Abstract
Anoxic sediments from Rotsee (Switzerland) were analyzed for the presence and diversity of methanogens by using molecular tools and for methanogenic activity by using radiotracer techniques, in addition to the measurement of chemical profiles. After PCR-assisted sequence retrieval of the 16S rRNA genes (16S rDNA) from the anoxic sediment of Rotsee, cloning, and sequencing, a phylogenetic analysis identified two clusters of sequences and four separated clones. The sequences in cluster 1 grouped with those of Methanosaeta spp., whereas the sequences in cluster 2 comprised the methanogenic endosymbiont of Plagiopyla nasuta. Discriminative oligonucleotide probes were constructed against both clusters and two of the separated clones. These probes were used subsequently for the analysis of indigenous methanogens in a core of the sediment, in addition to domain-specific probes against members of the domains Bacteria and Archaea and the fluorescent stain 4′,6-diamidino-2-phenylindole (DAPI), by fluorescent in situ hybridization. After DAPI staining, the highest microbial density was obtained in the upper sediment layer; this density decreased with depth from (1.01 ± 0.25) × 1010 to (2.62 ± 0.58) × 1010 cells per g of sediment (dry weight). This zone corresponded to that of highest metabolic activity, as indicated by the ammonia, alkalinity, and pH profiles, whereas the methane profile was constant. Probes Eub338 and Arch915 detected on average 16 and 6% of the DAPI-stained cells as members of the domains Bacteria and Archaea, respectively. Probe Rotcl1 identified on average 4% of the DAPI-stained cells as Methanosaeta spp., which were present throughout the whole core. In contrast, probe Rotcl2 identified only 0.7% of the DAPI-stained cells as relatives of the methanogenic endosymbiont of P. nasuta, which was present exclusively in the upper 2 cm of the sediment. Probes Rotp13 and Rotp17 did not detect any cells. The spatial distribution of the two methanogenic populations corresponded well to the methane production rates determined by incubation with either [14C]acetate or [14C]bicarbonate. Methanogenesis from acetate accounted for almost all of the total methane production, which concurs with the predominance of acetoclastic Methanosaeta spp. that represented on average 91% of the archaeal population. Significant hydrogenotrophic methanogenesis was found only in the organically enriched upper 2 cm of the sediment, where the probably hydrogenotrophic relatives of the methanogenic endosymbiont of P. nasuta, accounting on average for 7% of the archaeal population, were also detected.
Lake sediments are vertically structured ecosystems in which microbial activity is predominantly influenced by the periodic availability of detritic organic matter coming from the photic zone. Hence, the anaerobic mineralization of organic matter has a trigger function for microbial activity in the sediment and relies on the close interchange of chemical, physical, and biological processes. The investigation of anaerobic organic matter mineralization therefore requires combined analyses of physicochemical parameters and microbial community structure. Besides its dependence on environmental factors, the rate of organic matter mineralization relies on the balanced cooperation of different microbial populations within a complex community, i.e., fermentative and hydrolytic bacteria, acetogens, and methanogens (42, 57, 59). During the last two decades, Rotsee has been investigated intensively in order to assess the effect of organic input on the balanced function of the lake’s ecosystem. Due to a constant low-level input of organic material, a high methane production was observed and anaerobic microbial processes took place within the anaerobic sediment and the hypolimnion, which is below a water depth of 10 m during summer stratification (28, 47). Early studies focused on the sulfur cycle in the anaerobic hypolimnion as well as in surface sediments, where a sulfur reduction rate of 2 mmol per m2 of sediment per day was measured (28). Determination of the abundance of gene fragments encoding key enzymes was quantified by PCR in order to identify sulfate-reducing, denitrifying, and nitrogen-fixing bacteria (33). These analyses resulted in the detection of genes encoding assimilating sulfite reductase (dsrAB), nitrite reductase (nirS), and nitrogenase (nifD) in amounts of 108 to 109, 106 to 109, and 109 to 1010 gene copy numbers per g of sediment (wet weight), respectively. No data, however, are available on the methanogenic community that is responsible for methanogenesis as a final microbial step in organic matter degradation resulting in the production of methane. The productivity of methanogenic communities in lake sediments is dependent on the temperature regime, which may be between 4 and 23°C, as well as on the availability of substrates which are mainly H2-CO2 and acetate derived from mineralization processes (9, 12). Generally, an increase in temperature causes higher methane production rates, as well as a shift from acetoclastic methanogenesis, i.e., acetate-consuming processes, to hydrogenotrophic methanogenesis, i.e., H2-CO2-consuming pathways. Ex situ experiments with sediment slurries under psychrophilic conditions (4°C) showed a predominance of acetoclastic methanogenesis (44). In such systems, hydrogen derived from organic matter fermentation is consumed by acetogens (13). Hence, methane production from hydrogen under psychrophilic conditions is a two-step process via acetate (35). Furthermore, it has been shown that, at constant temperatures, the total methane production rate was positively correlated with the availability of organic matter (26). Predominance of hydrogenotrophic methanogenesis was favored by the availability of freshly sedimented organic matter (22), as well as by an increase in temperature up to 20°C (44). Although methanogens have been studied intensively by applying cultivation-dependent approaches (17, 58), there is still a lack of information about the in situ composition and activity of methanogenic communities under psychrophilic conditions, i.e., the natural situation of lake sediments in northern latitudes.
In recent years, in situ analysis of microbial communities based on 16S rRNA detection methods gained a broad acceptance in microbial ecology as reliable techniques that avoid certain biases associated with cultivation studies (36). Recent investigations applying molecular techniques have identified methanogens from sediment or peatland (22, 34) that resulted in the identification of methanogenic sequences which were closely related but not identical to reference sequences from cultured taxa, similar to the results of earlier reports (15, 19, 52). In addition, structural analyses of methanogenic communities were performed with artificial ecosystems such as anaerobic reactors or ex situ systems such as rice paddy microcosms (21, 38, 46).
Our study aimed to analyze the in situ distribution and activity of methanogens in the anoxic sediment of Rotsee by using a combination of in situ and ex situ molecular analyses, combined with ex situ determination of methanogenic activity and the measurement of chemical profiles. This investigation resulted in a vertical distribution pattern of two dominant methanogenic populations of acetoclastic and hydrogenotrophic methanogens identified by in situ hybridization. In addition, these results confirmed methane production rates measured ex situ with either acetate or H2-CO2, as well as chemical profiles describing the zones of highest microbial activity.
MATERIALS AND METHODS
Ex situ analysis of methane production rates.
For the ex situ analysis of methane production rates in the sediment of Rotsee, five sediment cores with a length of 50 cm and a diameter of 6.3 cm were retrieved from the deepest site (approximately 16 m) with a gravity corer in late summer (27). The upper 10 cm of each core was sectioned into 1-cm slices. Slices of all five replicate cores were pooled into one set of depth samples, subsequently divided into three replicates for each incubation treatment, and homogenized in a double volume of sterilized anaerobic hypolimnetic lake water to which sodium sulfide was added (0.5 g liter−1; final pH, 6.8). Aliquots of 12.5 ml of suspended sediment were transferred to 25-ml serum bottles, closed with butyl rubber stoppers, and subsequently flushed with N2 to a final pressure of 150 kPa. After preincubation for 4 days at the in situ lake temperature (6°C), either Na2[14C]acetic acid (Amersham, Rainham, United Kingdom) or NaH14CO3 (Amersham) was added to a final concentration of 7 or 40 μM, respectively. The background level of acetate has been measured by ion-exchange chromatography (3) and was between 32 μM (for core sections from 0 to 1 cm) and 15 μM (9 to 11 cm), the alkalinity was analyzed with the Aquamerk test kit (Merck, Dietikon, Switzerland) and was between 8 mM (0 to 1 cm) and 9 mM (9 to 11 cm). So, the resulting dilution of labeled acetate ranged from 4.5 times (0 to 1 cm) to 2.1 times (9 to 11 cm), and that of labeled bicarbonate ranged from 200 times (0 to 1 cm) to 225 times (9 to 11 cm), which was taken into account for calculating the methane production rates.
The production of [14C]methane was measured every 3 to 6 h on the first day of incubation and then daily over the next 3 days with a Tri-Carb 1600CA scintillation analyzer (Canberra Packard, Schlieren, Switzerland) as described previously, but with minor modifications (57). Briefly, 1 ml of the headspace was removed with a syringe, into which 1 ml of 2 M NaOH was drawn up previously. The syringe with the gas-NaOH mixture was shaken thoroughly to dissolve CO2. Subsequently, the gas phase was injected into scintillation vials containing a toluene-based cocktail for the analysis of gaseous samples (Toluene Scintillator; Canberra Packard), while the liquid phase was transferred to scintillation vials containing a cocktail for liquid samples (Emulsifier Scintillator Plus; Canberra Packard). Samples incubated without 14C-labeled substrates served as controls. The production of unlabeled methane was measured on a gas chromatograph (HRGC 51 60; Carlo Erba, Milan, Italy) equipped with a flame ionization detector (FID-40; Carlo Erba) and a megabore GS-Q column (J. & W. Scientific, Inc., Folsom, Calif.) with a length of 30 m. Hydrogen, with a flow rate of 4 ml min−1 at a temperature of 40°C, was used as the carrier gas. The headspace gas samples (0.2 ml) were injected with a Hamilton syringe and quantified with a calibration curve from a pure methane standard (1012.6 ppm; Carbagas, Bern, Switzerland).
Chemical analysis of in situ pore water composition.
The analysis of ions and gases in the pore water of the sediment and in the adjacent water column was based on the equilibrium diffusion technique (7). Dialysis plates 60 cm long and 1 cm thick (51) were mounted on a tripod and placed in the sediment at the deepest site of the lake (approximately 16 m) for 10 days in late summer. After retrieval of the dialysis plates from the sediments, pore water was obtained through septum stoppers at the sides of the plates by using syringes equipped with hypodermic needles. Alkalinity (acid-neutralizing capacity) in pore water was determined by end point titration to pH 4.3 with 0.1 M HCl (14). Ammonia was determined colorimetrically as described elsewhere (14). The pH was measured with a standard pH meter (Metrohm, Herisau, Switzerland) immediately after the pore water was sampled. For methane analysis, pore water was removed with a syringe immediately after retrieval of the dialysis plates and transferred through the rubber stoppers of 10-ml glass vials containing 2 ml of 1 M NaOH in order to inhibit bacterial growth. Methane in the headspace was subsequently analyzed by gas chromatography.
Ex situ analysis of Archaea based on 16S rRNA gene (16S rDNA) sequence retrieval.
For the ex situ analysis of Archaea, total nucleic acids were extracted from the upper 10 cm of a sediment core freshly collected in late summer. Cells were lyzed by a bead-beating procedure, followed by phenol-chloroform extraction for further purification of the nucleic acids (24). Archaea-specific PCR resulted in 825-bp-long fragments, which were subsequently cloned and sequenced (21). The phylogenetic position of the clones was determined after EMBL/GenBank database searches by using FASTA (Genetics Computer Group), and alignment of sequences of clones and selected Archaea was done by using the program CLUSTAL V (version 1.60) (23). A phylogenetic tree was calculated by the neighbor-joining method (41) in CLUSTAL V.
In situ analysis of microbial community based on in situ hybridization.
By using the ARB probe design program (49), probes Rotcl1 and Rotcl2, targeting the dominant methanogenic sequences retrieved from the sediment of Rotsee, as well as probes Rotp13 and Rotp17, targeting sequences in clones Rot13 and Rot17, respectively, were designed and labeled with the fluorescent dye Cy3 (Amersham) (Table 1). Probe specificity was tested and stringent hybridization conditions were established by in situ hybridization of pure culture strains, i.e., Methanosaeta concilii DSM2139, Methanobacterium thermoautotrophicum DSM2133, Methanospirillum hungatei DSM864, and Methanosarcina barkeri DSM800. In order to analyze microbial community structure in the sediment of Rotsee, the upper 10 cm of a sediment core retrieved in late summer was sectioned into 0.5-cm slices directly after sampling. Subsamples of approximately 0.5 g (fresh weight) were fixed immediately with 96% ethanol and stored at −20°C until used for in situ hybridization, which was performed as previously described, but the hybridization buffer contained 0.1% blocking reagent (Boehringer, Mannheim, Germany) (10). Probe specificity was adjusted by the addition of different concentrations of formamide to the hybridization buffer: 40% for Rotcl1, 30% for Eub338 (1) and Rotcl2, and 20% for Arch915 (48). Probes Rotp13 and Rotp17 were tested with formamide concentrations between 10 and 40%. No formamide was added for the hybridization with probes Eury498 and Cren499 (8).
TABLE 1.
Cy3-labeled oligonucleotide probes used for in situ hybridization analysis of microbial community in the sediment of Rotsee
| Probe | Microbial community or clone | 16S rRNA sequence location (positions) | Nucleotide sequence |
|---|---|---|---|
| Eub338 | Bacteria | 338–355 | 5′GCTGCCTCCCGTAGGAGT |
| Arch915 | Archaea | 915–934 | 5′GTGCTCCCCCGCCAATTCCT |
| Rotcl1 | Cluster related to Methanosaeta concilii | 644–663 | 5′CTCCCGGCCTCGAGCCAGAC |
| Rotcl2 | Cluster related to endosymbiont of P. nasuta | 648–665 | 5′CTCCCGATCCCAAGTCTTGC |
| Rotp13 | Clone Rot13 | 644–663 | 5′TTCCCGGTCTCAAGTCTTGC |
| Rotp17 | Clone Rot17 | 644–663 | 5′CTCTCCCGGTCCCTAGTC |
After hybridization and washing, slides were mounted with Citifluor (Canterbury, United Kingdom) solution and examined with a Zeiss Axiolab microscope fitted for epifluorescence with a high-pressure mercury bulb (50 W) and filter sets 02 (Zeiss; G365, FT395, LP420) and HQ-Cy3 (AHF Analysentechnik, Tübingen, Germany; G535/50, FT565, BP610/75). Microbial cells were counted at a magnification ×1,000. Forty fields covering an area of 0.01 mm2 that had been randomly selected from a sample distributed over two circular areas of 53 mm2 each were examined. Numbers are presented as means ± standard deviations (n = 40). The data were statistically analyzed by calculating a Pearson’s correlation between the depth and the mean cell counts obtained after hybridization with the respective oligonucleotide probes and 4′,6-diamidino-2-phenylindole (DAPI). The significance level was set at a P of <0.05.
Nucleotide sequence accession numbers.
Clones obtained from DNA extracted from the sediment of Rotsee were named Rot1 to Rot26, and sequences were deposited in the EMBL database under accession no. Y18080 to Y18095.
RESULTS AND DISCUSSION
Ex situ analysis of methane production rates.
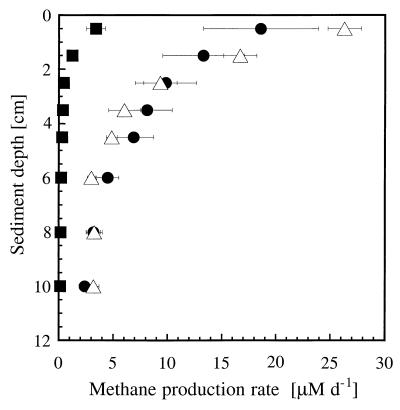
Methane production rates of either acetoclastic or hydrogenotrophic methanogens present in the sediment of Rotsee were measured after incorporation of either [14C]acetate or [14C]bicarbonate, respectively. The resulting methane production rates, and therefore the methanogenic activity, were highest in the upper sediment layers and decreased with depth. Methane produced from labeled acetate decreased with depth and ranged between 18.5 ± 5.3 and 2.4 ± 0.5 μM day−1, accounting for 70 to 75% of the totally formed methane, which was between 26.3 ± 1.5 and 3.2 ± 0.5 μM day−1 (Fig. 1). Methane production from labeled bicarbonate decreased from 3.4 ± 0.9 to 0.1 ± 0.02 μM day−1, representing between 3 and 13% of the totally formed methane. In paddy soils, methane production rates from acetate were found to range between 50 and 90%, whereas H2- CO2 was determined to be a precursor for 30 to 75% of totally produced methane in certain sediments (11, 45, 53). Bicarbonate either can be converted directly to methane by hydrogenotrophic methanogens or can follow a two-step process in which acetogens use bicarbonate to form acetate, which is then available as a substrate for acetoclastic methanogens (35). We assume that part of the methane produced comes directly from bicarbonate in the upper 2 cm of the sediment of Rotsee, since the measured total methane production could not be explained as a result of the acetate used as substrate alone. In addition, the environmental conditions in the upper layer of the sediment of Rotsee, which was highly enriched in organic matter, are comparable to those of studies where an increase in sedimentation rate caused a shift in the methanogenic community from acetoclastic methanogens to hydrogenotrophic methanogens (25, 45). Our results suggest that the dominant methane precursor in the sediment of Rotsee is acetate rather than bicarbonate, similar to other reports, although a small amount of methane is also produced directly from bicarbonate (11, 29, 30, 32, 40, 43, 50).
FIG. 1.
Methane production rate in sediment slurries without addition of 14C-labeled substrates (control) (▵) or with either [14C]bicarbonate (■) or [14C]acetate (●). d, day.
Chemical analysis of in situ pore water composition.
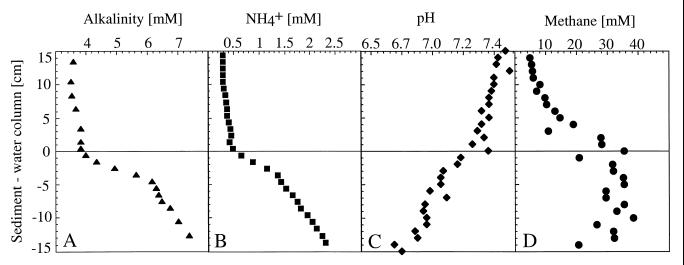
A clear zonal separation of the vertical profile of alkalinity could be observed in the sediment profile. Alkalinity (HCO3− concentration) was 3.6 mM in the water layer above the sediment, with a steep increase shown below the sediment-water interface. Within the upper 2 cm, a large alkalinity flux of 7.5 mmol m−2 day−1, which is caused by the degradation of freshly sedimented material, was calculated (Fig. 2A). This is in contrast to deeper and therefore older layers, which contained already degraded material. A concentration profile similar to that for alkalinity was obtained for ammonia (NH4+), resulting in a flux of 5.4 mmol of ammonia m−2 day−1 within the upper 2 cm of the sediment (Fig. 2B). The values of ammonia were constant at 0.3 mM in the mixed water layer and increased almost 10-fold to a depth of 8 cm. Ammonia and alkalinity profiles were stable within 3 to 4 cm just above the sediment. This indicated the existence of a stagnant water layer, where the prevented convection reduced the chemical interchange between sediment and water.
FIG. 2.
Depth profiles of alkalinity (A), ammonium (B), pH (C), and methane (D) analyzed in pore water from sediment and the adjacent water column.
In addition, the sediment composition showed remarkable differences within the first 2 cm. The upper layer was fluffier and different in color, and it originates from recently sedimented organic matter. The availability of easily degradable organic matter caused the high biological activity in this layer, which was indicated by the steep increase of ammonia and alkalinity values, as well as by the change of pH.
The pH in the hypolimnetic water was 7.5 and decreased steadily in the sediment pore water to 6.6 at a depth of 8 cm (Fig. 2C), due to mineralization processes. The subsequent release of CO2, nutrients, and protons to the pore water can increase the activity of hydrogenotrophic methanogens, since they use H2-CO2 as a substrate for the production of methane.
Methane concentrations in the sediment-water interface above the sediment were 5 to 6 mM and increased towards the sediment surface (Fig. 2D). In the first 25 cm of the sediment, methane concentrations were constant at 33 mM. By using a Bunsen coefficient of 0.043 for the methane solubility at 10°C (55), we obtained a clear indication that methane will form gas bubbles below a depth of 16 m. Methane transport in the form of bubbles explains the difference in the shapes of the ammonia and alkalinity profiles in comparison to the rather constant methane profile. In the water column above the sediment, the methane concentrations are close to saturation under the pressure conditions of a depth of 16 m. A lower estimate for the methane flux can be obtained by assuming only molecular diffusion according to Fick’s law. Using diffusion coefficients from Li and Gregory (31), we obtained a diffusive flux of 36 mmol m−2 day−1. However, due to the transport of methane in gas bubbles, the methane concentrations measured at a certain depth may have been produced in a deeper layer. Therefore, the measured methane profile did not clarify any differences in the activity of methanogens.
In addition, sulfate concentrations in the sediment of Rotsee were below 1 μM (data not shown), which can favor sulfate reducers to act as donors of net H2, which is then available for hydrogenotrophic methanogens (25).
Ex situ analysis of Archaea based on 16S rDNA retrieval.
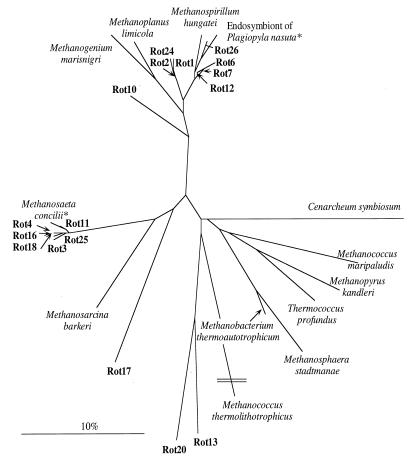
Seventeen 16S rDNA sequences were retrieved from the sediment of Rotsee. Comparative sequence analysis revealed that most of the sequences grouped into two clusters (Fig. 3). The first cluster comprised sequences of clones named Rot3, Rot4, Rot11, Rot16, Rot18, and Rot25, which were closely related to Methanosaeta concilii. The 16S rDNA sequence homologies between these clones and Methanosaeta concilii were above 97%, i.e., the sequences from the sediment of Rotsee represent a population of methanogens belonging at least to the genus Methanosaeta, if not to the species Methanosaeta concilii itself (2, 4, 56). Within the second cluster, sequences of clones Rot6, Rot7, Rot12, and Rot26 grouped together with those of the methanogenic endosymbiont of Plagiopyla nasuta, with similarity values between 95 and 98%. Only Rot10, R13, Rot17, and Rot20 did not belong to these two clusters, and Rot13 and Rot20 were identified as members of a novel euryarchaeotal lineage (rice cluster V) that had been detected previously only in rice paddy soil (20). The observed composition of methanogenic clones is like that reported for rice paddies: clones containing sequences closely related to Methanosaeta spp. are dominant, but sequences most like those of the endosymbiont of P. nasuta are also obtained (21). Sequences of methanogenic endosymbionts of P. nasuta have also been detected in peat soils (22). Whether the methanogens were really associated with protozoa, however, was not investigated.
FIG. 3.
Phylogenetic relationship of the 16S rRNA sequences retrieved from sediment of lake Rotsee (Rot1 to Rot26) with known methanogenic species. The scale bar represents 10% of the estimated substitutions per nucleotide position.
In situ analysis of microbial community based on in situ hybridization.
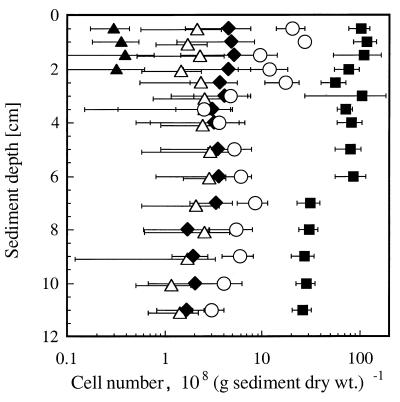
The highest density of DAPI-stained cells was detected within the upper 2 cm of the sediment core. It showed a continuous decrease with sediment depth (Fig. 4). The statistical analysis resulted in a significant correlation between depth and cell counts detected after in situ hybridization with probe Arch915 (r = 0.92, P < 0.001, n = 15) or probe Eub338 (r = 0.64, P < 0.01, n = 15) and total cell counts obtained after DAPI staining (r = 0.87, P < 0.001, n = 15). DAPI-stained cells decreased from (1.01 ± 0.25) × 1010 cells per g of sediment (dry weight) at the water-sediment interface to (2.62 ± 0.58) × 109 cells per g of sediment (dry weight) at a depth of 10 cm. Cells hybridizing to probe Eub338, which identified members of the domain Bacteria, represented the largest microbial group and accounted for 20 to 12% of the DAPI-stained cells [(2.05 ± 0.67) × 109 to (3.00 ± 1.05) × 108 cells per g of sediment (dry weight)]. Cells identified with probe Arch915, which targeted members of the domain Archaea, accounted for 5 to 7% of the DAPI-stained cells [(4.57 ± 2.97) × 108 to (1.69 ± 0.86) × 108 cells per g of sediment (dry weight)]. This result was in about the same range as has been reported for an alkaline aquifer where members of the domain Archaea represented between 1.8 and 2.5% of the detected cells (18). Probe Cren499, targeting members of the archaeal kingdom Crenarchaeota, did not result in counts, whereas probe Eury498, targeting members of the archaeal kingdom Euryarchaeota, identified similar numbers of cells as did probe Arch915 (data not shown). From this result, it was assumed that all detected Archaea belong to the kingdom Euryarchaeota.
FIG. 4.
Microbial community structure in anoxic sediment of Rotsee identified after in situ hybridization with probe Rotcl1 (▵), Rotcl2 (▴), Arch915 (⧫), and Eub338 (○). Total cell counts were determined after DAPI staining (■).
Probes Rotp13 and Rotp17 did not result in the identification of cells by in situ hybridization, even under nonstringent conditions, indicating that the abundance of retrieved clones did not reflect the abundance of methanogens in the sediment of Rotsee. Probe Rotcl1 targeting cluster 1 of retrieved sequences and including Methanosaeta concilii resulted in cell counts of 2 to 6% of the DAPI-stained cells [(2.16 ± 1.60) × 108 to (1.45 ± 0.77) × 108 cells per g of sediment (dry weight)] and identified 32 to 150% of the cells detected with probe Arch915. Identifying more cells with probe Rotcl1 than with Arch915 is not due to specificity problems of the gene probes used but is caused by the formation of cell aggregates. This problem could be overcome by neither chemical nor mechanical treatments in order to achieve a better dispersion of the cells. In addition, no significant correlation between depth and cell counts was obtained with probes Rotcl1 (n = 15) and Rotcl2 (n = 4). In contrast to probe Rotcl1, which identified the major methanogenic population, probe Rotcl2 identified a minor population present exclusively in the upper 2 cm of the core. Probe Rotcl2, identifying organisms of cluster 2 which are related to the methanogenic endosymbiont of P. nasuta, accounted for 0.3 to 1% of DAPI-stained cells [(3.01 ± 1.31) × 107 to (3.22 ± 2.86) × 107 cells per g of sediment (dry weight)] and for 7% of the cells identified with probe Arch915. In comparison to other data reporting on variable densities of methanogens in sediments between 104 and 109 cells per g of sediment (dry weight), the sediment of Rotsee is inhabited by a large methanogenic community (30, 39, 50, 53).
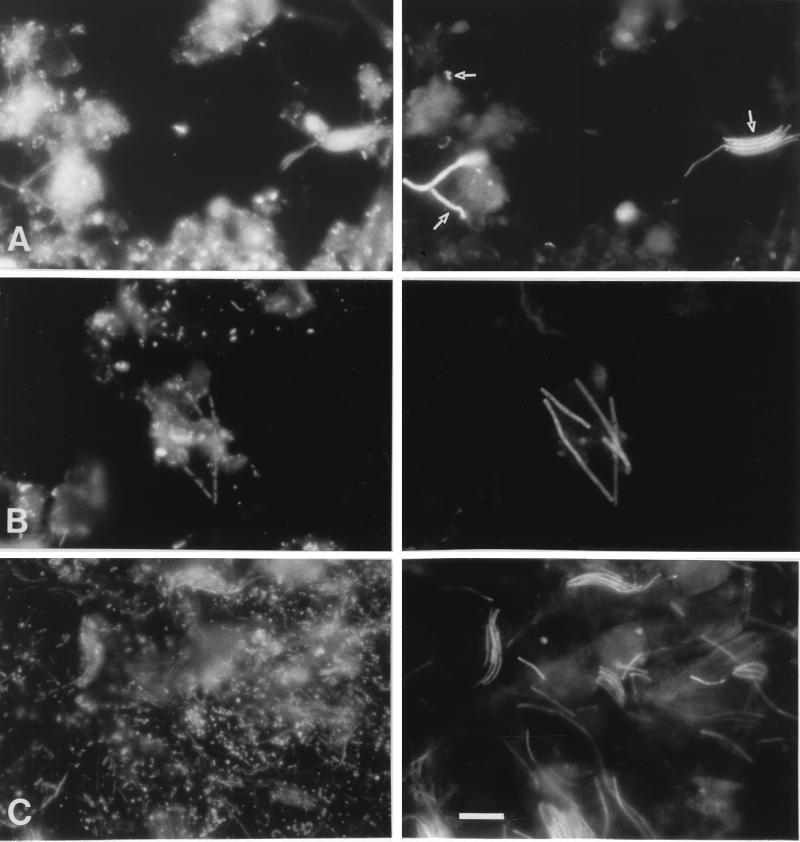
The fluorescence images obtained after in situ hybridization with probe Arch915 showed a variety of cell types, including cocci, thick filaments, and thin spiral-shaped cells (Fig. 5A), while probes Rotcl1 and Rotcl2 identified two distinct methanogenic populations, each represented by cells of the same morphology. Methanogens identified with probe Rotcl1 were rod shaped and aligned within a sheathed filament, similar to Methanosaeta spp., to which they cluster phylogenetically (Fig. 5B) (5). Cells detected by probe Rotcl2 showed a long, thin spiral shape, similar to Methanospirillum spp., bearing at maximum two subunits of longer rods (Fig. 5C) (37). Similar to cells with the latter morphology, methanogenic endosymbionts have been described as Methanospirillum sp.-like cells (54), despite the fact that endosymbiotic methanogens may have morphologies different from those of phylogenetically related free-living methanogens.
FIG. 5.
In situ hybridization of sediment samples from Rotsee with the Cy3-labeled oligonucleotide probes Arch915 (A, right panel), Rotcl1 (B, right panel), and Rotcl2 (C, right panel). The left panels show the corresponding epifluorescence micrographs after DAPI staining. Bar, 5 μm.
The ecological function of such a symbiotic association is more clear for the methanogens than for the respective hosts. Methanogens can consume H2 for energy production by intergenic H2 transfer from the host to the symbiont; these would be especially favorable in sulfate-rich habitats where methanogens are in competition with sulfate reducers. The positive effect on the host is less clear, despite the observation that certain ciliates, such as Plagiopyla frontata and Metopus contortus, grow faster while being associated with methanogenic symbionts (16). However, as mentioned above, this study did not focus on the detection of protozoa. Moreover, the sulfate concentrations in the anoxic sediment of Rotsee were very low, and therefore rather noncompetitive, although one may suggest that the sequences retrieved in the present study belong to a methanogenic species that is capable of living either free or in a symbiotic state, depending on the environmental conditions.
These results demonstrate the usefulness of a combined strategy using molecular, physiological, and chemical analyses in order to characterize the distribution and activity of methanogenic communities in an anoxic lake sediment. The ammonia and alkalinity profiles which indicate zones of highest microbial activity were found to correlate very well with the ex situ-measured methane production rates and with the vertical differences in density of the methanogenic community demonstrated by in situ hybridization. Moreover, the 16S rRNA-based molecular analyses could clearly identify a major and a minor methanogenic population simultaneously present in the upper layer, whereby in deeper layers, only the major population was active, showing slightly decreasing cell numbers with depth. The major population has been identified as Methanosaeta spp., whereas the minor population is phylogenetically related to the methanogenic endosymbiont of P. nasuta and is assumed to be hydrogenotrophic. This confirms the higher methane production rates from acetate relative to those from H2-CO2, since all members of the genus Methanosaeta utilize acetate exclusively as a substrate (6). In addition, this result strengthens the hypothesis that acetate is the predominant methane precursor in cold lake sediments. Moreover, the presence of hydrogenotrophic methanogens in the organically enriched upper layer emphasizes the ability of hydrogenotrophic methanogens to be active under psychrophilic conditions. However, still little is known about the in situ availability of methanogenic substrates under the influence of changing environmental conditions, which should be the focus of future studies.
ACKNOWLEDGMENTS
We thank Francisco Vazquez, Ruth Stierli, Christian Dinkel, and Erwin Grieder for their very helpful technical assistance during this project.
REFERENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ammann A A, Rüttimann T B. Simultaneous determination of small organic and inorganic anions in environmental water samples by ion-exchange chromatography. J Chromatogr. 1995;706:259–269. [Google Scholar]
- 4.Balch W E, Fox G E, Magrum L J, Woese C R, Wolfe R S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beveridge T J, Harris B J, Patel G B, Sprott G D. Cell division and filament splitting in Methanothrix concilii. Can J Microbiol. 1986;33:725–732. [Google Scholar]
- 6.Boone D R, Whitman W B, Rouvière P. Diversity and taxonomy of methanogens. In: Ferry J G, editor. Methanogenesis: ecology, physiology, biochemistry and Genetics. New York, N.Y: Chapman & Hall; 1993. pp. 35–80. [Google Scholar]
- 7.Brandl H, Hanselmann K W. Evaluation and application of dialysis porewater samplers for microbiological studies at sediment-water interfaces. Aquat Sci. 1991;53:55–73. [Google Scholar]
- 8.Burggraf S, Mayer T, Amann R, Schadhauser S, Woese C R, Stetter K O. Identifying members of the domain Archaea with rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1994;60:3112–3119. doi: 10.1128/aem.60.9.3112-3119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capone D G, Kiene R P. Comparison of microbial dynamics in marine and freshwater sediments: contrasts in anaerobic carbon catabolism. Limnol Oceanogr. 1988;33:725–749. [Google Scholar]
- 10.Chatzinotas, A., R.-A. Sandaa, W. Schönhuber, R. Amann, F. L. Daae, V. Torsvik, J. Zeyer, and D. Hahn. Analysis of broad-scale differences in microbial community structure in two pristine forest soils. Syst. Appl. Microbiol., in press. [DOI] [PubMed]
- 11.Chin K-J, Conrad R. Intermediary metabolism in methanogenic paddy soil and the influence of temperature. FEMS Microbiol Ecol. 1995;18:85–102. [Google Scholar]
- 12.Conrad R. Anaerobic hydrogen metabolism in aquatic sediments. In: Adams D D, Seitzinger S P, Crill P M, editors. Cycling of reduced gases in the hydrosphere. Stuttgart, Germany: Schweizerbart’sche Verlagsbuchhandlung; 1993. pp. 15–24. [Google Scholar]
- 13.Conrad R, Bak F, Seitz H J, Thebrath B, Mayer H P, Schütz H. Hydrogen turnover by psychrotrophic homoacetogenic and mesophilic methanogenic bacteria in anoxic paddy soil and lake sediment. FEMS Microbiol Ecol. 1989;62:285–294. [Google Scholar]
- 14.Deutsche Einheitsverfahren zur Wasser—Abwasser und Schlammuntersuchung (DEW). 1998. Wiley—VCH, Weinheim, Germany.
- 15.Embley T M, Finlay B J. The use of rRNA sequences to unravel the relationships between anaerobic ciliates and their methanogenic endosymbionts. Microbiology. 1994;140:225–235. doi: 10.1099/13500872-140-2-225. [DOI] [PubMed] [Google Scholar]
- 16.Fenchel T, Finlay B J. Endosymbiotic methanogenic bacteria in anaerobic ciliates: significance for the growth efficiency of the host. J Protozool. 1991;38:18–22. [Google Scholar]
- 17.Ferry J G. Methanogenesis. Vol. 1. New York, N.Y: Chapman & Hall; 1993. [Google Scholar]
- 18.Fry N K, Fredrickson J K, Fishbain S, Wagner M, Stahl D A. Population structure of microbial communities associated with two deep, anaerobic, alkaline aquifers. Appl Environ Microbiol. 1997;63:1498–1504. doi: 10.1128/aem.63.4.1498-1504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuhrman J A, McCallum K, Davis A A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 20.Großkopf, R., S. Stubner, and W. Liesack. Novel euryarchaeotal lineages detected in rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983–4989. [DOI] [PMC free article] [PubMed]
- 21.Großkopf R, Janssen P H, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hales B, Edwards C, Ritchie D A, Hall G, Pickup R W, Saunders J R. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol. 1996;62:668–675. doi: 10.1128/aem.62.2.668-675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins D G, Sharp P M. Clustal: a package for performing multiple alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 24.Hönerlage W, Hahn D, Zeyer J. Detection of mRNA of nprM in Bacillus megaterium ATCC 14581 grown in soil by whole-cell hybridization. Arch Microbiol. 1995;163:235–241. [Google Scholar]
- 25.Jones G J, Simon B M, Gardener S. Factors affecting methanogenesis and associated anaerobic processes in the sediments of a stratified eutrophic lake. J Gen Microbiol. 1982;128:1–11. [Google Scholar]
- 26.Kelly C A, Chynoweth D P. The contributions of temperature and of the input of organic matter in controlling rates of sediment methanogenesis. Limnol Oceanogr. 1981;26:891–897. [Google Scholar]
- 27.Kelts K, Briegel U, Ghilardi K, Hsü K. The limnogeology-ETH coring system. Schweiz Z Hydrol. 1986;1986:104–115. [Google Scholar]
- 28.Kohler H-P, Ahring B, Albella C, Ingvorsen K, Keweloh H, Laczko E, Stupperich E, Tomei F. Bacteriological studies on the sulfur cycle in the anaerobic part of the hypolimnion and in the surface sediments of Rotsee in Switzerland. FEMS Microbiol Lett. 1984;21:279–286. [Google Scholar]
- 29.Kotsyrbenko O R, Nozhevnikova A N, Zavarzin G A. Methanogenic degradation of organic matter by anaerobic bacteria at low temperature. Chemosphere. 1993;27:1745–1761. [Google Scholar]
- 30.Lay J-J, Miyahara T, Noike T. Methane release rate and methanogenic bacterial populations in lake sediments. Water Res. 1995;30:901–908. [Google Scholar]
- 31.Li Y H, Gregory S. Diffusion of ions in sea water and in deep-sea sediments. Geochim Cosmochim Acta. 1974;38:703–714. [Google Scholar]
- 32.Lovley D R, Klug M J. Methanogenesis from methanol and methylamines and acetogenesis from hydrogen and carbon dioxide in the sediments of a eutrophic lake. Appl Environ Microbiol. 1983;45:1310–1315. doi: 10.1128/aem.45.4.1310-1315.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller B, Wehrli B, Power M, van der Meer J R. Structure and activity of microbial communities in sediments. Chimia. 1997;51:878–883. [Google Scholar]
- 34.Munson M A, Nedwell D B, Embley T M. Phylogenetic diversity of Archaea in sediment samples from a coastal salt marsh. Appl Environ Microbiol. 1997;63:4729–4733. doi: 10.1128/aem.63.12.4729-4733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nozhevnikova A N, Holliger C, Ammann A, Zehnder A J B. Methanogenesis in sediments from deep lakes at different temperatures (2–70°C) Water Sci Technol. 1997;36:57–64. [Google Scholar]
- 36.Olsen G J, Lane D J, Giovannoni S J, Pace N R, Stahl D A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 37.Patel G B, Roth L A, van den Berg L, Clark D S. Characterization of a strain of Methanospirillum hungatei. Can J Microbiol. 1976;22:1404–1410. doi: 10.1139/m76-208. [DOI] [PubMed] [Google Scholar]
- 38.Raskin L, Rittmann B E, Stahl D A. Competition and coexistence of sulfate-reducing and methanogenic populations in anaerobic biofilms. Appl Environ Microbiol. 1996;62:3847–3857. doi: 10.1128/aem.62.10.3847-3857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothfuss F, Bender M, Conrad R. Survival and activity of bacteria in a deep, aged lake sediment (Lake Constance) Microb Ecol. 1997;33:69–77. doi: 10.1007/s002489900009. [DOI] [PubMed] [Google Scholar]
- 40.Rothfuss F, Conrad R. Thermodynamics of methanogenic intermediary metabolism in littoral sediment of Lake Constance. FEMS Microbiol Ecol. 1993;12:265–276. [Google Scholar]
- 41.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 42.Schink B. Principles and limits of anaerobic degradation. Environmental and technological aspects. In: Zehnder A J B, editor. Biology of anaerobic microorganisms. New York, N.Y: John Wiley & Sons, Inc.; 1988. pp. 771–846. [Google Scholar]
- 43.Schulz S, Conrad C. Effect of algal deposition on acetate and methane concentrations in the profundal sediment of a deep lake (Lake Constance) FEMS Microbiol Ecol. 1995;16:251–260. [Google Scholar]
- 44.Schulz S, Conrad R. Influence on temperature on pathways to methane production in the permanently cold profundal sediment of Lake Constance. FEMS Microbiol Ecol. 1996;20:1–14. [Google Scholar]
- 45.Schulz S, Matsuyama H, Conrad R. Temperature dependence of methane production from different precursors in a profundal sediment (Lake Constance) FEMS Microbiol Ecol. 1997;22:207–213. [Google Scholar]
- 46.Sørensen A H, Torsvik V L, Torsvik T, Poulsen L K, Ahring B K. Whole-cell hybridization of Methanosarcina cells with two new oligonucleotide probes. Appl Environ Microbiol. 1997;63:3043–3050. doi: 10.1128/aem.63.8.3043-3050.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stadelmann P. Stickstoffkreislauf und Primärproduktion im mesotrophen Vierwaldstättersee (Horwer Bucht) und im eutrophen Rotsee, mit besonderer Berücksichtigung des Nitrats als limitierender Faktor. Schweiz Z Hydrol. 1971;33:1–65. [Google Scholar]
- 48.Stahl D A, Amann R I. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 205–248. [Google Scholar]
- 49.Strunk O, Ludwig W. ARB: a software environment for sequence data. Munich, Germany: Technical University; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thebrath B, Rothfuss F, Whiticar M J, Conrad R. Methane production in littoral sediment of Lake Constance. FEMS Microbiol Ecol. 1993;102:279–289. [Google Scholar]
- 51.Urban N R, Dinkel C, Wehrli B. Solute transfer across the sediment surface of deep eutrophic lakes. I. Porewater profiles from dialysis samplers. Aquat Sci. 1997;59:1–25. [Google Scholar]
- 52.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal uncultured inhabitants of a well-studied thermal community. FEMS Microbiol Rev. 1990;75:105–116. doi: 10.1111/j.1574-6968.1990.tb04088.x. [DOI] [PubMed] [Google Scholar]
- 53.Ward T E, Frea J I. Sediment distribution of methanogenic bacteria in Lake Erie and Cleveland Harbor. Appl Environ Microbiol. 1980;39:597–603. doi: 10.1128/aem.39.3.597-603.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whatley J M. Bacteria and nuclei in Pelomyxa palustris: comments on the theory of serial endosymbiosis. New Phytol. 1976;76:111–120. [Google Scholar]
- 55.Wiesenburg A, Guinasso N L., Jr Equilibrium solubilities of methane, carbon monoxide and hydrogen in water and sea water. J Chem Eng Dat. 1979;24:356–360. [Google Scholar]
- 56.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zehnder A J B. Ecology of methane formation. In: Mitchell R, editor. Water pollution microbiology. New York, N.Y: John Wiley & Sons, Inc.; 1978. pp. 349–376. [Google Scholar]
- 58.Zehnder A J B. Biology of anaerobic microorganisms. New York, N.Y: John Wiley & Sons, Inc.; 1988. [Google Scholar]
- 59.Zeikus J G. Metabolism of one-carbon compounds by chemotrophic anaerobes. Adv Microb Physiol. 1983;24:215–299. doi: 10.1016/s0065-2911(08)60387-2. [DOI] [PubMed] [Google Scholar]