Abstract
Several studies have demonstrated impaired immune cell functions in chronic lymphocytic leukemia (CLL) patients, contributing to tumor evasion and disease progression. However, in CLL-like monoclonal B cell lymphocytosis (MBL) studies are scarce. Herein, we characterized the immune environment in 62 individuals with clinical MBL, 56 early stage CLL patients and 31 healthy controls. Gene expression arrays and qRT-PCR were performed on RNA from CD4+ peripheral blood cells; serum cytokines were measured by immunoassays; and HLA-DR expression on circulating monocytes as well as Th1, cytotoxic, exhausted and effector CD4+ T cells were characterized by flow cytometry. Besides, cell cultures of clonal B cells and CD14-enriched or depleted cell fractions were performed. Strikingly, MBL and early stage CLL differed in proinflammatory signatures. An increased inflammatory drive mainly orchestrated by monocytes was identified in MBL, which exhibited enhanced phagocytosis, pattern recognition receptors, IL8, HMGB1 and acute response signaling pathways, and increased proinflammatory cytokines (in particular IL8, IFNγ and TNFα). This inflammatory signature was diminished in early stage CLL (reduced IL8 and IFNγ levels, IL8 signaling pathway and monocytic HLA-DR expression compared to MBL), especially in those patients with mutations in IGHV genes. Additionally, CD4+ T cells of MBL and early stage CLL showed a similar upregulation of Th1 and cytotoxic genes, and expanded CXCR3+ and perforin+ CD4+ T cells as well as PD1+ CD4+ T cells compared to controls. Cell culture assays disclosed tumor-supporting effects of monocytes similarly observed in MBL and early stage CLL. These novel findings reveal differences in the inflammatory environment between MBL and CLL, highlighting an active role for antigen stimulation in the very early stages of the disease, potentially related to malignant B cell transformation.
Introduction
Several alterations have been described in T cells of patients with chronic lymphocytic leukemia (CLL), presumably a consequence of chronic exposure to tumor cells [1–8]. Additionally, an elevated number of monocytes has been detected in peripheral blood (PB) of patients with the disease, displaying deregulated genes involved in phagocytosis and inflammation [9]. Serum cytokine levels have also been described to be altered in CLL [10], and increased levels of some cytokines such as CCL3 or IL8 have been associated with a worse outcome [11, 12].
CLL-like monoclonal B cell lymphocytosis (MBL) is defined as the presence of a clonal population of B lymphocytes in PB (<5×109/l) having a phenotype consistent with CLL, without other clinical features of the disease. Its frequency increases with age, being present in >50% of people older than 90 years. Those cases with a clonal B cell count >0.5×109/l are referred to as clinical MBL and are considered a premalignant condition. They show a progression rate to CLL requiring therapy of 1–2%/year. Indeed, virtually all cases of CLL are preceded by an MBL stage [13–17].
The role of immune interactions in MBL remains uncharacterized. While B cell expansions in patients with advanced CLL are accompanied by immune suppression that restrains antitumor immunity, prior investigations have shown that T cell function in MBL is onlyss slightly deviated [1, 18]. Thus, although immunosuppressive features and T cell repertoire restrictions are already detectable in MBL and evolve toward a more suppressive profile across the disease stages [18–20], the functional immune cell fraction may play relevant roles in MBL dynamics and in clonal evolution leading to CLL. Understanding immune interactions in pre-leukemic individuals would also be relevant for deciphering molecular mechanisms underlying the natural history of the disease. In this study, we characterized PB CD4+ cells (including CD4+ T cells and monocytes) and serum samples of a large cohort of MBL and early stage CLL patients to elucidate more comprehensively these issues.
Material and Methods
Patients and samples
A total of 62 subjects with clinical CLL-like MBL and 56 untreated, early stage CLL patients (Binet A/Rai 0-I and <20×109 clonal B cells/l) were evaluated. Patients were cared for at Hospital del Mar (n=50) and Hospital Vall d’Hebron (n=11) in Barcelona (Spain) and at Northwell Health (n=57) in New York (USA). Additionally, 31 age-matched healthy subjects were studied as controls. No case had evidence of infection at sampling. The main characteristics of the entire cohort are summarized in Table 1. At the end of the study, all MBL cases remained asymptomatic and none progressed to CLL requiring therapy. The study was performed in accordance with National and International Guidelines (Professional Code of Conduct, Declaration of Helsinki) and approved by the Ethics Committee of Hospital del Mar, Barcelona (2011/4317/I). Fresh PB, cryopreserved PB mononuclear cells (PBMCs) and frozen serum samples were characterized by several methods (Supplementary Figure 1). Part of the samples were obtained from MARBiobanc (Barcelona). Data from Northwell Health were collected using REDCap [21].
Table 1.
Characteristics of the study cohort at the time of analysis and last follow-up.
| Control (n=31) | MBL (n=62) | CLL Binet A (n=56) | |
|---|---|---|---|
|
| |||
| Age | 64 (42–88) | 69 (45–89) | 64 (37–89) |
| Absolute lymphocyte count (×109 cells/l) | 1.7 (0.4–3.2) | 5.0 (0.9–10.1) | 13.2 (7.9–24.2) |
| Clonal B cell count (×109 cells/l) | NA* | 2.1 (0.5–4.8) | 10.2 (5.8–19.4) |
| Rai stage | |||
| 0 | NA | NA | 48 (85.7%) |
| I | 8 (14.3%) | ||
| Mutated IGHV | NA | 48/54 (88.9%) | 32/54 (59.3%) |
| Treated patients | NA | 0 | 11 (19.6%) |
| Deaths | NA | 3 (4.8%) | 6 (10.7%) |
| Follow-up from recruitment (months) | NA | 48 (0–117) | 67 (0–143) |
Values are given as median (range) or number (%). NA: not applicable.
Polyclonal B cell count: 0.07×109 cells/l (0.04–0.21).
Cell isolation and RNA extraction for gene expression analyses
Fresh PB was subjected to Ficoll density gradient centrifugation. Purified CD4+CD19− (total CD4+ cells, including CD4+ T cells and monocytes), CD4+CD14− (CD4+ T cells) and CD14+ cells (monocytes) were isolated from PBMCs based on positive selection methods employing immunomagnetic beads (CD19 Multisort Kit, CD19 MicroBeads, CD4 MicroBeads and CD14 MicroBeads) and autoMACS technology (Miltenyi Biotec, Bergisch Gladbach, Germany). A flow-chart describing all cell isolation procedures for gene expression analyses is shown in Supplementary Figure 2. To assess the purification process efficacy, 100,000 cells of each sample were analyzed by flow cytometry using anti-CD3 (APC), anti-CD4 (PE), anti-CD14 (FITC) and/or anti-CD45 (PerCP-Cy5.5) antibodies (BD Biosciences, San Jose, CA, USA). Purity of ≥90% was achieved in all samples, except for two MBL subjects in which purities of total CD4+ cells were 70%. Purified cells were stored at −80ºC in 1% BME RLT-plus buffer (QIAGEN, Venlo, The Netherlands). RNA was finally extracted following the RNeasy Plus Mini Kit protocol (QIAGEN).
Gene expression analysis by microarrays
RNA from total CD4+ cells was employed for gene expression arrays in 9 healthy controls, 15 MBL and 14 CLL cases. All samples used for microarray experiments had an RNA integrity number (RIN) >7. Briefly, 30 ng of total RNA were retrotranscribed into cDNA and amplified (Ovation® Pico WTA System V2, NUGEN, Redwood City, CA, USA). The cDNA product was purified (MinElute Reaction Cleanup Kit, QIAGEN), yield and purity measured, and biotinylated fragments were added to the hybridization cocktail, finally hybridized on GeneChip Human Gene 2.0 ST array (Affymetrix, Santa Clara, CA, USA) following manufacturer’s instructions. Data quality assessment was performed with the Expression Console (Affymetrix) and R (v3.1.1). A Robust Multi-array Average (RMA) algorithm, included in the aroma.affymetrix package, was used for data normalization [22]. To correct for batch effects, ComBat, implemented in the R package SVA, was employed [23]. Differential expression analysis was performed using limma R package [24]. P-values <0.05 together with a Fold Change |FC|>1.2 cut-off value were used to generate the list of differentially expressed genes. Gene annotations were obtained from the Affymetrix annotation file Human Gene 2.0 ST array (NetAffix version na34, hg19). UCSC database was employed (Feb. 2009 hg19, GRCh37) to complement Affymetrix annotations. Microarray data have been deposited in the Gene Expression Omnibus (GEO) database with series accession number GSE125791 [25]. A gene pathway analysis of the genetic signature of total CD4+ cells was accomplished using Ingenuity Pathway Analysis (IPA, QIAGEN). Finally, to have an insight into the differential contribution of monocytes and T cells in gene expression, the BioGPS tool (http://biogps.org) was employed.
Gene expression analysis by quantitative RT-PCR (qRT-PCR)
To confirm the gene expression levels detected by microarrays in total CD4+ cells, RNA from the same samples (excluding the two cases with purities <90%) and from an additional independent cohort consisting of 8 MBL and 5 CLL subjects (purity ≥90%, 49 cases in total) was subjected to qRT-PCR validation. To elucidate the different contributions of CD4+ T cells and monocytes in gene expression, RNA from additional CD14-depleted CD4+ cells (CD4+ T cells) and CD14+ cells (monocytes) was subjected to the same qRT-PCR validation panel (in 4 healthy and 8 MBL; and 3 healthy and 9 MBL individuals, respectively). This panel consisted of 13 selected genes (CCL2, CLU, CXCL5, DEFA1, FGFBP2, GZMH, ITGAM, NEXN, NKG7, PPBP, RAB31, TUBB1 and VSTM1; assays detailed in Supplementary Table 1) significantly upregulated by gene expression arrays and related to inflammation, Th1 cells and cytotoxic pathways. GUSB and GAPDH were employed as housekeeping genes. A total of 400 ng of RNA were analyzed in this customized gene panel, using Taqman Low Density Arrays (TLDA, Life Technologies, Foster City, CA) according to manufacturer’s instructions. For qRT-PCR data analysis, outlier replicates, defined by a SD >0.5 and a qRT-PCR threshold cycle (Ct) >40, were excluded. The ΔCt measures, defined as the mean of the expression of a studied gene minus the mean of the expression of the selected housekeeping genes, were compared.
Flow cytometry analysis
Cryopreserved PBMCs from 8 controls, 11 MBL and 10 CLL subjects were assessed. PBMCs were washed and labeled with the following antibodies: anti-CD45 (PerCP-Cy5.5), anti-CD14 (FITC), anti-CD3 (V450), anti-CD4 (PE), anti-CD4 (APC), anti-HLA-DR (PE-Cy7), anti-CXCR3 (PerCP-Cy5.5), anti-perforin (AF488), anti-granzyme B (FITC), anti-PD1 (PE) and anti-CD27 (PE). FVS510 was employed to discriminate viable from non-viable cells, and the Fixation/Permeabilization Solution Kit to stain intracellular molecules. Isotype controls (PE, PerCP-Cy5.5, PE-Cy7 and APC) and fluorescence-minus-one controls were used to correctly assess antigen expression. All reagents were purchased from BD Biosciences. Data acquisition was performed in a BD FACSCanto II cytometer and analyzed using the FACSDiva software (BD Biosciences). The gating strategy to define monocytes and CD4+ T cells and evaluate the expression of the antigens of interest is detailed in Figure 1.
Figure 1.
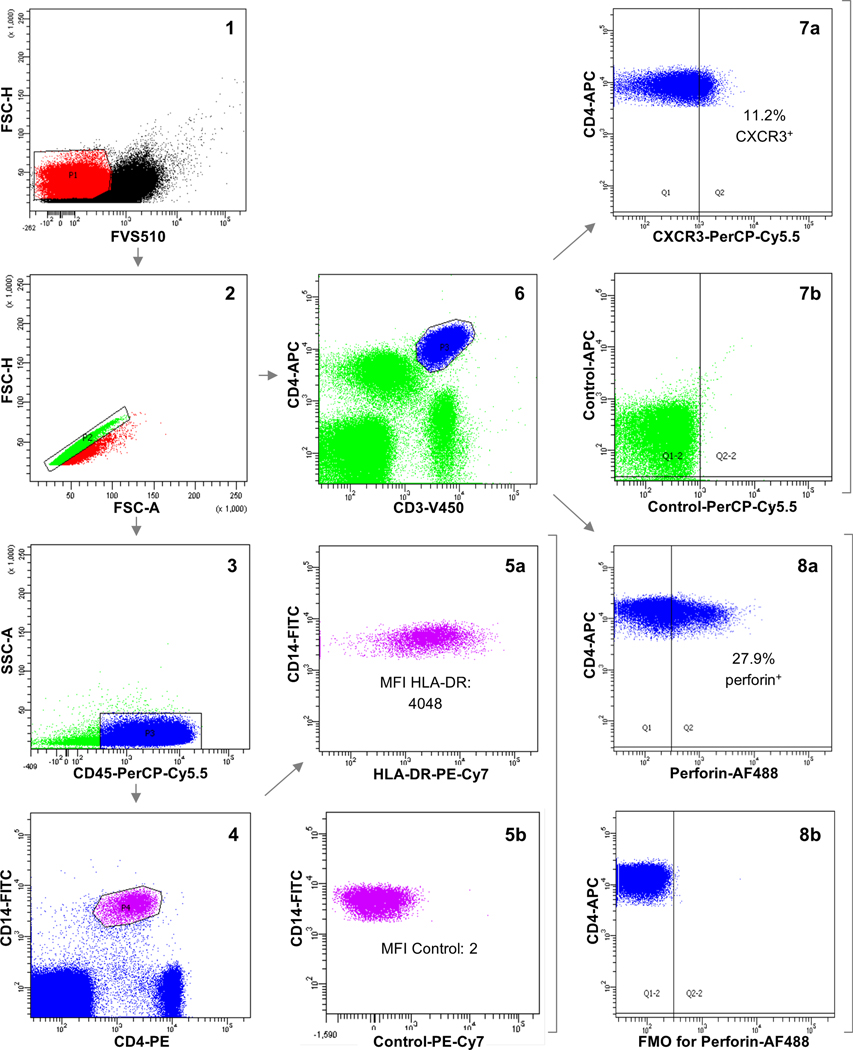
Flow cytometry gating strategy. FVS510 was used to select viable cells (1). After gating for singlets (2), CD45+CD14+CD4+ cells (monocytes) were selected (3 and 4) and linear mean fluorescence intensity (MFI) values of HLA-DR were registered (5a). The absence of fluorescence for PE-Cy7 on monocytes was confirmed employing isotype control (5b). On the other hand, CD3+CD4+ cells (CD4+ T cells) were gated (6). The percentage of CD4+ T cells that were positive for CXCR3 (7a) and PD1 or negative for CD27 was assessed employing isotype controls (7b), whereas the percentage of CD4+ T cells that were positive for perforin (8a) and granzyme B was evaluated using fluorescence-minus-one (FMO) controls (8b). In the plots, fluorescence intensity values are transformed into logarithmic scale.
Multiplex cytokine analysis
In 24 healthy subjects, 40 MBL and 44 CLL cases, serum levels of the following 20 cytokines were quantified: IL1β, IL2, IL4, IL5, IL6, IL8, IL10, IL12, IL15, IL17A, IFNα, IFNγ, TNFα, GM-CSF, CCL3, CCL4, CCL19, CXCL10 and CXCL11 using the U-PLEX Platform (Meso Scale Discovery, Rockville, MD, USA) and CXCL9 using Human CXCL9/MIG Quantikine ELISA Kit (R&D Systems, Minneapolis, MN, USA). All determinations were performed in duplicate and concentrations were reported in picograms per milliliter. To study the potential cytokine interactions, their respective coding genes were obtained and genomic data was analyzed using IPA.
Tumor cell survival analysis
Cryopreserved PBMCs from 6 MBL subjects and 5 CLL patients, all of them with mutated IGHV genes, were employed. Purified tumor B cells were isolated based on negative selection methods employing immunomagnetic beads (B-CLL Cell Isolation Kit) and autoMACS technology (Miltenyi Biotec). To assess the purification process efficacy, 100,000 cells of each sample were analyzed by flow cytometry using anti-CD33 (FITC), anti-CD4 (PE), anti-CD20 (PerCP-Cy5.5) and anti-CD5 (APC) antibodies (BD Biosciences). Purity of ≥90% of tumor B cells was achieved for all samples (median: 98.2%, range: 90.1–99.2). Additionally, CD14-enriched cell fractions and CD14-depleted PBMCs were obtained using CD14 MicroBeads (Miltenyi Biotec). The same flow cytometry panel was employed to evaluate the percentage of monocytes in the CD14-enriched cell fraction (median: 85.9%, range: 51.3–95.3), total PBMCs and CD14-depleted PBMCs (median: 4.7 vs. 0.9% respectively, P-value=0.003). A total of 1×105 tumor cells were cultured alone or in the presence of 6×105 cells of the CD14-enriched fraction, besides cell cultures of 1×106 total PBMCs and CD14-depleted PBMCs. Cells were resuspended in 200 μl of RPMI 1640 medium supplemented with 1% Fetal Bovine Serum (FBS), L-glutamine (4 mM), penicillin (100 U/ml) and streptomycin (100 μg/ml) in MW96 assay plates (Corning Inc., Corning, NY). Cells were then centrifuged at 200 x g for 4 minutes and cultured for 24 hours at 37ºC and 5% CO2. After that period, cells were washed and labeled with anti-CD33 (FITC), anti-CD4 (PE), anti-CD20 (PerCP-Cy5.5), anti-CD5 (APC) and FVS510, which allowed the identification of two subpopulations (viable and non-viable cells) within the tumor cell clone in all included patients, as shown in Supplementary Figure 3. Data acquisition was performed in a BD FACSCanto II cytometer and analyzed using FlowJo (FlowJo LLC, Ashland, OR). The effect of monocytes on tumor viability was assessed as the difference between the viability of tumor cells cultured with the CD14-enriched cell fraction minus the viability of tumor cells cultured alone.
Statistical analysis
The t-test and Mann-Whitney U test were used to estimate statistical significance of the differences identified between independent groups, whereas the results of the tumor cell survival analysis comparing different conditions within the same subject were evaluated using the Wilcoxon test. Linear regression analysis and Spearman correlations were calculated to evaluate the relationship between gene expression intensity and protein expression by flow cytometry. P-values <0.05 were considered significant. Statistical analyses were performed using R, SPSS v.22 and SAS v9.3.
Results
Increased inflammatory signature in MBL, mainly orchestrated by monocytes
The profiles identified by gene expression arrays in total CD4+ mononuclear cells were compared between MBL and controls. Considering only cases with purity ≥90%, a total of 1786 genes were differentially expressed in MBL (953 overexpressed and 833 underexpressed). The most differentially expressed genes are detailed in Supplementary Table 2. Significant overexpression in 11/13 gene determinations (85%) was corroborated by qRT-PCR analysis and was maintained when only IGHV mutated MBL subjects were considered (Supplementary Table 1).
A gene pathway analysis of the genetic signature observed in total CD4+ cells from MBL individuals was performed employing IPA, which recognized 1330 differentially expressed genes between MBL and controls. Significant canonical pathways predicted to be activated (Z-score ≥2) included several functions in which monocytes and macrophages are typically involved, such as Fcγ receptor-mediated phagocytosis, role of pattern recognition receptors (PRR) in pathogen recognition, acute phase response or TREM1 signaling pathways. Inflammasome, chemokine and IL8 signaling pathways were also predicted to be activated (Figure 2, Supplementary Tables 3 and 4). As expected, BioGPS analysis supported a myeloid contribution for the differentially expressed genes of these pathways (Supplementary Table 5). These results were maintained when only IGHV mutated MBL individuals were taken into account (Supplementary Table 4).
Figure 2.
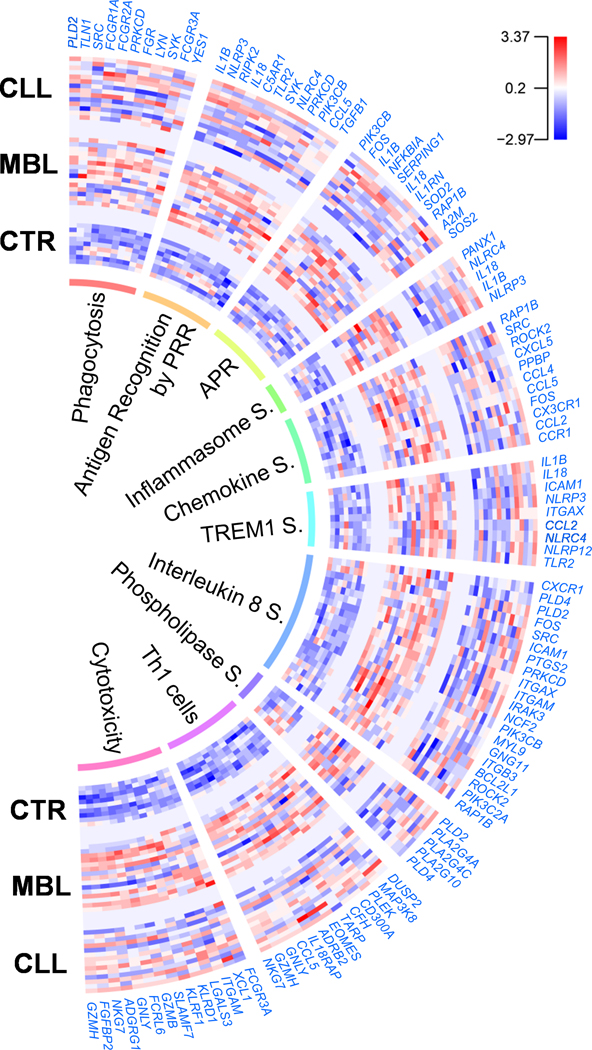
Differentially expressed genes and pathways in total CD4+ cells of MBL and CLL patients. Gene expression intensity is represented as log2 values centered and scaled. Detailed information of the enumerated genes is provided in Supplementary Table 3. CTR: Healthy controls; PRR: Pattern recognition receptors; APR: Acute phase response; S.: Signaling.
Additionally, an upstream analysis predicted some genes coding for receptors and co-receptors involved in pathogen recognition (TLR2, TLR7/8 or CD14) and proinflammatory cytokines (IFNG, TNF, IL1B, IL17A or IL18) as activated (Supplementary Table 6). Numerous functions related to cell migration and inflammation were also predicted to be activated (Supplementary Table 7).
This increased inflammatory drive detected in MBL was also supported by serum cytokine analysis, which showed that levels of 11/20 measured cytokines (IL5, IL6, IL8, IL10, IFNγ, TNFα, CCL3, CCL4, CXCL9, CXCL10 and CXCL11) were significantly higher in MBL compared to controls (Table 2). Of note, 5 of these (IL5, IL6, IL8, IFNγ and TNFα) are involved in HMGB1 signaling pathway, which induces the release of proinflammatory cytokines and was predicted to be activated by IPA (P-value<0.001, Z-score: 2.2). In line with the aforementioned, an upstream analysis predicted activation of Toll-like receptors (TLR) such as TLR3, TLR4 and TLR9 (P-value<0.001, Z-score: 2.4, 2.4 and 2, respectively), which can be stimulated through HMGB1 interaction. Notably, when we repeated the analysis considering IGHV mutational status (Supplementary Table 8), significantly increased levels of 10 and 4 cytokines were disclosed in MBL subjects with mutated and unmutated IGHV genes compared to controls, respectively. The small number of MBL cases with unmutated IGHV genes (n=4) may account for the reduced significant differences in this group, which actually displayed very similar high levels of those cytokines significantly increased in IGHV mutated MBL. Consequently, no significant differences were disclosed between both MBL groups.
Table 2.
Serum cytokine levels of healthy controls, MBL subjects and CLL patients.
| Cytokine | Control (n=24) | MBL (n=40) | CLL (n=44) | P-value | |||||
|---|---|---|---|---|---|---|---|---|---|
| Median | Range | Median | Range | Median | Range | MBL vs. Control | CLL vs. Control | CLL vs. MBL | |
|
| |||||||||
| IL1β | 0.26 | 0.07–5.85 | 0.35 | 0.07–124.27 | 0.47 | 0.07–114.13 | 0.475 | 0.061 | 0.219 |
| IL2 | 1.49 | 0.44–8.07 | 1.49 | 0.44–11.80 | 1.66 | 0.44–16.49 | 0.633 | 0.273 | 0.483 |
| IL4 | 0.21 | 0.06–0.79 | 0.21 | 0.06–40.67 | 0.08 | 0.06–1.34 | 0.875 | 0.270 | 0.210 |
| IL5 | 1.44 | 0.45–4.97 | 2.01 | 0.52–4.42 | 1.51 | 0.45–4.19 | 0.004 | 0.558 | 0.005 |
| IL6 | 2.04 | 1.16–151.09 | 3.62 | 1.16–1431.63 | 2.80 | 0.34–1689.05 | 0.021 | 0.080 | 0.495 |
| IL8 | 22.05 | 6.15–5216.03 | 238.01 | 7.21–6810.22 | 20.73 | 4.00–8807.27 | 0.005 | 0.980 | 0.019 |
| IL10 | 0.74 | 0.39–1.66 | 0.93 | 0.39–3.74 | 0.88 | 0.39–5.03 | 0.017 | 0.179 | 0.451 |
| IL12 | 1.07 | 0.60–3.57 | 1.07 | 0.60–18.42 | 1.07 | 0.60–3.08 | 0.299 | 0.358 | 0.824 |
| IL15 | 3.21 | 1.03–7.65 | 2.92 | 0.51–7.09 | 2.88 | 0.51–7.92 | 0.787 | 0.787 | 0.865 |
| IL17A | 2.51 | 0.51–13.14 | 2.52 | 0.51–21.39 | 2.52 | 0.51–16.69 | 0.199 | 0.196 | 0.895 |
| IFNα | 1.23 | 0.45–6.58 | 1.98 | 0.45–7.95 | 2.20 | 0.45–14.91 | 0.286 | 0.306 | 0.913 |
| IFNγ | 11.24 | 3.30–42.08 | 16.69 | 1.44–151.14 | 13.24 | 3.30–69.33 | 0.014 | 0.577 | 0.026 |
| TNFα | 2.25 | 0.50–29.74 | 4.99 | 1.30–69.19 | 3.30 | 0.70–28.21 | 0.015 | 0.140 | 0.149 |
| GM-CSF | 0.23 | 0.09–1.05 | 0.42 | 0.09–1.51 | 0.24 | 0.09–0.88 | 0.151 | 0.305 | 0.590 |
| CCL3 | 22.67 | 17.80–2925.30 | 46.00 | 19.05–2913.37 | 39.13 | 4.61–6427.70 | 0.001 | 0.070 | 0.395 |
| CCL4 | 114.53 | 56.66–1753.44 | 176.57 | 76.92–2358.09 | 151.61 | 29.75–2915.75 | 0.012 | 0.114 | 0.275 |
| CCL19 | 109.36 | 52.26–449.53 | 131.95 | 3.17–871.66 | 108.35 | 4.53–1561.50 | 0.071 | 0.908 | 0.078 |
| CXCL9 | 70.98 | 15.70–1044.45 | 165.53 | 15.70–1166.00 | 151.78 | 15.70–1982.50 | 0.003 | 0.001 | 0.964 |
| CXCL10 | 188.10 | 102.88–985.02 | 325.21 | 10.73–1904.78 | 230.79 | 5.25–735.04 | 0.016 | 0.581 | 0.029 |
| CXCL11 | 32.37 | 13.15–122.40 | 47.05 | 10.60–213.10 | 53.69 | 8.09–315.39 | 0.010 | 0.038 | 0.844 |
All determinations were performed in duplicate. Cytokine values are reported in picograms per milliliter (pg/ml). Significant P-values (<0.05) are highlighted in bold.
The inflammatory drive of MBL is decreased in early stage CLL, especially IGHV mutated
Next, gene expression profiles of total CD4+ mononuclear cells (purity ≥90%) were compared between CLL vs. controls and CLL vs. MBL. In all, 1039 genes were differentially expressed in the CLL vs. control comparison (460 overexpressed and 579 underexpressed) and 806 in CLL vs. MBL (398 overexpressed and 408 underexpressed). The most differentially expressed genes are detailed in Supplementary Table 2. The qRT-PCR analysis corroborated significant differential expression in 6 gene determinations (Supplementary Table 1).
IPA recognized 742 differentially expressed genes for CLL vs. control; however, significant activated canonical pathways were not detected. Some signs of functional immunity were predicted (such as CD28 or interferon activation and increased binding and phagocytosis of cells, Supplementary Table 9), although to a much lesser extent compared to MBL vs. control. This decreased inflammation in early stage CLL was also corroborated by cytokine analysis in serum, which only revealed 2 (CXCL9 and CXCL11) of the 20 measured cytokines to be significantly more elevated in CLL compared to controls (Table 2).
Concerning CLL vs. MBL, 598 differentially expressed genes were recognized by IPA, which showed phospholipases and IL8 signaling pathways as significantly decreased in CLL compared to MBL (Figure 2, Supplementary Tables 3 and 4), besides upstream inhibition of IFNG, IL5, TNF or TLR7 and decreased functions related to cell migration and lipid metabolism (Supplementary Table 10). The reduced immune activity observed in early stage CLL compared to MBL was specifically confirmed in circulating monocytes when HLA-DR expression was analyzed by flow cytometry. Thus, monocyte HLA-DR expression was significantly reduced in CLL compared to MBL, both when considering all patients or when restricting the analysis to IGHV mutated patients (Figure 3A, Supplementary Figure 4, Supplementary Table 11). Finally, serum levels of 4 cytokines (IL5, IL8, IFNγ and CXCL10) were significantly decreased in CLL compared to MBL (Table 2).
Figure 3.
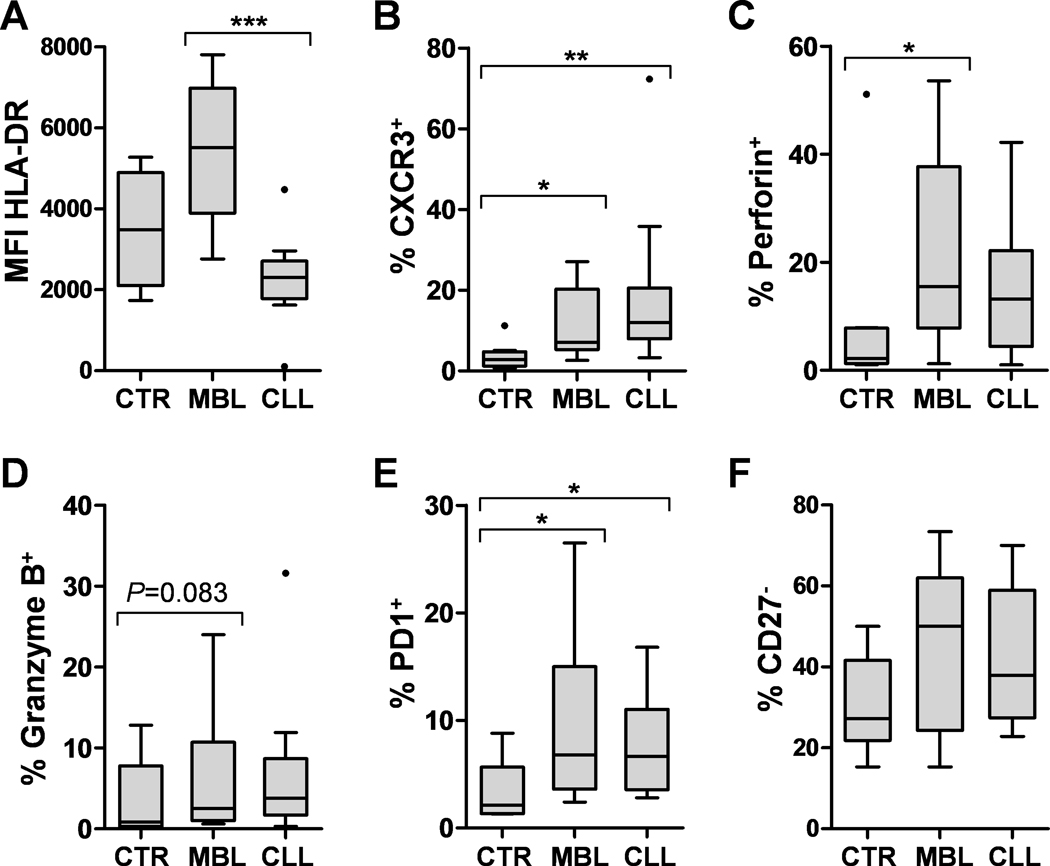
Flow cytometry analysis. MFI of HLA-DR was assessed on monocytes (A), and the percentages (%) of CD4+ T cells that were positive for CXCR3 (B), perforin (C), granzyme B (D), PD1 (E) or negative for CD27 (F) were registered. Significant P-values are indicated with * (<0.05), ** (<0.01) or *** (<0.001). CTR: Healthy controls.
Interestingly, when CLL cases were separated into two groups according to IGHV-mutation status, differences were underscored in MBL vs. CLL patients with mutated IGHV (M-CLL, <98% germline identity). In this sense, M-CLL cases displayed decreased levels of IL4, IL5, IL8, IFNγ, CCL3 and CXCL10 compared to MBL. Remarkably, 4 of these cytokines (IL4, IL5, IL8 and IFNγ) are involved in the proinflammatory HMGB1 pathway, which was predicted to be inhibited in M-CLL compared to MBL (P-value<0.001, Z-score:−2). Nonetheless, CLL patients with unmutated IGHV (U-CLL, ≥98% germline identity) only showed significant decreased levels of IL5 and CCL19 and increased levels of IL1β compared to MBL (Figure 4). Since levels of the aforementioned cytokines were similar between MBL subjects with mutated and unmutated IGHV genes, comparisons with CLL groups did not significantly differ when IGHV mutational status of MBL was considered, and the loss of statistical significance in some comparisons could be attributed to the more reduced number of cases in both MBL groups (Supplementary Table 8).
Figure 4.
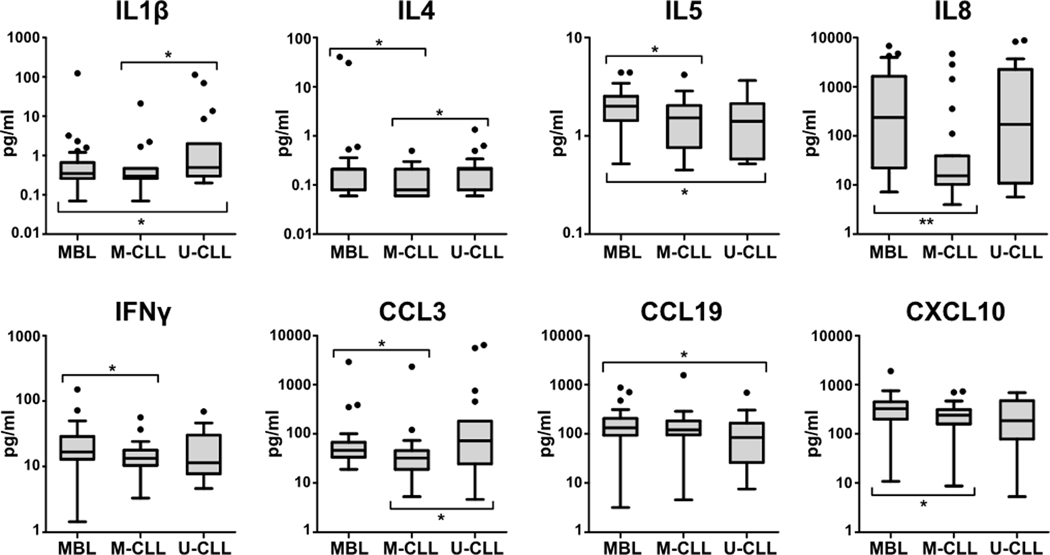
Serum cytokine levels in MBL and CLL patients with mutated IGHV (M-CLL) or unmutated IGHV (U-CLL). All determinations were performed in duplicate. Cytokine values (pg/ml) represented in the y-axis were transformed into logarithmic scale. Only those cytokines with significant differences in at least one comparison are shown. Significant P-values are indicated with * (<0.05) or ** (<0.01).
Similar increase of Th1 and cytotoxic CD4+ T cells in MBL and early stage CLL
A detailed analysis of the differentially expressed genes between groups also disclosed an upregulation of genes previously associated with Th1 cells [26] (DUSP2, MAP3K8, PLEK, CFH, TARP, IL18RAP, GNLY, GZMH or NKG7) in total CD4+ cells of both MBL and CLL compared to controls. Besides, genes related to cytotoxic pathways (GZMB, GZMH, GNLY, FGFBP2, FCRL6 or NKG7) were also upregulated in both entities compared to controls (Figure 2, Supplementary Table 3). If the two MBL cases with total CD4+ cell purity of 70% were considered, PRF1 (coding for perforin) was also significantly overexpressed in MBL vs. control (FC=1.64) and CLL vs. control (FC=1.55). BioGPS analysis pointed to a T cell contribution for most of the differentially expressed genes associated with Th1 cells and cytotoxicity, the latter only showing statistical differences for CD8+ T cells (Supplementary Table 5). This is not surprising considering that cytotoxic CD4+ T cells are closely related to CD8+ T cells. Nonetheless, when we analyzed RNA from CD14-depleted CD4+ cells (CD4+ T cells) and CD14+ cells (monocytes) of MBL subjects using the qRT-PCR panel previously described, a significant overexpression of cytotoxic genes (GZMH, FGFBP2 and NKG7) was only observed for CD4+ T cells (Supplementary Table 12), which confirms that CD4+ T cells account for the cytotoxic gene expression signature in MBL.
Flow cytometry analyses verified the similar increase of Th1 and cytotoxic CD4+ T cells in both entities. The percentages of CD4+ T cells that were positive for CXCR3, generally accepted as a marker of Th1 cells [27–28], and for the cytotoxic proteins perforin and granzyme B were higher in MBL and CLL subjects compared to controls. Moreover, increased percentages of PD1+ (exhausted) or CD27− (effector) CD4+ T cells were also detected in both entities. Nonetheless, statistical significance was only achieved for perforin (MBL vs. control) and for CXCR3 and PD1 (MBL and CLL vs. control), which was maintained when only IGHV mutated patients were considered (Figure 3B–F, Supplementary Table 11).
For those cases that were assessed by gene expression arrays and flow cytometry (n=13), a significantly positive correlation was observed between PRF1 and GZMB expression and the percentage of CD4+ T cells that were positive for perforin (ρ=0.62, P-value=0.024) and granzyme B (ρ=0.81, P-value=0.001), respectively.
Tumor-supporting effects of monocytes are similarly observed in MBL and early stage CLL
To have an insight into the possible functional consequences of the monocyte gene expression signature on tumor survival, different cell cultures were performed. After 24 hours, MBL and CLL subjects displayed a similar percentage of viable tumor cells (median: 64.6 vs. 68% respectively, P-value=0.931) and also a similar effect of monocytes on tumor viability (median increase: 10.1 vs. 9.7% respectively, P-value=0.537). Relating to the aforementioned finding, viability of tumor cells from MBL subjects was increased when they were cultured in the presence of the CD14-enriched cell fraction (median: 64.6 vs. 79.3%, P-value=0.046) and similar results were observed for CLL (median: 68 vs. 83.9%, P-value=0.043). In line with this, CD14+ cell depletion from PBMCs led to decreased tumor cell viability in CLL (median: 88.7 vs. 71.6%, P-value=0.043) and a similar trend was observed in MBL (median: 85.8 vs. 75.6%, P-value=0.075) (Figure 5).
Figure 5.
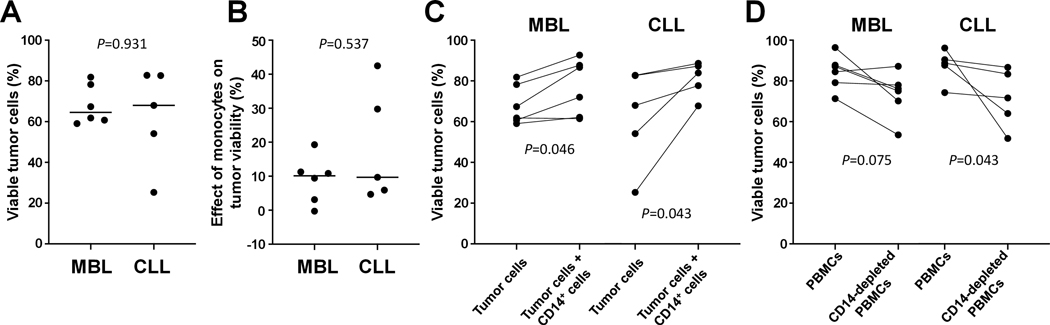
Tumor cell survival analysis. All cell cultures were incubated for 24h. To evaluate the viability of tumor cells, clonal B cells were cultured alone (A). The effect of monocytes on tumor viability was assessed as the viability of tumor cells cultured with CD14-enriched cells minus the viability of tumor cells cultured alone (B). For each subject, variations in tumor viability between the two previous conditions are shown (C), as well as the viability of tumor cells within total PBMCs or CD14-depleted PBMCs (D).
Discussion
Several studies in CLL have reported a dysfunctional microenvironment with tumor-supportive effects [4–9, 29, 30]. In the present study, signs of immune stimulation were found in MBL, which exhibited an increased inflammatory signature corroborated by different techniques. Notably, prior studies were mainly focused on more advanced stages of the disease, where independent malignant proliferation is accompanied by severe immunosuppresion. The lack of previous studies that comprehensively characterized the immune system in MBL, including molecular pathways of monocytes, may explain why this early inflammatory signature had not been revealed yet.
Remarkably, MBL displayed an activation of several functions relating to pathogen recognition by innate immune cells (e.g. monocytes). Among them, phagocytosis and stimulation of PRR such as TLR, which triggers the overexpression of proinflammatory genes [31], were highlighted. Similarly, a role for HMGB1 was noted, which can bind to PRR to mediate cytokine production [32]. Among these cytokines, IL8 is released and acts as an important mediator of the immune response. Of note, both IL8 serum levels and IL8 signaling pathway in total CD4+ cells were increased in our MBL cohort. Our results were also validated in an independent proteomic study, which showed especially increased serum levels of IL8 in MBL compared to controls, besides an activation of the HMGB1 signaling pathway (data not shown). This inflammatory axis of MBL could be initiated upon monocyte PRR stimulation by pathogens that may be related to the development of the disease. A role for self-antigens that can bind PRR and activate B cells was also reported [33]. In this sense, the maintenance of a proinflammatory environment, such as that identified here in pre-leukemic individuals, could contribute to neoplastic transformation. These results are in line with previous investigations suggesting a key role of self or foreign antigens in CLL development [2, 20, 34–36]. In this regard, the presence of immune stimulation was described several years before CLL diagnosis [37] and increased susceptibility to clonal expansions was associated with the development of the disease [38].
Strikingly, we also demonstrated that the aforementioned inflammatory drive was reduced in early stage CLL. Specifically, IPA predicted decreased IL8, IL5 and IFNγ signaling pathways in CLL compared to MBL, and the levels of these three cytokines were also significantly diminished in serum of CLL compared to MBL. The expression of HLA-DR on monocytes, which could be considered as a marker of immune activation since it is essential for antigen presentation that initiates a specific immune response, was also significantly decreased in CLL compared to MBL. These differences could be a consequence of a reduced function of pro-inflammatory monocytes and/or to a decreased percentage of these monocyte subsets (which express higher HLA-DR levels compared to classical monocytes) in CLL [39, 40]. Therefore, the distinct monocyte subsets should be isolated and further characterized to clarify their differential contribution in MBL and CLL pathogenesis. These as yet unknown findings, together with the gene expression and cytokine results showing augmented pro-inflammatory pathways in MBL, may reflect not only increased immunosuppression with tumor progression but also a stimulated immune system in MBL.
Additionally, the inflammatory signature of MBL subjects was especially reduced in M-CLL. In line with this, a different ongoing proteomics analysis predicted a significant inhibition of IL8 and HMGB1 signaling pathways in M-CLL compared to MBL, as demonstrated by cytokine immunoassays in the present study. The ongoing proteomic analyses also support that M-CLL patients rather than U-CLL may be responsible for the reduction of the inflammatory drive observed in MBL (unpublished results). Clonal cells observed in U-CLL patients are more dependent from environmental prosurvival signals [41], therefore interacting more actively with the immune environment than M-CLL clones. These interactions may imply cognate antigens presented by HLA molecules, which may account for our previous observations concerning a reduced monocyte HLA-DR expression in M-CLL. Nonetheless, MBL subjects with mutated IGHV genes significantly overexpressed monocyte HLA-DR compared to M-CLL, suggesting that another mechanism rather than the stimulation by the anergic and small B cell clone may explain monocyte activation in MBL. On the other hand, B cell receptor (BCR) signaling, which can stimulate cytokine production such as IL8 or CCL3 [42, 43], is more frequent in U-CLL. In our study, levels of both cytokines were significantly decreased in M-CLL compared to MBL, but similar increased levels were observed in U-CLL and in MBL subjects independently of their IGHV mutational status, suggesting that this increased inflammatory signature is MBL-specific. In this regard, BCR-dependent cytokine production by malignant cells may depend on their capacity to interact with the immune environment, which may be restricted in M-CLL and in the majority of MBL cases since 75–80% of them show mutated IGHV [15, 44]. Therefore, the increased inflammatory molecules observed in MBL may be actively produced by the immune environment rather than by the emerging and relatively small B cell clone. The gene expression results of circulating CD4+ cells of the present study showing increased inflammatory pathways also support this hypothesis.
We have also reported that co-cultures of clonal B cells from MBL patients with autologous CD14-enriched cells enhanced tumor cell viability. This finding suggests that the monocyte gene program and the related overexpressed inflammatory genes in MBL may not be globally a consequence of antitumor effects of monocytes. In this sense, tumor-supporting effects of the monocyte/macrophage lineage were previously reported in CLL [30, 45–47], and our current findings would extend this protective role to MBL. In addition, effects of monocytes on tumor survival were similarly observed in MBL and early stage CLL. Since the former exhibited increased inflammatory features, these may therefore be related to an increased antigen stimulation potentially associated with recent malignant transformation rather than to increased tumor-supporting effects of monocytes in MBL. However, the possibility that some of these inflammatory molecules act as tumor pro-survival signals cannot be discarded. It is also worth mentioning that in healthy conditions, macrophages promoted BCR-stimulated B cell proliferation [48] and monocytes exerted supporting effects on normal B cell survival [49]. In our experiments, monocytes from MBL and CLL patients increased normal B cell viability (median: 60% in total PBMCs vs. 40.7% in CD14-depleted PBMCs, P-value=0.043), which also suggests that supporting interactions between monocytes and B cells are not unique to tumor B cells. Therefore, culturing monocytes from healthy controls, MBL subjects and CLL patients together with the same tumor B cell clone would be helpful to definitely clarify if the differences in the inflammatory features that we observed among the three groups could be attributed to differences in tumor-supporting effects of monocytes.
Our findings also suggest that CD4+ T cells of MBL and early stage CLL are Th1 biased. We observed upregulated Th1 genes [26] and increased CXCR3+ CD4+ T cells, generally accepted as Th1 cells [27, 28]. Besides, MBL subjects displayed augmented serum levels of IFNγ, CXCL9–11, IL8 or TNFα, which are related to a Th1-monocytic response. As far as we know, this is the first time that a role for CD4+ Th1 cells is described in MBL, although prior investigations in CLL suggested a Th2 polarization [4, 50]. Nonetheless, other studies described a Th1 dominance in early stages of the disease [51], which is in accord with our results. Remarkably, Th1 cells can interact with monocytes/macrophages to induce a specific antitumor response in B cell tumors [52], and a recent study in CLL showed that activation of Th1 immunity was associated with clinical response to lenalidomide [53]. Tumoricidal effects of macrophages were also described in a mouse model of the disease [54]. Thus, the Th1-monocytic features that we observed in MBL could potentially restrain malignant expansions. However, when we cultured tumor cells together with the rest of PBMCs, increased tumor viabilities were observed compared to the same cell cultures without CD14+ cells, suggesting that interactions between T cells and monocytes may confer protective rather than antitumor roles in MBL and early stage CLL.
Besides, we identified increased CD4+ T cells with cytotoxic properties similarly in both MBL and early stage CLL. These cytotoxic features were specifically confirmed in CD14-depleted CD4+ T cells of MBL subjects by qRT-PCR, and in CD4+ T cells of MBL and CLL patients by flow cytometry. Additionally, our MBL group displayed the higher median percentage of effector CD27− CD4+ T cells. In line with this, a recent study demonstrated increased levels of CD27− CD4+ T cells mainly in MBL, and augmented CD27− granzyme B+ CD4+ T cells throughout all disease stages, including MBL [55]. Increased frequencies of CD4+ T cells expressing perforin had not been described in MBL before, although it is not surprising taking into account that perforin synergizes with granzymes in the same cytotoxic pathways. Other investigations also described cytotoxic CD4+ T cells in CLL [56], which were able to kill clonal B cells [57]. All these findings emphasize the non-exclusive role of the immune environment in supporting clonal B cell survival, although the global effect in MBL and early-stage CLL may rather be tumor-supporting, as suggested by the cell culture assays of this study. As previously highlighted, isolation of the different CD4+ T cell and monocyte subsets results mandatory to definitely clarify their specific effects on tumor development.
Taken together, we described an increased inflammatory response in MBL, mainly orchestrated by monocytes, that lessens in early stage CLL. The proinflammatory signature identified in MBL could represent an active role for self/foreign antigen stimulation in this pre-leukemic stage, potentially related to recent malignant transformation. Conversely, the possibility that these inflammatory molecules are associated with tumor-supporting effects of monocytes cannot be discarded. These findings offer novel insights into the very early stages of the disease, which will allow the development of future investigations addressed to completely understand its natural history and, ultimately, to develop pre-emptive strategies for CLL management.
Supplementary Material
Acknowledgements
The authors want to thank all patients and healthy volunteers for their participation in this study and Xavier Duran for statistical support. G. Blanco was funded by a grant from Fundació La Caixa and Fundación Española de Hematología y Hemoterapia (FEHH-Janssen). This work has been supported by the following grants: PI11/1621, PI15/437 and FEDER (PT13/0010/0005), Instituto de Salud Carlos III, Spanish Ministry of Economy and Competitiveness; “Xarxa de Bancs de tumors” sponsored by Pla Director d’Oncologia de Catalunya (XBTC); 2014/SGR585 and 2017/SGR437 from Generalitat de Catalunya.
Footnotes
Category for the Table of Contents
Immunobiology and Immunotherapy
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.te Raa GD, Tonino SH, Remmerswaal EB, et al. Chronic lymphocytic leukemia specific T cell subset alterations are clone-size dependent and not present in monoclonal B lymphocytosis. Leuk Lymphoma. 2012;53:2321–2325. [DOI] [PubMed] [Google Scholar]
- 2.Vardi A, Vlachonikola E, Karypidou M, et al. Restrictions in the T cell repertoire of chronic lymphocytic leukemia: high-throughput immunoprofiling supports selection by shared antigenic elements. Leukemia. 2017;31:1555–1561. [DOI] [PubMed] [Google Scholar]
- 3.Farace F, Orlanducci F, Dietrich PY, et al. T cell repertoire in patients with B chronic lymphocytic leukemia. Evidence for multiple in vivo T cell clonal expansions. J Immunol. 1994;153:4281–4290. [PubMed] [Google Scholar]
- 4.Görgün G, Holderried TA, Zahrieh D, Neuberg D, Gribben JG. Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Invest. 2005;115:1797–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramsay AG, Johnson AJ, Lee AM, et al. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest. 2008;118:2427–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramsay AG, Evans R, Kiaii S, Svensson L, Hogg N, Gribben JG. Chronic lymphocytic leukemia cells induce defective LFA-1-directed T cell motility by altering Rho GTPase signaling that is reversible with lenalidomide. Blood. 2013;121:2704–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riches JC, Davies JK, McClanahan F, et al. T cells from CLL patients exhibit features of T cell exhaustion but retain capacity for cytokine production. Blood. 2013;121:1612–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palma M, Gentilcore G, Heimersson K, et al. T cells in chronic lymphocytic leukemia display dysregulated expression of immune checkpoints and activation markers. Haematologica. 2017;102:562–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maffei R, Bulgarelli J, Fiorcari S, et al. The monocytic population in chronic lymphocytic leukemia shows altered composition and deregulation of genes involved in phagocytosis and inflammation. Haematologica. 2013;98:1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan XJ, Dozmorov I, Li W, et al. Identification of outcome-correlated cytokine clusters in chronic lymphocytic leukemia. Blood. 2011;118:5201–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sivina M, Hartmann E, Kipps TJ, et al. CCL3 (MIP-1α) plasma levels and the risk for disease progression in chronic lymphocytic leukemia. Blood. 2011;117:1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wierda WG, Johnson MM, Do KA, et al. Plasma interleukin 8 level predicts for survival in chronic lymphocytic leukaemia. Br J Haematol. 2003;120:452–456. [DOI] [PubMed] [Google Scholar]
- 13.Rawstron AC, Bennett FL, O’Connor SJ, et al. Monoclonal B lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008;359:575–583. [DOI] [PubMed] [Google Scholar]
- 14.Landgren O, Albitar M, Ma W, et al. B cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009;360:659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vardi A, Dagklis A, Scarfò L, et al. Immunogenetics shows that not all MBL are equal: the larger the clone, the more similar to CLL. Blood. 2013;121:4521–4528. [DOI] [PubMed] [Google Scholar]
- 16.Strati P, Shanafelt TD. Monoclonal B cell lymphocytosis and early stage chronic lymphocytic leukemia: diagnosis, natural history, and risk stratification. Blood. 2015;126:454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scarfò L, Dagklis A, Scielzo C, Fazi C, Ghia P. CLL-like monoclonal B cell lymphocytosis: are we all bound to have it? Semin Cancer Biol. 2010;20:384–390. [DOI] [PubMed] [Google Scholar]
- 18.Rissiek A, Schulze C, Bacher U, et al. Multidimensional scaling analysis identifies pathological and prognostically relevant profiles of circulating T cells in chronic lymphocytic leukemia. Int J Cancer. 2014;135:2370–2379. [DOI] [PubMed] [Google Scholar]
- 19.D’Arena G, Rossi G, Minervini MM, et al. Circulating regulatory T cells in “clinical” monoclonal B lymphocytosis. Int J Immunopathol Pharmacol. 2011;24:915–923. [DOI] [PubMed] [Google Scholar]
- 20.Blanco G, Vardi A, Puiggros A, et al. Restricted T cell receptor repertoire in CLL-like monoclonal B cell lymphocytosis and early stage CLL. OncoImmunology. 2018;7:e1432328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap) – A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. [DOI] [PubMed] [Google Scholar]
- 23.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. [DOI] [PubMed] [Google Scholar]
- 24.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gene expression Omnibus, a public functional genomics data repository supporting MIAME-compliant data submissions. Available from: http://www.ncbi.nlm.nih.gov/geo.
- 26.Ono C, Yu Z, Kasahara Y, Kikuchi Y, Ishii N, Tomita H. Fluorescently activated cell sorting followed by microarray profiling of helper T cell subtypes from human peripheral blood. PLoS One. 2014;9:e111405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonecchi R, Bianchi G, Bordignon PP, et al. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J Exp Med. 1998;187:129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin S, Rottman JB, Myers P, et al. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J Clin Invest. 1998;101:746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brusa D, Serra S, Coscia M, et al. The PD-1/PD-L1 axis contributes to T cell dysfunction in chronic lymphocytic leukemia. Haematologica. 2013;98:953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Attekum MHA, Terpstra S, Slinger E, et al. Macrophages confer survival signals via CCR1-dependent translational MCL-1 induction in chronic lymphocytic leukemia. Oncogene. 2017;36:3651–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stevens WB, Netea MG, Kater AP, van der Velden WJ. ‘Trained immunity’: consequences for lymphoid malignancies. Haematologica. 2016;101:1460–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Hreggvidsdottir HS, Palmblad K, et al. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc Natl Acad Sci U S A. 2010;107:11942–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suurmond J and Diamond B. Autoantibodies in systemic autoimmune diseases: specificity and pathogenicity. J Clin Invest. 2015;125:2194–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fais F, Ghiotto F, Hashimoto S, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102:1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ten Hacken E, Gounari M, Ghia P, Burger JA. The importance of B cell receptor isotypes and stereotypes in chronic lymphocytic leukemia. Leukemia. 2019;33:287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanemo Myhrinder A, Hellqvist E, Sidorova E, et al. A new perspective: molecular motifs on oxidized LDL, apoptotic cells, and bacteria are targets for chronic lymphocytic leukemia antibodies. Blood. 2008;111:3838–3848. [DOI] [PubMed] [Google Scholar]
- 37.Tsai HT, Caporaso NE, Kyle RA, et al. Evidence of serum immunoglobulin abnormalities up to 9.8 years before diagnosis of chronic lymphocytic leukemia: a prospective study. Blood. 2009;114:4928–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glenn MJ, Madsen MJ, Davis E, et al. Elevated IgM and abnormal free light chain ratio are increased in relatives from high-risk chronic lymphocytic leukemia pedigrees. Blood Cancer J. 2019;9:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas GD, Hamers AAJ, Nakao C, et al. Human blood monocyte subsets: A new gating strategy defined using cell surface markers identified by mass cytometry. Arterioscler Thromb Vasc Biol. 2017;37:1548–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee R, Barman PK, Thatoi PK et al. Non-classical monocytes display inflammatory features: Validation in sepsis and systemic lupus erythematous. Sci Rep. 2015;5:13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coscia M, Pantaleoni F, Riganti C, et al. IGHV unmutated CLL B cells are more prone to spontaneous apoptosis and subject to environmental prosurvival signals than mutated CLL B cells. Leukemia. 2011;25:828–837. [DOI] [PubMed] [Google Scholar]
- 42.Burger JA, Quiroga MP, Hartmann E, et al. High-level expression of the T cell chemokines CCL3 and CCL4 by chronic lymphocytic leukemia B cells in nurselike cell cocultures and after BCR stimulation. Blood. 2009;113:3050–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pede V, Rombout A, Vermeire J, et al. Expression of ZAP70 in chronic lymphocytic leukaemia activates NF-κB signalling. Br J Haematol. 2013;163:621–630. [DOI] [PubMed] [Google Scholar]
- 44.Morabito F, Mosca L, Cutrona G, et al. Clinical monoclonal B lymphocytosis versus Rai 0 chronic lymphocytic leukemia: A comparison of cellular, cytogenetic, molecular, and clinical features. Clin Cancer Res. 2013;19:5890–5900. [DOI] [PubMed] [Google Scholar]
- 45.Reinart N, Nguyen PH, Boucas J et al. Delayed development of chronic lymphocytic leukemia in the absence of macrophage migration inhibitory factor. Blood. 2013;121:812–821. [DOI] [PubMed] [Google Scholar]
- 46.Galletti G, Scielzo C, Barbaglio F et al. Targeting macrophages sensitizes chronic lymphocytic leukemia to apoptosis and inhibits disease progression. Cell Rep. 2016;14:1748–1760. [DOI] [PubMed] [Google Scholar]
- 47.Galletti G, Caligaris-Cappio F and Bertilaccio MTS. B cells and macrophages pursue a common path toward the development and progression of chronic lymphocytic leukemia. Leukemia. 2016;30:2293–2301. [DOI] [PubMed] [Google Scholar]
- 48.Craxton A, Magaletti D, Ryan EJ, Clark EA. Macrophage- and dendritic cell–dependent regulation of human B cell proliferation requires the TNF family ligand BAFF. Blood. 2003;101:4464–4471. [DOI] [PubMed] [Google Scholar]
- 49.Mueller CG, Boix C, Kwan WH, et al. Critical role of monocytes to support normal B cell and diffuse large B cell lymphoma survival and proliferation. J Leukoc Biol. 2007;82:567–575. [DOI] [PubMed] [Google Scholar]
- 50.Fayad L, Keating MJ, Reuben JM, et al. Interleukin-6 and interleukin-10 levels in chronic lymphocytic leukemia: correlation with phenotypic characteristics and outcome. Blood. 2001;97:256–263. [DOI] [PubMed] [Google Scholar]
- 51.Podhorecka M, Dmoszynska A, Rolinski J, Wasik E. T type 1/type 2 subsets balance in B cell chronic lymphocytic leukemia--the three-color flow cytometry analysis. Leuk Res. 2002;26:657–660. [DOI] [PubMed] [Google Scholar]
- 52.Haabeth OA, Lorvik KB, Hammarström C, et al. Inflammation driven by tumour-specific Th1 cells protects against B cell cancer. Nat Commun. 2011;2:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aue G, Sun C, Liu D, et al. Activation of Th1 immunity within the tumor microenvironment is associated with clinical response to lenalidomide in chronic lymphocytic leukemia. J Immunol. 2018;201:1967–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu QL, Buhtoiarov IN, Sondel PM, Rakhmilevich AL, Ranheim EA. Tumoricidal effects of activated macrophages in a mouse model of chronic lymphocytic leukemia. J Immunol. 2009;182:6771–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simões C, Silva I, Carvalho A, et al. Quantification and phenotypic characterization of peripheral blood Vδ1 +T cells in chronic lymphocytic leukemia and monoclonal B cell lymphocytosis. Cytometry B Clin Cytom. 2019;96:164–168. [DOI] [PubMed] [Google Scholar]
- 56.Porakishvili N, Roschupkina T, Kalber T, et al. Expansion of CD4+ T cells with a cytotoxic phenotype in patients with B-chronic lymphocytic leukaemia (B-CLL). Clin Exp Immunol. 2001;126:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Porakishvili N, Kardava L, Jewell AP, et al. Cytotoxic CD4+ T cells in patients with B cell chronic lymphocytic leukemia kill via a perforin-mediated pathway. Haematologica. 2004;89:435–443. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


