Abstract
Escherichia coli B has been engineered as a biocatalyst for the conversion of lignocellulose into ethanol. Previous research has demonstrated that derivatives of E. coli B can produce high levels of Erwinia chrysanthemi endoglucanase (encoded by celZ) as a periplasmic product and that this enzyme can function with commercial fungal cellulase to increase ethanol production. In this study, we have demonstrated two methods that improve celZ expression in E. coli B. Initially, with a low-copy-number vector, two E. coli glycolytic gene promoters (gap and eno) were tested and found to be less effective than the original celZ promoter. By screening 18,000 random fragments of Zymomonas mobilis DNA, a surrogate promoter was identified which increased celZ expression up to sixfold. With this promoter, large polar inclusion bodies were clearly evident in the periplasmic space. Sequencing revealed that the most active surrogate promoter is derived from five Sau3A1 fragments, one of which was previously sequenced in Z. mobilis. Visual inspection indicated that this DNA fragment contains at least five putative promoter regions, two of which were confirmed by primer extension analysis. Addition of the out genes from E. chrysanthemi EC16 caused a further increase in the production of active enzyme and facilitated secretion or release of over half of the activity into the extracellular environment. With the most active construct, of a total of 13,000 IU of active enzyme per liter of culture, 7,800 IU was in the supernatant. The total active endoglucanase was estimated to represent 4 to 6% of cellular protein.
The cost of cellulase enzymes is one of the principal economic bottlenecks for the commercialization of lignocellulosic biomass-to-ethanol technology (13, 16, 18, 29, 32). Three different approaches have been proposed to reduce the cost of cellulase for the bioethanol industry: (i) improved on-site production of cellulase (16, 32), (ii) optimized reconstitution of cellulase components from different sources into a more effective artificial cellulase system (12, 43, 44), and (iii) development of improved ethanologenic biocatalysts which supply a portion of the cellulase needed for the direct microbial conversion of cellulose into ethanol (6, 17). Our lab has focused on the latter approach.
Recombinant strains of Escherichia coli B have been engineered which produce ethanol efficiently from all sugar constituents of lignocellulose by using the Zymomonas mobilis genes encoding the pyruvate-to-ethanol pathway (pdc and adhB) (20, 30). Genes for cellulose hydrolysis are now being added (27, 47). The solubilization of cellulose involves three classes of enzymes: endoglucanase, exoglucanase, and β-glucosidase (32). The requirement for the addition of β-glucosidase has been eliminated by the functional insertion of genes for cellobiose utilization from a related organism, Klebsiella oxytoca M5A1. These K. oxytoca genes encode a phosphoenolpyruvate-dependent phosphotransferase system and phosphocellobiase (22).
Further improvement of cellulose utilization will require extracellular secretion or release of endoglucanase and exoglucanase activities. E. coli is very limited in its ability to secrete proteins into the extracellular environment (34). Recombinant proteins such as endoglucanases, which are secreted by their source organisms, are typically accumulated in the periplasmic space in E. coli (11, 26, 47). In related gram-negative bacteria, however, at least three different types of protein secretion systems have been described (type I, type II, and type III) (25, 34). Considerable progress has been made in our understanding of the type II secretion system (5, 14, 15, 23–25, 28, 34, 37), the most widely used system for protein secretion. Type II secretion involves a two-step process in which a pro-protein containing an N-terminal signal peptide is exported to the periplasm and processed into a mature enzyme by using a highly conserved Sec pathway. The mature enzyme is then secreted into the extracellular environment through the peptidoglycan and outer membrane. Seven type II secretion systems have been cloned from gram-negative bacteria (2, 15, 24, 28, 34, 35, 38, 41), and all contain 12 to 15 gene products which share homology. Although heterologous proteins are often exported to the periplasm by the Sec system, the genes which complete extracellular secretion are highly species specific. This is true even for closely related enteric bacteria such as Erwinia chrysanthemi and E. carotovora (15, 24, 35). Two type II secretion systems, encoded by pul genes from K. oxytoca and the out genes from E. chrysanthemi, have been reconstituted in K-12 strains of E. coli (15, 34). The E. chrysanthemi EC16 out gene products have been shown to secrete EC16 pectate lyase in recombinant E. coli (15). In EC16, this system also appears to facilitate the secretion of the endoglucanase EGZ (3).
Our laboratory has previously cloned celZ from E. chrysanthemi P86021 (4) and demonstrated that the encoded enzyme (EGZ) can function with commercial fungal cellulase to improve ethanol production (47). In these studies, EGZ was accumulated in the E. coli B periplasm and required cell disruption (solvents, detergent, or heat) for release. Total EGZ production was approximately 3 IU per ml of broth.
In this study, we have identified a strong promoter to enhance the expression of celZ endoglucanase in E. coli B and demonstrated that this enzyme can be effectively secreted (or released into the environment) by using the out genes from E. chrysanthemi EC16, thus eliminating the need for cell disruption.
MATERIALS AND METHODS
Organisms and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli DH5α was used as the host for plasmid construction. The celZ gene was previously cloned in our laboratory from E. chrysanthemi P86021 (4, 47). The out genes were cloned by He et al. (15) from E. chrysanthemi EC16.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Z. mobilis CP4 | Prototrophic | 31 |
| E. coli | ||
| DH5α | lacZ M15 recA | Bethesda Research Laboratory |
| B | Prototrophic | ATCC 11303 |
| HB101 | recA lacY recA | ATCC 37159 |
| Plasmids | ||
| pUC19 | bla cloning vector | New England Biolabs |
| pST76-K | kan low copy number, temperature sensitive | 33 |
| pRK2013 | kan mobilizing helper plasmid (mob+) | ATCC |
| pCPP2006 | Spr, ca. 40-kbp plasmid carrying the complete out genes from E. chrysanthemi EC16 | 15 |
| pLOI1620 | bla celZ | 4 |
| pLOI2164 | pLOI1620 with BamHI site removed (Klenow) | This study |
| pLOI2170 | NdeI-HindIII fragment (promoterless celZ) from pLOI2164 cloned into pUC19 | This study |
| pLOI2171 | BamHI-SphI fragment (promoterless celZ) from pLOI2170 cloned into pST76-K | This study |
| pLOI2173 | EcoRI-SphI fragment (celZ with native promoter) from pLOI2164 cloned into pST76-K | This study |
| pLOI2174 | EcoRI-BamHI fragment (gap promoter) cloned into pLOI2171 | This study |
| pLOI2175 | EcoRI-BamHI fragment (eno promoter) cloned into pLOI2171 | This study |
| pLOI2177 | Random Sau3A1 Z. mobilis DNA fragment cloned into pLOI2171 | This study |
| pLOI2178 | Random Sau3A1 Z. mobilis DNA fragment cloned into pLOI2171 | This study |
| pLOI2179 | Random Sau3A1 Z. mobilis DNA fragment cloned into pLOI2171 | This study |
| pLOI2180 | Random Sau3A1 Z. mobilis DNA fragment cloned into pLOI2171 | This study |
| pLOI2181 | Random Sau3A1 Z. mobilis DNA fragment cloned into pLOI2171 | This study |
| pLOI2182 | Random Sau3A1 Z. mobilis DNA fragment cloned into pLOI2171 | This study |
| pLOI2183 | Random Sau3A1 Z. mobilis DNA fragment cloned into pLOI2171 | This study |
| pLOI2184 | Random Sau3A1 Z. mobilis DNA fragment cloned into pLOI2171 | This study |
| pLOI2196 | pLOI2177 fused into pUC19 at the PstI site | This study |
| pLOI2197 | pLOI2180 fused into pUC19 at the PstI site | This study |
| pLOI2198 | pLOI2182 fused into pUC19 at the PstI site | This study |
| pLOI2199 | pLOI2183 fused into pUC19 at the PstI site | This study |
| pLOI2307 | EcoRI-SphI fragment from pLOI2183 cloned into pUC19 | This study |
Cultures were grown in Luria-Bertani broth (LB) (10 g of tryptone per liter [Difco], 5 g of yeast extract per liter [Difco], 5 g of sodium chloride per liter [40]) or on Luria agar (LB supplemented with 15 g of agar per liter). Carboxymethyl cellulose (CMC) plates (48) were used to screen clones for EGZ activity. Ampicillin (50 mg/liter), spectinomycin (100 g/liter), and kanamycin (50 g/liter) were added as appropriate for selection. Constructs containing plasmids with a temperature-conditional pSC101 replicon (33) were grown at 30°C. Unless stated otherwise, constructs with pUC-based plasmids were grown at 37°C.
Genetic methods.
E. chrysanthemi out genes (pCPP2006) were conjugated into E. coli by using pRK2013 for mobilization (10, 28). Standard methods were used for all plasmid constructions (1, 40). Small-scale plasmid isolation was performed by the TELT procedure (1). Large-scale plasmid isolation was performed with a Promega Wizard Kit. DNA fragments were isolated from gels with a Qiaquick Gel Extraction Kit from Qiagen. E. coli and Z. mobilis chromosomal DNAs were isolated as described by Cutting and Horn (8) and Yomano et al. (49), respectively. Purified chromosomal DNA from E. coli DH5α served as a template for the amplification of two glycolytic gene promoters by using the following primer pairs: gap promoter, 5′-CGAATTCCTGCCGAAGTTTATTAGCCA-3′ and 5′-AAGGATCCTTCCACCAGCTATTTGTTAGTGA-3′; and eno promoter, 5′-AGAATTCTGCCAGTTGGTTGACGATAG-3′ and 5′-CAGGATCCCCTCAAGTCACTAGTTAAACTG-3′.
DNA was sequenced by the dideoxy method by using a LI-COR model 4000-L DNA sequencer and fluorescent primers. For pST76-K-based plasmids, only one direction was sequenced with a T7 primer (5′-TAATACGACTCACTATAGGG-3′). For pUC18- or pUC19-based plasmids, sequencing was performed in forward (5′-CACGACGTTGTAAAACGAC-3′) and reverse (5′-ATAACAATTTCACACAGGA-3) directions. Extension reactions were performed by using a Perkin-Elmer GeneAmp PCR 9600 and a SequiTherm Long-Read Sequencing Kit-LC. Sequences were analyzed with the Wisconsin Genetics Computer Group software package (9).
Primer extension analysis of transcriptional initiation.
Promoter regions were identified by mapping the transcriptional start sites with a primer within the celZ gene. RNA was isolated from cells in late exponential phase by using a Qiagen RNeasy kit. Cells were treated with 400 μg of lysozyme per ml in TE containing 0.2 M sucrose (25°C, 5 min) prior to lysis. After ethanol precipitation, RNA was dissolved in 20 μl of Promega avian myeloblastosis virus (AMV) reverse transcriptase buffer (50 mM Tris-HCl, pH 8.3; 50 mM KCl; 10 mM MgCl2; 0.5 mM spermidine; 10 mM dithiothreitol). IRD41-labeled primer (5′-GACTGGATGGTTATCCGAATAAGAGAGAGG-3′; LI-COR, Inc.) was added. The sample was denatured at 80°C for 5 min, annealed at 55°C for 1 h, and purified by alcohol precipitation. Annealed samples were dissolved in 19 μl of AMV reverse transcriptase buffer containing 500 μM deoxynucleoside triphosphates and 10 U of AMV reverse transcriptase and then incubated for extension (1 h at 42°C). Products were treated with 0.5 μg of DNase-free RNase A per ml, precipitated, dissolved in loading buffer, and compared to parallel dideoxy sequences by using the LI-COR model 4000-L DNA sequencer.
Endoglucanase activity.
Expression of celZ was initially evaluated by using the Congo red procedure of Wood and Bhat (48). Recombinant clones were transferred to gridded CMC plates and incubated for 18 h at 30°C prior to staining. Endoglucanase-positive clones formed yellow zones on a red background. The diameters of these zones were recorded as a relative measure of celZ expression.
EGZ activity was also determined in vitro by using CMC as a substrate. Appropriate dilutions of cell-free culture broth (extracellular activity) or broth containing cells treated with ultrasound (total activity) were assayed at 35°C in 50 mM citrate buffer (pH 5.2) containing CMC (20 g/liter). Conditions for optimal enzyme release for 3- to 4-ml samples were determined to be four pulses at full power for 1 s each (model W-220F cell disruptor; Heat System-Ultrasonics, Inc., Plainview, N.Y.). Enzyme reactions were terminated by heating in a boiling water bath (10 min). Reducing sugars were measured by using the dinitrosalicylic acid reagent (48) with glucose as a standard. Enzyme activity (IU) is expressed as micromoles of reducing sugar released per minute or as a percentage of total activity. Results are averages of two or more determinations.
Ultrastructural analysis.
For ultrastructural analysis, fresh colonies from Luria agar plates were fixed in 2% glutaraldehyde in 0.2 M sodium cacodylate buffer (pH 7) and then in 1% osmium tetroxide, followed by treatment with 1% uranyl acetate in distilled water. Samples were dehydrated in ethanol and embedded in Spurr’s plastic (42). Ultrathin sections were examined with a Zeiss EM-10CA electron microscope.
RESULTS
Construction of a low-copy-number promoter probe vector with celZ as the reporter.
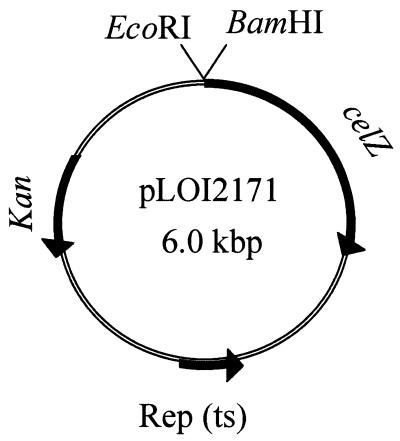
To facilitate the isolation of strong promoters, a low-copy-number vector was constructed with a pSC101 replicon and a BamHI site immediately preceding a promoterless celZ gene (pLOI2171). Plasmid pLOI1620 was used as a source of celZ and is a pUC18 derivative with expression from consecutive lac and celZ promoters. The BamHI site in this plasmid was eliminated by digestion and Klenow treatment (pLOI2164). The celZ gene was isolated as a promoterless NdeI fragment after Klenow treatment. The resulting blunt fragment was digested with HindIII to remove downstream DNA and then ligated into pUC19 (HindIII to HincII) to produce pLOI2170. In this plasmid, celZ is oriented opposite to the direction of lacZ transcription and was weakly expressed. The BamHI (amino terminus)-SphI (carboxyl terminus) fragment from pLOI2170 containing celZ was then cloned into the corresponding sites of pST76-K, a low-copy-number vector with a temperature-sensitive replicon, to produce pLOI2171 (Fig. 1). Expression of celZ in this vector was extremely low, facilitating its use as a probe for strong promoters.
FIG. 1.
Diagram illustrating plasmid pLOI2171, a low-copy-number promoter probe vector Abbreviations: Kan, kanamycin resistance gene; Rep (ts), temperature-sensitive pSC101 replicon.
Expression of celZ from E. coli gap and eno promoters.
Chromosomal DNA from DH5α was used as the template to amplify the gap and eno promoter regions by PCR. The resulting fragments of approximately 400 bp each were digested with EcoRI and BamHI and cloned into the corresponding sites in front of a promoterless celZ in pLOI2171 to produce pLOI2174 (gap promoter) and pLOI2175 (eno promoter). As a control, the EcoRI-SphI fragment from pLOI2164 containing the complete celZ gene and native E. chrysanthemi promoter was cloned into the corresponding sites of pST76-K to produce pLOI2173.
The three plasmids were transformed into strains B and DH5α to compare EGZ production. For both strains of E. coli, endoglucanase activities were lower on CMC plates with E. coli glycolytic promoters than with pLOI2173 containing the native E. chrysanthemi celZ promoter (Table 2). Assuming activity is related to the square of the radius of each zone (Fick’s law of diffusion), EGZ production with glycolytic promoters (pLOI2174 and pLOI2175) was estimated to be 33 to 65% lower than in the original construct.
TABLE 2.
Evaluation of promoter strength for celZ expression in E. coli by using CMC indicator plates
| Plasmids |
E. coli DH5α host
|
E. coli B host
|
||||
|---|---|---|---|---|---|---|
| No. of plasmidsa | CMC zone diam (mm)b | % Native promoter (100 · R2x/R2c)c | No. of plasmids | CMC zone diam (mm) | % Native promoter (100 · R2x/R2c) | |
| pLOI2171 (promoterless) | 1 | 0 | ||||
| pLOI2173 (native promoter) | 1 | 5.0 | 100 | 1 | 4.5 | 100 |
| pLOI2174 (gap promoter) | 1 | 4.0 | 77 | 1 | 3.5 | 60 |
| pLOI2175 (eno promoter) | 1 | 3.0 | 43 | 1 | 2.8 | 35 |
| Z. mobilis promoters | ||||||
| Group I | 5 | 13.0 | 676 | 4 | 10.8–11.3 | 570–625 |
| Group II | 14 | 9.0–11.0 | 324–484 | 17 | 9.0–10.5 | 445–545 |
| Group III | 56 | 6.0–9.0 | 144–324 | 54 | 5.0–8.8 | 125–375 |
The number of clones with the indicated range of activities.
The average size of the diameters from three CMC digestion zones.
R2x is the square of the radius of the clear zone with the test plasmid; R2c is the square of the radius of the clear zone for the control (pLOI2173).
Cloning Z. mobilis DNA fragments as promoters for celZ expression.
Previously, we have used random fragments of Z. mobilis DNA as an effective source of surrogate promoters for the high-level expression of heterologous genes in E. coli (7, 19). A similar approach was used to find promoters for Erwinia celZ expression. Z. mobilis chromosomal DNA was extensively digested with Sau3A1. The resulting fragments were ligated into pLOI2171 at the BamHI site and transformed into E. coli DH5α to generate a library of potential promoters. Resulting colonies were transferred to gridded CMC plates and stained for endoglucanase activity after incubation (Table 2). Approximately 20% of the 18,000 clones tested were CMC positive. The 75 clones which produced larger zones than the control, pLOI2173, were examined further by using E. coli B. In general, recombinants of DH5α produced larger zones than recombinants of strain B. However, the relative promoter strength in each host was similar for most clones. Based on zone size with CMC plates, four clones appeared to express celZ at approximately 6-fold-higher levels than the construct with the original E. chrysanthemi celZ gene (pLOI2173) and at 10-fold-higher levels than the E. coli glycolytic promoters.
Production and apparent secretion of endoglucanase.
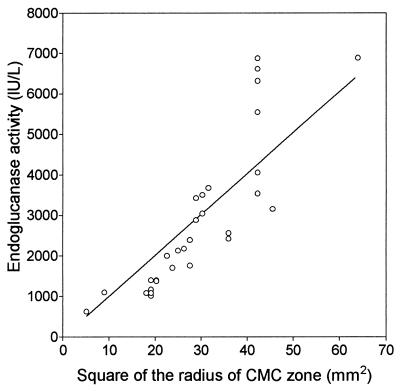
Eight plasmid derivatives of pST76-K (pLOI2177 to pLOI2184) were selected from group I and group II (Table 2) and assayed for total endoglucanase activity in strain B (Table 3). The four plasmids with the largest zones on CMC plates also had the highest endoglucanase activities (pLOI2177, pLOI2180, pLOI2182, and pLOI2183). The activities were approximately sixfold higher than that of the unmodified celZ (pLOI2173), which is in excellent agreement with our estimate by using the square of the radius of the cleared zone on CMC plates. Figure 2 shows a comparison of activity estimates from plates and in vitro enzyme assays for strain B containing a variety of different promoters, with and without the out genes. Although there is some scatter, a direct relationship is clearly evident, which validates the plate method for estimating relative activity. The original construct in pUC18, a high-copy-number plasmid, was also included for comparison (pLOI2164). This construct with consecutive lac and celZ promoters produced less EGZ activity than three of the low-copy-number plasmids with surrogate promoters (pLOI2177, pLOI2182, and pLOI2183). The DNA fragment containing celZ and the most effective surrogate promoter was isolated from pLOI2183 (EcoRI-SphI) and inserted into pUC19 with transcription oriented opposite to that of the lac promoter (pLOI2307). With this high-copy-number plasmid, endoglucanase activity was further increased twofold.
TABLE 3.
Comparison of promoters for EGZ production and secretion in E. coli B
| Plasmida | Without secretion genes
|
With secretion genes (pCPP2006)
|
||
|---|---|---|---|---|
| Total activity (IU/liter)b | Extracellularc activity (%) | Total activity (IU/liter) | Extracellularc activity (%) | |
| Low copy number | ||||
| pLOI2173 | 620 | 17 | 1,100 | 43 |
| pLOI2177 | 3,700 | 10 | 5,500 | 44 |
| pLOI2178 | 2,200 | 9 | 3,500 | 49 |
| pLOI2179 | 2,000 | 10 | 3,000 | 50 |
| pLOI2180 | 2,900 | 8 | 6,300 | 39 |
| pLOI2181 | 1,800 | 11 | 4,100 | 46 |
| pLOI2182 | 3,500 | 7 | 6,600 | 38 |
| pLOI2183 | 3,400 | 7 | 6,900 | 39 |
| pLOI2184 | 2,100 | 12 | 2,400 | 39 |
| High copy number | ||||
| pLOI2164 | 3,200 | 20 | 6,900 | 74 |
| pLOI2307 | 6,600 | 28 | 13,000 | 60 |
Plasmids pLOI2173 and pLOI2164 contain the celZ native promoter; pLOI2307 contains the strong promoter from pLOI2183. Plasmids pLOI2164 and pLOI2307 are pUC-based plasmids (high copy number). All other plasmids are derivatives of pST76-K (low copy number).
Endoglucanase activities were determined after 16 h of growth at 30°C.
Extracellular activity (secreted or released).
FIG. 2.
Relationship between cleared zones on CMC indicator plates and endoglucanase activity in disrupted cells.
Previous studies have shown that recombinant E. coli can secrete E. chrysanthemi EC16 pectate lyase (pelE) into the extracellular environment when provided with EC16 out genes (15). By using the same plasmid containing out genes (pCPP2006), we have investigated the secretion of EGZ in E. coli B (Table 3). With low-copy-number plasmids, 7 to 17% of the total EGZ was extracellular after 16 h of growth without plasmid pCPP2006. A larger fraction of EGZ (20 to 28%) was found in the extracellular broth with pUC-based plasmids than with the low-copy-number pST76-based plasmids containing the same promoters. Addition of pCPP2006 increased the total level of expression up to twofold and increased the fraction of extracellular enzyme (38 to 74%) approximately fourfold. The highest activity was produced by strain B(pLOI2307+pCPP2006): 13,000 IU of total endoglucanase per liter, of which 7,800 IU per liter was found in the cell-free supernatant.
Py et al. (36) reported a specific activity of 419 IU for pure EGZ enzyme (pH 7, 37°C). We have determined that EGZ was 25% more active under their assay conditions than under ours (pH 5.2, citrate buffer; 35°C). If a specific activity of 316 IU for pure enzyme at pH 5.2 (35°C) is assumed, cultures of E. coli B(pLOI2307+pCPP2006) produced approximately 41 mg of active EGZ per liter or 4 to 6% of total cellular protein.
Sequence of Z. mobilis fragment with strong promoter activity.
The sequences of the four strongest surrogate promoters (pLOI2177, pLOI2180, pLOI2182, and pLOI2183) were determined. To facilitate this process, each was fused with pUC19 at the PstI site. The resulting plasmids, pLOI2196, pLOI2197, pLOI2198, and pLOI2199, were produced at high copy numbers (ColE1 replicon) and could be sequenced in both directions by using M13 and T7 sequencing primers. All four plasmids contained identical pieces of Z. mobilis DNA and were siblings. Each was 1,417 bp in length and contained four internal Sau3A1 sites. DNA and translated protein sequences (six reading frames) of each piece were compared to the current database. Only one fragment exhibited a strong match in BLAST searches (281-bp internal fragment); it was 99% identical in DNA sequence to part of the Z. mobilis hpnB gene which is proposed to function in cell envelope biosynthesis (39). Primer extension analysis revealed both a single major start site, 67 bp upstream from the Sau3A1/BamHI junction site with celZ, and a second minor start site farther upstream (Fig. 3). Sequences in the −10 and −35 regions were compared to the conserved sequences for E. coli sigma factors (45, 46). The dominant promoter region (ca. 85% of the total start site) appears similar to a ς70 promoter, whereas the secondary promoter site resembles a ς38 promoter.
FIG. 3.
Partial nucleotide sequence of the Z. mobilis DNA fragment in pLOI2183 which served as a surrogate promoter. The full sequence has been assigned GenBank accession number AF109242. Sequence numbering began with the base adjacent to the upstream Sau3A1 site (GATC). Two transcriptional start sites were identified by primer extension analysis and are marked with a “#” symbol. Putative −35 and −10 regions are marked by double and single underlines, respectively. Vector and celZ DNA are shown as lowercase letters. The Shine-Dalgarno region and the celZ translational start are marked in boldface.
Periplasmic inclusion bodies.
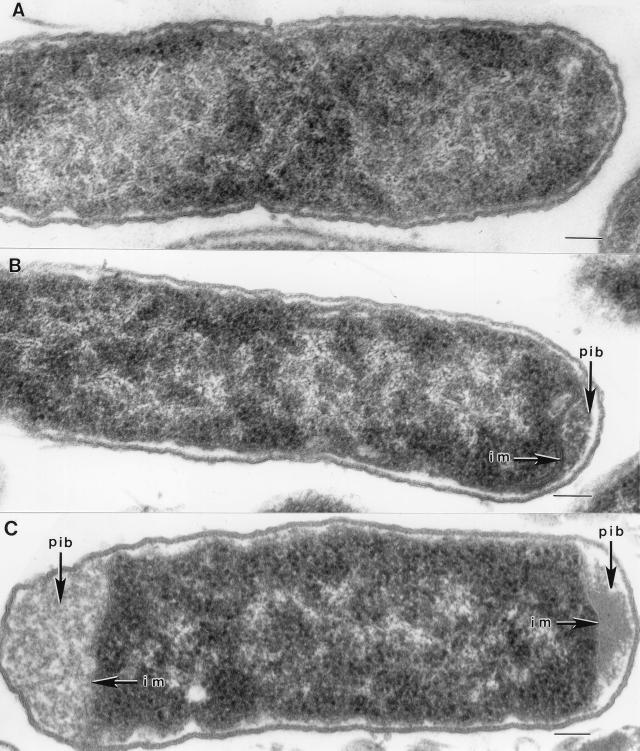
Little difference in cell morphology was observed between recombinants and the parental organism by light microscopy. Under the electron microscope, however, small polar inclusion bodies were clearly evident in the periplasm of strain B(pLOI2164) and are presumed to contain EGZ (Fig. 4). With strain B(pLOI2307), which produced twofold-higher activity, these polar bodies were very large and occupied up to 20% of the total cell volume. The large size of these polar bodies suggests that activity measurements may underestimate total EGZ production. Typically, polar inclusion bodies were smaller in constructs which also contained the out genes (not shown). No periplasmic inclusion bodies were evident in strain B(pUC19), which served as a control.
FIG. 4.
Electron micrographs of E. coli B cells harboring different plasmids. (A) pUC19 containing no visible periplasmic inclusion body. (B) pLOI2164 containing small periplasmic inclusion bodies. (C) pLOI2307 containing large periplasmic inclusion bodies. Abbreviations: im, inner membrane; pib, periplasmic inclusion body. Bars, 0.1 μm.
DISCUSSION
Despite extensive investigation of technology for the bioconversion of lignocellulose to fuel ethanol, the cost of cellulase is still one of the major barriers for commercialization (13, 16, 18, 29, 32). The development of inexpensive enzymatic methods for cellulose hydrolysis has great potential for improving the efficiency of substrate utilization and the economics of the process. Prior to fermentation, insoluble substrates such as cellulose must be converted into a soluble form which can be transported and metabolized. Fungal cellulase is commercially produced on a large scale and is used in food processing, detergents, and pulping (32). Although industrial experience with these enzymes makes it likely that they will also be used in cellulose-to-ethanol processes, it should be possible to develop ethanologenic biocatalysts that provide at least part of the required cellulase activities as a recombinant coproduct with ethanol. Ethanologenic derivatives of E. coli B have been previously engineered to metabolize cellobiose and cellotriose by inserting genes encoding the phosphoenolpyruvate-dependent phosphotransferase system and phospho-β-glucosidase from K. oxytoca M5A1 (22), eliminating a need for the least-stable component of fungal cellulase, β-glucosidase (27). However, enzymes which solubilize polymeric cellulose must be present in the extracellular environment to be effective.
Previous studies have identified E. chrysanthemi EGZ as an effective endoglucanase which can be used in combination with commercial fungal cellulase to improve ethanol production from cellulose (47). The gene encoding this enzyme was expressed well (ca. 3,000 IU per liter), but higher levels of expression would be desirable. The native E. chrysanthemi promoter was more effective in E. coli than either of two E. coli glycolytic promoters (gap and eno). However, Z. mobilis DNA served as an excellent source of surrogate promoters. Sau3A1 fragments with strong promoter activity were identified which provided a sixfold increase in EGZ production in a low-copy-number vector and a twofold increase in a high-copy-number vector. In both cases, most of the activity was cell associated (periplasmic, intracellular, or bound). Relative activity correlated with the size of periplasmic inclusion bodies. Based on activity, EGZ is calculated to represent 4 to 6% of the total cellular protein in the most active construct. However, this may be an underestimate of total EGZ protein since inclusion bodies represented approximately 20% of the total cell volume. It is possible that some of the proteins retained in the inclusion bodies are misfolded or partially degraded.
Native EGZ was secreted or released from the periplasm to the extracellular environment by the out gene products in E. chrysanthemi (21), and the out gene products have been previously demonstrated to secrete recombinant E. chrysanthemi pectate lyases in E. coli (15). Addition of the out genes increased the EGZ activity produced in all E. coli recombinants up to twofold and facilitated the apparent secretion of approximately one-half of the activity into the extracellular environment. The increase in EGZ activity could result from improved folding or reduced degradation. At this time, the basis for incomplete secretion or release remains unknown but may be growth associated. Preliminary experiments revealed that over 90% of the EGZ activity was extracellular in early exponential phase. The fraction of extracellular activity slowly declined and continued to decline during stationary phase, although total activity continued to increase. It is possible that the stability or synthesis of the Out proteins or Sec proteins limited EGZ export during the late-exponential and stationary phases, while the unregulated promoters driving EGZ production continued to function.
The ability of these E. coli strains to secrete or release high levels of EGZ demonstrates an approach which can be used to produce EGZ as a coproduct with ethanol in ethanologenic strains of E. coli B and eliminates the need for additional process steps for the release of endoglucanase activity (47). Although this approach may not be readily used with genes from other hosts, it does demonstrate the potential for E. coli to be further developed as a host for recombinant protein secretion. Considerable progress has been made in defining the basis for protein recognition and secretion in the analogous systems from seven different organisms (2, 5, 14, 15, 23–25, 28, 34, 35, 37, 38, 41). Structural information for extracellular secretion by type II systems does not appear to be localized within a particular region and may involve the entire structure of the secreted protein (14). Recognition of target proteins for secretion appears to reside in the outC and outD gene products (24). With continued progress in defining this relationship it should be possible to secrete large amounts of recombinant products for biotechnology applications by using E. coli as a host.
ACKNOWLEDGMENTS
We thank A. Collmer for sharing plasmid pCPP2006 containing the out genes from E. chrysanthemi EC16 and G. Posfai for sharing the low-copy-number vector, pST76-K.
This research was supported in part by grants from the U.S. Department of Agriculture, National Research Initiative (98-35504-6177), and the U.S. Department of Energy, Office of Basic Energy Science (DE-FG02-96ER20222).
Footnotes
Florida Agricultural Experiment Station journal series no. R-06614.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 2.Bally M, Filloux A, Akrim M, Ball G, Lazdunski A, Tommassen J. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol Microbiol. 1992;6:1121–1131. doi: 10.1111/j.1365-2958.1992.tb01550.x. [DOI] [PubMed] [Google Scholar]
- 3.Barras F, Gijsegem F, Chatterjee A K. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu Rev Phytopathol. 1994;32:201–234. [Google Scholar]
- 4.Beall D S. Genetic engineering of Erwinia for the conversion of biomass to ethanol. Ph.D. dissertation. Gainesville: University of Florida; 1995. [Google Scholar]
- 5.Bortoli-German I, Brun E, Py B, Chippaux M, Barras F. Periplasmic disulphide bond formation is essential for cellulase secretion by the plant pathogen Erwinia chrysanthemi. Mol Microbiol. 1994;11:545–553. doi: 10.1111/j.1365-2958.1994.tb00335.x. [DOI] [PubMed] [Google Scholar]
- 6.Burchardt G, Ingram L O. Conversion of xylan to ethanol by ethanologenic strains of Escherichia coli and Klebsiella oxytoca. Appl Environ Microbiol. 1992;58:1128–1133. doi: 10.1128/aem.58.4.1128-1133.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conway T, Osman Y A, Ingram L O. Gene expression in Zymomonas mobilis: promoter structure and identification of membrane anchor sequences forming functional LacZ′ fusion proteins. J Bacteriol. 1987;169:2327–2335. doi: 10.1128/jb.169.6.2327-2335.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutting S M, Horn P B V. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. New York, N.Y.: John Wiley & Sons, Inc.; 1990. pp. 61–74. [Google Scholar]
- 9.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figurski D, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilkes N R, Kilburn D G, Miller R C, Warren R A J. A mutant of Escherichia coli that leaks cellulase activity encoded by cloned cellulase genes from Cellulomonas fimi. BioTechniques. 1984;3:259–263. [Google Scholar]
- 12.Goyal A K, Wright R M, Hu P, Eveleigh D E. Proceedings of the Society of Automotive Engineering. Warrendale, Pa: SAE; 1992. Cellulase-producing organisms: development through recombinant DNA technology; pp. 761–769. [Google Scholar]
- 13.Hayn H, Steiner W, Klinger R, Steinmuller H, Sinner M, Esterbauer H. Basic research and pilot studies on the enzymatic conversion of lignocellulosics. In: Saddler J N, editor. Bioconversion of forest and agricultural residue. Biotechnology in agriculture series, no. 9. Wallingford, United Kingdom: C.A.B. International; 1993. pp. 33–72. [Google Scholar]
- 14.He S Y, Schoedel C, Chatterjee A K, Collmer A. Extracellular secretion of pectate lyase by the Erwinia chrysanthemi Out pathway is dependent upon Sec-mediated export across the inner membrane. J Bacteriol. 1991;173:4310–4317. doi: 10.1128/jb.173.14.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He S Y, Lindeberg M, Chatterjee A K, Collmer A. Cloned Erwinia chrysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc Natl Acad Sci USA. 1991;88:1079–1083. doi: 10.1073/pnas.88.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Himmel M E, Adney W S, Baker J O, Elander R, McMillan J D, Nieves R A, Sheehan J J, Thomas S R, Vinzant T B, Zhang M. Advanced bioethanol production technologies: a perspective. In: Saha B C, Woodward J, editors. Fuels and chemicals from biomass. ACS symposium series 666. Washington, D.C: American Chemical Society; 1997. pp. 2–45. [Google Scholar]
- 17.Hogsett D A, Ahn H J, Bernardez T D, South C R, Lynd L R. Direct microbial conversion: perspects, progress, and obstacles. Appl Biochem Biotechnol. 1992;35:527–542. [Google Scholar]
- 18.Ingram L O, Gomez P F, Lai X, Moniruzzaman M, Wood B E, Yomano L P, York S W. Metabolic engineering of bacteria for ethanol production. Biotechnol Bioeng. 1998;58:204–214. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<204::aid-bit13>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 19.Ingram L O, Conway T. Expression of different levels of ethanologenic enzymes from Zymomonas mobilis in recombinant strains of Escherichia coli. Appl Environ Microbiol. 1988;54:397–404. doi: 10.1128/aem.54.2.397-404.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingram L O, Conway T, Clark D P, Sewell G W, Preston J F. Genetic engineering of ethanol production in Escherichia coli. Appl Environ Microbiol. 1987;53:2420–2425. doi: 10.1128/aem.53.10.2420-2425.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotoujansky A. Molecular genetics of pathogenesis by soft-rot Erwinia. Annu Rev Phytopathol. 1987;25:405–430. [Google Scholar]
- 22.Lai X, Davis F C, Hespell R B, Ingram L O. Cloning of cellobiose phosphoenolpyruvate-dependent phosphotransferase genes: functional expression in recombinant Escherichia coli and identification of a putative binding region for disaccharides. Appl Environ Microbiol. 1997;63:355–363. doi: 10.1128/aem.63.2.355-363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindeberg M, Collmer A. Analysis of eight out genes in a cluster required for pectic enzyme secretion by Erwinia chrysanthemi: sequence comparison with secretion genes from other gram-negative bacteria. J Bacteriol. 1992;174:7385–7397. doi: 10.1128/jb.174.22.7385-7397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindeberg M, Salmond G P C, Collmer A. Complementation of deletion mutants in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologues reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via type II pathway. Mol Microbiol. 1996;20:175–190. doi: 10.1111/j.1365-2958.1996.tb02499.x. [DOI] [PubMed] [Google Scholar]
- 25.Lory S. Determinants of extracellular protein secretion in gram-negative bacteria. J Bacteriol. 1992;174:3423–3428. doi: 10.1128/jb.174.11.3423-3428.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Missiakas D, Raina S. Protein folding in the bacterial periplasm. J Bacteriol. 1997;179:2465–2471. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moniruzzaman M, Lai X, York S W, Ingram L O. Isolation and molecular characterization of high-performance cellobiose-fermenting spontaneous mutants of ethanologenic Escherichia coli KO11 containing the Klebsiella oxytoca casAB operon. Appl Environ Microbiol. 1997;63:4633–4637. doi: 10.1128/aem.63.12.4633-4637.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murata H, Fons M, Chatterjee A, Collmer A, Chatterjee A K. Characterization of transposon insertion out− mutants of Erwinia carotovora subsp. carotovora defective in enzyme export and of a DNA segment that complements out mutations in E. carotovora subsp. carotovora, E. carotovora subsp. atroseptica, and Erwinia chrysanthemi. J Bacteriol. 1990;172:2970–2978. doi: 10.1128/jb.172.6.2970-2978.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nieves R A, Ehrman C I, Elander R T, Himmel M E. Technical communication: survey and analysis of commercial cellulase preparations suitable for biomass conversion to ethanol. World J Microbiol Biotechnol. 1998;14:301–304. [Google Scholar]
- 30.Ohta K, Beall D S, Mejia J P, Shanmugan K T, Ingram L O. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl Environ Microbiol. 1991;57:893–900. doi: 10.1128/aem.57.4.893-900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osman Y A, Ingram L O. Mechanism of ethanol inhibition of fermentation in Zymomonas mobilis CP4. J Bacteriol. 1985;164:173–180. doi: 10.1128/jb.164.1.173-180.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Philippidis G P. Cellulase production technology: evaluation of current status. In: Himmel M E, Baker J O, Overend R P, editors. Enzymatic conversion of biomass for fuel production. ACS symposium series 566. Washington, D.C: American Chemical Society; 1994. pp. 188–217. [Google Scholar]
- 33.Posfai G, Koob M D, Kirkpatrick H A, Blattner F R. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J Bacteriol. 1997;179:4426–4428. doi: 10.1128/jb.179.13.4426-4428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Py B, Salmond G P C, Chippaux M, Barras F. Secretion of cellulase in Erwinia chrysanthemi and E. carotovora is species-specific. FEMS Microbiol Lett. 1991;79:315–322. [Google Scholar]
- 36.Py B, Bortoli-German I, Haiech J, Chippaux M, Barras F. Cellulase EGZ of Erwinia chrysanthemi: structural organization and importance of His98 and Glu33 residues for catalysis. Protein Eng. 1991;4:325–333. doi: 10.1093/protein/4.3.325. [DOI] [PubMed] [Google Scholar]
- 37.Py B, Chippaux M, Barras F. Mutagenesis of cellulase EGZ for studying the general protein secretory pathway in Erwinia chrysanthemi. Mol Microbiol. 1993;7:785–793. doi: 10.1111/j.1365-2958.1993.tb01169.x. [DOI] [PubMed] [Google Scholar]
- 38.Reeves P J, Whitcombe D, Wharam S, Gibson M, Allison G, Bunce N, Barallon R, Douglas P, Mulholland V, Stevens S, Walker D, Salmond G P C. Molecular cloning and characterization of 13 out genes from Erwinia carotovora subspecies carotovora: genes encoding members of a general secretion pathway (GSP) widespread in gram-negative bacteria. Mol Microbiol. 1993;8:443–456. doi: 10.1111/j.1365-2958.1993.tb01589.x. [DOI] [PubMed] [Google Scholar]
- 39.Reipen I G, Poralla K, Sahm H, Sprenger G A. Zymomonas mobilis squalene-hopene cyclase gene (shc): cloning, DNA sequence analysis, and expression in Escherichia coli. Microbiology. 1995;141:155–161. doi: 10.1099/00221287-141-1-155. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Sandkvist M, Michel L O, Hough L P, Morales V M, Bagdasarian M, Koomey M, Drita V J, Bagdasarian M. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spurr A R. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- 43.Thomas S R, Laymon R A, Chou Y C, Tucker M P, Vinzant T B, Adney W S, Baker J O, Nieves R A, Mielens J R, Himmel M E. Initial approaches to artificial cellulase systems for conversion of biomass to ethanol. In: Saddler J N, Himmel M E, editors. Enzymatic degradation of insoluble carbohydrates. ACS symposium series 618. Washington, D.C: American Chemical Society; 1995. pp. 208–236. [Google Scholar]
- 44.Walker L P, Belair C D, Wilson D B, Irwin D C. Engineering cellulase mixtures by varying the mole fraction of Thermomonospora fusca E5 and E3, Trichoderma reesei CBHI, and Caldocellum saccharolyticum β-glucosidase. Biotechnol Bioeng. 1993;42:1019–1028. doi: 10.1002/bit.260420902. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, Gralla J D. Multiple in vivo roles for the −12-region elements of sigma 54 promoters. J Bacteriol. 1998;180:5626–5631. doi: 10.1128/jb.180.21.5626-5631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wise A, Brems R, Ramakrishnan V, Villarejo M. Sequences in the −35 region of Escherichia coli rpoS-dependent genes promote transcription by EςS. J Bacteriol. 1996;178:2785–2793. doi: 10.1128/jb.178.10.2785-2793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood B E, Beall D S, Ingram L O. Production of recombinant bacterial endoglucanase as a co-product with ethanol during fermentation using derivatives of Escherichia coli KO11. Biotechnol Bioeng. 1997;55:547–555. doi: 10.1002/(SICI)1097-0290(19970805)55:3<547::AID-BIT12>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 48.Wood T M, Bhat K M. Methods for measuring cellulase activities. Methods Enzymol. 1988;160:87–112. [Google Scholar]
- 49.Yomano L P, Scopes R K, Ingram L O. Cloning, sequencing, and expression of the Zymomonas mobilis phosphoglycerate mutase gene (pgm) in Escherichia coli. J Bacteriol. 1993;175:3926–3933. doi: 10.1128/jb.175.13.3926-3933.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]