Abstract
The Omicron SARS-CoV-2 has several distinct sublineages, among which sublineage BA.1 is responsible for the initial Omicron surge and is now being replaced by BA.2 worldwide, whereas BA.3 is currently at a low frequency. The ongoing BA.1-to-BA.2 replacement underscores the importance to understand the cross-neutralization among the three Omicron sublineages. Here we test the neutralization of BA.1-infected human sera against BA.2, BA.3, and USA/WA1-2020 (a strain isolated in late January 2020). The BA.1-infected sera neutralize BA.1, BA.2, BA.3, and USA/WA1-2020 SARS-CoV-2s with geometric mean titers (GMTs) of 445, 107, 102, and 16, respectively. Thus, the neutralizing GMTs against heterologous BA.2, BA.3, and USA/WA1-2020 are 4.2-, 4.4-, and 28.4-fold lower than the GMT against homologous BA.1, respectively. These findings have implications in COVID-19 vaccine strategy.
Subject terms: SARS-CoV-2, Antibodies, Viral infection
It is important to understand the cross-neutralization among distinct SARS-CoV-2 Omicron sublineages. Zou et al. show that sera from Omicron BA.1-infected people are significantly weaker in neutralizing Omicron BA.2, BA.3, and USA-WA1/2020 than neutralizing BA.1.
Introduction
Since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late 2019, the virus has evolved to increase viral transmission and immune evasion. The World Health Organization (WHO) has so far designated 5 variants of concern (VOC), including Alpha, Beta, Gamma, Delta, and Omicron. At the time of submitting this paper, the newly emerged Omicron variant had 3 distinct sublineages: BA.1, BA.2, and BA.3. BA.1 was first identified in South Africa in November 2021. BA.1 and its derivative BA.1.1 (containing an extra R346K substitution in the spike of BA.1) caused the initial surges of Omicron around the world. Subsequently, the frequency of BA.2 increased steeply, replacing BA.1 in many parts of the world. In the USA, the frequency of BA.2 increased from 0.4% to 54.9% between 22 January 2022 and 24 March 2022 (https://covid.cdc.gov/covid-data-tracker/#variant-proportions). Compared with BA.1, BA.2 did not seem to cause more severe disease1, but may increase viral transmissible by ~30%2. As of 30 March 2022, the frequency BA.3 remained low in the GISAID database (https://www.gisaid.org/). All three sublineages of Omicron could significantly evade vaccine-elicited neutralization, among which BA.3 exhibited the greatest reduction3,4. In addition, Omicron BA.1 could efficiently evade non-Omicron SARS-CoV-2 infection-elicited neutralization5. The increased transmissibility and immune evasion of the Omicron variant may be responsible for the replacement of VOC from the previous Delta to the current Omicron. Many unvaccinated individuals were infected by BA.1 during the initial Omicron surge6. Therefore, in this work, we examine the cross-neutralization of BA.1 infection against BA.2, BA.3, and other variants. Such laboratory information is essential to guide vaccine strategy and public health policy.
Results and discussion
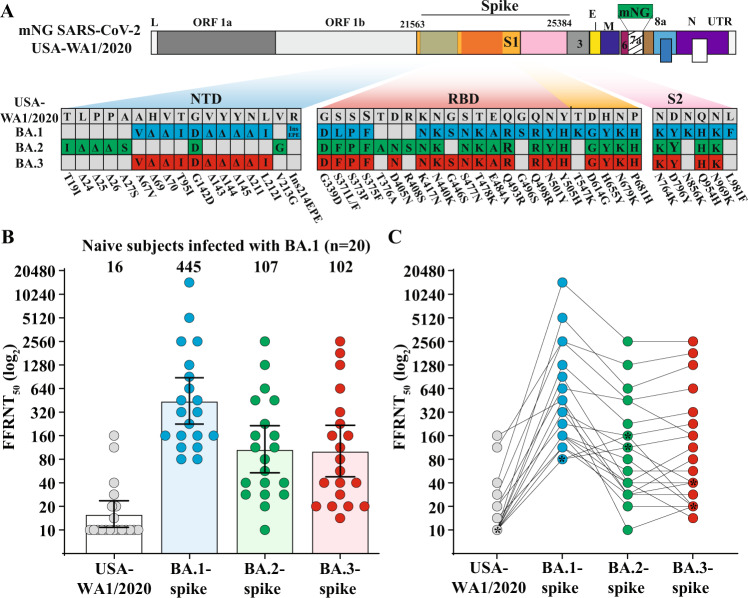
To examine the cross-neutralization among the three Omicron sublineages, we collected 20 human sera from unvaccinated patients who were infected with Omicron BA.1 (Table 1). The genotype of infecting virus was verified for each patient by Sanger sequencing. The sera were collected on day 8 to 62 after positive RT-PCR test. The serum panel was measured for neutralization against four recombinant SARS-CoV-2s (Fig. 1A): USA/WA1-2020 (wild-type) and three chimeric USA/WA1-2020 bearing the full-length spike protein from Omicron BA.1 (GISAID EPI_ISL_6640916), BA.2 (GISAID EPI_ISL_6795834.2), or BA.3 (GISAID EPI_ISL_7605591). The spike proteins of the three Omicron sublineages have distinct amino acid mutations, deletions, and insertions (Fig. 1A). To facilitate neutralization testing, an mNeonGreen (mNG) reporter was engineered into the four viruses, resulting in wild-type, BA.1-, BA.2-, and BA.3-spike mNG SARS-CoV-2s. The construction and characterization of the four mNG SARS-CoV-2s were recently reported4. Using an mNG-based fluorescent focus-reduction neutralization test (FFRNT), we determined the neutralizing geometric mean titers (GMTs) of the sera against wild-type, BA.1-, BA.2-, and BA.3-spike mNG SARS-CoV-2s to be 16, 445, 107, and 102, respectively (Fig. 1B). Thus, the neutralizing GMTs against heterologous BA.2-spike, BA.3-spike, and wild-type viruses were 4.2-, 4.4-, and 28.4-fold lower than the GMT against the homologous BA.1-spike virus, respectively (Fig. 1B). Consistently, all sera neutralized BA.1-spike virus at neutralizing titers of ≥80, whereas 13 out of 20 sera did not neutralize the wild-type USA/WA1-2020 (defined as 10 for plot and calculation purposes; Fig. 1B and Table 1). Notably, 2 sera neutralized BA.2-spike virus more efficiently than the BA.1-spike virus (indicated by symbol * in Fig. 1C). Collectively, the results support two conclusions. First, BA.1 infection elicited similar levels of cross-neutralization against BA.2 and BA.3, although at a decreased efficiency that was 4.2- to 4.4-fold lower than that against BA.1. This result is in contrast with the neutralization results from vaccinated sera (collected at 1 month after three doses of Pfizer/BioNTech’s BNT162b2 vaccine) which neutralized BA.1 and BA.2 much more efficiently than BA.34. Second, the neutralization of BA.1-infected sera against USA/WA1-2020 were 6.7- and 6.4-fold lower than that against Omicron BA.2 and BA.3, respectively. The results indicate the antigenic distinctions among different variant spikes, which must be carefully considered when deciding to switch the vaccine sequence to new variants7. If future variants are Omicron decedents, it would be conceptually attractive to switch vaccine sequence to an Omicron spike.
Table 1.
Serum information and FFRNT50 values.
| Serum ID | Age (years) | Gender (F/M) | Ethnicity | FFRNT50a | Serum collection time (days post positive RT-PCR test) | |||
|---|---|---|---|---|---|---|---|---|
| USA-WA1/2020 | BA.1-spike virus | BA.2-spike virus | BA.3-spike virus | |||||
| 1 | 21–30 | M | Hispanic | 10b | 80 | 160 | 20 | 26 |
| 2 | 41–50 | F | White | 10 | 80 | 113 | 40 | 33 |
| 3 | 81–90 | M | White | 14 | 113 | 40 | 14 | 21 |
| 4 | 11–20 | M | Black | 10 | 113 | 28 | 28 | 16 |
| 5 | 31–40 | F | Black | 10 | 160 | 40 | 40 | 16 |
| 6 | 21–30 | F | Black | 10 | 160 | 20 | 20 | 28 |
| 7 | 1–10 | F | Hispanic | 10 | 160 | 57 | 57 | 43 |
| 8 | 61–70 | M | Black | 10 | 160 | 80 | 20 | 40 |
| 9 | 81–90 | M | White | 20 | 226 | 160 | 226 | 40 |
| 10 | 1–10 | F | Hispanic | 10 | 320 | 28 | 40 | 26 |
| 11 | 1–10 | F | Hispanic | 10 | 320 | 113 | 160 | 56 |
| 12 | 51–60 | F | White | 10 | 453 | 28 | 80 | 35 |
| 13 | 71–80 | M | Hispanic | 160 | 453 | 10 | 20 | 17 |
| 14 | 21–30 | F | Hispanic | 20 | 640 | 453 | 160 | 62 |
| 15 | 51–60 | M | White | 10 | 905 | 40 | 113 | 29 |
| 16 | 71–80 | F | NA | 10 | 1280 | 226 | 320 | 8 |
| 17 | 71–80 | F | Hispanic | 28 | 2560 | 453 | 640 | 32 |
| 18 | 61–70 | M | Hispanic | 113 | 2560 | 1280 | 1810 | 15 |
| 19 | 71–80 | F | Hispanic | 10 | 5120 | 640 | 1280 | 16 |
| 20 | 81–90 | M | White | 40 | 14,482 | 2560 | 2560 | 13 |
| GMTc | – | – | – | 16 | 445 | 107 | 102 | 25 |
| 95% CId | – | – | – | 11–24 | 225–881 | 54–215 | 48–218 | 20–32 |
NA not available.
aIndividual FFRNT50 value is the geometric mean of duplicate FFRNT results.
bFFRNT50 of <20 was treated as 10 for plot purpose and statistical analysis.
cGeometric mean neutralizing titers (GMT).
d95% confidence interval (95% CI) for the GMT.
Fig. 1. Cross-neutralization of human sera from unvaccinated individuals who were infected with Omicron BA.1 SARS-CoV-2.
A Omicron BA.1-, BA.2-, and BA.3-spike mNG SARS-CoV-2s. The full-length spike gene from Omicron BA.1, BA.2, or BA.3 was engineered into an mNG USA-WA1/2020 SARS-CoV-2. The mNG gene was engineered at the open-reading-frame-7 of the viral genome. Amino acid mutations, deletions, and insertions (Ins) are indicated for BA.1, BA.2, and BA.3 spikes in reference to the USA-WA1/2020 spike. L leader sequence, ORF open reading frame, NTD N-terminal domain of S1, RBD receptor binding domain of S1, S spike glycoprotein, S1 N-terminal furin cleavage fragment of S, S2 C-terminal furin cleavage fragment of S, E envelope protein, M membrane protein, N nucleoprotein, UTR untranslated region. B Scatterplot of neutralization titers. A panel of 20 human sera collected from Omicron BA.1-infected individuals were tested for the 50% fluorescent focus-reduction neutralization titers (FFRNT50) against recombinant USA-WA1/2020 (gray circles), Omicron BA.1- (blue circles), BA.2- (green circles), and BA.3-spike (red circels) mNG SARS-CoV-2s. The neutralization titer for each virus was determined in duplicates. The serum information and FFRNT50 values are summarized in Table 1. Each data point represents the geometric mean FFRNT50 obtained with a serum specimen against the indicated virus. The bar heights and the numbers above indicate geometric mean titers (GMTs). Error bars indicate the 95% confidence interval (CI) of the GMTs. Data are presented as GMT with 95% CI. Statistical analysis was performed using the Wilcoxon matched-pairs signed-rank test. Two-tailed P values of the GMT against BA.1-spike and USA-WA1/2020, BA.2-spike, or BA.3 spikes are all <0.0001. C FFRNT50 values with connected lines for individual sera. Two sera exhibiting slightly higher FFRNT50s against BA.2 virus than that against BA.1-spike SARS-CoV-2 are indicated by symbol asterisk (serum ID 1 and 2 in Table 1).
There are two limitations of the serum specimens used in this study. First, the sample size of serum specimen was relatively small. Second, all sera were collected on days 8 to 62 after positive RT-PCR test. The immune status of these specimens were heterogenous, with some sera collected at an acute plasma blast stage and other sera collected at a convalescent IgG-dominant phase. Analysis of more specimens collected at later timepoints post-infection will substantiate the current observations.
Emerging evidence supports a vaccine booster strategy to minimize the health risk of the ongoing Omicron infection. First, 2 doses of BNT162b2 vaccine are inefficient to elicit robust neutralization against Omicron variant, whereas 3 doses of BNT162b2 produces robust neutralization against Omicron. Although Omicron-neutralizing activity remains robust for up to 4 months3, the durability of such neutralization beyond 4 months after dose 3 remains to be determined. The latter result, together with the real-world vaccine effectiveness, are required to guide the timing of dose 4 vaccine. Second, non-Omicron SARS-CoV-2 infection does not elicit robust neutralization against Omicron variant5, suggesting that previously infected individuals should be vaccinated to mitigate the health threat from Omicron. The cross-neutralization of BA.1-infected sera against BA.2 and BA.3 suggests the recent BA.1-infected individuals are likely to be protected against the ongoing BA.2 surge. Third, vaccine-mediated T cell immunity and non-neutralizing antibodies that mediate antibody-dependent cytotoxicity could also confer protection against severe COVID-19. After vaccination or infection, the majority of T cell epitopes are highly preserved against Omicron spikes8. In agreement with this notion, 3 doses of BNT162b2 conferred efficacy against Omicron disease, but the protection wanes over time, with overall efficacy remaining high up to 6 months after dose 39–13. The real-world vaccine effectiveness and laboratory studies will guide vaccine booster strategy to achieve optimal breadth and duration of protection.
Methods
Ethical statement
All virus work was performed in a biosafety level 3 (BSL-3) laboratory with redundant fans in the biosafety cabinets at The University of Texas Medical Branch at Galveston. All personnel wore powered air purifying respirators (Breathe Easy, 3 M) with Tyvek suits, aprons, booties, and double gloves.
The research protocol regarding the use of human serum specimens was reviewed and approved by the University of Texas Medical Branch (UTMB) Institutional Review Board (IRB number 20-0070). No informed consent was required since these deidentified sera were leftover specimens before being discarded. No diagnoses or treatment was involved either.
Cells
Vero E6 (ATCC® CRL-1586) were purchased from the American Type Culture Collection (ATCC, Bethesda, MD), and maintained in a high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS; HyClone Laboratories, South Logan, UT) and 1% penicillin/streptomycin at 37 °C with 5% CO2. All culture media and antibiotics were purchased from ThermoFisher Scientific (Waltham, MA). The cell line was tested negative for mycoplasma.
Recombinant Omicron spike mNG SARS-CoV-2s
The construction and characterization of recombinant Omicron BA.1-, BA.2-, and BA.3-spike mNG SARS-CoV-2s were recently reported4. The BA.1, BA.2, and BA.3 spike sequences were derived from GISAID EPI_ISL_6640916, EPI_ISL_6795834.2, and EPI_ISL_7605591, respectively. Passage 1 (P1) virus stocks were produced from infectious cDNA clones of corresponding viruses14,15. The P1 viruses were used for neutralization testing throughout the study. The spike gene from each P1 virus was sequenced to ensure no undesired mutations. Equivalent specific infectivities, defined by the genomic RNA-to-FFU (fluorescent focus-forming unit) ratios, were confirmed for individual recombinant P1 virus stocks, as previously reported4.
Serum specimens
The de-identified human sera from unvaccinated patients who were infected by Omicron sublineage BA.1 were heat-inactivated at 56 °C for 30 min before neutralization testing. The genotype of infecting virus was verified by the molecular tests with FDA’s Emergency Use Authorization and Sanger sequencing. The serum information is presented in Table 1.
Fluorescent focus reduction neutralization test
Neutralization titers of sera were measured by fluorescent focus reduction neutralization test (FFRNT) using the USA-WA1/2020, BA.1-, BA.2-, and BA.3-spike mNG SARS-CoV-2s. The FFRNT protocol was reported previously5. Briefly, Vero E6 cells were seeded to 96-well plates at 2.5×104 per well (Greiner Bio-one™). On the following day, heat-inactivated sera were 2-fold serially diluted in culture medium with the first dilution of 1:20 (final dilutions ranging from 1:20 to 1:20,480). The diluted serum was incubated with 100–150 FFUs of indicated mNG SARS-CoV-2s at 37 °C for 1 h. Afterwards, the serum-virus mixtures were loaded onto the pre-seeded Vero E6 cell monolayer in 96-well plates. After 1 h infection, the inoculum was aspirated and overlay medium (100 μl supplemented with 0.8% methylcellulose) was added to each well. After incubating the plates at 37 °C for 16–18 h, raw images of mNG foci were acquired using CytationTM 7 (BioTek) with Gene5 software. The foci in each well were counted and normalized to the no-serum-treated controls to calculate infection rates. The FFRNT50 value was defined as the minimal serum dilution that suppressed >50% of fluorescent foci. The neutralization titer of each serum was determined in duplicates, and the geometric mean was presented. Figures were initially plotted using GraphPad Prism 9.0 software, and assembled in Adobe Illustrator. FFRNT50 of <20 was treated as 10 for plot purpose and statistical analysis; in this way, we can differentiate between sera with FFRNT50 of 20 and sera with no neutralization activity at 1:20 dilution (treated as FFRNT50 of 10). Table 1 summarizes the FFRNT50 results.
Statistics
The nonparametric Wilcoxon matched-pairs signed-rank test was used to analyze the statistical significance in Fig. 1B.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank colleagues at the University of Texas Medical Branch (UTMB) for helpful discussions. We thank Michael L O’Rourke from the Information System Department at UTMB for assisting with electronic medical record systems. P.-Y.S. was supported by NIH grants HHSN272201600013C, U01AI151801, AI134907, AI145617, and UL1TR001439, and awards from the Sealy & Smith Foundation, the Kleberg Foundation, the John S. Dunn Foundation, the Amon G. Carter Foundation, the Gilson Longenbaugh Foundation, and the Summerfield Robert Foundation.
Source data
Author contributions
Conceptualization by X.X., P.R., and P.-Y.S.; Methodology by J.Z., C.K., M.L., X.X., P.R., and P.-Y.S; Investigation by J.Z., C.K., M.L., X.X., P.R., and P.-Y.S; Resources by C.K., M.L., X.X., P.R., and P.-Y.S; Data curation by J.Z., C.K., X.X., P.R., and P.-Y.S.; Writing-Original Draft by J.Z., H.X., X.X., R.R., and P.-Y.S; Writing-review & editing by X.X., P.R., and P.-Y.S.; Supervision by X.X., P.R., and P.-Y.S.; Funding acquisition by P.-Y.S.
Peer review
Peer review information
Nature Communications thanks Nicholas Wohlgemuth and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
The raw data that support the findings of this study are shown in the Source data file. Source data are provided with this paper.
Competing interests
X.X. and P.-Y.S. have filed a patent on the reverse genetic system. J.Z., C.K., X.X., and P.-Y.S. received compensation from Pfizer for COVID-19 vaccine development. Other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xuping Xie, Email: xuxie@utmb.edu.
Ping Ren, Email: piren@utmb.edu.
Pei-Yong Shi, Email: peshi@UTMB.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-30580-5.
References
- 1.Wolter N, et al. Clinical severity of Omicron sub-lineage BA.2 compared to BA.1 in South Africa. MedRxiv. 2022 doi: 10.1101/2022.02.17.22271030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyngse FP, et al. Transmission of SARS-CoV-2 omicron VOC subvariants BA.1 and BA.2: evidence from danish households. BioRxiv. 2022 doi: 10.1101/2022.01.28.22270044. [DOI] [Google Scholar]
- 3.Xia H, et al. Neutralization and durability of 2 or 3 doses of the BNT162b2 vaccine against Omicron SARS-CoV-2. Cell Host Microbe. 2022 doi: 10.1016/j.chom.2022.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurhade, C. et al. Neutralization of Omicron BA.1, BA.2, and BA.3 SARS-CoV-2 by 3 doses of BNT162b2 vaccine. BioRxivhttps://biorxiv.org/cgi/content/short/2022.2003.2024.485633v485631 (2022).
- 5.Zou J, et al. Neutralization against Omicron SARS-CoV-2 from previous non-Omicron infection. Nat. Commun. 2022;13:852. doi: 10.1038/s41467-022-28544-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amelia GJ. COVID-19 incidence and death rates among unvaccinated and fully vaccinated adults with and without booster doses during periods of delta and omicron variant emergence — 25 U.S. Jurisdictions, April 4–December 25, 2021. Morbidity Mortal. Wkly. Rep. 2022;71:132–138. doi: 10.15585/mmwr.mm7104e2. [DOI] [Google Scholar]
- 7.Liu Y, et al. Distinct neutralizing kinetics and magnitudes elicited by different SARS-CoV-2 variant spikes. bioRxiv. 2021 doi: 10.1101/2021.09.02.458740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redd, A. D. et al. Minimal crossover between mutations associated with omicron variant of SARS-CoV-2 and CD8(+) T-cell epitopes identified in covid-19 convalescent individuals. mBio10.1128/mbio.03617-21 (2022).
- 9.Ferdinands JM, et al. Waning 2-dose and 3-dose effectiveness of mRNA vaccines against COVID-19-associated emergency department and urgent care encounters and hospitalizations among adults during periods of delta and omicron variant predominance - VISION Network, 10 States, August 2021-January 2022. MMWR Morb. Mortal. Wkly Rep. 2022;71:255–263. doi: 10.15585/mmwr.mm7107e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chemaitelly H, Abu-Raddad LJ. Waning effectiveness of COVID-19 vaccines. Lancet. 2022;399:771–773. doi: 10.1016/S0140-6736(22)00277-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tartof, S. Y. et al. BNT162b2 (Pfizer–Biontech) mRNA COVID-19 Vaccine Against Omicron-Related Hospital and Emergency Department Admission in a Large US Health System: A Test-Negative Design. SSRNhttps://ssrn.com/abstract=4011905 or http://dx.doi.org/4011910.4012139/ssrn.4011905 (2022).
- 12.Andrews N, et al. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. New Engl. J. Med. 2022 doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agency, U. H. S. COVID-19 vaccine surveillance report – Week 9. 3 March 2022. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1058464/Vaccine-surveillance-report-week-1058469.pdf (2022).
- 14.Xie X, et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848 e843. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie X, et al. Engineering SARS-CoV-2 using a reverse genetic system. Nat. Protoc. 2021;16:1761–1784. doi: 10.1038/s41596-021-00491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data that support the findings of this study are shown in the Source data file. Source data are provided with this paper.