Abstract
We studied the expression of the genes encoding group I alcohol dehydrogenases (PsADH1 and PsADH2) in the xylose-fermenting yeast Pichia stipitis CBS 6054. The cells expressed PsADH1 approximately 10 times higher under oxygen-limited conditions than under fully aerobic conditions when cultivated on xylose. Transcripts of PsADH2 were not detectable under either aeration condition. We used a PsADH1::lacZ fusion to monitor PsADH1 expression and found that expression increased as oxygen decreased. The level of PsADH1 transcript was repressed about 10-fold in cells grown in the presence of heme under oxygen-limited conditions. Concomitantly with the induction of PsADH1, PsCYC1 expression was repressed. These results indicate that oxygen availability regulates PsADH1 expression and that regulation may be mediated by heme. The regulation of PsADH2 expression was also examined in other genetic backgrounds. Disruption of PsADH1 dramatically increased PsADH2 expression on nonfermentable carbon sources under fully aerobic conditions, indicating that the expression of PsADH2 is subject to feedback regulation under these conditions.
The fundamental mechanisms by which fermentation is regulated appear to differ profoundly in the glucose-fermenting yeast Saccharomyces cerevisiae and the xylose-fermenting yeast Pichia stipitis (22, 44). Even though the P. stipitis structural genes encoding alcohol dehydrogenase (ADH) and pyruvate decarboxylase (PDC) show significant sequence conservation with those of S. cerevisiae (9, 28, 32), their regulatory patterns differ. Oxygen availability is largely irrelevant to fermentative metabolism when S. cerevisiae is provided with excess glucose (24). Glucose induces high levels of fermentative S. cerevisiae ADH (ADH1) and represses oxidative ADH (ADH2), leading to ethanol production (3). In contrast, glucose does not induce fermentation in the Crabtree-negative yeast P. stipitis (6, 38). Efficient conversion of xylose to ethanol requires a limited amount of oxygen (12, 40), but ethanol accumulation does not occur when oxygen is freely available (15, 26, 39). As oxygen becomes limiting, P. stipitis PDC and ADH activities increase (16, 31), and malate dehydrogenase activity decreases (39).
The induction of fermentative enzymes by oxygen limitation also is observed in plants. Transcriptional regulation is implicated in the hypoxic synthesis of arabidopsis, barley, and maize ADH (1, 2, 8, 41). Similarly, in the filamentous fungus Aspergillus nidulans, ADH3 is specifically induced in response to periods of anaerobic stress, and its expression is regulated largely at the posttranscriptional level (23). Similar mechanisms may function in P. stipitis, because its primary phenomenological response to a lowered aeration rate is to increase fermentative enzymes. The mechanism and extent of this regulation, however, remain obscure, and the means by which its fermentation is regulated by oxygen have not been characterized in detail at the molecular level.
In a previous paper, (9) we described the isolation of two cytoplasmic ADH (PsADH) genes from P. stipitis. The results from gene disruption studies suggested that PsADH1 had both fermentative and respirative functions. However, the simultaneous presence of ethanol-producing and ethanol-oxidizing activities in the cytoplasm could result in a futile cycle. We hypothesized that oxygen-dependent regulation of PsADH expression could avoid futile cycling if respirative enzyme activities were repressed and fermentative enzyme activities were induced under oxygen-limited conditions.
Our objective in the present research was to elucidate the physiological signals regulating the expression of the PsADH gene(s). Transcription of PsADH1 appeared to involve regulation by oxygen. If P. stipitis requires oxygen for heme synthesis, as in other yeasts (48), the oxygen effect on transcription could be transduced through heme-dependent transcription factors (49). Because heme acts as a regulatory coeffector in the induction and repression of aerobic and hypoxic genes in S. cerevisiae (50), we hypothesized that it could be responsible for the repression of the fermentative gene, PsADH1. The results reported here show that heme represses expression of PsADH1 under fermentative conditions. Previous studies had shown that disruption of PsADH1 increased xylitol production on xylose, so we wanted to know how its disruption affected the levels of PsADH2 mRNA. Our results showed that PsADH2 transcription increases significantly in the Psadh1 disruptant. While posttranscriptional mechanisms possibly affect PsADH2 expression, feedback regulation of PsADH2 at the transcriptional level is most likely.
MATERIALS AND METHODS
Strains.
P. stipitis CBS 6054 was the ultimate origin of all yeast strains. P. stipitis PLU20 (ura3-3/ura3-3 Psleu2Δ-1/Psleu2Δ-1) was used as a recipient strain for transformations. Escherichia coli DH5α (Gibco BRL, Gaithersburg, Md.) [F− recA1 endA1 hsdR17 (rk− mk+) supE44 thi-1 gyrA relA1] was used for all recombinant DNA experiments that required a bacterial host.
Media and culture conditions.
Yeasts were routinely grown in yeast-peptone-dextrose (YPD) medium consisting of yeast extract (10 g/liter), peptone (20 g/liter), and glucose (20 g/liter). For cultivation of ura3 and leu2 auxotrophs, media were supplemented with 100 mg of uridine and 100 mg of leucine per liter, respectively. Induction studies were carried out in 1.7 g of yeast nitrogen base per liter without ammonium sulfate or amino acids (YNB; Difco, Detroit, Mich.), which was supplemented with Bacto Peptone (6.6 g/liter) plus urea (2.3 g/liter) (2× nitrogen) and 80 g of d-xylose or glucose per liter, plus leucine, as needed. P. stipitis CBS 6054 was grown in fermentative medium containing either 8% xylose or glucose under fully aerobic conditions (200 rpm in a baffled flask) until the cells reached an optical density at 600 nm (OD600) of between 0.8 and 1.0. Oxygen was then limited by centrifuging the cells and inoculating them at a density of 2.4 mg (dry wt)/ml in 50 ml of medium in a 125-ml Erlenmeyer flask shaken at 100 rpm. Cultures were incubated at 25°C. After the shift to oxygen-limited conditions, samples were harvested at various times to monitor the activity of ADH and the level of its mRNA. Comparisons made between samples by Northern blotting, zymogram analysis, and ADH activity were performed with samples prepared at the same time. Yeast transformants were selected on YNB plus 20 g of glucose per liter, without uracil or leucine, when URA3 or LEU2 was used as the selectable marker, respectively. For solid media, 20 g of agar per liter was added. E. coli cells were grown in Luria-Bertani medium (35) with 50 μg of ampicillin per ml in liquid media or 100 μg of ampicillin per ml in solid media.
Northern analyses.
Total RNA was extracted by lysing cells with glass beads in the presence of phenol and sodium dodecyl sulfate. Comparisons between samples by Northern analyses were performed with RNA samples prepared at the same time. Approximately 20 μg of total RNA from each sample was loaded in triplicate onto a 2.2 M formaldehyde–1.2% agarose gel and electrophoresed. After blotting to the positively charged nylon membrane, the blot was cut into two identical pieces and hybridized with radiolabeled PsADH1-, PsADH2-, and PsCYC1-specific oligonucleotides as probes: PsADH1, 5′-CTCGTCGGAGTGCTGGCAGAAT-3′; PsADH2, 5′-TTCGTGAGCAGTGACACAGTAC-3′; and PsCYC1, 5′-CTTGACCGGACTTTCTGCCCATG-3′. Oligonucleotide probes were γ-32P-end-labeled with T4 polynucleotide kinase (New England Biolabs., Beverly, Mass.) and purified with a Microspin G-25 column (Pharmacia Biotech, Piscataway, N.J.) to remove unincorporated radionucleotides. RNA concentrations were measured by OD260. Hybridizations were performed with excess probes. We normalized the amount of total RNA loaded into each lane by measuring the relative abundance of rRNA. rRNA levels were analyzed with a Macintosh One scanner with the public domain NIH image program V 1.61 (42). The mRNA levels in cells grown under different growth conditions were measured with a PhosphorImager system (Molecular Dynamics, Sunnyvale, Calif.) relative to those of the rRNAs.
Zymogram analysis.
Enzyme activity was visualized essentially as described by Dewey and Conklin (14). Cell extracts were prepared by vortexing cells with glass beads. Protein concentrations were determined by the method of Bradford (5) with the Bio-Rad protein assay as specified by the manufacturer. Proteins were separated on a 5% stacking gel and 10% separating gel by using Tris-HCl buffer (pH 8.3) in the gel and Tris-glycine buffer (pH 8.8) in the electrophoresis vessels, with an applied current of 20 mA/gel at 4°C overnight. Following electrophoresis, ADH enzyme activity was visualized by staining the gel for enzyme activity with a solution of 4.0 mg of phenazine methosulfate, 10.0 mg of nitroblue tetrazolium, 50 mg of NAD+, and 0.05 ml of ethanol dissolved in 50 ml of 0.1 M Tris-HCl buffer (pH 8.5). Controls were run in which ethanol or protein was omitted from the aforementioned procedure to test for nonspecific reduction of the tetrazolium dye.
Enzyme assays.
ADH activity was assayed according to the method of Bergmeyer (4). The reaction mixture contained 100 mM Tris-HCl buffer (pH 8.3), 2 mM NAD+, cell extract (100 to 200 μg of protein), and 0.8 M ethanol, in a total volume of 1 ml. The reaction was started by ethanol addition, and reduction of NAD+ was monitored by measuring the increase in OD340. The specific activity was expressed as micromoles of NADH produced per minute per milligram of protein at 25°C.
Plasmids and plasmid constructions.
Plasmids pBluescript KSII+ (Stratagene, La Jolla, Calif.) and pUC19 were used for cloning DNA fragments. As a reporter gene in P. stipitis, we used the lacZ gene of E. coli from pFusionator (43). The PsADH1 promoter-lacZ fusion gene was constructed as follows. The KpnI site in pUC19 was destroyed by treatment with T4 DNA polymerase and ligation reactions to form pUC19-kpn. A 3.0-kbp BamHI-BamHI fragment containing the lacZ gene was excised from pFusionator and cloned into the BamHI site of pUC19-kpn to form pGAL. The 598-bp PsADH1 promoter region and 10 codons of the PsADH1 coding region were amplified with restriction enzyme-tailed primers (top, PstI, 5′-AAAACTGCAGAACCGATCCGAGGGAAAAACCGGG-3′; bottom, KpnI, 5′-CGGGGTACCCCGACAACAGCCTTTTGAGTGG-3′) and cut with PstI and KpnI to be cloned into the corresponding sites of pGAL to form pAB. In the resulting construct, the BamHI site in front of the lacZ gene was destroyed. As a result, the PsADH1 promoter and 10 codons of PsADH1 coding region are fused in frame with the lacZ coding sequence. The desired fusion was verified by restriction enzyme digests and sequenced by using a specific oligonucleotide primer from the lacZ coding region. A 750-bp BglII-XbaI fragment containing the 3′ end of the PsADH2 gene was excised from pJY158 (9) and cloned into the unique BamHI site of pAB to form pABA. A 4.35-kbp PstI-XbaI (blunt) fragment containing the PsADH1 promoter-lacZ-PsADH2 transcription terminator fusion gene was isolated from pABA constructs and cloned into the XbaI (blunt) site of pJM6 (46), an autonomously replicating plasmid, to form pFUS. The resulting plasmid was digested with SmaI, and the linker (5′-TGCTCTAGAGCA-3′) containing an XbaI site was inserted to create an appropriate 5′-overhang restriction site in front of the PsADH1 promoter sequences for the nested deletions with exonuclease III (New England Biolabs). Oligonucleotides were synthesized by Genosys Biotechnologies, Inc. (The Woodlands, Tex.).
Yeast transformation.
Lithium acetate transformations of P. stipitis PLU20 (ura3-3/ura3-3 leu2Δ-1/leu2Δ-1) (29) were performed according to the method described by Ito et al. (21).
β-Galactosidase assays.
Expression of the PsADH1::lacZ fusion gene was monitored with an o-nitrophenyl-β-d-galactopyranoside (ONPG) assay of β-galactosidase activity. Transformed yeast strains were grown in the appropriate selective medium with 8% xylose under fully aerobic conditions and harvested at an OD600 of 1.0. One-half of the culture was collected and stored frozen at −70°C. The remaining half was washed once in water and suspended in 50 ml of minimal medium lacking uracil and containing 8% xylose with a starting OD600 of 10 (≈2.4 g of cells [dry wt]/liter). These cultures were grown for another 4 h at 25°C under oxygen-limited conditions and then collected and frozen at −70°C. Cells grown under aerobic and oxygen-limited conditions were thawed, suspended in 300 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol), and disrupted with glass beads (10). A 50-μl aliquot of extract was assayed with a reaction mixture containing 1 ml of Z buffer and 100 μl of ONPG (4 mg/ml; Sigma, St. Louis, Mo.). Assays were performed at 25°C. One β-galactosidase unit is defined as the amount of enzyme necessary to cause a change of [1.0 OD420 min−1 mg of total protein−1] × 1,000 (27). The total protein concentration was determined as described above. A minimum of four independent transformants were assayed for the fusion construct, and the values are the means of independent determinations from three different assays. The copy number of plasmids in transformants was determined by measuring the relative intensities of plasmid-borne and genomic DNA fragments in Southern blots with LacZ gene- or PsADH2 gene-specific oligonucleotides as probes, respectively. Radioactive signals were measured with the PhosphorImager system. The plasmid copy number in transformants was found to be constant under the different growth conditions employed in this study (standard errors were <20% [data not shown]).
RESULTS
PsADH expression.
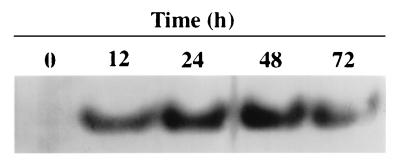
We analyzed the expression of the PsADH gene(s) in CBS 6054 following growth of the cells under fully aerobic and oxygen-limited conditions. A crude extract of cells grown on xylose under aerobic conditions and shifted to oxygen limitation showed a single major ADH activity in a zymogram analysis. Only one isozyme band was detectable in cells grown on xylose under oxygen-limited conditions (Fig. 1), even after xylose was exhausted from the medium and ethanol had begun to be utilized (72 h). Surprisingly, PsADH activity was not detectable when cells were grown on xylose under aerobic conditions, suggesting a negative effect of aerobiosis on the expression of the PsADH gene(s).
FIG. 1.
Zymogram analysis of ADH isozymes from P. stipitis CBS 6054 grown on 8% xylose under fully aerobic conditions (0 h) and after shifting to oxygen-limited conditions (12 to 72 h).
Regulation of PsADH1 expression and dependence on oxygen limitation.
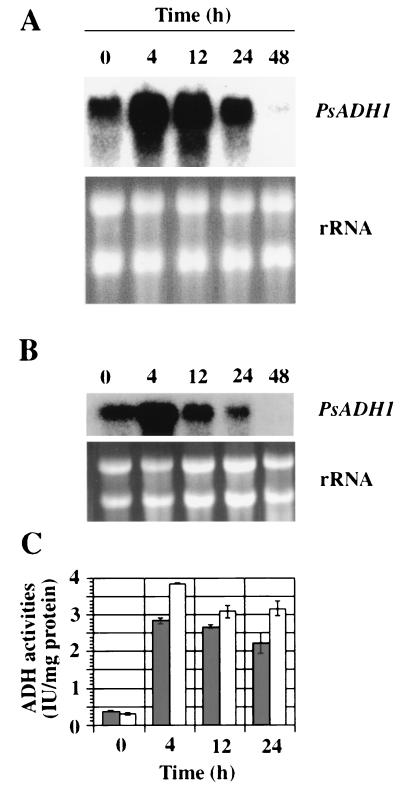
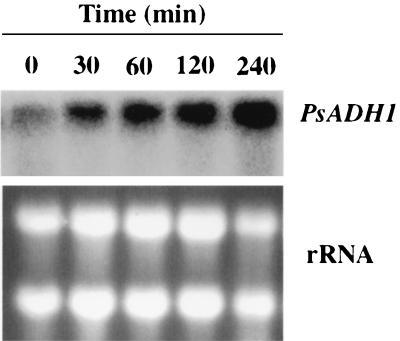
Total RNA was isolated from cells grown on xylose or glucose at various times following a shift from aerobic to oxygen-limited conditions. The levels of PsADH1 or PsADH2 mRNA relative to rRNAs were measured by quantitative Northern analyses with PsADH1- or PsADH2-specific oligonucleotides as probes. PsADH1 mRNA was induced following a shift to oxygen limitation (Fig. 2), which induced the ADH activity. No corresponding signals were detectable with the PsADH2 probe (data not shown). This difference is evidence that the induced ADH activity is PsADH1. No (or very low) PsADH activity was induced when we maintained cultures under fully aerobic conditions with either xylose or glucose as the carbon source. To maintain fully aerobic conditions, we kept cell densities low (less than 0.5 g/liter) and agitated the cells in baffled flasks at high speed (200 rpm). Even under such conditions, oxygen became limiting as cell densities increased, and ADH activity was induced. Small, but statistically significant, differences were noted in the titers of ADH activity in cells induced under oxygen limitation on xylose and glucose. Slightly higher levels of ADH activity were observed in cells grown on glucose (Fig. 2C). No statistically significant difference in the transcript levels could be observed with cells grown on these two carbon sources, but under both conditions, the transcript level of PsADH1 declined while ADH activity remained high. In a separate experiment, we studied the time course of induction of PsADH1 mRNA on xylose after shifting to oxygen limitation. The level of PsADH1 mRNA increased significantly within 30 min after shifting to oxygen limitation and continued to increase steadily up to 4 h (Fig. 3).
FIG. 2.
Induction of PsADH1 mRNA following a shift to oxygen-limited conditions. CBS 6054 cells were grown in either xylose (A) or glucose (B) under fully aerobic conditions and shifted to oxygen-limited conditions. Aliquots of cells were harvested at the indicated times, and total RNA was prepared for Northern analysis. Blots were probed with PsADH1. The positions of rRNA and PsADH1 mRNA are indicated. ADH activity (C) was also measured on samples prepared at the same time, as indicated. ■, xylose; □, glucose.
FIG. 3.
Kinetics of induction of PsADH1 mRNA level upon shift to oxygen-limited conditions. Cells were grown on xylose under aerobic conditions (repressed state) until they reached an OD600 of 1.0, collected by centrifugation, and shifted to oxygen-limited conditions by suspension in the same medium. Aliquots of cells were taken at the indicated times, and total RNA was prepared for Northern analysis. Blots were probed with PsADH1.
As is the case with PsADH1 mRNA, ADH-specific activities relative to cells grown on xylose or glucose under aerobic conditions were approximately 9- to 10-fold higher under oxygen-limited conditions than under fully aerobic conditions. However, the ADH-specific activity remained at induced levels, even after the PsADH1 mRNA level decreased (cf. Fig. 1 and 2).
We determined the level of β-galactosidase expressed from a PsADH1::lacZ fusion gene in order to compare the level of bona fide PsADH1 mRNA with the level of PsADH1 gene expression in cells grown on xylose following a shift from aerobic to oxygen-limited conditions. A fusion was constructed between the 598-bp PsADH1 promoter plus the coding region for the first 10 amino acids of PsADH1 and the coding region of the lacZ gene. The β-galactosidase activity reflects the transcriptional activity of the 598-bp PsADH1 promoter. The β-galactosidase activity was 2.5 ± 0.15 U under aerobic conditions and 25 ± 0.84 U under oxygen-limited conditions. Activity increased when the level of PsADH1 mRNA increased, suggesting that the oxygen-dependent regulation of PsADH1 involves control at the level of transcription. Because β-galactosidase is equally stable under aerobic and anaerobic conditions (18), the increased level of β-galactosidase following a shift to oxygen limitation reflects the transcriptional regulation of PsADH1.
Effect of heme addition on ADH1 expression in oxygen-limited cells.
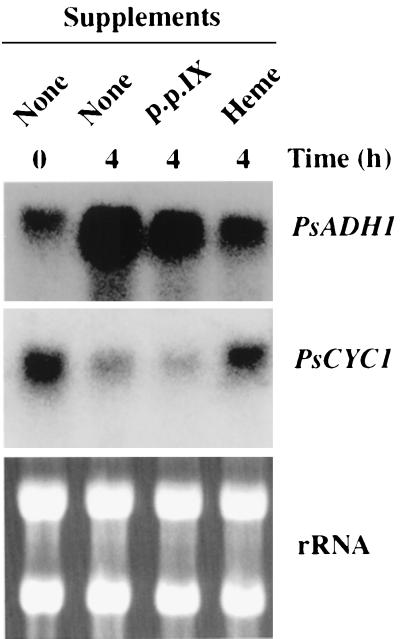
The regulation of PsADH1 transcription by oxygen could be direct or indirect. To determine whether heme affected the level of PsADH1 transcript, P. stipitis CBS 6054 was grown on xylose under fully aerobic conditions. Because the level of PsADH1 mRNA reached its maximum within 4 h after the shift to oxygen-limited conditions, we carried out a Northern blot analysis of the PsADH1 gene 4 h after adding heme and shifting the culture to oxygen-limited conditions. The transcription of PsADH1 was induced in untreated, oxygen-limited cells (Fig. 4, lane 2), but significantly less PsADH1 mRNA accumulated in oxygen-limited cells grown in the presence of heme (Fig. 4, lane 4). This result suggests that heme could be a negative coeffector of PsADH1 transcription. However, it is possible that heme depressed general mRNA synthesis. Therefore, as a control, we also analyzed the transcriptional level of PsCYC1, the P. stipitis gene that encodes cytochrome c, in the expectation that it would be induced under aerobic conditions (7, 17, 18, 20). The PsCYC1 transcript, which was repressed after the shift to the oxygen-limited conditions, was clearly present 4 h after the addition of heme (Fig. 4). This result showed that the addition of heme could maintain the transcription of PsCYC1 during this period and that heme was not toxic to the cells.
FIG. 4.
Effect of heme addition on PsADH1 and PsCYC1 expression in anaerobic cells. Cells were grown aerobically on xylose until they reached an OD600 of 1.0, collected by centrifugation, and shifted to oxygen-limited conditions by suspension in the same medium. After the cells had been shifted to oxygen limitation, heme (50 μg/ml) or protoporphyrin IX (p.p. IX [50 μg/ml]) was added as indicated, and growth was continued under oxygen-limited conditions for 4 h. The RNA blot was hybridized with PsADH1- or PsCYC1-specific oligonucleotides as probes.
A possible effect of heme precursors (protoporphyrin IX) was also tested because heme synthesis requires oxygen only at the penultimate step in the pathway (33), and several porphyrin precursors may occur in oxygen-limited cells and function as activators. Heme precursors (protoporphyrin IX) did not appear to function either as inhibitor or as activator. Therefore, it appears that heme is sufficient to inhibit transcription of PsADH1 under aerobic conditions.
Effect of PsADH1 disruption on PsADH2 expression.
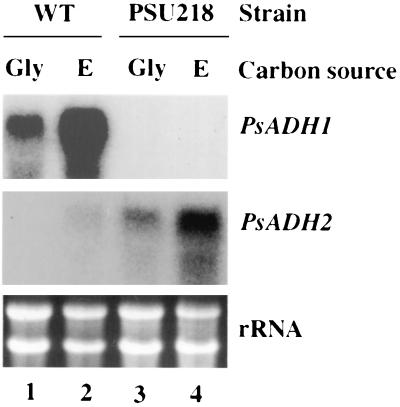
The PsADH2 gene was not, or at best was only poorly, expressed in the wild-type strain when it was grown under either fermentative or respirative conditions. To determine if a PsADH1 disruption affected the expression of PsADH2, we performed Northern analysis of PSU218, a strain in which Psadh1 is disrupted and in which xylose fermentation is impaired (9). The cells were grown on nonfermentable carbon sources under fully aerobic conditions to the early stationary phase and harvested for RNA preparation.
Disruption of PsADH1 caused a dramatic increase in PsADH2 expression in cells grown on a nonfermentable carbon source under aerobic conditions. PsADH2 was poorly expressed in wild-type yeast grown on either 2% ethanol or 3% glycerol (Fig. 5), but was expressed at high levels in PSU218 (psadh1/psadh1), suggesting that the expression of PsADH2 compensates—at least in part—for the missing PsADH1 activity under these conditions. PSU218 showed no apparent differences in its growth rate on nonfermentable carbon sources when compared with the parental strain (9).
FIG. 5.
Effect of PsADH1 disruption on expression of the PsADH2 gene during batch growth on nonfermentable carbon sources under fully aerobic conditions. Wild-type (WT) cells (lanes 1 and 2) and cells from PSU218 (lanes 3 and 4) carrying two disrupted Psadh1 alleles were grown in either glycerol (Gly [lanes 1 and 3]) or ethanol (E [lanes 2 and 4]) medium under aerobic conditions. Total RNA was extracted from these cultures for Northern analysis with PsADH1- or PsADH2-specific oligonucleotides as probes.
DISCUSSION
ADH activity is induced in P. stipitis under oxygen-limited conditions, and this activity corresponds to the product of PsADH1. The PsADH1 gene encodes a functional ADH protein and is inducible by oxygen limitation on xylose or glucose, and this induction is prevented by the addition of heme. This behavior markedly contrasts with that of the corresponding genes in S. cerevisiae (13) and Kluyveromyces lactis (30, 34, 37), but is itself not surprising; levels of various fermentative enzymes in P. stipitis were induced by growth on a fermentable carbon source under oxygen-limited conditions (31, 32, 38). Conditions that induce ADH activity also induce the transcription of PsADH1. Like the PsADH1 mRNA, ADH-specific activity relative to cells grown under aerobic conditions was approximately 10-fold higher under oxygen-limited conditions. These results are consistent with the hypothesis that the expression of PsADH1 is regulated at the transcriptional level. The ADH assay does not distinguish between PsADH1 and PsADH2, but because PsADH2 mRNA is not observed in the cells grown on xylose under oxygen-limited conditions, enzyme activity largely represents the translation product of PsADH1.
Regulation of PsADH1 transcript levels and of PsADH enzyme titers during oxygen-limited conditions is likely to be complex, and there is no a priori reason to expect them to change in parallel. As expected, the PsADH1 transcript levels rose initially, leading to enhanced synthesis of PsADH1 protein, which did not decline as rapidly as the corresponding transcript level. Presumably, this is because of its longer half-life or because of a delay between mRNA release and accumulation of active enzyme following protein synthesis. Note, however, that the existence of multiple, differentially expressed PsADH loci could account for the lack of an exact correlation between the PsADH1 mRNA level and ADH activities under the conditions employed. In our previously published studies, Southern blot analysis of P. stipitis genomic DNA revealed at least three loci with homology to S. cerevisiae ADH2 (9). Furthermore, the PsADH double disruptant, PLU1209, produces residual amount of ethanol from xylose under oxygen-limited conditions, raising the possibility that a third PsADH gene is expressed under these conditions.
The results observed through Northern analysis of PsADH1 expression agreed with the findings with the gene fusion construct. This similarity suggests that transcription plays an important role in regulating the expression of PsADH1. Therefore, PsADH1 expression appears to be responding to oxygen limitation, and it is regulated at the transcriptional level. However, it is not possible to rule out posttranscriptional mechanisms. It is not surprising to see the presence of PsADH1 mRNA in cells grown on glycerol under fully aerobic conditions, because the transcript level strongly increased in the presence of ethanol (Fig. 5), and PsADH1 has both fermentative and respirative functions (9).
Heme serves as the prosthetic group in oxygen-binding proteins such as catalases and cytochromes. Its function is intimately entwined with molecular oxygen, and its biosynthesis requires oxygen (49). Heme plays a regulatory role in many different processes in a wide variety of organisms, so it is not surprising that it serves as an intermediate in the signaling mechanism for oxygen levels in yeast cells. Because heme acts as a regulatory coeffector in the induction and repression of aerobic genes (45, 47), it is likely responsible for the simultaneous repression of the hypoxic gene PsADH1. A role for heme in transcriptional regulation of PsADH1 can be inferred from the observation that there were opposite concomitant changes in the level of PsCYC1 and PsADH1 transcripts when heme was added to the culture under oxygen-limited conditions.
Transcripts of PsADH2 were undetectable in cells grown on xylose under oxygen-limited conditions. Unexpectedly, the disruption of PsADH1 resulted in elevated expression of PsADH2 in mutant cells grown on either ethanol or glycerol. On the other hand, such an activation of PsADH2 expression did not seem to occur in cells grown on xylose under oxygen-limited conditions because the Psadh1 disruptant strain was not able to utilize xylose to produce ethanol or to contribute significantly to growth (9). Rather, the mutant produced large amounts of xylitol. At present, the role of PsADH2 and the basis for the physiological response of the cells are not completely understood. However, the mutational effect of Psadh1 on PsADH2 expression is reminiscent of feedback regulation of PDC expression in S. cerevisiae (19, 25, 36). In that instance, a signal for a PDC5 mRNA is only detectable in the pdc1 deletion mutant but not in the wild-type cells. Thus, in addition to oxygen-dependent regulation of PsADH1, expression of PsADH2 is subject to feedback regulation. The sharp increase of the PsADH2 transcript levels in the Psadh1 disruption mutant suggests that such regulation may occur at the transcriptional level. Alternatively, a posttranscriptional regulation similar to the mechanism described for the autoregulation of tubulin synthesis (11) may function.
Passoth et al. (32) recently reported cloning two genes for ADH from P. stipitis CBS 5774 by complementation of an S. cerevisiae Adh− mutant. The two genes that they described are virtually identical to the ones we reported earlier (9). However, Passoth et al. numbered them in the opposite order. PsADH1 in our nomenclature system codes for the principal ADH of P. stipitis. It is responsible for ethanol production under oxygen limitation. In this sense, it corresponds to the ADH1 of S. cerevisiae. Passoth et al. (32) also reported preliminary results showing that expression of only one of the two ADH genes could be detected under fermentative conditions. They detected expression of the gene corresponding to our PsADH2 only under aerobic conditions, and then only at a very low level. Our earlier investigations (9) showed that deletion of PsADH1 results in xylitol production under oxygen-limited conditions. Our present report shows that expression of PsADH2 increases significantly in the Δadh1 strain. We infer that PsADH1 has a higher affinity for NADH than PsADH2, but this hypothesis needs to be tested by assessing the biochemical properties of the native proteins.
Further investigations of the promoter region of PsADH1, along with site-specific base substitution and promoter reconstruction experiments, will help elucidate the mechanism of transcriptional regulation seen in P. stipitis. Because the physiological response of P. stipitis to oxygen has been well characterized in fermentation studies, the discovery that it has a system for regulating gene expression in response to oxygen will allow comparative studies of glycolytic regulation between P. stipitis and S. cerevisiae.
ACKNOWLEDGMENTS
We thank M. Culbertson for providing a pFusionator plasmid and B. Davis for cloning the PsCYC gene of P. stipitis.
This research was supported by National Renewable Energy Laboratory subcontract XAU-4-11193-02 and by USDA NRICGP grant no. 96-35500-3172.
REFERENCES
- 1.Andrews D L, Drew M C, Johnson J R, Cobb B G. The response of maize seedlings of different ages to hypoxic and anoxic stress. Plant Physiol. 1994;105:53–60. doi: 10.1104/pp.105.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey-Serres J, Freeling M. Hypoxic stress-induced changes in ribosomes of maize seedling roots. Plant Physiol. 1990;94:1237–1243. doi: 10.1104/pp.94.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennetzen J L, Hall B D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase I. J Biol Chem. 1982;257:3018–3025. [PubMed] [Google Scholar]
- 4.Bergmeyer H U. Methods of enzymatic analysis. 3rd ed. Vol. 11. Weinheim, Germany: Verlag Chemie; 1983. p. 139. [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Bruinenberg P M, de Bot P H M, van Dijken J P, Scheffers W A. NADH-linked aldose reductase: a key to anaerobic alcoholic fermentation of xylose by yeasts. Appl Microbiol Biotechnol. 1984;19:256–260. [Google Scholar]
- 7.Cerdan M E, Zitomer R S. Oxygen-dependent upstream activation sites of Saccharomyces cerevisiae cytochrome c genes are related forms of the same sequence. Mol Cell Biol. 1988;8:2275–2279. doi: 10.1128/mcb.8.6.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang C, Meyerowitz E M. Molecular cloning and DNA sequence of Arabidopsis thaliana alcohol dehydrogenase gene. Proc Natl Acad Sci USA. 1986;83:1408–1412. doi: 10.1073/pnas.83.5.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho J-Y, Jeffries T W. Pichia stipitis genes for alcohol dehydrogenase with fermentative and respirative functions. Appl Environ Microbiol. 1998;64:1350–1358. doi: 10.1128/aem.64.4.1350-1358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciriacy M. Genetics of alcohol dehydrogenase in Saccharomyces cerevisiae. Mutat Res. 1975;29:315–326. doi: 10.1007/BF02428119. [DOI] [PubMed] [Google Scholar]
- 11.Cleveland D W. Autoregulated instability of tubulin mRNAs: a novel eukaryotic regulatory mechanism. Trends Biochem Sci. 1988;13:339–343. doi: 10.1016/0968-0004(88)90103-x. [DOI] [PubMed] [Google Scholar]
- 12.Delgenes J P, Moletta R, Navarro J M. The effect of aeration on D-xylose fermentation by Pachysolen tannophilus, Pichia stipitis, Kluyveromyces marxianus and Candida shehatae. Biotechnol Lett. 1986;8:897–900. [Google Scholar]
- 13.Denis C L, Fergunson J, Young E T. mRNA levels for the fermentative alcohol dehydrogenase of Saccharomyces cerevisiae decrease upon growth on a nonfermentable carbon source. J Biol Chem. 1983;258:1165–1171. [PubMed] [Google Scholar]
- 14.Dewey N N, Conklin J L. Starch gel electrophoresis of lactic dehydrogenase from rat kidney. Proc Soc Exp Biol Med. 1960;105:492–494. doi: 10.3181/00379727-105-26153. [DOI] [PubMed] [Google Scholar]
- 15.du Preez J C. Process parameters and environmental factors affecting D-xylose fermentation by yeasts. Enzyme Microb Technol. 1994;16:944–956. [Google Scholar]
- 16.du Preez J C, van Driessel B, Prior B A. Effect of aerobiosis on fermentation and key enzyme levels during growth of Pichia stipitis, Candida shehatae and Candida tenuis on D-xylose. Arch Microbiol. 1989;152:143–147. [Google Scholar]
- 17.Freire-Picos M A, Hollenberg C P, Breunig K D, Cerdan M E. Regulation of cytochrome c expression in the aerobic respiratory yeast Kluyveromyces lactis. FEBS Lett. 1995;360:39–42. doi: 10.1016/0014-5793(95)00016-3. [DOI] [PubMed] [Google Scholar]
- 18.Guarente L, Mason T. Heme regulates transcription of the CYC1 gene of Saccharomyces cerevisiae via an upstream activation site. Cell. 1983;32:1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- 19.Hohmann S, Cederberg H. Autoregulation may control the expression of yeast pyruvate decarboxylase structural genes PDC1 and PDC5. Eur J Biochem. 1990;188:615–621. doi: 10.1111/j.1432-1033.1990.tb15442.x. [DOI] [PubMed] [Google Scholar]
- 20.Hörtner H G, Ammerer E, Hartter B, Hamilton J, Rytka T, Bilinski Ruis H. Regulation of synthesis of catalase and iso-1-cytochrome c in Saccharomyces cerevisiae by glucose, oxygen and heme. Eur J Biochem. 1982;128:179–184. doi: 10.1111/j.1432-1033.1982.tb06949.x. [DOI] [PubMed] [Google Scholar]
- 21.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeffries T W. Utilization of xylose by bacteria, yeasts, and fungi. Adv Biochem Eng Biotechnol. 1983;27:1–32. doi: 10.1007/BFb0009101. [DOI] [PubMed] [Google Scholar]
- 23.Kelly J M, Drysadale M R, Sealy-Lewis H M, Jones I G, Lockington R A. Alcohol dehydrogenase III in Aspergillus nidulans is anaerobically induced and post-transcriptionally regulated. Mol Gen Genet. 1990;222:323–328. doi: 10.1007/BF00633836. [DOI] [PubMed] [Google Scholar]
- 24.Lagunas R. Misconceptions about the energy metabolism of Saccharomyces cerevisiae. Yeast. 1986;2:221–228. doi: 10.1002/yea.320020403. [DOI] [PubMed] [Google Scholar]
- 25.Liesen T, Hollenberg C P, Heinisch J J. ERA, a novel cis-acting element required for autoregulation and ethanol repression of PDC1 transcription in Saccharomyces cerevisiae. Mol Microbiol. 1996;21:621–632. doi: 10.1111/j.1365-2958.1996.tb02570.x. [DOI] [PubMed] [Google Scholar]
- 26.Ligthelm M E, Prior B A, du Preez J C. The oxygen requirements of yeasts for the fermentation of D-xylose and D-glucose to ethanol. Appl Microbiol Biotechnol. 1988;28:63–68. [Google Scholar]
- 27.Lopes J M, Hirsch J P, Chorgo D A, Schulze K L, Henry S A. Analysis of sequences in the INO1 promoter that are involved in its regulation by phospholipid precursors. Nucleic Acids Res. 1991;19:1687–1693. doi: 10.1093/nar/19.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu P, Davis B P, Jeffries T W. Cloning and characterization of two pyruvate decarboxylase genes from Pichia stipitis CBS 6054. Appl Environ Microbiol. 1998;64:94–97. doi: 10.1128/aem.64.1.94-97.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu P, Hendrick J, Davis B P, Jeffries T W. Disruption of the β-isopropylmalate dehydrogenase gene LEU2 of Pichia stipitis with URA3 and recovery of the double auxotroph. Appl Microbiol Biotechnol. 1998;49:141–146. doi: 10.1007/s002530051150. [DOI] [PubMed] [Google Scholar]
- 30.Mazzoni C, Saliola M, Falcone C. Ethanol-induced and glucose-insensitive alcohol dehydrogenase activity in the yeast Kluyveromyces lactis. Mol Microbiol. 1992;6:2279–2286. doi: 10.1111/j.1365-2958.1992.tb01403.x. [DOI] [PubMed] [Google Scholar]
- 31.Passoth V, Zimmermann M, Klinner U. Peculiarities of the regulation of fermentation and respiration in the Crabtree-negative, xylose-fermenting yeast Pichia stipitis. Appl Biochem Biotechnol. 1996;57/58:201–212. doi: 10.1007/BF02941701. [DOI] [PubMed] [Google Scholar]
- 32.Passoth V, Schäfer B, Liebel B, Weierstall T, Klinner U. Molecular cloning of alcohol dehydrogenase genes of the yeast Pichia stipitis and identification of the fermentative ADH. Yeast. 1998;14:1311–1325. doi: 10.1002/(SICI)1097-0061(1998100)14:14<1311::AID-YEA315>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 33.Richter K, Ammerer G, Hartter E, Ruis H. The effect of δ-aminolevulinate on catalase T-messenger RNA levels in δ-aminolevulinate synthase-defective mutants of Saccharomyces cerevisiae. J Biol Chem. 1980;255:8019–8022. [PubMed] [Google Scholar]
- 34.Saliola M, Shuster J R, Falcone C. The alcohol dehydrogenase system in the yeast, Kluyveromyces lactis. Yeast. 1990;6:193–204. doi: 10.1002/yea.320060304. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Schaaff I, Green J B A, Gozalbo D, Hohmann S. A deletion of the PDC1 gene for pyruvate decarboxylase of yeast causes a different phenotype than previously isolated point mutations. Curr Genet. 1989;15:75–81. doi: 10.1007/BF00435452. [DOI] [PubMed] [Google Scholar]
- 37.Shain D H, Salvadore C, Denis C L. Evolution of the alcohol dehydrogenase ADH genes in yeast: characterization of a fourth ADH in Kluyveromyces lactis. Mol Gen Genet. 1992;232:479–488. doi: 10.1007/BF00266253. [DOI] [PubMed] [Google Scholar]
- 38.Skoog K, Hahn-Hägerdal B. Effect of oxygenation on xylose fermentation by Pichia stipitis. Appl Environ Microbiol. 1990;56:3389–3394. doi: 10.1128/aem.56.11.3389-3394.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skoog K, Jeppsson H, Hahn-Hägerdal B. The effect of oxygenation on glucose fermentation with Pichia stipitis. Appl Biochem Biotechnol. 1992;34/35:369–375. [Google Scholar]
- 40.Sliniger P J, Bothast R J, Okos M R, Ladisch M R. Comparative evaluation of ethanol production by xylose-fermenting yeasts presented high xylose concentrations. Biotechnol Lett. 1985;7:431–436. [Google Scholar]
- 41.Trick M, Dennis E S, Edwards K J R, Peacock W J. Molecular analysis of the alcohol dehydrogenase family of barley. Plant Mol Biol. 1988;211:147–160. doi: 10.1007/BF00015667. [DOI] [PubMed] [Google Scholar]
- 42.United States National Institutes of Health. 20 December 1996, posting date. [Online.] Image software V 1.61. http://rsb.info.nih.gov/nih-image/. [18 to 19 March 1999, last date accessed.]
- 43.Ursic D, DeMarini D J, Culbertson M R. Inactivation of the yeast Sen1 protein affects the localization of nucleolar proteins. Mol Gen Genet. 1995;249:571–584. doi: 10.1007/BF00418026. [DOI] [PubMed] [Google Scholar]
- 44.van Urk H, Voll W S L, Scheffers W A, van Dijken J P. Transient-state analysis of metabolic fluxes in Crabtree-positive and Crabtree-negative yeasts. Appl Environ Microbiol. 1990;56:281–287. doi: 10.1128/aem.56.1.281-287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verdiere J, Gaisne M, Labbe-Bois R. CYP1 HAP1 is a determinant effector of alternative expression of heme-dependent transcription in yeast. Mol Gen Genet. 1991;228:300–306. doi: 10.1007/BF00282480. [DOI] [PubMed] [Google Scholar]
- 46.Yang V W, Marks J A, Davis B P, Jeffries T W. High-efficiency transformation of Pichia stipitis based on its URA3 gene and a homologous autonomous replication sequence, ARS2. Appl Environ Microbiol. 1994;60:4245–4254. doi: 10.1128/aem.60.12.4245-4254.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zagorec M, Buhler J M, Treich I, Keng T, Guarente L, Labbe-Bois R. Isolation, sequence and regulation by oxygen of the yeast HEM13 gene coding for coproporphyrinogen oxidase. J Biol Chem. 1988;263:9718–9724. [PubMed] [Google Scholar]
- 48.Zagorec M, Labbe-Bois R. Negative control of yeast coproporphyrinogen oxidase synthesis by heme and oxygen. J Biol Chem. 1986;261:2506–2509. [PubMed] [Google Scholar]
- 49.Zitomer R S, Lowry C V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zitomer R S, Sellers J W, McCarter D W, Hastings G A, Wick P, Lowry C V. Elements involved in oxygen regulation of the Saccharomyces cerevisiae CYC7 gene. Mol Cell Biol. 1987;7:2212–2220. doi: 10.1128/mcb.7.6.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]