Abstract
Apolipoprotein E (APOE) allelic variation is associated with differences in overall circulating lipids and risks of major health outcomes. Lipid profiling provides the opportunity for a more detailed description of lipids that differ by APOE, to potentially inform therapeutic targets for mitigating higher morbidity and mortality associated with certain APOE genotypes. Here, we sought to identify lipids, lipid-like molecules, and important mediators of fatty acid metabolism that differ by APOE among 278 Black men ages 70–81. Using liquid chromatography-mass spectrometry methods, 222 plasma metabolites classified as lipids, lipid-like molecules, or essential in fatty acid metabolism were detected. We applied principal factor analyses to calculate a factor score for each main lipid category. APOE was categorized as ε4 carriers (n = 83; ε3ε4 or ε4ε4), ε2 carriers (n = 58; ε2ε3 or ε2ε2), or ε3 homozygotes (n = 137; ε3ε3). Using analysis of variance, the monoacylglycerol factor, cholesterol ester factor, the factor for triacylglycerols that consist mostly of polyunsaturated fatty acids, sphingosine, and free carnitine significantly differed by APOE (p < 0.05, false discovery rate < 0.30). The monoacylglycerol factor, cholesterol ester factor, and sphingosine were lower, whereas the factor for triacylglycerols that consisted mostly of polyunsaturated fatty acids was higher among ε2 carriers than remaining participants. Free carnitine was lower among ε4 carriers than ε3 homozygotes. Lower monoacylglycerols and cholesteryl esters and higher triacylglycerols that consist mostly of polyunsaturated fatty acids may be protective metabolic characteristics of APOE ε2 carriers, whereas lower carnitine may reflect altered mitochondrial functioning among ε4 carriers in this cohort of older Black men.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-021-00382-6.
Keywords: Lipids, Metabolites, Metabolism, Apolipoprotein E, APOE
Introduction
The apolipoprotein E (APOE) gene has three common alleles: ε2, ε3, and ε4. APOE encodes for the glycoprotein, apolipoprotein E, which combines with lipids to form lipoproteins that transport lipids through the bloodstream to different tissues for energy or storage [1]. APOE allelic variation causes apolipoprotein E to differ in lipid preferential binding; the ε4 allele causes apolipoprotein E to have a higher affinity for larger very low-density lipoprotein (VLDL) particles rich in triglycerides [2], whereas ε2 and ε3 alleles cause apolipoprotein E to have a higher affinity for smaller high-density lipoprotein (HDL) particles rich in phospholipids [2]. Another important distinction is the ε2 allele causes apolipoprotein E to have a 50 times lower affinity for binding to low-density lipoproteins (LDL) than the ε4 and ε3 alleles [3].
Differences in lipoprotein affinity by APOE result in differences in lipid metabolism and circulating lipids [2]. For example, the enhanced binding of apolipoprotein E to VLDL particles due to the ε4 allele results in altered lipid metabolism [4], characterized by increased liver uptake of LDL particles that paradoxically results in increased circulating LDL particles [5]. Specifically, ε4 carriers have higher triglycerides and LDL cholesterol, whereas ε2 carriers have higher triglycerides, but lower LDL cholesterol than ε3 homozygotes [6]. APOE ε4 carriers also have higher risks of Alzheimer’s disease [7, 8], cardiovascular disease [9, 10], and mortality [11], whereas ε2 carriers have lower risks of Alzheimer’s disease [12], cardiovascular disease [10], and mortality [11] when compared to ε3 homozygotes. U.S. frequencies for the ε2, ε3, and ε4 alleles are 0.07, 0.78, and 0.15, respectively [13], with a higher frequency of the ε4 “risk” allele among Black versus White older adults [14–18]. A more detailed description of lipids that differ by APOE could potentially inform novel therapeutic targets to mitigate morbidity and mortality related to APOE.
Metabolomics is the large-scale study of small molecules that are intermediates or end-products of cellular metabolism and typically include numerous lipids and lipid-like molecules. The Health, Aging, and Body Composition (Health ABC) study measured 350 plasma metabolites in a subset of 319 older Black men, of which more than 200 were classified as lipids or lipid-like molecules. Here, we sought to better characterize known differences in overall circulating lipids according to APOE by identifying lipids, lipid-like molecules, and important mediators of fatty acid metabolism that differ among older Black men who carry the APOE ε4 allele (n = 83; ε3ε4, ε4ε4), ε2 allele (n = 58; ε2ε3, ε2ε2), or two copies of the ε3 allele (n = 137; ε3ε3).
Methods
Cohort
The Health ABC study was a prospective cohort of 3075 Black and White men and women [19]. Participants were recruited from Pittsburgh, Pennsylvania and Memphis, Tennessee. White individuals were recruited from a random sample of Medicare beneficiaries and all age-eligible Black individuals were recruited. Eligible participants were ages 70–79 during recruitment (March 1997–July 1998) and self-reported no difficulty walking ¼ mile, climbing ten steps, or with basic activities of daily living. Ineligibility criteria included history of active cancer within 3 years or planning to move from the study area during the next 3 years. The study was approved by each sites’ Institutional Review Board. All participants provided written informed consent. All procedures were in accordance with institutional guidelines.
In 2016, a pilot study detected 350 metabolites in plasma collected at year 2 (1998–99) after an overnight fast of at least 8 h among a subset of 319 Black men, to better understand the influence of body composition on cellular metabolism [20]. The study was restricted to Black men due to limited funds and because U.S. Black individuals have a higher prevalence of aging-related metabolic diseases, but more muscle mass than U.S. White individuals [20]. The study was restricted to men to limit heterogeneity in body composition due to differences by sex.
APOE
Genotyping was performed at year 1 by the Center for Inherited Disease Research using Illumina Human1M-Duo BeadChip v3.3.7 system [21]. APOE was determined directly using polymerase chain reaction on coded DNA samples [11]. For analyses, APOE was categorized as ε4 carriers (ε3ε4 and ε4ε4; n = 83), ε2 carriers (ε2ε3 and ε2ε2; n = 58), or ε3 homozygotes (n = 137). Fourteen participants carrying the ε2ε4 alleles were examined but excluded from statistical tests due to the small number of participants. The proportions of APOE categories among the subset of Black men with metabolomics were almost identical to the proportions among the total sample of Health ABC Black men (Supplemental Table 1a versus 1b). The Health ABC Black men had higher proportions of ε2 carriers and ε4 carriers and a lower proportion of ε3 homozygotes than the White participants (Supplemental Table 1b).
Table 1.
Characteristicsa of Health ABC Black men by APOE genotype categories
| Mean (standard deviation) or frequency (percent) | APOE genotype categories | Overall p-valueb; pairwise comparisonsc | |||
|---|---|---|---|---|---|
| ε2 carriers (n = 58) | ε3 homozygotes (n = 137) | ε4 carriers (n = 83) | ε2ε4 carriers (n = 14) | ||
| Age | 75 (2.5) | 75 (2.9) | 75 (2.9) | 73 (2.1) | .99 |
| Pittsburgh site | 31 (53%) | 77 (56%) | 47 (57%) | 6 (43%) | .92 |
| More than high school education | 9 (16%) | 44 (32%) | 25 (30%) | 3 (21%) | .06 |
| Current smoker, year 1 | 10 (17%) | 23 (17%) | 17 (20%) | 5 (36%) | .78 |
| Height (cm), year 1 | 172 (7.2) | 174 (6.3) | 172 (7.5) | 171 (6.3) | .16 |
| Weight (kg) | 78 (15) | 83 (14) | 79 (15) | 80 (12) | .05 |
| Body mass index (kg/m2) | 26 (4.6) | 27 (4.5) | 27 (4.4) | 28 (3.8) | .18 |
| Waist circumference (cm), year 1 | 98 (13) | 101 (11) | 98 (13) | 98 (11) | .18 |
| Appendicular lean mass (kg/m2) | 8.2 (1.3) | 8.4 (1.1) | 8.2 (1.1) | 8.4 (1.1) | .32 |
| Body fat (%) | 27 (5.4) | 29 (5.6) | 28 (5.4) | 29 (6.3) | .25 |
| Daily dietary intake: | |||||
| Total calories (Kcal) | 2239 (1355) | 2145 (900) | 2309 (1168) | 2375 (1448) | .55 |
| Protein kilocalories (%) | 13 (2.7) | 14 (3.1) | 14 (3.6) | 16 (2.7) | .13 |
| Fat kilocalories (%) | 36 (7.6) | 35 (7.6) | 34 (7.0) | 35 (7.8) | .50 |
| Carbohydrates kilocalories (%) | 52 (8.9) | 52 (8.5) | 53 (10) | 50 (11) | .92 |
| Protein intake (g) | 73 (45) | 73 (32) | 82 (44) | 96 (66) | .25 |
| Fat intake (g) | 91 (63) | 84 (43) | 89 (50) | 95 (74) | .63 |
| Saturated fat intake (g) | 26 (18) | 24 (13) | 26 (15) | 27 (18) | .77 |
| Cholesterol intake (g) | 0.30 (0.2) | 0.27 (0.2) | 0.30 (0.2) | 0.37 (0.3) | .60 |
| Fiber intake (g) | 18 (11) | 18 (8.2) | 19 (10) | 17 (13) | .68 |
| Prevalent disease, year 1: | |||||
| Cardiovascular disease | 13 (22%) | 48 (35%) | 26 (31%) | 3 (21%) | .22 |
| Hypertension | 31 (56%) | 82 (62%) | 43 (54%) | 10 (77%) | .60 |
| Diabetes | 13 (22%) | 27 (20%) | 17 (20%) | 2 (14%) | .91 |
| Cancer | 4 (7%) | 17 (12%) | 11 (13%) | 0 | .46 |
| Number of rx medications | 3.4 (3.8) | 3.3 (2.9) | 3.1 (2.7) | 3.0 (2.4) | .82 |
| Taking lipid-lowering medication | 5 (9%) | 25 (18%) | 14 (16%) | 1 (7%) | .23 |
| Overall lipid profile: | |||||
| Total cholesterol | 183 (35) | 198 (36) | 200 (38) | 193 (30) | .01; ε2 < ε3, ε4 |
| Total cholesterol, year 1 | 175 (33) | 199 (37) | 199 (34) | 186 (20) | < .0001; ε2 < ε3, ε4 |
| LDL cholesterol, year 1 | 99 (33) | 124 (34) | 125 (33) | 106 (18) | < .0001; ε2 < ε3, ε4 |
| HDL cholesterol, year 1 | 53 (14) | 52 (14) | 51 (14) | 59 (19) | .81 |
| Triglycerides, year 1 | 120 (59) | 116 (60) | 116 (58) | 103 (44) | .92 |
| Interleukin-6 (pg/mL) | 3.7 (4.5) Med = 2.4 | 3.8 (3.6) Med = 2.8 | 3.8 (3.6) Med = 2.5 | 2.9 (1.8) Med = 2.3 | .69 |
| Creatinine (mg/dL), year 1 | 1.3 (0.3) | 1.2 (0.3) | 1.3 (0.4) | 1.2 (0.2) | .25 |
APOE, Apolipoprotein E genotype; Med, Median
aMeasured at year 2 unless stated otherwise
bExcluding n = 14 participants with APOE genotype ε2ε4
cε2 denotes ε2 carriers (ε2ε2 and ε2ε3); ε3 denotes ε3 homozygotes (ε3ε3), and ε4 denotes ε4 carriers (ε3ε4 and ε4ε4)
Metabolites
Metabolites were measured by the Broad Institute using plasma extracts collected at year 2 after an overnight fast of at least 8 h. These plasma samples had never been thawed and were stored at − 80 °C from collection (1998/1999) until metabolite profiling (2016). Three liquid chromatography-mass spectrometry (LC–MS) metabolite profiling methods were previously described in detail [22–24], measuring: (1) amines and polar metabolites; (2) central metabolites and polar metabolites, and (3) lipids [23]. Metabolite values were analyzed using TraceFinder (ThermoFisher Scientific, Waltham, MA, USA) and Progenesis QI (Nonlinear Dynamics, UK), confirmed manually using known standards, and if below the limit of quantitation (signal/noise < 10) were classified as unquantifiable [22]. Two pooled samples were run after every 20 samples, one to normalize the data, if necessary, and the other to assess quality of that normalization. The positive and negative ion mode detection methods used normalization to the nearest neighbor, whereas the lipid profiling method was not normalized. We found high reliability of known metabolites from sixteen blinded duplicates (intraclass correlation coefficient, median: 0.92; interquartile range: 0.81–0.97) [20]. Metabolite values were reported as LC–MS peak areas, which are proportional to concentrations [20, 23]. Metabolites were log-transformed and standardized to a mean of zero and standard deviation of one.
A total of 350 metabolites were detected using the three profiling methods [20]. For this report, our independent variable of interest, i.e., APOE, was a three-level categorical variable with a limited sample size, thus, to reduce the number of comparisons made, we focused on a subset of the 350 metabolites (Supplemental Table 2). We examined metabolites classified as lipids (e.g., triacylglycerols, cholesteryl esters, and sphingomyelins) or lipid-like molecules (bile acids, acylcarnitines) and metabolites that play a central role in fatty acid metabolism (e.g., free carnitine). Specifically, we examined a subset of 229 metabolites. Among the 229 metabolites, 189 were detected using the lipid profiling method, eight were detected using the polar metabolite profiling method with negative ion mode detection (i.e., five bile acids, two fatty acids, and one glycerophosphate), and 32 were detected using the polar metabolite profiling method with positive ion mode detection (i.e., 28 acetylcarnitine, free carnitine, two fatty acids, and one glycerophosphocholine).
Table 2.
Associations between lipids and APOE genotype among Health ABC Black men
| Mean (standard error) of APOE genotype categories | Overall p-valuea; pairwise comparisonsb | ||||
|---|---|---|---|---|---|
| ε2 carriers (n = 58) | ε3 homozygotes (n = 137) | ε4 carrier (n = 83) | ε2ε4 carriers (n = 14) | ||
| Monoacylglycerol (MAG) score | − 0.38 (0.10) | 0.19 (0.10) | 0.0007 (0.10) | − 0.24 (0.16) | 0.0005c; ε2 < ε4, ε3 |
| Cholesteryl ester (CE) score | − 0.38 (0.13) | 0.08 (0.09) | 0.14 (0.11) | − 0.04 (0.26) | 0.005c; ε2 < ε3, ε4 |
| Sphingosine | − 0.23 (0.13) | 0.18 (0.08) | − 0.06 (0.11) | − 0.49 (0.26) | .02c; ε2 < ε3 |
| Triacylglycerol (TAG) that consist mostly of polyunsaturated fatty acids score | 0.29 (0.13) | − 0.05 (0.09) | − 0.15 (0.11) | 0.17 (0.27) | 0.03c; ε2 > ε3, ε4 |
| Free carnitine | 0.06 (0.13) | 0.12 (0.09) | − 0.23 (0.11) | − 0.06 (0.27) | .04c; ε4 < ε3 |
| PE plasmalogen score | 0.23 (0.13) | − 0.02 (0.09) | − 0.17 (0.11) | 0.25 (0.27) | 0.07 |
| Lysophosphatidylethanolamine (LPE) score | 0.10 (0.12) | − 0.15 (0.09) | 0.10 (0.10) | 0.46 (0.26) | 0.11 |
| PS plasmalogen score | − 0.20 (0.13) | 0.10 (0.09) | − 0.04 (0.11) | 0.11 (0.27) | 0.16 |
| Sphingomyelin (SM) score | − 0.22 (0.13) | 0.05 (0.09) | 0.06 (0.11) | 0.12 (0.27) | 0.18 |
| PC plasmalogen score | 0.07 (0.13) | 0.04 (0.09) | − 0.18 (0.11) | 0.40 (0.27) | 0.22 |
| 3-methyladipate | 0.21 (0.16) | − 0.03 (0.08) | − 0.10 (0.11) | − 0.006 (0.25) | 0.28 |
| Phosphorylcholine (PC) score | − 0.19 (0.13) | 0.06 (0.09) | − 0.008 (0.11) | 0.24 (0.27) | 0.30 |
| Mevalonic acid | 0.11 (0.13) | − 0.09 (0.09) | 0.07 (0.11) | − 0.02 (0.27) | 0.32 |
| Lysophosphatidylcholine (LPC) score | 0.05 (0.13) | − 0.10 (0.09) | 0.06 (0.11) | 0.42 (0.27) | 0.47 |
| Alpha-glycerophosphocholine | 0.13 (0.13) | − 0.02 (0.09) | − 0.08 (0.11) | 0.12 (0.27) | 0.48 |
| Phosphatidylinositol (PI) score | − 0.12 (0.13) | 0.05 (0.09) | − 0.03 (0.11) | 0.15 (0.27) | 0.57 |
| Acylcarnitine score | 0.04 (0.13) | 0.05 (0.09) | − 0.09 (0.11) | − 0.11 (0.27) | 0.61 |
| Phosphatidylethanolamine (PE) score | 0.09 (0.13) | − 0.002 (0.09) | − 0.04 (0.11) | − 0.10 (0.27) | 0.73 |
| Bile acid score | − 0.01 (0.13) | − 0.0005 (0.09) | 0.09 (0.11) | − 0.49 (0.27) | 0.75 |
| Diacylglycerol (DAG) score | 0.08 (0.13) | 0.006 (0.08) | − 0.04 (0.11) | − 0.16 (0.27) | 0.78 |
| Alpha-glycerophosphate | 0.01 (0.13) | − 0.02 (0.09) | 0.06 (0.11) | − 0.18 (0.27) | 0.86 |
| C34:0 PS | − 0.09 (0.13) | − 0.002 (0.09) | − 0.02 (0.11) | 0.47 (0.27) | 0.87 |
| Adipate | 0.02 (0.13) | − 0.02 (0.09) | 0.03 (0.11) | − 0.11 (0.27) | 0.94 |
| Triacylglycerol (TAG) that do not consist mostly of polyunsaturated fatty acids score | 0.02 (0.13) | 0.006 (0.08) | − 0.002 (0.11) | − 0.14 (0.27) | 0.99 |
| Ceramide score | 0.0007 (0.13) | − 0.001 (0.09) | 0.004 (0.11) | − 0.01 (0.27) | 0.999 |
APOE, Apolipoprotein E; PE, phosphatidylethanolamine; PS, phosphatidylserine; PC, phosphorylcholine
aExcluding n = 14 participants with APOE genotype ε2ε4
bε2 denotes ε2 carriers (ε2ε2 and ε2ε3); ε3 denotes ε3 homozygotes (ε3ε3), and ε4 denotes ε4 carriers (ε3ε4 and ε4ε4)
cFalse discovery rate < 0.30
Among the subset of 229 metabolites, 206 were detected in all participants. Sixteen metabolites were detected in ≥ 80% of the cohort; missing values were assumed to be due to true values being below detectable limits [25] and for analyses, these missing values were replaced with half the minimum recorded value for the respective metabolite [25]. The remaining seven metabolites were excluded from analyses because they were detected in < 80% of participants [26]. Thus, our final analysis examined 222 metabolites by APOE (Supplemental Table 3).
Participant characteristics
Participants self-reported age, gender, Black race, highest level of education, and cigarette smoking habits. Height, weight, and waist circumference were recorded and body mass index was calculated. Usual diet during the past year was estimated using an interviewer-administered 108-item Food Frequency Questionnaire [27]. Using total body dual-energy x-ray absorptiometry (Hologic QDR 4500A; Hologic, Bedford, MA, USA), appendicular lean mass was estimated as the sum of bone-free lean mass in arms and legs divided by height2 and body fat was estimated relative to total mass. History or presence of cardiovascular disease, hypertension, diabetes, and cancer were based on self-report of a physician diagnosis or taking medication. Participants brought in all prescription medications used in the last 2 weeks for an inventory, and information on use of statins or other lipid-lowering medications was documented. Serum interleukin-6 and C-reactive protein and plasma total cholesterol were measured at year 2 by a core laboratory at Wake Forest University. Serum creatinine, cystatin C, and triglycerides and plasma total and HDL cholesterol were measured at year 1 by the Laboratory for Clinical Biochemistry Research at the University of Vermont. LDL cholesterol was calculated using the Friedewald equation [28].
Statistical analysis
Differences in demographics, behavioral factors, and markers of disease by APOE were tested using analysis of variance for normally distributed continuous measures, Kruskal–Wallis test for non-normally distributed continuous measures, and chi-square test for categorical measures. We applied principal factor analyses with varimax rotation in SAS 9.4 separately by lipid categories (see Supplemental Table 3 for lipids organized by categories). For each lipid category, we calculated subject-specific weighted factor scores from the first factor. For lipid categories that did not contain enough metabolites for a factor analysis or if the first factor had an eigenvalue less than one, then we examined these metabolites individually in our main analyses (e.g., sphingosine, free carnitine, and C34:0 PS). In our previous work on metabolomics of walking ability in older adults [29], we found that the direction of associations between triacylglycerols and walking ability differed based on the degree of fatty acid saturation. Thus, to account for this, we calculated separate factor scores for triacylglycerols that did versus did not consist mostly of polyunsaturated fatty acids.
We identified metabolite factor scores and individual metabolites that differed by APOE using analysis of variance, adjusting for unequal variances when necessary (p < 0.10 from Levene’s test for homogeneity). To account for multiple comparisons, we used a Benjamini–Hochberg correction [30] with 30% false discovery rate since this was a hypothesis-generating report [22, 31]. Pairwise comparisons by APOE were made when an overall difference in a metabolite was observed. As a sensitivity analysis, we examined differences in metabolite factor scores and individual metabolites by APOE when excluding participants taking lipid-lowering medication.
Results
Table 1 indicates minimal differences in study characteristics by APOE. Overall, 15% of the cohort was taking lipid-lowering medications, of which almost all of those participants were taking statins. Fewer ε2 carriers were taking lipid-lowering medications than ε3 homozygotes and ε4 carriers, though this difference was not statistically significant (p = 0.23). Total cholesterol was lower among ε2 carriers than ε3 homozygotes and ε4 carriers. HDL and LDL cholesterol and triglycerides were not measured at year 2 but were measured at year 1; ε2 carriers had lower LDL cholesterol, but there was no difference in HDL cholesterol nor triglycerides when compared to ε4 carriers or ε3 homozygotes.
Supplemental Table 4 includes the proportion of variance explained for each of the 17 metabolite factor scores. The two factor scores for triacylglycerols resulted in the highest proportion of variance explained, whereas the phosphatidylinositol factor score which consisted of five phosphatidylinositols had the lowest proportion of variance explained. Table 2 compares the 17 metabolite factor scores and the remaining log-transformed and standardized values for the eight individual metabolites by APOE. We found that the monoacylglycerol score, cholesteryl ester score, sphingosine, free carnitine, and the factor score for triacylglycerols that consist mostly of polyunsaturated fatty acids were significantly different by APOE (p < 0.05, false discovery rate < 0.30). When examining pairwise comparisons, we found ε2 carriers had lower average levels of the monoacylglycerol score, cholesteryl ester score, and sphingosine and higher average levels of the factor score for the triacylglycerols that consist mostly of polyunsaturated fatty acids when compared to remaining participants. Carnitine was significantly lower among ε4 carriers than ε3 homozygotes. We found consistent mean differences in the metabolite factor scores and individual metabolites by APOE when excluding participants taking lipid-lowering medication.
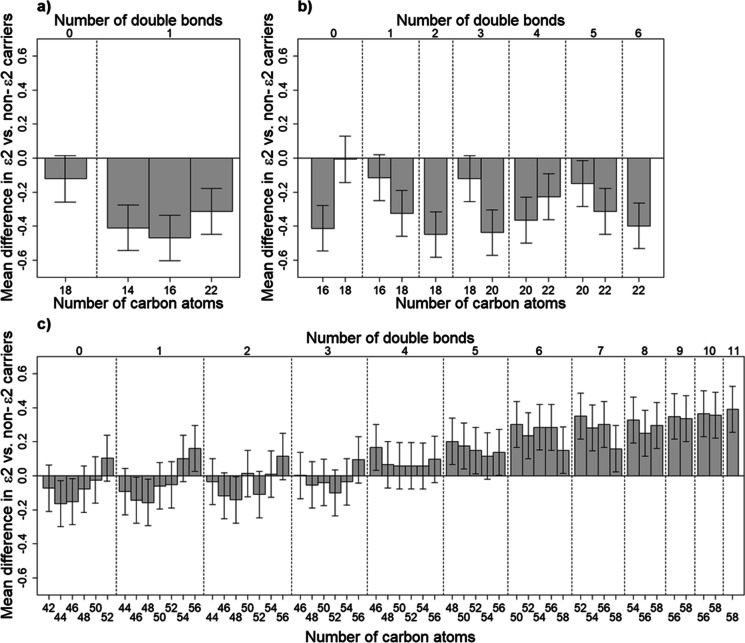
Supplemental Table 3 compares the average log-transformed and standardized values for all 222 individual metabolites by APOE. There were 23 metabolites that significantly differed by APOE (p < 0.05). Consistent with our main analysis using lipid-category factor scores, we found three monoacylglycerols, six cholesteryl esters, five triacylglycerols, sphingosine, and free carnitine differed significantly by APOE, in addition to four PE plasmalogens and one sphingomyelin (Supplemental Table 3). Among the triacylglycerols, we found only those consisting mostly of polyunsaturated fatty acids were significantly higher among ε2 carriers compared to ε3 homozygotes or ε4 carriers (Table 2 and Supplemental Table 3). To examine this pattern further, we plotted the mean difference among ε2 carriers versus non-ε2 carriers for all 55 triacylglycerols that were measured in the Health ABC study against the number of double bonds and the number of carbon atoms within each triacylglycerol. Figure 1C further illustrates that triacylglycerols that were higher among ε2 carriers tended to have five or more double bonds. We examined this same type of plot for monoacylglycerols and cholesteryl esters. Figure 1A and B illustrate that regardless of the number of double bonds and carbon atoms, all cholesteryl esters and monoacylglycerols tended to be lower among ε2 carriers versus non-ε2 carriers.
Fig. 1.
Standardized mean (± standard error) difference in apolipoprotein E (APOE) ε2 vs. non-ε2 carriers for all available A monoacylglycerols, B cholesteryl esters, and C triacylglycerols, sorted by number of double bonds and carbon atoms among Health ABC Black men
Discussion
APOE ε2 carriers were lower on cholesteryl esters, monoacylglycerols, and sphingosine and were higher on triacylglycerols that consisted mostly of polyunsaturated fatty acids than remaining participants. Carnitine was the only metabolite that differed significantly between ε4 carriers and ε3 homozygotes, which was lower among ε4 carriers. We found consistent mean differences in the metabolite factor scores and individual metabolites by APOE when excluding the small percentage of participants taking lipid-lowering medications.
Differences in metabolites by APOE were previously examined among Australian older adults who were either ε2 carriers or ε4 carriers and were matched on age, gender, and lipid-lowering medication to those who were ε3 homozygotes [32]. Similarly, differences in metabolites by APOE were mainly due to differences in ε2 carriers. Specifically, the Australian ε2 carriers versus ε3 homozygotes were higher on two lysophosphatidylcholines and four phosphatidylethanolamines [32]. Lack of overlap in metabolites that differed by APOE in the previous versus current study are likely due to differences in lipid profiling platforms and could partly be due to differences in demographics; our cohort consisted of U.S. Black men, whereas the previous report was an Australian population-based cohort of men and women, where Australia has a very small percentage of individuals of African ancestry [33]. There was also a higher proportion of participants taking lipid-lowering medication in the previous report than among the Health ABC Black men (55% versus 15%, respectively), and the previous report found no differences in total and LDL cholesterol by APOE. Future studies in older adult cohorts that use similar lipid profiling methods will be needed to replicate results.
Higher triglycerides have been reported among ε2 carriers versus ε3 homozygotes [6]. Using lipid profiling, we sought to identify triacylglycerols that potentially contribute to this known difference in overall triglycerides by APOE. Triacylglycerols contain three fatty acids, and in this report, triacylglycerols that consist mostly of polyunsaturated fatty acids were higher among ε2 carriers than ε3 homozygotes and ε4 carriers. A previous report examining fatty acid remodeling in cell membranes in response to different APOE isoforms [34] found that exposing human neuroblastoma cells to the APOE ε3 or ε4 isoforms resulted in decreased total polyunsaturated fatty acids and increased total saturated and monounsaturated fatty acids [34]. In addition, we previously found triacylglycerols consisting mostly of polyunsaturated fatty acids were higher among older adults with high versus low walking ability [29], and others have noted these metabolites associated with less insulin resistance [35], lower waist circumference [35], and lower type 2 diabetes risk [36]. Higher triacylglycerols consisting mostly of polyunsaturated fatty acids may indicate more optimal fats available to ε2 carriers and may be one mechanism contributing to lower morbidity and mortality among ε2 carriers, despite having higher overall triglycerides than ε3 homozygotes [11].
Triacylglycerols can be resynthesized or modified by the monoacylglycerol pathway [37]. Monoacylglycerols are classified based on fatty acid location; if the fatty acid is located on either end (i.e., 1- or 3-isomer), then it is an α-monoacylglycerol, whereas if the fatty acid is centered (i.e., 2-isomer), then it is a β-monoacylglycerol. The preferred substrates for the monoacylglycerol pathway are β-monoacylglycerols, which are the major end-product of dietary fat digestion in the small intestine and are directly converted to triacylglycerols by the monoacylglycerol pathway [37]. Monoacylglycerols are present in low levels and typically do not accumulate due to strong detergent properties [38]. Here, APOE ε2 carriers had lower levels of three of the four measured monoacylglycerols, whereas dietary fat intake did not differ. All monoacylglycerols measured in this report were α-monoacylglycerols, which can be formed by β-monoacylglycerols undergoing a random isomerize [39]. Though little else is known about α-monoacylglycerol, including what lower plasma α-monoacylglycerol among APOE e2 carriers may indicate and how lower levels relate to β-monoacylglycerols and triacylglycerols.
In addition to higher overall triglycerides, lower levels of total and LDL cholesterol have been reported among APOE ε2 carriers versus ε3 homozygotes [6]. Here, a cholesteryl ester factor score was significantly lower among ε2 carriers than ε4 carriers or ε3 homozygotes. Excess cholesterol is stored primarily as cholesteryl esters [40], which make up a major component of macrophage foam cells of atherosclerotic plaques [41]. For example, one study found total cholesteryl esters were 120 times higher in atherosclerotic plaques than in healthy radial arteries [42], with the majority of cholesteryl esters in plaques being C18:1 and C18:2 [42]. Here, both plasma C18:1 and C18:2 cholesteryl esters were significantly lower among ε2 carriers than ε4 carriers or ε3 homozygotes. Some reports have also found a protective association between the APOE ε2 allele and cardiovascular outcomes. For example, a meta-analysis of 121 studies with coronary outcomes and over 120,000 participants reported ε2 carriers had 20% lower odds of coronary disease than ε3 homozygotes [10]. Similarly, APOE ε2 carriers had a 35% lower risk of mortality due to cardiovascular disease than ε3 homozygotes among a large older adult cohort with a good representation of the ε2 allele and more than 18 years of follow-up [15]. Large prospective older adult cohorts with metabolomics and power to examine differences in incident cardiovascular disease risk by APOE will be able to further examine the protective effect of lower cholesteryl esters on cardiovascular outcomes among APOE ε2 carriers.
We found APOE ε4 carriers had lower levels of carnitine than ε3 homozygotes. Carnitine is a critical molecule in the mitochondrial metabolism of fatty acids. The most important function of carnitine is to transport long-chain fatty acids into the mitochondria [43] to be oxidized and used as an energy source for myocardium and skeletal muscle [38]. Carnitine can be synthesized in the body, but the majority comes from dietary animal-based sources, primarily from red meat and dairy products [38]. In a healthy individual, plasma carnitine and its acylcarnitine derivatives are tightly regulated within a fixed range, where altered levels, specifically lower carnitine, can be indicative of mitochondrial dysfunction [38, 43]. As a result, carnitine or acetyl-carnitine supplementation has been suggested as a potential therapy to combat aging-related decline in mitochondria [44]. Mitochondrial dysfunction is thought to be involved in the pathophysiology of several aging-related major health outcomes, including Alzheimer’s disease [45, 46] and cardiovascular disease [47]. Thus, mitochondrial dysfunction may partially explain the higher risk of morbidity and mortality associated with the APOE e4 allele and might be mitigated through carnitine supplementation. However, a potential undesirable effect of carnitine supplementation is an increase in trimethylamine oxide (TMAO) [48], a gut microbiota-dependent metabolite of carnitine, which is associated with cardiovascular disease [49]. In fact, supplementation with TMAO or TMAO-related metabolites (e.g., carnitine) promoted atherosclerosis in animal models [48, 49]. It remains to be determined whether a lower dose of carnitine can improve mitochondrial functioning, while reducing the risk of accelerated atherosclerosis formation.
Only one metabolite differed significantly between APOE ε4 carriers and ε3 homozygotes in this cohort of older Black men. This is likely due to our relatively small number of ε4 carriers, limiting power to detect smaller effect sizes. On the other hand, we identified several differences in metabolites between ε2 carriers versus remaining participants, despite an even smaller number of ε2 carriers than the number of ε4 carriers. This discrepancy is likely explained by the ε2 allele causing a 50 times lower affinity for apolipoprotein E to bind to LDL particles when compared to the ε3 and ε4 alleles [3]. This dramatic difference in lipoprotein affinity could result in much larger differences in circulating lipids among ε2 carriers, i.e., larger effect sizes that are easier to detect with smaller sample sizes. Multiple metabolites likely differ between APOE ε4 carriers and ε3 homozygotes that will need to be explored in a larger cohort.
In addition to the small number of ε4 carriers, we were limited by only having information on older Black men who were recruited to be non-disabled at baseline, it is unknown whether the identified metabolites that differed by APOE would generalize to all older adults, in particular to women, other racial/ethnic groups, or similarly aged older adults of poorer health than those meeting the eligibility criteria for this study. In addition, metabolite values were unitless peak areas, which limited the interpretability of the observed differences by APOE; for example, we were not able to assess whether the lower level of carnitine among ε4 carriers versus ε3 homozygotes was outside of a healthy range. Strengths of this work included the higher proportion of ε2 carriers when compared to U.S. frequencies, as well as a well-characterized cohort of older Black men with a large number of metabolites available from overnight-fasting plasma samples that had never been thawed previously.
The observed differences in lipids provide a better characterization of metabolic differences according to APOE in this cohort of older Black men. Lower levels of cholesteryl esters, monoacylglycerols, and sphingosine and higher levels of triacylglycerols that consist mostly of polyunsaturated fatty acids may be a protective profile of lipids among APOE ε2 carriers. Specifically, the higher triacylglycerols consisting mostly of polyunsaturated fatty acids among APOE ε2 carriers may indicate more optimal fats available in the periphery to be transported to tissues as an energy source and the lower cholesteryl esters potentially indicate less cholesterol available in the periphery to accumulate as atherosclerotic plaques. It remains to be determined whether this potentially protective profile of lipids contributes to the APOE ε2 allele-associated lower risks of Alzheimer’s disease, cardiovascular disease, and mortality. In addition, the observed lower circulating carnitine among APOE ε4 carriers versus ε3 homozygotes may potentially reflect lower mitochondrial functioning among ε4 carriers that might explain a portion of the higher risk of morbidity and mortality associated with the ε4 allele. A complete understanding of differences in lipid metabolism that occur as a result of APOE allelic variation could potentially indicate metabolic pathways involved in pathophysiology of multiple major health outcomes.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contribution
Health ABC study concept and design: ABN; metabolomics ancillary study: SCM; data analysis and manuscript writing: MMM; interpretation of data and manuscript editing and critical review: all authors.
Funding
This work was supported by National Institute on Aging Contracts N01-AG-6–2101, N01-AG-6–2103, and N01-AG-6–2106; National Institute on Aging Grant R01-AG028050, and National Institute of Nursing Research Grant R01-NR012459. This work was also supported in part by the Intramural Research Program of the National Institute on Aging. MMM is supported by the Cardiovascular Epidemiology training grant funded by the National Heart, Lung, and Blood Institute Grant T32-HL08382512 at the University of Pittsburgh.
Data availability
Data and material are available by submitting an analysis proposal at https://healthabc.nia.nih.gov/.
Code availability
Not applicable.
Declarations
Ethics approval
The Health ABC study was approved by the Institutional Review Board of each participating site.
Consent to participate
All Health ABC participants provided written informed consent.
Consent for publication
All authors and the Health ABC study approved this manuscript for publication.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Feingold KR, Grunfeld C. Introduction to lipids and lipoproteins. Endotext. 2018. https://www.ncbi.nlm.nih.gov/books/NBK305896/.
- 2.Mahley RW. Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders. J Mol Med (Berl) 2016;94:739–746. doi: 10.1007/s00109-016-1427-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10:333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Dhanasekaran P, Alexander ET, Rader DJ, Phillips MC, Lund-Katz S. Molecular mechanisms responsible for the differential effects of apoE3 and apoE4 on plasma lipoprotein–cholesterol levels. Arterioscler Thromb Vasc Biol. 2013;33:687–693. doi: 10.1161/ATVBAHA.112.301193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Martínez AB, Torres-Perez E, Devanney N, Del Moral R, Johnson LA, Arbones-Mainar JM. Beyond the CNS: the many peripheral roles of APOE. Neurobiol Dis. 2020;138:104809. doi: 10.1016/j.nbd.2020.104809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasmussen KL. Plasma levels of apolipoprotein E, APOE genotype and risk of dementia and ischemic heart disease: a review. Atherosclerosis. 2016;255:145–155. doi: 10.1016/j.atherosclerosis.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 7.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 8.Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, et al. Association of apolipoprotein E allele ϵ4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43:1467–1467. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 9.Song Y, Stampfer MJ, Liu S. Meta-analysis: apolipoprotein E genotypes and risk for coronary heart disease. Ann Intern Med. 2004;141:137–147. doi: 10.7326/0003-4819-141-2-200407200-00013. [DOI] [PubMed] [Google Scholar]
- 10.Bennet AM, Di Angelantonio E, Ye Z, Wensley F, Dahlin A, Ahlbom A, et al. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA. 2007;298:1300–1311. doi: 10.1001/jama.298.11.1300. [DOI] [PubMed] [Google Scholar]
- 11.Wolters FJ, Yang Q, Biggs ML, Jakobsdottir J, Li S, Evans DS, et al. The impact of APOE genotype on survival: results of 38,537 participants from six population-based cohorts (E2-CHARGE) PLoS ONE. 2019;14:e0219668. doi: 10.1371/journal.pone.0219668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, Jr, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7:180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 13.Raber J, Huang Y, Ashford JW. ApoE genotype accounts for the vast majority of AD risk and AD pathology. Neurobiol Aging. 2004;25:641–650. doi: 10.1016/j.neurobiolaging.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 14.Alzheimer’s Association. 2020 Alzheimer’s disease facts and figures. Special Report On the Front Lines: Primary Care Physicians and Alzheimer’s Care in America. 2020.
- 15.Rajan KB, Barnes LL, Wilson RS, McAninch EA, Weuve J, Sighoko D, et al. Racial differences in the association between apolipoprotein E risk alleles and overall and total cardiovascular mortality over 18 years. J Am Geriatr Soc. 2017;65:2425–2430. doi: 10.1111/jgs.15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitzpatrick AL, Kuller LH, Ives DG, Lopez OL, Jagust W, Breitner JC, et al. Incidence and prevalence of dementia in the Cardiovascular Health Study. J Am Geriatr Soc. 2004;52:195–204. doi: 10.1111/j.1532-5415.2004.52058.x. [DOI] [PubMed] [Google Scholar]
- 17.Evans DA, Bennett DA, Wilson RS, Bienias JL, Morris MC, Scherr PA, et al. Incidence of Alzheimer disease in a biracial urban community: relation to apolipoprotein E allele status. Arch Neurol. 2003;60:185–189. doi: 10.1001/archneur.60.2.185. [DOI] [PubMed] [Google Scholar]
- 18.Tang M, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, et al. The APOE-∊ 4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. 1998;279:751–755. doi: 10.1001/jama.279.10.751. [DOI] [PubMed] [Google Scholar]
- 19.Goodpaster BH, Carlson CL, Visser M, Kelley DE, Scherzinger A, Harris TB, et al. Attenuation of skeletal muscle and strength in the elderly: the Health ABC study. J Appl Physiol. 1985;2001(90):2157–2165. doi: 10.1152/jappl.2001.90.6.2157. [DOI] [PubMed] [Google Scholar]
- 20.Murphy RA, Moore SC, Playdon M, Meirelles O, Newman AB, Milijkovic I, et al. Metabolites associated with lean mass and adiposity in older black men. J Gerontol A Biol Sci Med Sci. 2017;72:1352–1359. doi: 10.1093/gerona/glw245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh J, Minster RL, Schupf N, Kraja A, Liu Y, Christensen K, et al. Genomewide association scan of a mortality associated endophenotype for a long and healthy life in the Long Life Family Study. J Gerontol A Biol Sci Med Sci. 2017;72:1411–1416. doi: 10.1093/gerona/glx011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marron MM, Harris TB, Boudreau RM, Clish CB, Moore SC, Murphy RA, et al. Metabolites associated with vigor to frailty among community-dwelling older black men. Metabolites. 2019;9:E83. doi: 10.3390/metabo9050083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Townsend MK, Clish CB, Kraft P, Wu C, Souza AL, Deik AA, et al. Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clin Chem. 2013;59:1657–1667. doi: 10.1373/clinchem.2012.199133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paynter NP, Balasubramanian R, Giulianini F, Wang DD, Tinker LF, Gopal S, et al. Metabolic predictors of incident coronary heart disease in women. Circulation. 2018;137:841–853. doi: 10.1161/CIRCULATIONAHA.117.029468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei R, Wang J, Su M, Jia E, Chen S, Chen T, et al. Missing value imputation approach for mass spectrometry-based metabolomics data. Sci Rep. 2018;8:663. doi: 10.1038/s41598-017-19120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houston DK, Nicklas BJ, Ding J, Harris TB, Tylavsky FA, Newman AB, et al. Dietary protein intake is associated with lean mass change in older, community-dwelling adults: the Health, Aging, and Body Composition (Health ABC) study. Am J Clin Nutr. 2008;87:150–155. doi: 10.1093/ajcn/87.1.150. [DOI] [PubMed] [Google Scholar]
- 28.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. doi: 10.1093/clinchem/18.6.499. [DOI] [PubMed] [Google Scholar]
- 29.Marron MM, Wendell SG, Boudreau RM, Clish CB, Santanasto AJ, Tseng GC, et al. Metabolites associated with walking ability among the oldest old from the CHS All Stars study. J Gerontol A Biol Sci Med Sci. 2020 doi: 10.1093/gerona/glaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc. 1995:289–300.
- 31.Diniz BS, Sibille E, Ding Y, Tseng G, Aizenstein HJ, Lotrich F, et al. Plasma biosignature and brain pathology related to persistent cognitive impairment in late-life depression. Mol Psychiatry. 2015;20:594–601. doi: 10.1038/mp.2014.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong MWK, Braidy N, Crawford J, Pickford R, Song F, Mather KA, et al. APOE genotype differentially modulates plasma lipids in healthy older individuals, with relevance to brain health. J Alzheimers Dis. 2019;72:703–716. doi: 10.3233/JAD-190524. [DOI] [PubMed] [Google Scholar]
- 33.Pietsch J. Race, ethnicity, and the participation gap: understanding Australia’s political complexion. University of Toronto Press; 2018.
- 34.Prasinou P, Dafnis I, Giacometti G, Ferreri C, Chroni A, Chatgilialoglu C. Fatty acid-based lipidomics and membrane remodeling induced by apoE3 and apoE4 in human neuroblastoma cells. Biochim Biophys Acta Biomembr. 2017;1859(10):1967–1973. doi: 10.1016/j.bbamem.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Kotronen A, Velagapudi VR, Yetukuri L, Westerbacka J, Bergholm R, Ekroos K, et al. Serum saturated fatty acids containing triacylglycerols are better markers of insulin resistance than total serum triacylglycerol concentrations. Diabetologia. 2009;52:684–690. doi: 10.1007/s00125-009-1282-2. [DOI] [PubMed] [Google Scholar]
- 36.Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121:1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gurr MI, Harwood JL, Frayn KN, Murphy DJ, Michell RH. Lipids: biochemistry, biotechnology and health. John Wiley & Sons; 2016.
- 38.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christie WW. Monoacylglycerols. The LipidWeb. 2019. https://www.lipidhome.co.uk/lipids/simple/mg/index.htm. Accessed 6 May 2020
- 40.Hinterwirth H, Stegemann C, Mayr M. Lipidomics: quest for molecular lipid biomarkers in cardiovascular disease. Circ Cardiovasc Genet. 2014;7:941–954. doi: 10.1161/CIRCGENETICS.114.000550. [DOI] [PubMed] [Google Scholar]
- 41.Ghosh S, Zhao B, Bie J, Song J. Macrophage cholesteryl ester mobilization and atherosclerosis. Vascul Pharmacol. 2010;52:1–10. doi: 10.1016/j.vph.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stegemann C, Drozdov I, Shalhoub J, Humphries J, Ladroue C, Didangelos A, et al. Comparative lipidomics profiling of human atherosclerotic plaques. Circ Cardiovasc Genet. 2011;4:232–242. doi: 10.1161/CIRCGENETICS.110.959098. [DOI] [PubMed] [Google Scholar]
- 43.Reuter SE, Evans AM. Carnitine and acylcarnitines: pharmacokinetic, pharmacological and clinical aspects. Clin Pharmacokinet. 2012;51:553–572. doi: 10.2165/11633940-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 44.Sharma S, Black SM. Carnitine homeostasis, mitochondrial function and cardiovascular disease. Drug Discov Today Dis Mech. 2009;6:e31–e39. doi: 10.1016/j.ddmec.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 46.Ising C, Venegas C, Zhang S, Scheiblich H, Schmidt SV, Vieira-Saecker A, et al. NLRP3 inflammasome activation drives tau pathology. Nature. 2019;575:669–673. doi: 10.1038/s41586-019-1769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siasos G, Tsigkou V, Kosmopoulos M, Theodosiadis D, Simantiris S, Tagkou NM, et al. Mitochondria and cardiovascular diseases—from pathophysiology to treatment. Ann Transl Med. 2018;6(12):256. doi: 10.21037/atm.2018.06.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallance H, Koochin A, Branov J, Rosen-Heath A, Bosdet T, Wang Z, et al. Marked elevation in plasma trimethylamine-N-oxide (TMAO) in patients with mitochondrial disorders treated with oral l-carnitine. Mol Genet Metab Rep. 2018;15:130–133. doi: 10.1016/j.ymgmr.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng S, Shah SH, Corwin EJ, Fiehn O, Fitzgerald RL, Gerszten RE, et al. Potential impact and study considerations of metabolomics in cardiovascular health and disease: a scientific statement from the American Heart Association. Circ Cardiovasc Genet. 2017;10:e000032. doi: 10.1161/HCG.0000000000000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and material are available by submitting an analysis proposal at https://healthabc.nia.nih.gov/.
Not applicable.