Abstract
Sequence homology between SARS-CoV-2 and common-cold human coronaviruses (HCoVs) raises the possibility that memory responses to prior HCoV infection can affect T cell response in COVID-19. We studied T cell responses to SARS-CoV-2 and HCoVs in convalescent COVID-19 donors and identified a highly conserved SARS-CoV-2 sequence, S811-831, with overlapping epitopes presented by common MHC class II proteins HLA-DQ5 and HLA-DP4. These epitopes are recognized by low-abundance CD4 T cells from convalescent COVID-19 donors, mRNA vaccine recipients, and uninfected donors. TCR sequencing revealed a diverse repertoire with public TCRs. T cell cross-reactivity is driven by the high conservation across human and animal coronaviruses of T cell contact residues in both HLA-DQ5 and HLA-DP4 binding frames, with distinct patterns of HCoV cross-reactivity explained by MHC class II binding preferences and substitutions at secondary TCR contact sites. These data highlight S811-831 as a highly conserved CD4 T cell epitope broadly recognized across human populations.
Keywords: heterologous immunity, seasonal coronavirus, T cell receptor repertoire, major histocompatibility complex, spike fusion peptide proximal region, CD4 T cells
Graphical abstract
Becerra-Artiles et al. study 2 partially overlapping CD4 T cell epitopes from the SARS-CoV-2 spike protein that are highly conserved across human and animal coronaviruses and are recognized by cross-reactive T cells present in most individuals. The molecular basis for the observed T cell cross-reactivity is discussed.
Introduction
Severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) infections result in coronavirus disease 2019 (COVID-19), a disease with a spectrum of clinical presentation from asymptomatic to severe disease and death, with most cases resulting in relatively mild symptoms (Wiersinga et al., 2020). Variations in the innate and adaptive host response to SARS-CoV-2 (Koch et al., 2021; Mallapaty, 2021) and genetic polymorphisms (Kousathanas et al., 2022; Soveg et al., 2021; Wickenhagen et al., 2021) play a critical role in the disparity of the clinical outcome. Studies also support a role for T cells (reviewed in Sette and Crotty, 2021). In severe cases, there are dysregulated T cell responses (Rydyznski Moderbacher et al., 2020), low CD4 and CD8 T cell counts are associated with severe disease (Chen et al., 2020; Du et al., 2020), and peak severity is inversely correlated with the frequency of SARS-CoV-2-specific interferon γ (IFN-γ)-producing CD8+ T cells (Rydyznski Moderbacher et al., 2020). In addition, early CD4 T cell responses are associated with mild disease (Peng et al., 2020; Tan et al., 2021).
Six other coronaviruses in addition to SARS-CoV-2 are known to infect humans: the highly pathogenic SARS-CoV and Middle Eastern respiratory syndrome (MERS)-CoV, which caused constrained outbreaks in 2002 and 2012, respectively, and the much less pathogenic α-coronaviruses 229E and NL63, and β-coronaviruses OC43 and HKU1, which probably emerged within the last few centuries, and now circulate seasonally and cause the common cold (Cui et al., 2019; Forni et al., 2017; Gaunt et al., 2010). Shortly after the discovery of SARS-CoV, it was shown that T cells from unexposed individuals recognize naturally processed and presented SARS antigens (Chen et al., 2005; Gioia et al., 2005; Yang et al., 2009). Serological cross-reactivity and sequence homology in major proteins between SARS and the human common cold coronaviruses (HCoVs) led to the suggestion that previous infections with HCoVs may have elicited the cross-reactive SARS-specific responses in unexposed individuals (Meyer et al., 2014), although some studies argued against this hypothesis (Chen et al., 2005; Yang et al., 2009). Immune cross-reactivity among the HCoVs has received attention recently with the spread of SARS-CoV-2 (Bonifacius et al., 2021; Lipsitch et al., 2020). Individuals with recent HCoV infection have been reported to have less severe COVID-19 (Sagar et al., 2021), although a similar study did not find that previous infection with HCoVs reduced the severity of COVID-19 (Gombar et al., 2021). Several studies have reported T cell responses to SARS-CoV-2 antigens in uninfected donors (reviewed in Grifoni et al., 2021), although the cross-reactive responses represent only a small fraction of the total response observed after SARS-CoV-2 infection (Le Bert et al., 2020; Tarke et al., 2021; Weiskopf et al., 2020). Recent evidence for a potentially protective role of preexisting SARS-CoV-2-specific T cells in COVID-19 comes from a study of people who had been exposed to SARS-CoV-2, but did not test positive for infection (i.e., undergoing abortive infections), who have early T cell responses to SARS-CoV-2 replication complex proteins that are highly conserved in coronaviruses and may have helped to prevent productive infection (Swadling et al., 2021). Preexisting memory T cells respond more quickly to spike (S) mRNA vaccine than do newly elicited T cells, with levels that correlate with antibodies specific for SARS-CoV-2 S protein, suggesting a possible supportive role for cross-reactive T cells in COVID-19 vaccination (Loyal et al., 2021).
To help clarify the role of cross-reactive T cell responses in COVID-19, we investigated SARS-CoV-2 S protein responses targeted by cross-reactive T cells isolated from previously uninfected donors and convalescent COVID-19 individuals. We used an unbiased screen to identify epitopes targeted by these cells. We systematically screened for T cells that were cross-reactive between SARS-CoV-2 and HCoV S epitopes in previously uninfected and convalescent COVID-19 donors. We identified a highly conserved immunodominant peptide broadly recognized by polyclonal and polyfunctional CD4 T cells. Two epitopes within this sequence are presented by HLA alleles common in populations worldwide. T cells that recognize this peptide respond to corresponding HCoV epitopes with similar avidity. The response is characterized by a broad repertoire of TCR, with subsets responding to different patterns of HCoV variations. The conserved sequence S811–831 (KPSKRSFIEDLLFNKVTLADA) can be used to follow cross-reactive responses to SARS-CoV-2 and may be a good candidate for consideration in pan-coronavirus vaccination strategies.
Results
T cell responses to coronavirus antigens in COVID-19 and uninfected donors
To characterize the cross-reactive T cell response to SARS-CoV-2 and HCoVs, we measured responses to overlapping peptide pools covering the S proteins of the four HCoVs and S, membrane (M), nucleoprotein (N), and envelope (E) proteins of SARS-CoV-2 using blood from recovered COVID-19 donors at convalescence and uninfected donors (both unexposed pre-pandemic donors sampled 2015–2018 and seronegative asymptomatic individuals sampled contemporaneously with the COVID-19 donors). We measured IFN-γ secretion in response to antigenic stimulation both ex vivo and in expanded cross-reactive T cells after a single in vitro stimulation with the four HCoV S peptide pools (S pools).
A representative COVID-19 donor (d0801) showed strong IFN-γ responses to peptide pools from SARS-CoV-2 S, M, and N but not E proteins (Figure 1A). Responses to HCoV S pools were weaker but clearly distinguishable from self-peptide and vehicle controls. Responses to HCoV S pools were expanded (∼27-fold) by in vitro stimulation (Figure 1B). Off-target expansion appeared to be minimal, as SARS-CoV-2 M, N, or E responses were not expanded. Responses to the SARS-CoV-2 S pool also were expanded by stimulation with the HCoV S pools (∼4-fold), indicating that a fraction of the SARS-CoV-2-responsive T cell population cross-reacts with HCoV homologs.
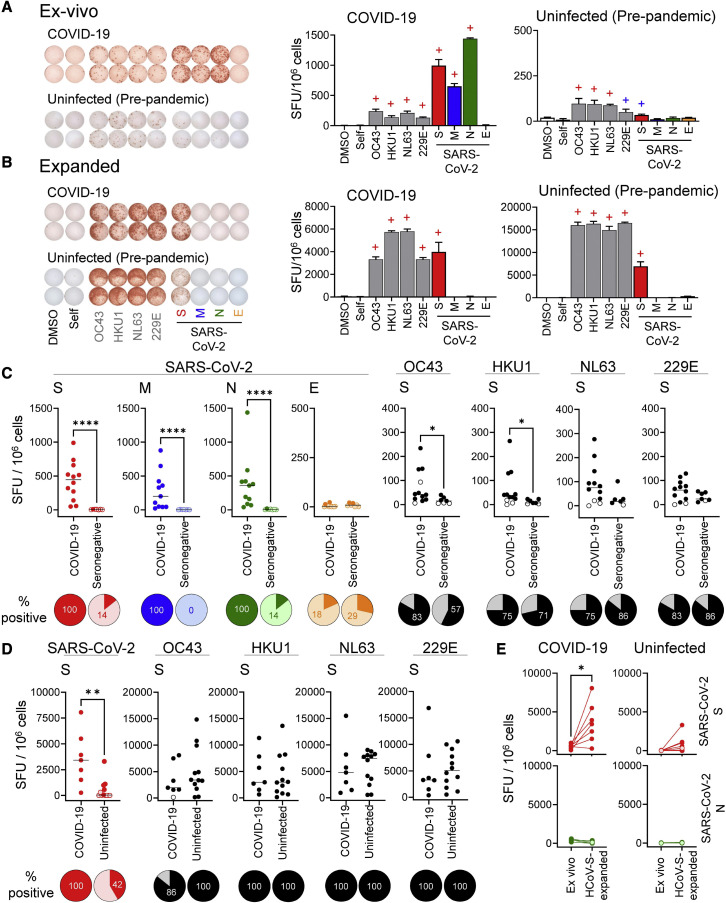
Figure 1.
Responses to coronavirus antigens in COVID-19 and uninfected donors
(A) Representative ex vivo responses for a COVID-19 donor and a pre-pandemic donor to S pools from OC43, HKU1, NL63, and 229E (gray), and S (red), M (blue), N (green), and E (orange) pools from SARS-CoV-2.
(B) Responses to re-stimulation after in vitro expansion with HCoV S pools in the same donors. IFN-γ ELISpot images and bar graphs (means ± standard deviations) are presented; +, positive responses by DFR1X (blue) or DFR2X (red) tests (Moodie et al., 2012).
(C) Summary of ex vivo responses in 12 COVID-19 donors at convalescence and 7 seronegative donors (pre-pandemic donors are shown in Figure S1).
(D) Summary of responses of HCoV-expanded T cells in 7 convalescent COVID-19 and 12 uninfected donors (both pre-pandemic and seronegative). (E) Responses to SARS-CoV-2 S or control N pools, before and after expansion with HCoV S pools, in convalescent COVID-19 and uninfected donors; paired t test: ∗p = 0.021.
For (C) and (D), Mann-Whitney test (∗∗p < 0.01; ∗∗∗∗p < 0.001); pies: percentage of positive responses (dark color) for each group/condition. For (C)–(E), positive responses by distribution free resampling (DFR) are indicated by dark-colored circles.
A pre-pandemic donor (L38) exhibited IFN-γ T cell responses to S pools from each of the four HCoVs and also from SARS-CoV-2 (Figure 1A). This donor was sampled before the emergence of SARS-CoV-2, and therefore the response to SARS-CoV-2 S suggests a possible cross-reactivity of T cells elicited by prior HCoV infection. To test this, we expanded T cells with HCoV S pools as just described (Figure 1B). SARS-CoV-2 S-specific responses expanded ∼90-fold after heterologous stimulation with the HCoV S pool, as were the HCoV-specific responses (∼51-fold). This indicates that some T cells from this unexposed donor responsive to HCoV homologs also are cross-reactive with SARS-CoV-2.
Similar responses were observed throughout the entire COVID-19 and uninfected study groups (Table S1). Ex vivo responses to SARS-CoV-2 S, M, and N pools were observed in all COVID-19 donors (Figure 1C, dark-colored circles). As previously observed (Le Bert et al., 2020; Mateus et al., 2020; Nelde et al., 2021; Tan et al., 2021), responses to SARS-CoV-2 antigens were seen in seronegative donors, with both the fraction of donors exhibiting positive responses and the numbers of responding T cells observed for these donors substantially lower than for COVID-19 donors (Figure 1C). Similar results were observed in pre-pandemic donors (Figure S1). Responses to the SARS-CoV-2 S pool in uninfected donors were relatively weak, and some positive responses may have been below the limit of detection in our ex vivo assay, although most donors responded to at least 1 of the HCoVs (Figures 1C and S1). Some of the COVID-19 donors appeared to have expanded responses to ≥1 HCoVs, with small but significant differences between COVID-19 and seronegative donors in response to OC43 and HKU1 S pools (Figure 1C).
After in vitro expansion, 42% of uninfected donors had IFN-γ responses for the SARS-CoV-2 S pool, compared with 7%–14% when tested ex vivo, and all but 1 donor was positive for at least 1 of the HCoVs (Figure 1D). T cells from COVID-19 donors responding to the SARS-CoV-2 S pool expanded on average 8-fold after stimulation with HCoV S pools, indicating that responding T cells recognize both SARS-CoV-2 and homologous HCoVs epitopes (Figure 1E, top). The expansion was specific for S antigens, as no expansion of N-specific responses was observed for either COVID-19 or uninfected donors (Figure 1E, bottom). T cell populations from uninfected donors responded to the S pool from SARS-CoV-2, and these responses also expanded after stimulation (average 66-fold increase) with homologous epitopes. Thus, both uninfected and COVID-19 donors exhibited T cell responses cross-reactive between corresponding HCoVs and SARS-CoV-2 antigens.
Identification of cross-reactive peptides
To identify epitopes recognized in these cross-reactive responses, we screened overlapping peptide libraries covering the SARS-CoV-2 S protein. We assessed responses in T cells from 3 COVID-19 donors at convalescence, enriching for cross-reactive populations by expansion in vitro with HCoV S pools. We used a pool-deconvolution approach to identify individual peptides. First, responses to peptides grouped into pools of 10 were measured (Figure 2A), and second, pools exhibiting positive responses were deconvoluted and retested to identify individual peptides (Figure 2B). In this manner, we identified 3 pairs of overlapping peptides: 163/164 (S811–831), recognized in 3 donors; 190/191 (S946–966), recognized in 2 donors; and 198/199 (S986–1,006), recognized by a single donor (Figure 2C).
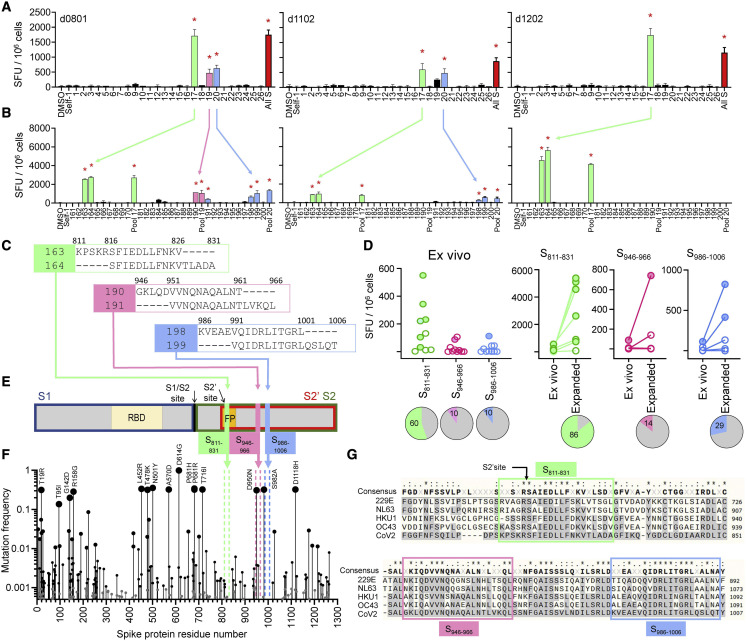
Figure 2.
Identification of cross-reactive peptides
(A) IFN-γ ELISpot responses of in vitro HCoV-expanded lines from 3 COVID-19 donors to re-stimulation with SARS-CoV-2 S overlapping peptide pools (pool number on x axis; all S, all S peptides; DMSO and Self-1, negative controls).
(B) Deconvolution of positive pools (peptide number on x axis; parent pool included).
(C) Amino acid sequences of candidate epitopes: S811–826/S816–831 (green), S946–961/S951–966 (pink), and S986–1,001/S991–1,006 (blue).
(D) Ex vivo responses to candidate epitopes in 10 COVID-19 donors, comparison of ex vivo and in vitro expanded responses (filled circles, positive; empty circles, negative by DFR2X). Pies: percentage of positive responses.
(E) Schematic of SARS-CoV-2 S protein with location of candidate epitopes. RBD, receptor binding domain; FP, fusion peptide; cleavage sites (S1/S2 and S2′) and cleavage products S1 (blue box), S2 (green box), and S2ʹ (red box).
(F) Mutation frequency is indicated by size of circles. Location of candidate epitopes in the protein shown by colored broken vertical lines. Common mutations are also indicated.
(G) Sequence alignment of S proteins from SARS-CoV-2 (bottom) and HCoVs (229E, NL63, HKU1, and OC43) in the region of the candidate epitopes (enclosed in boxes).
For (A) and (B), bar graphs: means ± standard deviations; red stars: positive responses by DFR2X.
We evaluated the responses to S811–831, S946–966, and S986–1,006 in additional convalescent COVID-19 donors. Of 10 donors analyzed, 6 showed ex vivo responses to S811–831, and 1 donor each recognized S946–966 and S986–1,006 (Figure 2D, left). After in vitro expansion with HCoV S pools, an additional donor was positive for peptide S811–831 (Figure 2D, right). Thus, S811–831 is recognized by a substantial fraction of COVID-19 donors across histocompatibility leukocyte antigen (HLA) types (Table S1).
All 3 cross-reactive candidate epitopes identified derive from the spike S2′ domain (Figure 2E). The S811–831 sequence contains the S2′ cleavage site and the fusion peptide (FP), critical for viral entry (Xia, 2021). S946–966 is in the first heptapeptide repeat (HP) and S986–1,006 is between HP1 and HP2. These regions are highly conserved among SARS-CoV-2 variants, including Delta (B1.617.2) and Omicron (B.1.1.529), with a mutation frequency <0.01 for most positions except S950 in S946–966 (Figure 2F). These regions also are highly conserved among the 4 HCoVs (Figure 2G). Overall, these results indicate that the S811–831 region is a broadly recognized immunogenic hotspot in which mutations are highly restricted.
Functional characteristics of cross-reactive T cell populations
To assess the functional characteristics of T cells responding to these peptides, we performed a phenotypic analysis of in vitro-expanded cross-reactive T cells (Figure 3 ). Most expanded cells are CD4+ (Figure 3B) with a predominantly effector-memory phenotype (Figure 3C). Intracellular cytokine secretion (ICS) assay showed that cells responding to re-stimulation with the SARS-CoV-2 S pool (CoV-2 S) or the individual peptides S811–826 and S816–831 were exclusively within the CD4+ T cell population, and produced mainly IFN-γ, some tumor necrosis factor α (TNF-α), very little interleukin-2 (IL-2), and mobilized CD107a to surface, suggesting a T helper 1 cell (Th1) population with cytotoxic potential (Figure 3D). Furthermore, the expanded cells were polyfunctional. Approximately half of the responding cells produced 1 or 2 cytokines and mobilized CD107a, although cells expressing only CD107a or only IFN-γ were frequent (Figure 3E). t-Distributed stochastic neighbor embedding (t-SNE) analysis of pooled samples reveals 2 major clusters (g1 and g2) along with some dispersed populations (Figure 3F). Cluster g1 includes cells producing high IFN-γ, TNF-α, and mobilizing CD107a. Cluster g2 includes cells that still mobilize CD107a but produce less IFN-γ and no TNF-α. Overall, responses to the SARS-CoV-2 S pool, S811–826, and S816–831 were similar. T cell responses to the other cross-reactive epitopes showed similar trends (Figures S2A and S2B). Since the responding T cells are mostly CD4+, we conclude that S811–831, S946–966, and S986–1,006 contain epitopes presented by major histocompatibility complex class II (MHC class II) proteins.
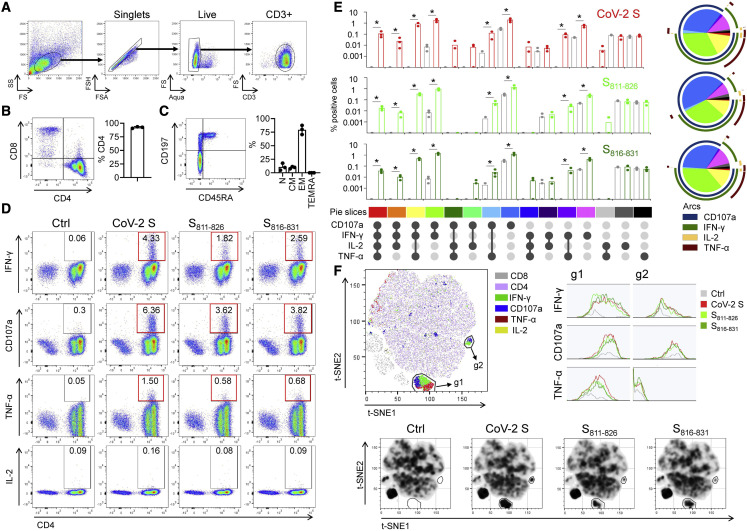
Figure 3.
Functional characterization of in vitro-expanded cross-reactive T cells
(A) Gating.
(B) Representative plot of CD4/CD8 distribution in the CD3+ population and percentage of CD4+ cells (bar graph) in cells expanded from 3 donors.
(C) Representative plot of CD45RA/CD197 in the CD4+ population and summary of the percentage of naive (N), central memory (CM), effector memory (EM), and EM re-expressing RA (TEMRA) populations in expanded cells (n = 3).
(D) Representative ICS plots for IFN-γ, TNF-α, and IL-2 production, and CD107a mobilization, in the CD3+ population after re-stimulation of expanded cells with SARS-CoV-2 S pool (CoV-2 S) or peptides S811–826 and S816–831; positive responses shown in red boxes (>3-fold background).
(E) Visualization of the polyfunctional response using Simplified Presentation of Incredibly Complex Evaluations (SPICE) (Roederer et al., 2011): bar graph (means ± standard deviations) for each stimulating antigen (red for CoV-2, light green for S811–826, dark green for S816–831) and comparison to control (gray) (p < 0.05, Wilcoxon rank-sum test); pie and arcs show the combined contribution of each marker (pie slice colors correspond to colors shown at the bottom of the bar graphs).
(F) t-SNE analysis of concatenated data from 3 donors for stimulation with same antigens, showing density plots for each condition. Two gates (g1 and g2) were drawn, indicating major differences among stimulated and unstimulated samples. Histograms show IFN-γ, TNF-α, and CD107a in each gate. Representative density plots for responses of d0801 are shown (see also Figure S2).
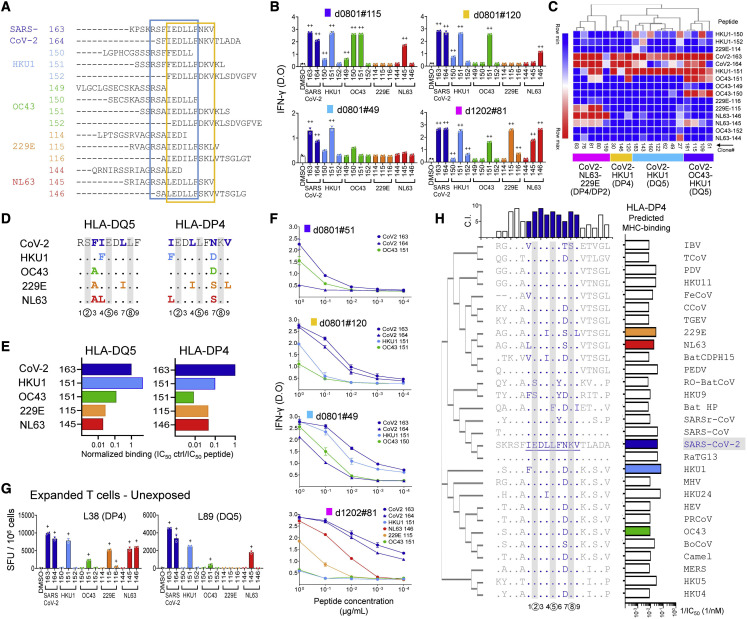
HLA restriction and epitope mapping
For epitope mapping studies, we focused on the broadly recognized S811–831. Because of the possibility that multiple epitopes and/or multiple MHC molecules may be involved, we generated a panel of T cell clones that recognize S811–831. Forty-nine clones, isolated using expanded cells from the same donors as investigated in Figure 2, were tested for HLA restriction and epitope recognition (Table S2). Donors d0801, d1102, and d1202 express a total of 15 different MHC class II alleles (Table S1), 9 of which are predicted to bind at least 1 epitope within S811–831 (Table S3). To define HLA restriction for these clones, we used partially HLA-matched antigen-presenting cells (APCs) (Figures 4A and 4C) and blocking experiments using antibodies to HLA-DR, -DQ, and -DP, and MHC class I (Figures 4B and 4D). For example, clone #143 from d0801 responded when S811–831 was presented by LG2 cells, which share DQ5 (DQA1∗01:01/DQB1∗05:01) and DP4 (DPA1∗01:03/DPB1∗04:01) with the donor (Figure 4E), but not when presented by 9068 cells, which share DR8 (DRB1∗08:01), nor with single HLA-transfected DP4 cells (MN605), which suggests a DQ5 restriction (Figure 4A, left panel). This was confirmed by blocking studies, in which inhibition was observed with anti-DQ, but not anti-DR, anti-DP, or anti-MHC class I (Figure 4B, left panel). Similarly, clone #83 from d1202 showed strong reactivity to 9068 cells, which was blocked by anti-DP antibody, suggesting restriction by DP2 (DPA1∗01:03 DPB1∗02:01), the DP allele shared between 9068 and the donor (Figures 4A and 4B, center panels). Clone #159 from d0801 showed reactivity to the DP4-transfected line and to a lesser extent to LG2 cells, which also express DP4; the blocking experiment confirmed the DP4 restriction (Figures 4A and 4B, right panels). In this way, 13 clones from d0801 were categorized as DQ5 restricted and 2 as DP4 restricted, while 3 of the clones from d1202 were categorized as DP2 restricted and 1 clone as DP4 restricted (Table S2). DP2 and DP4 are very similar proteins (Figure S3), and 2 clones were observed to recognize S811–831 presented by either allele (d0801#120 and #159; Table S2).
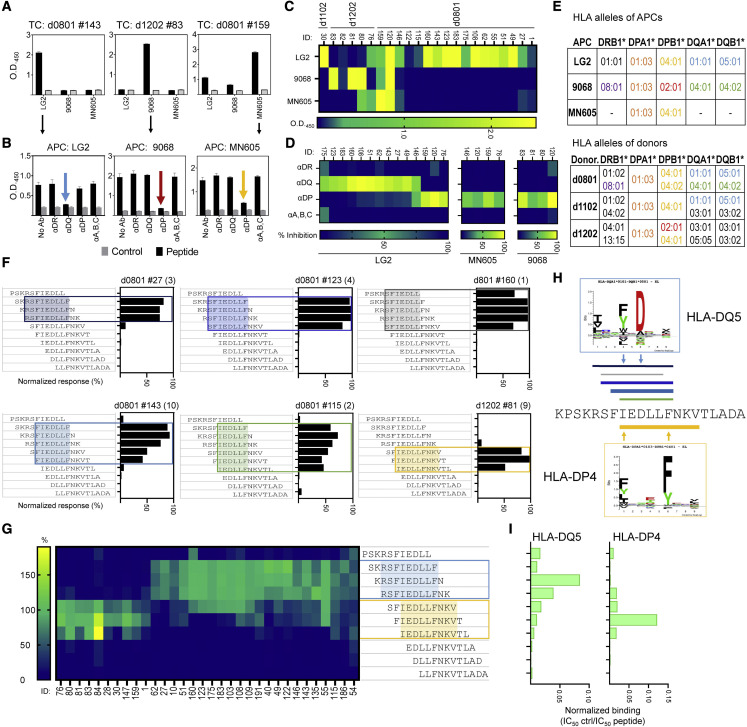
Figure 4.
HLA restriction and epitope mapping for S811–831
(A) IFN-γ responses (ELISA) of 3 representative T cell clones with confirmed reactivity to S811–831 presented by partially HLA-matched APCs (LG2, 9068, MN605); Bar graphs: means ± standard deviations, with responses to S811–831 in black and DMSO control in gray.
(B) Antigen presentation blocking assays using T cell clones presented in A, and antibodies to DR, DQ, DP, and MHC class I. Inhibition indicated by colored arrows. Bar graphs: means ± standard deviations.
(C) Summary of T cell clone responses (clone number on top of graph) to S811–831; color scale represents the ΔOD450 (peptide-DMSO).
(D) Summary of blocking assay; color scale represents the percentage of inhibition.
(E) HLA alleles expressed by APCs shared with donors originating the T cell clones.
(F) Responses to a set of 11-mer peptides covering S811–831; bars: response (%) of each truncated peptide relative to the full-length peptide. Representative clones for different reactivity patterns are shown; clone number on top; number of clones exhibiting a similar pattern in parentheses. Minimal sequence required to explain reactivity is highlighted.
(G) Summary of the 2 partially overlapping patterns observed in 32 clones (boxed in blue for DQ and yellow for DP).
(H) Location of minimal epitopes from (F), shown aligned with S811–831 (color of lines match color of boxes; thickness of line represents approximate frequency of the pattern). Binding motifs for DQ5 and DP4 are shown as sequence logos.
(I) Normalized binding (half-maximal inhibitory concentration[IC50]-positive control/IC50 peptide) for truncated peptides to purified DQ5 or DP4 (see also Figures S2, S3 and S4).
To map the precise epitopes recognized by the clones, we evaluated responses to a series of 11-mer peptides covering the S811–831 sequence. We identified 6 distinct patterns of minimal peptide sequences required to activate the 29 clones analyzed (Figure 4F; Table S2), which segregated into 2 main groups sharing similar reactivity (Figure 4G). One group, consisting of clones that mapped to DQ5 in the presentation/blocking experiments, responded to length variants of core epitope RSFIEDLLF (blue box in Figure 4G). The other group, consisting of clones that mapped to DP2 or DP4, responded to length variants of a different core epitope, IEDLLFNKV (yellow box in Figure 4G). DQ5-restricted clones recognized minimal peptide sequences 6–9 residues long, whereas the DP4-restricted clones recognized minimal peptide sequences 9 residues long (Figures 4F, shaded regions, and 4H, lines above and below sequence). These recognition patterns agree with the binding predictions for these specific alleles (Table S3), with minimal peptide sequences centered on the respective predicted core epitopes (Figure 4H).
We validated the tight binding of S811–831 to DQ5 and DP4 using purified proteins in competition binding assays (Figure S4) and confirmed the differential core epitope selection by DQ5 and DP4 using the same set of minimal-length peptides as was used to assess T cell recognition patterns (Figure 4I). Maximal binding to DQ5 was observed for peptides with the core epitope RSFIEDLLF, whereas maximal binding to DP4 was observed for peptides with the core epitope IEDLLFNKV. Thus, DQ5 and DP4 both bind epitopes within S811–831, but with a 3-residue register shift between the core epitopes, consistent with the recognition patterns observed for the DQ5-, DP2-, and DP4-restricted T cell clones (Figure 4H).
We used similar sets of overlapping truncated peptides to help map the specificity of T cell clones responding to S946–966 and S986–1,006 (Figure S2C). Combining patterns of responses to truncated peptides with binding predictions for HLA alleles present in responding donors (Table S3), we infer that clones recognizing these 2 regions likely are restricted by DR1 (DRB1∗01:02) and/or DR53 (DRB4∗01:03) (Figure S2D).
Cross-reactive recognition of S811–831 variants from circulating HCoVs
S811–831 is highly conserved across HCoVs. To determine the extent of T cell cross-reactivity between homologous epitopes, we tested responses of T cell clones from COVID-19 donors to overlapping HCoV peptides covering the SARS-CoV-2 S811–831 sequence (Figure 5A). All of the clones tested responded to at least 1 HCoV homolog (Table S2). Responses by 4 representative clones are shown in Figure 5B. Unsupervised hierarchical clustering of the responses of 17 clones to the homolog peptides revealed 4 major groups (Figure 5C), with preferences for homologs from different viruses, and DQ5 or DP2/DP4 restriction as identified previously. For instance, clones in the purple group, categorized as DQ5 restricted, show reactivity to SARS-CoV-2, OC43, and HKU1, while clones in the turquoise group, which is also DQ5 restricted, show reactivity only to SARS-CoV-2 and HKU1. Clones in the gold and magenta groups, categorized as DP4 or DP2, show reactivity to SARS-CoV-2 and HKU1 (gold) or to SARS-CoV-2, NL63, and 229E (magenta).
Figure 5.
Cross-reactive recognition of S811–831 homologs
(A) Sequences of SARS-CoV-2 S811–831 and corresponding HKU1, OC43, 229E, and NL63 peptides; boxes enclose the DQ5 (blue) and DP4 (yellow) 9-mer core epitopes.
(B) Responses of representative T cell clones to the peptides (IFN-γ ELISA). Bar graphs: means ± standard deviations)
(C) Hierarchical clustering of responses of 19 T cell clones to different homologs. Four major groups were defined: CoV2-OC43-HKU1/DQ5 (purple), CoV2-HKU1/DQ5 (cyan), CoV2-HKU1/DP4 (gold), and CoV2-NL63-229E/DP4-DP2 (magenta).
(D) Sequence alignment of DP4 and DQ5 core epitopes from SARS-CoV-2 and HCoVs. Numbers at bottom indicate peptide position, with T cell contacts encircled. Changes relative to SARS-CoV-2 shown in color; T cell contacts with gray bars.
(E) Normalized binding of homolog peptides relative to S811–831.
(F) Dose response of selected T cell clones to homolog peptides (IFN-γ ELISA). Symbols show means ± standard deviations.
(G) Responses of in vitro expanded cells from unexposed donors (expanded with SARS-CoV-2 peptides) to homolog peptides (IFN-γ ELISpot; +, positive response by DFR2X); relevant HLA shown in parentheses.
(H) Sequence alignment of S811–831 and positional homologs in 28 coronaviruses that sample the Orthocoronavirinae subfamily (Table S4). Complete SARS-CoV-2 sequence and differences for other viruses are shown. DP4 core epitope in blue, T cell contact positions with gray bars. Phylogenetic tree of the S proteins shown at left; conservation index (C.I.) for each position at top; predicted DP4 binding affinities at right (see also Figure S5).
These response patterns can be understood by considering the S811–831 homolog sequences in the various viruses. Alignment of SARS-CoV-2 and HCoV sequences in the region of the DQ5 and DP4 core epitopes shows that for both binding registers, the P2, P5, and P8 positions are 100% conserved (Figure 5D). These positions are located where the major T cell contacts are expected for conventionally oriented T cell receptors (TCRs) (Rossjohn et al., 2015; Stern and Wiley, 1994). Conservation of the key TCR contact residues helps to explain the overall high degree of SARS-CoV-2-HCoV cross-reactivity in these epitopes. Residues at other positions are less conserved in both DQ5 and DP4 registers, contributing to the differential MHC binding that we observed in the binding assays (Figure 5E). DQ5 showed a preference for the HKU1 homolog, which binds more strongly than SARS-CoV-2, with reduced binding to OC43 and substantially weaker binding to NL63 and 229E. For DP4, the strongest preference again was for HKU1, followed by 229E and NL63, with less binding for OC43. These patterns could be understood in terms of substitutions at the positions expected to bind into principal MHC side-chain binding pockets at P1, P4, P6, or P9, and minor pockets at P3 and P7. For example, the improved DQ5 binding of the HKU1 homolog relative to SARS-CoV-2 apparently is due to Phe at the P4 position, which is a preferred residue at the P4 position, and the only difference between SARS-CoV-2 and HKU1 core epitope sequences (Figure 4H). For DP4, weaker binding of the OC43 homolog appears to be due to an Asn-to-Asp substitution at the P7 position, which is partially compensated in the HKU1 homolog by Phe at the preferred DP4 P1 anchor residue position (Sidney et al., 2010b). These binding differences also help to explain the reactivity groups defined in Figure 5C. The purple and cyan groups, categorized as DQ5, show little or no reactivity with 229E and NL63, which exhibited minimal DQ5 binding. Similarly, the gold and magenta groups, categorized as DP4, exclude OC43, which was the worst DP4 binder. The restricted specificity for SARS-CoV-2 and HKU1 in the cyan group probably indicates an important TCR preference for Phe over Ala at the P3 position in the DQ5 register (Sidney et al., 2010a), and in the gold group, an important T cell contact at the Asn/Asp at P7 in the DP4 register.
Previous studies have reported low avidity for T cells cross-reactive between SARS-CoV-2 and HCoV homologs (Bacher et al., 2020; Dykema et al., 2021; Saini et al., 2021), although another study reported comparable responses for some epitopes (Mateus et al., 2020). We evaluated the dose response to SARS-CoV-2 and HCoV homologs for clones in each group. In most cases, the preferred HCoV homolog stimulated comparable IFN-γ secretion relative to the corresponding SARS-CoV-2 peptide over a wide range of concentrations (Figure 5F). For instance, for clone #51 in the purple group (DQ5, CoV2/OC43/HKU1), dose-dependent reactivities to S811–826 and OC43-151 peptides were similar. Likewise, for clone #49 in the cyan group (DQ5, CoV2/HKU1) dose-dependent reactivities for S811–826 and HKU1-151 also were similar. For clone #120 in the gold group (DP4, CoV2/HKU1) and clone #81 in the magenta group (DP4/DP2, CoV2/NL63/229E), reactivities for the targeted HKU1 and NL63 peptides were approximately 10-fold weaker than for SARS-CoV-2. We also tested cross-reactive T cells from unexposed donors obtained by expansion with S811–831. Patterns of cross-reactivity observed for these donors were similar to those defined for the clones from COVID-19 donors, with donor L38 exhibiting a pattern similar to the magenta group, and L89 similar to the gold group (Figure 5G).
S811–831 is highly conserved across human and animal coronaviruses
We performed an in silico analysis of S protein sequences from a variety of coronaviruses infecting animal species (Table S4) to explore S811–831, S946–966, and S986–1,006 conservation among potentially emergent viruses, and the prospects for these sequences as components of a pan-coronavirus vaccine. The S811–831 DP4 epitope was highly conserved, with predicted T cell contacts at positions P2, P5, and P8 almost invariant, and most species retaining high predicted MHC binding affinity (Figure 5H). The partially overlapping DQ5 epitope was similarly conserved, although in this case, substitutions clustered in MHC contact positions and in many cases strikingly reduced predicted MHC binding (Figure S5A). For S946–966, the tentatively assigned DR1/DR53 binding frame was less conserved, with approximately half of the sequences having substitutions at key predicted T cell contacts and several of the others having reduced predicted MHC binding (Figure S5B). In contrast, S986–1,006 was highly conserved, particularly in the tentatively assigned DR1 binding frame, with predicted T cell contacts invariant and MHC binding largely preserved across the sequences (Figure S5C). Thus, CD4 T cell responses to SARS-CoV-2 S811–831 and S986–1,006 would be expected to exhibit substantial cross-reactivity with many human and animal coronaviruses.
Broad recognition of S811–831 in the population
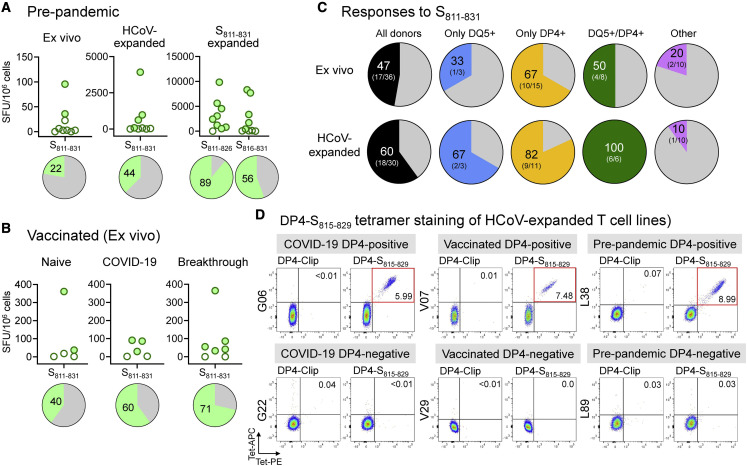
We evaluated the frequency of response to S811–831 in unexposed donors and donors receiving mRNA-based COVID-19 vaccines, directly ex vivo and after expansion in vitro. Of 9 donors analyzed, we found responses ex vivo in 2 donors; in cells expanded with HCoV S pools, we found responses in 4 donors, and in cells expanded with S811–831, we found responses in 8 donors (Figure 6A). In vaccinated individuals, we observed responses ex vivo in 10 of 17 donors tested, including those with no evidence of previous COVID-19 (2 positives of 5 tested), those who had COVID-19 and were later vaccinated (3 positives of 5 tested), and those who were vaccinated and later got COVID-19 (5 positives of 7 tested) (Figure 6B). As noted in Figure 2D, 6 of 10 convalescent COVID-19 donors exhibited responses to S811–831 ex vivo. Thus, the S811–831 epitope is recognized broadly in unexposed, mRNA-vaccinated, and COVID-19 donors.
Figure 6.
Broad recognition of S811–831 in the population
(A) Responses (IFN-γ ELISpot) to S811–831 in 9 pre-pandemic donors, ex vivo and after in vitro expansion with HCoV S pools or SARS-CoV-2 S811–831.
(B) Ex vivo responses to S811–831 in vaccinated donors (naive: no previous COVID-19; COVID-19, previous COVID-19; breakthrough, COVID-19 after vaccination). Pies show percentage of positive responses in (A) and (B).
(C) Responses to S811–831ex vivo and in HCoV-expanded cells for donors categorized according to DQ5 and DP4 status: all donors: donors regardless of DQ5/DP4 status; only DQ5: express DQ5 but not DP4; only DP4: express DP4 but not DQ5; DQ5/DP4: express both alleles; other: express neither allele. Percentage of donors with a positive response and number of positive and total donors are shown.
(D) Representative DP4-S815–829 double-tetramer staining (PE and APC) of HCoV-expanded T cells from COVID-19, vaccine recipients, and unexposed donors in CD4+ population, for donors expressing DP4 (top) or not expressing DP4 (bottom). DP4-Clip tetramers used as controls (see also Figures S6 and S7).
We analyzed T cell responses to S811–831 for all donors tested considering their DQ5 and DP4 status (Figure 6C). For those expressing only DQ5 but not DP4, 33% responded to S811–831 ex vivo and 67% after in vitro expansion. For those expressing only DP4 but not DQ5, the percentage responding was substantially larger—67% ex vivo and 82% after expansion. In this analysis, we also included 3 donors with the closely related DP4 variant DPA1∗01:03/DPB1∗04:02 (DP402) and 6 donors expressing both DP4 and DP402 variants. The amino acid differences between DP4 and DP402 are buried underneath the peptide largely away from peptide binding pockets (Figure S3) and are known to have little effect on peptide binding specificity (Castelli et al., 2002). Thus, S811–831 is broadly recognized in individuals expressing DQ5 or DP4/402 alleles.
The IFN-γ ELISpot results for the S811–831-specific responses reported in Figure 2D for COVID-19 convalescent donors, and in Figure 6B for vaccinated donors, corresponds to approximately 0.017% (0.003%–0.055%, n = 16) of the total T cell populations present in these donors. Most of the responding T cells are CD4, based on the preponderance of these cells observed by ICS in T cell lines after expansion (Figure 3B). For pre-pandemic donors (Figure 6A), this fraction was lower, at 0.007% (0.004%–0.010%), and with only 2 donors showing positive responses. Overall, this confirms that S811–831 is a broadly-recognized immunodominant epitope, although the frequency of cells to S811–831 varies greatly between individuals.
Tetramer staining
We investigated the use of MHC class II tetramers to follow cross-reactive T cell populations recognizing S811–831. We focused on DP4 because of the high prevalence of this allele across most human population groups (Castelli et al., 2002; Sidney et al., 2010b). The tetramer carries a 15-mer peptide centered around the DP4 epitope (RSFIEDLLFNKVTLA S815–829; core epitope underlined). DP4-restricted T cell clones were recognized as a distinct population strongly staining with both phycoerythin (PE)- and APC-labeled tetramers, while DP2- and DQ5-restricted clones, as well as a negative control DP4-Clip tetramer, exhibited no detectable staining (Figure S6A). T cell lines from DP4+ donors expanded in vitro using HCoV S pools exhibited populations staining strongly as compared to negative controls, including lines from convalescent COVID-19 (G06), vaccinated (V07), and pre-pandemic (L38) donors (Figure 6D). For L38, we evaluated DP4-S815–829 tetramer staining in resting unstimulated peripheral blood mononuclear cells (PBMCs) and observed a small population visible at ∼5-fold increased abundance over the background (Figure S6B). Expanded T cell lines from unexposed and vaccinated DP402 donors also could be detected (Figure S6C). Finally, we evaluated DP4-S815–829 tetramer staining for T cell populations expanded in vitro using S811–831 or HCoV homologs of that epitope (Figure S7A). Tetramer+ populations were observed for T cell lines expanded with each of the homologs, with the relative size of the populations matching ELISpot results on these same lines (Figure S7B) and consistent with the patterns of HCoV reactivity observed in Figure 5. These results confirm DP4 presentation of S811–831 as identified in cellular and biochemical studies and validate the use of the DP4-S815–829 tetramer in detecting SARS-CoV-2 and HCoV-cross-reactive T cell populations.
TCRs
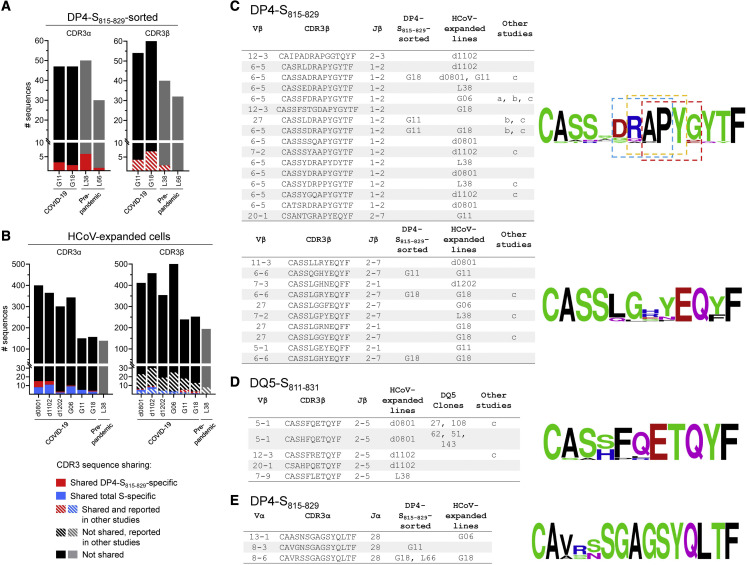
To further characterize the cross-reactive response, we analyzed TCR repertoires specific for S811–831 and other epitopes within the S protein. To identify S811–831 TCR-α and TCR-β sequences, we sorted tetramer+ cells from 2 COVID-19 and 2 pre-pandemic donors, after in vitro expansion with S811–831. A total of 172 TCR-α and 184 TCR-β unique sequences were identified (Table S5). For each donor, a few of the sequences were observed in at least 1 other donor, or in previous studies of COVID-19-associated TCR-β repertoires (Figure 7A; Table S5). To extend these results to additional cross-reactive specificities beyond DP4-S811–831, we analyzed bulk TCR-α and TCR-β repertoires from 6 COVID-19 donors and 1 pre-pandemic donor after expansion with HCoV S pools. A total of 1,796 TCR-α and 2,368 TCR-β unique sequences were identified in these polyclonal lines (Table S5). As before, several of the sequences were shared between donors or were previously observed in other studies (Figure 7B). Several sequences were observed in both tetramer-sorted and polyclonal lines from the same donors (Table S5).
Figure 7.
Analysis of cross-reactive TCR repertoires from COVID-19 and pre-pandemic donors
(A) Summary of CDR3α and CDR3β clonotypes identified in DP4-S815-829 tetramer-sorted cells. Sequences shared among different donors shown in red, hatched lines show sequences also reported in other studies. (B) Same as previous panel, but for clonotypes identified in polyclonal lines expanded with HCoV S pool. Sequences shared among different donors shown in blue; hatched lines show sequences reported by others.
(C) Selected TCR-β DP4-specific convergence groups (CGs) identified in DP4-S815–829-tetramer sorted samples, HCoV-expanded polyclonal lines, and in other studies: a=(Nolan et al., 2020); b=(Low et al., 2021); c=(Dykema et al., 2021). Sequence logos associated with the two CGs (scores 2.8E-14 and 4.4E-14) are shown at right. Three sequence motifs identified in the first CG are boxed (Motifs: RAPY [p<0.001], APYG [p<0.001], DRAP [p<0.001]). CGs identified using GLIPH (Glanville et al., 2017).
(D) TCR-β DQ5-specific CG identified in HCoV-expanded polyclonal lines, DQ5-restricted T cell clones, and reported in other studies. CG score: 2.5E-15. CGs identified using GLIPH (Glanville et al., 2017)
(E) TCR-α DP4-specific cluster identified in DP4- S815-829-tetramer-sorted samples and HCoV-expanded polyclonal lines. Sequence cluster identified using TCRdist (Dash et al., 2017).
To identify sequence motifs that could be shared more broadly among donors than exact CDR3 sequence matches, we used clustering algorithms that group TCR-α or TCR-β by shared specificity (Dash et al., 2017; Glanville et al., 2017). Analysis of the pooled TCR-β sequences from polyclonal lines, T cell clones, and DP4-S815–829-sorted cells revealed 117 TCR convergence groups (CGs), some of which were observed across multiple donors (Table S6). For instance, 2 TCR-β CGs associated with DP4-S815–829 were observed in multiple donors from this and other studies (Figure 7C). Likewise, 1 TCR-β CG associated with DQ5 was observed in polyclonal lines from 3 donors and DQ5-restricted clones (Figure 7D). One TCR-α cluster observed in 4 donors in both DP4-S815–829-sorted cells and polyclonal lines was associated with DP4 (Figure 7E). In summary, we found a highly diverse repertoire of TCRs recognizing S811–831 in the context of DP4 and other HLA alleles, with little sharing of either TRAV/TRBV usage or CDR3 sequences among donors, although a few low-frequency public TCR-α and TCR-β clonotypes and convergence groups were identified.
Discussion
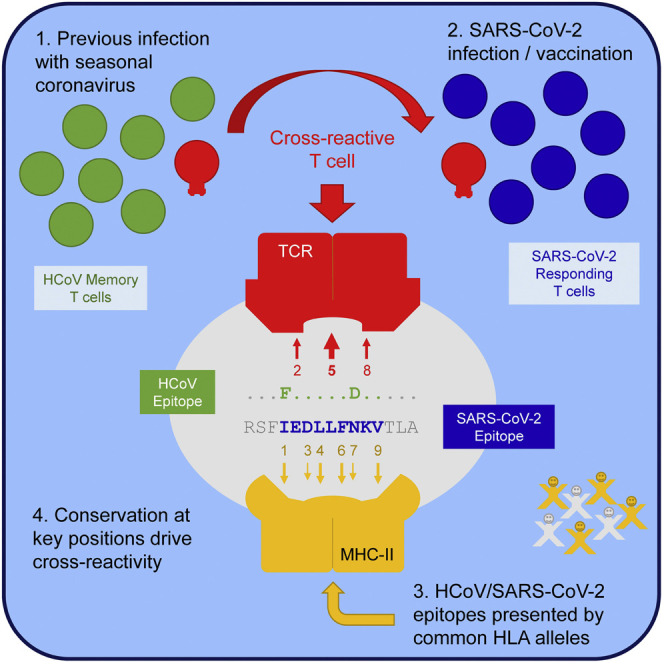
We studied T cell cross-reactivity between SARS-CoV-2 and the 4 seasonal HCoVs by measuring responses to S protein in samples from convalescent COVID-19 donors, vaccine recipients, and individuals not exposed to SARS-CoV-2. We challenged T cells from SARS-CoV-2-exposed donors with HCoV S peptide pools, and vice versa. We made particular use of T cell lines from SARS-CoV-2 donors expanded in vitro by stimulation with HCoV S peptides, which enriches for cross-reactive T cells. We identified peptides recognized by these cross-reactive T cells and found that 1 of these sequences, S811–831, dominated the cross-reactive response, and was recognized in most of the donors tested. The S811–831 region, located proximal to the S2′ cleavage site at the start of the fusion peptide, is highly conserved across SARS-CoV-2 variants, and human and animal coronaviruses, presumably because processing at the spike S2/S2′ junction is necessary to release the fusion peptide essential for viral entry and membrane fusion. We used a panel of T cell clones to identify minimal epitopes and presenting HLA alleles, and identified 2 major reactivity patterns, with an N-terminal epitope RSFIEDLLF S815–823 presented by DQ5, and a partially overlapping C-terminal epitope IEDLLFNKV S818–826 presented by DP2 and DP4. T cells recognizing these epitopes were highly cross-reactive for corresponding HCoV sequences.
Most of the donors tested recognized S811–831, including severe and mild COVID-19 donors, individuals receiving mRNA COVID-19 vaccines, and previously unexposed donors, although most unexposed donors required in vitro expansion to detect cells to S811–831. The broad recognition of these epitopes is driven by the prevalence of the presenting HLA alleles, particularly DP4, which is the most common allele worldwide (Castelli et al., 2002; Sidney et al., 2010b). The same epitope was presented by DP4 and closely related DP2 and DP402, which share identical DPα subunits, similar DPβ subunits each with only 4 substitutions, and similar peptide-binding motifs (Figure S3C). These alleles cover a large fraction of many human populations worldwide (Figure S3E).
The key to this cross-reactive recognition seems to be the remarkable conservation across coronaviruses of amino acids at expected T cell contact positions for both the DQ5 and DP4 binding frames. The selection for DP4 and DQ5 as preferred presenting elements for cross-reactive recognition of SARS-CoV-2 and HCoV homologs may be related to their particular binding motifs, which accommodate peptide sequence variability while still presenting identical residues for TCR recognition. Other alleles that present epitopes in different binding frames would not have such conserved T cell contact cell residues. For example, a SARS-CoV-2 peptide overlapping S811–831 was identified in a CD4 T cell epitope screen (Verhagen et al., 2021) and as a naturally processed epitope derived from SARS-CoV-2 S protein (Parker et al., 2021), where it was predicted to be presented by DR3 and DR4, respectively. For these alleles, however, the expected T cell contact positions are not conserved.
We observed that T cells cross-reactive for both SARS-CoV-2 and seasonal HCoVs comprise a small fraction of the overall response in both pre-pandemic and COVID-19 donors. The responses were characterized by a highly diverse TCR repertoire, mainly specific to individual donors, although a few public clonotypes were present across donors with varying abundance. There seems to be a large and highly diverse repertoire of cross-reactive cells available for expansion. Why cross-reactive T cells present before SARS-CoV-2 infection do not expand and dominate the overall response is not clear.
Immune responses to S811–831 have been observed in other studies (Deng et al., 2021; Dykema et al., 2021; Low et al., 2021; Loyal et al., 2021; Mateus et al., 2020; Saini et al., 2021; Tarke et al., 2021; Woldemeskel et al., 2021). An overlapping epitope accessible in the pre-fusion conformation was recognized by antibodies from COVID-19 donors as one of the most highly recognized linear epitopes and antibodies recognizing this sequence were detected in both COVID-19 and unexposed donors (Poh et al., 2020; Shrock et al., 2020; Voss et al., 2021). Several studies of CD4 and CD8 T cell responses in SARS-CoV-2 donors, including unbiased epitope screens as well as those based on MHC-binding predictions, identified peptides overlapping the S811–831 sequence, among many others (Deng et al., 2021; Dykema et al., 2021; Low et al., 2021; Loyal et al., 2021; Saini et al., 2021; Tarke et al., 2021; Woldemeskel et al., 2021). A peptide overlapping the S811–831 sequence was found among MHC class II-bound peptides eluted from human monocyte-derived dendritic cells (DCs) pulsed in vitro with S protein, showing that this epitope is presented after antigen processing and presentation in a natural context, although in that study the presenting HLA alleles were not assigned (Knierman et al., 2020). S811–831 also has been investigated previously in the context of cross-reactivity between SARS-CoV-2 and HCoVs. Mateus et al. (2020) identified an overlapping epitope in 1 of 9 peptides for which cross-reactive T cell responses were validated for SARS-CoV-2 and HCoV variants. Loyal et al. (2021) observed T cell populations responding to 2 overlapping peptides from the same region that we report here, in a study of SARS-CoV-2 epitopes recognized in uninfected donors, where they were shown to also contribute to the initial response to primary SARS-CoV-2 infection. An epitope overlapping with S811–831 was 1 of 2 highlighted in a study by Low et al. (2021), who mapped the specificity, HLA restriction, and HCoV cross-reactivity of T cell clones from COVID-19 and unexposed donors. The same study reported T cell responses to a 20-mer peptide that overlaps S986–1,006 (Low et al., 2021), another of the cross-reactive epitopes reported here. Cross-reactive epitopes partially overlapping with S811–831, S946–966, and S986–1,006 were also reported recently (Johansson et al., 2021). Dykema et al. (2021) mapped peptide reactivity for TCRs overrepresented in cross-reactive, in vitro-expanded CD4 T cell lines, and identified a sequence from the S811–831 region recognized by TCR transfectants that also recognized the NL63 homolog. We systematically evaluated which epitopes dominated the SARS-CoV-2-HCoV cross-reactive T cell response and validated a proposed DP4 restriction through direct MHC-peptide binding and minimal peptide mapping studies; we also identified DQ5 as a presenting molecule that recognizes a register-shifted epitope, and we identified patterns of cross-reactivity with the various HCoV homologs.
In conclusion, S811-831 is a pan-coronavirus epitope that dominates the cross-reactive T cell response to the S protein after SARS-CoV-2 infection. The epitope is highly conserved across human coronaviruses, with T cell contact positions invariant in each of 2 partially overlapping MHC class II binding frames. Most people have CD4 T cells responding to this epitope before SARS-CoV-2 infection, because of its robust presentation by common HLA alleles and the seasonal prevalence of infection by HCoVs. Responding T cells appear to be functionally competent and are strongly expanded ex vivo by cross-stimulation, but they do not dominate the primary response after natural infection, at least as assessed 3–9 months post-infection. The S811–831 sequence, completely conserved in SARS-CoV-2 variants, including Delta and Omicron, may be useful in studies relating preexisting HCoV immunity to COVID-19 severity or incidence and could be considered for inclusion in pan-coronavirus vaccination strategies.
Limitations of the study
We mapped the specificity of the cross-reactive response by following IFN-γ-secreting cells, but non-IFN-γ-secreting populations could also contribute to the response. In expanded T cells, we observed higher frequencies of T cells staining with DP4-S815–829 tetramer than responding to the same peptide in IFN-γ ELISpot assays, indicating that some T cells can recognize the epitope but not secrete IFN-γ. We observed the cross-reactive T cell response to involve mostly CD4 T cells. This may be due to in vitro culture conditions that favor CD4 over CD8 T cell populations, the use of relatively long peptides that favor presentation by MHC class II molecules (Fiore-Gartland et al., 2016), or an intrinsic bias of cross-reactive T cells because of the different patterns of peptide-MHC-TCR interaction for MHC class I and class II proteins (Stern and Wiley, 1994). We were not able to confidently estimate the fraction of the overall S-specific CD4 T cell response that is represented by S811–831. We calculated the fraction of T cells secreting IFN-γ in response to S811–831 relative to those responding to the entire S pool (0.47 ± 0.47, n = 17), but this likely overestimates the contribution of S811–831 because of competition within the pool for antigen presentation (Fiore-Gartland et al., 2016). In addition to S-specific responses, COVID-19 donors also respond to other antigens (Tarke et al., 2021), but these were not considered in our study. We studied a relatively small group of 27 individuals exposed to SARS-CoV-2 antigens by infection or vaccination, mostly older than 40 years of age. Younger individuals with more frequent previous exposures to HCoVs may show a different response pattern. Our initial screen for immunodominant epitopes involved only 3 donors, all of whom recognized S811–831, but other immunodominant cross-reactive epitopes may have escaped our attention, including those recognized by other MHC proteins. For all of the donors, previous HCoV infection was inferred; we did not attempt to determine which donors were exposed previously to which of the HCoVs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| LB3.1 | Produced in-house | n/a |
| SPVL-3 | Produced in-house | n/a |
| B7/21 | Produced in-house | n/a |
| mouse anti-human CD107a-PE-CF594 (H4A3) | BD Biosciences | 562628 |
| mouse anti-human CD3-APC-H7 (SK7) | BD Biosciences | 560176 |
| mouse anti-human CD4-PerCPCy5.5 (RPA-T4) | BD Biosciences | 560650 |
| mouse anti-human CD8-APC-R700 (RPA-T8) | BD Biosciences | 565166 |
| mouse anti-human CD14-BV510 (MφP9) | BD Biosciences | 563079 |
| mouse anti-human CD19-BV510 (SJ25C1) | BD Biosciences | 562947 |
| mouse anti-human CD56-BV510 (NCAM16.2) | BD Biosciences | 563041 |
| mouse anti-human IFN-γ-V450 (B27) | BD Biosciences | 560371 |
| mouse anti-human TNF-α-PE-Cy7 (MAb11) | BD Biosciences | 557647 |
| mouse anti-human IL-2-BV650 (5344.111) | BD Biosciences | 563467 |
| mouse anti-human CD45RA-BV650 (HI100) | BD Biosciences | 563963 |
| mouse anti-human CD197-PB (G043H7) | Biolegend | 353209 |
| Human Ig | Millipore Sigma | I2511 |
| Biological samples | ||
| Leukopacks from healthy donors | New York Biologics Inc. | https://www.newyorkbiologics.com/ |
| Blood from COVID-19 donors | Recruited as part of the study | UMass Chan Medical School IRB H00020145 |
| Blood from healthy donors | Recruited as part of the study | UMass Chan Medical School IRB I-306-19 |
| Chemicals, peptides, and recombinant proteins | ||
| Human AB+ serum | GeminiBio | 100-512 |
| Fetal Bovine Serum | R&D Systems | S11550 |
| OpTmizer CTS T cell Expansion Medium | Gibco | A1048501 |
| Penicillin Streptomycin | Gibco | 15140-122 |
| GlutaMAX | Gibco | 35050-061 |
| MEM Non-Essential Amino Acids | Gibco | 11140-050 |
| Sodium Piruvate | Gibco | 11360-070 |
| 2-Mercaptoethanol | Gibco | 21985-023 |
| RPMI 1640 | Gibco | 22400-089 |
| Ficoll-Paque | Cytiva | 17144003 |
| DPBS | Corning | 21031-CV |
| PBS 10X | Fisher Scientific | BP3994 |
| Dimethyl Sulfoxide | Gibco | 21985-023 |
| N,N-Dimethylformamide | Millipore Sigma | D4551 |
| Tween 20 | Fisher Scientific | BP337-100 |
| 3-Amino--9-Ethylcarbazole | AlfaAesar | B22529 |
| Acetic Acid, Glacial | Fisher Scientific | A38S-500 |
| Hydrogen Peroxide 30% (W/W) in H2O | Millipore Sigma | H1009-5ML |
| PHA-M | Gibco | 10576015 |
| PHA-P | Sigma | L9017 |
| RPMI w/o phenol red | Gibco | 11835-030 |
| Golgi Plug | BD Biosciences | 51-2301KZ |
| Golgi Stop | BD Biosciences | 51-2092KZ |
| BD Perm/Wash | BD Biosciences | 51-2091KZ |
| BD Cytofix/Cytoperm | BD Biosciences | 51-2090KZ |
| Live/Dead Fixable Aqua Dead Cell Stain Kit | Invitrogen, Life Technologies | L34966 |
| RNeasy Mini | QIAGEN | 74104 |
| RNeasy Micro | QIAGEN | 74004 |
| Ultrapure water | Invitrogen, Life Technologies | 10977-015 |
| RNAse inhibitor 40 U/uL | Takara Bio USA, Inc | 2313A |
| SMARTScribe reverse transcriptase 100U/mL | Takara Bio USA, Inc | 639536 |
| Uracil DNA glycosylase (UDG) 5000 U/mL | New England Biolabs | M0280S |
| Betaine 5 M | Affymetrix | 77507 |
| MgCl2 1 M | Invitrogen, Life Technologies | AM9530G |
| AMPure XP beads | Beckman Coulter | A63881 |
| KOD Hot Start DNA polymerase | Novagen, Millipore Sigma | 71086-3 |
| Ethanol, molecular biology grade | Fisher Bioreagents | BP2818 |
| QIAQuick gel extraction kit | QIAGEN | 28706 |
| Isopropanol, molecular biology grade | Fisher Scientific | BP2618 |
| CEF Control Peptide Pool | AnaSpec | AS-61036-05 |
| SARS-Related Coronavirus 2 Spike (S) Glycoprotein | BEI Resources | NR-52402 |
| SARS-Related Coronavirus 2 Membrane (M) Protein | BEI Resources | NR-52403 |
| SARS-Related Coronavirus 2 Nucleocapsid (N) Protein | BEI Resources | NR-52404 |
| SARS-Related Coronavirus 2 Envelope (E) Protein | BEI Resources | NR-52405 |
| PeptMix HCoV-OC43 Spike Glycoprotein | JPT Peptide Technologies GmbH | PM-OC43-S-1 |
| PepMix™ HCoV-229E Spike Glycoprotein | JPT Peptide Technologies GmbH | PM-229E-S-1 |
| PepMix™ HCoV-NL63 Spike Glycoprotein | JPT Peptide Technologies GmbH | PM-NL63-S-1 |
| PepMix™ HCoV- HKU1 (isolate N1) Spike Glycoprotein | JPT Peptide Technologies GmbH | PM-HKU1-S-1 |
| HCoV-OC43 Spike Glycoprotein | 21st Century Biochemicals, Inc. | Custom |
| HCoV-229E Spike Glycoprotein | 21st Century Biochemicals, Inc. | Custom |
| HCoV-NL63 Spike Glycoprotein | 21st Century Biochemicals, Inc. | Custom |
| HCoV-HKU1 (isolate N1) Spike Glycoprotein | 21st Century Biochemicals, Inc. | Custom |
| SARS-CoV-2 Spike | 21st Century Biochemicals, Inc. | Custom |
| SARS-CoV-2 163 | 21st Century Biochemicals, Inc. | Custom |
| SARS-CoV-2 164 | 21st Century Biochemicals, Inc. | Custom |
| SARS-CoV-2 190 | 21st Century Biochemicals, Inc. | Custom |
| SARS-CoV-2 191 | 21st Century Biochemicals, Inc. | Custom |
| SARS-CoV-2 198 | 21st Century Biochemicals, Inc. | Custom |
| SARS-CoV-2 199 | 21st Century Biochemicals, Inc. | Custom |
| SARS-CoV-2 163-164 truncated | 21st Century Biochemicals, Inc. | Custom |
| SARS-CoV-2 190-191 truncated | 21st Century Biochemicals, Inc. | Custom |
| SARS-CoV-2 198-199 truncated | 21st Century Biochemicals, Inc. | Custom |
| DR4 reporter | 21st Century Biochemicals, Inc. | DR4 reporter |
| DQ5 reporter | 21st Century Biochemicals, Inc. | DQ5 reporter |
| DR1 reporter | 21st Century Biochemicals, Inc. | DR1 reporter |
| Self-1 Peptides pool | 21st Century Biochemicals, Inc. | Custom; Becerra-Artiles, et al. 2019 |
| Human recombinant IL-2 (Proleukin) | Prometheus Laboratories Inc. | NDC 65483-116-07 |
| DP4-163/164-PE | NIH Tetramer Facility | Custom made |
| DP4-163/164-APC | NIH Tetramer Facility | Custom made |
| DP4-CLIP-PE | NIH Tetramer Facility | Custom made |
| DP4-CLIP-APC | NIH Tetramer Facility | Custom made |
| Critical commercial assays | ||
| human IFN-gamma ELISPOT Kit | Invitrogen | 88-7386-88 |
| human IFN-gamma ELISA kit | Invitrogen | 88-7316-88 |
| Protrans HLA typing kits | Protrans Medizinische Diagnostische Produkte GmbH | https://www.protrans.info/nano.cms/en/products/MainCatID/9/ |
| Experimental models: Cell lines | ||
| 9069, BMG | IHWG | 9068 |
| M12C3-DPA1∗0103/DPB1∗0401 (MN605) | S. Kent, UMass | n/a |
| LG2 | IPD-IMGT/HLA | 10984 |
| Oligonucleotides | ||
| TrueSeq R1-UMI: CTACACGACGCTCTT CCGATCTNNNNUNNNNUNNNNUCTTr GrGrGrGrG |
Integrated DNA Technologies | Custom |
| 2nd strand: TCTTTCCCTACACGACGCT CTTCCGATCT |
Integrated DNA Technologies | Custom |
| 5′ RACE forward primer with P5 TrueSeq adapter: AATGATACGGCGACCACCGAG ATCTACACTCTTTCCCTACACGACGCTC TTCCGATCT |
Integrated DNA Technologies | Custom |
| hTRAC_RT8: GGCAGACAGACTTGTCACTG | Integrated DNA Technologies | Custom |
| hTRAC_RT1: CAGAATCCTTACTTTGTGACAC | Integrated DNA Technologies | Custom |
| hTRAC_RT2: GTCTAGCACAGTTTTGTCTG | Integrated DNA Technologies | Custom |
| hTRAC_RT3: CTGTTGCTCTTGAAGTCCATAG | Integrated DNA Technologies | Custom |
| hTRAC_RT4: GTTGAAGGCGTTTGCACATG | Integrated DNA Technologies | Custom |
| hTRAC_RT5: GGTGTCTTCTGGAATAATGC | Integrated DNA Technologies | Custom |
| hTRAC_RT6: GAACCCAATCACTGACAGG | Integrated DNA Technologies | Custom |
| hTRAC_RT7: CACTTTCAGGAGGAGGATTC | Integrated DNA Technologies | Custom |
| hTRAC_RT9: GCGTCATGAGCAGATTAAAC | Integrated DNA Technologies | Custom |
| hTCRAC_Nested with TrueSeq R2: AGTTCA GACGTGTGCTCTTCCGATCtNNNNGAGTCT CTCAGCTGGTACACGGCAGGG |
Integrated DNA Technologies | Custom |
| hTRBC_RT8: CACACCAGTGTGGCCTTTTG | Integrated DNA Technologies | Custom |
| hTRBC_RT1: CTCCTTCCCATTCACCCAC | Integrated DNA Technologies | Custom |
| hTRBC_RT2: GCAGTATCTGGAGTCATTGAG | Integrated DNA Technologies | Custom |
| hTRBC_RT3: CTTGACAGCGGAAGTGGTTG | Integrated DNA Technologies | Custom |
| hTRBC_RT4: CACTCGTCATTCTCCGAGAG | Integrated DNA Technologies | Custom |
| hTRBC_RT5: GTTTGGCCCTATCCTGGGTC | Integrated DNA Technologies | Custom |
| hTRBC_RT6: CTTTCTCTTGACCATGGCCATC | Integrated DNA Technologies | Custom |
| hTRBC_RT9: CATAGAGGATGGTGGCAGAC | Integrated DNA Technologies | Custom |
| hTCRBC_Nested with TrueSeq R2: AGTTCAG ACGTGTGCTCTTCCGATCtNNNNCTCAAACA CAGCGACCTCGGGTGGGAAC |
Integrated DNA Technologies | Custom |
| Barcodes (i7 index): CAAGCAGAAGACGGCAT ACGAgatxxxxxxGTGACTGGAGTTCAGACGTG TGCTCTT |
Integrated DNA Technologies | Custom |
| P2: CAAGCAGAAGACGGCATACGA | Integrated DNA Technologies | Custom |
| Software and algorithms | ||
| GraphPad Prism 9 | GraphPad Software LLC. | https://www.graphpad.com/scientific-software/prism/ |
| RunDFR Web Tool | FRED HUTCH | https://rundfr.fredhutch.org/ |
| ImmunoSpot ver 7 Professional DC | CTL ImmunoSpot | https://immunospot.com |
| FLOWJO ver. 10.7.1 | FLOWJO LLC | https://www.flowjo.com/ |
| NetMHCIIpan – MHC peptide binding prediction version 4.0 | DTU Health Tech | https://services.healthtech.dtu.dk/service.php?NetMHCIIpan-4.0 |
| Morpheus (Broad) – Matrix Visualization and Analysis: | Broad Institute | https://software.broadinstitute.org/morpheus/ |
| CoV-GLUE-Viz – GISAID visualization server version 1.1.108 | MRC-University of Glasgow Centre for Virus Research | http://cov-glue-viz.cvr.gla.ac.uk |
| Immune Epitope Database (IEDB) | IEDB/NIAID | http://www.iedb.org/ |
| Spice Data Mining & Visualization Software for Multicolor Flow Cytometry | NIH | https://niaid.github.io/spice/ |
| IPD-IMGT/HLA Database | HLA Informatics Group Anthony Nolan Research Institute | https://www.ebi.ac.uk/ipd/imgt/hla/ |
| WebLogo 3 | University of California, Berkeley | https://weblogo.berkeley.edu/ |
| Virus Pathogen Database and Analysis Resource (ViPR) | NIAID | http://www.viprbrc.org/ |
| SnapGene Viewer 5.3.1 | Insightful Science | snapgene.com |
| Clustal Omega v 1.2.4 | EMBL/EBI | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| AL2CO | Sequence conservation analysis server | http://prodata.swmed.edu/al2co/al2co.php |
| Other | ||
| 96-well Filtration Plates Immobilon-P membrane | Millipore Sigma | MSIPS4W10 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact Lawrence J. Stern (lawrence.stern@umassmed.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Blood, PBMC, and HLA typing
Whole blood from COVID-19 convalescent donors, healthy donors, or vaccine recipients was collected under a protocol approved by the UMass Chan Medical School Institutional Review Board of the University of Massachusetts and informed consent was obtained from all subjects. In total, samples from 34 donors were collected, with a median age of 56 (range 27–71) y/o, and 16 females/18 males. Demographic data are summarized in Table S1. Leukopaks were obtained from New York Biologics, Inc. (Southampton, NY). Peripheral blood mononuclear cells (PBMC) were isolated using Ficoll-Paque (Cytiva, Marlborough, MA) density gradient centrifugation and used fresh (COVID-19 and unexposed donors), or frozen (pre-pandemic donors) until use. The HLA class II haplotype of pre-pandemic donors was determined using the Protrans HLA typing kits (Protrans Medizinische Diagnostische Produkte GmbH, Hockenheim, Germany) or The Sequencing Center (Fort Collins, CO); for other donors, HLA typing was performed using a Nanopore protocol (Stockton et al., 2020) or by the Histocompatibility Laboratory at UMass Memorial Medical Center (Worcester, MA).
Method details
Generation of peptide-expanded T cells
Peptide-pool or individual peptide expanded T cell lines were generated for each donor by a single in vitro expansion of freshly isolated or frozen PBMC (2 × 106 cells in 1 mL in a 24 well plate) with a final concentration of 1 μg/mL of peptide. As antigens were used individual peptides, or peptides covering the entire SARS-CoV-2 spike protein in a single pool or pools of 10 peptides, or peptides pools covering the entire spike proteins of OC43, HKU1, NL63, and 229E. Cells were maintained in complete CRPMI (RPMI 1640 supplemented with 10% AB + human serum (GeminiBio, West Sacramento, CA), 50 μM beta-mercaptoethanol, 1 mM non-essential amino acids, 1 mM sodium pyruvate, and 100 U/mL penicillin and 100 mg/mL streptomycin (all Gibco, Grand Island, NY)). After 3 days, cultures were supplemented with recombinant human IL-2 (Proleukin, Prometheus, San Diego, CA) at a final concentration of 100 U/mL. During the following 2-15 days, one-half of the medium was replaced with fresh CRPMI supplemented with 100 U/mL IL-2 every 3 days. When cultures reached confluence, cells were resuspended, one-half of the culture transferred to another well, and fresh CRPMI+100 U/mL IL-2 was added to replenish the original volume.
ELISpot assay
IFN-γ ELISpots were performed using Human IFN gamma ELISpot KIT (Invitrogen, San Diego, CA) and MultiScreen Immobilon-P 96 well filtration plates (EMD Millipore, Burlington, MA), following the manufacturer’s instructions. Assays were performed in CST™ OpTmizer™ T cell medium (Gibco, Grand Island, NY). Peptides or peptides pools were used at a final concentration of 1 μg/mL per peptide; as negative controls were used DMSO (DMSO, Fisher Scientific, Hampton, NH) and a pool of human self-peptides (Self-1, (Becerra-Artiles et al., 2019)), and PHA-M (Gibco, Grand Island, NY) was used as a positive control. For ex vivo assays, PBMC were incubated with peptides or controls for ∼24-48 hours. We used 2 × 105 per well for fresh samples from COVID-19, seronegative, and vaccinated donors, or 5 × 105 per well for frozen samples from pre-pandemic donors. For assays with cells expanded in vitro, 2-5x104 cells per well were incubated with an equal number of autologous irradiated PBMC in the presence of peptides or controls for ∼18 hours. Two to four wells of each peptide, pool of peptides, or PHA-M, and at least 6 wells for DMSO were usually tested. Secreted IFN-γ was detected following the manufacturer’s protocol. Plates were analyzed using the CTL ImmunoSpot Image Analyzer (ImmunoSpot, Cleveland, OH) and ImmunoSpot 7 software. For estimation of the overall T cell response due to S811-831, we considered that T cells represent 30% of PBMC.
Intracellular cytokine secretion assay (ICS)
ICS was performed using in vitro expanded T cells as previously described (Becerra-Artiles et al., 2019) with minor modifications. Briefly, autologous irradiated PBMC were resuspended in CRPMI (w/o phenol red) +10% fetal bovine serum (FBS, R&D Systems) containing 1 μg/mL of each peptide and incubated overnight. On the day of the assay, T cell lines were collected, washed, and resuspended in the same medium and added to the pulsed PBMC (1:1 ratio); at this time, anti-CD107a-CF594 was added, along with brefeldin A and monesin at the suggested concentrations (Golgi plug/Golgi stop, BD Biosciences, San Jose, CA). After 6 hours of incubation, cells were collected, washed, and stained using a standard protocol, which included: staining for dead cells with Live/Dead Fixable Aqua Dead Cell Stain Kit™ (Life Technologies, Thermo Fisher Scientific, Waltham, MA); blocking of Fc receptors with human Ig (Sigma-Aldrich, St. Louis, MO); surface staining with mouse anti-human CD3-APC-H7, CD4-PerCPCy5.5, CD8-APC-R700, CD14-BV510, CD19-BV510, CD56-BV510; fixation and permeabilization using BD Cytofix/Cytoperm™; and intracellular staining with mouse anti-human IFN-γ-V450, TNF-α-PE-Cy7, IL-2-BV650, (all from BD Biosciences, San Jose, CA). Data were acquired using a BD LRSII flow cytometer equipped with BD FACSDiva software (BD Biosciences, San Jose, CA) and analyzed using FlowJo v.10.7 (FlowJo, LLC, Ashland, OR). The gating strategy consisted in selecting lymphocytes and single cells, followed by discarding cells in the dump channel (dead, CD14+, CD19+, and CD56+ cells), and selecting CD3+ cells in the resulting population. Polyfunctional analysis was performed in FlowJo, defining Boolean combinatorial gates for all the markers in the CD3+/CD4+/CD8- population. These results were visualized in SPICE software v6.0 (Roederer et al., 2011). t-SNE analysis was done in concatenated samples (control, SARS-CoV-2 S pool, peptide S811-826, and peptide S816-831) from 3 donors using the available plugin in FlowJo.
Partially-match HLA cell lines
EBV-transformed LG2 cell line (10984, IPD-IMGT/HLA), 9068 cell line (BM9, IHWG), and mouse DP4-transfected MN605 cell line (M12C3-DPA1∗0103/DPB1∗0401; (Williams et al., 2018); kindly provided by Dr. S. Kent, UMMS), were maintained in RPMI 1640 medium supplemented with L-glutamine (2 mM), penicillin (100 U/mL), streptomycin (100 μg/mL) and 10% FBS at 37°C/5% CO2.
Isolation of T cell clones
T cell clones were isolated by limiting dilution (∼1 cell per well) using as feeder cells a pool of irradiated heterologous PBMC in CRPMI medium supplemented with PHA-P (Gibco, Grand Island, NY) at 1:500 and 100 U/mL IL-2. After 12-14 days of incubation wells with cell growth were screened for responses to S811-831, S946-966, and S986-1006 by IFN-γ ELISA assay. (Invitrogen, San Diego, CA) and following the manufacturer’s protocol. Absorbance at 450 nm was acquired in a BMG plate POLARstar Optima plate reader (Offenburg, Germany). Positive responses were assessed using a cutoff value of 2-times over background + 3-times the standard deviation of background. Sixty-seven T cell clones with the highest specific signal were selected for further analysis.
T cell clones stimulation and blocking assays
T cell clones (5 × 104 cells per well) were incubated in CRPMI+10% FBS with an equal number of irradiated partially-match HLA cell lines pulsed with peptides (candidate epitopes, truncated 11-mers, HCoV homologs) at 1 μg/mL; DMSO was used as control. For dose-response experiments, 10-fold dilution series (1-10−4 μg/mL) of the peptides were used. Supernatants were collected after 24 hours. Duplicated wells for antigens and 6 wells for negative controls were used. Secreted IFN-γ was measured using ELISA assay as described above. For blocking of antigen-stimulation assays, in-house produced antibodies to HLA-DR (LB3.1), HLA-DQ (SPVL-3), HLA-DP (B7/21), or HLA-ABC (w6/32), were added at a final concentration of 10 μg/mL.
Peptide binding assay
A fluorescence polarization (FP) competition binding assay similar to published ones (Jurewicz et al., 2019; Yin and Stern, 2014) was used to measure spike peptide binding. Soluble DP4 (HLA-DPA1∗01:03/DPB1∗04:01) with a covalently-linked Clip peptide was prepared essentially as described (Willis et al., 2021), as were DQ5-Clip (HLA-DQA∗01:01/DQB1∗05:01) (Jiang et al., 2019) and peptide-exchange catalyst HLA-DM (Busch et al., 1998). Human oxytocinase EKKYFAATQFEPLAARL and influenza nucleoprotein AAHSKAFEDLRVSSY peptides were labeled with Alexa Fluor 488 (Alexa488) tetrafluorophenyl ester (Invitrogen, Carlsbad, CA) and used as probe peptides for DP4 and DQ5 binding. Binding reactions were carried out at 37°C in 100 mM sodium citrate, 50 mM sodium chloride, 0.1% octyl β-D-glucopyranoside, 5 mM ethylenediaminetetraacetic acid, 0.1% sodium azide, 0.2 mM iodoacetic acid, 1 mM dithiothreitol as described, but with 1 U/μg thrombin added to cleave the Clip linker. Thrombin was inactivated after 3 hrs of reaction using 0.1 mM phenylmethanesulfonyl fluoride, and the reaction was continued for 21 hours before FP measurement using a Victor X5 Multilabel plate reader (PerkinElmer, Shelton, CT). DP4-Clip (500 nM) and DQ5-Clip (300 nM) concentrations were selected to provide 50% maximum binding of 25 nM probe peptide in the presence of 500 nM DM. Binding reactions also contained serial dilutions of test peptides with 5-fold dilutions. IC50 values were determined as described (Yin and Stern, 2014).
Tetramer staining
DP4-S815-829 PE and APC tetramers were obtained from the NIH Tetramer Core Facility (Emory University, Atlanta, GA). Cells were collected, washed, and stained using a standard protocol which included: staining for dead cells with Live/Dead Fixable Aqua Dead Cell Stain Kit™ (Life Technologies, Thermo Fisher Scientific, Waltham, MA); blocking of Fc receptors with human Ig (Sigma-Aldrich, St. Louis, MO); staining with the mix of DP4-PE and APC tetramers (final concentration of 4 μg/mL each) at 37°C for 2 hours; surface staining antibodies CD3-APC-H7, CD4-PerCP-Cy5.5, CD8-APC-R700, CD14-BV510, CD19-BV510, CD56-BV510 were added for the last 20 minutes of incubation, followed by washes and resuspension in buffer for data acquisition. Data were acquired using a BD LRSII flow cytometer equipped with BD FACSDiva software (BD Biosciences, San Jose, CA) and analyzed using FlowJo v. 10.7 (FlowJo, LLC, Ashland, OR). The gating strategy consisted in selecting lymphocytes and single cells, followed by discarding cells in the dump channel (dead, CD14+, CD19+, and CD56+ cells), CD3+/CD4+ cells, and assessing the double-staining PE/APC in this population.
Sorting of DP4-S815-829 cells
For tetramer-sorting, T cells were expanded in vitro with S811-831 as indicated previously. After 2 weeks, cells were collected, washed, and stained using the procedure described before. Cell populations in the PE+/APC+ gate were sorted using a BD FACS Aria Cell Sorter at The University of Massachusetts Medical School Flow Cytometry Core Facility. Sorted cells were washed and frozen at −80°C until use.
TCR repertoire analysis
RNA was prepared from HCoV S pool-expanded lines, T cell clones, or DP4-S815-829-sorted cells (0.5-1 × 106 cells) using RNeasy Mini or RNeasy Micro kits from (QIAGEN, Germantown, MD), following user’s manual instructions. Cells were usually frozen in RLT buffer at the time of collection. RNA quality and concentration were assessed using the Fragment Analyzer Service at The University of Massachusetts Molecular Biology Core Labs. RNA with RQN above 7.2 was used for sequencing. We used an in-house RACE (Rapid Amplification of cDNA Ends) approach with template-switch effect, adapted from Turchaninova et al. (Turchaninova et al., 2016). Reverse transcription was performed using ∼100 ng of RNA and 1 μM primers recognizing the constant region of TCRA or TCRB (Integrated DNA Technologies, IDT, Coralville, IA), and annealed for 3 minutes at 72°C. A reaction mix was added to a final concentration of 1 μM UMI/R1 oligo (IDT), 5 U/μL SMARTScribe reverse transcriptase, 0.5 mM dNTP, 2 U/μL RNAse inhibitor (all Takara Bio USA, Inc, Mountain View, CA), 5 mM DTT (Invitrogen), 1 M betaine (Affymetrix), 6 mM MgCl2 (Invitrogen, Thermo Fisher Scientific). Samples were incubated at 42°C for 90 minutes followed by 10 cycles of 50°C/42°C for 2 minutes each, with final incubation at 70°C for 15 minutes. Excess oligo was removed by incubating at 37°C for 40 minutes with 214 U/mL Uracil DNA glycosylase (New England Biolabs, Ipswich, MA). cDNA was purified using AMPure XP Beads (Beckman Coulter, Brea, CA) following the manufacturer’s instructions. Four PCR reactions were performed to add TrueSeq R2, P5, and P7 sequences, and i7 indices. All reactions were performed at a final concentration of 0.2 μM primers (IDT), 0.02 U/μL KOD Hot Start DNA Polymerase, 0.2 mM dNTP, 1.5 mM MgSO4 (all Novagen/Millipore Sigma, Burlington, MA). All primers sequences are shown in STAR Methods. First PCR utilizes purified cDNA, and 2nd strand and RT8 primers; second PCR utilizes purified product from previous PCR, and 2nd strand and nested primers; third PCR utilizes purified product from 2nd PCR, and 5′RACE and barcodes with i7 index primers; fourth PCR utilizes purified product from 3rd PCR, and P1 and P2 primers. Cycling conditions for PCR1: 95°C for 2 minutes; 10 cycles of 95°C for 20 seconds, 70°C for 10 seconds (-1°C per cycle), 70°C for 30 seconds; 15 cycles of 95°C for 20 seconds, 60°C for 10 seconds, 70°C for 30 seconds; final extension at 70°C for 3.5 minutes. PCR2-3: 95°C for 2 minutes; 8 cycles of 95°C for 20 seconds, 60°C for 10 seconds, 70°C for 30 seconds; final extension at 70°C for 3.5 minutes. PCR4: 95°C for 2 minutes; 7 cycles of 95°C for 20 seconds, 60°C for 10 seconds, 70°C for 30 seconds; final extension at 70°C for 3.5 minutes. PCR products from PCR1-3 were purified using AMPure XP magnetic beads, and the final PCR product was purified using the QIAquick Gel Extraction kit (QIAGEN, Germantown, MD); TCRA and TCRB libraries were quantified using the Fragment Analyzer Service. TCR Sequencing was performed at The University of Massachusetts Deep Sequencing Core. Equimolar concentrations of 8–12 libraries were mixed and sequenced in an Illumina MiSeq System (250 × 250 paired-end reads). Data ere de-multiplexed and single FASTQ files generated. These files were processed using MIGEC v1.2.9 pipeline: Checkout-batch, Histogram, and Assemble-batch (Shugay et al., 2014); followed by MiXCR v3.0.13: analyze amplicon pipeline (Bolotin et al., 2015). Further data analysis included VDJTools, for gene usage and statistics (Shugay et al., 2014); and GLIPH (Glanville et al., 2017), and TCRDist (Dash et al., 2017), for clustering and to find TCRs with shared motifs and convergence groups.
Peptides and HLA binding predictions
Peptides for these studies were obtained from 21st Century Biochemicals (Marlborough, MA), BEI Resources (Manassas, VA), and JPT (Berlin, Germany). Peptide sequences are shown in Table S7. HLA-peptide binding prediction was performed with NetMHCIIpan v4.0 server (Reynisson et al., 2020). Sequence logo of predicted motifs obtained using Motif Viewer in NetMHCIIpan v4.0 server.
Conservation analysis
We selected twenty-nine coronaviruses, including viruses in the alpha, beta, gamma, and delta genera. Selected viruses infect a variety of animal species, including humans. Sequence alignment and phylogenetic tree (Neighbor-joining tree without distance corrections) of S proteins from selected viruses were generated using Clustal Omega v1.2.4 (Goujon et al., 2010). Conservation indices for each position of the alignment were calculated using the AL2CO algorithm (Pei and Grishin, 2001) using the alignment previously generated and the default settings.
Quantification and statistical analysis
Statistical analyses were performed in GraphPad Prism v9.2.0. Comparisons between groups were done using Mann-Whitney tests or paired t-tests. ELISpot statistical analysis was performed using a distribution-free resampling (DFR) algorithm described by Moodie et al. (Moodie et al., 2012), available as an online resource at https://rundfr.fredhutch.org, which tests null hypotheses of equal (DFR1x) or less than two-fold difference (DFR2x) between background and experimental sample means.
Acknowledgments
We deeply acknowledge the blood donors, without whom this study would not have been possible. R.W.F. passed away during the course of this work. We thank the UMass Memorial Medical Center Histocompatibility Laboratory and Jeff Bailey (Brown University) for HLA typing; Mollie Jurewicz for help with the TCR repertoire methodology; Manuel Garber for assistance with the TCR repertoire analysis, and Ann M Rothstein for helpful discussions. We acknowledge the assistance of the UMass Chan Medical School Clinical Research Center, Flow Cytometry Core, and Deep Sequencing Core, and we thank the NIH Tetramer Core Facility (contract no. 75N93020D00005) for providing DP4-S815–829 tetramers. This work was supported by grants from the Massachusetts Consortium Pathogen Readiness MassCPR (to A.M.M.) and the UMass COVID-19 Pandemic Fund (to L.J.S.) and NIH-127869 (to L.J.S.).
Author contributions
J.M.C.-C., A.B.-A., and L.J.S. conceived the analysis. R.W.F., M.C., and A.M.M. recruited donors and participated in the study design. J.C., C.F., J.M.C.-C., and A.B.-A. assisted with blood processing. A.B.-A. and J.M.C.-C. performed all of the T cell studies. C.F. and A.M.M. measured the antibody responses. L.L. and G.C.W. prepared the recombinant proteins, and P.P.N. performed the binding assays. A.B.-A., J.M.C.-C., and L.J.S. analyzed the T cell data, HCoV homologies, peptide binding patterns, and TCR repertoires. A.B.-A., J.M.C.-C., and L.J.S. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: May 27, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110952.
Supplemental information
Data and code availability
Additional Supplemental Items are available from Mendeley Data at https://data.mendeley.com/datasets/dfhdfm8gmy/1.
This paper does not report original code.
References
- Bacher P., Rosati E., Esser D., Martini G.R., Saggau C., Schiminsky E., Dargvainiene J., Schroder I., Wieters I., Khodamoradi Y., et al. Low-avidity CD4(+) T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity. 2020;53:1258–1271.e5. doi: 10.1016/j.immuni.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra-Artiles A., Cruz J., Leszyk J.D., Sidney J., Sette A., Shaffer S.A., Stern L.J. Naturally processed HLA-DR3-restricted HHV-6B peptides are recognized broadly with polyfunctional and cytotoxic CD4 T-cell responses. Eur. J. Immunol. 2019;49:1167–1185. doi: 10.1002/eji.201948126. [DOI] [PubMed] [Google Scholar]
- Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., Chudakov D.M. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- Bonifacius A., Tischer-Zimmermann S., Dragon A.C., Gussarow D., Vogel A., Krettek U., Godecke N., Yilmaz M., Kraft A.R.M., Hoeper M.M., et al. COVID-19 immune signatures reveal stable antiviral T cell function despite declining humoral responses. Immunity. 2021;54:340–354.e6. doi: 10.1016/j.immuni.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch R., Doebele R.C., von Scheven E., Fahrni J., Mellins E.D. Aberrant intermolecular disulfide bonding in a mutant HLA-DM molecule: implications for assembly, maturation, and function. J. Immunol. 1998;160:734–743. [PubMed] [Google Scholar]
- Castelli F.A., Buhot C., Sanson A., Zarour H., Pouvelle-Moratille S., Nonn C., Gahery-Segard H., Guillet J.G., Menez A., Georges B., Maillere B. HLA-DP4, the most frequent HLA II molecule, defines a new supertype of peptide-binding specificity. J. Immunol. 2002;169:6928–6934. doi: 10.4049/jimmunol.169.12.6928. [DOI] [PubMed] [Google Scholar]
- Chen H., Hou J., Jiang X., Ma S., Meng M., Wang B., Zhang M., Zhang M., Tang X., Zhang F., et al. Response of memory CD8+ T cells to severe acute respiratory syndrome (SARS) coronavirus in recovered SARS patients and healthy individuals. J. Immunol. 2005;175:591–598. doi: 10.4049/jimmunol.175.1.591. [DOI] [PubMed] [Google Scholar]
- Chen J., Qi T., Liu L., Ling Y., Qian Z., Li T., Li F., Xu Q., Zhang Y., Xu S., et al. Clinical progression of patients with COVID-19 in Shanghai, China. J. Infect. 2020;80:e1–e6. doi: 10.1016/j.jinf.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash P., Fiore-Gartland A.J., Hertz T., Wang G.C., Sharma S., Souquette A., Crawford J.C., Clemens E.B., Nguyen T.H.O., Kedzierska K., et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017;547:89–93. doi: 10.1038/nature22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Pan J., Qiu M., Mao L., Wang Z., Zhu G., Gao L., Su J., Hu Y., Luo O.J., et al. Identification of HLA-A2 restricted CD8(+) T cell epitopes in SARS-CoV-2 structural proteins. J. Leukoc. Biol. 2021;110:1171–1180. doi: 10.1002/JLB.4MA0621-020R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du R.H., Liang L.R., Yang C.Q., Wang W., Cao T.Z., Li M., Guo G.Y., Du J., Zheng C.L., Zhu Q., et al. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur. Respir. J. 2020;55:2000524. doi: 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykema A.G., Zhang B., Woldemeskel B.A., Garliss C.C., Cheung L.S., Choudhury D., Zhang J., Aparicio L., Bom S., Rashid R., et al. Functional characterization of CD4+ T cell receptors crossreactive for SARS-CoV-2 and endemic coronaviruses. J. Clin. Invest. 2021;131:e146922. doi: 10.1172/JCI146922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore-Gartland A., Manso B.A., Friedrich D.P., Gabriel E.E., Finak G., Moodie Z., Hertz T., De Rosa S.C., Frahm N., Gilbert P.B., et al. Pooled-peptide epitope mapping strategies are efficient and highly sensitive: an evaluation of methods for identifying human T cell epitope specificities in large-scale HIV vaccine efficacy trials. PLoS One. 2016;11:e0147812. doi: 10.1371/journal.pone.0147812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt E.R., Hardie A., Claas E.C.J., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioia C., Horejsh D., Agrati C., Martini F., Capobianchi M.R., Ippolito G., Poccia F. T-Cell response profiling to biological threat agents including the SARS coronavirus. Int. J. Immunopathol. Pharmacol. 2005;18:525–530. doi: 10.1177/039463200501800312. [DOI] [PubMed] [Google Scholar]
- Glanville J., Huang H., Nau A., Hatton O., Wagar L.E., Rubelt F., Ji X., Han A., Krams S.M., Pettus C., et al. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547:94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombar S., Bergquist T., Pejaver V., Hammarlund N.E., Murugesan K., Mooney S., Shah N., Pinsky B.A., Banaei N. SARS-CoV-2 infection and COVID-19 severity in individuals with prior seasonal coronavirus infection. Diagn. Microbiol. Infect. Dis. 2021;100:115338. doi: 10.1016/j.diagmicrobio.2021.115338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Vita R., Peters B., Crotty S., Weiskopf D., Sette A. SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe. 2021;29:1076–1092. doi: 10.1016/j.chom.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Birtley J.R., Hung S.C., Wang W., Chiou S.H., Macaubas C., Kornum B., Tian L., Huang H., Adler L., et al. In vivo clonal expansion and phenotypes of hypocretin-specific CD4(+) T cells in narcolepsy patients and controls. Nat. Commun. 2019;10:5247. doi: 10.1038/s41467-019-13234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A.M., Malhotra U., Kim Y.G., Gomez R., Krist M.P., Wald A., Koelle D.M., Kwok W.W. Cross-reactive and mono-reactive SARS-CoV-2 CD4+ T cells in prepandemic and COVID-19 convalescent individuals. PLoS Pathog. 2021;17:e1010203. doi: 10.1371/journal.ppat.1010203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurewicz M.M., Willis R.A., Ramachandiran V., Altman J.D., Stern L.J. MHC-I peptide binding activity assessed by exchange after cleavage of peptide covalently linked to β2-microglobulin. Anal. Biochem. 2019;584:113328. doi: 10.1016/j.ab.2019.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knierman M.D., Lannan M.B., Spindler L.J., McMillian C.L., Konrad R.J., Siegel R.W. The human leukocyte antigen class II immunopeptidome of the SARS-CoV-2 spike glycoprotein. Cell Rep. 2020;33:108454. doi: 10.1016/j.celrep.2020.108454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C.M., Prigge A.D., Anekalla K.R., Shukla A., Do Umehara H.C., Setar L., Chavez J., Abdala-Valencia H., Politanska Y., Markov N.S., et al. Age-related differences in the nasal mucosal immune response to SARS-CoV-2. Am. J. Respir. Cell Mol. Biol. 2021;66:206–222. doi: 10.1165/rcmb.2021-0292OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A., Pairo-Castineira E., Rawlik K., Stuckey A., Odhams C.A., Walker S., Russell C.D., Malinauskas T., Wu Y., Millar J., et al. Whole genome sequencing reveals host factors underlying critical Covid-19. Nature. 2022 doi: 10.1038/s41586-022-04576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Lipsitch M., Grad Y.H., Sette A., Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low J.S., Vaqueirinho D., Mele F., Foglierini M., Jerak J., Perotti M., Jarrossay D., Jovic S., Perez L., Cacciatore R., et al. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science. 2021;372:1336–1341. doi: 10.1126/science.abg8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyal L., Braun J., Henze L., Kruse B., Dingeldey M., Reimer U., Kern F., Schwarz T., Mangold M., Unger C., et al. Cross-reactive CD4(+) T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science. 2021;374:eabh1823. doi: 10.1126/science.abh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. Kids and COVID: why young immune systems are still on top. Nature. 2021;597:166–168. doi: 10.1038/d41586-021-02423-8. [DOI] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B., Drosten C., Muller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodie Z., Price L., Janetzki S., Britten C.M. Response determination criteria for ELISPOT: toward a standard that can be applied across laboratories. Methods Mol. Biol. 2012;792:185–196. doi: 10.1007/978-1-61779-325-7_15. [DOI] [PubMed] [Google Scholar]
- Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lubke M., Bauer J., Rieth J., Wacker M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- Nolan S., Vignali M., Klinger M., Dines J.N., Kaplan I.M., Svejnoha E., Craft T., Boland K., Pesesky M., Gittelman R.M., et al. A large-scale database of T-cell receptor beta (TCRβ) sequences and binding associations from natural and synthetic exposure to SARS-CoV-2. Res. Sq. 2020 doi: 10.21203/rs.3.rs-51964/v1. [DOI] [Google Scholar]
- Parker R., Partridge T., Wormald C., Kawahara R., Stalls V., Aggelakopoulou M., Parker J., Powell Doherty R., Ariosa Morejon Y., Lee E., et al. Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells. Cell Rep. 2021;35:109179. doi: 10.1016/j.celrep.2021.109179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J., Grishin N.V. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17:700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh C.M., Carissimo G., Wang B., Amrun S.N., Lee C.Y.P., Chee R.S.L., Fong S.W., Yeo N.K.W., Lee W.H., Torres-Ruesta A., et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020;11:2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M., Nozzi J.L., Nason M.C. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry. 2011;79A:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossjohn J., Gras S., Miles J.J., Turner S.J., Godfrey D.I., McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 2021;131:e143380. doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S.K., Hersby D.S., Tamhane T., Povlsen H.R., Hernandez S.P.A., Nielsen M., Gang A.O., Hadrup S.R. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8(+) T cell activation in COVID-19 patients. Sci. Immunol. 2021;6:eabf7550. doi: 10.1126/sciimmunol.abf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugay M., Britanova O.V., Merzlyak E.M., Turchaninova M.A., Mamedov I.Z., Tuganbaev T.R., Bolotin D.A., Staroverov D.B., Putintseva E.V., Plevova K., et al. Towards error-free profiling of immune repertoires. Nat. Methods. 2014;11:653–655. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- Sidney J., Steen A., Moore C., Ngo S., Chung J., Peters B., Sette A. Divergent motifs but overlapping binding repertoires of six HLA-DQ molecules frequently expressed in the worldwide human population. J. Immunol. 2010;185:4189–4198. doi: 10.4049/jimmunol.1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J., Steen A., Moore C., Ngo S., Chung J., Peters B., Sette A. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J. Immunol. 2010;184:2492–2503. doi: 10.4049/jimmunol.0903655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soveg F.W., Schwerk J., Gokhale N.S., Cerosaletti K., Smith J.R., Pairo-Castineira E., Kell A.M., Forero A., Zaver S.A., Esser-Nobis K., et al. Endomembrane targeting of human OAS1 p46 augments antiviral activity. Elife. 2021;10:e71047. doi: 10.7554/eLife.71047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern L.J., Wiley D.C. Antigenic peptide binding by class I and class II histocompatibility proteins. Structure. 1994;2:245–251. doi: 10.1016/s0969-2126(00)00026-5. [DOI] [PubMed] [Google Scholar]
- Stockton J.D., Nieto T., Wroe E., Poles A., Inston N., Briggs D., Beggs A.D. Rapid, highly accurate and cost-effective open-source simultaneous complete HLA typing and phasing of class I and II alleles using nanopore sequencing. HLA. 2020;96:163–178. doi: 10.1111/tan.13926. [DOI] [PubMed] [Google Scholar]
- Swadling L., Diniz M.O., Schmidt N.M., Amin O.E., Chandran A., Shaw E., Pade C., Gibbons J.M., Le Bert N., Tan A.T., et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature. 2021;601:110–117. doi: 10.1038/s41586-021-04186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.C.S., Owen C.J., Tham C.Y.L., Bertoletti A., van Dorp L., Balloux F. Pre-existing T cell-mediated cross-reactivity to SARS-CoV-2 cannot solely be explained by prior exposure to endemic human coronaviruses. Infect. Genet. Evol. 2021;95:105075. doi: 10.1016/j.meegid.2021.105075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchaninova M.A., Davydov A., Britanova O.V., Shugay M., Bikos V., Egorov E.S., Kirgizova V.I., Merzlyak E.M., Staroverov D.B., Bolotin D.A., et al. High-quality full-length immunoglobulin profiling with unique molecular barcoding. Nat. Protoc. 2016;11:1599–1616. doi: 10.1038/nprot.2016.093. [DOI] [PubMed] [Google Scholar]
- Verhagen J., van der Meijden E.D., Lang V., Kremer A.E., Volkl S., Mackensen A., Aigner M., Kremer A.N. Human CD4(+) T cells specific for dominant epitopes of SARS-CoV-2 Spike and Nucleocapsid proteins with therapeutic potential. Clin. Exp. Immunol. 2021;205:363–378. doi: 10.1111/cei.13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss C., Esmail S., Liu X., Knauer M.J., Ackloo S., Kaneko T., Lowes L., Stogios P., Seitova A., Hutchinson A., et al. Epitope-specific antibody responses differentiate COVID-19 outcomes and variants of concern. JCI Insight. 2021;6:e148855. doi: 10.1172/jci.insight.148855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickenhagen A., Sugrue E., Lytras S., Kuchi S., Noerenberg M., Turnbull M.L., Loney C., Herder V., Allan J., Jarmson I., et al. ISARIC4C Investigators A prenylated dsRNA sensor protects against severe COVID-19. Science. 2021;374:eabj3624. doi: 10.1126/science.abj3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a Review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Williams T., Krovi H.S., Landry L.G., Crawford F., Jin N., Hohenstein A., DeNicola M.E., Michels A.W., Davidson H.W., Kent S.C., et al. Development of T cell lines sensitive to antigen stimulation. J. Immunol. Methods. 2018;462:65–73. doi: 10.1016/j.jim.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis R.A., Ramachandiran V., Shires J.C., Bai G., Jeter K., Bell D.L., Han L., Kazarian T., Ugwu K.C., Laur O., et al. Production of class II MHC proteins in lentiviral vector-transduced HEK-293t cells for tetramer staining reagents. Curr. Protoc. 2021;1:e36. doi: 10.1002/cpz1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemeskel B.A., Garliss C.C., Blankson J.N. mRNA vaccine-elicited severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–Specific T cells persist at 6 Months and recognize the delta variant. Clin. Infect. Dis. 2021:ciab915. doi: 10.1093/cid/ciab915. [DOI] [PubMed] [Google Scholar]
- Xia X. Domains and functions of spike protein in Sars-Cov-2 in the context of vaccine design. Viruses. 2021;13:109. doi: 10.3390/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., James E., Roti M., Huston L., Gebe J.A., Kwok W.W. Searching immunodominant epitopes prior to epidemic: HLA class II-restricted SARS-CoV spike protein epitopes in unexposed individuals. Int. Immunol. 2009;21:63–71. doi: 10.1093/intimm/dxn124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Stern L.J. Measurement of peptide binding to MHC class II molecules by fluorescence polarization. Curr. Protoc. Immunol. 2014;106:5 10 11–15 10 12. doi: 10.1002/0471142735.im0510s106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional Supplemental Items are available from Mendeley Data at https://data.mendeley.com/datasets/dfhdfm8gmy/1.
This paper does not report original code.