Abstract
Multiple COVID-19 vaccines, representing diverse vaccine platforms, successfully protect against symptomatic COVID-19 cases and deaths. Head-to-head comparisons of T cell, B cell, and antibody responses to diverse vaccines in humans are likely to be informative for understanding protective immunity against COVID-19, with particular interest in immune memory. Here, SARS-CoV-2-spike-specific immune responses to Moderna mRNA-1273, Pfizer/BioNTech BNT162b2, Janssen Ad26.COV2.S, and Novavax NVX-CoV2373 were examined longitudinally for 6 months 100% of individuals made memory CD4+ T cells, with cTfh and CD4-CTL highly represented after mRNA or NVX-CoV2373 vaccination. mRNA vaccines and Ad26.COV2.S induced comparable CD8+ T cell frequencies, though only detectable in 60–67% of subjects at 6 months. A differentiating feature of Ad26.COV2.S immunization was a high frequency of CXCR3+ memory B cells. mRNA vaccinees had substantial declines in antibodies, while memory T and B cells were comparatively stable. These results may also be relevant for insights against other pathogens.
Keywords: COVID-19 vaccine, SARS-COV2, humoral immunity, cellular immunity, immune memory
Graphical abstract
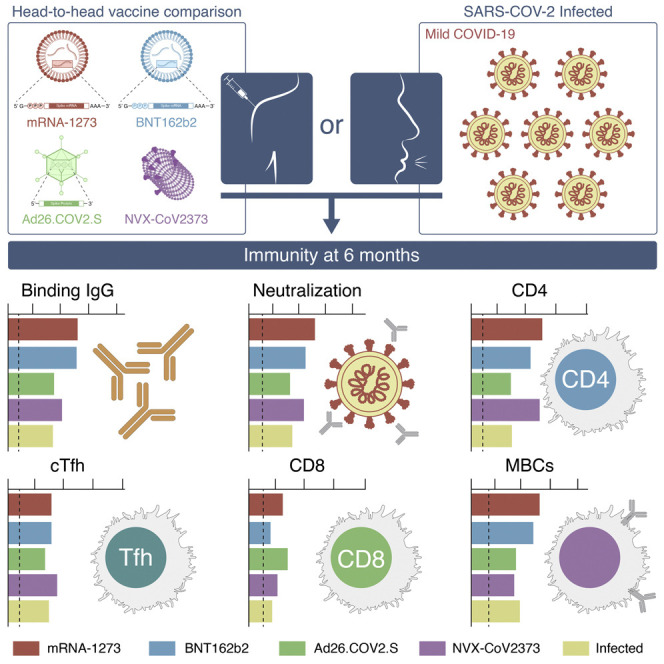
Longitudinal analysis of T cell, B cell, and antibody response to four different COVID-19 vaccines in humans, representing three different vaccine technologies, identifies different quantities and quality of CD4+ T cell, CD8+ T cell, and antibody response elicited by mRNA-1273, BNT162b2, Ad26.COV2.S, and NVX-CoV2373. Antibody levels decline, while memory T cells and B cells were comparatively stable.
Introduction
The response to the SARS-CoV-2 pandemic has relied in large part on the development, testing, and deployments of vaccines. In a short time, several different vaccine platforms have been developed and, after establishing their safety and efficacy, deployed for use in a large number of individuals. In the USA, two different mRNA vaccines (Moderna mRNA-1273 [Jackson et al., 2020], and Pfizer/BioNTech BNT162b2 [Vogel et al., 2021; Walsh et al., 2020] and a viral vector-based vaccine [Janssen/J&J Ad26.COV2.S] [Sadoff et al., 2021] have been widely used. The recombinant protein-based adjuvanted vaccine Novavax NVX-CoV2373 completed successful Phase 3 efficacy clinical trials in the USA, Mexico, and the UK (Dunkle et al., 2021; Heath et al., 2021) and is approved for use or expected to be approved for use in several different countries (Novavax, 2022). These four vaccines are representatives of the three main vaccine platforms in use for the prevention of COVID-19, namely mRNA, viral vector, and recombinant protein plus adjuvant (Pollard and Bijker, 2021).
In Phase 3 trials, these vaccines proved remarkably effective with early vaccine efficacy (VE) of 95% for BNT162b2 (Thomas et al., 2021), 94% for mRNA-1273 (Baden et al., 2021), and 90% for NVX-CoV2373 (Dunkle et al., 2021; Heath et al., 2021) against COVID-19 cases. A single dose of Ad26.COV2.S was associated with 67% VE overall and 70% in the USA (Sadoff et al., 2021). 6-month efficacy data for BNT162b2 and mRNA-1273 were 91 and 93% against COVID-19 cases (Thomas et al., 2021). Population-based "real world" studies of COVID-19 VE have provided additional insights, including comparisons between vaccines. VE wanes against symptomatic COVID-19 over time (Leon et al., 2022; Lin et al., 2022; Pilishvili et al., 2021; Rosenberg et al., 2022; Tartof et al., 2021). In one large study, VE against symptomatic COVID-19 for BNT162b2 and mRNA-1273 decreased to 67 and 75% at 5–7 months (Rosenberg et al., 2022). 1-dose Ad26.COV2.S VE started lower and also declined (Rosenberg et al., 2022). Comparable findings were made in multiple studies of populations using BNT162b2, mRNA-1273, and Ad26.COV2.S (Leon et al., 2022; Lin et al., 2022; Mallapaty et al., 2021; Rosenberg et al., 2022; Tartof et al., 2021). If any detectable SARS-CoV-2 infection is considered, as opposed to symptomatic disease, lower VE is observed for all vaccines (Nordstrom et al., 2022; Pouwels et al., 2021). Higher VE against hospitalization is observed for all COVID-19 vaccines, with somewhat lower hospitalization VE for Ad26.COV2.S compared to the mRNA vaccines (e.g. 82 vs. 94% [Rosenberg et al., 2022]). Notably, in multiple large "real world" studies, VE against hospitalization was stable over time in contrast to VE against infections (Tartof et al., 2021), potentially indicating distinct immunological mechanisms of action contributing to protection against hospitalization compared to detectable infection (Sette and Crotty, 2021).
Antibodies have been established as a clear correlate of protection against infection over the first months post-vaccination (Gilbert et al., 2022; Khoury et al., 2021), but several lines of evidence also suggest important contributions from T and B cell memory responses in protective immunity (Sette and Crotty, 2021), with neutralizing antibodies playing a dominant role in prevention of infection, while cellular immunity might be key to modulate disease severity and resolve infection (Kedzierska, 2022). Overall, available data suggest that coordinated functions of different branches of adaptive immunity may provide multiple mechanisms of protective immunity against COVID-19.
Differences between VE of COVID-19 vaccines suggest that the different vaccines might generate differential immune memory. Comparisons of immunogenicity and immune memory of different COVID-19 vaccines have been limited, hampered by multiple challenges. First, side-by-side comparisons with standardized cellular assays are often lacking. Standardized binding antibody and neutralizing antibody quantitation is possible via the use of WHO international standards (Mattiuzzo, 2020). However, CD4+ T cell, CD8+ T cell, and memory B cell assays all use live cells and complex reagents, which are far less amenable to cross-laboratory comparisons, and thus memory T and B cell measurements within the same study are required for quantitative comparisons. This is highlighted by the initial discordant findings regarding CD8+ T cell responses to COVID-19 mRNA vaccines, with early reports suggesting quite different CD8+ T cell response rates to BNT162b2 compared to mRNA-1273 (Corbett et al., 2020; Jackson et al., 2020; Sahin et al., 2021). Second, longitudinal studies with cryopreserved PBMCs are needed to directly determine kinetics of vaccine-specific immune memory in humans. Additionally, few studies have assessed antibody, CD4+ T cell, CD8+ T cell, and memory B cell vaccine responses simultaneously in the same individuals.
The massive COVID-19 immunization campaigns represent a unique opportunity to comprehensively collect and analyze immune responses in a longitudinal fashion for individuals immunized in the same year and having no prior immunity. The present study was designed to establish the magnitude and duration of vaccine-induced immune memory with four different vaccine platforms. A direct, side-by-side, comprehensive evaluation of effector and memory immune responses induced by different vaccine platforms is important to advance our understanding of the protection afforded by the various COVID-19 vaccines, as well as understand fundamental differences in immunogenicity and immune memory to mRNA, adenoviral vector, and recombinant protein vaccine platforms in humans. Here, we compare the immune responses induced by three different vaccine platforms, namely two different mRNA vaccines (Moderna mRNA-1273 and Pfizer/BioNTech BNT162b2), a viral vector-based vaccine (Janssen Ad26.COV2.S) and the protein-based adjuvanted vaccine Novavax NVX-CoV2373. The inclusion of NVX-CoV2373 was of particular interest for head-to-head comparisons of immune memory between a more conventional recombinant protein vaccine and mRNA and viral vectors. We additionally compared their immune memory to natural infection for binding antibodies, neutralizing antibodies, spike-specific CD4+ T cells, spike-specific CD8+ T cells, and spike- and RBD-specific memory B cells. To the best of our knowledge, this is the most comprehensive side-by-side evaluation of the kinetics of immune memory to these four different vaccine platforms.
Results
COVID-19 vaccine cohorts
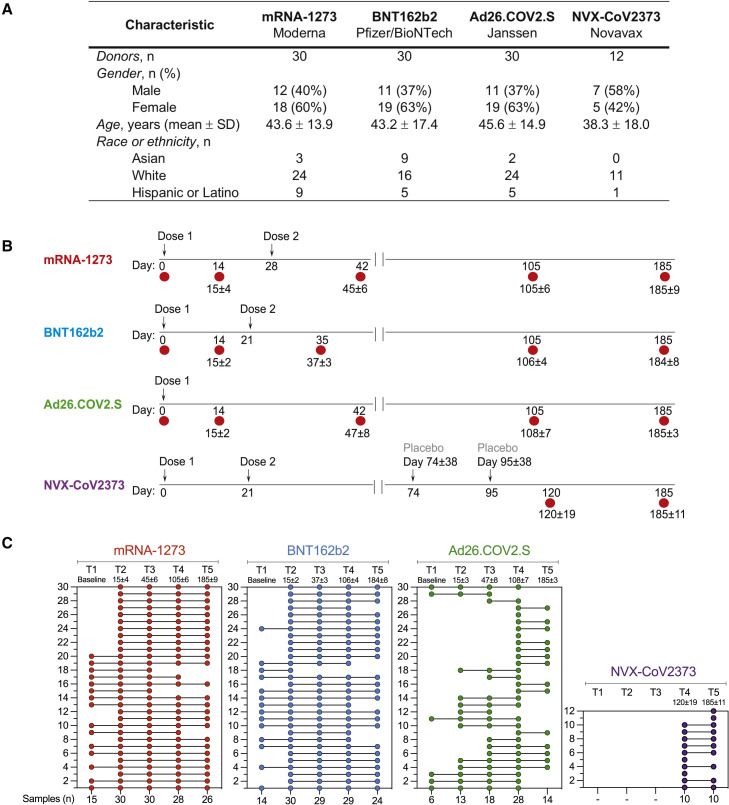
To compare the development of immune memory, we enrolled subjects who were either planning or had received immunization with mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 vaccine. Characteristics of the donor cohorts are shown in Figure 1 A. All four vaccine groups were similar in their distribution of gender, age, and race or ethnicity. Blood donations were obtained at multiple time points, and both plasma and peripheral blood mononuclear cells (PBMC) were preserved. For example, sampling time points for mRNA-1273 were pre-vaccination (T1), and then four sampling time points after immunization (T2 to T5), counting days after the first immunization: T2 at 15 ± 4 days, T3 at 45 ± 6 days, T4 at 3.5 months (105 ± 6 days), and T5 at 6 months (185 ± 9 days) (Figures 1B and 1C). Both cohorts of mRNA vaccinees (mRNA-1273, BNT162b2) received two doses of vaccine, approximately 28 and 21 days apart, respectively. Ad26.COV2.S was authorized as a 1-dose vaccine and thus blood donation timepoints were based on the initial immunization date. For NVX-CoV2373, volunteers were recruited locally who had participated in an NVX-CoV2373 efficacy trial of two intramuscular 5 μg doses of NVX-CoV2373 plus adjuvant 21 days apart (Dunkle et al., 2021). The NVX-CoV2373 trial was structured such that donors initially received two doses of placebo or vaccine in a blinded manner and were then provided two doses of the opposite (vaccine or placebo), such that all participants were vaccinated (Clinicaltrials.gov, 2022) (Figure 1B). To measure possible exposure to natural SARS-CoV-2 infection, IgG levels against the Nucleocapsid (N) protein were measured in each vaccinee (Figure S1 A. See method details for exclusion criteria).
Figure 1.
COVID-19 vaccine recipient cohorts
(A) Donor cohort characteristics.
(B) The timeline of immunizations and bleeds for each vaccine is shown. Arrows indicate immunizations, red circles indicate bleeds, and the numbers below red circles indicate the days after the first-immunization. See method details for detailed information.
(C) Subjects received mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 vaccine and donated blood at different times post-vaccination. Each COVID-19 vaccine cohort is color-coded: mRNA-1273 (red), BNT162b2 (blue), Ad26.COV2.S (green), or NVX-CoV2373 (purple). The first column displays the number of donors included in each vaccine cohort, and the bottom row shows the number of samples collected for each time point.
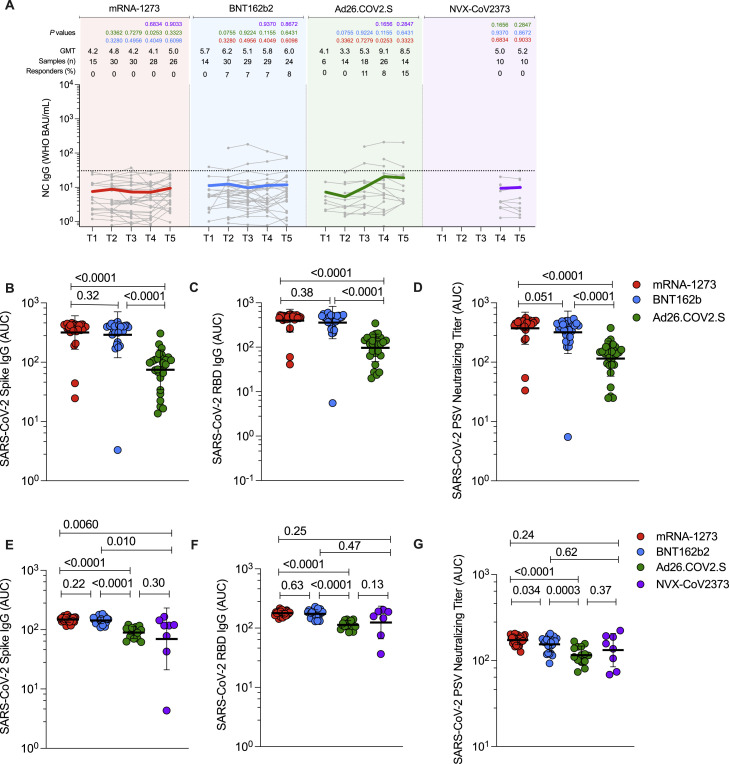
Figure S1.
Antibodies elicited by mRNA-1273, BNT162b2, Ad26.COV2.S, and NVX-CoV2373 COVID-19 vaccine platforms. Related to Figure 2
(A) Comparison of longitudinal SARS-CoV-2 spike Nucleocapsid (NC) levels from all donors to the mRNA-1273 (red), BNT162b2 (blue), Ad26.COV2.S (green) and NVX-CoV2373 (purple) over 6 months. Individual subjects are show as gray symbols with connecting lines for longitudinal samples. Geometric means of overall responses are shown in thick colored lines. The dotted line indicates the limit of quantification (LOQ). LOQ was established on the basis of pre-vaccinated samples (timepoint 1) and set as the titer at which 95% of pre-vaccinated samples (T1) fell below the dotted line. p values on the top show the differences between each time point and vaccine between the different vaccines, color-coded per comparison based on the vaccine compared. NS, non-significant; GMT, Geometric mean titers.
(B–D) (B) Comparison of area under the curve (AUC) for spike IgG, (C) RBD IgG, and (D) PSV neutralization titers across the full 6-month window for mRNA-1273, BNT162b2, and Ad26.COV2.S. Statistical analysis by Mann-Whitney t-test. Data are represented as geometric mean ± geometric SD.
(E–G) (E) Comparison of area under the curve (AUC) for spike IgG, (F) RBD IgG, and (G) PSV neutralization titers across the 3.5 to 6-month window for mRNA-1273, BNT162b2, Ad26.COV2.S, and NVX-COV2373. Statistical analysis by Mann-Whitney t-test. Data are represented as geometric mean ± geometric SD.
Spike antibody magnitude and durability elicited by different vaccine platforms
For all donors at all available time points, SARS-CoV-2 spike antibodies (Figure 2 A), receptor-binding domain (RBD) antibodies (Figure 2B), N antibodies (Figure S1A), and SARS-CoV-2 pseudovirus (PSV) neutralization titers (Figure 2C) were determined, for a total of 1,408 measurements from 352 samples. Binding antibody titers and PSV neutralization titers were quantified based on a WHO standard.
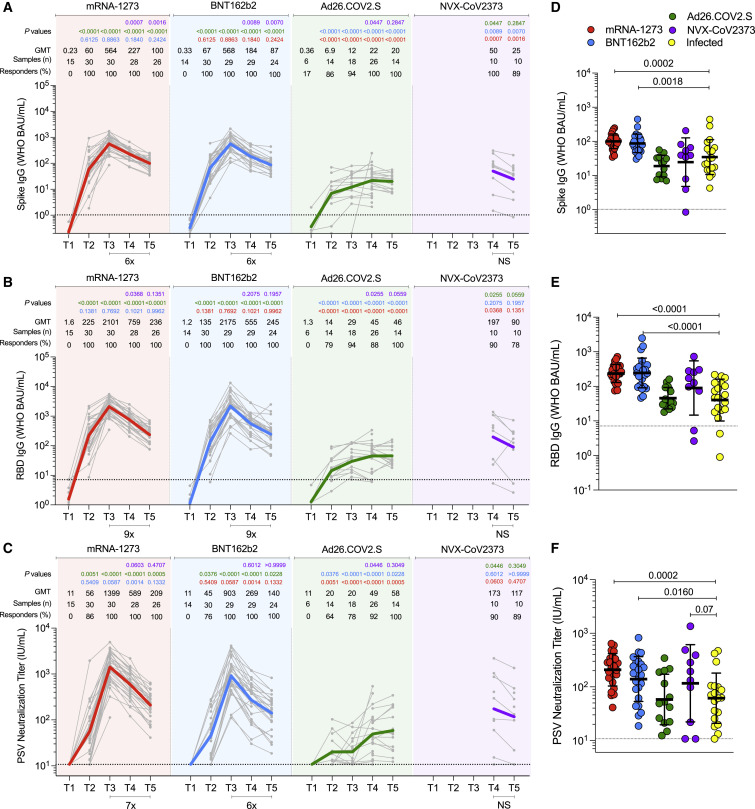
Figure 2.
Antibodies elicited by mRNA-1273, BNT162b2, Ad26.COV2.S, and NVX-CoV2373 COVID-19 vaccine platforms
(A–C) (A) Comparison of longitudinal SARS-CoV-2 spike IgG levels, (B) SARS-CoV-2 RBD IgG levels, and (C) SARS-CoV-2 pseudovirus neutralizing titers (PSV) from all donors to the mRNA-1273 (red), BNT162b2 (blue), Ad26.COV2.S (green) and NVX-CoV2373 (purple) COVID-19 vaccines over 6 months. Individual subjects are show as gray symbols with connecting lines for longitudinal samples. Geometric means are shown in thick colored lines. Dotted lines indicate the limit of quantification (LOQ). p values show differences between each time point between the different vaccines, color-coded per comparison based on the vaccine compared. NS, non-significant; GMT, geometric mean titers. Bottom bars indicate fold changes between two time points.
(D–F) (D) Comparison of spike IgG, (E) RBD IgG, and (F) PSV neutralization titers at 185 ± 6 days post-vaccination to SARS-CoV-2-infected individuals at 170 to 195 days post-symptom onset. Statistical analysis by Mann-Whitney t-test. Data are represented as geometric mean ± geometric SD. See also Figure S1.
For mRNA-1273, after first dose immunization, 100% of vaccinees had detectable spike IgG and RBD IgG titers (Figures 2A and 2B). 86% of vaccinees had detectable neutralizing antibody titers after the first dose (Figure 2C). These early findings are consistent with a large mRNA-1273 clinical trial cohort that measured serology at early time points (100% positive for RBD IgG and spike IgG, 82% positive for neutralizing antibody (Gilbert et al., 2022). After the second immunization, antibody levels of both spike and RBD IgG were boosted 9-fold (Figures 2A and 2B) and neutralizing antibody titers were boosted 25-fold (GMT 1,399) (Figure 2C). 100% of mRNA-1273 recipients remained positive for spike IgG, RBD IgG, and neutralizing antibodies at 6-month post-vaccination (T5). From peak (T3) to 6-month (T5), GMTs of spike IgG decreased 6-fold, RBD IgG decreased 9-fold, and neutralizing antibodies decreased 7-fold.
For BNT162b2, after first dose immunization, 100% of vaccinees had detectable Spike IgG and RBD IgG titers (Figures 2A and 2B). 76% of vaccinees had detectable neutralizing antibodies after the first dose, which was slightly lower than the 86% with mRNA-1273 (Figure 2C). After the second immunization, spike and RBD IgG were boosted nine- to 16-fold (Figures 2A and 2B), and neutralizing antibody titers were boosted 20-fold (GMT 903) (Figure 2C). 100% of BNT162b2 recipients remained positive for spike IgG, RBD IgG, and neutralizing antibodies at 6-month post-immunization (Figures 2A–2C). From peak (T3) to 6-month (T5), GMT of spike IgG, RBD IgG, and neutralizing antibody titers decreased by 6-fold, 9-fold, and 6-fold, respectively. These antibody declines after BNT162b2 immunization were comparable with declines after mRNA-1273 immunization (Figures 2A–2C). Neutralizing antibody titers in BNT162b2 recipients were lower than mRNA-1273 recipients by 1.6-fold (p = 0.059), 2.2-fold (p = 0.0014), and 1.5-fold (p = 0.13), at the T3, T4, and T5 time points, respectively. Neutralizing antibody titers trended lower in BNT162b2 than mRNA-1273 recipients when assessed in aggregate across the entire 6-month time period (area under curve [AUC], p = 0.051, Figures S1B-D).
For Ad26.COV2.S 1-dose immunization, 86% of vaccinees had detectable Spike IgG and 79% RBD IgG at T2 (Figures 2A and 2B). 64% of vaccinees had detectable neutralizing antibodies at T2, which was somewhat lower than the 86% with mRNA-1273 and 76% with BNT162b2 (Figure 2C). Ad26.COV2.S antibody binding and neutralization titers gradually increased over time, with 100% of recipients having detectable Spike IgG, RBD IgG, and neutralizing antibodies at 6-month post-immunization. Ad26.COV2.S neutralizing antibody titers peaked at T5 (GMT 58), but that peak was still 24-fold lower than the mRNA-1273 peak (GMT 1,399) and 16-fold lower than the BNT162b2 peak (GMT 903). At 6-month post-immunization, Ad26.COV2.S neutralizing antibody titers were 3.6-fold lower than mRNA-1273 and 2.4-fold lower than BNT162b2 (Figure 2C). Over the entire 6-month time period, Ad26.COV2.S spike IgG, RBD IgG, and neutralizing antibody titers were significantly lower than mRNA vaccine recipients (p < 0.0001 mRNA-1273, p < 0.0001 BNT162b2. Figures S1B–S1D).
For NVX-CoV2373, antibody titers were available for 3.5 and 6 months. Spike and RBD IgG titers were substantial at 3.5 months post-vaccination and were marginally (not significantly) decreased at T5 (Figures 2A and 2B). Neutralizing antibody titers were comparable at both timepoints (Figure 2C). At 6-month post-immunization, NVX-CoV2373 neutralizing antibody titers (GMT 152) were 2.6-fold higher than Ad26.COV2.S (GMT 58), and were comparable to mRNA-1273 (GMT 209) and BNT162b2 (GMT 140). Considering the 3.5-month to 6-month period in aggregate, RBD IgG and neutralizing antibody titers in NVX-CoV2373 recipients were comparable to both mRNA vaccines (Figures S1F-G). Spike IgG decay rates at the final time point were comparable for NVX-CoV2373, mRNA-1273, and BNT162b2 (t 1/2 were 52, 68, and 69 days respectively. 95% Cls: 37–88, 59–81, and 60–82 days).
Lastly, antibody titers at 6 months were compared to SARS-CoV-2 infected subjects (Figures 2D–2F) who were enrolled for a previously reported study (Mateus et al., 2021). The previously infected individuals were selected randomly. Recipients of the mRNA vaccines (mRNA-1273 and BNT162b2) had 4.5-fold higher spike IgG (Figures 2D), 6.4-fold higher RBD IgG (Figure 2E), and 3.4-fold higher neutralizing antibody titers (Figure 2F) compared to previously infected subjects. Antibody titers from NVX-CoV2373 recipients also trended higher than SARS-CoV-2 infected subjects (Figures 2D–2F). Antibody titers from Ad26.COV2.S were similar to titers from SARS-CoV-2 infected subjects (Figures 2D–2F).
Overall, antibody titers were significantly higher for mRNA recipients than Ad26.COV2.S recipients. Recipients of NVX-Co2373 immunization also had higher peak antibody titers than recipients of Ad26.COV2.S. Antibody titers to mRNA-1273, BNT162b2, and Ad26.COV2.S changed substantially over the 6+ months of observation, with different patterns seen for the mRNA versus adenoviral vector platforms.
Spike-specific CD4+ T cell memory elicited by four different vaccines
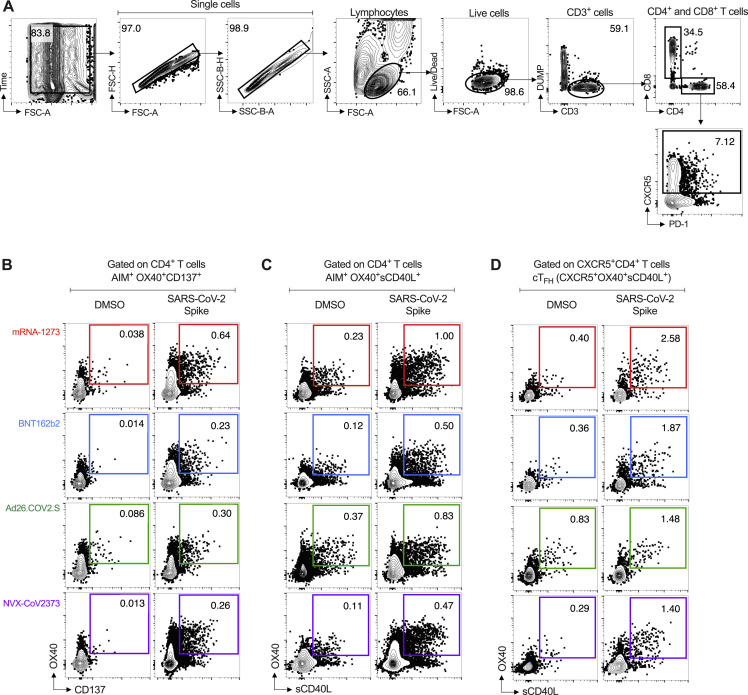
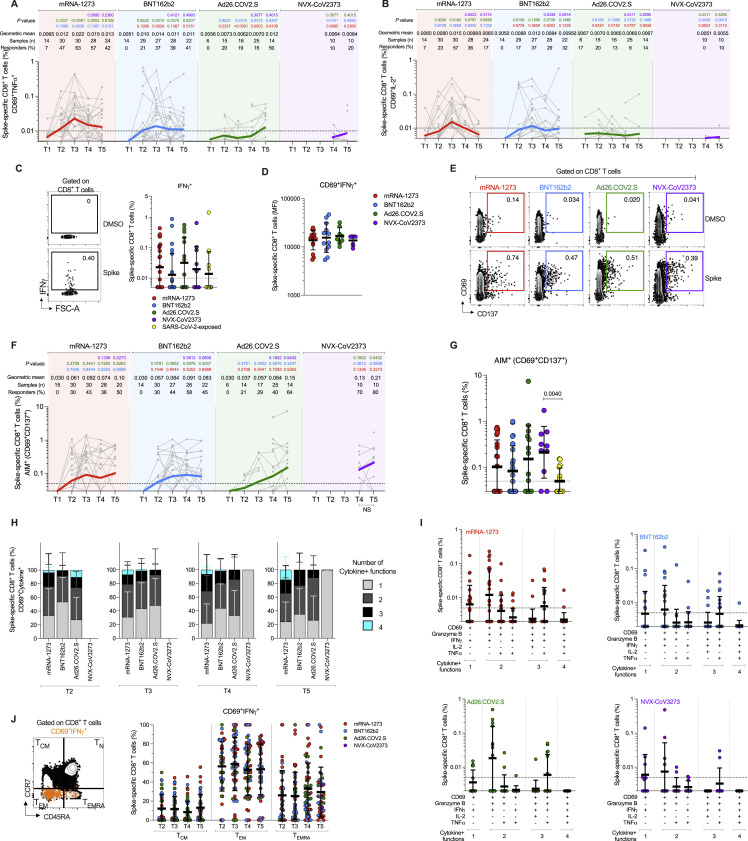
SARS-CoV-2 spike-specific CD4+ T cell responses were measured for all donors at all available timepoints utilizing two previously described flow cytometry activation-induced marker (AIM) assays (OX40+CD137+ and OX40+ surface CD40L+ [sCD40L]) (Figure 3 A, 3B, and S2) and separate intracellular staining (ICS) for cytokines (IFNγ, TNFα, IL-2), granzyme B (GzB), and intracellular CD40L (iCD40L) (Figure 4 ). SARS-CoV-2 spike-specific circulating follicular helper T (cTfh) cells were measured at all time points (Figures 3C and S2D), as this subpopulation of CD4+ T cells is crucial for supporting antibody responses following vaccination (Crotty, 2019; Lederer et al., 2022; Mudd et al., 2022).
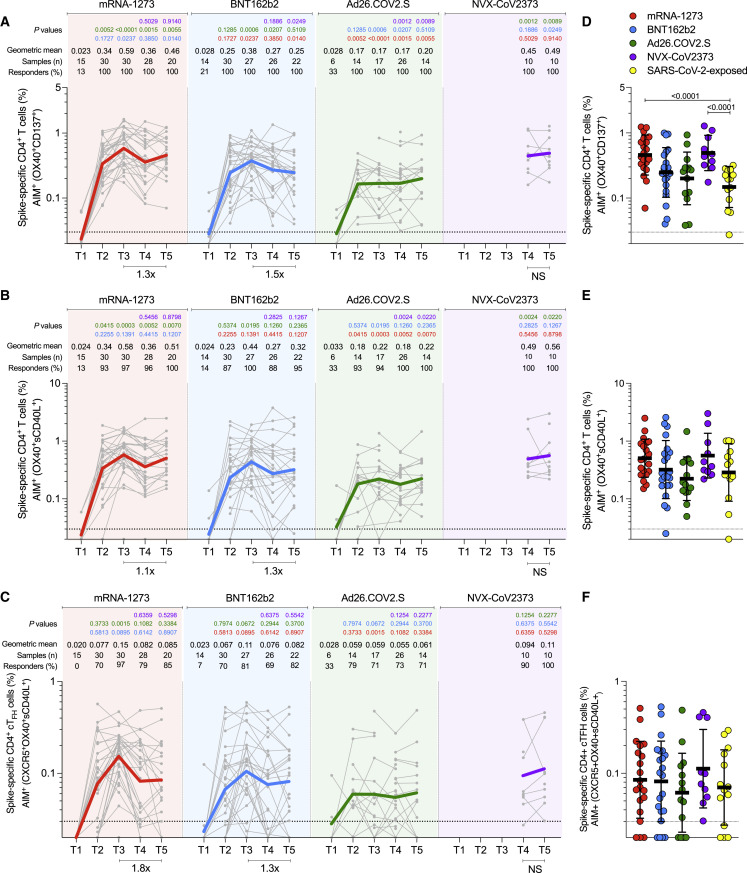
Figure 3.
Acute and memory CD4+ T cell responses after mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 immunization
(A) Longitudinal spike-specific CD4+ T cell responses induced by four different COVID-19 vaccines measured by OX40+CD137+ AIM+ after spike megapool (MP) stimulation. See Figure S2B for the representative gating strategy of OX40+CD137+ AIM+ cells.
(B) Longitudinal spike-specific CD4+ T cell responses induced by four different COVID-19 vaccines measured by OX40+ surface CD40L+ AIM after spike megapool (MP) stimulation. See Figure S2C for the representative gating strategy of OX40+sCD40L+AIM+ cells.
(C) Longitudinal spike-specific circulating T follicular helper cells (cTfh) induced by COVID-19 vaccines. Spike-specific cTfh cells (CXCR5+OX40+sCD40L+, as % of CD4+ T cells) after stimulation with spike MP. See Figure S2D for the representative gating strategy of cTfh cells.
(D–F) Comparison of spike-specific CD4+ T cells by OX40+CD137+ AIM+ (D), OX40+sCD40L+AIM+ (E), and cTfh (F) between COVID-19 vaccinees at 185 ± 6 days post-vaccination and SARS-CoV-2-exposed subjects 170 to 195 days PSO. Data are represented as geometric mean ± geometric SD.
The dotted black line indicates the limit of quantification (LOQ). The color-coded bold lines in (A), (B), and (C) represent the geometric mean each time post-vaccination. Background-subtracted and log data analyzed. p values on the top in (A), (B), and (C) show the differences between each time point in the different vaccines and are color-coded as per Figure 1. Bottom bars in (A), (B), and (C) show fold-changes between T3 and T5. Data were analyzed for statistical significance using the Mann-Whitney test ([A–F]). NS, non-significant.
See also Figure S2.
Figure S2.
Representative gating strategy for T cell analysis. Related to Figure 3, Figure 4, Figure 5
(A) Representative strategies to define CD3+CD4+ and CD3+CD8+ cells by AIM and ICS assays.
(B-C) Representative gating strategy of spike-specific AIM+ CD4+ T cells induced by mRNA-1273, BNT162b2, Ad26.COV2.S, and NVX-CoV2373 COVID-19 vaccine platforms. Spike-specific CD4+ T cells were measured by Activation-Induced Makers (AIM) assay: AIM+ OX40+ and CD137+ (B) and AIM+ OX40+ and surface CD40L+ (C).
(D) Representative gating strategy of spike-specific circulating follicular helper T cells (cTFH) induced by mRNA-1273, BNT162b2, Ad26.COV2.S, and NVX-CoV2373 COVID-19 vaccine platforms.
mRNA-1273 (red), BNT162b2 (blue), Ad26.COV2.S (green), and NVX-CoV2373 (purple).
Figure 4.
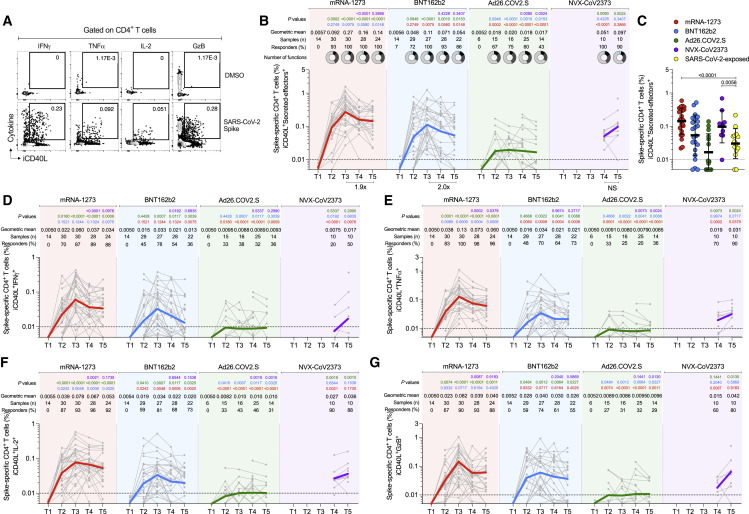
Cytokine memory CD4+ T cell responses after mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 immunization
(A) Representative gating strategy of spike-specific CD4+ T cells expressing iCD40L+ producing cytokines or Granzyme B (GzB) detected in COVID-19 vaccine platforms at T3. Secreted-effector+ CD4+ T cell responses were quantified by expressing iCD40L+ along with the production of IFNγ, TNFα, IL-2, and/or GzB after stimulation with spike MP.
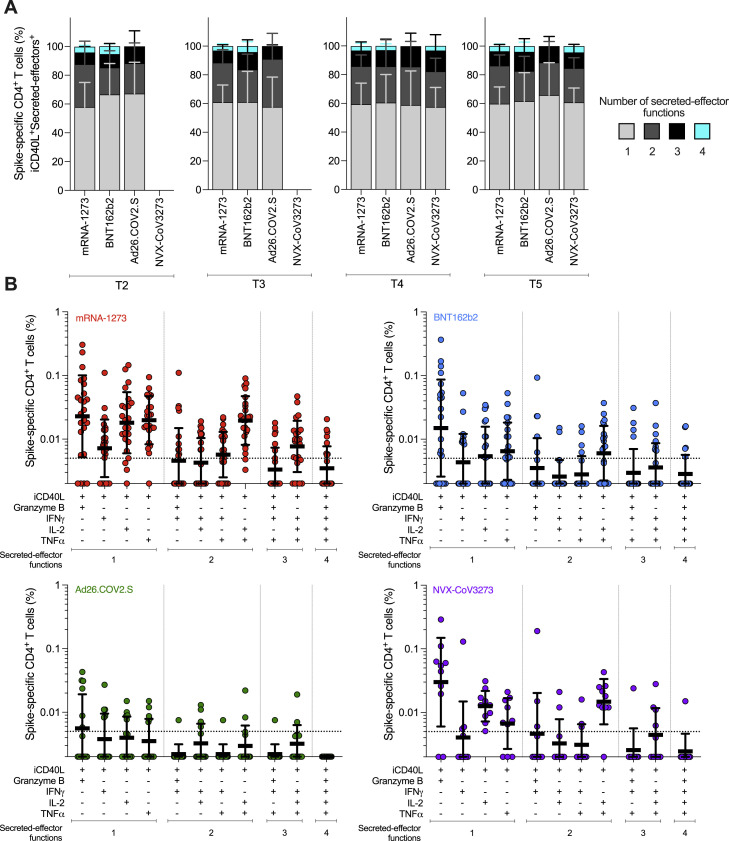
(B) Spike-specific CD4+ T cells measured by ICS. Expressing iCD40L and producing IFNγ, TNFα, IL-2, or GzB (Secreted-effector+ = ICS+). Donut charts depict the proportions of multifunctional secreted effector profiles among the spike-specific ICS+ CD4+ T cells: 1 (light gray), 2 (dark gray), 3 (black), and 4 (turquoise) functions.
(C) Comparison of spike-specific CD4+ T cells measured by ICS between COVID-19 vaccinees at 185 ± 6 days post-vaccination and SARS-CoV-2-exposed subjects 170 to 195 days PSO. Data are represented as geometric mean ± geometric SD.
(D–G) Spike-specific CD4+ T cells expressing iCD40L+ and producing IFNγ (B), TNFα (C), IL-2 (D), or GzB (E) from COVID-19 vaccinees evaluated at T1, T2, T3, T4, and T5.
The dotted black line indicates the limit of quantification (LOQ). The color-coded bold lines in (B and D–G) represent the Geometric mean in each time post-vaccination. Background-subtracted and log data analyzed. p values on the top in (B and D–G) show the differences between each time point in the different vaccines and are color coded as follows: mRNA-1273 (red), BNT162b2 (blue), Ad26.COV2.S (green), or NVX-CoV2373 (purple). Data were analyzed for statistical significance using the Mann-Whitney test ([B–G]). T1, Baseline; T2, 15 ± 3 days; T3, 42 ± 7 days; T4, 108 ± 9 days; T5, 185 ± 8 days. See also Figures S2 and S3.
In response to a single dose of the mRNA-1273 vaccine (T2), a majority of subjects developed a spike-specific CD4+ T cell response as measured by both AIM+ (Figures 3A-B) and iCD40L+ secreted-effector+ (ICS+) CD4+ T cells (Figure 4A and 4B). Spike-specific CD4+ T cell responses peaked after the second mRNA-1273 vaccination (100% responders, T3) and were well maintained out to 6 months post-vaccination, with only a 1.1- to 1.9-fold reduction in AIM+ or ICS+ CD4+ T cells, respectively (Figures 3A, 3B, 4 A and 4B). mRNA-1273 vaccination induced spike-specific cTfh cells in most donors after the first dose, which peaked after the second dose (97%, T3), and memory cTfh cells were maintained out to 6 months post-vaccination with only a 1.8-fold change from peak (T3 to T5, Figure 3C). Memory cTfh cells represented 27% of the spike-specific memory CD4+ T cells, on average.
Vaccination with BNT162b2 induced spike-specific AIM+ and ICS+ CD4+ T cells after the first vaccination (T2), with peak responses after the second immunization (T3). However, peak responses to BNT162b2 vaccination were significantly lower than mRNA-1273 peak vaccine responses both by AIM and ICS (1.5-fold lower, p = 0.023; and 2.5-fold lower, p = 0.0075. Figures 3A and 4). Memory CD4+ T cells were detectable in 86–100% of BNT162b2 vaccinees at 6 months after immunization, but the memory CD4+ T cell frequencies were significantly lower than for mRNA-1273 (1.8-fold lower by AIM, p = 0.014 and 2.6-fold lower by ICS, p = 0.0148. Figures 3A and 4). Spike-specific memory cTfh cell frequencies were comparable between BNT162b2 and mRNA-1273 vaccination (Figure 3C).
Both mRNA-1273 and BNT162b2 vaccination induced ICS+ spike-specific memory CD4+ T cells, including iCD40L+IFNγ+, iCD40L+TNFα+, and iCD40L+IL-2+ cells, detectable out to 6 months post-vaccination. mRNA-1273 vaccinees had significantly higher frequencies of TNFα+ and IL-2+ CD4+ T cells at all timepoints and higher levels of IFNγ+ memory CD4+ T cells at 6 months relative to BNT162b2 vaccinees (Figure 4). GzB+ CD4+ T cells (iCD40L+GzB+) were assessed as indicators of CD4+ cytotoxic T lymphocytes (CD4-CTL). Interestingly, both mRNA vaccines generated GzB+ CD4+ T cells as a significant fraction of the overall spike-specific CD4+ T cell response (Figures 4G and 4S3). Multifunctional spike-specific CD4+ T cells were observed after the first dose of either mRNA-1273 or BNT162b2, and multifunctionality was stably maintained out to 6 months (Figures 4B and 4S3).
For the Ad26.COV2.S vaccine, spike-specific CD4+ T cell responses were detectable in a majority of individuals and were largely stable out to 6 months post-vaccination (71–100% of individuals with spike-specific CD4+ T cells by AIM assays; 43% with spike-specific CD4+ T cells by ICS (Figures 3A, 3B, and 4). cTfh cells were detectable in the majority of individuals (Figure 3C). Peak CD4+ T cell responses were lower to Ad26.COV2.S than either of the mRNA vaccines. Peak AIM+ CD4+ T cells to Ad26.COV2.S were 2.2- to 2.4-fold lower than BNT162b2 and 3.4- to 3.2-fold lower than mRNA-1273 peak responses (Figures 3A and 3B). Peak spike-specific ICS+ CD4+ T cell responses to Ad26.COV2.S were 6.1-fold lower than BNT162b2 and 15-fold lower than mRNA-1273 (Figure 4B). Both mRNA vaccines generated significantly higher peak frequencies of IFNγ+ CD4+ T cells than Ad26.COV2.S vaccination (iCD40L+IFNγ+, mRNA1273 p < 0.0001, BNT162b2 p = 0.007), and mRNA-1273 vaccinees had significantly higher IFNγ+ spike-specific memory CD4+ T cells than Ad26.COV2.S at 6 months post-vaccination (p = 0.0009, Figure 4D). The mRNA vaccines also induced significantly more CD4-CTLs at peak than Ad26.COV2.S (mRNA1273 p < 0.0001, BNT162b2 p = 0.0012, Figure 4G), and the CD4-CTLs induced by the mRNA vaccines were more sustained as memory cells at the 6-month memory timepoint relative to Ad26.COV2.S (Figure 4G). Spike-specific CD4+ T cells induced by Ad26.COV2.S had less multifunctionality at all time points relative to both mRNA vaccines (>3 functions, T3 p = 0.023, T4 p = 0.017, T5 p = 0.023; Figures 4B and S3A). Overall, memory CD4+ T cell frequencies were lower after Ad26.COV2.S immunization compared to mRNA vaccines, assessed as total spike-specific memory (AIM+), cTfh memory, IFNγ+ memory, CD4-CTL memory, or memory CD4+ T cell multifunctionality.
Figure S3.
Multifunctional spike-specific CD4+ T cells expressing iCD40L+ in subjects vaccinated with the mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 COVID-19 vaccines. Related to Figure 4
(A) Comparison of multifunctional profiles of spike-specific CD4+ T cells iCD40L+Secreted-effector+ in subjects vaccinated with the mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 COVID-19 vaccine at T2, T3, T4, and T5. The blue, green, yellow, and red colors in the stacked bar charts depict the production of one, two, three, and four Secreted-effector+ functions, respectively. Data were analyzed for statistical significance using the Kruskal-Wallis (KW) test and Dunn’s post-test for multiple comparisons.
(B) Predominant multifunctional profiles of spike-specific CD4+ T cells expressing iCD40L with one, two, three, and four Secreted-effector+ functions were analyzed in subjects vaccinated with the mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 COVID-19 vaccine at 6 months post-vaccination (T5). Boolean analysis was carried out to define the functional profiles and the analysis included GzB, IFNγ, IL-2, and TNFα gated on CD3+CD4+ cells expressing iCD40L (See Figure 4). Each Secreted-effector+ profile combination was considered positive with >0.005% and an SI > 2 for CD4+ T cells. The dotted line indicates the limit of quantification (LOQ). The bars show the Geometric mean and geometric SD of the spike-specific CD4+ T cells iCD40L+.
For the NVX-CoV2373 vaccine, 100% of immunized individuals developed spike-specific memory CD4+ T cells detected by both AIM and ICS assays (Figures 3A, 3B, and 4). All NVX-CoV2373 immunized individuals had spike-specific memory cTfh cells (Figure 3C). Memory CD4+ T cell responses to NVX-CoV2373 were comparable in magnitude to the mRNA vaccines by AIM (Figures 3A and 3B). At 6 months post-vaccination, AIM+ memory CD4+ T cell frequencies appear to have stabilized, with no discernible half-life for all four vaccines. By ICS, NVX-CoV2373 responses 6 months post-vaccination were comparable to BNT162b2 (NVX-CoV2373 geomean 0.074%, BNT162b2 geomean 0.059%), and significantly higher than the Ad26.COV2.S vaccine (Ad26.COV2.S geomean 0.015%, p = 0.0057. Figure 4B). NVX-CoV2373 induced multifunctional memory spike-specific CD4+ T cells comparably to both mRNA vaccines (T4 and T5, Figures 4B and S3A), with a shift in the relative abundance of IL-2+ cells over IFNγ+ memory CD4+ T cells observed for NVX-CoV2373 (Figures 4D and 4F).
Spike-specific CD4+ T cell responses in COVID-19 recovered individuals were assessed to compare infection-induced versus vaccine-elicited T cell memory (Figures 3D–3F and 4C). Spike-specific CD4+ T cell memory at 6 months post-vaccination in mRNA-1273 and NVX-CoV2373 vaccinees was significantly higher than for COVID-19 recovered individuals, both by AIM and ICS (Figures 3D and 4C). The frequencies of memory CD4+ T cells generated by BNT162b2 and Ad26.COV.2S were not significantly different than SARS-CoV-2 infection (Figures 3D and 4C). Memory cTfh cell frequencies were similar between all four vaccines and infection (Figure 3F). Overall, all four of the COVID-19 vaccines generated memory CD4+ T cells in the majority of vaccinated individuals, with representation of both Th1 (IFNγ+) and Tfh memory, with memory CD4-CTL also generated by mRNA and NVX-CoV2373 vaccines. Additionally, the magnitude of spike-specific CD4+ T cell memory was generally higher for mRNA vaccines and NVX-CoV2373 than seen in COVID-19 recovered individuals.
Spike-specific CD8+ T cells elicited by four different vaccines
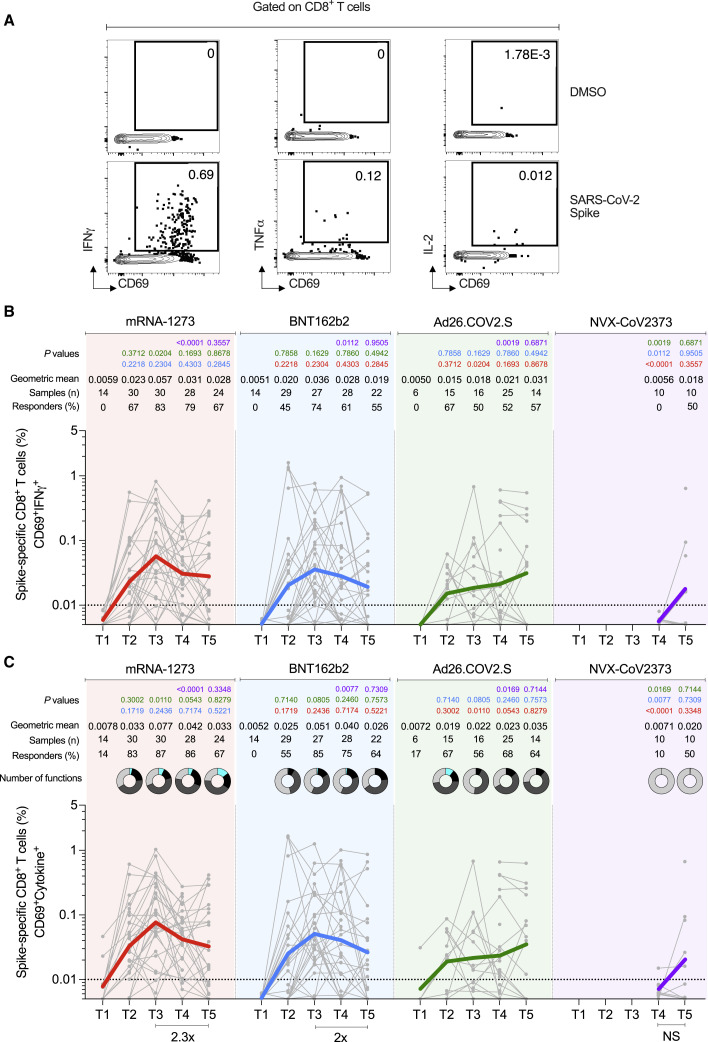
SARS-CoV-2 spike-specific CD8+ T cells were measured by ICS at all time points to identify IFNγ, TNFα, or IL-2 producing cells (CD69+ cytokine+ gating = “ICS+”. Figures 5 and S4 ) for all vaccine modalities. Spike-specific CD8+ T cells were also measured by AIM (CD69+CD137+, Figures S4E–S4G).
Figure 5.
Acute and memory CD8+ T cell responses after mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 immunization
(A) Representative gating of spike-specific CD8+ T cells. Cytokine-producing (“cytokine+”) CD8+ T cells were quantified as CD69+ along with IFNγ, TNFα, or IL-2 expression after stimulation with spike MP.
(B) Longitudinal quantitation of CD69+IFNγ+ spike-specific CD8+ T cells. See Figures S4A and S4B for TNFα and IL-2, and Figures S4H and S4I for additional analysis.
(C) Longitudinal quantitation of cytokine+ spike-specific CD8+ T cells. CD8+ T cells were quantified as CD69+ along with IFNγ, TNFα, or IL-2 expression after stimulation with spike MP. Bottom bars show fold-changes between T3 and T5. The donut charts depict the proportions of multifunctional cytokine+ profiles of the spike-specific CD8+ T cells, including IFNγ, TNFα, or IL-2 and GzB: 1 (light gray), 2 (dark gray), 3 (black), and 4 (turquoise) functions.
The dotted black line indicates the limit of quantification (LOQ). Graphs are color-coded as per Figure 1. Background-subtracted and log data analyzed. Data were analyzed for statistical significance using the Mann-Whitney test ([B], [C]). See Figure S4 for additional analysis on spike-specific CD8+ T cells.
See also Figures S2 and S4.
Figure S4.
Additional analyses in spike-specific CD8+ T cells in subjects vaccinated with the mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 COVID-19 vaccines. Related to Figure 5
(A-B) Spike-specific CD8+ T cells expressing CD69+ and producing TNFα (A) and IL-2 (B) from COVID-19 vaccinees at T1, T2, T3, T4, and T5.
(C) Comparison of spike-specific IFNγ+ CD8+ T cell responses between COVID-19 vaccinees at 185 ± 6 days post-vaccination and SARS-CoV-2-exposed subjects 170 to 195 days PSO. For this analysis, IFNγ-producing CD8+ T cells were gated based on total CD8+ T cells (no CD69 gating), as no CD69 marker was available for the samples from the previously SARS-CoV-2-infected subjects. Representative gating strategy of spike-specific CD8+ T cells producing IFNγ detected in COVID-19 vaccine platforms (Left panel).
(D) Spike-specific CD8+ T cells expressing CD69+ and producing IFNγ from COVID-19 vaccinees at 6 months post-vaccination (T5). Median fluorescence intensity (MFI) levels of IFNγ were evaluated on COVID-19 vaccinees with a positive IFNγ response at T5 (See Figure 5).
(E-F) Longitudinal spike-specific CD8+ CD69+CD137+ AIM+ T cell responses induced by COVID-19 vaccines at T1, T2, T3, T4, and T5. Representative gating strategy of spike-specific AIM+ CD8+ T cells (E) and spike-specific CD8+ T cells were measured by AIM assay: AIM+ CD69+ and CD137+ after stimulation with spike MP (F).
(G) Comparison of spike-specific CD8+ T cell responses (AIM+ CD69+CD137+) between COVID-19 vaccinees at 185 ± 6 days post-vaccination and SARS-CoV-2-exposed subjects 170 to 195 days PSO.
(H) Comparison of multifunctional profiles of spike-specific CD8+ T cells CD69+Cytokine+ in subjects vaccinated with the mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 COVID-19 vaccine at T2, T3, T4, and T5.
(I) Predominant multifunctional profiles of spike-specific CD8+ T cells expressing CD69 with one, two, three, and four functions analyzed in subjects vaccinated with the mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 COVID-19 vaccines at 6 months post-vaccination (T5). Boolean analysis was carried out to define the functional profiles and the analysis included GzB, IFNγ, IL-2, and/or TNFα gated on CD3+CD8+ cells expressing CD69 (See Figure 5). Each Cytokine+ profile combination was considered positive with >0.005% and an SI > 2 for CD8+ T cells.
(J) Longitudinal analysis of the memory subsets evaluated on spike-specific CD8+ T cells expressing CD69+ and producing IFNγ in subjects vaccinated with the mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 COVID-19 vaccines. Representative gating strategy of the memory subsets evaluated on CD8+ T cells. The memory subsets were defined based on the expression of CCR7 and CD45RA: central memory (TCM, CCR7+CD45RA−), effector memory (TEM, CCR7-CD45RA−), and terminally differentiated effector cells (TEMRA, CCR7-CD45RA+) (Left Panel).
The dotted black line indicates the limit of quantification (LOQ). The color-coded bold lines (A, B, and F) represent the Geometric mean each time post-vaccination. Lines in C, D, G, and I represent the Geometric mean and Geometric SD, or in H and J represent the Mean and SD Background-subtracted and log data analyzed. p values on the top (A, B, and F) show the differences between each time point in the different vaccines. Data were analyzed for statistical significance using the Mann-Whitney test [(A, B, F)]. T1, Baseline; T2, 15 ± 3 days; T3, 42 ± 7 days; T4, 108 ± 9 days; T5, 185 ± 8 days.
For the mRNA-1273 vaccine, 83% of vaccinees had detectable spike-specific CD8+ T cell responses after the first immunization (Figure 5 C). ICS+ CD8+ T cell response rates peaked after the second immunization (87% T3 responders, Figure 5C). Spike-specific memory CD8+ T cells were largely maintained out to 6 months after mRNA-1273 vaccination (67% responders, Figure 5C), with only a 2.3-fold decline in geomean frequency from the peak (0.077%–0.033%, Figure 5C). Both acute and memory CD8+ T cell responses were dominated by IFNγ-producing cells (Figures 5B, 5C, S4A, S4B and S4I), the majority of which co-expressed GzB (Figure S4I). The majority of the memory spike-specific CD8+ T cells exhibited an effector memory (TEM) surface phenotype (Figure S4J).
For the BNT162b2 vaccine, IFNγ+ and total ICS+ CD8+ T cell responses also peaked after the second immunization (T3 73 and 85% responders, respectively Figures 5B and 5C). Memory CD8+ T cells were maintained out to 6 months after BNT162b2 vaccination (64% responders, Figure 5C), with only a 2-fold decline in geomean frequency (Figure 5C). Multifunctional spike-specific memory CD8+ T cells were more common in mRNA-1273 compared to BNT162b2 vaccinees (Figures 5C, S4H, and S4I), with the responses dominated by IFNγ+ cells (Figures 5C, S4H, and S4I). Overall, spike-specific CD8+ T cell acute and memory responses to BNT162b2 were similar to mRNA-1273 but slightly lower in frequency and multifunctionality.
The fraction of CD8+ T cell responders to Ad26.COV2.S was lower than both mRNA vaccines (67% compared to 87 and 85%, Figure 5C). Nevertheless, Ad26.COV2.S spike-specific CD8+ T cell frequencies were relatively stable through 6 months post-vaccination (Figures 5B, 5C, S4A, and S4B) and geomean frequencies of memory CD8+ T cells after Ad26.COV2.S vaccination were comparable to both mRNA vaccines at 6 months (Figures 5B, 5C, S4A, and S4B). The estimated t 1/2 for memory CD8+ T cells at 6 months post-mRNA1273 or Ad26.COV2.S was greater than 1 year (t 1/2 449 and 381 days, respectively. 95% Cls: 101 to days, and 89 to days, respectively).
For the NVX-CoV2373 vaccine, spike-specific ICS+ memory CD8+ T cells were observed in 10%–50% of donors (T4 and T5, Figure 5C). There were minimal multifunctional CD8+ T cells (Figures 5C, S4H, and S4I).
Overall, memory CD8+ T cell frequencies and response rates were similar between mRNA-1273, BNT162b2, and Ad26.COV2.S immunizations. Low but detectable memory CD8+ T cells were observed in some individuals after NVX-CoV2373 immunization. CD8+ T cell responses to all COVID-19 vaccines were dominated by IFNγ-producing cells. No differences in IFNγ MFI were observed between memory CD8+ T cells generated for each of the vaccines (Figure S4D). All vaccines elicited IFNγ+ memory CD8+ cells (Figure S4C) and AIM+ memory CD8+ cells (Figure S4G) at frequencies comparable to, or slightly higher than, frequencies observed in SARS-CoV-2 recovered individuals at six months (Figures S4C and S4G).
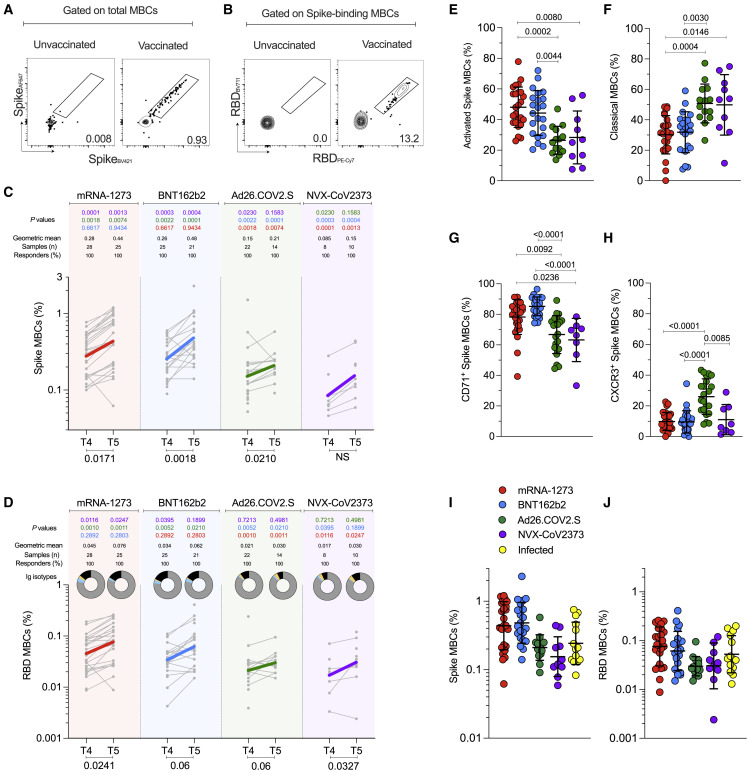
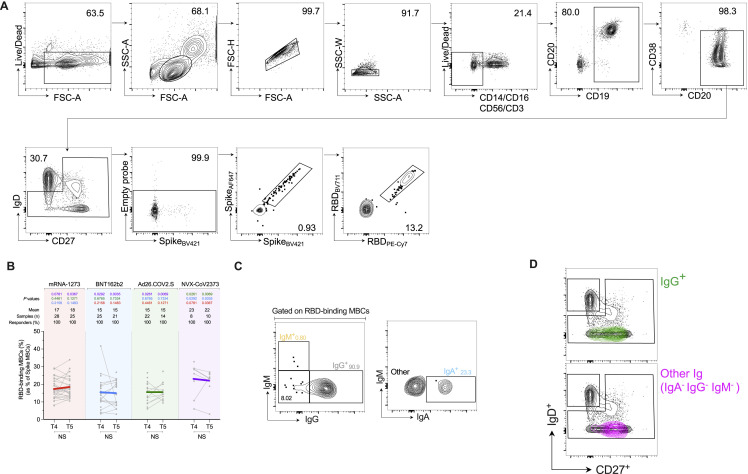
Spike- and RBD-specific B cell memory to four COVID-19 vaccines
Next, we sought to characterize and compare the development of B cell memory across the four different COVID-19 vaccines. For that, we utilized spike and RDB probes to identify, quantify and phenotypically characterize memory B cells from vaccinated subjects at 3.5 (T4) and 6 months (T5) after immunization (Figures 6A, 6B, and S5A). Spike-specific and RBD-specific memory B cells were detected in all vaccinated subjects at 6 months (Figures 6C and 6D). RBD-specific memory B cells comprised 15 to 20% of the spike-specific memory B cell population, on average (Figure S5 B). Immunization with mRNA-1273 or BNT162b2 led to higher frequencies of spike-specific and RBD-specific memory B cells compared to Ad26.COV2.S and NVX-CoV2373 at 3.5 and 6 months (each p < 0.01. Figures 6C and 6D).
Figure 6.
SARS-CoV-2-specific memory B cells to mRNA-1273, BNT162b2, Ad26.COV2.S, and NVX-CoV2373 vaccines
(A and B) Representative gating strategy for (A) spike-binding and (B) RBD-binding memory B cells (“MBCs”) (See also Figure S5).
(C and D) Frequency of (C) spike-binding and (D) RBD-binding MBCs from total MBCs elicited after 3.5 and 6 months. Limit of detection = 0.0017. RBD donut graphs represent isotype distribution; IgG (gray), IgA (blue), IgM (yellow), and other (black).
(E and F) Proportion of spike-binding MBCs with (E) activated (CD21−CD27+) and (F) classical (CD21+CD27+) phenotypes at 6 months. Data are represented as mean ± SD.
(G and H) Proportion of spike-binding MBCs expressing (G) CD71 or (H) CXCR3 at 3.5 months. Data are represented as mean ± SD.
(I and J) Comparisons between vaccinees and SARS-CoV-2-infected individuals for (I) Spike-binding MBCs and (J) RBD-binding MBCs at 6 months. Data are represented as geometric mean ± geometric SD.
The vaccines are color-coded as per Figure 2. The color-coded bold lines in (C) and (D) represent the geometric mean at each time post-vaccination. Bottom bars show T4 to T5 statistics. Data were analyzed for statistical significance using the Mann-Whitney test ([C], [D]), Kruskal-Wallis (KW) test and Dunn’s post-test for multiple comparisons ([E], [F], [G], [H], [I], [J]0. NS, non-significant. See also Figures S5 and S6.
Figure S5.
Identification of SARS-CoV-2-binding MBCs. Related to Figure 6
(A) Flow cytometry gating strategy for identification of spike and RBD-binding MBCs.
(B) Frequency of RBD MBCs gated from spike MBCs. p values on the top show the differences between each time point in the different vaccines. The bottom bars show T4 to T5 statistics.
(C) Representative gating strategy of Ig isotypes gated on RBD-binding MBCs. Colors reflect the Ig isotype distribution displayed in the donut graphs in Figure 6D.
(D) Overlay of Ig isotypes from RBD MBCs onto CD27 and IgD flow cytometry plots. The “other” RBD MBCs were predominantly IgDneg and CD27+, similarly to the IgG+ RBD MBCs. Since most of the RBD MBCs were IgG+, the phenotypic similarity of the “other” RBD MBCs suggests they may be IgG isotype MBCs that did not sufficiently bind to the anti-IgG reagent.
Data were analyzed for statistical significance using the Mann-Whitney test [(B)].
Memory B cell responses to the four vaccines did not exhibit the same kinetics as the antibody responses. The frequency of spike-specific memory B cells increased over time, (mRNA-1273, p = 0.017; BNT162b2, p = 0.0018, Ad26.COV2.S, p = 0.021. Figure 6C). RBD-specific memory B cell frequencies increased at 6 months after mRNA-1273 (1.7-fold, p = 0.024), BNT162b2 (2.2-fold, p = 0.06), Ad26.COV2.S (2.1-fold, p = 0.06), and NVX-CoV2373 (3.05-fold, p = 0.033) (Figure 6D).
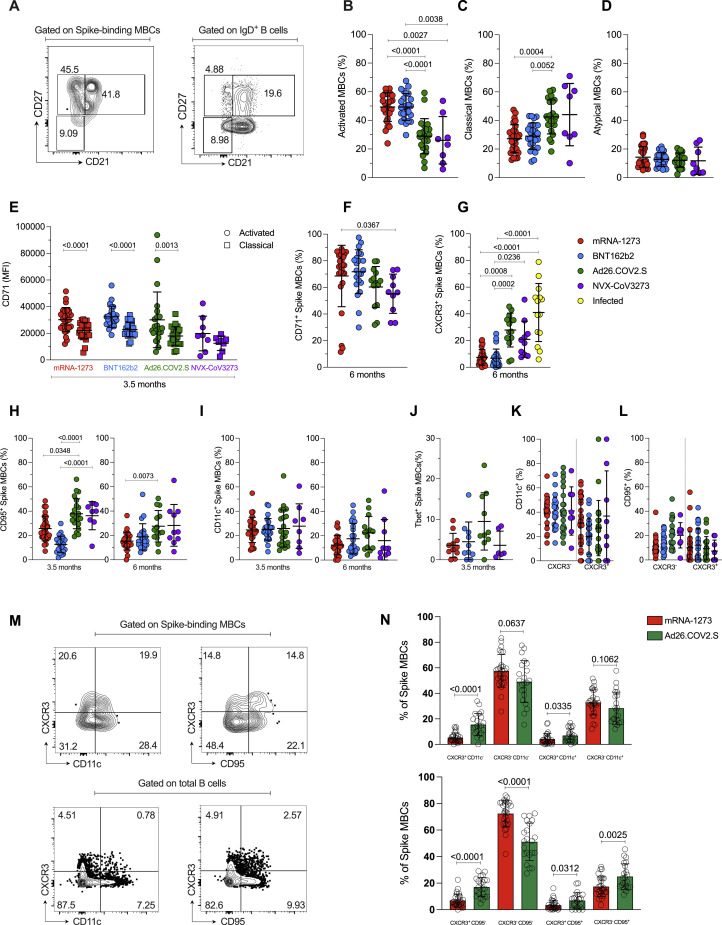
RBD-specific memory B cell isotypes were mostly comparable among the different vaccines, with an average distribution of 83% IgG, 2.5% IgM, and 2.2% IgA at 6 months (donut graphs, Figures 6D and S5C); however, IgA+ RBD-specific memory B cells were higher at 3.5 months in mRNA vaccinees compared to Ad26.COV2.S (mRNA-1273 p = 0.003. BNT162b2 p = 0.04. Figure 6D). Phenotypically, activated memory B cells (CD21−CD27+) comprised 49% of spike-specific memory B cells after mRNA vaccination (Figure S6 B), which was significantly higher than observed for Ad26.COV2.S or NVX-CoV2373 (mRNA-1273 or BNT162b2 vs. Ad26.COV2.S, p < 0.0001. mRNA-1273 vs. NVX-CoV2373, p = 0.0027; BNT162b2 vs. NVX-CoV2373 p = 0.0038 (Figure S6B), and these differences persisted at 6 months (Figure 6E). Reciprocally, the representation of classical memory B cells (CD21+CD27+) was lower in response to mRNA vaccines (Figures 6F and S6C). The frequency of atypical memory B cells (CD21−CD27−) was low and comparable among vaccines (Figure S6D). To further qualitatively compare memory B cells across vaccine platforms, we assessed CD71, CXCR3, CD95, and CD11c expression on spike-specific memory B cells. No differences in CD11c were observed (Figure S6I). CD71+ memory B cells were more common at 3.5 months in response to mRNA vaccines than Ad26.COV2.S or NVX-CoV2373 (T4 Figure 6G), with higher expression on activated memory B cells (Figure S6E). Considering that CD71 is a proliferation marker of B cells, this may reflect greater continuing production of memory B cells in response to mRNA vaccines at 3.5 months compared to Ad26.COV2.S and NVX-CoV2373 vaccines. At 6 months, the frequency of CD71+ spike-specific memory B cells remained elevated for mRNA-1273 (Figure S6F).
Figure S6.
Phenotypic characterization of SARS-CoV-2-specific MBCs elicited by COVID-19 vaccines. Related to Figure 6
(A) Representative gating strategy of spike MBCs with a classical (CD21+CD27+), activated (CD21−CD27+) and atypical (CD21−CD27−) phenotype. Control gating on IgD+ B cells is shown for comparison.
(B-D) Freq. of (B) activated (CD21−CD27+), (C) classical (CD21+CD27+) and (D) atypical (CD21− CD27−) spike MBCs at 3.5 months.
(E) CD71 expression by activated and classical spike-binding MBCs, at 3.5 months.
(F-G) Frequency of (F) CD71+ and (G) CXCR3+ spike-binding MBCs, at 6 months.
(H) Frequency of CD95+ cells among spike MBCs at 3.5 (left) and 6 (right) months.
(I) Frequency of CD11c+ cells among spike MBCs at 3.5 (left) and 6 (right) months.
(J) Frequency of Tbet+ cells among spike MBCs at 3.5 months.
(K-L) Frequency of CD11c+ or CD95+ cells among CXCR3+ or CXCR3- spike MBCs at 3.5 months.
(M) Representative gating to evaluate co-expression of CXCR3 and CD95 or CD11c.
(N) Example of co-expression of CXCR3 and CD95 or CD11c in response to mRNA-1273 or Ad26.COV2.S vaccines.
Data were analyzed for statistical significance using Kruskal-Wallis (KW) test [(B), (C), (D), (E), (F), (G), (H), (I), (J), (K), (L)], Mann-Whitney test [(N)], Wilcoxon matched-pairs signed rank test [(E)]. Data are represented as mean ± SD.
CXCR3+ spike-specific memory B cell frequencies were substantially higher in response to Ad26.COV2.S compared to the other vaccine platforms (mRNA-1273 p < 0.001, BNT162b2 p < 0.001, NVX-CoV2373 p = 0.008. Figure 6H) and remained elevated at 6 months (Figure S6G). In previously infected individuals, ∼40% of spike-binding memory B cells were CXCR3+ at 6 months post-symptom onset (PSO), which was significantly higher than the ∼7% CXCR3+ observed from mRNA vaccinees at 6 months (Figure S6G). Since CXCR3 expression during viral responses can be driven by the transcription factor Tbet, we assessed the frequency of Tbet+ spike-specific memory B cells. There was a trend toward higher Tbet expression in Ad26.COV2.S vaccinees (Figure S6J). Atypical memory B cells or “age-associated B cells” are often Tbet+ and express CXCR3 (Rubtsova et al., 2015). However, atypical memory B cells are rare in response to these COVID-19 vaccines by the CD21−CD27− cell definition (Figure S6D). Furthermore, while CD95+ spike-specific memory B cells were increased in Ad26.COV2.S vaccinees (Figure S6H), CD11c or CD95 expression was not selectively observed on CXCR3+ compared to CXCR3- spike-binding memory B cells (Figure S6K and S6L); and, by performing quadrant gating, the majority of the spike-specific memory B cell enrichment in Ad26.COV2.S vaccinees was CXCR3+ CD95− CD11c− cells (Figure S6M and S6N). In sum, using multiple approaches to characterize the CXCR3+ memory B cells, and to classify atypical B cells, our data suggest that atypical memory B cells are uncommon in response to any of the COVID-19 vaccines analyzed here, whereas CXCR3+ memory B cells are substantially generated in response to Ad26.COV2.S immunization or SARS-CoV-2 infection.
Lastly, the frequencies of spike-specific and RBD-specific memory B cells at 6 months post-vaccination were comparable to the frequencies found in previously infected subjects at 6 months (Figures 6I and 6J), indicating robust memory B cell development in response to each of the four COVID-19 vaccines.
Multiparametric comparisons across vaccine platforms
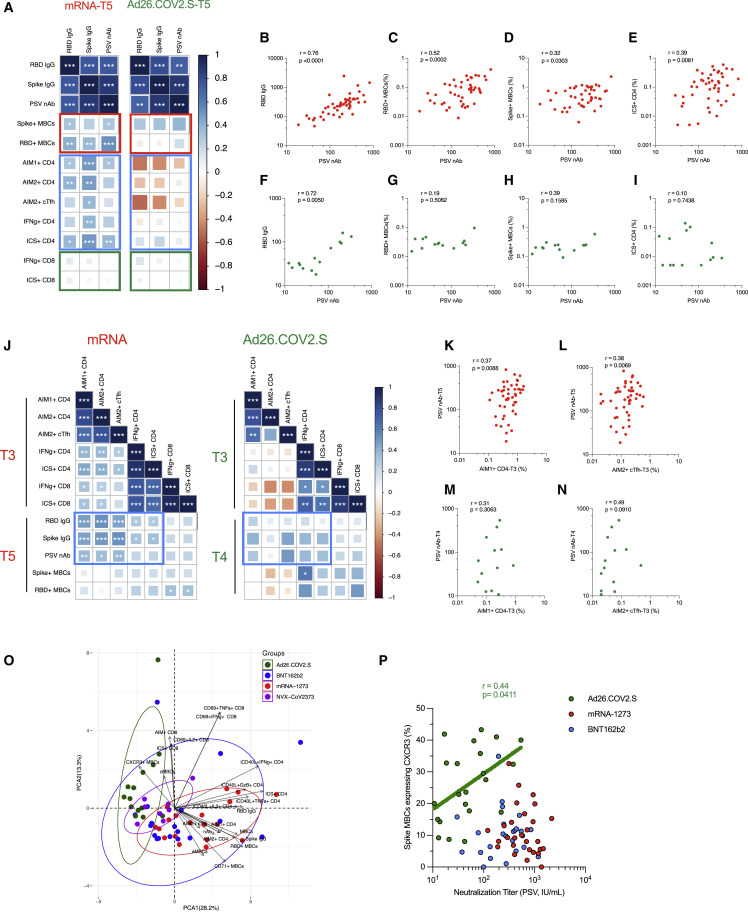
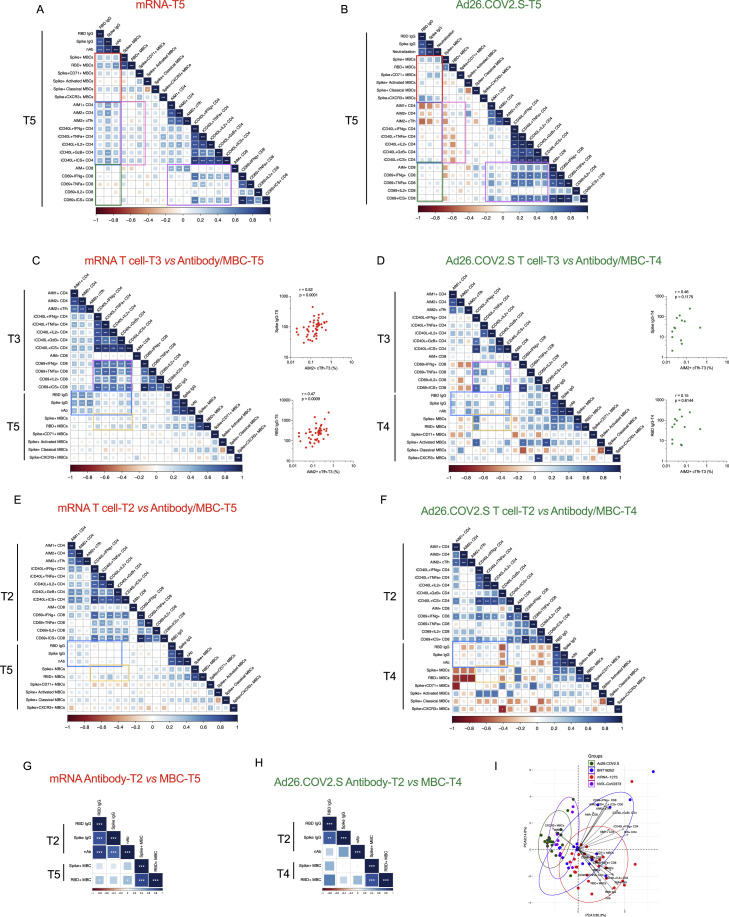
We performed multiparametric analyses, utilizing both correlation matrixes and principal component analysis (PCA) to assess the relative immunogenicity of the four vaccines (Figures 7 and S7 ). Considering all parameters of vaccine antigen-specific immune responses at 6 months after mRNA (mRNA-1273 and BNT162b2) or Ad26.COV2.S vaccination (Figures S7A and S7B), we observed strong correlations between spike IgG, RBD IgG, and neutralizing antibody titers (Figures 7A, 7B, and 7F). Neutralizing antibody titers correlated with spike-specific and RBD-specific memory B cells for mRNA vaccinees at 6 months (Figures 7A, 7C, and 7D). Antibody levels and memory CD4+ T cells were significantly associated in mRNA vaccinees by multiple metrics (Figures 7A and 7E). In contrast, no relationship was observed between antibodies and memory CD8+ T cells (Figure 7A). Memory CD4+ T cells and CD8+ T cells were significantly cross-correlated in mRNA vaccinees (Figure S7A). For Ad26.COV2.S vaccination, no significant correlations were detected at 6 months between antibodies, memory B cells, memory CD4+ T cells, or memory CD8+ T cells, which may be related to the smaller cohort size (Figures 7A and 7G–7I).
Figure 7.
Vaccine-specific immunological correlations analyses
(A) Correlation matrix of T5 (6-month) samples, plotted as mRNA (mRNA-1273 and BNT162b2) and Ad26.COV2.S COVID-19 vaccines. The red rectangle indicates the association between antibody and MBC; the blue rectangle indicates the association between antibody and CD4+ T cells; the green rectangle indicates the association between antibody and CD8+ T cells. Spearman rank-order correlation values (r) are shown from red (−1.0) to blue (1.0); r values are indicated by color and square size. p values are indicated by white asterisks as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. MBCs indicates memory B cell, AIM1 indicates OX40+CD137+, AIM2 indicates OX40+sCD40L+, nAb indicates neutralization antibody.
(B–I) The association of indicated parameters shown by scatterplot. Red indicated mRNA, green indicated Ad26.COV2.S. Spearman rank-order correlation values (r) and p values were shown.
(J) Correlation matrix of CD4+ and CD8+ T cell data from the early time point with MBCs and antibody data from the late timepoint. The blue rectangle indicates the association between CD4+ T cell and antibody. Spearman rank-order correlation values (r) are shown from red (−1.0) to blue (1.0); r values are indicated by color and square size. p values are indicated by white asterisks as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. T4 MBC and antibody data were preferred for Ad26.COV2.S due to fewer T5 paired samples.
(K–N) The association of indicated parameters shown by scatterplot. Red indicated mRNA, green indicated Ad26.COV2.S. Spearman rank-order correlation values (r) and p values were shown.
(O) Principal component analysis (PCA) representation of mRNA-1273 (n = 19), BNT162b2 (n = 14), Ad26.COV2.S (n = 14), and NVX-Cov-2373 (n = 10) on the basis of all parameters obtained 6-month post-vaccination. Only paired subjects were used for the PCA analysis. Arrows indicated the prominent immunological distinguishing features. Ellipse represents the clustering of each vaccine. Red indicates mRANA-1273, blue indicates BNT162b2, and green indicates Ad26.COV2.S. MBCs indicates spike-specific memory B cell, cMBCs indicates spike-specific classical MBCs, aMBCs indicates spike-specific activated MBCs, AIM1+ indicates OX40+CD137+, AIM2+ indicates OX40+CD40L+, nAb indicates neutralization antibody.
(P) Spearman rank-order correlation between PSV neutralization titers and frequency of spike MBCs expressing CXCR3 at 3.5 months after vaccination. Background-subtracted and log data analyzed. Only Ad26.COV2.S shows a positive correlation and spearman rank-order correlation values (r) and p values are shown as green. Linear regression analysis of Ad26.COV2.S is shown for visual reference. See also Figure S7.
Figure S7.
Vaccine-specific correlation analyses. Related to Figure 7
(A-B) Correlation matrix of T5 (6-month) samples, plotted as mRNA (mRNA-1273 and BNT162b2) and Ad26.COV2.S COVID-19 vaccines. The red rectangle indicated the association between antibody and memory B cells; the blue rectangle indicated the association between antibody and CD4+ T cell; the green rectangle indicated the association between antibody and CD8+ T cell; the pink rectangle indicated the association between CD4 T cells and memory B cell; the purple rectangle indicated the association between CD4 T cells and CD8 T cells. Spearman rank-order correlation values (r) are shown from red (−1.0) to blue (1.0); r values are indicated by color and square size. p values are indicated by white asterisks as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(C-F) Correlation matrix of CD4+ and CD8+ T cell data from the early time point T3 (C-D) or T2 (E-F) with memory B cell and antibody data from the late timepoint. The blue rectangle indicates the association between CD4+ T cell and antibody; the orange rectangle indicated the association between CD4 T cells and memory B cell. Spearman rank-order correlation values (r) are shown from red (−1.0) to blue (1.0); r values are indicated by color and square size. p values are indicated by white asterisks as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. The T4 MBC and antibody data were preferred for Ad26.COV2.S due to fewer T5 paired samples. The association of spike IgG or RBD IgG with AIM2+cTfh were also shown by scatterplot (C-D). Red indicated mRNA, green indicated Ad26.COV2.S. Spearman rank-order correlation values (r) and p values were shown.
(G-H) Correlation matrix of antibody data from the T2 time point with memory B cell data from the late timepoint. Spearman rank-order correlation values (r) are shown from red (−1.0) to blue (1.0); r values are indicated by color and square size. p values are indicated by white asterisks as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. The T4 MBC data was preferred for Ad26.COV2.S due to fewer T5 paired samples.
(I) Principal component analysis (PCA) representation of mRNA-1273 (n = 28), BNT162b2 (n = 19), Ad26.COV2.S (n = 20), and NVX-Cov-2373 (n = 8) on the basis of all parameters obtained 3.5-month post-vaccination. Only paired subjects were used for the PCA analysis. Arrows indicated the prominent immunological distinguishing features. Ellipse represented the clustering of each vaccine. Red indicated mRANA-1273, blue indicated BNT162b2, and green indicated Ad26.COV2.S. MBCs indicates spike-specific memory B cell, cMBCs indicates spike-specific classical memory B cell, aMBCs indicates spike-specific activated memory B cell, AIM1+ indicates OX40+CD137+, AIM2+ indicates OX40+CD40L+, nAb indicates neutralization antibody.
Next, we tested for relationships between early immune responses and immune memory (Figures 7J–7N and S7C–S7H). Peak post-second mRNA immunization cTfh CD4+ T cells were strongly associated with 6-month antibody levels (Figures 7J–7L, S7C, and S7D), providing an early indicator of long-term humoral immunity. Early RBD IgG titers after the first mRNA immunization were positively associated with 6-month RBD-specific memory B cell frequencies (Figures S7G and S7H). For both mRNA and Ad26.COV2.S, peak ICS+ CD4+ and CD8+ T cell responses significantly cross-correlated (T3, Figure 7J). Overall, these observations suggest that early peak CD4+ T cells responses had a lasting effect on the humoral response.
PCA mapping was performed using 3.5-month (Figure S7I) and 6-month (Figure 7O) post-vaccination data. PCA discriminated mRNA-1273 and Ad26.COV2.S, indicating these two vaccines generated distinct immunological profiles (Figure 7O). BNT162b2 largely developed the same profile as mRNA-1273 but with more heterogeneity. NVX-CoV2373 generated an immune memory profile overlapping with that of mRNA and adenoviral vectors (Figure 7O). Prominent immunological features distinguishing between mRNA and Ad26.COV2.S were CXCR3+ spike-specific memory B cells, ICS+ memory CD4+ T cells, CD71+ memory B cells, and spike IgG (Figures 7O and S7I). Notably, neutralizing antibody titers and CXCR3+ spike-specific memory B cells were correlated for Ad26.COV2.S vaccinees (r = 0.44, p = 0.04) but not mRNA vaccinees (mRNA-1273, p = 0.25. BNT162b2, p = 0.79. Figure 7P), corroborating the immunologically distinct outcomes. Overall, substantial relationships were observed between multiple components of immune memory for these COVID-19 vaccines, with distinct immune memory profiles for different vaccine platforms.
Discussion
COVID-19 vaccines have achieved extraordinary success in protection from infection and disease, yet some limitations exist, including differences in VE between vaccines and waning of protection against infection over a period of several months. Here, diverse metrics of adaptive responses were measured to mRNA-1273, BNT162b2, Ad26.COV2.S, and NVX-CoV2373, with implications for understanding the protection against COVID-19 associated with each of the vaccines. A strength of this study is that the samples from different vaccine platforms were obtained from the same blood processing facility, from the same geographical location, and were analyzed concomitantly, utilizing the same experimental platform.
In the present study, antibody responses were detected in 100% of individuals. At 6 months post-immunization, the neutralizing antibody titer hierarchy between the vaccines was mRNA-1273∼BNT162b2∼NVX-CoV2373 > Ad26.COV2.S. These serological data are consistent with previous reports for single vaccines (Atmar et al., 2022; Doria-Rose et al., 2021; Goel et al., 2021; Naranbhai et al., 2021a; Pajon et al., 2022; Pegu et al., 2021), and serological comparisons between vaccines (Barouch et al., 2021; Carreno et al., 2022; Dashdorj et al., 2021; Lafon et al., 2022; Naranbhai et al., 2021b; Self et al., 2021), though in much larger serological studies ∼2-fold higher neutralizing antibody titers were discerned with mRNA-1273 compared to BNT162b2 (Atmar et al., 2022; Steensels et al., 2021). Comparisons of NVX-CoV2373 antibody responses compared to other vaccines after 6 months have been very limited. Here we observed that NVX-CoV2373 neutralizing antibody titers were comparable to that of BNT162b2 and only moderately lower than mRNA-1273.
In this side-by-side comparative study, spike-specific CD4+ T cell responses were detected in 100% of individuals to all four vaccines. While neutralizing antibody kinetics were different between mRNA and viral vector vaccines, the CD4+ T cell response kinetics were similar. The hierarchy of the magnitude of the memory CD4+ T cells was mRNA-1273 > BNT162b2∼NVX-CoV2373 > Ad26.COV2.S. These overall findings are consistent with previous reports on COVID-19 vaccine T cell responses (Barouch et al., 2021; Goel et al., 2021; Guerrera et al., 2021; Khoo et al., 2022; Liu et al., 2022; Mateus et al., 2021; Rodda et al., 2022; Tarke et al., 2022), but the analysis reported herein extensively expand these observations, including four different vaccines representing three different vaccine platforms, and with longitudinal data and single-cell cytokine expression resolution providing insights regarding CD4+ T cell subpopulations between the vaccines. Interestingly, multifunctional CD4+ T cells were observed most frequently after mRNA-1273 immunization, and GzB+ early CD4-CTLs represented a substantial fraction of the memory CD4+ T cells after mRNA-1273, BNT162b2, or NVX-CoV2373 vaccination. cTfh memory cells were represented as a substantial fraction of CD4+ T cell memory for each of the four vaccines, consistent with these vaccine platforms being selected for their ability to induce antibody responses. Memory CD4+ T cell responses were also compared to infected individuals, demonstrated that each vaccine was successful in generating circulating spike-specific CD4+ T cell memory frequencies similar to or higher than SARS-CoV-2 infection, though of course infection also generates responses to other viral antigens.
The two mRNA vaccines and Ad26.COV2.S induced comparable acute and memory CD8+ T cell frequencies. These data are broadly consistent with previous reports for mRNA vaccines or adenoviral vectors (Goel et al., 2021; Guerrera et al., 2021; Keeton et al., 2022; Mateus et al., 2021; Tarke et al., 2022), with the exception being represented by reduced cytokine-expressing CD8+ T cells detected after mRNA vaccinations when using a 6- to 8-h assay (Atmar et al., 2022; Collier et al., 2021), compared to the overnight stimulation used here. As expected for a protein-based vaccine, IFNγ+ memory CD8+ T cell frequencies after NVX-CoV2373 were lower than the other vaccine platforms assessed, but it was notable that NVX-CoV2373 generated spike-specific CD8+ T cell memory in a significant fraction of individuals, and the IFNγ+ spike-specific memory CD8+ T cell frequencies were similar at 6 months between NVX-CoV2373 immunizations and SARS-CoV-2 infection.
Spike- and RBD-specific memory B cell responses were detected in all individuals to each of the four vaccines. While neutralizing antibody titers declined over time in mRNA vaccinees, the frequency of spike-specific memory B cells increased over time. These divergent antibody and memory B cell kinetics were also observed in SARS-CoV-2 infection (Dan et al., 2021). The mRNA vaccine data are comparable to Goel et al. (2021), but memory B cell data and kinetics for the Ad26.COV2.S or NVX-CoV2373 vaccines have not previously been available. At 6 months post-immunization, the spike-specific memory B cell hierarchy was mRNA1273∼BNT162b2 > Ad26.COV2.S > NVX-CoV2373. One of the most differentiating features of Ad26.COV2.S immunization observed here was the high frequency of CXCR3+ memory B cells. CXCR3+ memory B cells were correlated with neutralizing antibody titers after Ad26.COV2.S immunization, but not mRNA immunization, suggesting a specific functional role in viral vector B cell responses. CXCR3 expression was present on most memory B cells after SARS-CoV-2 infection (Fig. S6G). CXCR3 expression on memory B cells has been found to be important for mucosal immunity in two mouse models (Oh et al., 2019, 2021). CXCR3 ligands are also expressed in infection-induced IFNγ inflammation in non-mucosal sites.
Between the mRNA vaccines, mRNA-1273 elicited more immune memory than BNT162b2. Vaccine dose and timing are likely explanations for the observed differences. mRNA-1273 contains 100 μg mRNA, while BNT162b2 contains 30 μg. In a clinical trial of mRNA-1273, a 25 μg dose elicited somewhat lower neutralizing antibody and memory CD4+ T cell responses than the 100 μg dose (Mateus et al., 2021), and the differences between 25 and 100 μg mRNA-1273 were comparable to the differences observed here between 100 μg mRNA-1273 and 30 μg BNT162b2. The time interval between the first and second mRNA dose may also contribute to the differences in the frequencies and phenotypes of memory T cells between mRNA1273 and BNT162b2. Memory T cells with high proliferative potential are typically not generated until several weeks after a first immunization (Sallusto et al., 2010); thus, a second immunization prior to maximal establishment of memory T cells after the first dose may contribute to sub-optimal T cell memory later. A longer time interval likely also contributes to higher quality memory B cell responses, as observed for SARS-CoV-2 infection and vaccines (Cho et al., 2021; Muecksch et al., 2022), and other contexts (Lee et al., 2021).
Across the antigen-specific immune metrics assessed, mRNA vaccines were consistently the most immunogenic, with levels higher than or equal to that of Ad26.COV2.S and NVX-CoV2373 vaccines for each immune response. NVX-CoV2373 elicited CD4+ T cell memory and neutralizing antibody titers comparably to the mRNA vaccines. The responses induced by the Ad26.COV2.S were generally lower but relatively stable. The mRNA vaccine platforms were associated with substantial declines in neutralizing antibody titers over 6 months, while memory CD4+ T cells, memory CD8+ T cells, and memory B cells exhibited small reductions (T cells) or increases (B cells). These observations appear to be consistent with the relatively high degree of protection maintained against hospitalizations with COVID-19 after these vaccines over 6 months, and the differential VE reported between mRNA COVID-19 vaccines and Ad26.COV2.S. These results of detailed immunological evaluations, coupled with analyses of VE data published for the various vaccine platforms, may also be relevant for other vaccine efforts.
Limitations of study
We did not evaluate recognition of variants, as this was evaluated in independent studies from our laboratories and others (Flemming, 2022; Gao et al., 2022; GeurtsvanKessel et al., 2022; Keeton et al., 2021; Tarke et al., 2021, 2022). The current study did not evaluate responses elicited by other vaccine platforms (AstraZeneca, Coronavac, Sinopharm, Sputnik) commonly utilized in other regions because samples from individuals vaccinated with these platforms were not available to us.
Quantification and statistical analysis
Cytometry data was analyzed using FlowJo 10.8.1. Statistical analyses were performed in GraphPad Prism 9.3.0, unless otherwise stated. The statistical details of the experiments are provided in the respective figure legends. Data plotted in linear scale were expressed as Mean ± SD. Data plotted in logarithmic scales were expressed as Geometric Mean ± Geometric SD. Mann–Whitney U or Wilcoxon tests were applied for unpaired or paired comparisons, respectively. Kruskal– Wallis and Dunn’s post-test were also applied for multiple comparisons in vaccine cohorts. Details pertaining to significance are also noted in the respective legends.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 11A9 (BUV496) [anti-CCR6] | BD Biosciences | Cat# 356,920; RRID: AB_2833076 |
| J252D4 (BV421) [anti-CXCR5] | BioLegend | Cat# 356,920; RRID: AB_2562303 |
| G025H7 (BV605) [anti-CXCR3] | BioLegend | Cat# 353,728; RRID: AB_2563157 |
| G043H7 (BV711) [anti-CCR7] | BioLegend | Cat# 353,228; RRID: AB_2563865 |
| G043H7 (PE/Cyanine7) [anti-CCR7] | BioLegend | Cat# 353,226; RRID: AB_11126145 |
| HB14 [anti-CD40] | Miltenyi Biotec | Cat# 130-108-041; RRID: AB_2660897 |
| UCHT1 (BUV395) [anti-CD3] | BD Biosciences | Cat# 563,546; RRID: AB_2744387 |
| SK7 (PerCP) [anti-CD3] | BioLegend | Cat# 344,814; RRID: AB_10639948 |
| SK1 (BUV805) [anti-CD8] | BD Biosciences | Cat# 612,889; RRID: AB_2833078 |
| 3G8 (BV510) [anti-CD16] | BioLegend | Cat# 302,048; RRID: AB_2562085 |
| 3G8 (PerCP) [anti-CD16] | BioLegend | Cat# 302,030; RRID: AB_940380 |
| 63D3 (BV510) [anti-CD14] | BioLegend | Cat# 367,124; RRID: AB_2716229 |
| 63D3 (PerCP) [anti-CD14] | BioLegend | Cat# 367,152; RRID: AB_2876693 |
| 2H7 (BV510) [anti-CD20] | BioLegend | Cat# 302,340; RRID: AB_2561941 |
| HI100 (BV570) [anti-CD45RA] | BioLegend | Cat# 304,132; RRID: AB_2563813 |
| SK3 (cFluor548) [anti-CD4] | Cytek | Cat# R7-20043 |
| G46-6 (APC-R700) [anti-HLA-DR] | BD Biosciences | Cat# 565,127; RRID: AB_2732055 |
| DX2 (BB700) [anti-CD95] | BD Biosciences | Cat# 566,542; RRID: AB_2869780 |
| DX2 (BUV737) [anti-CD95] | BD Biosciences | Cat# 612,790; RRID: AB_2870117 |
| HB-7 (BV650) [anti-CD38] | BioLegend | Cat# 356,620; RRID: AB_2566233 |
| EH12.2H7 (BV785) [anti-PD-1] | BioLegend | Cat# 329,930; RRID: AB_2563443 |
| FN50 (FITC) [anti-CD69] | BioLegend | Cat# 310,904; RRID: AB_314839 |
| 24-31 (PE/Dazzle594) [anti-CD154/CD40L] | BioLegend | Cat# 310,840; RRID: AB_2566245 |
| 24-31 (PerCP-eFluor710) [anti-CD154/CD40L] | Thermo Fisher Scientific | Cat# 6-1548-42; RRID: AB_10670357 |
| 4b4-1 (BUV737) [anti-CD137] | BD Bioscience | Cat# 741,861; RRID: AB_2871191 |
| Ber-Act35 (APC) [anti-CD134/OX40] | BioLegend | Cat# 350,008; RRID: AB_10719958 |
| 4S.B3 (FITC) [anti-IFNγ] | eBioscience | Cat# 11-7319-82; RRID: AB_465415 |
| MP4-25D2 (BUV737) [anti-IL-4] | BD Bioscience | Cat# 612,835; RRID: AB_2870157 |
| BL168 (BV785) [anti-IL17] | BioLegend | Cat# 512,338; RRID: AB_2566765 |
| MQ1-17H12 (BB700) [anti-IL-2] | BD Bioscience | Cat# 566,405; RRID: AB_2744488 |
| JES3-19F1 (PE/Dazzle594) [anti-IL10] | BioLegend | Cat# 506,812; RRID: AB_2632783 |
| Mab11 (eFluor450) [anti-TNFα] | Thermo Fisher Scientific | Cat# 48-7349-42; RRID: AB_2043889 |
| GB11 (Alexa Fluor 647) [anti-Granzyme B] | BD Bioscience | Cat# 560,212; RRID:AB_11154033 |
| Hu Fc Block Pure Fc1.3216 | BD Bioscience | Cat# 564,220; RRID: AB_2869554 |
| SJ25C1 (BUV563) [anti-CD19] | BD Biosciences | Cat# #612916; RRID: AB_2870201 |
| 1C6 (BUV805) [anti-CXCR3] | BD Biosciences | Cat# 612,790: RRID: AB_2871338 |
| IA6-2 (Pacific Blue) [anti-IgD] | BioLegend | Cat# 348,224; RRID:AB_2561597 |
| MHM-88 (BV570) [anti-IgM] | BioLegend | Cat# 314,517; RRID: AB_10913816) |
| M-T271 (BB515) [anti-CD27] | BD Biosciences | Cat# 564,642; RRID: AB_2744354 |
| IS11-8E10 (Vio Bright) [anti-IgA] | Miltenyi Biotec | Cat# 130-113-480; RRID:AB_2734076 |
| HCD56 (PerCP) [anti-CD56] | BioLegend | Cat# 318,342; RRID: AB_2561865 |
| M1310G05 (PerCP/Cyanine5.5) [anti-IgG] | BioLegend | Cat# 410,710; RRID: AB_2565788 |
| CY1G4 (PE/Dazzle594) [anti-CD71] | BioLegend | Cat# 334,120; RRID: AB_2734335 |
| 3.9 (PE/Cyanine5) [anti CD11c] | BioLegend | Cat# 301,610; RRID: AB_493578 |
| Bu31 (Alexa Fluor 700) [anti-CD21] | BioLegend | Cat# 354,918; RRID: AB_2750239 |
| HIT2 (APC/Fire810) [anti-CD38] | BioLegend | Cat# 303,550; RRID: AB_2860784 |
| HP6043 (Peroxidase) [anti-IgG] | Hybridoma Reagent Lab | Cat# HP6043-HRP |
| Biological samples | ||
| COVID-19 vaccinee donor blood samples | LJI Clinical Core | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Fixable Live/Dead Blue | Thermo Fischer Scientific | Cat# L34962 |
| SARS-CoV-2 Spike MP | Grifoni et al, 2020 | N/A |
| Brilliant Staining Buffer Plus | BD Biosciences | Cat# 566,385 RRID: AB_2869761 |
| Brilliant Stain Buffer | BD Biosciences | Cat# 566,349 RRID: AB_2869750 |
| SARS-CoV-2 Spike protein | Acro Biosystems | Cat# SPN-C82E9 |
| SARS-CoV-2 Spike protein Receptor-Binding Domain (RBD) | BioLegend | Cat# 793,906 |
| Alexa Fluor 647 Streptavidin | Thermo Fisher Scientific | Cat# S21374 |
| BV421 Streptavidin | BioLegend | Cat# 405,225 |
| BV711 Streptavidin | BioLegend | Cat# 405,241 |
| PE-Cy7 Streptavidin | BioLegend | Cat# 405,206 |
| PE-Cy5.5 Streptavidin | Thermo Fisher Scientific | Cat# SA1018 |
| Biotin | Avidity | Cat# Bir500A |
| Bacterial and virus strains | ||
| rVSV-SARS-CoV-2 | This study | N/A |
| Experimental models: Cell lines | ||
| VERO cells | ATCC | ATCC CCL-81 |
| HEK293T cells | ATCC | ATCC CRL-3216 |
| Software and algorithms | ||
| Flowjo 10.8.1 | FlowJo, LLC | www.flowjo.com |
| GraphPad Prism 9.3.0 | GraphPad | www.graphpad.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Shane Crotty (shane@lji.org).
Materials availability
Upon specific request and execution of a material transfer agreement (MTA) to the lead contact or to Daniela Weiskopf, aliquots of the peptide pools utilized in this study will be made available. Limitations might be applied to the availability of peptide reagents due to cost, quantity, demand, and availability.
Data and code availability
All the data generated in this study are available in the published article and summarized in the corresponding tables, figures and supplemental materials.
Experimental model and subject details
Human sample donors
A total of 354 peripheral blood samples were obtained from 102 participants who received either the Moderna mRNA-1273 (n = 30), Pfizer/BioNTech BNT162b2 (n = 30), Janssen Ad26.COV2.S (n = 30) and Novavax NVX-CoV2373 vaccines (n = 12), according to the approved dose schedule. Blood sample collection schedules are shown in Figure 1B. For baseline determinations, for a subset of donors, blood samples were collected before vaccination (T1). T2 was 2 weeks (targeting 15 ± 3 days) after the first immunization. T3 was 2 weeks after the second immunization, or approximately day 45 after the first immunization for the 1-dose Ad26.COV2.S. T4 was 3.5 months after the first dose. T5 was 6 months after the first dose. An overview of all samples analyzed in this study is provided in Figure 1C.
Both cohorts of mRNA vaccinees (mRNA-1273, Pfizer/BioNTech BNT162b2) received two doses of the vaccine (28 and 21 days apart, respectively). In the case of the Novavax NVX-CoV2373, we advertised locally to recruit subjects who had participated in an investigational NVX-CoV2373 trial conducted in the San Diego region, where two intramuscular 5-μg doses of NVX-CoV2373 or placebo were administered 21 days apart (Clinicaltrials.gov). The NVX-CoV2373 trial was structured such that donors initially received two doses of placebo or vaccine in a blinded manner and were then provided two doses of the opposite (vaccine or placebo), such that all participants were vaccinated. The gap between dose two and dose three for the LJI recruited cohort was 53 ± 38 days (Figure 1B); participants were blinded to their immunization regimen, and LJI had no information on which group the participants were in. All experiments performed at the La Jolla Institute (LJI) were approved by the institutional review boards (IRB) of the La Jolla Institute (IRB#: VD-214).
To avoid any batch effect all of the samples and cells in this study were collected, processed and stored by the clinical core at LJI following the same standard operating procedures. Experimental characterization of antibody and B cell responses were carried out by one person, utilizing the exact same protocol and same experimental equipment. T cell assay sample batching and standardization is described in sections below. After all experiments were run, experimentalists were unblinded to allow of vaccine specific analysis and data interpretation.
To compare levels of immune memory responses induced by any of the vaccine platforms to immune memory responses induced by infection with SARS-CoV-2, samples were used from individuals that experienced infection with SARS-CoV-2, originally reported in (Mateus et al., 2021). We matched the 7 months (209 days) post-vaccination samples with samples from convalescent donors collected on average 181 days (range 170–195) post symptoms onset (PSOB cell experiments were repeated for nine donors of this cohort and new five donors. The five new donors were selected randomly based to match the timepoint post symptom onset of the other samples. Seropositivity against SARS-CoV-2 was confirmed by ELISA, as described below. At the time of enrollment, all COVID-19 convalescent donors provided informed consent to participate in the present and future studies.
Exclusion criteria
Before analyzing the entire dataset for our cohort, we generated exclusion criteria as follows: subjects who tested positive for RBD and neutralization antibodies at baseline were excluded (one subject, mRNA-1273); subjects with no baseline sample available and whose RBD and neutralization antibody reached the peak after first-dose immunization (indicative of memory from previous infection) and were nucleocapsid (NC) antibody-positive were also excluded as previously infected subjects (one subject, mRNA-1273). In addition, any time points following a confirmed COVID-19 booster immunization were excluded for any subject (five subjects, BNT162b2, two Ad26.COV2.S, two NVX-CoV2373).
Method details
Peripheral blood mononuclear cells (PBMCs) and plasma isolation.
Whole blood samples from subjects vaccinated with the mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 COVID-19 vaccine and convalescent samples after COVID-19 infection were collected at La Jolla Institute in heparin-coated blood bags and centrifuged for 15 min at 803 g to separate the cellular fraction and plasma. Blood samples were collected at the times described above. The plasma was then carefully removed from the cell pellet and stored at minus 20 °C. PBMCs were isolated by density-gradient sedimentation using Ficoll-Paque (Lymphoprep, Nycomed Pharma, Oslo, Norway) as previously described (Dan et al., 2021; Grifoni et al., 2020; Mateus et al., 2020, 2021; Rydyznski Moderbacher et al., 2020). Isolated PBMCs were cryopreserved in cell recovery media containing 10% Dimethyl sulfoxide (DMSO) (Gibco), supplemented with 10% heat-inactivated fetal bovine serum (FBS; Hyclone Laboratories, Logan UT), and stored in liquid nitrogen until used in the assays. Plasma samples were used for antibody measurements by ELISA and PSV neutralization assay and PBMC samples were used for flow cytometry in the T cell and B cell assays.
SARS-CoV-2 ELISAs
The SARS-CoV-2 ELISAs have been described previously (Dan et al., 2021; Grifoni et al., 2020; Mateus et al., 2021; Rydyznski Moderbacher et al., 2020). Briefly, 96-well half-area plates (ThermoFisher 3690) were coated with 1 g/mL of antigen and incubated at 4 °C overnight. Antigens included recombinant SARS-CoV-2 RBD protein and spike protein, both obtained from the Saphire laboratory at LJI, and recombinant nucleocapsid protein (GenScript Z03488). The next day, plates were blocked with 3% milk in PBS (PBS) containing 0.05% Tween 20 for 1.5 h at room temperature. Plasma was heat-inactivated at 56 °C for 30 to 60 min. Plasma was diluted in 1% milk containing 0.05% Tween 20 in PBS starting at a 1:3 dilution followed by serial dilutions by three and incubated for 1.5 h at room temperature. Plates were washed five times with 0.05% PBS-Tween-20. Secondary antibodies were diluted in 1% milk containing 0.05% Tween 20 in PBS. Anti-human IgG peroxidase antibody produced in goat (Sigma A6029) was used at a 1:5,000 dilution. Plates were read on Spectramax Plate Reader at 450 nm, and data analysis was performed using SoftMax Pro.
End-point titers were plotted for each sample, using background-subtracted data. Negative and positive controls were used to standardize each assay and normalize across experiments. A positive control standard was created by pooling plasma from six convalescent COVID-19 donors to normalize between experiments. The limit of detection (LOD) was defined as 1:3 of IgG. The limit of quantification (LOQ) for COVID-19 vaccinated individuals were established based on pre-vaccinated individuals (timepoint 1) and set as the titer at which 95% of pre-vaccinated samples (T1) fell below the dotted line (Figures 2A and 2B). Titers, LOD, and LOQ were calibrated to the WHO International Reference Panel for anti-SARS-CoV-2 spike, RBD, and nucleocapsid binding antibody units per milliliter (WHO BAU/mL). For Spike IgG, the LOD was 0.20 with a LOQ of 1.024 (Figures 2A and 2D). For RBD IgG, the LOD was 0.83 with a LOQ of 7.12 (Figures 2B and 2E). For NC IgG, the LOD was 0.68 with a LOQ of 30.48 (Figure S1A).
For comparison among mRNA-1273, BNT162b, and Ad26.COV2.S over the entire 6 + month time period, log10 transformed end-point titers (WHO BAU/mL) were used to generate area under the curve (AUC) for each donor (Figures S1B-D). Donors with only one timepoint excluded. If there was no (T1), T1 was set as the LOD ET (BAU/mL). Correction factors for AUCs were determined by the number of time points and normalized to compare donor to donor. For comparison among mRNA-1273, BNT162b, Ad26.COV2.S, and NVZ-CoV2373 over the 3.5 months–6 months period, log10 transformed end-point titers (WHO BAU/mL) were used to generate area under the curve (AUC) for each donor (Figures S1E-G). Donors with only one timepoint excluded. Kruskal-Wallis tests for AUC were <0.0001 for Figures S1B-G. Comparison between different vaccines were made by Mann-Whitney. Values plotted show GMT with GM SD
To calculate the spike IgG half-life (t 1/2) for the memory time point, a simple linear regression was performed using log2-transformed T4-T5 paired data. The mean and 95% Cl of t 1/2 were described in the Results section.
Pseudovirus (PSV) Neutralization Assay
The PSV neutralization assays in samples from vaccinated subjects were performed as previously described (Dan et al., 2021; Grifoni et al., 2020; Mateus et al., 2021; Rydyznski Moderbacher et al., 2020). Briefly, 2.5×104 VERO cells (ATCC, Cat. No. CCL-81) were seeded in clear flat-bottom 96-well plates (Thermo Scientific, Cat. No. 165305) to produce a monolayer at the time of infection. Recombinant SARS-CoV-2-S-D614G pseudotyped VSV-ΔG-GFP were generated by transfecting HEK293T cells (ATCC, Cat. No. CRL-321) with plasmid phCMV3-SARS-CoV2-Spike kindly provided by Dr. E. Saphire and then infecting with VSV-ΔG-GFP. Pre-titrated rVSV-SARS-CoV-2-S-D614G was incubated with serially diluted human heat-inactivated plasma at 37 °C for 1–1.5 h before addition to confluent VERO cell monolayers. Cells were incubated for 16 h at 37 °C in 5% CO2 then fixed in 4% paraformaldehyde in PBS pH 7.4 (Santa Cruz, Cat. No. sc-281692) with 10 μg/mL of Hoechst (Thermo Scientific, Cat. No. 62249), and imaged using a CellInsight CX5 imager to quantify the total number of cells and infected GFP expressing cells to determine the percentage of infection. Neutralization titers or inhibition dose 50 (ID50) were calculated using the One-Site Fit Log IC50 model in Prism 9.3 (GraphPad). As internal quality control to define the variation inter-assay, a pooled plasma (secondary standard) from 10 donors who received the mRNA-1273 vaccine was included across the PSV neutralization assays. Samples that did not reach 50% inhibition at the lowest serum dilution of 1:20 were considered as non-neutralizing and the values were set to 19. PSV neutralization titers were done with two replicates per experiment. We included the WHO International Reference Panel for anti-SARS-CoV-2 immunoglobulin (20/268) to calibrate our PSV neutralization titers. The WHO IU calibrated neutralization ID50 (cID50-IU/mL) was graphed in figures. The limit of detection was calculated as 10.73 IU/mL.
T cell experiments
All T cell assays were carried out by two independent investigators (JMT and CMR) on the same model analyzer (Cytek Aurora). To ensure comparability between results, all samples were distributed in a blinded fashion and subjects which received different vaccines were distributed between the batches. All timepoints (T1-T5) from a given donor were run in the same experiment, including a positive and negative control for each donor. Furthermore, each independent experiment included a sample of the same control donor, with known SARS-CoV-2 specific CD4 and CD8 T cell reactivity. Deviation from the expected reactivity resulted in repetition of the experiment. Details of the T cell assays are provided below. After all experiments were run, experimentalists were unblinded to allow of vaccine specific analysis and data interpretation.
Spike megapool (spike MP)
We have previously developed the MP approach to allow simultaneous testing of a large number of epitopes, as reported previously (Grifoni et al., 2020; Mateus et al., 2020; Rydyznski Moderbacher et al., 2020). According to this approach, large numbers of different epitopes are solubilized, pooled, and re-lyophilized to avoid cell toxicity problems associated with high concentrations of DMSO typically encountered when single pre-solubilized epitopes are pooled (Grifoni et al., 2020; Mateus et al., 2020; Rydyznski Moderbacher et al., 2020). Here, were used for ex vivo stimulation of PBMCs for flow cytometry an MP to evaluate the antigen-specific T cell response against SARS-CoV-2 spike. We used a Spike MP of 253 overlapping peptides spanning the entire sequence of the Spike protein. As this peptide pool consists of peptides with a length of 15 amino acids, both CD4+ and CD8+ T cells have the capacity to recognize this MP, as described previously (Dan et al., 2021; Mateus et al., 2021).
Activation-induced markers (AIM) assay
The AIM assays in samples from subjects vaccinated with mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 COVID-19 vaccine were performed as previously described (Dan et al., 2021; Grifoni et al., 2020; Mateus et al., 2020, 2021; Rydyznski Moderbacher et al., 2020).
Spike-specific CD4+ T cells were measured as a percentage of OX40+CD137+ AIM+ or OX40+sCD40L+ AIM+ and spike-specific CD8+ T cells were measured as a percentage of CD69+CD137+ AIM+ CD8+ T cells after stimulation of PBMCs from subjects vaccinated with the Spike MP. Also, spike-specific circulating T follicular helper (cTfh) cells (CXCR5+OX40+CD40L+, as a percentage of CD4+ T cells) were defined by the AIM assay. Briefly, prior to the addition of the Spike MP, PBMCs were blocked at 37 °C for 15 min with 0.5 μg/mL anti-CD40 mAb (Miltenyi Biotec). Then, cells were incubated at 37 °C for 24 h in the presence of fluorescently labeled chemokine receptor antibodies (anti-CCR6, CXCR5, CXCR3, and CCR7) and the Spike MP (1 μg/mL) in 96-wells U-bottom plates, as previously described (Mateus et al., 2021; Rydyznski Moderbacher et al., 2020). In addition, PBMCs were incubated with an equimolar amount of DMSO as negative control and with phytohemagglutinin (5 μg/mL) (PHA, Roche) as a positive control. For the surface stain, 1×106 PBMCs were resuspended in PBS, incubated with BD human FC block (BD Biosciences, San Diego, CA) and the LIVE/DEAD marker in the dark for 15 min and washed with PBS. Then, the antibody mix containing the rest of the surface antibodies was added directly to cells and incubated for 60 min at 4 °C in the dark. Following surface staining, cells were then washed twice with PBS containing 3% FBS (FACS buffer). All samples were acquired on a Cytek Aurora (Cytek Biosciences, Fremont, CA). A list of antibodies used in this panel can be found in table S1 and a representative gating strategy of spike-specific CD4+ and CD8+ T cells using the AIM assay is shown in Figure S2, respectively.
Spike-specific CD4+ and CD8+ T cells were measured as background (DMSO) subtracted data, with a minimal DMSO level set to 0.005%. Response >0.02% and a stimulation index (SI) > 2 for CD4+ and >0.03% and SI > 3 for CD8+ T cells were considered positive. The limit of quantification (LOQ) for antigen-specific CD4+ T cell responses (0.03%) and antigen-specific CD8+ T cell responses (0.05%) was calculated using the median 2-fold SD of all negative controls.
Intracellular cytokine staining (ICS) assay
The ICS assays in samples from subjects vaccinated with mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 COVID-19 vaccine were performed as previously described (Mateus et al., 2021; Rydyznski Moderbacher et al., 2020).
Prior to the addition of the Spike MP, PBMC were blocked at 37 °C for 15 min with 0.5 μg/mL anti-CD40 mAb, as previously described (Mateus et al., 2021; Rydyznski Moderbacher et al., 2020). PBMCs were cultured in the presence of the Spike MP (1 μg/mL) for 20 h at 37 °C in 96-wells U-bottom plates. In addition, cells were incubated with an equimolar amount of DMSO as a negative control. After 20 h, Golgi-Plug and Golgi-Stop were added to the culture for 4 h along with the anti-CD69 Ab. Cells were then washed, incubated with BD human FC block, and stained with the LIVE/DEAD marker as described above. Then, cells were washed and surface stained for 30 min at 4 °C in the dark and fixed with 1% of paraformaldehyde (Sigma-Aldrich, St. Louis, MO). Subsequently, cells were permeated and stained with intracellular antibodies for 30 min at room temperature in the dark. All samples were acquired on a Cytek Aurora. Antibodies used in the ICS assay are listed in table S2 and a representative gating strategy of spike-specific CD4+ and CD8+ T cells producing IFNγ, TNFα, IL-2 and/or GzB using the ICS assay is shown in Figures 4A and 5A.
To define the spike-specific T cells by the ICS assay, we gated the cytokine- or GzB-producing cells together with the expression of iCD40L or CD69 on CD4+ or CD8+ T cells, respectively (Figures 4A and 5A). Then, a Boolean analysis was performed to define the multifunctional profiles on FlowJo 10.8.1. The overall response to spike, denoted as Secreted-effector+ (IFNγ, TNFα, IL-2, and/or GzB) or Cytokine+ (IFNγ, TNFα, and/or IL-2), was defined as the sum of the background-subtracted responses to each combination of individual cytokines or GzB. The total spike-specific CD4+ and CD8+ T cells producing IFNγ, TNFα, IL-2, and/or GzB are shown in Figures 4 and 5 and Figure S3, Figure S4. To define the multifunctional profiles of spike-specific T cells, all positive background-subtracted data (>0.005% and an SI > 2 for CD4+ T cells and CD8+ T cells) was aggregated into a combined sum of antigen-specific CD4+ or CD8+ T cells based on the number of functions. Values higher than the LOQ (0.01%) were considered for the analysis of the multifunctional spike-specific T cell responses. The average of the relative CD4+ and CD8+ T cell responses was calculated per donor and visit to define the proportion of multifunctional spike-specific T cell responses with one, two, three, and four functions (Figures 4B, 5C, S3A-B and S4H-I).
To calculate the ICS+ CD8+ T cell and AIM+ CD4+ T cell half-life (t 1/2) for the memory time point, a simple linear regression was performed using log2-transformed T4-T5 paired data. The mean and 95% Cl of t 1/2 were described in the Result section.
Detection of SARS-CoV-2-specific memory B cells
Detection of SARS-CoV-2-specific memory B cells (MBCs) in samples from subjects vaccinated with mRNA-1273, BNT162b2, Ad26.COV2.S, or NVX-CoV2373 COVID-19 vaccine was performed using B cell probes as previously described (Dan et al., 2021) Biotinylated full-length SARS-CoV-2 Spike protein was purchased from Acro Biosystems and SARS-CoV-2 Spike protein Receptor-Binding Domain (RBD) was purchased from BioLegend.
To enhance specificity, identification of both spike- and RBD-specific MBCs was performed using two fluorochromes for each protein. Thus, the biotinylated SARS-CoV-2 spike was incubated with either Alexa Fluor 647 or BV421 at a 20:1 ratio (∼6:1 M ratio) for 1 h at 4 °C. Biotinylated RBD was conjugated with BV711 or PE-Cy7 at a 2.2:1 ratio (∼4:1 M ratio). Streptavidin PE-Cy5.5 was used as a decoy probe to minimize background by eliminating SARS-CoV-2 nonspecific streptavidin-binding B cells. Then, 9×106 PBMCs were placed in U-bottom 96 well plates and stained with a solution consisting of 5 μM of biotin to avoid cross-reactivity among probes, 20 ng of decoy probe, 211 ng of Spike and 31.25 ng of RBD per sample, diluted in Brilliant Buffer and incubated for 1 h at 4°C, protected from light. After washing with PBS, cells were incubated with surface antibodies (table S3) diluted in Brilliant Staining Buffer for 30 min at 4 °C in dark. Viability staining was performed using Live/Dead Fixable Blue Stain Kit diluted at 1:200 in PBS and incubation at 4 °C for 30 min. For T-bet staining, cells were fixed and permeabilized in 100 uL of eBioscience Intracellular Fixation & Permeabilization Buffer Set (Thermo Fisher Scientific, catalog no. 88-8824-00) for 30 min at 4 °C. Cells were then washed with 1X BD Perm/Wash buffer (BD Biosciences, catalog no. 554723) and stained for T-bet in 50 uL of eBioscience Permeabilization Buffer (Thermo Fisher Scientific, catalog no. 00-8333-56). Cells were subsequently washed twice in 1X BD Perm/Wash and resuspended in FACS buffer before acquisition. The acquisition was performed using Cytek Aurora. The frequency of antigen-specific MBCs was expressed as a percentage of total memory B cells (Singlets, Lymphocytes, Live, CD3– CD14– CD16– CD56–CD19+ CD20+ CD38int/– IgD– and/or CD27+). For every experiment, PBMCs from a known positive control (COVID-19 convalescent subject) and an unexposed subject were included to ensure consistent sensitivity and specificity of the assay. Limit of detection was calculated as median + 2x SD of [1/(number of total B cells recorded)]
Correspondence: Correlation and principal component analysis (PCA)
Correlograms plotting the Spearman rank correlation coefficient (r) between all paired parameters were created with the corrplot package (v0.84) running in Rstudio (1.1.456) as previously described (Rydyznski Moderbacher et al., 2020). Spearman rank two-tailed p values were calculated using corr.mtest and graphed based on ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. The codes used are:
M = cor(DataFrame, method = "spearman", use = "pairwise.complete.obs")
MP = cor.mtest(DataFrame, method = "spearman", use = "pairwise.complete.obs", conf.level = 0.95, exact = FALSE)
corrplot(M, p.mat = MP$p, method = 'square', tl.col = "black", tl.cex = 0.7, tl.srt = 45, cl.align = "l", type = 'lower', sig.level = c(0.001, 0.01, 0.05), pch.cex = 0.7, insig = 'label_sig', pch.col = 'white')
The “DataFrame” is the data from each correlation matrix shown in Figure 6, collected and organized in spreadsheet.
The codes for PCA analysis are as follows:
res.pca = PCA(na.omit(MP), scale = TRUE)
fviz_eig(res.pca, addlabels = TRUE)
fviz_pca_biplot(res.pca, label = "var", labelsize = 3, repel = TRUE, geom.ind = "point", pointsize = 4, col.ind = Group$Vaccine, palette = c("darkgreen", "blue", "red", "purple"), col.var = "black", alpha.var = 0.5, addEllipses = TRUE, ellipse.alpha = 0, select.var = list(name = c("RBD IgG", "Spike IgG", "nAbs", "MBC", "aMBC", "cMBC", "CXCR3+ MBC", "AIM2+ CD4", "ICS + CD4", "ICS + CD8″)), ellipse.level = 0.8, legend.title = "Groups", invisible = "quali", title = "").
Acknowledgments
We thank Gina Levi and the LJI clinical core for assistance in sample coordination and blood processing. This project has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under grant CCHI AI142742 (S.C. and A.S.), Contract No. 75N9301900065 (A.S. and D.W.), and U01 CA260541-01 (D.W.). This work was additionally supported in part by LJI Institutional Funds and the NIAID under K08 award AI135078 (J.M.D.).
Author contributions
Conceptualization: D.W., A.S., S.C.; Methodology: Z.Z., J.M., J.M.D., C.H.C., C.R.M., R.I.G., F.H.C., J.C., E.A.E., B.G., N.I.B., D.W., A.G., A.T.; Formal analysis: J.M., J.M.D., Z.Z., C.H.C., C.R.M., R.I.G., F.H.C., B.G., N.I.B., D.W.; Investigation: Z.Z., J.M., J.M.D., C.H.C., C.R.M., J.C., E.A.E., B.G., N.I.B., D.W.; Project administration A.F., A.S., S.C., D.W.; Funding acquisition: J.M.D., A.S., S.C., D.W.; Writing: Z.Z., J.M., C.H.C., D.W., A.S., S.C.; Supervision: D.W., A.S., S.C.
Declaration of interests
A.S. is a consultant for Gritstone Bio, Flow Pharma, ImmunoScape, Avalia, Moderna, Fortress, Repertoire, Gerson Lehrman Group, RiverVest, MedaCorp, and Guggenheim. S.C. has consulted for GSK, JP Morgan, Citi, Morgan Stanley, Avalia NZ, Nutcracker Therapeutics, University of California, California State Universities, United Airlines, Adagio, and Roche. LJI has filed for patent protection for various aspects of T cell epitope and vaccine design work. All other authors declare no conflict of interest.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. We worked to ensure that the study questionnaires were prepared in an inclusive way. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper received support from a program designed to increase minority representation in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: May 27, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2022.05.022.
Supplemental information
References
- Atmar R.L., Lyke K.E., Deming M.E., Jackson L.A., Branche A.R., El Sahly H.M., Rostad C.A., Martin J.M., Johnston C., Rupp R.E., et al. Homologous and heterologous Covid-19 booster vaccinations. N. Engl. J. Med. 2022 doi: 10.1056/NEJMoa2116414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch D.H., Stephenson K.E., Sadoff J., Yu J., Chang A., Gebre M., McMahan K., Liu J., Chandrashekar A., Patel S., et al. Durable humoral and cellular immune responses 8 Months after Ad26.COV2.S vaccination. N. Engl. J. Med. 2021;385:951–953. doi: 10.1056/NEJMc2108829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., Sominsky L.A., Clark J.J., Adelsberg D.C., Bielak D.A., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- Cho A., Muecksch F., Schaefer-Babajew D., Wang Z., Finkin S., Gaebler C., Ramos V., Cipolla M., Mendoza P., Agudelo M., et al. Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination. Nature. 2021;600:517–522. doi: 10.1038/s41586-021-04060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinicaltrials.gov, (2022). A Study to Evaluate the Efficacy, Immune Response, and Safety of a COVID-19 Vaccine in Adults ≥ 18 Years with a Pediatric Expansion in Adolescents (12 to < 18 Years) at Risk for SARS-CoV-2. https://clinicaltrials.gov/ct2/show/NCT04611802
- Collier A.Y., Yu J., McMahan K., Liu J., Chandrashekar A., Maron J.S., Atyeo C., Martinez D.R., Ansel J.L., Aguayo R., et al. Differential kinetics of immune responses elicited by Covid-19 vaccines. N. Engl. J. Med. 2021;385:2010–2012. doi: 10.1056/NEJMc2115596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O'Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. T follicular helper cell Biology: a decade of Discovery and diseases. Immunity. 2019;50:1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashdorj N.J., Wirz O.F., Roltgen K., Haraguchi E., Buzzanco A.S., 3rd, Sibai M., Wang H., Miller J.A., Solis D., Sahoo M.K., et al. Direct comparison of antibody responses to four SARS-CoV-2 vaccines in Mongolia. Cell Host Microbe. 2021;29:1738–1743.e1734. doi: 10.1016/j.chom.2021.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N., Suthar M.S., Makowski M., O'Connell S., McDermott A.B., Flach B., Ledgerwood J.E., Mascola J.R., Graham B.S., Lin B.C., et al. Antibody persistence through 6 Months after the second dose of mRNA-1273 vaccine for Covid-19. N. Engl. J. Med. 2021;384:2259–2261. doi: 10.1056/NEJMc2103916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle L.M., Kotloff K.L., Gay C.L., Anez G., Adelglass J.M., Barrat Hernandez A.Q., Harper W.L., Duncanson D.M., McArthur M.A., Florescu D.F., et al. Efficacy and safety of NVX-CoV2373 in Adults in the United States and Mexico. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2116185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming A. Cross reactive T cells hold up against Omicron. Nat. Rev. Immunol. 2022 doi: 10.1038/s41577-022-00690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Cai C., Grifoni A., Muller T.R., Niessl J., Olofsson A., Humbert M., Hansson L., Osterborg A., Bergman P., et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 2022 doi: 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Geers D., Schmitz K.S., Mykytyn A.Z., Lamers M.M., Bogers S., Scherbeijn S., Gommers L., Sablerolles R.S.G., Nieuwkoop N.N., et al. Divergent SARS CoV-2 Omicron-reactive T- and B cell responses in COVID-19 vaccine recipients. Sci Immunol. 2022:eabo2202. doi: 10.1126/sciimmunol.abo2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell responses to SARS-CoV-2 Coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e1415. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrera G., Picozza M., D'Orso S., Placido R., Pirronello M., Verdiani A., Termine A., Fabrizio C., Giannessi F., Sambucci M., et al. BNT162b2 vaccination induces durable SARS-CoV-2-specific T cells with a stem cell memory phenotype. Sci Immunol. 2021;6:eabl5344. doi: 10.1126/sciimmunol.abl5344. [DOI] [PubMed] [Google Scholar]
- Heath P.T., Galiza E.P., Baxter D.N., Boffito M., Browne D., Burns F., Chadwick D.R., Clark R., Cosgrove C., Galloway J., et al. Safety and efficacy of NVX-CoV2373 Covid-19 vaccine. N. Engl. J. Med. 2021;385:1172–1183. doi: 10.1056/NEJMoa2107659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA vaccine against SARS-CoV-2 - Preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzierska K. Count on us: T cells in SARS-CoV-2 infection and vaccination. Cell Rep Med. 2022 doi: 10.1016/j.xcrm.2022.100562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton R., Richardson S.I., Moyo-Gwete T., Hermanus T., Tincho M.B., Benede N., Manamela N.P., Baguma R., Makhado Z., Ngomti A., et al. Prior infection with SARS-CoV-2 boosts and broadens Ad26.COV2.S immunogenicity in a variant-dependent manner. Cell Host Microbe. 2021;29:1611–1619.e1615. doi: 10.1016/j.chom.2021.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton R., Tincho M.B., Ngomti A., Baguma R., Benede N., Suzuki A., Khan K., Cele S., Bernstein M., Karim F., et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature. 2022 doi: 10.1038/s41586-022-04460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo N.K.H., Lim J.M.E., Gill U.S., de Alwis R., Tan N., Toh J.Z.N., Abbott J.E., Usai C., Ooi E.E., Low J.G.H., et al. Differential immunogenicity of homologous versus heterologous boost in Ad26.COV2.S vaccine recipients. Med (N Y) 2022;3:104–118.e104. doi: 10.1016/j.medj.2021.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Lafon E., Jager M., Bauer A., Reindl M., Bellmann-Weiler R., Wilflingseder D., Lass-Florl C., Posch W. Comparative analyses of IgG/IgA neutralizing effects induced by three COVID-19 vaccines against variants of concern. J. Allergy Clin. Immunol. 2022;149:1242–1252.e1212. doi: 10.1016/j.jaci.2022.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer K., Bettini E., Parvathaneni K., Painter M.M., Agarwal D., Lundgreen K.A., Weirick M., Muralidharan K., Castano D., Goel R.R., et al. Germinal center responses to SARS-CoV-2 mRNA vaccines in healthy and immunocompromised individuals. Cell. 2022 doi: 10.1016/j.cell.2022.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Sutton H., Cottrell C.A., Phung I., Ozorowski G., Sewall L.M., Nedellec R., Nakao C., Silva M., Richey S.T., et al. Long-lasting germinal center responses to a priming immunization with continuous proliferation and somatic mutation. bioRxiv. 2021 doi: 10.1101/2021.12.20.473537. 2021.2012.2020.473537. [DOI] [Google Scholar]
- Leon T.M., Dorabawila V., Nelson L., Lutterloh E., Bauer U.E., Backenson B., Bassett M.T., Henry H., Bregman B., Midgley C.M., et al. COVID-19 cases and hospitalizations by COVID-19 vaccination Status and previous COVID-19 Diagnosis - California and New York, may-November 2021. MMWR Morb. Mortal. Wkly. Rep. 2022;71:125–131. doi: 10.15585/mmwr.mm7104e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D.Y., Gu Y., Wheeler B., Young H., Holloway S., Sunny S.K., Moore Z., Zeng D. Effectiveness of Covid-19 vaccines over a 9-month period in North Carolina. N. Engl. J. Med. 2022 doi: 10.1056/NEJMoa2117128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Chandrashekar A., Sellers D., Barrett J., Jacob-Dolan C., Lifton M., McMahan K., Sciacca M., VanWyk H., Wu C., et al. Vaccines Elicit highly Conserved cellular immunity to SARS-CoV-2 Omicron. Nature. 2022 doi: 10.1038/s41586-022-04465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S., Callaway E., Kozlov M., Ledford H., Pickrell J., Van Noorden R. How COVID vaccines shaped 2021 in eight powerful charts. Nature. 2021;600:580–583. doi: 10.1038/d41586-021-03686-x. [DOI] [PubMed] [Google Scholar]
- Mateus J., Dan J.M., Zhang Z., Rydyznski Moderbacher C., Lammers M., Goodwin B., Sette A., Crotty S., Weiskopf D. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science. 2021;374:eabj9853. doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiuzzo G. 2020. Establishment of the WHO International Standard and Reference Panel for Anti-SARS-CoV-2 Antibody. [Google Scholar]
- Mudd P.A., Minervina A.A., Pogorelyy M.V., Turner J.S., Kim W., Kalaidina E., Petersen J., Schmitz A.J., Lei T., Haile A., et al. SARS-CoV-2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell. 2022;185:603–613. doi: 10.1016/j.cell.2021.12.026. e615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muecksch F., Wang Z., Cho A., Gaebler C., Tanfous T.B., DaSilva J., Bednarski E., Ramos V., Zong S., Johnson B., et al. Increased potency and Breadth of SARS-CoV-2 neutralizing antibodies after a Third mRNA vaccine dose. bioRxiv. 2022 doi: 10.1101/2022.02.14.480394. 2022.2002.2014.480394. [DOI] [Google Scholar]
- Naranbhai V., Garcia-Beltran W.F., Chang C.C., Mairena C.B., Thierauf J.C., Kirkpatrick G., Onozato M.L., Cheng J., St Denis K.J., Lam E.C., et al. 2021. Comparative Immunogenicity and Effectiveness of mRNA-1273, BNT162b2 and Ad26.COV2.S COVID-19 Vaccines. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranbhai V., Garcia-Beltran W.F., Chang C.C., Mairena C.B., Thierauf J.C., Kirkpatrick G., Onozato M.L., Cheng J., St Denis K.J., Lam E.C., et al. Comparative immunogenicity and effectiveness of mRNA-1273, BNT162b2 and Ad26.COV2.S COVID-19 vaccines. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiab593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom P., Ballin M., Nordstrom A. Risk of infection, hospitalisation, and death up to 9 months after a second dose of COVID-19 vaccine: a retrospective, total population cohort study in Sweden. Lancet. 2022;399:814–823. doi: 10.1016/S0140-6736(22)00089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novavax . 2022. Coronavirus Vaccine Candidate Updates. [Google Scholar]
- Oh J.E., Iijima N., Song E., Lu P., Klein J., Jiang R., Kleinstein S.H., Iwasaki A. Migrant memory B cells secrete luminal antibody in the vagina. Nature. 2019;571:122–126. doi: 10.1038/s41586-019-1285-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J.E., Song E., Moriyama M., Wong P., Zhang S., Jiang R., Strohmeier S., Kleinstein S.H., Krammer F., Iwasaki A. Intranasal priming induces local lung-resident B cell populations that secrete protective mucosal antiviral IgA. Science Immunology. 2021;6:eabj5129. doi: 10.1126/sciimmunol.abj5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajon R., Doria-Rose N.A., Shen X., Schmidt S.D., O'Dell S., McDanal C., Feng W., Tong J., Eaton A., Maglinao M., et al. SARS-CoV-2 Omicron variant neutralization after mRNA-1273 booster vaccination. N. Engl. J. Med. 2022 doi: 10.1056/NEJMc2119912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegu A., O'Connell S.E., Schmidt S.D., O'Dell S., Talana C.A., Lai L., Albert J., Anderson E., Bennett H., Corbett K.S., et al. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science. 2021;373:1372–1377. doi: 10.1126/science.abj4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilishvili T., Gierke R., Fleming-Dutra K.E., Farrar J.L., Mohr N.M., Talan D.A., Krishnadasan A., Harland K.K., Smithline H.A., Hou P.C., et al. Effectiveness of mRNA Covid-19 vaccine among U.S. Health Care Personnel. N. Engl. J. Med. 2021;385:e90. doi: 10.1056/NEJMoa2106599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard A.J., Bijker E.M. A guide to vaccinology: from basic principles to new developments. Nat. Rev. Immunol. 2021;21:83–100. doi: 10.1038/s41577-020-00479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouwels K.B., Pritchard E., Matthews P.C., Stoesser N., Eyre D.W., Vihta K.D., House T., Hay J., Bell J.I., Newton J.N., et al. Effect of Delta variant on viral burden and vaccine effectiveness against new SARS-CoV-2 infections in the UK. Nat. Med. 2021;27:2127–2135. doi: 10.1038/s41591-021-01548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda L.B., Morawski P.A., Pruner K.B., Fahning M.L., Howard C.A., Franko N., Logue J., Eggenberger J., Stokes C., Golez I., et al. Imprinted SARS-CoV-2-specific memory lymphocytes define hybrid immunity. 2022. Cell. [DOI] [PMC free article] [PubMed]
- Rosenberg E.S., Dorabawila V., Easton D., Bauer U.E., Kumar J., Hoen R., Hoefer D., Wu M., Lutterloh E., Conroy M.B., et al. Covid-19 vaccine effectiveness in New York state. N. Engl. J. Med. 2022;386:116–127. doi: 10.1056/NEJMoa2116063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsova K., Rubtsov A.V., Cancro M.P., Marrack P. Age-associated B cells: a T-bet-dependent effector with roles in protective and pathogenic immunity. J. Immunol. 2015;195:1933–1937. doi: 10.4049/jimmunol.1501209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012. doi: 10.1016/j.cell.2020.09.038. e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff J., Gray G., Vandebosch A., Cardenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., Quandt J., Bidmon N., Ulges A., Baum A., et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature. 2021;595:572–577. doi: 10.1038/s41586-021-03653-6. [DOI] [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A., Araki K., Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self W.H., Tenforde M.W., Rhoads J.P., Gaglani M., Ginde A.A., Douin D.J., Olson S.M., Talbot H.K., Casey J.D., Mohr N.M., et al. Comparative effectiveness of Moderna, Pfizer-BioNTech, and Janssen (Johnson & Johnson) vaccines in preventing COVID-19 hospitalizations among Adults without Immunocompromising Conditions - United States, March-August 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:1337–1343. doi: 10.15585/mmwr.mm7038e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensels D., Pierlet N., Penders J., Mesotten D., Heylen L. Comparison of SARS-CoV-2 antibody response following vaccination with BNT162b2 and mRNA-1273. JAMA. 2021;326:1533–1535. doi: 10.1001/jama.2021.15125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022 doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., Wang E., et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartof S.Y., Slezak J.M., Fischer H., Hong V., Ackerson B.K., Ranasinghe O.N., Frankland T.B., Ogun O.A., Zamparo J.M., Gray S., et al. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet. 2021;398:1407–1416. doi: 10.1016/S0140-6736(21)02183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S.J., Moreira E.D., Jr., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Polack F.P., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine through 6 Months. N. Engl. J. Med. 2021;385:1761–1773. doi: 10.1056/NEJMoa2110345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Guler A., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine Candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated in this study are available in the published article and summarized in the corresponding tables, figures and supplemental materials.