Abstract
Two genetic variants in strong linkage disequilibrium (rs9536314 and rs9527025) in the Klotho (KL) gene, encoding a transmembrane protein, implicated in longevity and associated with brain resilience during normal aging, were recently shown to be associated with Alzheimer disease (AD) risk in cognitively normal participants who are APOE ε4 carriers. Specifically, the participants heterozygous for this variant (KL-SVHET+) showed lower risk of developing AD. Furthermore, a neuroprotective effect of KL-VSHET+ has been suggested against amyloid burden for cognitively normal participants, potentially mediated via the regulation of redox pathways. However, inconsistent associations and a smaller sample size of existing studies pose significant hurdles in drawing definitive conclusions. Here, we performed a well-powered association analysis between KL-VSHET+ and five different AD endophenotypes; brain amyloidosis measured by positron emission tomography (PET) scans (n = 5,541) or cerebrospinal fluid Aβ42 levels (CSF; n = 5,093), as well as biomarkers associated with tau pathology: the CSF Tau (n = 5,127), phosphorylated Tau (pTau181; n = 4,778) and inflammation: CSF soluble triggering receptor expressed on myeloid cells 2 (sTREM2; n = 2,123) levels. Our results found nominally significant associations of KL-VSHET+ status with biomarkers for brain amyloidosis (e.g., CSF Aβ positivity; odds ratio [OR] = 0.67 [95% CI, 0.55–0.78], β = 0.72, p = 0.007) and tau pathology (e.g., biomarker positivity for CSF Tau; OR = 0.39 [95% CI, 0.19–0.77], β = -0.94, p = 0.007, and pTau; OR = 0.50 [95% CI, 0.27–0.96], β = -0.68, p = 0.04) in cognitively normal participants, 60–80 years old, who are APOE e4-carriers. Our work supports previous findings, suggesting that the KL-VSHET+ on an APOE ε4 genotype background may modulate Aβ and tau pathology, thereby lowering the intensity of neurodegeneration and incidence of cognitive decline in older controls susceptible to AD.
Introduction
Alzheimer disease (AD), the most common form of dementia, affects about 30% of those aged over 85 years [1]. AD is classified as a neurodegenerative disease, affecting brain integrity and function, eventually resulting in progressive deterioration of cognitive capabilities [2]. Besides aging, a strong genetic risk factor for developing AD is the epsilon 4 allele of the apolipoprotein E gene (APOE ε4) [3, 4]. As such, participants carrying one or two APOE ε4 alleles are significantly overrepresented among persons diagnosed with AD, in comparison to non-carriers [5, 6]. This particular genetic variant has been shown to be associated with cognitive decline [7] and reduced mean age at onset even within families with late onset AD [8]. Further, APOE ε4 homozygosity among cognitively normal participants is associated with earlier and more abundant Aβ deposition [9–12], earlier pre-clinical memory decline [13], and an increased incidence of conversion to dementia [9], in comparison to APOE ε4 heterozygotes and noncarriers. These observations suggest that APOE ε4 influences the very core of AD pathophysiology, in large part due to its key role in fostering cerebral Aβ pathology. Therefore, identifying genetic factors that interact with APOE ε4 genotype to reduce Aβ burden and, eventually, a participant’s risk for developing AD, may inspire novel strategies for preventing or halting the progression of AD and reveal novel targets for effective therapeutic interventions.
One such genetic factor that has been recently reported and showed a protective effect among cognitively normal APOE ε4 carriers is polymorphism on Klotho (KL) gene [6, 14, 15]. KL is a transmembrane protein that is cleaved by α- and β-secretases and shed into CSF and plasma, where it acts as a signaling molecule and longevity factor [16, 17] that promotes neuronal functions and brain resilience during aging [18–20]. In humans, two KL gene variants (rs9536314 for p.F352V and rs9527025 for p.C370S) exist in strong linkage disequilibrium and segregate together as a functional haplotype called KL-VS. Interestingly, heterozygosity for the KL-VS haplotype (KL-VSHET+) has been associated with higher serum concentrations of KL [18, 21] which, in turn, has been reported to promote healthy brain aging and protect synaptic functions in comparison to participants who carry two copies of the KL-VS haplotype (KL-VSHET-) [20, 22]. Even though KL-VSHET+ is associated with better cognitive health and longevity among those aging normally, there exist no clear indication of its involvement in protection against aging-associated neurodegenerative disorders, such as AD.
Identification of genetic risk factors for AD based on clinical diagnosis poses several challenges. A clinical diagnosis of AD relies in part on evidence of cognitive decline using standard cognitive tests that might be influenced by factors unrelated to disease (e.g., anxiety, education, and general test-taking ability of the participant [23]). In addition, other salient factors that can play an important role include variability in the cognitive measures [24, 25], over-reliance on normative cut-off scores to diagnose dementia, and practice effect [24, 26]. A complementary approach to classical case-control studies involves the use of more robust and stable measures (endophenotypes) that support a diagnosis of AD, such as cerebrospinal fluid (CSF) biomarkers and Aβ burden assessed by positron emission tomography (PET). This approach can increase statistical power to identify AD genetic risk factors, discover novel associations, and understand their impact on the brain [27]. For example, by using brain endophenotypes, researchers have identified novel protective genetic variants in TMEM106B and MS4A genes, associated with neuroprotection (high neuronal proportion) in AD [28], and increased CSF soluble triggering receptor expressed on myeloid cells 2 (sTREM2) concentrations with reduced AD risk, respectively [29].
In the spectrum of AD pathology and different genetic factors that exert a protective effect in the context of disease onset and/or progression, KL-VS appears to be a compelling candidate due to its implication in promoting longevity and cognitive resilience during aging [18, 20, 22]. Interestingly, two recent studies evaluating the protective effect of KL-VSHET+ against AD pathology in cognitively normal participants [15, 30] provided contradictory evidence. The first study [15] focused on 309 late-middle-aged adults (mean age 61 years) and found KL-VSHET+ to be associated with reduced Aβ aggregation, suggesting its protective effect against APOE ε4-linked pathways to disease onset in AD. The second study [30] analyzed data from 581 adults (mean age 71 years) and found no significant associations between KL-VSHET+ and cognitive decline, independent of the APOE ε4 genotype, suggesting no modifying effect of KL-VSHET+ on Aβ aggregation and APOE ε4-driven cognitive decline in preclinical AD. Furthermore, a study assessing the association between Klotho KL-VS haplotype and cognition using data from the population-based Heinz Nixdorf Recall Study in 1812 subjects (55–87 years) suggested a slightly lower cognitive performance in KL-VSHET+ subjects [31]. However, a recent large-scale meta-analysis [6] that focused on cognitively normal participants in the age range of 60–80 years revealed a 30% reduction in AD risk for participants who are APOE ε4 carriers and KL-VSHET+. They also observed a significant associations between KL-VSHET+ and higher Aβ42 in CSF (p = 0.03) and between KL-VSHET+ and lower Aβ on PET scans (p = 0.04), modulated by APOE ε4 status. Focusing on tau pathology, a recent study showed KL-VSHET+ to be associated with both lower levels and slower rate of change in amyloid-related increase of tau-PET accumulation in asymptomatic and symptomatic elderly participants, supporting a protective effect of KL-VSHET+ on the primary AD pathologies [32]. Due to contradictory outcomes from the existing reports and their relatively small sample sizes, we performed a systematic evaluation of association between KL-VSHET+ and multiple well-established AD endophenotypes to evaluate whether it has a protective effect on AD pathology in asymptomatic elderly participants. Here, we have performed a well-powered association analysis between KL-VSHET+ and five different AD endophenotypes: Aβ assessed by PET scans (n = 5,541) and CSF (n = 5,093), as well as the CSF Tau (n = 5,127), phosphorylated Tau (pTau181; n = 4,778) and sTREM2 (n = 2,123). In line with previous studies, we performed APOE ε4- and age-stratified (60–80 years) analyses to determine if there is any association between KL-VSHET+ and Aβ aggregation that is modulated by APOE ε4 status. In addition, we also evaluated if there is any association between KL-VSHET+ and other AD endophenotypes that include Tau, pTau, and sTREM2 measured by CSF. Briefly, in the case of APOE ε4-carriers, we found significant associations between KL-VSHET+ and biomarkers for brain amyloidosis (CSF Aβ42; p = 0.007) and tau pathology (CSF Tau; p = 0.007, and pTau; p = 0.04). As evident from the observed P-values, the detected associations are nominally significant and would likely fail multiple test correction, indicating the need for validating these findings in studies with even larger sample sizes before drawing definitive conclusions. Nevertheless, these findings suggest that KL-VSHET+ exerts an APOE ε4-genotype dependent protective effect on CSF Tau and pTau concentrations and on subsequent cognitive decline in older cognitively normal participants susceptible to AD.
Methods
Study samples and phenotype processing
Written consent was obtained for all participants. This study was approved the Washington University Human study committee. IRB approval #: 201109148.
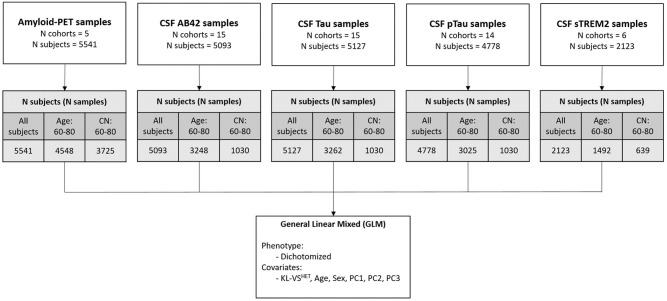
For this study, we collected data from 17 different AD-related cohorts. Participants were enrolled in the Memory and Aging Project (MAP) at the Knight Alzheimer’s Disease Research Center (Knight-ADRC), Alzheimer’s Disease Neuroimaging Initiative (ADNI, adni.loni.usc.edu), BIOCARD, the Dominantly Inherited Alzheimer Network (DIAN), HB, Hospital Sant Pau (Lleo), London, MOLI, Pau, Mayo Clinic (Mayo), SWEDEN, UPENN, UW, Parkinson’s Progression Markers Initiative (PPMI), Anti-Amyloid Treatment in Asymptomatic Alzheimer’s Disease (A4), and ADNI Department of Defense (ADNIDOD) studies. Total sample size was 9,526 (S1 Table in S2 File). Collection of genotype data, PET image processing, and CSF data processing for each cohort are described in detail in the respective studies [10, 11, 28, 29, 33, 34]. We analyzed the association between KL-VSHET+ and five different AD endophenotypes (Table 1) that served as biomarkers for brain amyloidosis (Aβ pathology assessed by amyloid-PET [n = 5,541] and Aβ42 measured from CSF [n = 5,093]), tau pathology (Tau [n = 5,127] and pTau181 [n = 4,778] from CSF), and inflammation (sTREM2 levels from CSF [n = 2,123]). A schematic overview of the analyses conducted and the datasets used is provided in Fig 1.
Table 1. Demographics of analyzed Alzheimer’s disease (AD) endophenotypes.
| Total | Amyloid-PET | Aβ42 | Tau | pTau181 | sTREM2 | |
|---|---|---|---|---|---|---|
| Sample size | 9,526 | 5,541 | 5,093 | 5,127 | 4,778 | 2,123 |
| Female (%) | 51.86 | 54.54 | 49.30 | 49.19 | 48.68 | 50.31 |
| Age, mean (SD) | 68.93 (11.13) | 69.53 (10.73) | 67.04 (13.27) | 67.15 (13.30) | 66.93 (13.43) | 68.17 (12.13) |
| APOE ε4+ (%) | 39.03 | 37.39 | 40.74 | 41.08 | 39.47 | 42.11 |
| Biomarker, mean (SD) | 0.028 (0.02) | 0.045 (1.03) | -0.0018 (1) | 0.027 (1) | 0.02 (1) | 0.05 (0.98) |
| Klotho-VSHET+ (%) | 25.76 | 25.99 | 25.31 | 25.16 | 25.09 | 25.20 |
| Cases | 3,109 | 1,090 | 2,424 | 2,443 | 2,297 | 1,074 |
| Controls | 5,286 | 4,117 | 1,584 | 1,589 | 1,582 | 879 |
Demographics of participants at the time of amyloid PET imaging and CSF sampling. This table summarizes basic demographic information of participants included in the analysis. For each modality, we report percentage of females, mean age of the participants and standard deviation (SD) in the age, percentage of APOE ε4-carriers (APOE ε4+) participants, mean value of the endophenotypic biomarker and its SD, percentage of KL-VS heterozygous (KL-VSHET+) participants, and number of cases and controls. Samples with missing case/controls status were also considered in the ‘all participants’ analysis. To normalize endophenotypes across different cohorts, we converted different amyloid imaging measures (e.g., Centiloid, PiB, and AV45) into log-normalized z-score using “scale” function in base R. Phenotype from each cohort was normalized individually to account for within cohort variation. These AD endophenotypes are used for checking their association with KL-VSHET+. Abbreviations: PET, positron emission tomography; Aβ, β-amyloid; pTau, phosphorylated tau; soluble triggering receptor expressed on myeloid cells 2, sTREM2; sd, standard deviation; KL-VS, Klotho-VS; Het+, heterozygosity.
Fig 1. Schematic overview of datasets and performed analysis.
Number of participants in each modality were stratified into three categories: 1) All of the participants; 2) Age: 60–80, participants aged 60 to 80 years; 3) CN: 60–80, cognitively normal participants aged 60 to 80 years. Association between KL-VSHET and endophenotypes were assessed using generalized linear mixed (logistic regression) model for dichotomized phenotype. Age, sex, and first three genetic PCs were used as covariates in an APOE ε4-stratified analysis. Abbreviations: PET, positron emission tomography; N, number of; CSF, cerebrospinal fluid; Aβ, β-amyloid; pTau, phosphorylated tau181; soluble triggering receptor expressed on myeloid cells 2, sTREM2; CN, cognitively normal; KL, Klotho; Het, heterozygous; PC, principal component.
Briefly, participants were diagnosed as cognitively normal (controls) or AD (cases), based on the clinical dementia rating (CDR) that was available for 88% of the total dataset. The CDR is a five-point scaling system that describes the overall dementia severity for each participant (no dementia = 0, very mild = 0.5, mild = 1, moderate = 2, and severe = 3). Participants with CDR = 0 were categorized as controls, and those with CDR > 0 were defined as cases. Any participant who was missing information about age, sex, KL-VSHET+, APOE ε4 genotype, or genetic principal components (PCs) was excluded from the analysis. Following this rationale, we considered 3,725 cognitively normal participants assessed by amyloid-PET and 1,030 cognitive normal participants measured by CSF (Aβ42, Tau, and pTau) and 639 participants with CSF sTREM2 levels. Similarly, the number of clinically defined AD participants assessed from amyloid-PET, CSF Aβ42, Tau, pTau, and sTREM2 were 1,090, 2,424, 2,443, 2,297, and 1,074, respectively.
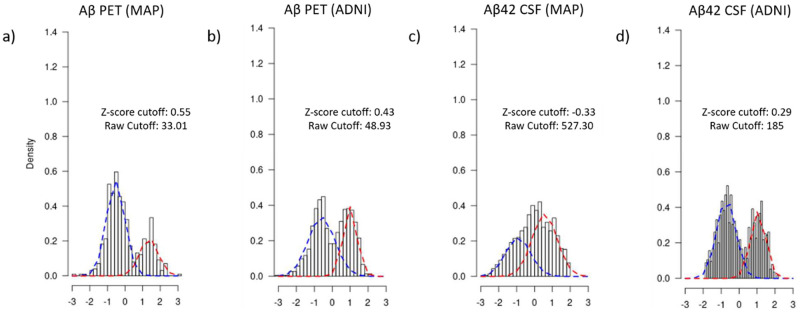
For each cohort, amyloid PET images were normalized to their reference cerebellar regions to obtain standardized uptake value ratios (SUVR) in a composite of cortical brain areas. The normalized z-scores were calculated for each endophenotype using the mean and standard deviation (SD) units across each cohort and applied to the entire endophenotype. These normalized z-scores were used for dichotomizing each endophenotype into biomarker positive (case) and negative (control), as previously described [11]. Briefly, defining biomarker positivity and negativity requires the selection of a cut-point. We and others [11, 35, 36] have demonstrated that it is possible to use Gaussian mixture model (GMM) to statistically infer that cut-off. We overlapped the distributions of quantitative z-scores from cases and controls for each endophenotype and employed a GMM that relies on hierarchical model-based agglomerative clustering to get votes for defining a cut-point for dichotomization. We used Mclust function from “mclust” R package (version 5.4.6) for dichotomizing all quantitative endophenotypes separately. The empirical dichotomization cutoffs obtained using this approach appeared consistent with existing literature. For instance, our model suggested the cut-point of 527 pg/mL for Aβ42 from CSF in MAP cohort which is between 500 pg/mL [37] to 518 ng/l [35] depending on the study. A density plot defining the dichotomization cutoffs for Aβ assessed by PET scan and Aβ42 from CSF in MAP and ADNI cohorts is shown in Fig 2. Further details about the empirical cutoffs derived from z-scores and their corresponding raw values are provided in S2 Table in S2 File. The dichotomized (biomarker positive/negative) endophenotypic status was used as a response variable to assess its association with Klotho heterozygosity.
Fig 2. Cutoffs for dichotomizing different AD endophenotypes across MAP and ADNI cohorts.
A density plot defining the dichotomization cutoffs for Aβ assessed by PET scan and Aβ42 from CSF in MAP and ADNI cohorts. The distribution of z-score for cases and controls is shown by red and blue dotted lines, respectively. The cut-off point where both these distributions overlap was selected as the dichotomization threshold for each endophenotype. The dichotomization was performed on normalized z-scores for each endophenotype. However, the corresponding raw score for each dichotomization threshold is also labelled in the plot. Abbreviations: Aβ, β-amyloid; PET, positron emission tomography; MAP, Memory and Aging Project; ADNI, Alzheimer’s Disease Neuroimaging Initiative; CSF; Cerebrospinal fluid.
Genotyping, quality checks, imputation, and population structure
We applied stringent quality control (QC) steps to process the genotyping array and sequencing data. We used the threshold of 98% for removing single nucleotide polymorphisms (SNPs) and participants with low call rate. Autosomal SNPs that were not in the Hardy-Weinberg equilibrium (P < 1×10−6) were also removed. Duplication and relatedness of participants were estimated from identity-by-descent (IBD) analysis carried out in Plink version 1.9 [38]. In case of related participants (Pihat ≥0.25), the samples from MAP or with a higher number of variants that passed the QC were prioritized. For phasing and imputation, we used The 1000 Genomes Project Phase 3 data (October 2014), SHAPEIT v2.r837 [39], and IMPUTE2 v2.3.2 [40]. We used imputed probability score < 0.90 and ≥0.90 as thresholds for missing and fully observed participant genotypes, respectively. Genotyped and imputed variants with MAF < 0.02 or IMPUTE2 information score < 0.30 were discarded. Principle component analysis (PCA) was performed on the genotype data to obtain genetic PCs that capture population substructure (S2 Fig in S1 File). To obtain the largest and most homogeneous pool of population, only European participants were considered (S3 Fig in S1 File) for the subsequent statistical analyses.
Statistical analyses
Statistical analyses and data visualization were performed in Plink version 1.9 [38] and R version 3.5.2 [41]. We performed association analyses of KL-VSHET+ status with different AD endophenotypes from PET scan (Aβ) and CSF (Aβ42, Tau, pTau, and TREM2). The associations between the biomarker positivity and KL-VSHET+ were tested using logistic regression model. The implementation of regression model from the base R [41] “stats” package was used for the evaluation of association and the outcomes measurements were adjusted for sex, age, and first three genetic PCs. For the Aβ levels measured by PET scan, we considered the age at scan and in the case CSF biomarker levels, age at lumbar puncture. Furthermore, associations were evaluated across three different strata: (1) all participants (AD and controls); (2) those aged 60 to 80 years (AD and controls); and (3) only cognitively normal participants aged 60 to 80 years. All association analyses were stratified by APOE ε4 status: APOE ε4 carriers (APOE- 24, 34, and 44) and APOE ε4 non-carriers (APOE- 22, 32, and 33). Associations were deemed significant at a threshold of p < 0.05.
Results
The primary aim of this study was to determine whether there is a significant association between KL-VSHET+ and AD endophenotypes in cognitively normal participants who are APOE ε4-carriers. In addition, we also assessed the KL-VS effects in participants with AD dementia. To accomplish this goal, we analyzed genetic data and five different AD endophenotypes from 9,526 participants obtained through 17 different AD-related cohorts. In line with existing studies [6, 15], our findings validate the protective effect of KL-VSHET+ against AD in cognitively normal participants who are APOE ε4 carriers.
Association between KL-VSHET+ status and brain amyloidosis measured by PET scan and CSF
We evaluated the association of KL-VSHET+ status with Aβ pathology as measured by amyloid-PET and CSF Aβ42 for 5,541 and 5,093 participants, respectively (Table 1). Although associations were evaluated for three different age ranges (S3 Table in S2 File), our main focus was cognitively normal participants who are 60 to 80 years old (Table 2). We focused on this age range to be consistent with existing studies that report a pronounced effect of APOE ε4 positivity on AD dementia risk between age 60 to 80 years in comparison to older participants (≥80 years) [6, 42]. For Aβ levels measured by PET, we observed that KL-VSHET+ status was more prevalent in Aβ biomarker negative participants in comparison to the positive ones, but there was no significant association within any age group or APOE ε4 strata (S3 Table in S2 File). On the other hand, we found a significant association between KL-VSHET+ status and CSF Aβ biomarker positivity in cognitively normal APOE ε4-carrier participants who are 60 to 80 years old (Table 2; OR = 0.67 [95% CI, 0.55–0.78], β = 0.72, p = 0.007). We also observed a significant association for APOE ε4 non-carriers; however, the effect size and the strength of the association was lower in this group (OR = 0.61 [95% CI, 0.51–0.70], β = 0.46, p = 0.03) than the APOE ε4-carriers (Table 2). Taken together, we were able to replicate the previously reported associative findings [6, 42] between increased Aβ42 CSF levels and KL-VSHET+ in a larger sample group (S1 Fig in S1 File).
Table 2. Genetic association of KL-VSHET+ with AD endophenotypes in cognitively normal participants aged 60–80 years, stratified by APOE ε4 status.
| Modality | Group | CN participants | Odds ratio | Estimate | P value |
|---|---|---|---|---|---|
| (KL-VSHET+ %) | |||||
| Amyloid PET | APOE4+ | 1328 (27.5) | 0.94 | -0.07 | 0.61 |
| APOE4- | 2397 (26.5) | 0.99 | -0.01 | 0.90 | |
| Aβ42 | APOE4+ | 308 (26.3) | 0.67 | 0.72 | 0.007 |
| APOE4- | 722 (26.3) | 0.61 | 0.46 | 0.03 | |
| Tau | APOE4+ | 308 (26.9) | 0.39 | -0.94 | 0.007 |
| APOE4- | 722 (26.5) | 0.85 | -0.16 | 0.49 | |
| pTau | APOE4+ | 308 (26.9) | 0.50 | -0.68 | 0.04 |
| APOE4- | 722 (26.3) | 0.89 | -0.11 | 0.61 | |
| sTREM2 | APOE4+ | 199 (31.2) | 1.08 | 0.08 | 0.80 |
| APOE4- | 440 (25.2) | 1.20 | 0.18 | 0.43 |
Association between KL-VSHET+ and different dichotomized AD endophenotypes were assessed using logistic regression model. We used dichotomized endophenotype as the response variable, whereas, age, sex, and first three genetic PCs were used as covariates in an APOE4-stratified analysis. Significant associations are represented by bold P-values. Abbreviations: KL-VSHET+, Klotho-VS heterozygous; CN, cognitively normal; AD, Alzheimer’s disease; Std. Error, Standard error; %, percentage; Aβ, β-amyloid; pTau, phosphorylated tau181; soluble triggering receptor expressed on myeloid cells 2, sTREM2.
APOE ε4-related alteration in tau pathology varies by KL-VSHET+ status
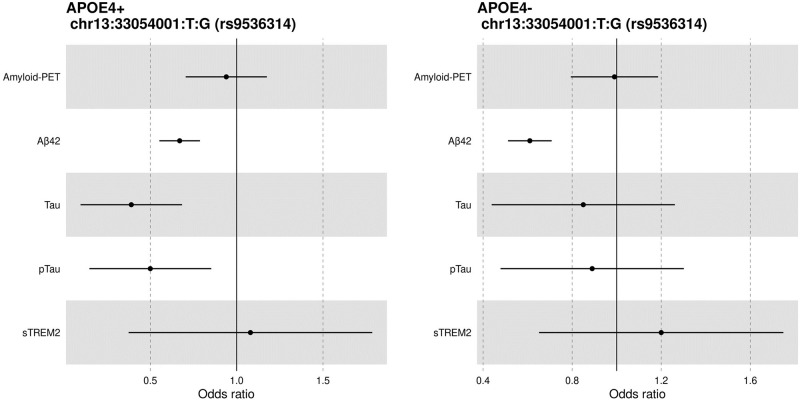
Similar to Aβ association analyses, we also determined if there was an age- and APOE ε4-dependent association of KL-VSHET+ status with dichotomized CSF Tau and pTau biomarker positivity. In the age range of 60 to 80 years, we observed significant association between KL-VSHET+ status and CSF Tau (OR = 0.39 [95% CI, 0.20–0.77], p = 0.007), and pTau (OR = 0.50 [95% CI, 0.27–0.96], p = 0.04) levels in elderly (60–80 years) cognitively normal APOE ε4-carrier participants (Table 2). In both cases, the KL-VSHET+ was associated with biomarker negative status (e.g., lower CSF Tau or pTau levels which are associated with lower AD risk (β = -0.94 and -0.68 for Tau and pTau, respectively). Regarding the effect size, we observed an almost 6-fold decrease for both Tau and pTau levels in APOE ε4-carriers as compared to the non-carriers, suggestive of a more pronounced protective effect for participants carrying one copy of KL-VS haplotype. As expected, a consistent negative association was observed for both modalities when effect sizes were represented in the form of a Forest plot (Fig 3). While we observed similar trends in the effect of KL-VSHET+ on tau pathology biomarker negativity for cognitively normal participants who are APOE ε4 non-carriers (β = -0.16 and -0.11 for Tau and pTau, respectively), the association was not deemed significant (p = 0.49 and 0.61 for Tau and pTau, respectively).
Fig 3. Forest plot of odds ratio (OR) for KL-VSHET+ association with dichotomized AD endophenotypes in 60–80 year cognitively normal participants, stratified by APOE ε4 status.
A significant association was detected between KL-VSHET+ and dichotomized Aβ, Tau, and pTau CSF levels. In case of Aβ, the associations were deemed significant across both APOE ε4 strata, whereas for Tau and pTau, associations were observed only in APOE ε4-carriers, representing an exclusive protective effect of KL-VSHET+ for the cognitively normal participants aged 60 to 80 years and carrying APOE ε4. Abbreviations: APOE4+, Apolipoprotein E4 positive; APOE4-, Apolipoprotein E4 negative; PET, positron emission tomography; Aβ, β-amyloid; pTau, phosphorylated tau181; soluble triggering receptor expressed on myeloid cells 2, sTREM2.
To further elucidate the link between APOE ε4 and KL-VSHET+, we performed the association analysis including an interaction effect for the presence or absence of APOE ε4 and the status of each biomarker (positive or negative). Results did not showed any significant association across any endophenotype (S4 Table in S2 File), emphasizing the APOE ε4-dependent association of KL-VSHET+ and different AD endophenotypes. In addition, we also assessed associations between APOE ε4 and AD pathologies in the group with 0 and 2 copies of KL-VS haplotype (KL-VSHET-). We observed no significant association across any AD endophenotypes (S5 Table in S2 File), confirming the existing hypothesis that suggests the neuroprotective effect of KL heterozygosity (KL-VSHET+) but not the homozygosity (KL-VSHET-) [20, 22].
Direct effect of KL-VSHET+ status on inflammation-specific biomarker
For sTREM2 CSF levels, we did not observe any significant association between this inflammation biomarker and KL-VSHET+ status across any participant stratification, regardless of the age group and APOE ε4 status. Nevertheless, for cognitively normal participants aged 60 to 80 years, we observed that KL-VSHET+ status is associated with increased sTREM2 CSF levels, which represents the positivity of inflammation-specific biomarker, but association was not deemed significant in APOE ε4-carriers (OR = 1.08 [95% CI, 0.58–2], β = 0.08, p = 0.80) as well as non-carriers (OR = 1.20 [95% CI, 0.77–1.86], β = 0.18, p = 0.43).
Sensitivity analysis result: Robustness of the associations between KL-VSHET+ status and brain amyloidosis to the APOE ε4 status, age, and sex
In order to estimate whether the associations between KL-VSHET+ and amyloidosis were confounded by uneven sample size of APOE ε4-carriers and non-carriers as well as by differences in the sex and age of the participant groups, the association analyses were repeated with equal numbers of APOE ε4-carriers (N = 308) and non-carriers (N = 308) matched for age and sex. As in the full-sample analyses for cognitively normal participants aged 60 to 80, these smaller, balanced analyses revealed that KL-VSHET+ was consistently associated with CSF Aβ biomarker positivity among APOE ε4-carriers (OR = 0.68 [95% CI, 0.56–0.78], β = 0.75, p = 0.005; S6 Table in S2 File) and among APOE ε4 non-carriers (OR = 0.69 [95% CI, 0.51–0.79], β = 0.70, p = 0.034). Likewise, similar trends were observed between the full-sample and smaller, balanced analyses for amyloid imaging, CSF Tau, pTau181, and sTREM2 (S6 Table in S2 File). In these corrections for class imbalance of APOE ε4-carriers and non-carriers, as well as for males and females of same age, the direction of effect remains the same for each endophenotype, and the strength of association becomes more profound (lower p-values). Taken together, these results suggest that observed associations between KL heterozygous cognitively normal participants and different AD endophenotypes (e.g. tau and pTau) are independent of the distribution of APOE ε4 carriage status, age, and sex of participants.
Cognitively normal participants (aged 60 to 80) drive association between KL-VSHET + status and AD endophenotypes
We also conducted APOE ε4-stratified association analyses between KL-VSHET+ status and biomarkers for brain amyloidosis (Aβ from PET and CSF), tau-related pathology (CSF Tau and pTau), and inflammation (CSF sTREM2) for all participants, regardless of their age and case-control status. In these larger inclusive analyses, no significant association was observed between KL-VSHET+ and any of the five endophenotypes (S3 Table in S2 File). Similarly, we also performed the same analyses but restricting to participants between 60 and 80 years of age, regardless of their case-control status; even in that case, no significant association was detected across any endophenotype (S3 Table in S2 File). Only when participants were restricted to the age range 60–80 and cognitive normalcy were significant association detected between KL-VSHET+ status and CSF Aβ, Tau, and pTau levels (S3 Table in S2 File). Notably, these findings suggest that the nearly-significant associations observed in the more inclusive analyses (e.g. for Aβ42) were mainly driven by the cognitively normal participants who are 60 to 80 years old.
Discussion
The role of Klotho protein as a longevity factor is widely recognized [16, 17]. There has been an increasing amount of evidence supporting the relationship between KL-VSHET+ and preserved brain integrity and cognitive performance during normal aging [18–20]. In this study, we examined the association of KL-VSHET+ status with five different AD-related endophenotypes that serve as biomarkers for brain amyloidosis (Aβ levels measured from CSF and amyloid PET), tau pathology (CSF Tau and pTau), and inflammation (sTREM2). To our knowledge, we have analyzed the largest sample size of AD endophenotypic data for evaluating its association with KL-VSHET+; this approach is instrumental to discern the potential protective effect of this heterozygous genetic variant for AD in cognitively normal APOE ε4-carriers. Our results showed that KL-VSHET+ status was associated with CSF Aβ42, Tau and pTau biomarker negativity in participants who are cognitively normal APOE ε4-carriers within an age range of 60 to 80 years. This finding suggests that KL-VSHET+ status reduces the risk of subsequent AD dementia among APOE ε4-carriers by lowering the AD pathology burden [6, 42].
We were able to replicate the findings by Belloy et al. [6]; that is, KL-VSHET+ status was significantly associated with increased CSF Aβ42 levels (Aβ biomarker positivity) for cognitively normal participants aged 60 to 80 years who are APOE ε4-carriers (OR = 0.67 [95% CI, 0.55–0.78], β = 0.72, p = 0.007). Further, our analyses also found this association to be significant among APOE ε4 non-carriers (OR = 0.61 [95% CI, 0.51–0.70], β = 0.46, p = 0.03), with similar (overlapping 95% CI) effect sizes (Table 2). Although no significant association was observed with amyloid PET, the detected trend towards a negative association suggests that KL-VSHET+ may protect against AD by reducing deposits of Aβ that are capable of binding amyloid PET tracers. Consistently, studies have shown a very high concordance between CSF Aβ42 and amyloid PET [43], but with a proportion of individuals with discordant results (CSF+/PET-); such individuals may represent the earliest stages of AD neuropathologic change, when low CSF Aβ levels appear to coincide with early amyloid deposition, but amyloid deposits have not yet accrued sufficiently to reach threshold for amyloid-PET tracer binding, and neurodegeneration has not yet begun [44, 45]. In support of this interpretation, a previous study investigating longitudinal differences in cognition between participants without dementia with different CSF and PET profiles found no memory decline in concordant-negative (CSF−/PET−) and discordant (CSF+/PET−) groups, in contrast to the concordant-positive (CSF+/PET+) group that deteriorated over time [46]. Furthermore, Palmqvist et al. [44] reported similar results, when they analyzed 437 non-demented participants from ADNI whose results from amyloid PET scans and CSF Aβ measurements showed that CSF Aβ levels become abnormal in the earliest stages of AD, before amyloid PET and before neurodegeneration starts.
We also investigated whether KL-VSHET+ status is significantly associated with Tau and pTau levels in CSF. We found that KL-VSHET+ status was significantly associated with decreased levels of CSF Tau (OR = 0.39 [95% CI, 0.20–0.77], β = -0.94, p = 0.007) and pTau (OR = 0.50 [95% CI, 0.27–0.96], β = -0.68, p = 0.04) i.e., CSF Tau and pTau biomarker negativity, in participants who are APOE ε4-carriers and 60 to 80 years old. Interestingly, APOE ε4 non-carriers showed similar negative trends, but the associations were not significant for Tau (OR = 0.85 [95% CI, 0.54–1.35], β = -0.16, p = 0.49) or pTau (OR = 0.89 [95% CI, 0.57–1.39], β = -0.11, p = 0.61). This indicates that KL-VSHET+ status interaction with pathological aspects of AD are more profound among APOE ε4-carriers, such as Aβ and Tau accumulation during the pre-clinical phase of the disease [47, 48]. Although we did not find a protective effect of KL-VSHET+ in individuals with AD dementia, a recent study reported that KL-VSHET+ attenuated the association between higher amyloid PET and higher increases in tau PET accumulation [32]. Reasons for the discrepancy could be that here we did not investigate interaction effects of KL-VSHET+ and amyloid on tau pathology, and second, we did not investigate tau PET, which assesses fibrillar tau deposits, whereas CSF p-tau181 appears to represent one or more earlier phenomena that do not closely correlate with neurofibrillary tangle burden. Indeed, previous studies have also reported a significant association between KL-VS heterozygosity and reduced tau accumulation and lower memory impairment in elderly humans at risk of AD dementia [32, 49]. However, in a mouse model of AD that was used to examine the neuroprotective effects of Klotho protein against neuronal damage associated with oxidative stress and neurodegeneration, no changes in Tau phosphorylation were observed in the presence of Klotho [50]. Unlike Tau and pTau association with KL-VSHET+ status, we observed a positive association with CSF levels of sTREM2 (inflammation biomarker). The observed increase in CSF sTREM2 levels was not significantly associated with KL-VSHET+ status for either APOE ε4-carriers (OR = 1.08 [95% CI, 0.59–2], β = 0.08, p = 0.80) or non-carriers (OR = 1.20 [95% CI, 0.77–1.86], β = 0.18, p = 0.43). Interestingly, recent studies [29, 34, 51, 52] have shown that higher sTREM2 levels are associated with lower AD risk and slower progression. Therefore, the observed positive association suggests that the protective effect of the KL-VSHET+ might be mediated by higher CSF sTREM2 levels. However, this hypothesis will need to be validated in studies with larger sample size for CSF sTREM2 levels.
Taken together, the observed significant associations between KL-VSHET+ status and biomarkers for brain amyloidosis (CSF Aβ positivity) and tau pathology (CSF Tau and pTau negativity) are suggestive of neuroprotective effect of KL-VSHET+ against age-related biomarker, biomolecular, and cognitive alterations that confer risk for AD. Our results further strengthen the findings of a recent meta-analysis including 25 independent studies, showing that APOE ε4-carriers who were also KL-VSHET+, were at a reduced risk for the combined outcome of conversion to mild cognitive impairment (MCI) or AD [6]. Besides, several other studies that evaluated the association of KL-VSHET+ status with different cognitive measures in control participants did not consider interactions with APOE ε4 but did observe protective associations that were more pronounced closer to 80 years of age [18, 53, 54].
Notably, we assessed the associations between KL-VSHET+ status and AD endophenotypes across three age strata: all of the participants (AD and controls); only those aged 60 to 80 years (AD and controls); and only cognitively normal participants aged 60 to 80 years (S3 Table in S2 File). Owing to a higher genetic risk for AD attributable to APOE ε4 in individuals who are 60 to 80 years old [55–57] and an existing study that hypothesized protective association of KL-VSHET+ status to be strongest in APOE ε4 carriers who are 60 to 80 years old [6], the a priori focus of the current study was also at this particular age range. Although we observed similar associative trends most of the time, it was interesting to see how the effects became apparent when restricting analyses to the cognitively normal participants in the age range of 60 to 80 years. In all of the cases, no significant associations were observed between KL-VSHET+ status and AD endophenotypes while considering all of the AD and cognitively normal participants, or all of the AD and cognitively normal participants within age range of 60–80. However, pronounced effects and associations were apparent for Aβ, Tau, and pTau levels from CSF, while considering cognitively normal participants who are APOE ε4-carriers and KL-VSHET+. These findings suggest that the cognitively normal participants group, aged 60 to 80 years, mainly drove the outcome in our analyses, further strengthening the existing hypothesis that KL-VS heterozygous genotype is favorable for better health and cognition in older people [18, 53, 58]. Importantly, we also observed that associations between KL heterozygous cognitively normal participants and different AD endophenotypes are robust to uneven sample size of APOE ε4-carriers and non-carriers as well as differences in the sex and age of the participants (S6 Table in S2 File). Although, the observed findings appear consistent with our initial hypothesis and confirm existing literature [6, 42], the detected associations are nominally significant and would likely fail multiple test correction due to limited sample size. Therefore, additional studies are required to investigate the associations between KL-VSHET+ and AD endophenotypes with relatively larger sample size to draw definitive conclusions.
The exact mechanism underlying the KL-VSHET+ interaction with APOE ε4 and modulation of Aβ, Tau, and pTau burden is yet unknown. However, it is logical to postulate that KL-VSHET+ may confer resilience by increasing the serum level of circulating Klotho protein [18, 21] or by changing its function. In animal mouse models, elevated klotho levels have led to an extended lifespan [17], enhanced cognition [19] and increased resilience to AD-related toxicity [58]. Other studies in humans indicated that KL-VSHET+ status has protective effects against brain aging and cognitive decline [21, 59], suggestive of its protective association against AD. Our findings also suggest that middle-aged APOE ε4-carriers who are KL-VSHET+ might show resilience to age-induced cognitive and tau changes. Interestingly, we have observed an age-specific association between KL-VSHET+ and AD endophenotypes, which is in line with existing studies reporting a specific time window for the effect of KL-VS polymorphism [20, 59].
To conclude, our work contributes to the existing literature by demonstrating that the protective effects of KL-VSHET+ extend to AD-related Aβ, Tau, and pTau endophenotypes and deficits in memory and executive function in cognitively normal APOE ε4-carriers who are at risk for developing AD. One promising research avenue for the future studies could be to assess whether Klotho protein levels in the CSF or serum/plasma of participants associate with measures of preclinical and symptomatic AD.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
We thank all the participants and their families, as well as the many involved institutions and their staff. Data used in preparation of this article were also obtained from the Dominantly Inherited Alzheimer Network (DIAN) and Alzheimer’s Disease Neuroimaging Initiative (ADNI) consortiums (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provide data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Data Availability
All relevant data are within the paper and its Supporting information files.
Funding Statement
This work was supported by grants from the National Institutes of Health (R01AG044546, P01AG003991, RF1AG053303, R01AG058501, U01AG058922, RF1AG058501 and R01AG057777), the Alzheimer Association (NIRG-11-200110, BAND-14-338165, AARG-16-441560 and BFG-15-362540). This work was supported by access to equipment made possible by the Hope Center for Neurological Disorders, and the Departments of Neurology and Psychiatry at Washington University School of Medicine. The recruitment and clinical characterization of research participants at Washington University were supported by NIH P50 AG05681, P01 AG03991, and P01 AG026276. HZ is a Wallenberg Scholar supported by grants from the Swedish Research Council (#2018-02532), the European Research Council (#681712), Swedish State Support for Clinical Research (#ALFGBG-720931), the Alzheimer Drug Discovery Foundation (ADDF), USA (#201809-2016862), the AD Strategic Fund and the Alzheimer’s Association (#ADSF-21-831376-C, #ADSF-21-831381-C and #ADSF-21-831377-C), the Olav Thon Foundation, the Erling-Persson Family Foundation, Stiftelsen för Gamla Tjänarinnor, Hjärnfonden, Sweden (#FO2019-0228), the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 860197 (MIRIADE), and the UK Dementia Research Institute at UCL. KB is supported by the Swedish Research Council (#2017-00915), the Alzheimer Drug Discovery Foundation (ADDF), USA (#RDAPB-201809-2016615), the Swedish Alzheimer Foundation (#AF-742881), Hjärnfonden, Sweden (#FO2017-0243), the Swedish state under the agreement between the Swedish government and the County Councils, the ALF-agreement (#ALFGBG-715986), the European Union Joint Program for Neurodegenerative Disorders (JPND2019-466-236), the National Institute of Health (NIH), USA, (grant #1R01AG068398-01), and the Alzheimer’s Association 2021 Zenith Award (ZEN-21-848495).
References
- 1.Ferri CP, Prince M, Brayne C, Brodaty H, Fratiglioni L, Ganguli M, et al. Global prevalence of dementia: A Delphi consensus study. Lancet. 2005. doi: 10.1016/S0140-6736(05)67889-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Budson AE, Solomon PR. New diagnostic criteria for Alzheimer’s disease and mild cognitive impairment for the practical neurologist. Practical Neurology. 2012. doi: 10.1136/practneurol-2011-000145 [DOI] [PubMed] [Google Scholar]
- 3.Roses AD. Apolipoprotein E alleles as risk factors in Alzheimer’s disease. Annual Review of Medicine. 1996. doi: 10.1146/annurev.med.47.1.387 [DOI] [PubMed] [Google Scholar]
- 4.Brousseau T, Legrain S, Berr C, Gourlet V, Vidal O, Amouyel P. Confirmation of the epsilon 4 allele of the apolipoprotein E gene as a risk factor for late-onset Alzheimer’s disease. Neurology. 1994. doi: 10.1212/wnl.44.2.342 [DOI] [PubMed] [Google Scholar]
- 5.Polvikoski T, Sulkava R, Haltia M, Kainulainen K, Vuorio A, Verkkoniemi A, et al. Apolipoprotein E, Dementia, and Cortical Deposition of β-Amyloid Protein. N Engl J Med. 1995. doi: 10.1056/nejm199511093331902 [DOI] [PubMed] [Google Scholar]
- 6.Belloy ME, Napolioni V, Han SS, Le Guen Y, Greicius MD. Association of Klotho -VS Heterozygosity with Risk of Alzheimer Disease in Individuals Who Carry APOE4. JAMA Neurol. 2020. doi: 10.1001/jamaneurol.2020.0414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosentino S, Scarmeas N, Helzner E, Glymour MM, Brandt J, Albert M, et al. APOE ε4 allele predicts faster cognitive decline in mild Alzheimer disease. Neurology. 2008. doi: 10.1212/01.wnl.0000304038.37421.cc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993. doi: 10.1126/science.8346443 [DOI] [PubMed] [Google Scholar]
- 9.Vlassenko AG, Mintun MA, Xiong C, Sheline YI, Goate AM, Benzinger TLS, et al. Amyloid-beta plaque growth in cognitively normal adults: Longitudinal [11C]Pittsburgh compound B data. Ann Neurol. 2011. doi: 10.1002/ana.22608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maxwell TJ, Corcoran C, Del-Aguila JL, Budde JP, Deming Y, Cruchaga C, et al. Genome-wide association study for variants that modulate relationships between cerebrospinal fluid amyloid-beta 42, tau, and p-tau levels. Alzheimer’s Res Ther. 2018. doi: 10.1186/s13195-018-0410-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deming Y, Li Z, Kapoor M, Harari O, Del-Aguila JL, Black K, et al. Genome-wide association study identifies four novel loci associated with Alzheimer’s endophenotypes and disease modifiers. Acta Neuropathol. 2017. doi: 10.1007/s00401-017-1685-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan Q, Nho K, Del-Aguilla J, Wang X, Risacher SL, Fan KH, et al. Genome-wide association study of brain amyloid deposition as measured by Pittsburgh Compound-B (PiB)-PET imaging. Mol Psychiatry. 2021;26: 309–321. doi: 10.1038/s41380-018-0246-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caselli RJ, Graff-Radford NR, Reiman EM, Weaver A, Osborne D, Lucas J, et al. Preclinical memory decline in cognitively normal apolipoprotein E-ε4 homozygotes. Neurology. 1999. doi: 10.1212/wnl.53.1.201 [DOI] [PubMed] [Google Scholar]
- 14.Belloy ME, Eger SJ, Le Guen Y, Napolioni V, Deters KD, Yang HS, et al. KL*VS heterozygosity reduces brain amyloid in asymptomatic at-risk APOE*4 carriers. Neurobiol Aging. 2021. doi: 10.1016/j.neurobiolaging.2021.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson CM, Schultz SA, Oh JM, Darst BF, Ma Y, Norton D, et al. KLOTHO heterozygosity attenuates APOE4-related amyloid burden in preclinical AD. Neurology. 2019. doi: 10.1212/WNL.0000000000007323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Château MT, Araiz C, Descamps S, Galas S. Klotho interferes with a novel FGF-signalling pathway and insulin/Igf-like signalling to improve longevity and stress resistance in Caenorhabditis elegans. Aging (Albany NY). 2010. doi: 10.18632/aging.100195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, et al. Physiology: Suppression of aging in mice by the hormone Klotho. Science. 2005. doi: 10.1126/science.1112766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubal DB, Yokoyama JS, Zhu L, Broestl L, Worden K, Wang D, et al. Life Extension Factor Klotho Enhances Cognition. Cell Rep. 2014. doi: 10.1016/j.celrep.2014.03.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leon J, Moreno AJ, Garay BI, Chalkley RJ, Burlingame AL, Wang D, et al. Peripheral Elevation of a Klotho Fragment Enhances Brain Function and Resilience in Young, Aging, and α-Synuclein Transgenic Mice. Cell Rep. 2017. doi: 10.1016/j.celrep.2017.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arking DE, Krebsova A, Macek M, Macek M, Arking A, Mian IS, et al. Association of human aging with a functional variant of klotho. Proc Natl Acad Sci U S A. 2002. doi: 10.1073/pnas.022484299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokoyama JS, Sturm VE, Bonham LW, Klein E, Arfanakis K, Yu L, et al. Variation in longevity gene KLOTHO is associated with greater cortical volumes. Ann Clin Transl Neurol. 2015. doi: 10.1002/acn3.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arking DE, Atzmon G, Arking A, Barzilai N, Dietz HC. Association between a functional variant of the KLOTHO gene and high-density lipoprotein cholesterol, blood pressure, stroke, and longevity. Circ Res. 2005. doi: 10.1161/01.RES.0000157171.04054.30 [DOI] [PubMed] [Google Scholar]
- 23.Braskie MN, Ringman JM, Thompson PM. Neuroimaging measures as endophenotypes in Alzheimer’s disease. International Journal of Alzheimer’s Disease. 2011. doi: 10.4061/2011/490140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Storandt M, Morris JC. Ascertainment bias in the clinical diagnosis of Alzheimer disease. Arch Neurol. 2010. doi: 10.1001/archneurol.2010.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sliwinski MJ, Smyth JM, Hofer SM, Stawski RS. Intraindividual coupling of daily stress and cognition. Psychol Aging. 2006. doi: 10.1037/0882-7974.21.3.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassenstab J, Ruvolo D, Jasielec M, Xiong C, Grant E, Morris JC. Absence of practice effects in preclinical Alzheimer’s disease. Neuropsychology. 2015. doi: 10.1037/neu0000208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farias FHG, Benitez BA, Cruchaga C. Quantitative endophenotypes as an alternative approach to understanding genetic risk in neurodegenerative diseases. Neurobiology of Disease. 2021. doi: 10.1016/j.nbd.2020.105247 [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Farias FHG, Dube U, Del-Aguila JL, Mihindukulasuriya KA, Fernandez MV, et al. The TMEM106B FTLD-protective variant, rs1990621, is also associated with increased neuronal proportion. Acta Neuropathol. 2020. doi: 10.1007/s00401-019-02066-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deming Y, Filipello F, Cignarella F, Cantoni C, Hsu S, Mikesell R, et al. The MS4A gene cluster is a key modulator of soluble TREM2 and Alzheimer’s disease risk. Sci Transl Med. 2019. doi: 10.1126/scitranslmed.aau2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porter T, Burnham SC, Milicic L, Savage G, Maruff P, Lim YY, et al. Klotho allele status is not associated with Aβ and APOE ε4–related cognitive decline in preclinical Alzheimer’s disease. Neurobiol Aging. 2019. doi: 10.1016/j.neurobiolaging.2018.12.014 [DOI] [PubMed] [Google Scholar]
- 31.Müller BW, Hinney A, Scherbaum N, Weimar C, Kleinschnitz C, Peters T, et al. Klotho KL-VS haplotype does not improve cognition in a population-based sample of adults age 55–87 years. Sci Rep. 2021. doi: 10.1038/s41598-021-93211-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neitzel J, Franzmeier N, Rubinski A, Dichgans M, Brendel M, Weiner M, et al. KL-VS heterozygosity is associated with lower amyloid-dependent tau accumulation and memory impairment in Alzheimer’s disease. Nat Commun. 2021. doi: 10.1038/s41467-021-23755-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dube U, Del-Aguila JL, Li Z, Budde JP, Jiang S, Hsu S, et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci. 2019. doi: 10.1038/s41593-019-0501-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suárez-Calvet M, Morenas-Rodríguez E, Kleinberger G, Schlepckow K, Caballero MÁA, Franzmeier N, et al. Early increase of CSF sTREM2 in Alzheimer’s disease is associated with tau related-neurodegeneration but not with amyloid-β pathology. Mol Neurodegener. 2019. doi: 10.1186/s13024-018-0301-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartlett JW, Frost C, Mattsson N, Skillbäck T, Blennow K, Zetterberg H, et al. Determining cut-points for Alzheimer’s disease biomarkers: statistical issues, methods and challenges. Biomark Med. 2012. doi: 10.2217/bmm.12.49 [DOI] [PubMed] [Google Scholar]
- 36.Guvakova MA. Improving patient classification and biomarker assessment using Gaussian Mixture Models and Bayes’ rule. Oncoscience. 2019. doi: 10.18632/oncoscience.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ingber AP, Hassenstab J, Fagan AM, Benzinger TLS, Grant EA, Holtzman DM, et al. Cerebrospinal Fluid Biomarkers and Reserve Variables as Predictors of Future “Non-Cognitive” Outcomes of Alzheimer’s Disease. J Alzheimer’s Dis. 2016. doi: 10.3233/JAD-150478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delaneau O, Marchini J, McVeanh GA, Donnelly P, Lunter G, Marchini JL, et al. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 2014. doi: 10.1038/ncomms4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012. doi: 10.1038/ng.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Team RC. R core team (2014). R A Lang Environ Stat Comput R Found Stat Comput Vienna, Austria URL http://www.R-project.org. 2014.
- 42.Lo MT, Kauppi K, Fan CC, Sanyal N, Reas ET, Sundar VS, et al. Identification of genetic heterogeneity of Alzheimer’s disease across age. Neurobiol Aging. 2019. doi: 10.1016/j.neurobiolaging.2019.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blennow K, Mattsson N, Schöll M, Hansson O, Zetterberg H. Amyloid biomarkers in Alzheimer’s disease. Trends Pharmacol Sci. 2015. doi: 10.1016/j.tips.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 44.Palmqvist S, Mattsson N, Hansson O. Cerebrospinal fluid analysis detects cerebral amyloid-β accumulation earlier than positron emission tomography. Brain. 2016. doi: 10.1093/brain/aww015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vlassenko AG, McCue L, Jasielec MS, Su Y, Gordon BA, Xiong C, et al. Imaging and cerebrospinal fluid biomarkers in early preclinical alzheimer disease. Ann Neurol. 2016. 10.1002/ana.24719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Wilde A, Reimand J, Teunissen CE, Zwan M, Windhorst AD, Boellaard R, et al. Discordant amyloid-β PET and CSF biomarkers and its clinical consequences. Alzheimer’s Res Ther. 2019. doi: 10.1186/s13195-019-0532-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caselli RJ, Reiman EM. Characterizing the preclinical stages of Alzheimer’s disease and the prospect of presymptomatic intervention. Journal of Alzheimer’s Disease. 2013. doi: 10.3233/JAD-2012-129026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jadhav S, Avila J, Schöll M, Kovacs GG, Kövari E, Skrabana R, et al. A walk through tau therapeutic strategies. Acta neuropathologica communications. 2019. doi: 10.1186/s40478-019-0664-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Driscoll I, Ma Y, Gallagher CL, Johnson SC, Asthana S, Hermann BP, et al. Age-related tau burden and cognitive deficits are attenuated in Klotho KL-vs heterozygotes. J Alzheimer’s Dis. 2021. doi: 10.3233/JAD-200944 [DOI] [PubMed] [Google Scholar]
- 50.Zeldich E, Di Chen C, Colvin TA, Bove-Fenderson EA, Liang J, Tucker Zhou TB, et al. The neuroprotective effect of Klotho is mediated via regulation of members of the redox system. J Biol Chem. 2014. doi: 10.1074/jbc.M114.567321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ewers M, Biechele G, Suárez-Calvet M, Sacher C, Blume T, Morenas-Rodriguez E, et al. Higher CSF sTREM2 and microglia activation are associated with slower rates of beta-amyloid accumulation. EMBO Mol Med. 2020. 10.15252/emmm.202012308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ewers M, Franzmeier N, Suárez-Calvet M, Morenas-Rodriguez E, Caballero MAA, Kleinberger G, et al. Increased soluble TREM2 in cerebrospinal fluid is associated with reduced cognitive and clinical decline in Alzheimer’s disease. Sci Transl Med. 2019. doi: 10.1126/scitranslmed.aav6221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Vries CF, Staff RT, Harris SE, Chapko D, Williams DS, Reichert P, et al. Klotho, APOEε4, cognitive ability, brain size, atrophy, and survival: a study in the Aberdeen Birth Cohort of 1936. Neurobiol Aging. 2017. 10.1016/j.neurobiolaging.2017.02.019 [DOI] [PubMed] [Google Scholar]
- 54.Deary IJ, Harris SE, Fox HC, Hayward C, Wright AF, Starr JM, et al. KLOTHO genotype and cognitive ability in childhood and old age in the same individuals. Neurosci Lett. 2005. 10.1016/j.neulet.2004.12.005 [DOI] [PubMed] [Google Scholar]
- 55.Neu SC, Pa J, Kukull W, Beekly D, Kuzma A, Gangadharan P, et al. Apolipoprotein E genotype and sex risk factors for Alzheimer disease: A meta-analysis. JAMA Neurol. 2017. doi: 10.1001/jamaneurol.2017.2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997. doi: 10.1001/jama.1997.03550160069041 [DOI] [PubMed] [Google Scholar]
- 57.Bickeböller H, Campion D, Brice A, Amouyel P, Hannequin D, Didierjean O, et al. Apolipoprotein E and Alzheimer disease: Genotype-specific risks by age and sex. Am J Hum Genet. 1997. [PMC free article] [PubMed] [Google Scholar]
- 58.Dubal DB, Zhu L, Sanchez PE, Worden K, Broestl L, Johnson E, et al. Life extension factor klotho prevents mortality and enhances cognition in hAPP transgenic mice. J Neurosci. 2015. doi: 10.1523/JNEUROSCI.5791-12.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Invidia L, Salvioli S, Altilia S, Pierini M, Panourgia MP, Monti D, et al. The frequency of Klotho KL-VS polymorphism in a large Italian population, from young subjects to centenarians, suggests the presence of specific time windows for its effect. Biogerontology. 2010. doi: 10.1007/s10522-009-9229-z [DOI] [PubMed] [Google Scholar]