SUMMARY
We describe the investigation of two temporally coincident illness clusters involving salmonella and Staphylococcus aureus in two states. Cases were defined as gastrointestinal illness following two meal events. Investigators interviewed ill persons. Stool, food and environmental samples underwent pathogen testing. Alabama: Eighty cases were identified. Median time from meal to illness was 5·8 h. Salmonella Heidelberg was identified from 27 of 28 stool specimens tested, and coagulase-positive S. aureus was isolated from three of 16 ill persons. Environmental investigation indicated that food handling deficiencies occurred. Colorado: Seven cases were identified. Median time from meal to illness was 4·5 h. Five persons were hospitalised, four of whom were admitted to the intensive care unit. Salmonella Heidelberg was identified in six of seven stool specimens and coagulase-positive S. aureus in three of six tested. No single food item was implicated in either outbreak. These two outbreaks were linked to infection with Salmonella Heidelberg, but additional factors, such as dual aetiology that included S. aureus or the dose of salmonella ingested may have contributed to the short incubation periods and high illness severity. The outbreaks underscore the importance of measures to prevent foodborne illness through appropriate washing, handling, preparation and storage of food.
Key words: Epidemiology, food-borne infections, outbreaks, public health, water-borne infections
INTRODUCTION
Non-typhoidal salmonella (NTS) is a common cause of foodborne infection in the United States (US), causing 26% of foodborne illness outbreaks [1] and resulting in over 1 million illnesses, nearly 20 000 hospitalisations, and close to 400 deaths each year [2]. Although salmonella can be transmitted through a variety of mechanisms, the predominant mode of transmission is consumption of contaminated food [3]. The incubation period following exposure is commonly reported as 6–72 h, but is usually 12–36 h [4], and infection commonly manifests as acute enterocolitis. In the US, approximately 27% of laboratory-confirmed foodborne NTS infections result in hospitalisation, and case fatality is estimated at <0·5% [2]. Several studies have noted associations between larger infectious doses and shorter incubation period and/or increased severity of illness [5–9].
In contrast, only 3% of foodborne illness outbreaks in the US can be attributed to Staphylococcus aureus enterotoxin [1]. S. aureus causes foodborne intoxication through production of toxin in foods that have been inadequately heated or refrigerated [10, 11]. Once S. aureus enterotoxin is produced, heating can kill the bacteria but does not eliminate the heat-stable toxin. Symptoms include profuse vomiting and diarrhoea, and incubation periods are generally shorter than bacterial enteritides, ranging from 30 min to 8 h, but usually 2–4 h [10]. To attribute an outbreak to S. aureus intoxication, pre-formed toxin must be detected in food that was consumed, large numbers of S. aureus (>105 organisms per gram) must be recovered from epidemiologically implicated food, or matching S. aureus clinical isolates from two or more patient specimens must be recovered. Approximately 6% of laboratory-confirmed foodborne S. aureus intoxications result in hospitalisation, and case fatality is <0·1% [2].
On 8 July 2013, the Alabama Department of Public Health (ADPH) was notified by Sumter County health officials of a cluster of gastrointestinal illnesses among persons who shared a meal 2 days earlier at a church following a funeral service. On 16 July 2013, Colorado's Tri-County Health Department (TCHD) received a call from a local hospital about a cluster of illnesses among persons who shared a single home-prepared meal. In this report, we describe these two illness clusters and the investigations into their causes.
METHODS
Cases were defined as onset of gastrointestinal illness within 3 days of attending the meal events described above in Alabama (AL) or Colorado (CO). ADPH defined a case as gastrointestinal illness in a person who ate lunch at the church on 6 July and became ill within 72 h, and TCHD defined a case as gastrointestinal illness in a person who ate the home-prepared meal on 13 July 2013 in Aurora, CO and became ill within 72 h. State and local investigators interviewed event attendees to identify exposures potentially related to illness using standard and outbreak-specific questionnaires. Environmental investigations were conducted following the two events, including interviews with food preparers. In AL, environmental sampling was conducted, retail samples were collected from a local grocery store, and leftover food from the event was obtained from the homes of ill persons.
The Bureau of Clinical Laboratories (BCL) in AL and the Colorado Department of Public Health and Environment (CDPHE) performed testing on stool and blood samples related to the two events. Food samples from CO were not tested, given food samples collected (from trash) were infested with maggots and hence not suitable for testing. BCL tested clinical specimens and food for salmonella, shigella, Shiga toxin-producing Escherichia coli (STEC) and norovirus; salmonella isolates were serotyped using conventional techniques [12] and further subtyped by pulsed-field gel electrophoresis (PFGE) using XbaI as the primary restriction enzyme and, for several isolates, BlnI as the secondary enzyme [13]. PFGE patterns were analysed and compared with others in the National PulseNet database. CDPHE cultured clinical specimens for salmonella, campylobacter, STEC and S. aureus. Antimicrobial susceptibility testing (AST) was completed on select salmonella isolates by CDPHE.
Clinical specimens, bacterial isolates, food samples and environmental samples from AL and clinical specimens from CO underwent additional testing at laboratories at the Centers for Disease Control and Prevention (CDC). Clinical specimens, food and environmental samples were cultured for salmonella (standard identification methods); selected stool specimens were also cultured for Bacillus cereus [14], campylobacter, shigella, S. aureus (standard identification methods), STEC [15] and enterotoxigenic E. coli (ETEC) [16]. Isolates were tested by a rapid latex agglutination test for identification of S. aureus (Staphaurex®, Remel, Lenexa, KS, USA), for coagulase reactivity using the direct tube method (BD BBL Coagulase Plasma with EDTA, BD, Franklin Lakes, NJ, USA), and by polymerase chain reaction (PCR) for S. aureus enterotoxin (SET) genes (sea, b, c, d, e, h) [17]. PFGE was performed on the S. aureus isolates recovered using the SmaI restriction enzyme as described previously [18]. PFGE patterns were compared with the S. aureus library of strain types at the CDC using BioNumerics version 6·6. Isolates were assigned to pulsed-field types (PFTs) at 80% relatedness and to pulsed-field patterns at 95% relatedness by use of Dice coefficients [18]. Isolates were tested by PCR for B. cereus enterotoxin genes (ces, nhe, hbl, cytK), and STEC virulence genes (stx1, stx2, eae, ehxA). Food samples were tested for S. aureus (Tecra SIDVIA72, 3 M, St. Paul, MN, USA; Oxoid SET-RPLA, Thermo Fisher Scientific, Waltham, MA, USA) and B. cereus enterotoxins [14] (Tecra BDEVIA48, 3 M, St. Paul, MN, USA; Oxoid BCET-RPLA, Thermo Fisher Scientific), and stools were tested for Clostridium perfringens enterotoxin (CPE; Oxoid PET-RPLA, Thermo Fisher Scientific). Seven salmonella isolates recovered from food were analysed by a PulseNet standardised PFGE protocol with XbaI and BlnI restriction enzymes, analysed using BioNumerics software, and patterns were compared to others in the National PulseNet database. Subtyping was conducted through PulseNet, the national molecular subtyping network for foodborne disease surveillance. AST was conducted on three salmonella isolates recovered from stools using the National Antimicrobial Resistance Monitoring System (NARMS) standard panel [19]. Pure cultures were tested for susceptibility to amoxicillin/clavulanate, ampicillin, cefoxitin, ceftiofur, ceftriaxone, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfisoxazole, tetracycline and trimethoprim/sulfamethoxazole. Whole genome sequencing (WGS) and analysis was conducted by CDC on a total of 16 salmonella isolates, 10 from the AL cluster (five food and five clinical) and six clinical isolates from CO cases, to determine genetic relatedness of salmonella isolated in the two illness clusters. Isolates were prepared for WGS according to the PulseNet MiSeq protocol (https://www.cdc.gov/pulsenet/pathogens/wgs.html). As previously reported, metagenomic sequences were obtained from 11 specimens (six from AL and five from CO cases) and analysed for salmonella and S. aureus signatures, and for microbial community composition [20].
To help determine whether the AL and CO illness clusters had a common source, state and federal regulatory agencies conducted a traceback investigation of selected food items served at the two events.
RESULTS
AL investigation
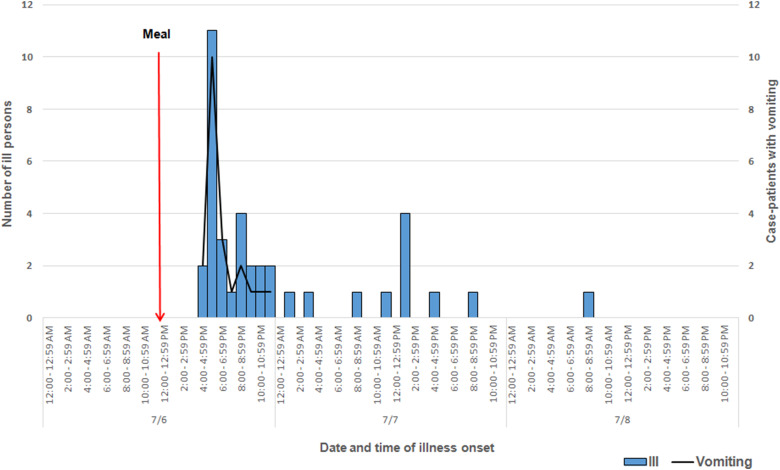
Among approximately 100 attendees of the church funeral service, 80 cases were identified. Among 38 ill persons interviewed, the median age was 35 years (range 4–75), 14 ill persons (37%) were male, 30 ill persons (79%) were hospitalised and one died. Median time from meal to illness onset was 5·8 h (range 2·5–42·5 h; see Fig. 1). Symptoms reported included diarrhoea (38; 100%), abdominal pain (36; 95%), nausea (32; 84%), objective fever >101°Fahrenheit (31; 82%) and vomiting (29; 76%; see Table 1). Of note, the proportion of ill persons with incubations <8 h who reported vomiting (16 of 17; 94%) was significantly greater than the proportion of ill persons with incubations ⩾8 h who reported vomiting (13 of 21; 62%; Fisher's exact test P-value 0·023). Food consumed by ill persons included fried chicken (21; 55% of ill persons consumed) and baked chicken (18; 47%), pulled pork (12; 32%), green beans (23; 61%), potato salad (27; 71%), macaroni and cheese (29; 76%) and cake (23; 61%). Analysis indicated that individuals who ate green beans were 27% more likely to become ill than individuals who did not eat green beans (RR = 1·27, P-value = 0·04). However, only 61% of the ill reported eating green beans. While analysis suggests that individuals who ate macaroni and cheese may have been 29% more likely to become ill than individuals who did not (RR = 1·29, P-value = 0·06), statistical significance was not reached. Over 90% of attendees who reported eating each food item became ill; no single food item was implicated. The person who died was found deceased 3 days after the meal; friends stated that he had complained of gastrointestinal symptoms the day after the church lunch.
Fig. 1.
Epidemic curve depicting date and time of symptom onset (N = 38) involved in an outbreak at a church meal event, all ill and case-patients reporting vomiting – Alabama, July 2013.
Table 1.
Summary of demographic and clinical characteristics of interviewed individuals involved in an outbreak at a church meal event – Alabama, July 2013
| Characteristic | Ill (N = 38) | Not ill (N = 4) |
|---|---|---|
| Age (years) | ||
| Mean (s.d.) | 35·8 (21·5) | 65·0 (7·1) |
| Range | 4–75 | 60–70 |
| Median | 35 | 65 |
| Gender, male (%) | 14 (36·8) | 1 (25·0) |
| Race (%) | ||
| African American | 30 (78·9) | 4 (100·0) |
| White/Caucasian | 2 (5·3) | – |
| Ethnicity, not Hispanic (%) | 35 (92·1) | 4 (100·0) |
| Incubation period (in hours) | ||
| Mean (s.d.) | 10·5 (9·7) | |
| Range | 2·5–42·5 | |
| Median | 5·8 | |
| Symptoms (yes), (%) | ||
| Diarrhoea | 38 (100·0) | |
| Abdominal pain | 36 (94·7) | |
| Nausea | 32 (84·2) | |
| Fever | 31 (81·6) | |
| Headache | 30 (78·9) | |
| Myalgia/muscle aches | 30 (78·9) | |
| Chills | 30 (78·9) | |
| Vomiting | 29 (76·3) | |
| Lethargy | 28 (73·7) | |
| Backache | 23 (60·5) | |
| Gas | 15 (39·5) | |
| Outcome (%) | ||
| Visited ER/physician | 38 (100·0) | |
| Hospitalised | 30 (78·9) | |
| Died | 1 (2·6) |
ADPH identified Salmonella serovar Heidelberg from 27 of 28 stool specimens tested. All 27 isolates yielded indistinguishable XbaI PFGE patterns (JF6X01·0022) and six of six isolates subtyped using a second enzyme had BlnI pattern JF6A26·0013. ADPH noted heavy growth in 23 of the stool specimens. Three stool isolates were susceptible to all antimicrobials on the NARMS panel. Coagulase-positive S. aureus carrying staphylococcal enterotoxin (SET) genes was isolated from stool samples from three (19%) of 16 ill persons; however, the three isolates represented two different SET genotypes (sec and sed). The sed-positive stool isolate was PFT USA 800 with PFGE pattern SAUSMA.0011. The two sec-positive isolates, from two individuals who shared a surname, matched by PFGE and were PFT USA 600 with PFGE pattern SAUSSMA.0036. Salmonella Heidelberg was also recovered from these stools. Cultures were negative for STEC, ETEC, shigella, campylobacter and B. cereus, and stools also tested negative for CPEs. Four of five blood samples tested by ADPH grew out Salmonella Heidelberg, as did one of six tested by CDC.
All seven isolates from food (baked chicken, pulled pork, green beans, potato salad) were confirmed as PFGE pattern combination JF6X01·0022/JF6A26·0013, indistinguishable from the stool isolates tested by ADPH. Coagulase-positive S. aureus carrying the sed toxin gene and SED toxin was detected in a cooked chicken sample from an attendee's leftover food plate. Likewise, an environmental swab from the counter on which the potato dish was prepared (prep counter) taken from the church kitchen yielded sed-positive S. aureus. However, PFGE subtyping on the sed-positive S. aureus isolates (from the chicken and the prep counter) revealed two PFTs, USA100 and USA800, with PFGE patterns SAURSMA.0166 and SAURSMA.0179. The stool isolate carrying the sed gene (mentioned above) did not match these environmental isolates.
The environmental assessment indicated that multiple food handling issues occurred during the meal preparation process. Prior to baking, raw chicken pieces were washed twice in the kitchen sink, where they were subsequently seasoned. After baking, the chicken was placed on a table in front of an air conditioning unit to cool for approximately 1·5 h prior to refrigeration. Cooked potatoes were drained in the same sink where raw chicken was earlier washed and seasoned. An inspection conducted on 9 July 2013 found the refrigerator in which food was stored prior to serving had a temperature at 46°Fahrenheit.
CO investigation
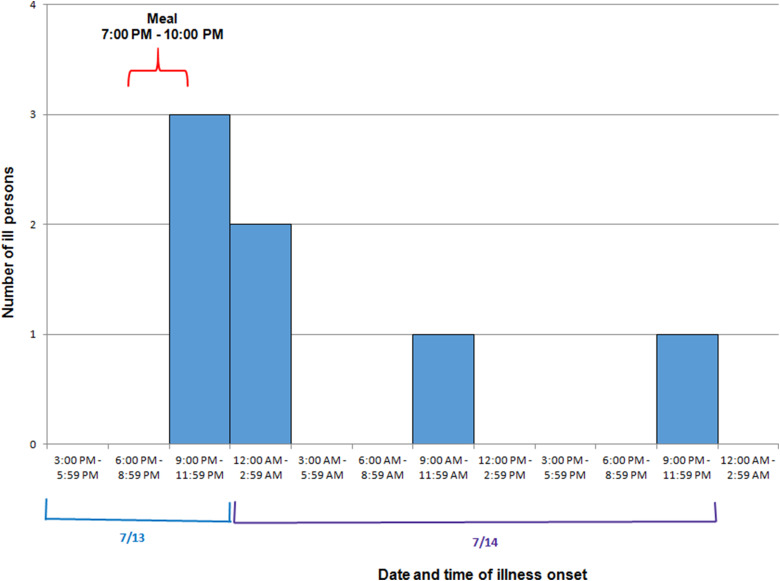
Among eight attendees, a total of seven cases were identified. Ages of ill persons ranged from 18 to 57 years with a mean of 45 years, and four were female (57%). Median time from meal to illness onset was 4·5 h (range 2·5–25 h; see Fig. 2). Five persons (71%) were admitted to the hospital. Of those, four persons were diagnosed with acute renal failure upon admission and four were admitted to the intensive care unit. One person was seen in the emergency department and discharged. One ill person who was not hospitalised died within 42–45 h after developing symptoms. Symptoms included diarrhoea (six of six; 100%), fever (five of five; 100%), headache (five of five; 100%), abdominal cramps (five of five; 100%) and vomiting (three of five; 60%; see Table 2).
Fig. 2.
Epidemic curve depicting date and time of symptom onset (N = 7) involved in an outbreak at a group meal event – Colorado, July 2013.
Table 2.
Summary of demographic and clinical characteristics of interviewed individuals involved in an outbreak at a group meal event – Colorado, July 2013
| Characteristic | Ill (N = 7*) | Number of case-patients for whom information is available |
|---|---|---|
| Age (years) | ||
| Mean | 45 | 7 |
| Range | 18–57 | 7 |
| Median | 55 | 7 |
| Gender, male (%) | 3 (42·9) | 7 |
| Race (%) | ||
| African American | 3 (60·0) | 5 |
| Mixed race (African American and Pacific Islander) | 2 (40·0) | 5 |
| Incubation period (in hours) | ||
| Mean | 8 | 7 |
| Range | 2·5–25 | 7 |
| Median | 5 | 7 |
| Symptoms (yes), (%) | ||
| Diarrhoea | 6 (100·0) | 6 |
| Bloody stool | 2 (40·0) | 5 |
| Abdominal cramps | 5 (100·0) | 5 |
| Fever | 5 (100·0) | 5 |
| Headache | 5 (100·0) | 5 |
| Vomiting | 3 (60·0) | 5 |
| Outcome (%) | ||
| Visited ER/physician | 6 (85·7) | 7 |
| Hospitalised | 5 (71·4) | 7 |
| Died | 1 (14·3) | 7 |
All interviewed persons had been ill.
In six of seven ill persons, Salmonella Heidelberg with PFGE-XbaI pattern JF6X01·0022, the same XbaI pattern as in the AL cluster was found, but the BlnI pattern, JF6A26·0001, differed by two bands from that in the AL cluster. AST testing was performed on three isolates and all were susceptible to all antimicrobials on the NARMS panel. Coagulase-positive S. aureus was isolated from the stool of three of six (50%) of the ill persons; two of these specimens also yielded Salmonella Heidelberg. Only one of four S. aureus stool isolates tested from the three persons was found to carry an SET gene (sea); the isolate with the sea toxin gene was from stool that was positive for Salmonella Heidelberg. The sea-positive isolate was S. aureus group D with PFGE pattern SAUSSMA.0253. The other three share PFGE pattern SAURSMA.0047 and were PFT USA 300; two of these three were from the same case-patient. Blood samples from two case-patients tested by CDPHE and CDC grew out Salmonella Heidelberg. These blood samples were from the deceased case-patient and a case-patient who was hospitalised for 12 days, including ICU admission. Two of the four persons with renal failure had S. aureus isolated from stool, and the sea toxin gene was from one of their stool samples. None of the S. aureus isolates matched the AL isolates by PFGE.
Gumbo and white rice were consumed by all seven ill persons; the only meal attendee who did not become ill denied consuming gumbo or white rice. The gumbo consisted of king crab, snow crab, shrimp, Andouille sausage, chicken, onions, bell peppers, chicken broth and spices. Interview of the meal preparer revealed that she boiled a whole, frozen chicken 2 days before serving. After cooking, the chicken was left on the counter to cool, then deboned and refrigerated. The next day, the chicken was combined with other ingredients, boiled in a large pot, then left to cool again for several hours prior to refrigeration. The following day, the gumbo was reheated to boiling prior to serving and rice was prepared on the day it was served. The meal preparer did not report any gastrointestinal illness in the week prior to preparing the meal, nor did any household members. Based on the investigation, no single ingredient could be implicated.
Traceback and WGS
Traceback of the chicken used at the two events was performed by the US Department of Agriculture's Food Safety and Inspection Service (USDA-FSIS). USDA-FSIS identified two different slaughter facilities for chicken used in AL and CO. WGS of 11 Salmonella Heidelberg isolates from ill persons in each of the outbreaks showed that the salmonella from the AL and CO illness clusters were genetically distinct; genomes from the two outbreaks were 105–116 single nucleotide polymorphisms different from each other. Metagenomic analysis also detected and differentiated salmonella signatures from the two outbreaks (two specimens from CO and three from AL) and identified distinct microbial community compositions (data not shown). Signatures of S. aureus, including multiple toxin-encoding genes, were detected in two samples from CO [20].
DISCUSSION
These two geographically distinct, temporally coincident outbreaks were caused by infection with Salmonella Heidelberg. Illnesses in these outbreaks occurred after an unusually short incubation period for salmonellosis; hospitalisation frequency was also higher than for typical outbreaks of salmonella infections. Although these outbreaks occurred only a week apart and the Salmonella Heidelberg had the same XbaI pattern, the epidemiological investigation identified no food source common to both events, suggesting that the outbreaks were unrelated. Further subtyping using PFGE with a second enzyme (BlnI) and WGS and metagenomic analysis also support the conclusion that these two outbreaks were not linked to a single common source. The biologic mechanism responsible for the shorter incubation period and higher illness severity in these two clusters is unknown. Of note, while renal dysfunction has been observed in 36% of patients infected with salmonella [21], four of seven ill persons (57%) involved in the CO outbreak had renal failure.
A dual aetiology might explain the unusual characteristics of the two outbreaks. Few reports have been published on outbreaks caused by a combination of bacterial infection and intoxication, due to bacteria capable of expressing enterotoxins [22]. Further, little is known about the physiological response to simultaneous infection and intoxication, such as Salmonella Heidelberg infection and S. aureus enterotoxins. SETs are pyrogenic toxins that cause immunosuppression, a superantigen effect [23–26], by altering T-cell regulation, and this property may have allowed salmonella to cause more severe disease and reduce the incubation period of the bacteria in the host. One food sample from AL did contain evidence of contamination with staphylococcal enterotoxin. Further, S. aureus itself is well known to cause Staphylococcus-associated glomerulonephritis, an immune complex-mediated renal disease [27]; a high proportion of case-patients in CO had renal failure. Additionally, from the AL outbreak, the proportion of ill persons with incubations <8 h who reported vomiting, a common symptom of S. aureus intoxication, was significantly greater than the proportion of ill persons with incubations ⩾ 8 h who reported vomiting, suggesting the plausibility of S. aureus playing a role. However, S. aureus is at times found in the stools of healthy people, and the S. aureus isolated from clinical specimens in this investigation represented more than one distinct enterotoxin gene. Further, the renal failure could have been due to profound dehydration alone. The evidence collected during this investigation was insufficient to determine whether coincident S. aureus intoxication with Salmonella Heidelberg infection was responsible for the short incubation periods, high hospitalisation frequency, or high frequency of renal failure. Nonetheless, the paucity of S. aureus culture evidence from patient samples does not rule out intoxication with SETs, which are heat-stable pre-formed toxins that may cause illness in fully cooked foods if present in sufficient quantity. This explanation would require multiple modes of contamination, as Salmonella Heidelberg was clearly viable in both outbreaks.
The dose of salmonella ingested by ill persons may have also contributed to the short incubation periods and high illness severity seen in both outbreaks. In prior salmonella outbreaks, persons self-reporting consumption of larger amounts of contaminated foods were associated with shorter median incubation periods [6, 7, 9], more severe symptoms [6, 9], higher frequency of hospitalisation [5, 7, 8] and longer duration of illness [6]. The long duration food left at unsafe temperatures at these events may have led to an unusually high dose of salmonella being present in the foods consumed.
Underlying health conditions of individuals who fell ill may have further contributed to the unusual profile of these outbreaks. Information on underlying health conditions was not collected.
Worthy of additional mention is the emetic illness caused by B. cereus, associated with a heat-stable peptide toxin. The emetic syndrome is characterised mainly by vomiting 0·5–6 h after contaminated food is ingested, and has been linked to foods including rice, noodles, pasta and pastry [28]. Its symptom profile and incubation period are identical to that of S. aureus intoxication. While investigation findings did not suggest a B. cereus aetiology, its involvement cannot be ruled out.
To facilitate a timely response, investigators will frequently characterise foodborne outbreaks based on the suspected food vehicle, incubation period, duration and clinical profile before results of laboratory testing are available [29]. When illness onset is <12 h after a suspected meal, investigators will often consider intoxication with preformed toxins of bacteria such as S. aureus and B. cereus; toxin-producing bacteria such as C. perfringens, Vibrio parahaemolyticus and ETEC; and chemical and heavy metal ingestion [30]. This investigation suggests that salmonella, particularly at higher ingested loads, should also be considered as an aetiologic agent when incubation periods are short.
Because it is not unusual to find salmonella or S. aureus on some food items, food safety measures should be taken to minimise the likelihood that they will cause illness [31, 32]. Consumers should cook meat, poultry and eggs thoroughly and avoid cross-contaminating foods by washing hands, utensils and cutting boards after they have been in contact with raw meat or poultry and before they touch other foods. Leftovers should be refrigerated promptly, and refrigerator temperatures should be kept below 40°Fahrenheit [33, 34]. Produce that is not pre-washed should be rinsed, but meats should not. Washing raw meat and poultry can spread bacteria because their juices may splash. Food preparers should routinely wash their hands with soap and water before preparing food. When foodborne illnesses are suspected, they should be reported immediately to the appropriate local health department. The AL and CO outbreaks reiterate the importance of measures to prevent foodborne illness through appropriate washing, handling, preparation and storage of food [31, 32], and highlight the severe illness that can result from infection with enteric pathogens such as salmonella.
ACKNOWLEDGMENTS
The authors thank Alabama Department of Public Health; Tri-County Health Department, Colorado; Colorado Department of Public Health and Environment; Alabama Bureau of Clinical Laboratories; CDC Enteric Diseases Laboratory Branch. This work was supported by the Centers for Disease Control and Prevention; the Alabama Department of Public Health; the Tri-County Health Department, Colorado; and the Colorado Department of Public Health and Environment.
ETHICAL STANDARDS
This work does not include human or animal experimentation.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Gould LH, et al. Surveillance for foodborne disease outbreaks – United States, 1998–2008. Morbidity and Mortality Weekly Report 2013; 62: 1–34. [PubMed] [Google Scholar]
- 2.Scallan E, et al. Foodborne illness acquired in the United States – major pathogens. Emerging Infectious Diseases 2011; 17: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anon. Salmonellosis. In Heymann DL, ed. Control of Communicable Diseases Manual, 19th edn. Washington DC, USA: American Public Health Association, 2008, pp. 534–539. [Google Scholar]
- 4.Hale CR, et al. Estimates of enteric illness attributable to contact with animals and their environments in the United States. Clinical Infectious Diseases 2012; 54: S472–S479. [DOI] [PubMed] [Google Scholar]
- 5.Rejnmark L, et al. Impact of infecting dose on severity of disease in an outbreak of food-borne Salmonella enteritidis. Scandinavian Journal of Infectious Diseases 1997; 29: 37–40. [DOI] [PubMed] [Google Scholar]
- 6.Mintz ED, et al. Dose-response effects in an outbreak of Salmonella enteritidis. Epidemiology and Infection 1994; 112: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glynn JR, Palmer SR. Incubation period, severity of disease, and infecting dose: evidence from a salmonella outbreak. American Journal of Epidemiology 1992; 136: 1369–1377. [DOI] [PubMed] [Google Scholar]
- 8.Glynn JR, Bradley DJ. The relationship between infecting dose and severity of disease in reported outbreaks of Samonella infections. Epidemiology and Infection 1992; 109: 371–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor DN, et al. An outbreak of salmonellosis associated with a fatality in a healthy child: a large dose and severe illness. American Journal of Epidemiology 1984; 119: 907–912. [DOI] [PubMed] [Google Scholar]
- 10.Anon. Staphylococcal food intoxication. In: Heymann DL, ed. Control of Communicable Diseases Manual, 19th edn. Washington, DC, USA: American Public Health Association, 2008, pp. 240–242. [Google Scholar]
- 11.Bennett SD, Walsh KA, Gould LH. Foodborne disease outbreaks caused by Bacillus cereus, Clostridium perfringens, and Staphylococcus aureus—United States, 1998–2008. Clinical Infectious Diseases 2013; 57: 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenner FW, McWhorter-Murlin AC. Identification and Serotyping of Salmonella. Atlanta, GA, USA: Centers for Disease Control and Prevention, 1998. [Google Scholar]
- 13.Ribot EM, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, salmonella, and shigella for PulseNet. Foodborne Pathogens and Disease 2006; 3: 59–67. [DOI] [PubMed] [Google Scholar]
- 14.Schoeni JL, Wong AC. Bacillus cereus food poisoning and its toxins. Journal of Food Protection 2005; 68: 636–648. [DOI] [PubMed] [Google Scholar]
- 15.Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. Journal of Clinical Microbiology 1998; 36: 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultsz C, et al. Detection of enterotoxigenic Escherichia coli in stool samples by using nonradioactively labeled oligonucleotide DNA probes and PCR. Journal of Clinical Microbiology 1994; 32: 2393–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moran GJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. New England Journal of Medicine 2006; 355: 666–674. [DOI] [PubMed] [Google Scholar]
- 18.McDougal LK, et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. Journal of Clinical Microbiology 2003; 41: 5113–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Surveillance Report for 2014 (Final Report). Atlanta, Georgia, USA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, 2016. [Google Scholar]
- 20.Huang AD, et al. Metagenomics of two severe foodborne outbreaks provides diagnostic signatures and signs of coinfection not attainable by traditional methods. Applied and Environmental Microbiology 2017; 83: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doorn KJ, Pierard D, Spapen H. Acute renal dysfunction in salmonella gastroenteritidis. Journal of Clinical Gastroenterology 2006; 40: 910–912. [DOI] [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention. Foodborne outbreak online database (http://wwwn.cdc.gov/foodborneoutbreaks). Accessed 8 October 2016.
- 23.Le Loir Y, Baron F, Gautier M. Staphylococcus aureus and food poisoning. Genetics and Molecular Research 2003; 2: 63–76. [PubMed] [Google Scholar]
- 24.Argudín MÁ, Mendoza MC, Rodicio MR. Food poisoning and Staphylococcus aureus enterotoxins. Toxins (Basel) 2010; 2: 1751–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortega E, et al. Multiple roles of Staphylococcus aureus enterotoxins: pathogenicity, superantigenic activity, and correlation to antibiotic resistance. Toxins (Basel) 2010; 2: 2117–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulhankova K, King J, Salgado-Pabón W. Staphylococcal toxic shock syndrome: superantigen-mediated enhancement of endotoxin shock and adaptive immune suppression. Immunologic Research 2014; 59: 182–187. [DOI] [PubMed] [Google Scholar]
- 27.Nadasdy T, Hebert LA. Infection-related glomerulonephritis: understanding mechanisms. Seminars in Nephrology 2011; 31: 369–375. [DOI] [PubMed] [Google Scholar]
- 28.Ehling-Schulz M, Fricker M, Scherer S. Bacillus cereus, the causative agent of an emetic type of food-borne illness. Molecular Nutrition and Food Research 2004; 48: 479–487. [DOI] [PubMed] [Google Scholar]
- 29.Hedberg CW, et al. The use of clinical profiles in the investigation of foodborne outbreaks in restaurants: United States, 1982–1997. Epidemiology and Infection 2008; 136: 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers HS, et al. Short incubation foodborne outbreaks [abstract 61453]. Presented at: the 131st Annual Meeting of American Public Health Association, 2003. (https://www.aphl.org/conferences/proceedings/Documents/2006_2nd_Annual_National_Foodborne_Epidemiologists_Meeting/30_Rogers_Etiologies.pdf). Accessed 9 December 2016.
- 31.United States Department of Agriculture Food Safety Inspection Service. Check your steps: food safe families (http://www.fsis.usda.gov/wps/portal/fsis/topics/food-safety-education/teach-others/fsis-educational-campaigns/check-your-steps). Accessed 8 October 2016.
- 32.United States Department of Agriculture Food Safety Inspection Service. Washing food: does it promote food safety? (http://www.fsis.usda.gov/wps/portal/fsis/topics/food-safety-education/get-answers/food-safety-fact-sheets/safe-food-handling/w85ashing-food-does-it-promote-ood-safety/washing-food). Accessed 8 October 2016.
- 33.Centers for Disease Control and Prevention. Be food safe: protect yourself from food poisoning, 2013. (http://www.cdc.gov/Features/befoodsafe/) Accessed 8 October 2016.
- 34.Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) (http://www.cdc.gov/narms/). Accessed 8 October 2016.