Peptide Polybia-CP Identifies Activity Determinants and Generates Synthetic Therapeutic Candidates. Commun. Biol. 2018, 1 (1), 221. 10.1038/s42003-018-0224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M. D. T.; Voskian S.; Brown P.; Liu A.; Lu T. K.; Hatton T. A.; De La Fuente-Nunez C. Coatable and Resistance-Proof Ionic Liquid for Pathogen Eradication. ACS Nano 2021, 15 (1), 966–978. 10.1021/acsnano.0c07642. [DOI] [PubMed] [Google Scholar]
- Becerra S. C.; Roy D. C.; Sanchez C. J.; Christy R. J.; Burmeister D. M. An Optimized Staining Technique for the Detection of Gram Positive and Gram Negative Bacteria within Tissue. BMC Res. Notes 2016, 9 (1), 216. 10.1186/s13104-016-1902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero M.; Escámez M. J.; García M.; Duarte B.; Holguín A.; Retamosa L.; Jorcano J. L.; Río M. d.; Larcher F. In Vitro and In Vivo Wound Healing-Promoting Activities of Human Cathelicidin LL-37. J. Invest. Dermatol. 2008, 128 (1), 223–236. 10.1038/sj.jid.5701043. [DOI] [PubMed] [Google Scholar]
- Xu H.; Yan F.; Monson E. E.; Kopelman R. Room-Temperature Preparation and Characterization of Poly(Ethylene Glycol)-Coated Silica Nanoparticles for Biomedical Applications. J. Biomed. Mater. Res. - Part A 2003, 66 (4), 870–879. 10.1002/jbm.a.10057. [DOI] [PubMed] [Google Scholar]
- Cesaro A.; Torres M.; de la Fuente-Nunez C.. Methods for the Design and Characterization of Peptide Antibiotics. In Methods in Enzymology; Academic Press: Cambridge, USA, 2022; 10.1016/bs.mie.2021.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.