Graphical abstract
Keywords: COVID-19, SARS-CoV-2, Intranasal vaccine, Influenza vector, Cell-mediated immunity
Abstract
Remarkable progress has been made in developing intramuscular vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); however, they are limited with respect to eliciting local immunity in the respiratory tract, which is the primary infection site for SARS-CoV-2. To overcome the limitations of intramuscular vaccines, we constructed a nasal vaccine candidate based on an influenza vector by inserting a gene encoding the receptor-binding domain (RBD) of the spike protein of SARS-CoV-2, named CA4-dNS1-nCoV-RBD (dNS1-RBD). A preclinical study showed that in hamsters challenged 1 d after single-dose vaccination or 9 months after booster vaccination, dNS1-RBD largely mitigated lung pathology, with no loss of body weight. Moreover, such cellular immunity is relatively unimpaired for the most concerning SARS-CoV-2 variants, especially for the latest Omicron variant. In addition, this vaccine also provides cross-protection against H1N1 and H5N1 influenza viruses. The protective immune mechanism of dNS1-RBD could be attributed to the innate immune response in the nasal epithelium, local RBD-specific T cell response in the lung, and RBD-specific IgA and IgG response. Thus, this study demonstrates that the intranasally delivered dNS1-RBD vaccine candidate may offer an important addition to the fight against the ongoing coronavirus disease 2019 pandemic and influenza infection, compensating limitations of current intramuscular vaccines.
1. Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has had an immeasurable impact on health, economy, and social stability worldwide. The rapid development of multiple COVID-19 vaccines has been an incredible scientific achievement [1]. Multiple vaccines based on traditional or modern platform technologies have demonstrated high effectiveness for preventing severe COVID-19, hospitalization, and death in clinical trials as well as in the real world for at least several months [2], [3], [4], [5], enabling widespread vaccine administration to curb the COVID-19 pandemic globally. Nonetheless, the effectiveness of current vaccines in interrupting human-to-human transmission and for mild or asymptomatic patients has been well below expectations, especially for variants with stronger transmissibility and antigenic changes, such as the delta variant. Indeed, the number of newly confirmed cases is increasing rapidly again even in countries with extremely high levels of vaccine coverage [6], [7], [8], [9], [10]. Thus, it is imperative to continue developing new COVID-19 vaccines using different vaccine strategies.
To date, COVID-19 vaccines approved for use by different regulatory authorities, including mRNA vaccines, inactivated vaccines, recombinant adenovirus vaccines, and recombinant protein vaccines, are all administered through traditional muscle injection, which are commonly limited for their ability to induce mucosal immunity and local immunity [11], [12], [13], [14]. While some countries with sufficient vaccine supplies have been achieving the potential “herd” immunity [15], breakthrough infections are common among vaccinated people. Importantly, the majority of children are not among the vaccinated groups. With countries reopening borders for international travelers and the increasing emergence of variants of concern, epidemics with high transmission among specific groups of people will become very common. Solutions in response to the evolving COVID-19 pandemic are imminently needed. Given the predominant respiratory tropism of SARS-CoV-2 and the evidence that intranasal live attenuated influenza vaccine (LAIV) has equivalent and even improved efficacy compared with that of inactivated influenza vaccine (IIV) [16], [17], several vaccine candidates intended to be delivered by intranasal administration or inhalation are under development, and some of them have shown potential in animal models and early phase clinical trials [18], [19]. To our knowledge, night intranasally delivered COVID-19 vaccines have been tested in clinical trials globally, seven of which are based on virus vectors, including adenovirus, respiratory syncytial virus, and influenza virus [12], [20], [21]. These intranasal vaccines have shown the potential to elicit mucosal IgA and CD8+ T cell-mediated immune responses in the respiratory tract as well as serum IgG responses, resulting in a more efficient reduction of virus replication and shedding in both the lungs and the nasal passages than intramuscular vaccination [12], [22], [23].
Here, we present data demonstrating the rapid (1 d), prolonged (9 months), and broad protection of and comprehensive innate and adaptive immune responses to an intranasally delivered COVID-19 vaccine based on the LAIV vector in animal models. This vaccine candidate has been shown to be well tolerated and immunogenic in Chinese adults [24], and a global multicenter phase III clinical trial is being initiated and ongoing now.
2. Materials and methods
2.1. Cell cultures
All cell lines were obtained from the American Type Culture Collection (ATCC). Human embryonic kidney cells (293T), African green monkey kidney epithelial cells (Vero E6), and Madin-Darby canine kidney cells (MDCK) were maintained in Dulbecco’s modified Eagle’s medium (DMEM)-high glucose (Sigma Aldrich, St. Louis, USA) supplemented with 10% low endotoxin fetal bovine serum (FBS) (Cegrogen Biotech, Stadtallendorf, Germany) and penicillin–streptomycin.
2.2. Construction of plasmids
The receptor-binding domain (RBD) segment of SARS-CoV-2 (GenBank No. MN908947) was codon optimized for eukaryotic expression system and constructed by overlapping primers with the B2M signal peptide at the 5′ end and the foldon motif with the V5 tag at the 3′ end. The sequence encoding the RBD segment was then cloned into the NS1 deletion plasmid pHW2000-DelNS1 as described previously [25].
2.3. Generation and passage of dNS1-RBD viruses
Eight pHW2000 plasmids containing the DelNS1 segment and the other seven influenza virus genomic segments, together with an NS1 expression plasmid, pCX-CA04-NS1-Flag, which was derived from the parental influenza virus A/California/04/2009 (H1N1) (GenBank No. MN371610.1-371617.1), were transfected into 293T cells and incubated overnight at 37 °C. The DNA mixture was removed, and Opti-MEM supplemented with 1 μg/mL 6-(1-tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK)-treated trypsin (Sigma, St. Louis, USA) was added. Viral supernatant was collected 72 h later, designated dNS1-RBD passage 0 virus, and was subsequently passaged in MDCK cells at 33 °C. The supernatant was harvested 48 h post transfection when most of the cells showed signs of cytopathic effect (CPE). Infectious virus titers (plaque forming unit (PFU)/mL) were determined by plaque assay on MDCK cells.
For the rescued viruses, deletion of the NS1 gene and insertion of the RBD gene were confirmed by reverse transcription- polymerase chain reaction (RT-PCR) using NS-specific and RBD-specific primers. Total RNA from virus supernatants was extracted using the PureLink™ Viral RNA/DNA Mini Kit (Thermo Fisher, Waltham, USA) according to the manufacturer’s protocol and then converted to cDNA by SuperScript® III Reverse Transcriptase (Invitrogen, Carlsbad, USA). The cDNA was then subjected to RT-PCR using primers and probes that are specific to the target sequence (RBD forward-5'-ACATTGGCCACCATGTTCACTGTAGAAAAAGGAAT-3'; RBD reverse-5’-AATGTGTCAATTTCAACTTCGGCTATATTCCGGAA-3’; NS forward-5'-CCGAAGTTGGGGGGGAGCAAAAGCAGGGTGACAAAAACATA-3'; NS reverse-5’-GATAAAAAACACCCTTGTTTCTACTAATAACCCGGCGGCC-3’, respectively). RT-PCR was performed under the following reaction conditions: 94 °C for 2 min, followed by 35 cycles of 98 °C for 15 s, 55 °C for 30 s and 68 °C for 90 s, and 68 °C for 10 min. The presence of inserted sequences in generated vaccine virus was further confirmed by Sanger sequencing.
2.4. Growth kinetics
MDCK cells seeded in 24-well plates were infected with viruses at the indicated multiplicity of infection (MOI). After 1 h adsorption, the viral supernatant was removed, and the cells were washed twice with phosphate-buffered saline (PBS). DMEM containing 1 μg/mL TPCK-treated trypsin was added, and the cells were incubated at the indicated temperature. Supernatants were collected at different time points, and titers were determined by plaque assay.
2.5. Plaque assay for dNS1-RBD viruses
Viruses were 10-fold serially diluted, added to confluent MDCK cells in 6-well plates, and then incubated at 37 °C for 1 h. The supernatant was removed, and the cells were washed twice with PBS and then overlaid with 1% minimum essential medium (MEM) agarose containing 1 μg/mL TPCK-treated trypsin. The plates were incubated at 33 °C for 72 h and then fixed with 4% PBS-buffered formaldehyde solution for at least 1 h. Plaques were visualized by staining with 1% crystal violet solution.
2.6. Western blot
MDCK cells were cultured and infected with dNS1-RBD virus as described above. 36 h later, cell lysates were harvested using modified NEP cell lysis buffer. Proteins were separated on a 10% gel, and then following transfer, blots were incubated with an anti-influenza A nucleoprotein (NP) protein antibody 19C10 generated by our laboratory (1:1000) and anti-V5 tag antibody (Thermo,1:5000), and visualized with horseradish peroxidase (HRP)-conjugated anti-mouse IgG (Invitrogen, 1:5000).
2.7. Immunofluorescence imaging
For direct visualization of the expression of hemagglutinin (HA) and RBD, MDCK cells were seeded at 2 × 104 cells per well in CellCarrier-96 Black plates and then infected with dNS1-RBD, CA04-dNS1, and CA04 WT at an MOI of 1. PBS was used as a negative control. After 72 h, the cells were fixed with 2% paraformaldehyde in PBS for 15 min in the dark. The cells were then permeated by the addition of 0.3% Triton X-100 in PBS (PBST) for 10 min at room temperature and blocked with 2% bovine serum albumin (BSA). The plates were incubated with a DyLight 488-labeled mAb against 6G9-488 (anti-HA, 1:100 dilution) and DyLight 650-labeled mAb against R4D11 (anti-RBD, 1:100 dilution) generated by our laboratory at 37 °C for 60 min, and the assay plates were washed three times with PBS. Cell nuclei were labeled with 4′, 6-diamidino-2-phenylindole (DAPI). The images were acquired on an Opera Phenix using a 63 × water immersion objective.
2.8. Vaccine formulation
The vaccine dNS1-RBD was prepared on a large scale at Beijing Wantai Biological Pharmacy Enterprise Co., Ltd., Beijing, China. After rounds of passage and amplification with the cell factory based on the MDCK cell line, the viruses were further purified through the process step by step including ultrafiltration, size-exclusion chromatography, nuclease treatment, and then ion-exchange chromatography to confirm the exclusion of exogenous factors. Purified dNS1-RBD virions were then mixed with virus protectants, which contained carbohydrates, amino acids, human albumin, etc., and were preserved at −15 °C. Based on the enzyme-linked immunosorbent assay (ELISA) results using a sandwich assay with anti-RBD monoclonal antibodies on both sides (Wantai) and plaque assay results, serial passages 1 to 10 of purified vaccines were confirmed to be stable under current vaccine manufacturing conditions. The purified vaccines were further used in this study for comprehensive evaluation. The CA04-dNS1 virus was also prepared in the same way as dNS1-RBD virus for further evaluation. To further eliminate the placebo effect for preparing vaccine in clinical trial, the virus protectant produced on a large scale was used as placebo control in this study.
2.9. Ethics statements
All animals involved in this study were housed and cared for in an Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited facility. All experimental procedures with mice, ferrets, and hamsters were conducted according to Chinese animal use guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC) of Xiamen University (XMULAC20200232). The hamster studies were performed in an animal biosafety level 3 (ABSL-3) laboratory affiliated to the State Key Laboratory of Emerging Infectious Diseases, The University of Hong Kong.
2.10. Vaccine safety evaluation
The safety of the potential SARS-CoV-2 vaccine, dNS1-RBD was evaluated in BALB/c mice and ferrets. BALB/c mice were intranasally inoculated 50 μL (105–107 PFU/mL) of dNS1-RBD and CA04-WT under isoflurane anesthesia and monitored daily for morbidity and mortality for 14 d pos inoculation. Vaccines were concentrated at first by 100 kD limitation Sartorius Vivaspin centrifugal concentrators and then diluted in 1640 media to a final 50 μL volume and administered bilaterally for BALB/c mice. Animals that lost more than 25% of their initial body weight were euthanized in accordance with our animal ethics protocol. Ferret studies were performed at JOINN Labs (Suzhou, China). Two groups of ferrets (5 female and 5 male ferrets in the vaccine group and 3 female and 3 male ferrets in the control group) were immunized intranasally with a single-dose 1 × 106 PFU of dNS1-RBD and CA04-WT virus respectively diluted in 1640 media to a final 500 μL volume. Datasets of the safety-related parameters were collected during and after immunization, including clinical observations, body weight, and body temperatures. Viral loads were detected for throat swabs and nasal washes collected at days −1, 1, 3, 5, and 7 post inoculation by RT-PCR. Histopathological evaluations in lungs from two groups of ferrets were conducted at day 8. Lung tissues were collected and stained with hematoxylin and eosin. Six to eight-week-old, female BALB/c mice and five to six-month-old, male and female ferrets were used throughout this study.
2.11. Immunization and infection of mice
BALB/c mice were immunized intranasally with 50 μL (1 × 106 PFU/mL) of the vaccine per dose prepared as indicated above under isoflurane anesthesia, while the control group was administered CA04-WT or CA04-dNS1 virus. For antibody response evaluation, all groups of BALB/c mice (6 animals in each group) were vaccinated with a prime-boost regimen (days 0 and 14), and blood was collected via retro-orbital bleeding before each immunization and 14 d after the second injection, followed by a binding assay to analyze vaccine immunogenicity.
For innate immune response analyses, C57BL/6 mice (5 animals in each group) were vaccinated with a single dose and sacrificed 1 d after vaccination. For cellular immune response analyses of PBMCs, splenic lymphocytes, pulmonary lymphocytes, and lymph node cells, C57BL/6 mice (6–8 weeks old) were immunized intranasally with 50 μL (1 × 106 PFU/mL) of the vaccine by the one-dose or two-dose regimen as described above (10 animals in each group). Then, splenic lymphocytes, pulmonary lymphocytes, and lymph node cells (6 animals in each group) were collected on day 28 of a prime-boost regimen with a 2-week interval for intracellular cytokine staining (ICS) measurements.
For protective efficacy evaluation against influenza, CA04-WT was treated with 0.03% formaldehyde solution as inactivated vaccine control and further used for vaccination by intramuscular injection. In influenza virus challenge studies, BALB/c mice (5 animals in each group) were inoculated with a lethal dose of various subtypes of influenza virus via the intranasal route. In brief, 14 d after two doses of 50 μL (1 × 106 PFU/mL) of dNS1-RBD, CA04-dNS1, and inactivated CA04-WT virus, serum and pulmonary lymphocyte were collected for antibody assay and T cell response evaluation, respectively. Immunized mice were further anesthetized and inoculated with 50 μL vaccine containing 25 MLD50 influenza virus which was adapted to the mouse model. The mice were observed daily for mortality and morbidity, and body weight was measured for up to 14 d after infection. Vaccinated mice were sacrificed for virological analysis on day 5 after the virus challenge. The virus challenge studies were performed in an animal biosafety level 2 (ABSL-2) facility.
2.12. Immunization and infection of hamsters
Hamsters (male:female = 1:1) were vaccinated with the indicated amount of the vaccine. All hamsters received 100 μL (1 × 106 PFU/mL) of vaccine per dose via the intranasal route. At the indicated time after vaccination, the hamsters were further evaluated by direct contact challenge of SARS-CoV-2. Three strains were used in this study: the prototype-like virus AP8 (hCoV-19/China/AP8/2020; GISAID accession No. EPI_ISL_1655937), the B.1.351 variant AP100 (hCoV-19/China/AP100/2021; GISAID accession No. EPI_ISL_2779638) and B.1.1.529 variant strain (share an identical sequence with EPI_ISL_8182026) were passaged on Vero cells (#CCL-81, ATCC). For prototype-like virus and B.1.351 variant, contact-transmission challenges of SARS-CoV-2 were used in the study. Virus-carrying hamsters (donors) were pre-infected via inoculation of 1 × 103 PFU of SARS-CoV-2 through the intranasal route. Each donor was transferred to a new cage and cohoused with four vaccinated or control animals. One day after cohousing, donors were isolated from the cage, and the other hamsters were further observed. For B.1.1.529 variant challenge, hamsters were inoculated directly with 1 × 105 PFU of SARS-CoV-2 virus through the intranasal nasal route under anesthesia. The hamsters were fed a daily food amount of 7 g per 100 g of body weight. The weight changes and typical symptoms (piloerection, hunched back, and abdominal respiration) in hamsters were recorded daily after virus inoculation or contact. Hamsters were sacrificed for tissue pathological and virological analyses on day 5 after the virus challenge. The virus challenge studies were performed in an animal biosafety level 3 (ABSL-3) facility.
2.13. Anti-RBD IgA measurements
Bronchoalveolar lavage (BAL) was collected on control-infected and vaccine-infected mice. Mice were euthanized, and a short needle insulin syringe (BD, Franklin Lakes, USA) was inserted gently into the lumen of the exposed trachea. The lungs were then lavaged with two separate 1-mL washes of sterile normal saline. The RBD-specific IgA titer of BAL samples was next evaluated by ELISA as described above with goat anti-mouse IgA alpha chain-HRP (Abcam, Cambridge, UK, 1:3000).
2.14. Anti-RBD and anti-NP IgG measurements
RBD-specific antibody titers in serum samples collected from immunized animals with 50 μL (1 × 106 PFU/mL) of vaccine were determined by indirect ELISA. Ninety-six-well microtiter plates were coated with 100 ng of purified NP protein of influenza A virus A/WSN/33(H1N1) derived from Xiamen Wantai Biological Pharmacy Enterprise Co. and 200 ng of purified RBD protein which was generated and expressed in 293F from the codon-optimized RBD sequence of SARS-CoV-2 spike protein (GenBank accession number MN908947) individually at 4 °C overnight and blocked with 2% BSA for 2 h at 37 °C. Diluted sera (1:100) were successively diluted in a 2-fold series and applied to each well for 1 h at 37 °C, followed by incubation with goat anti-mouse, anti-hamster, or anti-human antibodies conjugated with HRP for 1 h at 37 °C after 3 washes. The plate was developed using TMB, followed by the addition of 2 mol/L H2SO4 to stop the reaction, and read at 450/630 nm by an ELISA plate reader for final data acquisition.
2.15. ELISPOT assay
ELISPOT assays were performed using mouse Interferon-γ (IFN-γ)ELISPOT plates (DAKEWE, Shenzhen, China). Ninety-six-well ELISPOT plates pre-coated with capture antibody were blocked with RPMI-1640 for 10 min at room temperature. Briefly, a total of 106 cells per well from C57BL/6 mouse spleen, lymph nodes, lung, or peripheral blood mononuclear cells (PBMCs) immunized with 50 μL (1 × 106 PFU/mL) of vaccine were plated into each well and stimulated for 20 h with pooled peptides of RBD of wild type SARS-CoV-2 or variants (15-mer peptide with 11 amino acids overlap, cover the RBD305-547, Genscript) and pooled peptides of influenza A virus (mix peptides reported by the related reports [26], [27]). The segmented RBD peptides pool (1–6, 0.667 μg/mL each) and individual peptide (1.334 μg/mL) were used in T cell epitope identification. The spots were developed based on the manufacturer’s instructions. PBS and cell stimulation cocktails from the kit were used as negative and positive controls, respectively. Spots were scanned and quantified by an ImmunoSpot Cellular Technology Limited reader. Spot-forming units (SFUs) per million cells were calculated by subtracting the negative control wells.
2.16. Intracellular cytokine staining assay
The expression of phenotypic markers, activation markers, and cytokines was evaluated using flow cytometry for T cells, B cells, and monocytes/macrophages in single-cell suspensions from tissues. The cells were stained with murine antibodies for phenotype and activation (CD4 [clone GK1.5, APC/Cy7], CD8 [clone 53–6.7, PerCP/Cy5.5], CD11b [clone M1/70, PE], CD11c [clone N418, BV421], CD49b [clone dx5, FITC], MHC2 [clone M5/114.15.2, APC], Ly-6C [clone HK1.4, APC-Cy7], Ly-6G [clone 1A8, BV605], CD62L [clone MEL-14, APC-Cy7], CD103 [clone 2E7, PE], CD69 [clone H1.2F3, BV421], CD44 [clone IM7, APC], CD80 [clone 16-10A1, FITC], and CD86 [clone GL-1, PE-Cy7]), and cytokine expression (IL4 [clone 11B11, BV421], IL2 [clone JES6-5H4, PE], and IFN-γ [clone XMG1.2, APC]), and a LIVE/DEAD® Fixable Aqua Dead Cell Stain Kit was also used. For RBD-specific T cell assays, each sample was stimulated with pooled spike peptides (1 μg/well) in a U-bottom plate and incubated at 37 °C for 18 h. After incubation, 0.12 μL of protein transport inhibitors (BD GolgiPlugTM, BD Biosciences) in 20 μL of 10% FBS/RPMI 1640 medium was added to each well, and the plate was incubated at 37 °C for 6 h. Then, the cells were washed once with 2% FBS/PBS and further stained with labeled antibodies. After incubation at 4 °C for 30 min, the cells were washed once with 2% FBS/RPMI 1640 medium and passed through a 0.22 μm filter. The cells were transferred to 5-mL round-bottom tubes and analyzed by a BD LSRFORTESSA X-20 system. The data were analyzed by FlowJo V10.6.0 (Tree Star Inc., Ashland, USA) and GraphPad Prism 9 (GraphPad Software Inc., San Diego, USA).
2.17. Measurements of cytokine and chemokine levels
Lung homogenate samples were prepared for analysis with ProcartaPlex Multiplex Immunoassay, a mouse cytokine/chemokine magnetic bead panel (36-plex, Thermo Fisher), following kit-specific protocols. Analytes were quantified using a Magpix analytical test instrument using a standard curve derived from recombinant cytokine and chemokine standards, which utilizes xMAP technology (Luminex Co., Austin, USA) and xPONENT 4.2 software (Luminex). The results were expressed as ng/mL.
2.18. SARS-CoV-2, influenza A virus, and dNS1-RBD RNA quantification
Viral RNA levels in the lungs of challenged hamsters were detected by quantitative RT-PCR. Briefly, for quantification of viral levels and gene expression after challenge or passage experiments, RNA was extracted from homogenized organs or cultured cells using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Hamster tissue samples were homogenized by TissueLyser II (Qiagen) in 1 mL of PBS. Subsequently, viral RNA quantification was conducted using a SARS-CoV-2 RT-PCR Kit (Wantai) by measuring the copy numbers of the N gene. Quantification of IAV replication was measured with primers targeting M mRNA while CA4-dNS1-nCoV-RBD was quantified with primers targeting the RBD and NS genes.
2.19. SARS-CoV-2 titration assay
Live virus titers in homogenized lung tissues and cell cultures were measured by the standard TCID50 method in Vero E6 cells seeded in 96-well plates. In brief, the samples were serially diluted, added to the 96-well plates, and incubated with the Vero E6 cells for 1 h. 3 d after incubation, the cytopathic effects were observed and used to calculate the viral titers.
2.20. Histopathology
The lung tissues from challenged hamsters were fixed with 10% formalin for 48 h, embedded in paraffin and sectioned. Next, the fixed lung sections were subjected to hematoxylin and eosin (H&E) staining. Whole-slide images of the lung sections were captured by an EVOS M7000 Images System (Thermo Fisher). The standards for the pathological score of lung tissues in this study are adapted and optimized from a recent study of SARS-CoV-2 infection in the hamster model [28]. In brief, the H&E staining result of the whole lung tissue was analyzed for its severity of pathological change. The pathological score includes: (a) alveolar septum thickening and consolidation; (b) hemorrhage, exudation, pulmonary edema, and mucous; (c) recruitment and infiltration of inflammatory immune cells. For each issue, score related to the severity (0, no pathological change was observed; 1, moderate pathological change; 2, mild pathological change; 3, indicates severe pathological change; and 4, very severe pathological change). In conclusion, scores of such three issues were added as the comprehensive lung pathological score of lung tissue.
2.21. Statistical analysis
Statistical significance was assigned when P values were <0.05 using GraphPad Prism 9.0. Viral titers and RBD-specific IgG titers were analyzed after log transformation. The bars in this study represent the mean ± standard deviation (SD) or median (interquartile range, IQR) according to data distribution. The number of animals and independent experiments that were performed are indicated in the figure legends. Student’s t-test (two groups) or one-way analysis of variance (ANOVA) (three or more groups) was used for comparison of normally distributed continuous variables. For nonnormally distributed continuous variable comparisons, the Mann–Whitney U test (two groups) or Kruskal–Wallis test (three or more groups) was used. Two-way repeated-measures ANOVA was adopted for repeated data comparison. For multiple comparisons of three or more groups, Dunnett’s multiple comparison test was used.
3. Results
3.1. Construction and pathogenic analysis of the dNS1-RBD vaccine candidate
The vaccine candidate CA4-dNS1-nCoV-RBD (dNS1-RBD) was constructed by inserting a gene encoding the RBD of the spike protein of the SARS-CoV-2 prototype strain into the previously reported NS1-deleted backbone of H1N1 influenza virus California/4/2009 (CA04-dNS1) [24] (Fig. 1 a). We compared the growth kinetics of dNS1-RBD with those of the wild-type A/California/04/2009 (H1N1) parental virus (CA04-WT) and its NS1-deleted version (CA04-dNS1) in MDCK cells. As expected, the replication of dNS1-RBD was significantly suppressed at 37 °C and 39 °C compared with that at 33 °C due to the existence of temperature-sensitive mutations in the CA04-dNS1 vector (Fig. 1b), which is a desirable feature for reducing the risk of influenza-associated adverse reactions in the lung. In line with the above results, both dNS1-RBD and CA04-WT could replicate effectively in the nasal turbinate and throat in ferrets and only ferrets in the CA04-WT group still showed viral shedding in the nasal turbinate and throat at 7 d post nasal inoculation, in contrast to few of the ten ferrets in the dNS1-RBD group (Fig. S1a and b online). The expression of the RBD and HA antigen in dNS1-RBD-infected MDCK cells was visualized using confocal analysis and further confirmed by Western blot (Fig. 1c and d). Evaluated by ten continuous passages, the genetic and expression stability of the RBD fragment of dNS1-RBD in the MDCK cell culture system seemed acceptable for large-scale production (Fig. 1e).
Fig. 1.
Construction and characterization of a recombinant LAIV-based SARS-CoV-2 vaccine. (a) Construction of an mRNA-encoding plasmid that transcribes DelNS1 with RBD-inserted mRNA. (b) Replication efficiency of the dNS1-RBD, CA04-dNS1, and CA04-WT viruses varied with 33 °C, 37 °C, and 39 °C conditions in MDCK cells. Data are shown as mean ± SD of results from three independent experiments. Analysis was performed by two-way repeated-measures ANOVA and ordinary one-way ANOVA multiple comparison. Significance was set at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (c) Confocal analysis of the RBD and HA protein expressed by the influenza vector in MDCK cells. The co-expression of RBD and HA could be detected only for dNS1-RBD. MDCK cells were fixed 72 h after infection. Red fluorescence indicates the RBD; green fluorescence indicates HA. (d) Immunoblot analysis of RBD and NP expression in denatured and nondenatured cell lysate samples 36 h after infection with dNS1-RBD (1), CA04-dNS1 (2), and CA04-WT (3). Most of the secreted RBD protein for dNS1-RBD formed an RBD trimer, with RBD rarely existing in the dimer form. (e) Plaque assay and sandwich ELISA analysis of RBD expression were performed on the virus supernatant harvested from serial passages 1 to 10 of dNS1-RBD. Significance was determined by one-way ANOVA with the Kruskal–Wallis test. ns, not significant.
Intranasal inoculation in BALB/c mice and ferrets confirmed the obvious attenuation of dNS1-RBD compared to the parental CA04-WT virus (Figs. S1 and S2 online). Mice inoculated with 5 × 103–5 × 105 PFU of the parental CA04-WT virus succumbed to infection after seven d, whereas mice inoculated with dNS1-RBD continued to maintain their body weight and health (Fig. S2a, b online). Likewise, for ferrets, which are highly susceptible to influenza virus infection, inoculation with 1 × 106 PFU of CA04-WT but not dNS1-RBD resulted in obvious influenza-like symptoms, with fever, weight loss, and pathological injury in lung tissues (Fig. S1 online). In summary, a recombinant LAIV stably expressing the SARS-CoV-2 RBD with remarkably less virulence than its parental influenza virus was generated.
3.2. Rapid, prolonged, and broad protection against infection with the prototype, Beta variant, and Omicron variant of SARS-CoV-2 in hamsters after intranasal immunization with dNS1-RBD
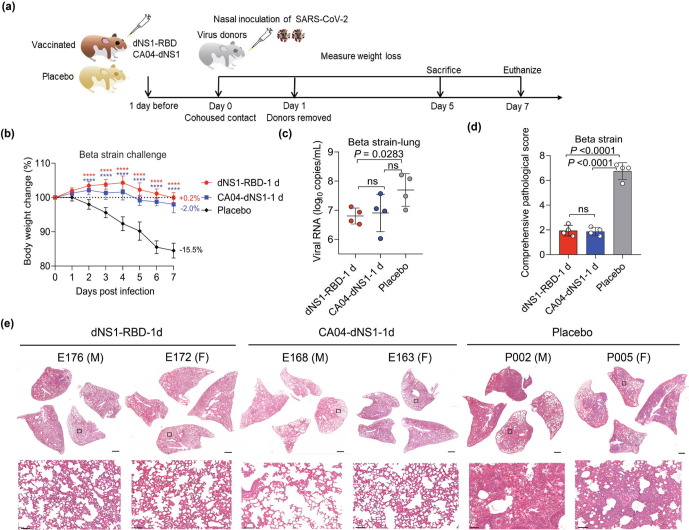
For understanding the protective effects provided by intranasal immunization, we validated the efficacy of intranasal immunization with dNS1-RBD in the interanimal transmission model in golden Syrian hamsters to mimic the predominant natural route of SARS-CoV-2 infection at 1 d after single-dose immunization and 3 or further more months after two doses of dNS1-RBD (prime and boost regimen with a 14-d interval). The model is preferred because it has been demonstrated to be sensitive to SARS-CoV-2 infection and associated COVID-19-like lung damage and can support efficient viral transmission from inoculated hamsters to naïve hamsters by direct contact and via aerosols [29], [30], [31]. Hamsters vaccinated with dNS1-RBD and CA04-dNS1 were infected through cohousing with donor hamsters infected by the Beta variant for rapid protective effects evaluation (Fig. 2 a). The placebo hamsters showed continuous body weight loss beginning 1 d post infection (dpi), with maximal weight loss at 7 dpi (mean: −15.5% for the Beta variant challenge group); In contrast, weight loss was not obvious in animals of all vaccine groups (mean: +0.2% for dNS1-RBD group and −2.0% for CA04-dNS1) (Fig. 2b). Lung damage at 5 dpi was quantitatively measured using a comprehensive pathological scoring system. Animals in the placebo groups had significantly higher pathological scores than those in both vaccine groups (Fig. 2d). The pathological histology analysis of lung tissues (Fig. 2e) taken at 5 dpi showed that vaccinated hamsters with dNS1-RBD and CA04-dNS1 were largely protected from lung damage caused by infection with the SARS-CoV-2 Beta variant, with minimal, if any, focal histopathological changes in the lung lobes, which analyzed in detail described above. In contrast, hamsters in placebo groups developed severe lung pathology with consolidated pathological lesions and severe or intensive interstitial pneumonia characterized by inflammatory cell infiltration in a focally diffuse or multifocal distribution. On average, 30% to 50% of the alveolar septa of these placebo animals became thicker, resembling findings in patients with severe COVID-19 bronchopneumonia [28], [29]. Additionally, at 5 dpi, the viral loads in the lung tissue of vaccinated hamsters were slightly lower than those in the lung tissue of placebo controls (Fig. 2c). Interestingly, dNS1-RBD and CA04-dNS1 seem to induce similar protection against SARS-CoV-2, demonstrating that this rapid protective immune could be attributed to the nonspecific immune induced by CA04-dNS1 vector.
Fig. 2.
Rapid protective efficacy of dNS1-RBD and CA04-dNS1 against SARS-CoV-2 challenge in Syrian hamsters one d after vaccination. (a) The hamsters received a single dose of dNS1-RBD, CA04-dNS1 control, and placebo control 1 d before virus challenge were evaluated with co-housed transmission mode. Virus-carrying hamsters (donors) were pre-infected via inoculation of 1 × 103 PFU of SARS-CoV-2 of Beta strain through the intranasal route. Each donor was then transferred to a new cage and cohoused with four vaccinated or placebo control animals. 1 d after cohousing, donors were isolated from the cage, and the other hamsters were further observed. (b) Changes in the body weights of hamsters were recorded following cohousing exposure. The average weight loss of each group at 7 dpi is indicated as a colored number. Data are shown as mean ± SD. Significant differences compared to the placebo group were analyzed using two-way repeated-measures ANOVA with Dunnett’s multiple comparisons test. (c) Viral loads of lung tissue obtained at 5 dpi from hamsters challenged by Beta strain were determined by RT-PCR. Data are shown as mean ± SD; significance was determined by ordinary one-way ANOVA multiple comparison. Symbols represent individual hamsters. (d) Comprehensive pathological scores of the hamster lungs. Scores were determined based on the severity and percentage of injured areas for the whole lung tissue collected from the indicated animal. Significance compared to the placebo group was determined by ordinary one-way ANOVA multiple comparison. (e) H&E staining of lung sections from tested hamsters collected on day 5 after cohousing exposure. Views of the whole lung lobes (4 independent sections) are presented in the above panel (scale bars, 1 mm), and the areas in the small black boxes are enlarged in the lower panel (scale bars, 100 μm).
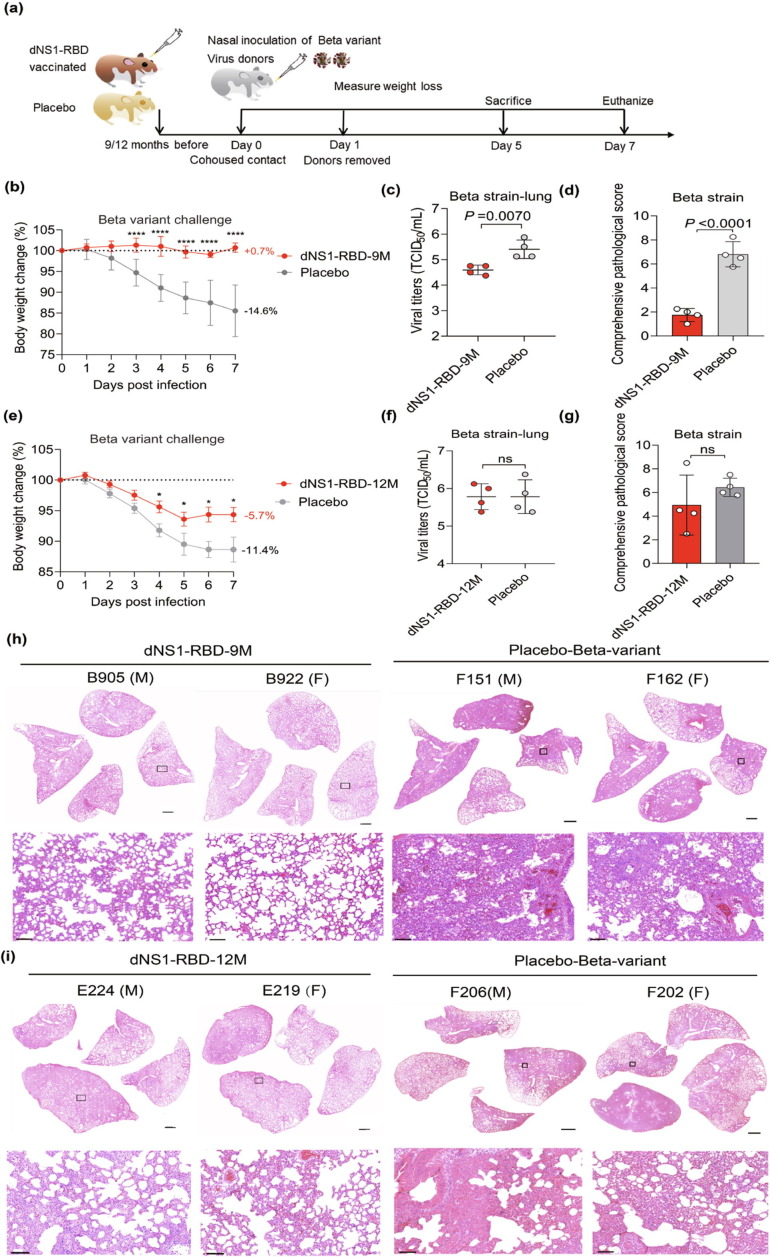
Having observed rapid protective efficacy of dNS1-RBD against infection by Beta variant, we further evaluated its prolonged protection (Fig. 3 a). Upon Beta variant infection for hamsters at 9 months post vaccination, while the placebo groups lost an average of 14.6% of body weight by 7 d post infection (Fig. 3b), the vast majority of dNS1-RBD immunized hamsters were still well protected with slight histological changes with significant reduction of viral loads (Fig. 3c, d, and h). We next assessed the prolonged protective efficacy of dNS1-RBD against Beta variant when hamsters were inoculated 12 months before challenge. Remarkably, the weight loss was diminished in vaccinated groups compared to placebo control hamsters (5.7% vs 11.4%, P = 0.0127) (Fig. 3e) and the pathological images change to a lesser degree with mild perivascular and alveolar infiltration in medium areas in 75% (3/4) animals (Fig. 3g, i), such as the E223 hamsters showed consolidated pathological lesions in all four lung sections, suggesting the protective immune response induced by dNS1-RBD proved to decline but last at least one year in hamster models.
Fig. 3.
Prolonged protective efficacy of dNS1-RBD against SARS-CoV-2 challenge in Syrian hamsters co-housed transmission model after two-dose vaccination. (a) The hamsters after two-dose vaccination were challenged by Beta strain of SARS-CoV-2 at 9 months and even 12 months after vaccination respectively. The hamsters treated with virus protectant was used as placebo controls. (b, e) Changes in the body weights of hamsters were recorded following cohousing exposure. The average weight loss of each group at 7 dpi is indicated as a colored number. Data are shown as mean ± SD. Significant differences compared to the placebo group were analyzed using two-way repeated-measures ANOVA with Dunnett’s multiple comparisons test. (c, f) Viral loads of lung tissue obtained at 5 dpi from hamsters challenged by Beta strain were determined by TCID50 assay. Data are shown as mean ± SD; significance was determined by two-tailed Student’s t-test. Symbols represent individual hamsters. (d, g) Comprehensive pathological scores of the hamster lungs. Scores were determined based on the severity and percentage of injured areas for the whole lung tissue collected from the indicated animal. Significance compared to the placebo group was determined by two-tailed Student’s t-test. (h, i) H&E staining of lung sections from tested hamsters collected on day 5 after cohousing exposure. Views of the whole lung lobes (4 independent sections) are presented in the above panel (scale bars, 1 mm), and the areas in the small black boxes are enlarged in the lower panel (scale bars, 100 μm).
When the Omicron variant outbreaks 6 months ago, more data are needed to understand the vaccine effectiveness against Omicron in this predominant trend. Indeed, while the attenuated replication and pathogenicity of Omicron variant for wild-type hamsters, infection by Omicron variant caused milder body weight loss, clinical disease, and lung pathology change compared to other variants of concern [32], [33]. Thus, the vaccinated hamsters were evaluated for Omicron variant challenge by intranasal inoculation directly, compared to the contact-transmission challenge for prototype strain (Fig. 4 a). For hamsters at 3 months post prime-boost vaccination, the placebo hamsters against prototype strain showed obvious weight loss (mean: −10.7%) with pulmonary edema and hemorrhage (Fig. 4b, h), while infection by Omicron variant reached the clinical endpoint at 5 dpi and cause less body weight loss (mean: −1.9%) with fewer pathological change in the alveoli and bronchial epithelia (Fig. 4e, i). In contrast, dNS1-RBD completely protected the animals from morbidity with no weight loss observed against both prototype strain and Omicron variant, and keep lungs normal, or near to normal with no more signs of bronchopneumonia after prototype strain and Omicron variant infection. In line with the protective results above, the immunized hamsters by dNS1-RBD show slight reduction in viral loads but mitigate the disease development to minimize the pathological change against both prototype and Omicron strain, suggesting that the dNS1-RBD might be still effective for new variants in future. (Fig. 4c, d, f, and g).
Fig. 4.
Broad protective efficacy of dNS1-RBD against SARS-CoV-2 challenge in Syrian hamsters co-housed transmission model after two-dose vaccination. (a) The hamsters after two-dose vaccination were challenged by prototype and Omicron strain of SARS-CoV-2 at 3 months after vaccination respectively. The hamsters treated with virus protectant were used as placebo controls. (b, e) Changes in the body weights of hamsters were recorded following cohousing exposure. The average weight loss of each group at 7 dpi is indicated as a colored number. Data are shown as mean ± SD. Significant differences compared to the placebo group were analyzed using two-way repeated-measures ANOVA with Dunnett’s multiple comparisons test. (c, f) Viral loads of lung tissue obtained at 5 dpi from hamsters challenged were determined by TCID50 assay. Data are shown as mean ± SD; significance was determined by two-tailed Student’s t-test. Symbols represent individual hamsters. (d, g) Comprehensive pathological scores of the hamster lungs. Scores were determined based on the severity and percentage of injured areas for the whole lung tissue collected from the indicated animal. Significance compared to the placebo group was determined by two-tailed Student’s t-test. (h, i) H&E staining of lung sections from tested hamsters collected on day 5 after cohousing exposure. Views of the whole lung lobes (4 independent sections) are presented in the above panel (scale bars, 1 mm), and the areas in the small black boxes are enlarged in the lower panel (scale bars, 100 μm).
Taken together, these results demonstrated that dNS1-RBD vaccination could efficiently block the pathogenicity of homogeneous and heterogeneous SARS-CoV-2 infection in golden Syrian hamsters in the direct contact model in the short and long term.
3.3. Intranasal inoculation of dNS1-RBD promotes comprehensive local immunity in the respiratory tract
It is well recognized that at least several days or weeks are needed before protective adaptive immunity is adequately activated. To understand the mechanism of the rapid and robust protection induced by intranasal administration of dNS1-RBD, the levels of innate immune response biomarkers in the respiratory tract of BALB/c mice after intranasal administration of dNS1-RBD were compared to those in placebo controls and animals infected with CA04-WT and CA04-dNS1 virus (Fig. 5 a, b). The results showed that the levels of the proinflammatory cytokines and chemokines interleukin (IL)-6, IL-1α, IFN-γ, IFN-α, monocyte chemoattractant protein (MCP)-1, interferon-inducible protein (IP)-10, macrophage inflammatory protein (MIP)-2α, MIP-1β, granulocyte–macrophage colony-stimulating factor (GM-CSF), and tumor necrosis factor (TNF)-α, which are linked to the activation of innate immunity against respiratory viruses, were significantly elevated in lung tissue of mice 24 h post immunization for both CA04-dNS1 and dNS1-RBD compared to CA04-WT. At 3 or 5 d post inoculation, only CA04-WT group maintained activated state for cytokines and even climbed to peak levels at 5 d post inoculation, which might cause cytokine storms. In the meantime, CA04-dNS1 and dNS1-RBD were found as similar innate immune response kinetics and only induced moderate and rapid anti-viral immune response at 1 d post vaccination with few cytokines detectable at 3 or 5 d post inoculation, which might contribute to rapid protection against SARS-CoV-2. As they were treated with a nasal spray vaccine, dNS1-RBD-vaccinated animals were expected to produce robust and RBD-specific cell-mediated immunity (CMI) compared to CA04-dNS1 (Fig. 5c), and had a significantly greater number of RBD-specific immune cells within the respiratory system than among PBMCs or lymphocytes from the spleen and cervical lymph nodes (Fig. 5d) after prime-boost immunization, which suggested that the CMI response induced by dNS1-RBD is local and intensive in the respiratory tract. In particular, the RBD-specific cellular immune response was 22 times higher than that in PBMCs (Fig. 5d), which poses a challenge in evaluating the immune response of this vaccine based on PBMC test results in clinical trials.
Fig. 5.
The profile of immune responses in the respiratory tract and blood induced by intranasal administration of dNS1-RBD in mice. (a, b) Lung homogenates from BALB/c mice vaccinated with dNS1-RBD, CA04-dNS1 and CA04-WT were assayed for cytokine and chemokine expression levels by ProcartaPlex immunoassays. (a) Heat maps showing the scaled expression level of 36 immune response genes on days 1, 3, and 5 after vaccination, normalized to the relative value of placebo controls. (b) Expression levels of 10 cytokines and chemokine that were significantly reduced in dNS1-RBD and CA04-dNS1 immunized mice 1 d after vaccination were analyzed. The data are expressed as ng/mL. (c) Pulmonary lymphocytes collected 7 d after immunization by dNS1-RBD and CA04-dNS1 were subjected to IFN-γ ELISPOT assays. (d) Lymphocytes in the peripheral blood, spleen, lung, and lymph nodes collected 14 d after booster immunization at day 14 were subjected to IFN-γ ELISPOT assays. (e) IFN-γ ELISPOT assays for pulmonary lymphocytes from C57BL/6 mice vaccinated intranasally with two doses, with a booster at day 14 to assess T cell response kinetics at the indicated time points. Data are the median with IQR; ns, not significant (P > 0.05); significance was determined by one-way ANOVA with the Kruskal–Wallis test. (f) The responses were assessed under pre-stimulation of various peptide pools covering the spike of SARS-CoV-2 variants. (g) TRM markers expressed in pulmonary T cells 2 weeks after booster vaccination at a 14-d interval. TRM markers expressed in pulmonary T cells were detected in mice boosted with an extra 3rd inoculation 3 months after booster vaccination with a 14-d interval. (h) RBD-specific IgA levels in BAL fluid and IgG levels in serum were measured by ELISA for BALB/c mice vaccinated twice by dNS1-RBD and CA04-dNS1 at days 0 and 14. Data for antibody analysis are presented as the geometric mean with the geometric SD from four independent experiments. LLOD-lower limit of detection. Data are shown as mean ± SD; significance was determined by ordinary one-way ANOVA multiple comparison (b–f) or two-tailed Student’s t-test (g, h). ns, not significant.
For RBD-specific T cell activation and proliferation, the CMI response reached a peak at 7 d after a single-dose intranasal administration, with more rapid and robust response dynamics compared to those of the humoral response, and fell to a moderate level at 42 d following the prime-boost regimen with a 2-week interval (Fig. 5e). Although the CMI response progressively waned, the specific T cell response from 9/10 animals was detectable at 3 months by IFN-γ ELISPOT after booster immunization, with 6/10 animals further proven to be positive at 6 months. In addition to the longevity of vaccine-induced immunity, a substantial number of CD4+ and CD8+ T cells in the lungs of mice vaccinated with dNS1-RBD showed upregulated expression of the tissue-resident memory (TRM) marker CD69, while dNS1-RBD-generated CD8+ TRM cells also expressed the canonical CD8+ TRM marker CD103 (Fig. 5g and Fig. S3 online), indicating that vaccination with dNS1-RBD generated lung-resident memory RBD-specific CD4+ and CD8+ TRM populations. Three months post 2nd vaccination with a 14-d interval, activation and proliferation of memory CD69+CD103+ TRM cells could be detected 7 d after the boost inoculation.
As a recent study showed that SARS-CoV-2 variants (B.1.1.7 in the UK, B.1.351 in South Africa, B.1.525 in Nigeria, P1 in Brazil, and especially Omicron) are relatively resistant to serum from individuals who have recovered from COVID-19 or serum from individuals who have been vaccinated against SARS-CoV-2 [34], we used peptides covering the RBD with key mutations from the major variants (including Alpha, Beta, Gamma, Delta, Kappa, Eta, Iota, Lambda, Mu, Omicron BA.1, and Omicron BA.2) and prototype strains to stimulate lymphocytes and found similar RBD-specific T cell responses in the lungs from vaccinated mice, especially for the latest BA.1 and BA.2, suggesting that the key mutants are still covered by the dNS1-RBD vaccine (Fig. 5f).
Notably, bioinformatics analyses suggest that the majority of CD4+ and CD8+ T cells epitopes are unaffected by mutations, regardless of whether early or late variants were considered including Omicron [35], in line with the above T cell response which can cross-recognize variants from Alpha to Omicron induced by dNS1-RBD. To identify the T cell epitopes of dNS1-RBD, an IFN-γ ELISPOT assay was performed by stimulating the pulmonary lymphocyte obtained from immunized mice 7 d after single immunization with 6 segmented peptide pools covering the RBD region (Table S1 online). After rounds of screening, the level of response provoked by treatment with the RBD539-546 peptide was similar to the SFU values stimulated with the entire RBD peptide pool, while only RBD509-519 of the other 56 individual RBD peptides induced the production of IFN-γ responses (Fig. S4a–d and Table S2 online). In sum, these epitope mapping data showed the preservation of T cell recognition of Omicron RBD (BA.1 and BA.2) was 100% (16/16) (Fig. S4e online) and suggested that dNS1-RBD appears to elicit more broad RBD-specific T cell response, which is related to the protective efficacy against Omicron in hamster challenge model.
In-depth profiling of the T cell compartment by intracellular cytokine staining confirmed a significant increase in RBD-specific IFN-γ+ effector memory T cells in the lung, spleen, and cervical lymph nodes (Fig. S5b online), RBD-specific TNF-α+ CD8+ T cells in the lung and spleen (Fig. S5c online) and RBD-specific IL-2+CD8+ T cells in the lung (Fig. S5d online) from immunized mice in comparison with those from mice in the control group upon ex vivo stimulation with pools of overlapping 15-mer RBD peptides. Significant enrichment of other subpopulations, such as IL-2+, IFN-γ+, and TNF-α-expressing CD4+ T lymphocytes, was not observed. The robust production of IFN-γ from CD8+ T cells indicated a favorable immune response with both antiviral and immune-augmenting properties, suggesting the induction of a Th1-biased cellular immune response and the potential safety of this vaccine.
Serum samples and bronchoalveolar lavage fluid (BAL) were also collected 14 d after primary or booster immunization, and RBD-specific sIgA or IgG responses were evaluated by ELISA (Fig. 5h). The levels of RBD-specific sIgA and IgG titers increased significantly for dNS1-RBD groups but not CA04-dNS1 groups after boost immunization and peaked at 28 d post immunization, with all mice seroconverting. Whereas vaccines can induce the production of moderate levels of RBD-specific sIgA and IgG, the neutralizing activity of the induced antibodies was below the limit of detection.
Overall, these data suggest that dNS1-RBD vaccination rapidly elicits vigorous and long-lived local innate and adaptive immune responses in the local respiratory tract that confer protection against SARS-CoV-2 infection.
3.4. Broad protection of dNS1-RBD against influenza in BALB/c mice
To confirm the protective ability of dNS1-RBD vaccine in influenza virus infection, mice were immunized with 50 μL (1 × 106 PFU/mL) of dNS1-RBD and CA04-dNS1 viruses via the nasal route and inactivated CA04-WT virus intramuscularly and boosted 14 d later (Fig. 6 a). NP-specific IgG and Flu-specific cell-mediated immune (CMI) response for pulmonary lymphocyte by IFN-γ ELISPOT was detected at 14 d after boost immunization, and showed that dNS1-RBD and CA04-dNS1 virus could induce Flu-specific moderate IgG level and robust T cell response in lung tissue compared to CA04-WT vaccine (Fig. 6b, c). To further assess whether the dNS1-RBD vaccine can provide protection against homologous and heterologous influenza virus challenges, two mouse-adapted (MA) H1N1 and H5N1 viruses were used to evaluate the prophylactic efficacy. The results showed that the dNS1-RBD and CA04-dNS1 virus vaccination provided broad and considerable prophylactic activity in mice against lethal dose of both representative influenza A viruses compared to CA04-WT group. Immunization with either vaccine or CA04-WT fully protected against lethal challenge with mouse-adapted H1N1 virus and no virus was detected in the lung tissues of mice from vaccinated groups at day 5 post infection (Fig. 6d–f). Interestingly, immunization with dNS1-RBD and CA04-dNS1 virus also protected against H5N1 virus challenge compared to CA04-WT group, with mice displaying only slight body weight loss during the first 5 d of infection, followed by full recovery (Fig. 6g, h). Consistent with this body weight loss, virus titer in the lungs of mice immunized with dNS1-RBD and CA04-dNS1 virus were significantly lower than CA04-WT group for H5N1 virus challenge (Fig. 6i), further demonstrating that dNS1-RBD and CA04-dNS1 virus provide better cross-protective activity than CA04-WT virus.
Fig. 6.
Immunogenicity and protective efficacy against influenza virus of dNS1-RBD in BALB/c mice. (a) Scheme of vaccination and challenge. Six-week-old BALB/c female mice were immunized dNS1-RBD, CA04-dNS1, and CA04-WT via intranasal route, and the mice treated with virus protectant were used as placebo controls. On day 30 post immunization, mice were challenged with 25 MLD50 of A/California/04/2009 (H1N1) and A/Qinghai/15/2005 (H5N1). (b) Antibody response in sera of immunized mice at 14 d after boosting was evaluated. (c) Cellular immune responses for IFN-γ in the lung were assessed at 14 d after boosting for pulmonary lymphocyte by stimulation with pooled peptides of influenza A virus for 24 h. (d–i) Survival curves and body weight changes for BALB/c mice (n = 5 per group) against lethal challenge with 25 MLD50 of A/California/04/2009 (H1N1) (d, e) and A/Qinghai/15/2005 (H5N1) (g, h) are shown. Viral loads of lung tissue obtained at 5 dpi challenged by the H1N1 (f) or H5N1 (i) were determined by TCID50 assay. All data are shown as mean ± SD from four independent experiments; error bars reflect SD. Significant differences compared to the placebo group were analyzed using two-way repeated-measures ANOVA with Dunnett’s multiple comparisons test (e, h) or by ordinary one-way ANOVA multiple comparison (b, c, f, and i).
Thus, the dNS1-RBD vaccine appears to be a promising candidate for the development of universal vaccines against influenza virus and SARS-CoV-2 infection.
4. Discussion and conclusion
To date, all COVID-19 vaccines approved are administered by intramuscular injection to elicit the production of primarily serum neutralizing antibodies and systemic T cell responses to fight against SARS-CoV-2 infection [6]. However, intramuscular vaccines induce poor local immunity in the respiratory tract, which is the primary infection site for SARS-CoV-2 [18]. It is evident that these vaccines are protective against severe diseases even in children [36], however, breakthrough infections among vaccinated individuals are common [8], [9], [37], [38]. How to achieve more effective prevention of infection or transmission has become extremely important in the ongoing response to the COVID-19 pandemic.
One solution is to enhance the local immunity in the respiratory tract. Cold-adapted, live attenuated intranasal influenza vaccines have been used for more than a decade and shown to be effective in seasonal influenza, in particular among young children [39]. Based on this concept, we have developed a live attenuated influenza vector (dNS1) by deleting viral immune modulator, the NS1 protein, from viral genome and identified adaptative mutations to support virus replication in eggs or MDCK cells which are commonly used for vaccine production. Using this dNS1 vector, we inserted the RBD gene of SARS-CoV-2 into the deleted NS1 site and made an influenza viral vector vaccine for COVID-19 (dNS1-RBD). This vaccine system has a few unique advantages which are immunogenic due to the lack of the NS1 which is a strong immune antagonist; it is extremely safe for use in all age groups; similar to the intranasal influenza vaccines, it is used intranasally to specifically induce mucosal immunity in the respiratory tract.
Our data showed that intranasal immunization of this dNS1-RBD vaccine can induce rapid protective, long-lasting and broad immunities in hamsters when immunized hamsters were challenged 1 d after single-dose vaccination or 9 months after booster vaccination. The protective immune response largely mitigated the lung pathology, with no apparent loss of body weight, caused by either the prototype-like strain or Beta variant and Omicron variant, suggesting cross-protective properties of this vaccine. This study demonstrates that nasal vaccines may offer an attractive alternative in fighting against the COVID-19 pandemic.
What is special about this vaccine is that it is effective in preventing the pathological changes caused by COVID-19 without high levels of neutralizing IgG antibodies, which is different from traditional vaccines mainly based on neutralizing antibodies. We believe that there are at least three aspects of the protective immune mechanism based on the current data. (i) Previous studies have reported that LAIVs induce the innate immune response in the nasal epithelium in animals, which not only is crucial for viral clearance and attenuation but also may play an important role in the induction of a protective immune response [40]. In this study, we also observed the activation and secretion of antiviral cytokines and chemokines in lung tissue from vaccinated mice and correlated their production with rapid protection in hamsters. (ii) We believe that robust and local RBD-specific T cell responses should contribute to providing effective protection against SARS-CoV-2 infection [41]. Considering resident memory CD8+ T cells, which are thought to provide long-lasting and broad-spectrum immune protection for LAIVs[42], our data suggest that dNS1-RBD has the potential to confer long-lasting protective immunity, particularly around the bronchoalveolar space and lungs. Consistently, the hamster challenge results showed that dNS1-RBD conferred persistent protection against both the prototype-like strain and beta variant at 9 months after vaccination. (iii) Regarding the humoral immune response, RBD-specific IgA in BAL fluid and IgG in serum could be detected at a moderate level, which may effectively inhibit SARS-CoV-2 in the upper airways and nasal passages [43]. Certainly, the translational gap for animal to human cannot be ignored, because the human immune system is more sophisticated and the local environment of the nasal or respiratory tract is “messy” rather than “naïve”. That could be factors influencing the immune response induced by our vaccine with possible pre-existing immune. However, the phase I/II clinical trial of the vaccine showed that pre-existing H1N1 antibody has no negative effect on the T cell response for our vaccine. In any cases, the efficacy of dNS1-RBD will be confirmed in the near future by the results of the ongoing phase III trial.
LAIVs for intranasal administration were first licensed in the Soviet Union in 1987, the US in 2003, and Europe in 2012, and have a proven record of efficacy over decades of use [38], [44], [45]. However, the immune response to LAIVs is multifaceted and does not necessarily involve a serum antibody response; LAIVs have been licensed based on efficacy trials that measure protection rather than correlates of protection. A human challenge trial of LAIVs (FluMist®) also suggested that a low antibody response was not directly associated with low protective efficacy. In that study, the virus challenge results indicated that the LAIV had the equivalent and even improved efficacy of trivalent inactivated vaccine (TIV), while a higher seroresponse rate was induced by the TIV [17]. In general, the induction of the production of mucosal antibodies and a local T cell response by FluMist® was similar to those induced by dNS1-RBD in adults [24]. As we can find in COVID-19 vaccine tracker and landscape from WHO, there are other influenza-based vaccines for COVID-19 using different strategies in pre-clinical trials. Indeed, the attenuated virus based on NS1 modification mutants, which is a virulence element and the critical factor for regulation of the host immune response, might not merely act as an attenuation strategy, but exhibit more potent and long-lasting immunity compared to cold-adapted influenza virus through activating multidimensional immune responses by NS1 gene deletion.
Data from three earlier-phase clinical trials involving 1084 naïve adults showed that dNS1-RBD is very well tolerated and immunogenic in inducing the production of mucosal IgA, systemic T cell responses, and IgG against the RBD of SARS-CoV-2 [24]. Undoubtedly, a phase III clinical trial conducted in COVID-19 epidemic countries is essential to finally determine the efficacies of dNS1-RBD against pivotal clinical outcomes associated with SARS-CoV-2 infection in humans, which is expected to be initiated soon in multi-international sites (ChiCTR2100051391).
Thus, dNS1-RBD, an intranasally delivered vaccine candidate based on a LAIV vector, is unique for offering very rapid and prolonged broad protection against SARS-CoV-2 infection by inducing comprehensive local immune responses in the respiratory tract and might be a very promising vaccine that could fill the gap of current intramuscular vaccines. Further studies should be conducted to understand the unique immune activation and protection mechanism of intranasal immunization dNS1-RBD for SARS-CoV-2 in humans. Furthermore, our proposed flu-based vaccine can also offer protection against seasonal human influenza viruses, representing a potential approach to fight against a possible “twin-demic” of COVID-19 and flu in the future.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
Acknowledgments
This work was supported by the National Program on Key Research Project of China (2020YFC0842600), the National Natural Science Foundation of China (82041038, 81871651, and 81991491), the Major Science and Technology Program of Fujian Province (2020YZ014001), and the Natural Science Foundation of Fujian Province (2021J02006).
Author contributions
Junyu Chen, Pui Wang, Lunzhi Yuan, Liang Zhang, Limin Zhang, Hui Zhao, Congjie Chen, Xijing Wang, Jinle Han, Yaode Chen, Jizong Jia, Zhen Lu, Junping Hong, Zicen Lu, Qian Wang, Rirong Chen, Ruoyao Qi, Jian Ma, Min Zhou, Huan Yu, Chunlan Zhuang, Xiaohui Liu, Qiangyuan Han, and Guosong Wang performed experiments and analyzed data. Xiangzhong Ye, Changgui Li, Tianying Zhang, Jun Zhang, Huachen Zhu, Yixin Chen, Honglin Chen, and Ningshao Xia designed and supervised the study. Changgui Li, Tianying Zhang, Jun Zhang, Huachen Zhu, Yixin Chen, Honglin Chen, and Ningshao Xia wrote the manuscript. Yingying Su, Quan Yuan, Tong Cheng, and Ting Wu participated in the design and result analysis.
Biographies

Junyu Chen is currently a post-doctoral fellow at the State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics, School of Public Health, Xiamen University. He received his Ph.D. degree from the School of Life Sciences, Xiamen University in 2019. His research interest focuses on the development of intranasal COVID-19 vaccine and universal influenza vaccine.

Pui Wang got his Ph.D. degree from the Department of Microbiology, The University of Hong Kong. His research interest includes the development of live attenuated influenza vaccine and molecular mechanisms of virus/host interaction.

Lunzhi Yuan obtained his Ph.D. degree from Xiamen University in 2019. He is currently a post-doctoral fellow at the School of Public Health, Xiamen University. His research interest focuses on animal models of emerging pathogens and infectious diseases, development of vaccines and antiviral drugs, and stem cell-derived organ regeneration.

Liang Zhang is a Ph.D. candidate at the State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics, National Institute of Diagnostics and Vaccine Development in Infectious Diseases, Xiamen University. His research interest focuses on the mechanism of vaccine immune response and respiratory mucosal immunity.

Limin Zhang is a Ph.D. candidate at the Department of Biochemistry and Molecular Biology, School of Life Sciences, Xiamen University. Her research interest focuses on the development of influenza virus vaccine.

Hui Zhao is a professor at the National Institute for Food and Drug Control, China. She received her Ph.D. degree from the Chinese Academy of Medical Sciences & Peking Union Medical College in 2009. Her research interest focuses on the quality control of vaccines and advanced techniques for the evaluation of vaccines.

Congjie Chen is a Ph.D. candidate at the State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics; National Institute of Diagnostics and Vaccine Development in Infectious Diseases, School of Life Sciences, Xiamen University. His research focuses on live-attenuated influenza and the Intranasal SARS-CoV-2 vaccine.

Xijing Wang is a Ph.D. candidate at the National Institute of Diagnostics and Vaccine Development in Infectious Diseases, School of Life Sciences, Xiamen University. Her research interest focuses on virus infection and the immunology mechanism of vaccines.

Jinle Han got her master's degree from Harbin Normal University and now works at Beijing Wantai Biological Pharmacy Enterprise Co., Ltd. Her interest focuses on the vaccine research and development.

Yaode Chen is a graduate student at the National Institute of Diagnostics and Vaccine Development in Infectious Diseases, School of Public Health, Xiamen University. His major research focuses on the attenuated influenza virus vector vaccine.

Ting Wu is a professor at the National Institute of Diagnostics and Vaccine Development in Infectious Disease, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics, Xiamen University. She received her Ph.D. degree from the School of Pharmacy, Fudan University in 2000 and joined the School of Public Health, Xiamen University in 2003. Her research interest focuses on the vaccine clinical trials and epidemiological studies of infectious diseases.

Xiangzhong Ye got his master’s degree from Beijing Institute of Biotechnology and now works at Beijing Wantai Biological Pharmacy Enterprise Co., Ltd. His research interest focuses on vaccine R & D and industrialization research.

Tianying Zhang is an assistant professor at the School of Public Health, Xiamen University. He received his Ph.D. degree from the School of Life Sciences, Xiamen University in 2016. His research interest focuses on the vaccine development and viral immunology.

Changgui Li is a member of the Expert Commission of Chinese Pharmacopoeia. He got his Ph.D. degree from Fourth Military Medical University in 2007. He is a professor at National Institutes for Food and Drug Control, China. His research interest focuses on the quality control of vaccines and advanced techniques for the evaluation of vaccines.

Jun Zhang is a professor at the National Institute of Diagnostics and Vaccine Development in Infectious Disease, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics, School of Public Health, Xiamen University. His research interest focuses on the development of vaccines and diagnostic reagents against infectious pathogens, including the hepatitis E vaccine, HPV16/18 vaccine, acquired immunodeficiency syndrome, epidemiological studies of infectious diseases, and so on.

Huachen Zhu is an associate professor at The University of Hong Kong and an adjunct professor at Shantou University. She also serves as associate director of the Guangdong-Hong Kong Joint Laboratory of Emerging Infectious Diseases and the Key Laboratory of Ministry of Education (China) for International Collaboration in Virology and Emerging Infectious Diseases. Her research interest focuses on mechanisms that lead to the virus emergence at the human and animal interface, and control measures for the emerging viral diseases.

Yixin Chen is an associate professor at the National Institute of Diagnostics and Vaccine Development in Infectious Disease, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics, Xiamen University. He earned his Ph.D. degree from the School of Life Sciences, Xiamen University in 2009 and stayed as an assistant professor. His research interest focuses on the vaccines, therapeutic agents, and diagnostic kits against COVID-19, influenza, Epstein-Barr virus (EBV), hand-foot-mouth disease, herpesvirus-associated diseases, and oncolytic virus.

Honglin Chen is a professor at the Department of Microbiology, The University of Hong Kong. He also serves as the coordinator in operation in the State Key Laboratory for Emerging Infectious Diseases at The University of Hong Kong. His research interest focuses on molecular mechanisms for host restriction and cross-species transmission of RNA viruses, and regulation of EBV gene expression and cell signaling in EBV-associated cancers.

Ningshao Xia is a professor, director of the National Institute of Diagnostics and Vaccine Development in Infectious Disease, and director of State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics, Xiamen University. His research interest focuses on biomedical development including vaccines, diagnostics, and therapeutics that are related to infectious diseases, cancers, and so on.
Footnotes
Supplementary materials to this article can be found online at https://doi.org/10.1016/j.scib.2022.05.018.
Appendix A. Supplementary materials
The following are the Supplementary data to this article:
References
- 1.Mathieu E., Ritchie H., Ortiz-Ospina E., et al. A global database of COVID-19 vaccinations. Nat Hum Behav. 2021;5:947–953. doi: 10.1038/s41562-021-01122-8. [DOI] [PubMed] [Google Scholar]
- 2.Al Kaabi N., Zhang Y., Xia S., et al. Effect of 2 inactivated SARS-CoV-2 vaccines on symptomatic COVID-19 infection in adults: a randomized clinical trial. JAMA. 2021;326:35–45. doi: 10.1001/jama.2021.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jara A., Undurraga E.A., González C., et al. Effectiveness of an inactivated SARS-CoV-2 vaccine in chile. N Engl J Med. 2021;385:875–884. doi: 10.1056/NEJMoa2107715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baden L.R., El Sahly H.M., Essink B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2020;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu T., Dai J., Yang Z., et al. Inactivated SARS-CoV-2 vaccine does not influence the profile of prothrombotic antibody nor increase the risk of thrombosis in a prospective chinese cohort. Sci Bull. 2021;66:2312–2319. doi: 10.1016/j.scib.2021.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tregoning J.S., Flight K.E., Higham S.L., et al. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat Rev Immunol. 2021;21:626–636. doi: 10.1038/s41577-021-00592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadoff J., Gray G., Vandebosch A., et al. Safety and efficacy of single-dose Ad26.Cov2.S vaccine against COVID-19. N Engl J Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinde V., Bhikha S., Hoosain Z., et al. Efficacy of NVX-CoV2373 COVID-19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384:1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madhi S.A., Baillie V., Cutland C.L., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez Bernal J., Andrews N., Gower C., et al. Effectiveness of COVID-19 vaccines against the B.1.617.2 (delta) variant. N Engl J Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou D., Chan J.F.-W., Zhou B., et al. Robust SARS-CoV-2 infection in nasal turbinates after treatment with systemic neutralizing antibodies. Cell Host Microbe. 2021;29:551–563. doi: 10.1016/j.chom.2021.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassan A.O., Kafai N.M., Dmitriev I.P., et al. A single-dose intranasal chad vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183:169–184. doi: 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bleier B.S., Ramanathan M., Lane A.P. COVID-19 vaccines may not prevent nasal SARS-CoV-2 infection and asymptomatic transmission. Otolaryng Head Neck. 2020;164:305–307. doi: 10.1177/0194599820982633. [DOI] [PubMed] [Google Scholar]
- 14.Mercado N.B., Zahn R., Wegmann F., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kustin T., Harel N., Finkel U., et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2-mRNA-vaccinated individuals. Nat Med. 2021;27:1379–1384. doi: 10.1038/s41591-021-01413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohn K.G.I., Smith I., Sjursen H., et al. Immune responses after live attenuated influenza vaccination. Hum Vacc Immunother. 2018;14:571–578. doi: 10.1080/21645515.2017.1377376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treanor J.J., Kotloff K., Betts R.F., et al. Evaluation of trivalent, live, cold-adapted (CAIV-T) and inactivated (TIV) influenza vaccines in prevention of virus infection and illness following challenge of adults with wild-type influenza A (H1N1), A (H3N2), and B viruses. Vaccine. 1999;18:899–906. doi: 10.1016/s0264-410x(99)00334-5. [DOI] [PubMed] [Google Scholar]
- 18.Lund F.E., Randall T.D. Scent of a vaccine. Science. 2021;373:397–399. doi: 10.1126/science.abg9857. [DOI] [PubMed] [Google Scholar]
- 19.Wu S., Huang J., Zhang Z., et al. Safety, tolerability, and immunogenicity of an aerosolised adenovirus type-5 vector-based COVID-19 vaccine (Ad5-nCoV) in adults: preliminary report of an open-label and randomised phase 1 clinical trial. Lancet Infect Dis. 2021;21:1654–1664. doi: 10.1016/S1473-3099(21)00396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King R.G., Silva-Sanchez A., Peel J.N., et al. Single-dose intranasal administration of AdCOVID elicits systemic and mucosal immunity against SARS-CoV-2 and fully protects mice from lethal challenge. Vaccines. 2021;9:881. doi: 10.3390/vaccines9080881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tioni MF, Jordan R, Pena AS, et al. One mucosal administration of a live attenuated recombinant COVID-19 vaccine protects nonhuman primates from SARS-CoV-2. bioRxiv, 2021, 2021.07.16.452733. [DOI] [PMC free article] [PubMed]
- 22.Bricker T.L., Darling T.L., Hassan A.O., et al. A single intranasal or intramuscular immunization with chimpanzee adenovirus-vectored SARS-CoV-2 vaccine protects against pneumonia in hamsters. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Doremalen N., Purushotham Jyothi N., Schulz Jonathan E., et al. Intranasal ChAdOx1 nCoV-19/AZD1222 vaccination reduces viral shedding after SARS-CoV-2 D614G challenge in preclinical models. Sci Transl Med. 2021;13:eabh0755. doi: 10.1126/scitranslmed.abh0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu FC, Zhuang CL, Chu K, et al. Safety and immunogenicity of a live-attenuated influenza virus vector-based intranasal SARS-CoV-2 vaccine in adults: randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Respir Med. 2022 doi: 10.1016/S2213-2600(22)00131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P., Zheng M., Lau S.-Y., et al. Generation of DelNS1 influenza viruses: a strategy for optimizing live attenuated influenza vaccines. mBio. 2019;10 doi: 10.1128/mBio.02180-19. e02180–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Assarsson E., Bui H.-H., Sidney J., et al. Immunomic analysis of the repertoire of T-cell specificities for influenza a virus in humans. J Virol. 2008;82:12241. doi: 10.1128/JVI.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkinson T.M., Li C.K.F., Chui C.S.C., et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 28.Imai M., Iwatsuki-Horimoto K., Hatta M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci USA. 2020;117:16587. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan L., Zhu H., Zhou M., et al. Gender associates with both susceptibility to infection and pathogenesis of SARS-CoV-2 in syrian hamster. Signal Transduct Target Ther. 2021;6:136. doi: 10.1038/s41392-021-00552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sia S.F., Yan L.-M., Chin A.W.H., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bi Z., Hong W., Yang J., et al. Animal models for SARS-CoV-2 infection and pathology. MedComm. 2021;2:548–568. doi: 10.1002/mco2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He X., Hong W., Pan X., et al. SARS-CoV-2 omicron variant: characteristics and prevention. MedComm. 2021;2:838–845. doi: 10.1002/mco2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halfmann P.J., Iida S., Iwatsuki-Horimoto K., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cele S., Gazy I., Jackson L., et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarke A., Coelho C.H., Zhang Z., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from alpha to Omicron. Cell. 2022;185:847–859. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu L., Zhang C., Wu J., et al. Strong immunity against COVID-19 in the early two years of age links to frequent immunization of routine vaccines. Sci Bull. 2020;65:2057–2060. doi: 10.1016/j.scib.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Supasa P., Zhou D., Dejnirattisai W., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alter G., Yu J., Liu J., et al. Immunogenicity of ad26.Cov2.S vaccine against SARS-CoV-2 variants in humans. Nature. 2021;596:268–272. doi: 10.1038/s41586-021-03681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caspard H., Heikkinen T., Belshe R.B., et al. A systematic review of the efficacy of live attenuated influenza vaccine upon revaccination of children. Hum Vacc Immunother. 2016;12:1721–1727. doi: 10.1080/21645515.2015.1115164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forero A., Fenstermacher K., Wohlgemuth N., et al. Evaluation of the innate immune responses to influenza and live-attenuated influenza vaccine infection in primary differentiated human nasal epithelial cells. Vaccine. 2017;35:6112–6121. doi: 10.1016/j.vaccine.2017.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poon Maya ML, Rybkina K, Kato Y, et al. SARS-CoV-2 infection generates tissue-localized immunological memory in humans. Sci Immunol 2021;6:eabl9105 [DOI] [PMC free article] [PubMed]
- 42.Pizzolla A., Wakim L.M. Memory T cell dynamics in the lung during influenza virus infection. The J Immunol. 2019;202:374. doi: 10.4049/jimmunol.1800979. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z., Lorenzi J.C.C. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci Transl Med. 2021;373:397–399. doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monto A.S., Ohmit S.E., Petrie J.G., et al. Comparative efficacy of inactivated and live attenuated influenza vaccines. N Engl J Med. 2009;361:1260–1267. doi: 10.1056/NEJMoa0808652. [DOI] [PubMed] [Google Scholar]
- 45.Nichol K.L., Mendelman P.M., Mallon K.P., et al. Effectiveness of live, attenuated intranasal influenza virus vaccine in healthy, working adults: a randomized controlled trial. JAMA. 1999;282:137–144. doi: 10.1001/jama.282.2.137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.