Abstract
Gasdermin proteins form large membrane pores in human cells that release immune cytokines and induce lytic cell death. Gasdermin pore formation is triggered by caspase-mediated cleavage during inflammasome signaling and is critical for defense against pathogens and cancer. We discovered gasdermin homologs encoded in bacteria that defended against phages and executed cell death. Structures of bacterial gasdermins revealed a conserved pore-forming domain that was stabilized in the inactive state with a buried lipid modification. Bacterial gasdermins were activated by dedicated caspase-like proteases that catalyzed site-specific cleavage and the removal of an inhibitory C-terminal peptide. Release of autoinhibition induced the assembly of large and heterogeneous pores that disrupted membrane integrity. Thus, pyroptosis is an ancient form of regulated cell death shared between bacteria and animals.
One-Sentence Summary:
Bacteria encode gasdermins that are activated by dedicated proteases, defend from phage, and induce cell death.
In mammals, gasdermin proteins execute pyroptotic cell death by oligomerizing into membrane pores that release inflammatory cytokines and induce cell lysis. The human genome encodes six gasdermin proteins (GSDMA to GSDME and pejvakin) including the prototypical member GSDMD (1–3). Gasdermin activation requires caspase- or granzyme-mediated cleavage of an inter-domain linker that liberates a lipophilic N-terminal domain (NTD) from a large inhibitory C-terminal domain (CTD) (4–6). Proteolysis enables gasdermin NTD oligomerization and the formation of membrane pores important for innate immunity in mammals and primitive eukaryotes (7–9). Recent structural analyses have explaind a mechanism of gasdermin pore formation (5, 6, 10, 11), but the evolutionary origin and biological roles of diverse gasdermin proteins remain unknown (12).
While analyzing bacterial anti-phage defense islands we identified uncharacterized genes with predicted homology to mammalian gasdermins (Table S1). Sequence analysis revealed 50 bacterial gasdermins (bGSDMs) that form a clade distinct from that of eukaryotic homologs (Fig. 1A and fig. S1) (7, 9, 13). We determined crystal structures of bGSDMs from Bradyrhizobium tropiciagri and Vitiosangium sp., which revealed that bGSDMs each adopt a shared overall architecture that exhibits notable homology to the mammalian gasdermin NTD (fig. S2B and Table S2), including conservation of a twisted central anti-parallel β sheet and the shared placement of connecting helices and strands throughout the periphery (Fig. 1B–C).
Fig. 1. Structures of bGSDMs reveal homology with mammalian cell death effectors.
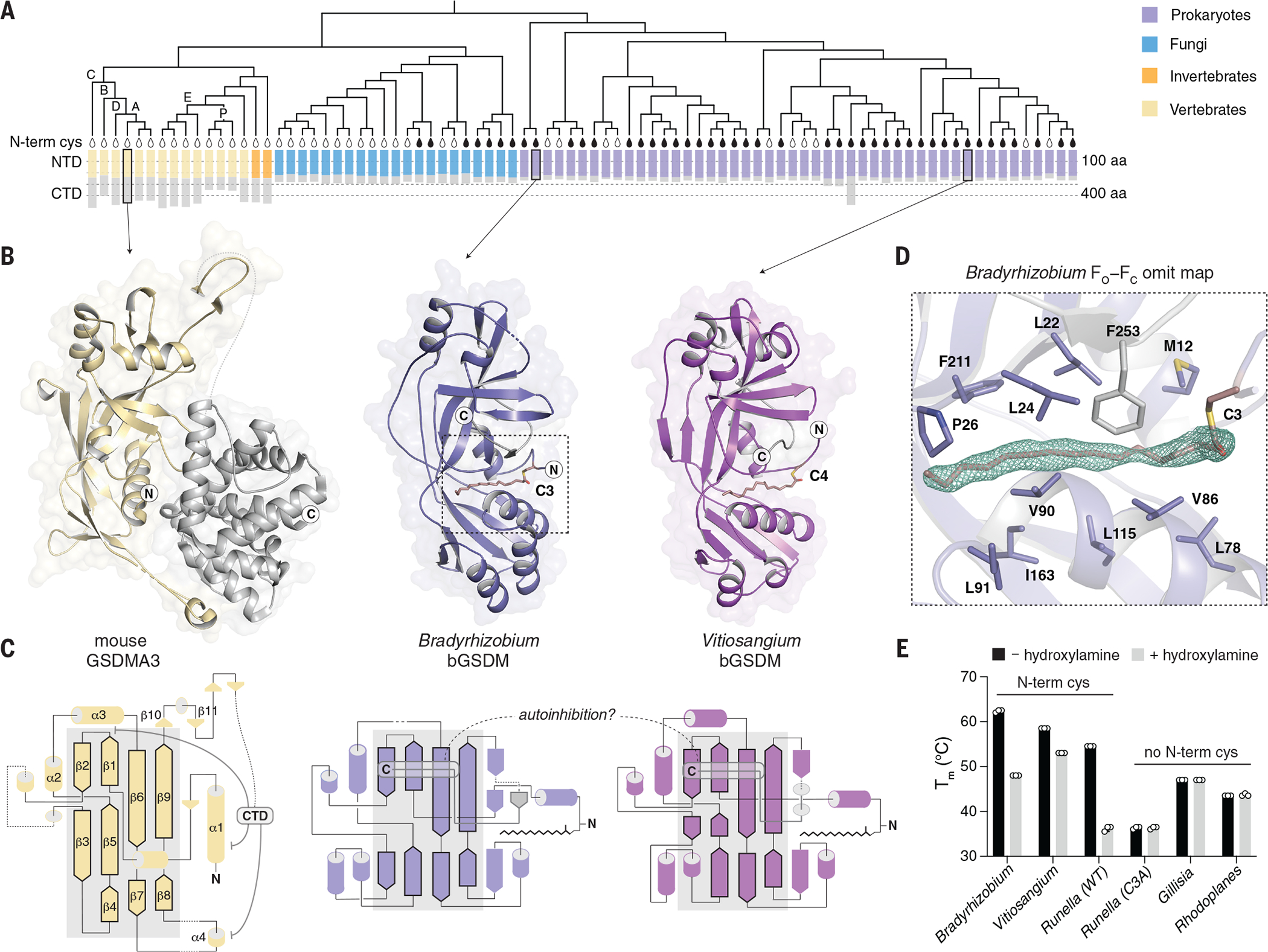
(A) Gasdermin phylogenetic tree. The sizes of the gasdermin NTDs and CTDs are depicted. Vertebrate gasdermins are labeled with single letters indicating human gasdermins GSDMA to GSDME (“A” to “E”), and “P” depicts pejvakin. The black teardrop indicates a conserved N-terminal cysteine(N-termin cys). A representative set of 20 fungal gasdermins are included in the tree. aa, amino acid. (B) Crystal structures of bGSDMs from species of the genera Bradyrhizobium and Vitiosangium. bGSDM structures reveal homology to the NTD of mammalian gasdermins in an inactive conformation including mouse GSDMA3 (Protein Data Bank ID 5B5R). (C) Gasdermin topology diagrams indicate a conserved central core of the bacterial and mammalian NTD. bGSDMs notably lack the CTD required for autoinhibition of mammalian gasdermins and instead encode a short C-terminal peptide. (D) Simulated annealing FO−FC omit map (contoured at 3.0 σ) from the Bradyrhizobium bGSDM fit with a palmitoyl modification at C3. Omit map is shown as green mesh and select residues forming a hydrophobic pocket around the palmitoyl group are indicated. (E) Melting temperatures (Tm) of bGSDMs with and without N-terminal cysteines, as determined with thermofluor assays. Data are the mean and standard deviation of three technical replicates and are representative of three independent experiments. WT, wild-type. Single-letter abbreviation for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
The structures revealed complete absence of the large α-helical CTD required to maintain mammalian gasdermins in an autoinhibited state (Fig. 1). Though lacking the CTD, the bGSDM structures adopted the same conformation as the inactive mammalian gasdermin complex (Fig. 1B–C). In the inactive structure of mammalian GSDMA3, the NTD forms two interfaces with the CTD that mediate autoinhibition, with the primary interface at the α1 helix and the β1–β2 hairpin (Fig. 1C) (5, 11). Cleavage of GSDMA3 results in NTD activation through the lengthening of strands β3, β5, β7, and β9 and oligomerization of ~27 protomers into a membrane-spanning pore (2, 5, 6). Both the Bradyrhizobium and Vitiosangium bGSDM structures contained strands equivalent to GSDMA3 β1 to β2 and β6 to β9, but in bGSDMs, a short C-terminal peptide wrapped around the twisted β-sheet core and terminated across strand β2 to stabilize the inactivated state (Fig. 1B–C).
While building the bGSDM atomic models, we observed a snakelike density protruding from the Bradyrhizobium cysteine C3 sidechain. The density occupies a hydrophobic tunnel across the protein that is capped by F25 from the C-terminal peptide. In the 1.5 Å Bradyrhizobium bGSDM electron density map, the density could be assigned as a 16-carbon palmitoyl thioester (Fig. 1D and fig. S2C). We confirmed bGSDM palmitoylation with mass spectrometry and found that a cysteine at this position is conserved in gasdermins across most bacteria and some fungi (Fig. 1A and fig. S3A–B). The presence of the palmitoyl in a hydrophobic cavity suggests that bGSDM palmitoylation occurs through autocatalysis (14). Palmitoylation contributes to stability of the inactive state protein (Fig. 1E), and modeling suggests substantial reorganization of residues along the hydrophobic tunnel during bGSDM activation (Fig. 1D and fig. S2C–D) (6).
The majority of bGSDMs (43 of 50) are genomically encoded next to one or more genes with a predicted protease domain (Fig. 2A and fig. S5A–C, Table S1). In most cases, the associated proteases are caspase-like peptidases including peptidase C14 (Pfam database ID PF00656) and CHAT (Pfam ID PF12770) proteases (Fig. 2B and fig. S5A). Fungal gasdermins are also commonly encoded next to protease domain-containing genes (40 of 52) (Table S3 and fig. S5B) and are activated through proteolysis (13). bGSDM-protease systems are found in diverse bacteria and archaea, as well as in metagenomic samples of prokaryotic origin (fig. S5D, Table S4). Analysis of the bGSDM-associated proteases revealed that they are fused to divergent repeat or NACHT domains frequently involved in pathogen recognition and inflammasome function in human innate immunity (Fig. 2B and fig. S5C) (15). bGSDM genes are occasionally encoded near known immune defense systems (Fig. 2A and fig. S7A, Tables S1, S4), so we tested bGSDM systems for a role in anti-phage defense. bGSDM systems evolutionarily distant from the model organism Escherichia coli exhibited no discernible phage restriction (fig. S6). However, a four gene operon from Lysobacter enzymogenes exhibited robust defense against coliphages T4, T5, and T6 (Fig. 2C–D and fig. S6B–C). Deletion of the bGSDM gene from the Lysobacter operon abolished protection (Fig. 2C–D and fig. S6C). Thus, the bGSDM is essential for defense.
Fig. 2. bGSDMs are associated with proteases, defend from phages, and execute cell death.
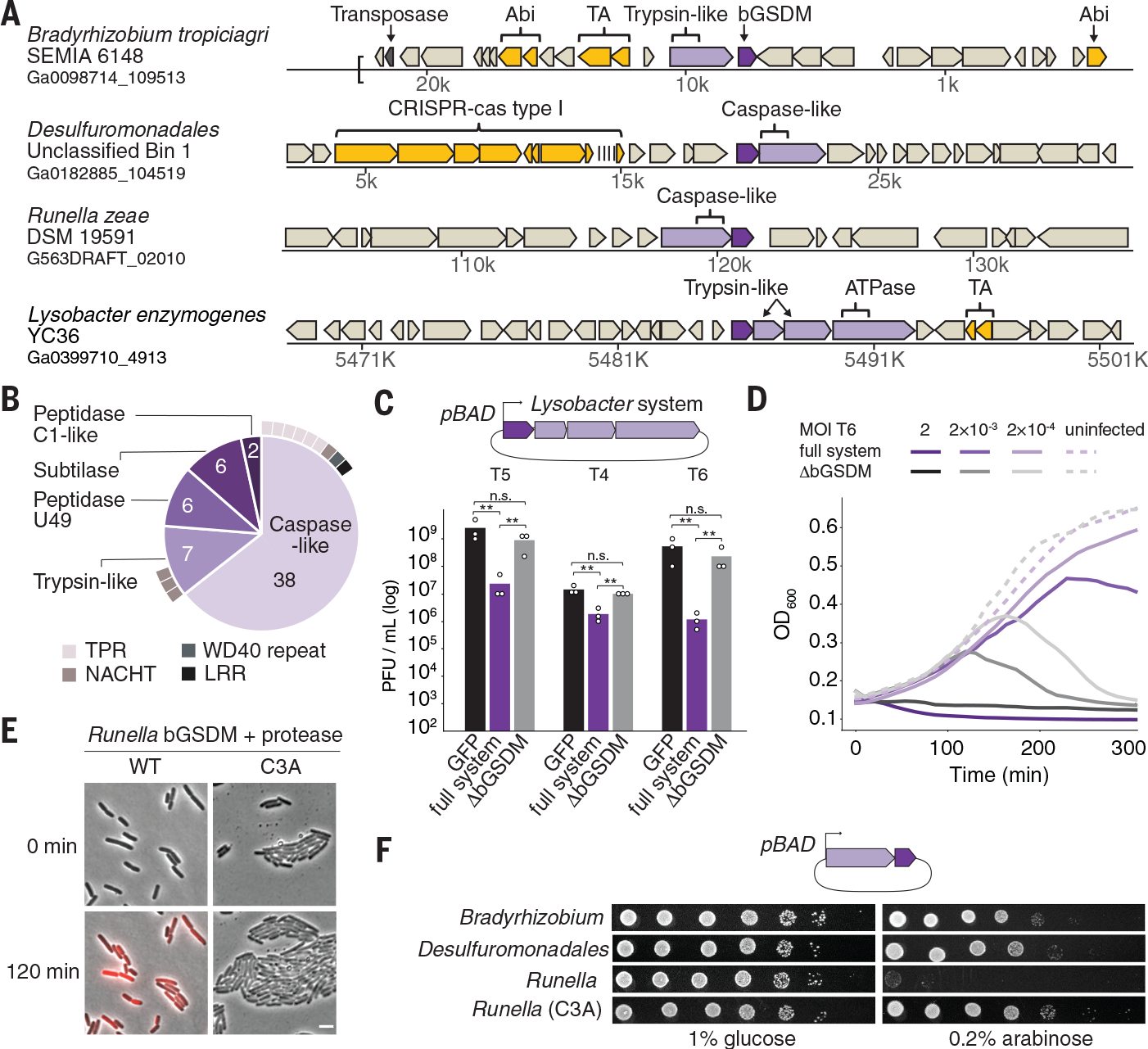
(A) Representative instances of bGSDMs and associated proteases, in their genomic neighborhoods. Genes known to be involved in anti-phage defense are shown in yellow. TA, toxin-antitoxin; Abi, abortive infection; ATPase, adenosine triphosphatase. (B) Types of proteases found adjacent to bGSDMs (n = 59). Some bGSDMs appear with more than one adjacent protease. Caspase-like proteases include peptidase C14 (n = 15) and CHAT (n = 23). Cases in which the protease gene also encodes an additional domain are indicated. TPR, tetratricopeptide repeat; LRR, leucine-rich repeat. (C) A bGSDM-containing operon protects against phages. The efficiency of plating of phages on E. coli MG1655 cells expressing the Lysobacter bGSDM WT or mutated operon is shown. Data represent plaque-forming units (PFU) per milliliter and are the averages of three independent replicates, with individual data points overlaid. GFP represents a control strain. Statistical significance was determined by a one-way analysis of variance (ANOVA) and Tukey multiple comparison test. Not significant (n.s.) ≥ 0.05, **P = 0.001 to 0.01. (D) Growth of liquid cultures of E. coli expressing the WT and mutated Lysobacter bGSDM operons. Cells were infected with phage T6. For each experiment, data represent one out of three biological replicates (replicates are shown in Fig. S6). OD600, optical density at 600 nm. (E) The Runella bGSDM operon causes cell death. E. coli DH5α cells expressing the Runella protease and WT or C3A mutated bGSDM were examined by time lapse microscopy. Overlay images from PI (red) and phase contrast of cells captured at the start of the experiment and after 120 min of incubation are shown. Scale bar, 2 μm. (F) bGSDM operons are toxic. Cells encoding protease and WT or mutated bGDSM were plated in 10-fold serial dilution on LB-agar in conditions that repress operon expression (1% glucose) or induce expression (0.2% arabinose).
Expression of some of the bGSDM-protease systems in E. coli induced potent cellular toxicity in the absence of phage infection (Fig. 2E–F, fig. S7B–C, and Table S5). Particularly strong toxicity was observed for a Runella system, which required bGSDM palmitoylation (Fig. 2E and fig. S7B–C). Time-lapse microscopy in the presence of propidium iodide (PI) showed that cells expressing the Runella system ceased dividing and lost membrane integrity, which suggests that bGSDM activation induces membrane disruption (Fig. 2E–F and fig. S7D, Movies S1–S2). Mutation of the predicted Runella caspase-like protease active site residues H796 and C840 ablated all cellular toxicity (Fig. 3A). The Runella bGSDM and protease only induced cellular toxicity when expressed together, which suggests that the protease targets bGSDM during system activation (Fig. 3A and fig. S8A). In fact, a mutation that disrupted the active site of the second trypsin-like protease in the Lysobacter bGSDM system abolished anti-phage defense (fig. S8B).
Fig. 3. bGSDMs are activated by proteolytic cleavage.
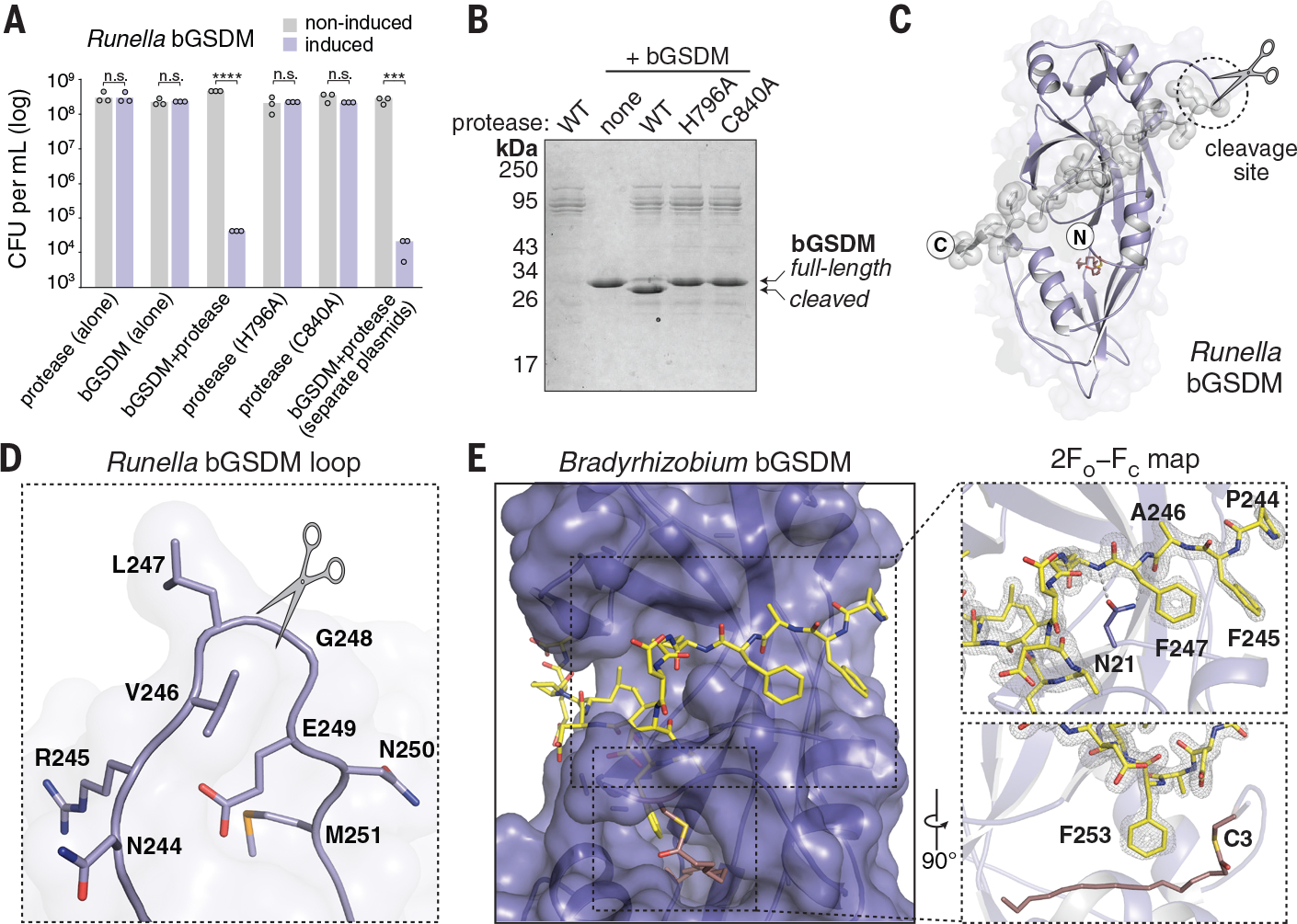
(A) Toxicity of Runella bGSDM in vivo requires the associated protease. Bacteria expressing WT and mutated versions of the Runella bGSDM–protease operon were grown on LB-agar in conditions that repress or induce expression. Data represent colony-forming units (CFU) per milliliter, and bar graphs represent an average of three independent replicates, with individual data points overlaid. Asterisks indicate statistically significant differences compared with the respective noninduced control using two-sided t-test. n.s. ≥ 0.05; ***P = 0.0001–0.001; ****P < 0.0001. (B) Runella bGSDM cleavage by its associated protease is dependent on catalytic histidine and residues in vitro. 15% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) were run after cleavage at room temperature for 18 h and visualized by Coomassie staining. (C) The Runella bGSDM crystal structure and protease cleavage site. The Runella bGSDM structure is shown in lavender with the last 21 amino acids highlighted as gray spheres. (D) Close-up view of the Runella bGSDM cleavage site wherein cleavage occurs after the P1 L247 residue. (E) Structural overview of the Bradyrhizobium CTD and autoinhibitory interactions. The bGSDM is colored purple except for its last 16 residues, which are colored yellow. Insets show interactions of F245 and F247 adjacent to D21 of the N-terminal β sheet (top) and F253 and the palmitoyl modification at C3 (bottom). The 2FO−FC (contoured at 1.5 σ) map is shown as gray mesh fit to the last 16 residues.
We focused on the Runella system and reconstituted cleavage with purified components (Fig. 3B and fig. S8A–F). Co-incubation with the protease resulted in specific bGSDM cleavage and formation of a lower molecular weight Runella bGSDM species (fig. S8A). Cleavage requires the protease catalytic residues but not bGSDM palmitoylation (Fig. 3B and fig. S8D–E). Using mass spectrometry, we determined that the Runella bGSDM cleavage site occurs after the P1 residue L247 (fig. S9A–B). A 2.9 Å structure of the Runella bGSDM (Table S2) revealed that cleavage occurs in a loop that immediately precedes the C-terminal peptide (Fig. 3C–D). Packing in the Runella bGSDM crystal lattice additionally indicates an ability of the peptide to dissociate from the bGSDM face which supports release after cleavage (fig. S10A).
Analysis of the high-resolution Bradyrhizobium bGSDM structure explains how the C-terminal peptide restrains the bGSDM core (Fig. 3E). Bradyrhizobium bGSDM F245 and F247 lay along the surface formed between the mammalian β9 strand and the α1 helix equivalent positions and are further supported with contacts between N21 and the peptide backbone. A Bradyrhizobium-specific β-strand from N21 to L24 extends off the β9 strand equivalent and is supported by a short parallel β strand from F253 to D255. Bradyrhizobium bGSDM F253 latches over the palmitoyl modification, with similar hydrophobic contacts also observed in the Runella and Vitiosangium structures (fig. S10A–B). The Bradyrhizobium bGSDM C-terminal peptide terminates below the strand equivalent to β2 and is supported by hydrogen bonds from R27 to the L256 backbone and N29 to E258. Truncation of the C-terminal peptide in the Runella, Bradyrhizobium, or Vitiosangium bGSDM constructs led to arrested cell growth which confirms that the C-terminal peptide is required to maintain the bGSDM autoinhibition (fig. S11A–B).
We next used mutagenesis of the Runella bGSDM system to define the specificity of proteolytic cleavage and bGSDM activation. In vitro, the L247 P1 position was essential for cleavage, and proteolysis was inhibited by mutations that disrupt the P1′ glycine and the P4, P3, P2 and P3′ residues (fig. S11C–D). Likewise, mutations that disrupt the P1 and P1′ positions eliminated toxicity in vivo (fig. S11E–F). The Runella protease was not capable of activating divergent bGSDMs engineered to contain the Runella cleavage loop, which suggests that additional contacts specify bGSDM recognition (fig. S11G–H). Thus, like their mammalian homologs, bGSDMs are cell death effectors activated by proteolytic cleavage.
To determine whether activated bGSDMs associate with bacterial membranes, we fused green fluorescent protein (GFP) to the N-terminus of the Runella bGSDM and visualized expression in E. coli. Upon co-expression with the Runella protease, GFP-bGSDM coalesced into membrane-associated puncta and induced cellular toxicity (Fig. 4A and fig. S12A–C). Transmission electron microscopy analysis of E. coli expressing the active Runella bGSDM system revealed clear disruption of membrane integrity (fig. S13A–C). In vitro reconstituted Runella bGSDM activity demonstrated that cleaved Runella bGSDMs permeabilized liposomes and caused rapid release of the internal contents (Fig. 4B and fig. S14A–B). Protease active-site or bGSDM cleavage-site mutations disrupted all liposome permeabilization, which confirms that proteolysis is essential for bGSDM activation (Fig. 4B and fig. S14B). Blocking bGSDM palmitoylation with mutation of residue C3 reduced but did not abolish liposome leakage or membrane-associated puncta formation in cells (Fig. 4B and fig. S12A). Likewise, a C7A mutation to the putative Lysobacter bGSDM palmitoylation site was not sufficient to abolish anti-phage defense (fig. S8B), which suggests that lipid-modification supports but is not required for membrane permeabilization.
Fig. 4. Cleaved bGSDMs form membrane pores to elicit cell death.
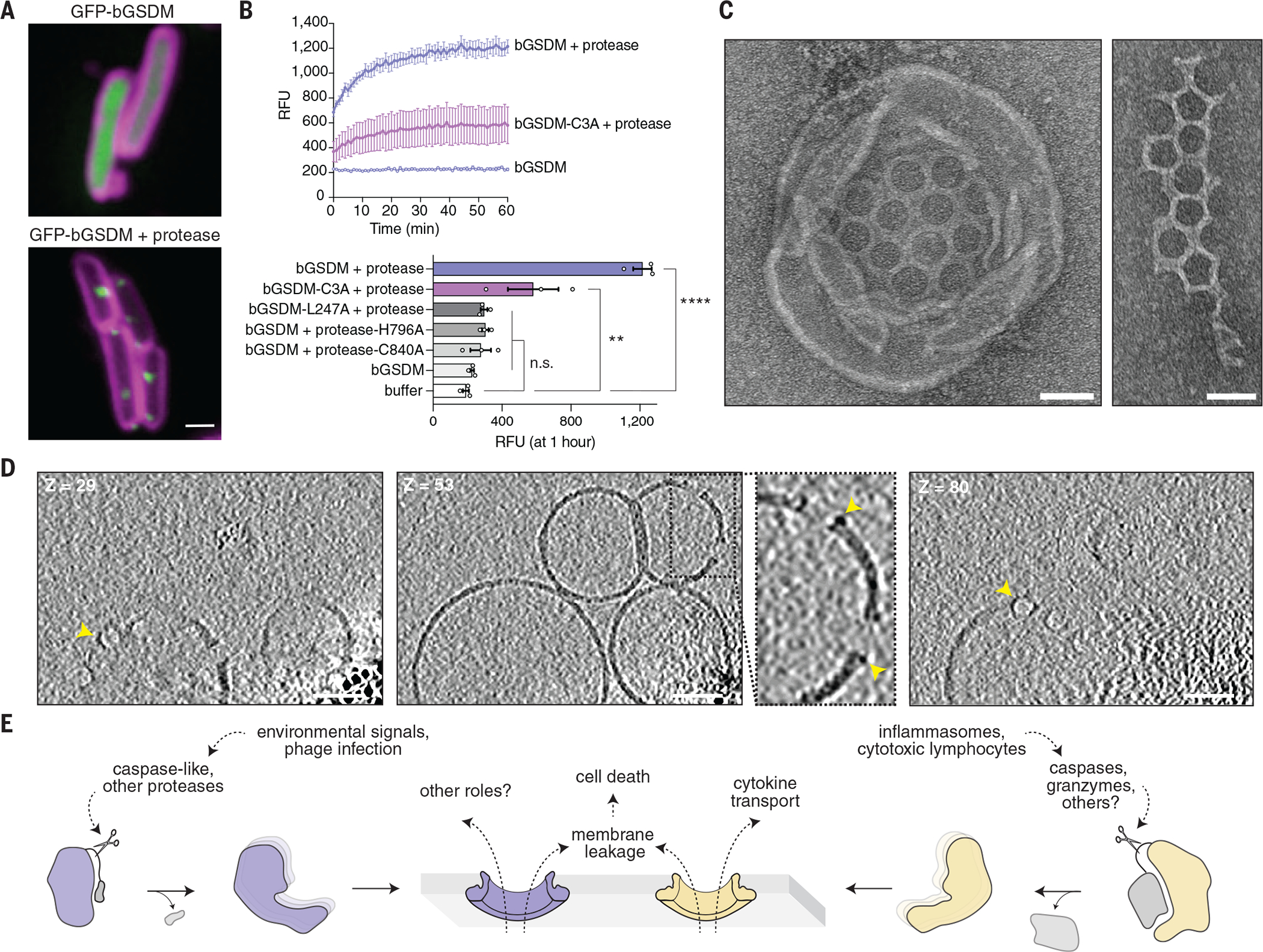
(A) GFP was fused to the N-terminus of the Runella bGSDM. Cells expressing GFP-bGSDM alone (top) or with the caspase-like protease (bottom) are shown. GFP is colored green. Membrane dye (FM4-64) is in magenta. Scale bar, 1 μm. (B) Cleaved Runella gasdermin permeabilizes liposome membranes. Relative fluorescence units (RFU) were measured continuously from cleavage reactions of dioleoylphosphatidylcholine (DOPC) liposomes loaded with TbCl3 with an external solution containing 20 μM dipicolonic acid (DPA). The top plot represents an example of time-course liposome leakage, whereas the bottom bar chart shows values for each condition at 60 min. Error bars represent the SEM of three technical replicates and statistical significance was determined by one-way ANOVA and Tukey multiple comparison test. n.s. ≥ 0.05; **P = 0.001–0.01; ****P < 0.0001. (C) Negative stain electron microscopy of Runella gasdermin pores in DOPC liposomes (left) and in mesh-like arrays (right). Scale bars, 50 nm. (D) Slices from representative tomogram (1 of 10) of Runella gasdermin pores in DOPC liposomes, at three different depths (Z). Yellow arrowheads indicate pores inserted within the liposome membrane. Scale bars, 50 nm. (E) Model of pyroptosis for bGSDMs and mammalian gasdermins.
To compare the bGSDM pore with its mammalian counterparts, we used electron microscopy to visualize Runella bGSDM cleavage reactions and liposomes (Fig. 4C and fig. S15A–C). bGSDM pores were observed within liposomes and as fragmented mesh-like arrays. Cryo-electron microscopy (cryo-EM) and two-dimensional (2D) classification analysis of detergent-solubilized complexes revealed that Runella bGSDM forms a ring-like pore that exhibits a width of ~50 Å and an inner diameter ranging from 200 to 300 Å (fig. S17A–D). Runella bGSDM pores within liposomes measured ~240 to 330 Å, modestly larger than the 135 to 215 Å mammalian gasdermin pores (fig. S17A–D) (6, 10, 16). We also reconstituted cleavage of a bGSDM from a metagenomic Bacteroidetes scaffold and observed smaller 130 to 190 Å pores within liposomes, which suggests heterogeneity in the architecture of diverse bGSDM pores (fig. S18A–D and fig. S19A–C). Cryo-electron tomography (cryo-ET) tilt series reconstructions of the pore-liposome assemblies confirmed that bGSDM pores span the liposomal surface to disrupt membrane integrity (Fig. 4D, fig. S20, and Movie S3–S4).
Our results support a model for gasdermin pore formation and effector function that has notable parallels between bacteria and mammals (Fig. 4E). bGSDM systems can exert antiphage defense, and the fusion of bGSDM-associated proteases with NACHT and repeat domains suggests that similar to inflammasome sensors in mammals, foreign pathogen recognition may control the initiation of gasdermin cleavage (Fig. 2B–D) (15, 17). In both mammalian gasdermin and bGSDM systems, proteolytic cleavage after the lipophilic NTD releases gasdermin inhibition. The notably short C-terminal peptide responsible for bGSDM inhibition suggests the possibility that short-form eukaryotic gasdermins including pejvakin may undergo activation through a similar mechanism. Furthermore, widespread palmitoylation of bGSDMs indicates that cysteine modifications are a conserved mechanism for regulating gasdermin pores (18). The size distribution of pores from Runella and Bacteroidetes species might suggest that bGSDM pores, like those in mammals, could be customized for the secretion of certain molecules (10). Defining the cues that activate bGSDM systems will provide insight into their roles in prokaryotic biology and the origins of pyroptotic cell death.
Supplementary Material
Acknowledgements:
The authors thank members of the Kranzusch and Sorek laboratories for helpful discussions. Mass spectrometry was performed at the Biopolymers and Proteomics Core Facility at the Koch Institute of MIT, the Taplin Mass Spectrometry Facility at Harvard Medical School, and the Weizmann De Botton Protein Profiling Institute. We thank W. Shih’s laboratory for training and use of the JEOL-1400 electron microscope, the Harvard Center for Cryo-Electron Microscopy (HC2EM), the HMS Electron Microscopy Facility, M. Eck for sharing computational resources, and the SBGrid consortium for computational support. We thank J. Leitz and A. Brunger for sharing scripts for cryo-ET reconstruction.
Funding:
This study was supported by the Pew Biomedical Scholars Program (P.J.K.), the Burroughs Wellcome Fund PATH award (P.J.K.)
Mathers Foundation (P.J.K.), the Parker Institute for Cancer Immunotherapy (P.J.K.), the European Research Council grant ERC-CoG 681203 (R.S.), the Israel Science Foundation grant ISF 296/21 (R.S.), the Ernest and Bonnie Beutler Research Program of Excellence in Genomic Medicine (R.S.), the Minerva Foundation and Federal German Ministry for Education and Research (R.S.), the Knell Family Center for Microbiology (R.S.), the Yotam project and the Weizmann Institute Sustainability And Energy Research Initiative (R.S.), the Dr. Barry Sherman Institute for Medicinal Chemistry (R.S.), the National Institute of Health Cancer Immunology training grant T32CA207021 (A.G.J.), a Life Science Research Foundation postdoctoral fellowship of the Open Philanthropy Project (A.G.J.), a Minerva Foundation postdoctoral fellowship (T.W.), and a Herchel Smith Graduate Research Fellowship (B.D.-L.).
Footnotes
Data and Materials Availability:
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 7N50 (Bradyrhizobium bGSDM), 7N51 (Vitiosangium bGSDM), and 7N52 (Runella bGSDM). Correspondence and requests for other materials should be addressed to PJK or RS.
References and Notes
- 1.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, Dixit VM, Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 526, 666–671 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F, Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 526, 660–665 (2015). [DOI] [PubMed] [Google Scholar]
- 3.He WT, Wan H, Hu L, Chen P, Wang X, Huang Z, Yang ZH, Zhong CQ, Han J, Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 25, 1285–1298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J, Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 535, 153–158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang DC, Shao F, Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 535, 111–116 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Ruan J, Xia S, Liu X, Lieberman J, Wu H, Cryo-EM structure of the gasdermin A3 membrane pore. Nature. 557, 62–67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang S, Zhou Z, Sun Y, Zhang T, Sun L, Coral gasdermin triggers pyroptosis. Sci. Immunol. 5 (2020), doi:eabd2591. [DOI] [PubMed] [Google Scholar]
- 8.Lieberman J, Wu H, Kagan JC, Gasdermin D activity in inflammation and host defense. Sci. Immunol. 4, eaav1447 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daskalov A, Mitchell PS, Sandstrom A, Vance RE, Glass NL, Molecular characterization of a fungal gasdermin-like protein. Proc. Natl. Acad. Sci. U. S. A. 117, 18600–18607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia S, Zhang Z, Magupalli VG, Pablo JL, Dong Y, Vora SM, Wang L, Fu TM, Jacobson MP, Greka A, Lieberman J, Ruan J, Wu H, Gasdermin D pore structure reveals preferential release of mature interleukin-1. Nature. 593, 607–611 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z, Wang C, Yang J, Zhou B, Yang R, Ramachandran R, Abbott DW, Xiao TS, Crystal Structures of the Full-Length Murine and Human Gasdermin D Reveal Mechanisms of Autoinhibition, Lipid Binding, and Oligomerization. Immunity. 51, 43–49.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broz P, Pelegrín P, Shao F, The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 20, 143–157 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Clavé C, Dyrka W, Granger-Farbos A, Pinson B, Saupe SJ, Daskalov A, Fungal gasdermin-like proteins are controlled by proteolytic cleavage. UPDATE IF POSSIBLE bioRxiv (2021), doi: 10.1101/2021.06.03.446900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kümmel D, Heinemann U, Veit M, Unique self-palmitoylation activity of the transport protein particle component Bet3: A mechanism required for protein stability. Proc. Natl. Acad. Sci. U. S. A. 103, 12701–12706 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi M, Zhang P, Vora SM, Wu H, Higher-order assemblies in innate immune and inflammatory signaling: A general principle in cell biology. Curr. Opin. Cell Biol. 63, 194–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen JM, de Jong MF, Wu Q, Zhang L, Heisler DB, Alto LT, Alto NM, Pathogenic ubiquitination of GSDMB inhibits NK cell bactericidal functions. Cell, 1–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur G, Burroughs AM, Iyer LM, Aravind L, Highly-regulated, diversifying NTP-dependent biological conflict systems with implications for the emergence of multicellularity. Elife. 9, 1–45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphries F, Shmuel-Galia L, Ketelut-Carneiro N, Li S, Wang B, Nemmara VV, Wilson R, Jiang Z, Khalighinejad F, Muneeruddin K, Shaffer SA, Dutta R, Ionete C, Pesiridis S, Yang S, Thompson PR, Fitzgerald KA, Succination inactivates gasdermin D and blocks pyroptosis. Science. 369, 1633–1637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen I-MA, Chu K, Palaniappan K, Pillay M, Ratner A, Huang J, Huntemann M, Varghese N, White JR, Seshadri R, Smirnova T, Kirton E, Jungbluth SP, Woyke T, Eloe-Fadrosh EA, Ivanova NN, Kyrpides NC, IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millman A, Melamed S, Amitai G, Sorek R, Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat. Microbiol. 5, 1608–1615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7 (2011), doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Söding J, Protein homology detection by HMM-HMM comparison. Bioinformatics. 21, 951–60 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V, A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Pei J, V Grishin N, PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol. Biol. 1079, 263–71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG, Clustal W and Clustal X version 2.0. Bioinformatics. 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Letunic I, Bork P, Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh K, Rozewicki J, Yamada KD, MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R, IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Ostell J, Pruitt KD, Schuler GD, Shumway M, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E, Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 36, D13–D21 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinegger M, Söding J, MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W, Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD, Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K, Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC, MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karplus PA, Diederichs K, Linking crystallographic model and data quality. Science. 336, 1030–1033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss MS, Global indicators of X-ray data quality. J. Appl. Crystallogr. 34, 130–135 (2001). [Google Scholar]
- 38.Käll L, Storey JD, Noble WS, Non-parametric estimation of posterior error probabilities associated with peptides identified by tandem mass spectrometry. Bioinformatics. 24, 42–48 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nesvizhskii AI, Keller A, Kolker E, Aebersold R, A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Mazzocco A, Waddell TE, Lingohr E, Johnson RP, Enumeration of bacteriophages using the small drop plaque assay system. Methods Mol. Biol. 501, 81–5 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M, Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 10, 1794–1805 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Schorb M, Haberbosch I, Hagen WJH, Schwab Y, Mastronarde DN, Software tools for automated transmission electron microscopy. Nat. Methods. 16, 471–477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA, MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheres SHW, RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner T, Merino F, Stabrin M, Moriya T, Antoni C, Apelbaum A, Hagel P, Sitsel O, Raisch T, Prumbaum D, Quentin D, Roderer D, Tacke S, Siebolds B, Schubert E, Shaikh TR, Lill P, Gatsogiannis C, Raunser S, SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mastronarde DN, Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120, 343–52 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 7N50 (Bradyrhizobium bGSDM), 7N51 (Vitiosangium bGSDM), and 7N52 (Runella bGSDM). Correspondence and requests for other materials should be addressed to PJK or RS.


