Abstract
Background
Rickettsia africae is a tick-borne bacterium that causes African tick-bite fever (ATBF) in humans. In southern Africa, the tick Amblyomma hebraeum serves as the primary vector and reservoir for R. africae and transmits the bacterium during any life stage. Previous research has shown that even when malaria has been dramatically reduced, unexplained acute febrile illnesses persist and may be explained by the serological evidence of rickettsiae in humans.
Methodology/Principal findings
We collected 12,711 questing Amblyomma larvae across multiple land use types in a savanna landscape in Eswatini. Our results show that host-seeking Amblyomma larvae are abundant across both space and time, with no significant difference in density by land use or season. We investigated the entomological risk (density of infected larvae) of ATBF from A. hebraeum larvae by testing over 1,600 individual larvae for the presence of R. africae using a novel multiplex qPCR assay. We found an infection prevalence of 64.9% (95% CI: 62.1–67.6%) with no land use type significantly impacting prevalence during the dry season of 2018. The mean density of infected larvae was 57.3 individuals per 100m2 (95% CI: 49–65 individuals per 100m2).
Conclusions
Collectively, our results demonstrate R. africae infected A. hebraeum larvae, the most common tick species and life stage to bite humans in southern Africa, are ubiquitous in the savanna landscape of this region. Increased awareness of rickettsial diseases is warranted for policymakers, scientists, clinicians, and patients. Early detection of disease via increased clinician awareness and rapid diagnostics will improve patient outcomes for travelers and residents of this region.
Author summary
African tick-bite fever (ATBF) is a widespread, endemic tick-borne rickettsiosis caused by the bacterium Rickettsia africae. ATBF is second only to malaria as the cause of acute febrile illness in travelers returning from sub-Saharan Africa, and R. africae seropositivity rate (which is indicative of past infection) is high in indigenous populations from rural areas. The primary vector of R. africae in southern Africa is the South African bont tick, Amblyomma hebraeum. This tick is a highly aggressive human biter during the larval life stage. In this study, we quantified the density of infected A. hebraeum larvae across the lowveld savanna in Eswatini. We found over half of tested A. hebraeum larvae were infected with R. africae, and the density of infected larvae can be high in multiple land uses, including wildlife preserves, ranches, and community grazing lands. This high likelihood of a bite by an infected A. hebraeum in the savanna of Eswatini suggests ATBF poses a significant risk to travelers and may contribute to unexplained acute febrile diseases in the region.
Introduction
Rickettsial diseases are vector-borne bacterial infections that cause acute febrile illness, and they are among the most common emerging or re-emerging zoonoses globally [1]. Rickettsial diseases are prevalent in tropical and subtropical regions of the world and disproportionately affect those living in poverty with limited access to modern health care [2]. For decades, rickettsial diseases have been an overlooked cause of morbidity, mortality, and economic losses in marginalized populations [3]. Despite the increasing awareness of the importance of rickettsial diseases, there remains a lack of information on the prevalence of rickettsiae in vector species across all infective life stages.
African tick-bite fever (ATBF), caused by the bacterium Rickettsia africae, is a neglected rickettsial disease transmitted by ticks in sub-Saharan Africa. Rickettsia africae belongs to the spotted-fever group (SFG) Rickettsia, which comprises a diverse group of pathogenic bacteria that can cause spotted fevers in humans. Rickettsia africae is predominately transmitted by the ticks, Amblyomma variegatum and Amblyomma hebraeum [4]. Both tick species can maintain rickettsiae within the tick population by vertical transmission of the bacteria from mother to egg [5,6]. Therefore, the distribution of the tick vectors determines the distribution of the R. africae pathogen. The distribution of A. variegatum spans western, central, and eastern parts of sub-Saharan Africa and the West Indies, whereas A. hebraeum is only in southern Africa [7].
ATBF is a neglected disease that can cause illness in travelers and rural inhabitants bitten by ticks in endemic regions [4,8,9]. Rickettsial infections are second only to malaria as the cause of acute febrile illnesses among travelers to sub-Saharan Africa [8]. Reports of ATBF in local populations are limited [4,9]. Still, studies that measure the seroprevalence of past Rickettsia infection indicate rates can reach 60–90% across the geographic range (central Africa: [10,11]; western Africa: [12]; southern Africa: [13,14]). Living and working in rural settings where cattle and Amblyomma ticks are present is linked with a heightened risk of R. africae infection. Other outdoor activities associated with vector contact, such as game hunting, can also increase risk [15,16]. The contribution of rickettsioses to undiagnosed acute febrile illnesses remains unknown due to the lack of laboratory diagnostic tests in rural and developing regions and the low reliability of test results even when the test is available [17]. In an area where malaria, the most common diagnosis for febrile patients in low-resource health care settings [18,19], has been dramatically reduced, unexplained acute febrile illnesses persist and may be explained by the serological evidence of rickettsiae in humans [20].
Amblyomma hebraeum larvae pose a significant risk of parasitism to humans as they are abundant and aggressive. A study in South Africa found A. hebraeum larvae were five times more abundant in the vegetation, over two times more abundant on human clothing, and inflicted more bites to humans than any other tick species or life stage [21]. Fully engorged females can lay between 6 and 18 thousand eggs [22], resulting in thousands of larvae available to attach to hosts. Larvae are small and may go undetected on the human body, resulting in longer attachment times. This extended time may lead to an increased potential for pathogen transmission, as the probability of transmission increases with time of attachment [23].
Members of SFG Rickettsia are relatively unique in their efficient transovarial transmission (passage from the adult female through the ovaries to the unfed larvae of the next generation). Amblyomma hebraeum and A. variegatum, naturally infected with R. africae, can maintain the bacterium through trans-stadial and transovarial transmission for multiple generations [5,6] as the majority of egg masses from R. africae-infected A. hebraeum females contain at least one R. africae positive egg [24]. The combination of high transovarial rates in transmission studies and high prevalence of infected ticks of all life stages from field collections has led to the conclusion that R. africae may be an endosymbiont of these tick vectors [25].
Previous studies on rickettsiae in southern African ticks have documented the presence of R. africae in A. hebraeum adults and nymphs [26,27]. Rickettsia africae infection in A. hebraeum is high, typically ranging from 60–80% [26–30]. Previous studies testing for R. africae in any region have either not included larvae [31,32] or failed to identify Amblyomma larvae to the species level [30,33]. Given that several species of Amblyomma ticks co-occur with A. hebraeum [34] and Amblyomma species can differ in R. africae prevalence [33,35], differentiating Amblyomma larvae to species level before pathogen analysis is essential for accurate estimates of infection prevalence.
This study investigated the entomological risk [36] of ATBF from A. hebraeum larvae across multiple land use types in a savanna landscape. We estimated entomological risk as the density of infected larvae (DIL) or probability of an infected A. hebraeum bite in multiple rural land use types in Eswatini. As A. hebraeum larvae are small, abundant, and aggressive ticks that can spread ATBF to humans, assessment of pathogen prevalence is a crucial step towards understanding the risk of rickettsial diseases in endemic areas. The entomological risk may differ by land use because microhabitat and host community composition, both of which are influenced by land use [37], can impact larval tick survival, development, and activity [38]. Identifying hazards across multiple land uses will aid in designing disease prevention measures through vector control and education awareness programs, as people’s interaction with the landscape can differ due to their occupation, recreational activities, and economic status.
Methods
Study area and sampling design
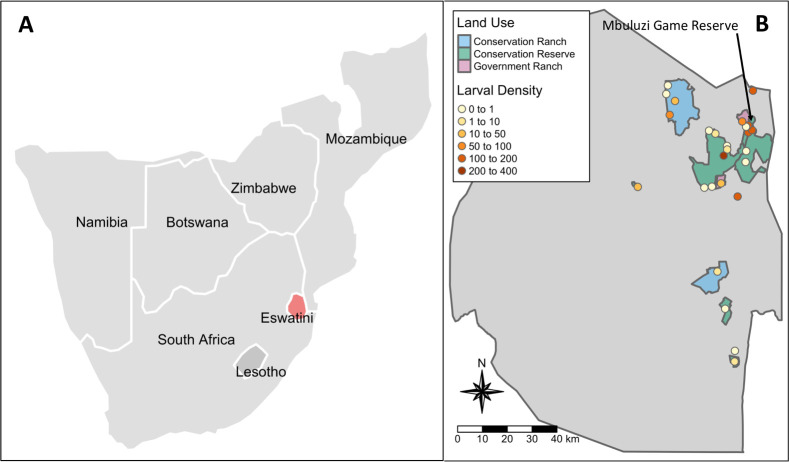
We conducted this study in the lowveld region of Eswatini. This region experiences mild, dry winters (8–26°C; 0-50mm) and hot, wet summers (15–33°C; 200-500mm) [39–41]. To quantify entomological risk over space, we selected 26 sites representing four common land uses of lowveld savanna grasslands in Eswatini (wildlife conservation reserves, cattle ranches, conservation ranches that maintain a mixture of cattle and game animals, and community Swazi Nation Lands near rural settlements often used for livestock grazing). We sampled all 26 sites during the dry season of 2018. All life stages of ticks were abundant in savanna grasslands, but not in agricultural areas, during the dry season [27,38]. To quantify entomological risk over time, we also sampled a wildlife conservation reserve, Mbuluzi Nature Reserve, five times over two years. We sampled two or three plots per site, where each site included 2 to 6 100-meter transects of drag sampling. We used a tape measure to mark straight transects and maintain 10 meters between transects within a plot. We accounted for differences in sampling effort per site in our density estimates. We selected the temporal sampling intervals to represent the dry and wet seasons in Eswatini.
We obtained research permits to collect ticks inside wildlife conservation reserves from the Eswatini National Trust Commission. We received verbal permission from landowners to collect ticks on government-owned cattle ranches, conservation ranches, and community Swazi Nation Lands. Permission to import dead, unengorged ticks to the United States was granted by the United States Department of Agriculture, the United States Centers for Disease Control and Prevention, and the United States Fish and Wildlife Service.
Our tick sampling technique has been previously described [27]. In short, to mimic how humans may encounter host-seeking ticks, we sampled ticks by dragging a 1m2 white flannel cloth across the vegetation allowing ticks to attach to the cloth as it passed. We inspected the cloth every 10 meters and spent a maximum of two minutes transferring ticks directly into 90% molecular-grade ethanol. Samples from unique transects and sampling sessions were collected and stored in individually labeled vials. We established a maximum time of two minutes to collect ticks from the cloth due to the large numbers of larvae present at some of the sites. We collected a representative sample of larvae into ethanol along the transect within a reasonable time using this method. We removed all larvae remaining on the cloth at the end of a transect using a lint roller. The larvae collected in vials were identified morphologically to the genus-level using taxonomic keys [7,42] and stored at -20°C. We counted the total number of larvae collected on lint rollers and extrapolated the total number belonging to each genus from the proportion of larvae collected into a vial from the identical transect and sampling session.
Sample size determination for pathogen screening
To determine the number of individual Amblyomma larvae to extract per site and sampling session, we calculated sample size using the formula:
where Z = value from standard normal distribution corresponding to desired confidence level, P = proportion of infected larvae, and e = desired precision or half desired confidence level width [43]. We used an estimated pathogen prevalence of 50% as a recent study from the same region found 19 out of 38 A. hebraeum nymphs were positive for R. africae [27]. This value is conservative because, for prevalence studies, the appropriate sample size is maximal when the true prevalence is 50% and decreases as the true prevalence approaches 0% or 100%. We selected the desired precision of 0.1 with a 95% confidence level. We determined that the appropriate sample size was n = 97 per site. To represent and replicate all land use types, we selected four sites in wildlife conservation reserves, two in cattle ranches, two in conservation ranches, and three in community lands, all sampled in the dry season of 2018, for pathogen screening. To quantify R. africae prevalence over time, we screened at least 97 individual Amblyomma larvae collected from Mbuluzi Game Reserve over each sampling session (dry seasons of 2017, 2018, 2019, and wet seasons of 2017/18 and 2018/19).
Molecular analyses
Primer and probe design
To determine the presence of Rickettsia africae in Amblyomma hebraeum larvae, we designed a multiplex qPCR assay to include R. africae specific primers and probe to target a 119 bp segment of the ompB gene [25], A. hebraeum specific primers and probe to target a 151 bp segment of the CO1 gene, and Ixodidae tick primers and probe to target a 150 bp segment of the 16S gene. We used the 16S primers and probe as endogenous controls for each PCR reaction. The primer-probe combination specifically amplifying R. africae DNA was obtained from a previously published qPCR assay [25]. To design a primer-probe combination to detect A. hebraeum DNA, we generated potential primer-probe combinations using CO1 gene sequence of A. hebraeum (KY457513). Candidate primer-probe sets were then evaluated for specificity to A. hebraeum and exclusion of co-occurring Amblyomma species using available CO1 sequences (Amblyomma exornatum: MN150166; Amblyomma marmoreum: KY457516; Amblyomma tholloni: KT307493; Amblyomma transversale: MN150172; Amblyomma variegatum: GU062743). To design a primer-probe combination for use as an extraction control, we generated potential primer-probe combinations by using the 16S gene of 14 tick species (S1 Table). The forward primer region was manually edited to include mixed nucleotides and enable detection of DNA from all hard tick genera tested. We generated all primer-probe combinations using the PrimerQuest Tool from Integrated DNA Technologies. We used the following criteria for identifying candidate primer-probe sets: optimal primer melting temperature of 61°C, optimal probe melting temperature of 68°C, and an amplicon size ranging from 100–150 bp.
DNA extraction and pathogen detection
We performed all molecular analyses at the Molecular Ecology Laboratory at the University of Florida, FL, USA. We performed 1637 individual extractions of whole Amblyomma larvae using an adapted protocol from the Gentra Puregene Tissue Kit (Qiagen). Before extraction, larvae were rinsed in diH2O and 70% ethanol and dried. We made a single cut across the scutum using a sterile scalpel to preserve the tick exoskeleton for further morphological identification.
qPCR was performed in a 10 μL reaction volume using 2x QuantiTect Multiplex PCR Master Mix and 2 μL of template DNA (Table 1). All reaction plates included one positive control of A. hebraeum DNA, a three-point standard curve of R. africae plasmid positive controls at 103, 105, and 107 copies per reaction, and one negative control (molecular grade water). We performed the real-time PCR-amplification assays using an Applied Biosystems QuantStudio 5 Real-Time PCR system (Thermo Fisher Scientific).
Table 1. The primers, probes, and thermocycling conditionsa that were used to detect Rickettsia africae, Rickettsia, and tick DNA in a novel multiplex qPCR assay for this study.
| Target organism | Target gene | Primer name | Primer orientation | 5’-3’ sequence | Reference | Concentration (nM) |
|---|---|---|---|---|---|---|
| Tick | 16S | Tick-F2 | Forward | CTCTAGGGATAACAGCGTWATAWT | This study | 600 |
| Tick-R | Reverse | GTCTGAACTCAGATCAAGTAGG | 600 | |||
| Tick-P1 | Probe | 6-FAM/TGCGACCTC/ZEN/GATGTTGGATTAGGA/3’IowaBlack | 250 | |||
| Amblyomma hebraeum | CO1 | Aheb_F | Forward | CATCATAATTGGCGGGTTTG | This study | 400 |
| Aheb_R | Reverse | AGTAAACAAAGAGATGGTGGTA | 400 | |||
| Aheb_P | Probe | Cy5/TGACTAGTT/TAO/CCAATTATGCTAGGTGCCC/3’IowaBlack | 250 | |||
| Rickettsia africae | ompB | Raf1797F | Forward | TTGGAGCTAATAATAAAACTCTTGGAC | [25] | 100 |
| Raf1915R | Reverse | GAATTGTACTGCACCGTTATTTCC | 100 | |||
| Raf1879P | Probe | HEX/CGCGATGTTAATAGCAACATCACC GCCACTATCGCG/Black Hole Quencher | 250 |
aThermocycling conditions were as follows: initial denaturation of 95°C for 15 minutes followed by 40 cycles of 95°C for 45 seconds and 60°C for 90 seconds.
To evaluate the specificity of the R. africae primer-probe set, we screened DNA from previously identified samples of R. africae, Rickettsia amblyommatis, Rickettsia belli, Candidatus Rickettsia barbariae, Rickettsia conorii, Rickettsia massiliae, Rickettsia parkeri, and Rickettsia rhipicephali. Furthermore, we used conventional PCR (cPCR) to amplify the ompB gene of 18 randomly selected positive samples as previously described [27]. To evaluate the specificity of the A. hebraeum primer-probe set, we screened DNA from previously identified ticks, including five species of Amblyomma (Amblyomma americanum, A. hebraeum, Amblyomma maculatum, Amblyomma marmoreum, and Amblyomma rotumdatum), Dermacentor variabilis, Haemaphysalis elliptica, Ixodes scapularis, and six species of Rhipicephalus (Rhipicephalus appendiculatus, Rhipicephalus decoloratus, Rhipicephalus maculatus, Rhipicephalus microplus, Rhipicephalus sanguineus, Rhipicephalus simus). We used cPCR to amplify the CO1 gene of 13 randomly selected individuals positive for the A. hebraeum probe and seven randomly selected individuals negative for the A. hebraeum probe using previously described methods [27]. To further characterize the Amblyomma larvae negative for the A. hebraeum probe, we used cPCR to amplify the 12S gene and ITS2 gene of two randomly selected individuals using previously described methods [27]. All cPCR amplicons were purified using Exo-SAP (New England Biolabs) and sent for sequencing with the same primers (Eurofins Genomics). We assembled sequences with Geneious Prime 2021.1.1.
To evaluate the sensitivity of the detection of R. africae in the qPCR assay, we obtained a synthetic gene strand spanning the R. africae ompB target region (Eurofins Genomics). We used the restriction enzyme EcoRI (New England BioLabs) to linearize the plasmid. Ten-fold serial dilutions ranging from 1 x 107 copies to 1 copy R. africae ompB DNA in triplicate were amplified using the previously mentioned qPCR conditions. We identified the detection limit as the lowest concentration of DNA with detectable fluorescence above the background prior to cycle 40. We calculated goodness of fit (R2) for the standard curve and reaction efficiency (E) based on the slope of the standard curve.
Data analyses
Density of larvae
To calculate the density of questing Amblyomma larvae (hereafter: DOL) per 100m2 for each site, we divided the number of collected Amblyomma larvae on a transect by the total distance sampled in m2 and multiplied by 100. The mean and variance of DOL were then calculated at the site level using the transect-level densities. To assess the influence of the number of unique transects on DOL, we evaluated the association between the number of unique transects per site and the mean DOL and variance of DOL using the Kruskal-Wallis test.
To investigate the relationship between DOL with a categorical variable representing land use (conservation reserve, cattle ranch, conservation ranch, community land), we used the DOL for each site collected during the dry season of 2018. We used negative binomial generalized linear mixed models to account for overdispersion in the count data. We modeled log-transformed DOL with land use as a fixed variable and location (i.e., name of the specific reserve, as more than one sampling site in some conservation areas) as a random variable. We checked model fit by simulating the residuals from the model and quantifying deviation. The null model and land use model were ranked using Akaike Information Criterion (AIC) and compared by computing the analysis of variance.
We obtained monthly precipitation data from the Eswatini Meteorological Services weather station located within the study area (Tabankulu) from 2016 to 2019. We calculated the total wet season (October to March) and the total dry season (April to September) precipitation for each year over the four years. To test for seasonal variability (wet vs. dry) in DOL, we conducted a two-sample t-test. Then, we used a simple linear regression to test if the current season total precipitation, the total precipitation 12 months prior, or the total precipitation 24 months prior significantly predicted DOL per sampling session in Mbuluzi Game Reserve.
Prevalence of R. africae and density of infected larvae
We removed all individual larval samples with a cycling threshold (CT) > 40 for the tick endogenous control from further analysis. To calculate the infection prevalence (hereafter: LIP) of A. hebraeum larvae with R. africae, we included only samples with a CT < 38 for the A. hebraeum qPCR probe. LIPs with 95% exact binomial confidence intervals (95%CI) were calculated at the site-level using Fisher’s exact test [44]. Using the Kruskal-Wallis test, we evaluated the association between the number of transects from which A. hebraeum larvae were collected per site and LIP. We also used a simple linear regression to test if DOL significantly predicted LIP.
We calculated the density of infected larvae (hereafter: DIL) by multiplying the DOL and LIP for each site. We used binomial and negative binomial generalized linear models to investigate the relationship of both LIP and DIL with land use. Model fit was checked by simulating the residuals from the model and quantifying deviation. The null model and land use model were ranked using AIC. To test for seasonal variability (wet vs. dry) in LIP and DIL, we conducted a two-sample t-test. We used a simple linear regression to test if the current season total precipitation, the total precipitation six months prior, or the total precipitation 12 months prior significantly predicted LIP and DIL. We performed all statistical analyses using the R software version 4.0.5 [45] and implemented models using the package GLMMadpative [46].
Results
Density of larvae
We collected 12,711 Amblyomma larvae across 19 of the 26 sites sampled during the dry season of 2018. We determined the apparent proportion of A. hebraeum larvae out of all Amblyomma larvae based on a subset of our dataset set to be 97%. No Amblyomma larvae were detected in seven sites. The average DOL across all sites during the dry season of 2018 was 51 ± 16 individuals per 100m2 (Fig 1). The highest average DOL from a single site was 333 individuals per 100m2 in Hlane Royal National Park conservation reserve (S2 Table). We observed no significant difference in DOL among land use categories, as the null model outperformed the model including land use (null model AIC = 111.0; LU model AIC = 114.7; anova p-value = 0.5) (S3 Table). Neither the mean nor variance of Amblyomma larval density were significantly associated with the number unique transects sampled per site (mean DOL: H(5) = 3.6, p = 0.6; variance DOL: H(5) = 3.9, p = 0.6).
Fig 1. Study area.
(A) Map of southern Africa with Eswatini indicated in red and (B) map of Eswatini indicating land use and the density of Amblyomma larvae per 100m2 at sampling sites during the dry season of 2018. Sampling sites not located within a conservation reserve, cattle ranch, or conservation ranch were in community lands. Made with Natural Earth (https://www.naturalearthdata.com.
DOL varied seasonally and annually over the five sampling sessions spanning three dry seasons and two wet seasons in Mbuluzi Game Reserve. We observed higher DOL in the wet seasons than dry seasons, but this difference was not statistically significant (mean dry = 73 ± 23 per 100m2; mean wet = 182 ± 90 per 100m2; t-test: t(4) = -1.2; p = 0.43). The wet season of 2015/16 received approximately 62% less rainfall than an average wet season. The simple linear regression of the mean precipitation 24 months prior and DOL was statistically significant (ß = 0.74; R2 = 0.88; F(1,3) = 21.5; p = 0.02) (S4 Table). The high DOL in the wet season of 2018/19 drove this relationship. Neither the linear regression testing precipitation 12 months prior nor current precipitation significantly predicted DOL, but both showed positive relationships with wetter years resulting in higher DOL.
qPCR assay validation
The tick primer-probe set successfully amplified DNA from all tick species tested. The A. hebraeum primer-probe set only amplified DNA from A. hebraeum. We observed no amplification for any other tick species, including the other species of Amblyomma. The R. africae specific primer-probe set amplified R. africae DNA, but not any other species of Rickettsia DNA tested. The limit of detection for R. africae primer-probe set in the multiplex assay was 1000 copies of the linearized DNA plasmid. The standard curve had a goodness of fit of R2 = 0.998 and reaction efficiency of E = 58.5%.
Molecular identification of Amblyomma larvae
1558 out of 1605 Amblyomma larvae included in molecular analyses were identified as A. hebraeum using the multiplex qPCR assay. We validated the identification of 13 randomly selected A. hebraeum individuals by sequencing the CO1 gene and finding 100% similarity (644 out of 644 base pairs) to A. hebraeum [KY457513]. From the subset of Amblyomma larvae that did not fluoresce the A. hebraeum probe, we sequenced the CO1 gene from 7 randomly selected individuals and found all identical and had a 95.2% similarity (613 out of 644 base pairs) to Amblyomma marmoreum [KY457515]. The 12S and ITS2 genes from two individuals of Amblyomma larvae had 96.5% similarity (360 out of 373 base pairs) to A. marmoreum [KY457515] and 99.9% similarity (1023 out of 1024 base pairs) to A. marmoreum [KY457491], respectively. We archived our Amblyomma marmoreum sequences to GenBank under the accession numbers: OM315185 (CO1), OM320577 (12S), and OM320578 (ITS2).
R. africae infection prevalence in Amblyomma larvae
We quantified LIP at 11 sites and found a study-wide infection prevalence of 64.9% (95% CI: 62.1–67.6%) for R. africae in A. hebraeum larvae. The highest LIP from a single site was 99% (95% CI: 94.6–100%) in a conservation ranch, and the lowest LIP was 1% (95% CI: 0–5.5%) in community lands near the rural settlement of Lomahasha (S2 Table). Across all sites, there was no significant relationship between DOL and LIP (R2 = 0.22, F(1,9) = 2.6, p = 0.14) and LIP was not significantly associated with land use as the null model outperformed the model including land use (null model AIC = 111.8; LU model AIC = 123.8) (S5 Table). Neither the mean nor variance of LIP were significantly associated with the number of unique transects from which we collected and tested larvae from each plot (mean LIP: H(7) = 4.9, p = 0.7; variance LIP: H(6) = 5.9, p = 0.4).
In Mbuluzi Game Reserve, LIP ranged from 51.6% (95% CI: 44.8–58.3%) during the dry season of 2018 to 90.5% (95% CI: 82.8–95.6%) in the wet season of 2018/19 (S6 Table). Overall, we observed no significant difference in LIP between the two seasons (mean dry = 73.8% ± 11.3%; mean wet = 73.6% ± 17.0%; t-test: t(4) = 0.01; p = 0.99). We identified one out of 46 A. marmoreum larvae to be positive for R. africae (both tick and R. africae identification confirmed by conventional PCR and sequencing) for a prevalence of 2.2% (95% CI: 0.1–11.5%).
To further validate the specificity of our assay, we submitted 18 randomly selected samples positive for R. africae for Sanger sequencing using the conventional genus-wide Rickettsia ompB PCR assay. 17 of the 18 samples [OL505022] had 100% similarity (829 out of 829 base pairs) to R. africae [CP001612], and one sample [OL505023] with two base pairs differences had 99.8% similarity to R. africae [CP001612].
Density of infected larvae
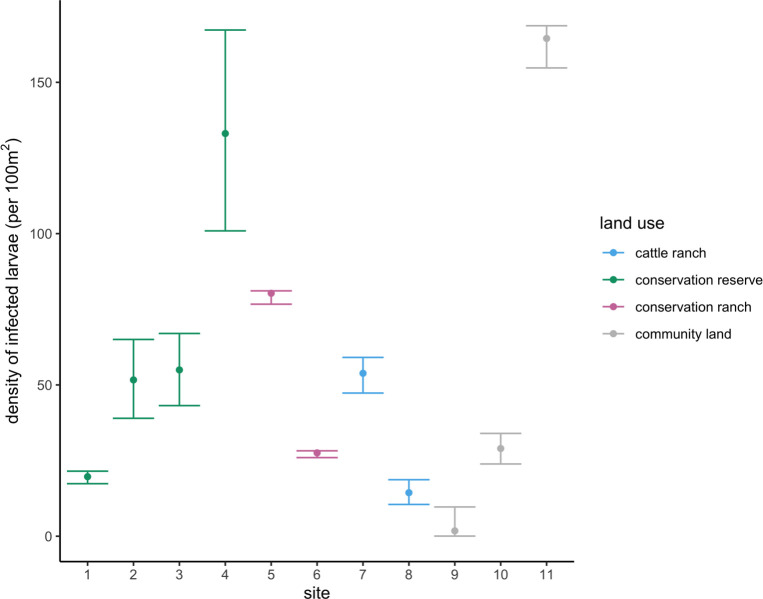
The average DIL across all sites during the 2018 dry season was 57.3 (95% CI: 49–65) individuals per 100m2 (S2 Table). The highest DIL from a single site was 165 (95% CI: 154–169) individuals per 100m2 in community lands near the rural settlement Sitsatsaweni (Fig 2). DIL was not significantly associated with land use as the null model outperformed the model that included land use (null AIC = 42.5; LU model AIC = 48.1) (S7 Table). In Mbuluzi Game Reserve, DIL ranged from 31.3 (95% CI: 28–33) individuals per 100m2 during the 2019 dry season to 246 (95% CI: 225–260) individuals per 100m2 during the 2018/19 wet season (S6 Table). We observed higher DIL in the wet seasons than in the dry seasons, but this difference was not statistically significant (mean dry = 48.4 ± 8.7 per 100m2; mean wet = 149 ± 97.2 per 100m2; t-test: t(4) = -1.0; p = 0.49). The simple linear regression testing if the mean precipitation 24 months prior predicted DIL was statistically significant (ß = 0.71; R2 = 0.88; F(1,3) = 21.7; p = 0.02) (S4 Table). Neither the linear regression testing precipitation 12 months prior nor current precipitation significantly predicted DIL, but both showed positive relationships with wetter years resulting in higher DIL.
Fig 2. Density of Amblyomma hebraeum larvae infected with Rickettsia africae at the 11 sampling sites surveyed in the dry season of 2018, from which we estimated infection prevalence.
There was no significant difference by land use category.
Discussion
This study detected large numbers of A. hebraeum larvae and a high prevalence of R. africae DNA in A. hebraeum larval ticks collected in Eswatini. We demonstrated that A. hebraeum larvae are abundant, ubiquitous ticks and act as the primary vector of R. africae in southern Africa. These results illustrate that these aggressive host-seeking larvae are widespread in the lowveld savanna of Eswatini and, due to the high infection rate, likely play a significant role in human infection with R. africae.
We observed a large variance in the density of Amblyomma larvae across our study due to the patchy distribution of larval ticks across space. Despite this variability, we found ticks in all land use types (conservation areas, ranches, and community lands) with high densities of Amblyomma larvae infected with R. africae. The high densities of infected ticks suggest significant entomological risk of ATBF regardless of land use. The variability of the density of infected larvae was driven by the variability in the density of larvae, as larval infection prevalence was high over both space and time. Amblyomma hebraeum represented the vast majority (~95%) of our Amblyomma larvae included in molecular analysis. Overall, the high contact rate with infected larval A. hebraeum reported in this study shows that R. africae transmission to humans may be an undiagnosed public health issue in Eswatini.
Apart from traveler cases, there are limited studies on the epidemiology of R. africae infections. To date, there are no published records describing disease incidence of ATBF or seroprevalence of R. africae in local populations from Eswatini. However, the seroprevalence of past infection with SFG Rickettsia in people from a rural area in South Africa, approximately 100 km away from Eswatini, was 63–92% [13]. While the morbidity and mortality caused by R. africae in communities across sub-Saharan Africa are unknown, cases of SFG Rickettsia can be fatal [47]. Elsewhere in sub-Saharan Africa, a study from Tanzania found 57 out of 641 (8.9%) patients with fever and paired serology had R. africae infection [9]. Rickettsial infections are a reality in sub-Saharan Africa, but the limited surveillance for nonmalarial febrile illnesses severely limits our understanding of the epidemiology and impact of diseases caused by R. africae and other SFG Rickettsia.
The prevalence of R. africae in Amblyomma ticks is consistent with previous studies from southern Africa. While most of these studies only tested the infection rate for the adult life stage, our prevalence range of 52–90% in larvae is comparable to that found in adults (60–80%) [24,28–30,48]. This result provides evidence that the proportion of infected individuals remains relatively constant over both life stages and generation, suggesting efficient transstadial and transovarial transmission in nature. The larval sampling and calculation of larval infection prevalence across time were limited to sites within a single conservation area. While this sampling scheme limits the interpretation of our data, we provide empirical evidence that infected A. hebraeum ticks are searching for hosts during both the wet and dry seasons, indicating a continuous risk of transmission throughout the year in this study region. Ultimately, this result supports the role of A. hebraeum ticks as the main reservoir of R. africae in southern Africa.
For this study, we selected sites in savanna grasses where ticks could be present in the vegetation, and human activity could result in exposure to ticks. While we tested an adequate number of individual samples for each site to achieve a 95% confidence interval within +/- 0.1 of the prevalence estimate, more sampling locations may have provided additional power to differentiate the density of larvae or the infection prevalence of larvae between land use types. Despite these limitations, we found large numbers of A. hebraeum larvae and a high infection prevalence of R. africae across the study area and duration.
The sampling associated with this study found seven different tick species collected as adults or nymphs [27]. Tick species richness was greatest in wildlife conservation areas (n = 7) and lowest in cattle-only ranches and community lands (n = 4). Tick-borne pathogen testing of adults and nymphs detected Anaplasma, Ehrlichia, Rickettsia, Babesia, Heptatozoon, and Theileria DNA. Detected Rickettsia species included three described species (R. africae, Rickettsia conorii, and Rickettsia massiliae), one candidate species (Ca. Rickettsia barbariae), and three undescribed genotypes of Rickettsia. Rickettsia africae was detected in A. hebraeum, with 19/38 (50%) of nymphs and 1/12 (8.3%) of adults testing positive [27].
The novel multiplex qPCR assay designed in this study demonstrated high specificity as there was no cross-reactivity of the R. africae primer-probe set with other species of Rickettsia. The A. hebraeum primer-probe set also did not cross-react with other tick species. The sensitivity and reaction efficiency of the assay fell below desired levels for diagnostic assays [49]. The less-than-ideal sensitivity of the assay may have resulted in false negatives in this study. The low sensitivity means that even more than the detected 65% of A. hebraeum larvae may indeed be infected with R. africae and further increase the risk of transmission. Nonetheless, the assay was able to identify the high prevalence of R. africae in our samples.
This study’s collection included the second published record of A. marmoreum in Eswatini and the first A. marmoreum larvae from the vegetation. These samples also included the first documentation of R. africae infection from A. marmoreum. While A. marmoreum has a lower infection prevalence of R. africae than A. hebraeum, our results show that larval A. marmoreum could play a role as a vector of ATBF as larvae are known to attach to humans [21] and can be abundant in the vegetation [50].
Previous studies found the peak of A. hebraeum larval activity in vegetation in the Eastern Cape to the south of Eswatini and Kruger National Park, South Africa to the north of Eswatini is highest during the wet season (November to January) and is lowest during the dry season (June to October) [51,52]. While our study did find the density of Amblyomma larvae to be highest during the wet season, the regional drought that affected the region in 2015 and 2016 may have influenced the relationship. A long-term (~13 years) study on the density of questing ticks also observed lower numbers of questing A. hebraeum larvae in the years immediately following a drought [51]. Our tick density and rainfall observations 24 months before sampling were strongly correlated, while current rainfall and rainfall 12 months prior were only weakly correlated. This relationship suggests that there may be a multi-year lag in tick population recovery after extreme weather events. Furthermore, seasonal rainfall may not be the primary determinant of questing ticks, and climate-induced variation in host populations may also have influenced the observed relationship. Over time, the variability in tick numbers may reflect the effect of climatic conditions on free-living ticks and their hosts.
Conclusion
The findings from this study add further evidence that A. hebraeum is an abundant tick species, and the high percentage of infected larval ticks with R. africae increases the probability an infected tick will bite humans. This work, along with the documentation of high seroprevalence in humans [10–14], adds to the evidence that rickettsiosis contributes to febrile bacterial disease in Africa and the impact of its neglect on patient health globally is potentially substantial. There is a need to increase awareness of the importance of the larval life stage of A. hebraeum in the transmission of ATBF to people in southern Africa, including tourists who engage in outdoor activities, locals who work in conservation areas and cattle ranches, and local people who live in rural settings with either domestic or wild animals near their home. Overall, rickettsioses, specifically African tick-bite fever, warrant increased recognition by policymakers, funders, healthcare workers, and scientists.
Supporting information
(DOCX)
For land use, wildlife refers to wildlife conservation areas, and mixed refers to conservation lands with both cattle and wildlife. Mean DOL = mean density of larvae per 100m2. No. tested = the number of individual larvae screened for R. africae using the multiplex qPCR. No. positive = the number of individual larvae positive for R. africae. LIP = larval infection prevalence. DIL = density of infected larvae per 100m2.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We would like to thank all the landowners who granted permission to work on their respective properties and the numerous field assistants who aided in collecting this field data. We would like to thank Zoe White for her technical advice in developing laboratory assays. We also thank the staff of Savanna Research Center and All Out Africa for their logistical support in Eswatini.
Data Availability
Data available at Open Science Framework. https://doi.org/10.17605/OSF.IO/CG7MVhttps://osf.io/cg7mv/.
Funding Statement
KJL was supported by the National Science Foundation Graduate Research Fellowship under grant no. DGE-1842473 and the University of Florida’s Center for African Studies’ Summer Pre-Dissertation Research Award. This study was funded by NSF IRES grant (no. 1459882) (to SMW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Walker DH, Ismail N. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nature Reviews Microbiology. 2008;6(5):375–86. doi: 10.1038/nrmicro1866 [DOI] [PubMed] [Google Scholar]
- 2.Xu G, Walker DH, Jupiter D, Melby PC, Arcari CM. A review of the global epidemiology of scrub typhus. Plos Neglected Tropical Diseases. 2017;11(11). doi: 10.1371/journal.pntd.0006062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salje J, Weitzel T, Newton PN, Varghese GM, Day N. Rickettsial infections: A blind spot in our view of neglected tropical diseases. Plos Neglected Tropical Diseases. 2021;15(5). doi: 10.1371/journal.pntd.0009353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jensenius M, Fournier PE, Kelly P, Myrvang B, Raoult D. African tick bite fever. Lancet Infectious Diseases. 2003;3(9):557–64. doi: 10.1016/s1473-3099(03)00739-4 [DOI] [PubMed] [Google Scholar]
- 5.Socolovschi C, Huynh TP, Davoust B, Gomez J, Raoult D, Parola P. Transovarial and trans-stadial transmission of Rickettsiae africae in Amblyomma variegatum ticks. Clinical Microbiology and Infection. 2009;15:317–8. doi: 10.1111/j.1469-0691.2008.02278.x [DOI] [PubMed] [Google Scholar]
- 6.Kelly PJ, Mason PR. Transmission of a spotted-fever group Rickettsia by Amblyomma hebraeum (Acari, Ixodidae). Journal of Medical Entomology. 1991;28(5):598–600. doi: 10.1093/jmedent/28.5.598 [DOI] [PubMed] [Google Scholar]
- 7.Walker AR. Ticks of domestic animals in Africa: a guide to identification of species: Bioscience Reports Edinburgh; 2003. [Google Scholar]
- 8.Freedman DO, Weld LH, Kozarsky PE, Fisk T, Robins R, von Sonnenburg F, et al. Spectrum of disease and relation to place of exposure among ill returned travelers. New England Journal of Medicine. 2006;354(2):119–30. doi: 10.1056/NEJMoa051331 [DOI] [PubMed] [Google Scholar]
- 9.Pisharody S, Rubach MP, Carugati M, Nicholson WL, Perniciaro JL, Biggs HM, et al. Incidence Estimates of Acute Q Fever and Spotted Fever Group Rickettsioses, Kilimanjaro, Tanzania, from 2007 to 2008 and from 2012 to 2014. American Journal of Tropical Medicine and Hygiene. 2022;106(2):494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupont HT, Cornet JP, Raoult D. Identification of Rickettsiae from ticks collected in the Central African Republic using the polymerase chain reaction. American Journal of Tropical Medicine and Hygiene. 1994;50(3):373–80. doi: 10.4269/ajtmh.1994.50.373 [DOI] [PubMed] [Google Scholar]
- 11.Ndip LM, Bouyer DH, Da Rosa A, Titanji VPK, Tesh RB, Walker DH. Acute spotted fever rickettsiosis among febrile patients, Cameroon. Emerging Infectious Diseases. 2004;10(3):432–7. doi: 10.3201/eid1003.020713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niang M, Parola P, Tissot-Dupont H, Baidi L, Brouqui P, Raoult D. Prevalence of antibodies to Rickettsia conorii, Rickettsia africae, Rickettsia typhi and Coxiella burnetii in Mauritania. European Journal of Epidemiology. 1998;14(8):817–8. doi: 10.1023/a:1007571412030 [DOI] [PubMed] [Google Scholar]
- 13.Simpson GJG, Quan V, Frean J, Knobel DL, Rossouw J, Weyer J, et al. Prevalence of selected zoonotic diseases and risk factors at a human-wildlife-livestock interface in Mpumalanga Province, South Africa. Vector-Borne and Zoonotic Diseases. 2018;18(6):303–10. doi: 10.1089/vbz.2017.2158 [DOI] [PubMed] [Google Scholar]
- 14.Noden BH, Tshavuka FI, van der Colf BE, Chipare I, Wilkinson R. Exposure and risk gactors to Coxiella burnetii, Spotted Fever Group and Typhus Group Rickettsiae, and Bartonella henselae among volunteer blood donors in Namibia. Plos One. 2014;9(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensenius M, Fournier PE, Vene S, Hoel T, Hasle G, Henriksen AZ, et al. African tick bite fever in travelers to rural sub-equatorial Africa. Clinical Infectious Diseases. 2003;36(11):1411–7. doi: 10.1086/375083 [DOI] [PubMed] [Google Scholar]
- 16.Ndip LM, Biswas HH, Nfonsam LE, LeBreton M, Ndip RN, Bissong MA, et al. Risk factors for African tick-bite fever in rural Central Africa. American Journal of Tropical Medicine and Hygiene. 2011;84(4):608–13. doi: 10.4269/ajtmh.2011.10-0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trataris-Rebisz AN, Rossouw J, Markotter W, Frean JA, Weyer J. Spotted fever rickettsiosis in South Africa: Evaluation of laboratory diagnostic capacity and inter-laboratory comparison of serological testing. Samj South African Medical Journal. 2019;109(4):223–6. doi: 10.7196/SAMJ.2019.v109i4.13788 [DOI] [PubMed] [Google Scholar]
- 18.Stoler J, Awandare GA. Febrile illness diagnostics and the malaria-industrial complex: a socio-environmental perspective. Bmc Infectious Diseases. 2016;16. doi: 10.1186/s12879-016-1338-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blair PW, Lamorde M, Dumler JS. Rickettsioses and Q fever in Tanzania: Estimating the burden of pervasive and neglected causes of severe febrile illness in sub-Saharan Africa. American Journal of Tropical Medicine and Hygiene. 2022;106(2):371–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsi TE, Hsiao SW, Minahan NT, Yen TY, Carvalho AVD, Raoult D, et al. Seroepidemiological and molecular investigation of spotted fever group rickettsiae and Coxiella burnetii in Sao Tome Island: A One Health approach. Transboundary and Emerging Diseases. 2020;67:36–43. doi: 10.1111/tbed.13191 [DOI] [PubMed] [Google Scholar]
- 21.Horak IG, Fourie LJ, Heyne H, Walker JG, Needham GR. Ixodid ticks feeding on humans in South Africa: with notes on preferred hosts, geographic distribution, seasonal occurrence and transmission of pathogens. Experimental and Applied Acarology. 2002;27(1–2):113–36. doi: 10.1023/a:1021587001198 [DOI] [PubMed] [Google Scholar]
- 22.Norval R. The life cycle of Amblyomma hebraeum Koch, 1844 (Acarina: Ixodidae). Journal of the Entomological Society of Sout hern Africa 1974. p. 357–67. [Google Scholar]
- 23.Eisen L. Pathogen transmission in relation to duration of attachment by Ixodes scapularis ticks. Ticks and Tick-Borne Diseases. 2018;9(3):535–42. doi: 10.1016/j.ttbdis.2018.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazhetese E. Investigating Rickettsia africae infection in Amblyomma hebraeum ticks in Mnisi, Bushbuckridge Municipality, South Africa: University of Pretoria; 2019. [DOI] [PubMed] [Google Scholar]
- 25.Maina AN, Jiang J, Omulo SA, Cutler SJ, Ade F, Ogola E, et al. High prevalence of Rickettsia africae variants in Amblyomma variegatum ticks from domestic mammals in rural western Kenya: Implications for human health. Vector-Borne and Zoonotic Diseases. 2014;14(10):693–702. doi: 10.1089/vbz.2014.1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halajian A, Palomar AM, Portillo A, Heyne H, Romero L, Oteo JA. Detection of zoonotic agents and a new Rickettsia strain in ticks from donkeys from South Africa: Implications for travel medicine. Travel Medicine and Infectious Disease. 2018;26:43–50. doi: 10.1016/j.tmaid.2018.10.007 [DOI] [PubMed] [Google Scholar]
- 27.Ledger KJ, Beati L, Wisely SM. Survey of Ticks and Tick-Borne Rickettsial and Protozoan Pathogens in Eswatini. Pathogens. 2021;10(8). doi: 10.3390/pathogens10081043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pillay AD, Mukaratirwa S. Genetic diversity of Rickettsia africae isolates from Amblyomma hebraeum and blood from cattle in the Eastern Cape province of South Africa. Experimental and Applied Acarology. 2020;82(4):529–41. doi: 10.1007/s10493-020-00555-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beati L, Kelly PJ, Matthewman LA, Mason PR, Raoult D. Prevalence of Rickettsia-like organisms and spotted-fever group Rickettsiae in ticks (Acari, Ixodidae) from Zimbabwe. Journal of Medical Entomology. 1995;32(6):787–92. doi: 10.1093/jmedent/32.6.787 [DOI] [PubMed] [Google Scholar]
- 30.Magaia V, Taviani E, Cangi N, Neves L. Molecular detection of Rickettsia africae in Amblyomma ticks collected in cattle from Southern and Central Mozambique. Journal of Infection in Developing Countries. 2020;14(6):614–22. doi: 10.3855/jidc.11625 [DOI] [PubMed] [Google Scholar]
- 31.Lucy M, Fokam EB, Bouyer DH, Roland N, Titanji VPK, Walker DH, et al. Detection of Rickettsia africae in patients and ticks along the coastal region of Cameroon. American Journal of Tropical Medicine and Hygiene. 2004;71(3):363–6. [PubMed] [Google Scholar]
- 32.Moumouni PFA, Terkawi MA, Jirapattharasate C, Cao S, Liu MM, Nakao R, et al. Molecular detection of spotted fever group rickettsiae in Amblyomma variegatum ticks from Benin. Ticks and Tick-Borne Diseases. 2016;7(5):828–33. doi: 10.1016/j.ttbdis.2016.03.016 [DOI] [PubMed] [Google Scholar]
- 33.Kumsa B, Socolovschi C, Raoult D, Parola P. Spotted fever group rickettsiae in ixodid ticks in Oromia, Ethiopia. Ticks and Tick-Borne Diseases. 2015;6(1):8–15. doi: 10.1016/j.ttbdis.2014.08.001 [DOI] [PubMed] [Google Scholar]
- 34.Horak IG, Heyne H, Williams R, Gallivan GJ, Spickett AM, Bezuidenhout JD, et al. The Ixodid Ticks (Acari: Ixodidae) of Southern Africa: Springer; 2018. [Google Scholar]
- 35.Mura A, Socolovschi C, Ginesta J, Lafrance B, Magnan S, Rolain JM, et al. Molecular detection of spotted fever group rickettsiae in ticks from Ethiopia and Chad. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2008;102(9):945–9. doi: 10.1016/j.trstmh.2008.03.015 [DOI] [PubMed] [Google Scholar]
- 36.Mather TN, Nicholson MC, Donnelly EF, Matyas BT. Entomologic index for human risk of Lyme disease. American Journal of Epidemiology. 1996;144(11):1066–9. doi: 10.1093/oxfordjournals.aje.a008879 [DOI] [PubMed] [Google Scholar]
- 37.Diuk-Wasser MA, VanAcker MC, Fernandez MP. Impact of Land Use Changes and Habitat Fragmentation on the Eco-epidemiology of Tick-Borne Diseases. Journal of Medical Entomology. 2021;58(4):1546–64. doi: 10.1093/jme/tjaa209 [DOI] [PubMed] [Google Scholar]
- 38.Ledger KJ, Keenan RM, Sayler KA, Wisely SM. Multi-scale patterns of tick occupancy and abundance across an agricultural landscape in southern Africa. PloS one. 2019;14(9):e0222879. doi: 10.1371/journal.pone.0222879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goudie A. The atlas of Swaziland: Swaziland National Trust Commission; 1983. [Google Scholar]
- 40.Abidoye BO, Odusola AF. Climate Change and Economic Growth in Africa: An Econometric Analysis. Journal of African Economies. 2015;24(2):277–301. [Google Scholar]
- 41.Sam AG, Abidoye BO, Mashaba S. Climate change and household welfare in sub-Saharan Africa: empirical evidence from Swaziland. Food Security. 2021;13(2):439–55. [Google Scholar]
- 42.Walker JB, Keirans JE, Horak IG. The genus Rhipicephalus (Acari, Ixodidae): a guide to the brown ticks of the world: Cambridge University Press; 2005. [Google Scholar]
- 43.Humphry RW, Cameron A, Gunn GJ. A practical approach to calculate sample size for herd prevalence surveys. Preventive Veterinary Medicine. 2004;65(3–4):173–88. doi: 10.1016/j.prevetmed.2004.07.003 [DOI] [PubMed] [Google Scholar]
- 44.Collett D. Modelling Binary Data (2nd ed): Chapman and Hall/CRC.; 2003. [Google Scholar]
- 45.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: 2021. [Google Scholar]
- 46.Rizopoulos D. GLMMadaptive: Generalized Linear Mixed Models using Adaptive Gaussian Quadrature. https://drizopoulos.github.io/GLMMadaptive/, https://github.com/drizopoulos/GLMMadaptive2021. [Google Scholar]
- 47.Rutherford JS, Macaluso KR, Smith N, Zaki SR, Paddock CD, Davis J, et al. Fatal spotted fever rickettsiosis, Kenya. Emerging Infectious Diseases. 2004;10(5):910–3. doi: 10.3201/eid1005.030537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandara S. The distribution of Amblyomma variegatum and A. hebraeum in Zimbabwe and their infection with Ehrlichia ruminantium and Rickettsia africae. Onderstepoort, South Africa: University of Pretoria; 2018. [Google Scholar]
- 49.Kralik P, Ricchi M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Frontiers in Microbiology. 2017;8. doi: 10.3389/fmicb.2017.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallivan GJ, Spickett A, Heyne H, Spickett AM, Horak IG. The dynamics of questing ticks collected for 164 consecutive months off the vegetation of two landscape zones in the Kruger National Park (1988–2002). Part III. The less commonly collected species. Onderstepoort Journal of Veterinary Research. 2011;78(1):27–35. doi: 10.4102/ojvr.v78i1.41 [DOI] [PubMed] [Google Scholar]
- 51.Horak IG, Gallivan GJ, Spickett AM. The dynamics of questing ticks collected for 164 consecutive months off the vegetation of two landscape zones in the Kruger National Park (1988–2002). Part I. Total ticks, Amblyomma hebraeum and Rhipicephalus decoloratus. Onderstepoort Journal of Veterinary Research. 2011;78(1):8–17. doi: 10.4102/ojvr.v78i1.32 [DOI] [PubMed] [Google Scholar]
- 52.Norval R. Studies on the ecology of the tick Amblyomma hebraeum Koch in the Eastern Cape Province of South Africa. I. distribution and seasonal activity. The Journal of Parasitology 1977. p. 734–9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
For land use, wildlife refers to wildlife conservation areas, and mixed refers to conservation lands with both cattle and wildlife. Mean DOL = mean density of larvae per 100m2. No. tested = the number of individual larvae screened for R. africae using the multiplex qPCR. No. positive = the number of individual larvae positive for R. africae. LIP = larval infection prevalence. DIL = density of infected larvae per 100m2.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Data available at Open Science Framework. https://doi.org/10.17605/OSF.IO/CG7MVhttps://osf.io/cg7mv/.