Abstract
Even though substantial progress has been made in the treatment of hepatitis C virus (HCV) infection, viral resistance and relapse still occur in some patients and additional therapeutic approaches may ultimately be needed should viral resistance become more prevalent. Microtubules play important roles in several HCV life cycle events, including cell attachment, entry, cellular transportation, morphogenesis and progeny secretion steps. Therefore, it was hypothesized that microtubular inhibition might be a novel approach for the treatment of HCV infection. Here, the inhibitory effects of our recently developed microtubule inhibitors were studied in the HCV replicon luciferase reporter system and the infectious system. In addition, the combination responses of microtubule inhibitors with daclatasvir, which is a clinically used HCV NS5A inhibitor, were also evaluated. Our results indicated that microtubule targeting had activity against HCV replication and showed synergistic effect with a current clinical drug.
Keywords: Hepatitis C virus, Viral resistance, Microtubule inhibitor, Combinational therapy
Highlights
-
•
Microtubule inhibition affects HCV replication.
-
•
Compound 9f displays time and concentration dependent inhibitory activities against HCV production.
-
•
Combination of compound 9f with Daclatasvir shows modest synergistic effects against HCV replication.
1. Introduction
Since the identification of hepatitis C virus (HCV) in 1989 [1], its contribution to chronic liver diseases including cirrhosis and hepatocellular carcinoma has become increasingly apparent [2,3]. Therapeutic treatment of HCV infection have progressed from interferon alpha to well tolerated oral direct acting antivirals (DAAs) [4,5] and significantly increased our ability to induce sustained virologic response (SVR, defined as undetectable HCV RNA after cessation of therapy) [6]. Currently, agents directed at the viral protease, NS5A protein, and NS5B RNA-dependent RNA polymerase are in widespread clinical use [[7], [8], [9], [10], [11]]. Daclatasvir plus sofosbuvir based regimens are highly effective in patients with chronic HCV genotype 1, 3 or 4 infection, and when patients have advanced liver disease, post-transplant recurrence and HIV-1 co-infection [12].
Despite the advances, there are still up to 5% of patients do not achieve SVR with contemporary combinations of DAAs [13,14]. Previously existing resistance mutations may contribute to these failures [15,16]. Host directed (antiviral) therapy (HDT) targeting cellular functions have been suggested as an alternative approach with advantages of broader spectrum antiviral activities and a lower likelihood of emergence of viral mutants with reduced drug susceptibility [17,18]. Microtubules, as a key compartment of cellular cytoskeleton [[19], [20], [21]], also serve in the crucial cargo transport mechanisms for a number of viruses [[22], [23], [24], [25]]. Microtubule aggregation has been observed in HCV infected hepatocytes [[26], [27], [28], [29]] and Kunjin virus infected cells [30,31]. However, there are only handfuls of studies investigating the roles of microtubule targeting agents in virus life cycle [30,[32], [33], [34]]. Colchicine, nocodazole and vinblastine sulfate were reported to decrease HCV replication levels in a dose dependent manner [25,32]. Our laboratories have recently designed and synthesized a series of small molecules that inhibit microtubule polymerization [35]. Here, we examined their effects on interrupting HCV life cycle in both subgenomic replicon cells and infectious particles generating Huh7.5.1 cells and investigated their synergistic activities in combination with daclatasvir. Because of the small amount of HDT antiviral drugs undergoing clinical trials so far, the combination of microtubule inhibitors with DAAs should be prioritized for further investigation and could become an effective therapeutic intervention strategy for HCV infection.
2. Material and methods
2.1. Cells and cell culture
The BM4-5 Feo subgenomic replicon cells, stably transfected with a genotype 1b replicon (BM45-Feo) plasmid with a neomycin-luciferase fusion as described elsewhere [5,36], are kind gifts of Dr. Aleem Siddiqui (University of California, San Diego). Cells were maintained in DMEM high glucose medium supplemented with 10% fetal bovine serum, 100 mg/ml streptomycin, 100 units/ml penicillin and 500 μg/ml Geneticin. The Huh7.5.1 cells were grown in DMEM high glucose medium supplemented with 10% fetal bovine serum, 100 mg/ml streptomycin and 100 units/ml penicillin.
2.2. Virus
The cell culture generated JFH-1 D183 HCV infectious particles (HCVcc) were kind gift from Dr. Aleem Siddiqui in University of California, San Diego. They were harvested from RNA transfection as described [37]. New virus supernatant was collected and aliquot after infection of Huh7.5.1 cells with JFH-1 D183 virus for 8 days [38]. The infectivity of newly generated supernatant was determined by focus forming unit assays. Briefly, supernatant containing HCV particles were 10-fold series diluted by DMEM culture medium and used to infect Huh7.5.1 cells in 96 well plates. After 72 h, cells were fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.1% Triton X-100. Intracellular virus was detected by immunofluorescence using a sheep anti-NS5A IgG [28] as primary antibody and anti-sheep IgG, Alexa Fluor 594 conjugated, as secondary antibody. NS5A positive foci were counted according to previous description [39]. Infectivity titer was calculated and expressed as focus-forming units per milliliter of supernatant (FFU/mL).
2.3. Compounds tested
Ledipasvir and daclatasvir were purchased from Fisher Scientific. Compounds 27a, 27b, 9f, 19a, 31b and 31d were synthesized as described before [35]. In our previously published results of the in vitro tumor proliferation assay, compounds 27a and 27b were most potent while compounds 19a and 9f were moderate and compounds 31b and 31d least potent. These six compounds were thus chosen for this anti-HCV study as they represent a wide spectrum of potency in the tumor cell proliferation assay.
2.4. Antiviral assay against HCV replicon
Following trypsinization, 2 104 replicon cells/well were seeded into 96-well plates and incubated overnight. Compounds were added to selected wells to achieve various concentrations in the same cell culture medium. Plates were maintained in the cell culture incubator for 24 h or 48 h. Luciferase activity was measured by the Bright-Glo™ Luciferase Assay System (E2610, Promega) and luminescence signals were recorded by microplate luminometer (VERITAS™ microplate luminometer, Turner Biosystems), as previously described [40].
2.5. Antiviral assay against HCVcc particles
Huh7.5.1 cells were trypsinized, 1.5 105 cells/well were seeded into 12-well plate and incubated overnight. Cells were infected with JFH-1 D183 virus at MOI 0.01. After infection at 37 °C for 3 h, cells were washed twice with PBS. Various concentrations of compounds diluted by the same culture media were added. After incubation for 24 h or 48 h, intracellular HCV RNA levels were determined by real-time quantitative PCR.
2.6. Cell viability assays
Compound toxicities were detected by CellTiter-Blue Cell Viability Assay (G8081, Promega). After 24 h or 48 h treatment periods of different concentrations of compounds, 20 μl/well of CellTiter-Blue Regent was added and mixed well. The cell plates were incubated at 37 °C for 1–4 h. After shaking plates for 10 s, fluorescence at 560/590 nm were recorded.
2.7. Analyses of drug synergism
Various concentrations of daclatasvir and compound 9f were added to BM4-5 Feo cells. The combination effects of daclatasivir with compounds 9f were evaluated by two softwares. One is Combenefit software (Cancer Research UK Cambridge Institute; Cambridge, UK) [41] using the classical Loewe's synergy reference model [[42], [43], [44]], another is CumpuSyn software which calculates the combination index based on median-effect principle of the mass-action law [45,46].
2.8. Real-time quantitative PCR assay (RT-qPCR)
Intracellular total RNA was extracted following instructions of RNeasy mini kit (74104, Qiangen). Reverse transcriptions were conducted using Superscript™ VILO Matster Mix (11755050, Thermo Fisher). HCV RNA was quantified by realtime quantative PCR using Ssoadvanced universal SYBR Green Supermix (Bio-rad). The JFH-1 primers are, forward 5’-TCTGCGGAACCGGTGAGTA-3’, reverse 5’-TCAGGCAGTACCACAAGGC-3’. Housekeeping gene GAPDH were used as control for RNA base levels, primers are, forward 5’- GTCTCCTCTGACTTCAACAGCG-3’, reverse 5’ –ACCACCCTGTTGCTGTAGCCAA-3’.
2.9. Immunofluorescence assay
Cells seeded on coverslips were washed with PBS twice and fixed with 4% PFA for 15 min at room temperature. Then cells were permeabilized with 0.1% Triton X-100 for 10 min and blocked with 5% BSA in PBS for 1 h at room temperature. Primary antibodies diluted by same blocking buffer were added and incubated at 4 °C overnight. After three times wash with PBS, secondary antibodies diluted by the same buffer were added and incubated for 1 h at room temperature. After three times wash, coverslips were mounted by mounting medium which contains DAPI. Immunofluorescence photos were captured by Leica SP8 Confocal with Lightning Deconvolution microscope.
3. Results
3.1. Microtubule inhibitors block HCV replication in BM45-Feo system
Colchicine and compounds 27a, 27b, 19a, 9f, 31b and 31d were tested on BM45-Feo replicon cells which allow for ongoing and persistent HCV subgenomic RNA synthesis. Daclatasvir and ledipasvir were used as positive controls (compounds structures were shown in Fig. 1A). After cells were treated with various concentrations of different compounds for 24 h or 48 h respectively, HCV replication level was quantified by luciferase activity. Meanwhile, the compound's cytotoxicity was determined by cell-titer blue assays to exclude the possibilities of HCV production reduction caused by cell death. As shown in Table 1, daclatasvir and ledipasvir displayed picomolar IC50 values in both time treatments, especially low IC50 values after 48 h of incubation. No obvious toxicity was observed during both time periods. Our 6 compounds (27a, 27b, 19a, 9f, 31b and 31d) and colchicine each have a nanomolar IC50 value for decreasing HCV replication in the replicon system at both time points, but concentrations at which toxicity is observed (TC50) are substantially higher. All of them barely showed toxicities at concentrations below 10 μM at 24 h, although toxicities increased at 48 h. Correspondingly, over this time interval, therapeutic indices (defined as the TC50/IC50 ratio) at 48 h decreased greatly because of cumulative cellular toxicities induced by microtubular inhibition. By 48 h, therapeutic indices of colchicine, compounds 9f and 19a remained above 100 while therapeutic indices of other compounds were substantially lower.
Fig. 1.
(A) Compounds structure. Compounds 27a, 27b, 9f, 19a, 31b and 31d are our recently developed microtubule inhibitors which target the colchicine binding site. Daclatasvir and ledipasvir are clinically used drugs which target HCV NS5A. (B) Immunofluorescence images of FFU assays to determine infectivity titer. Huh7.5.1 cells were infected by various dilutions of cell culture medium (a, mock infected), JFH-1 D183 virus stock (b) and newly produced viral supernatant (c) separately. After 72 h infection, Huh7.5.1 cells were fixed and stained for NS5A (red) and nuclei (blue). Clusters of 2–50 NS5A positive cells were considered as foci. (C) Quantitative real time PCR (qRT-PCR) results (left panel) and toxicity assay results (right panel) of daclatasvir and compound 9f at 24 h and 48 h. HCV RNA was relatively quantified by normalization to untreated control. GAPDH was used as housekeeping gene to guarantee the same base level. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Inhibitory activities and toxicities of microtubule inhibitors, daclatasvir and ledipasvir in BM45-Feo replicon cells.
| Compound | 24 h (nM, mean SD) |
48 h (nM, mean SD) |
||
|---|---|---|---|---|
| IC50a | TC50b | IC50a | TC50b | |
| Colchicine | 57.49 18.56 | >10000 | 4.14 1.29 | 78.19 45.58 |
| 27a | 15.51 12.01 | >10000 | 33.88 13.45 | 128.04 94.46 |
| 27b | 68.90 42.42 | >10000 | 33.98 11.90 | 119.93 46.70 |
| 9f | 20.1 13.81 | >10000 | 13.37 2.86 | 4804.67 1175.25 |
| 19a | 130.8 6.56 | >10000 | 57.41 36.76 | 4868.25 1885.84 |
| 31b | 112.46 97.93 | >10000 | 389.97 86.79 | 2794.33 1066.43 |
| 31d | 754.43 428.06 | >10000 | 294.87 9.31 | 6233.33 2091.71 |
| Daclatasvir | 0.14 0.16 | >10000 | 0.038 0.0087 | >10000 |
| Ledipasvir | 0.08 0.03 | >10000 | 0.0097 0.0021 | >10000 |
IC50: The concentration of a compound required for 50% inhibition of HCV replication.
TC50: The concentration of a compound required to cause 50% cell death.
3.2. Compound 9f inhibits HCV replication in HCVcc system
We next examined whether compound 9f had inhibitory effect on HCVcc replication. JFH-1 D183 virus was obtained by infecting Huh7.5.1 cells with the virus stock which was a gift of Dr. Sddiqui. As shown in Fig. 1B, the newly produced virus has higher efficiency of infection. After infection with JFH-1 D183 virus, Huh7.5.1 cells were incubated with various concentrations of compound 9f and daclatasvir. Intracellular virus RNA was analyzed by realtime quantitative PCR. As displayed in Fig. 1C, compound 9f exhibited better inhibitory effects after 48 h incubation time than 24 h, with IC50s at 26 nM and 1318 nM respectively, which suggested the long-lasting role of microtubule in the whole HCV life cycle. The toxicities of compound 9f were higher at 48 h than 24 h, with TC50s at 677 nM and 13053 nM. Nevertheless, compound 9f inhibited HCV replication at concentrations of no cytotoxicity, which means that the reduction of HCV RNA was not attributed to cell death. These results therefore indicated the potential value of microtubule inhibitors as anti-HCV agents.
3.3. Combination studies of daclatasvir with compound 9f
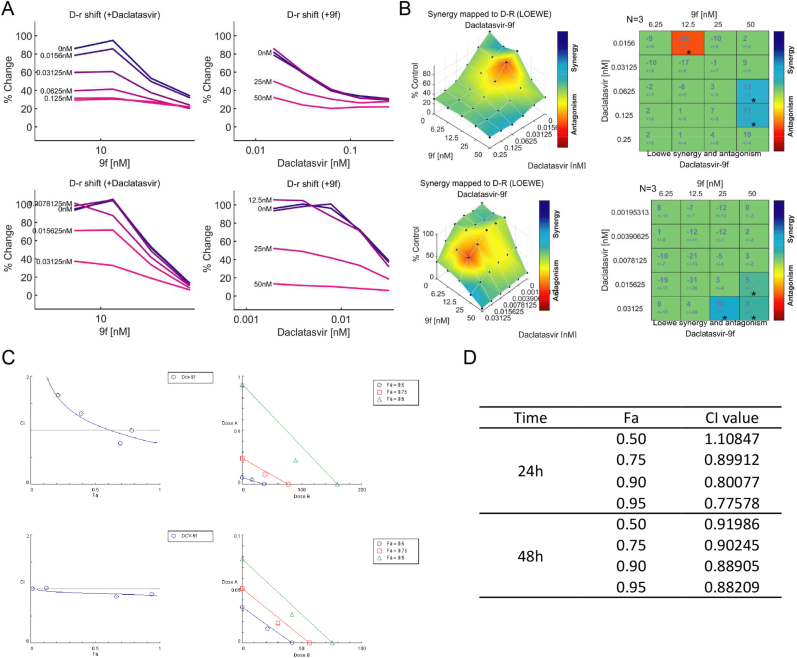
Having demonstrated the inhibitory effects of compound 9f, we investigated if the combination of daclatasvir and compound 9f would exhibit stronger activities because of their different affected targets. We performed the combination studies in the BM4-5 Feo replicon cells at treatment times of 24 h and 48 h, respectively. A range of concentrations of each compound, above and below their IC50 values, were added to the cells and the combined effects were examined by luciferase activities. The combination data for each experiment was analyzed by two softwares. One is Combenefit software which uses Loewe drug synergy model [41]. As shown in Fig. 2A, the addition of compound 9f or daclatasvir both shifted the inhibitory curve of the single agent. Compound 9f displayed larger fold change of the curve of daclatasvir at 48 h. Fig. 2B plotted synergy mapped surface of the combination groups. The main part indicates their additive or synergistic effects. Some part indicated antagonistic effect, however it could be false negative because of the low inhibition effects at low concentrations. Fig. 2B also showed calculated synergy score table, which also demonstrated that daclatasvir could additively or synergistically interact with compound 9f. Another software we used was Compusyn. It is based on median effect equation which was derived from mass action law principle, according to Ting-chao Chou's theory [45]. Fig. 2C left panel presents the Fa-CI plots which quantificationally defined synergism (CI < 1), additivity (CI = 1) and antagonism (CI > 1). The combination group had lower CI values at 48 h than 24 h. Fig. 2C right panel are isobolograms, where if the middle combination data point falls below the line it represents synergism, on the line represents additivity and above the line represents antagonism. Fig. 2D shows the fitted CI values at different Fa levels of the combination group. Taken together, these results consistently suggested that combination of daclatasvir with compound 9f had stronger synergistic effects at 48 h than 24 h.
Fig. 2.
(A)(B) Combenefit software plotted the combinational responses of daclatasvir with compound 9f using Loewe model. Upper panel, 24 h. Lower panel, 48h. (A) D-r shift map when added with a range of concentrations of different compounds. (B) Synergy surface map (left panel) and Matrix score table (right panel), N means the number of replicates, the number under the score is standard deviation, stars indicate the level of significance (*P < 0.05, **P < 0.001, ***P < 0.0001). (C)(D) Compusyn software plotted the combinational responses of daclatasvir with compound 9f using median effect equation. Upper panel, 24 h. Lower panel, 48 h. (C) Fa-CI plot (left panel), synergism (CI < 1), additivity (CI = 1), antagonism (CI > 1). Classic isobologram (right panel), synergism (middle combination data point below the line), additivity (middle combination data point on the line), antagonism (middle combination data point above the line). (D) CI data for compounds combination group at different Fa levels.
3.4. Immunofluorescence studies of daclatasvir and compound 9f
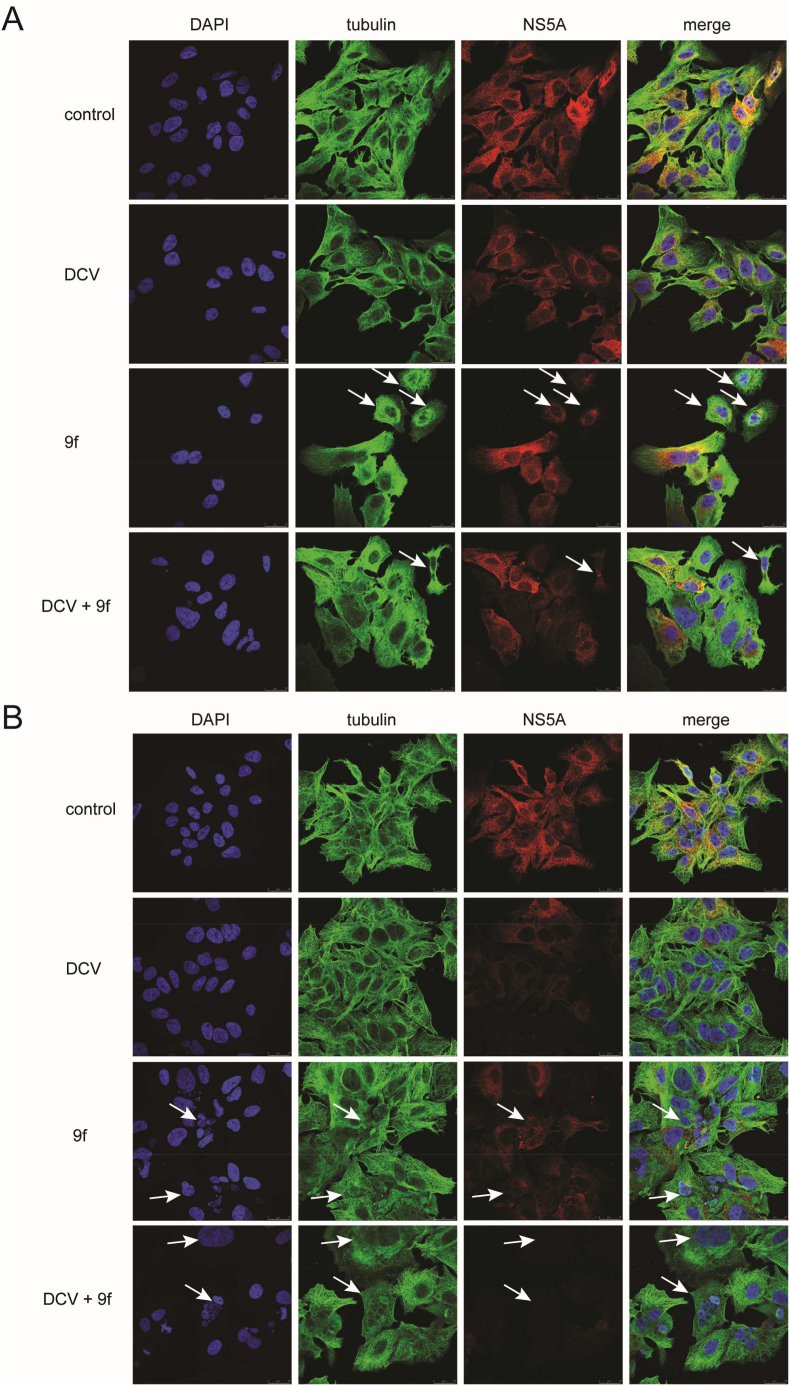
To examine whether the mechanism of action of compound 9f is related to its effect on microtubules, we performed the immunofluorescence experiment on BM4-5 Feo cells at 24 h and 48 h. As demonstrated in Fig. 3, the white arrows show that compound 9f induced abnormal distributions of microtubules. Accordingly, these cells exhibited reduced signals of HCV NS5A. Interestingly, when cells were treated with compound 9f combined with daclatasvir, more cells showed abnormal microtubule and these cells displayed stronger attenuated activities of HCV NS5A. The results in 48 h (Fig. 3B) were more distinguishable. Compound 9f caused increased abnormal microtubules in more cells, thus they showed weaker signals of HCV NS5A. Remarkably, when cells were treated with compound 9f combined with daclatasvir for 48 h, their signals of HCV NS5A became almost undetectable. All together, these data suggested that the inhibitory mechanism of compound 9f on HCV correlated with its effect on microtubules, and that daclatasvir could enhance the interruption on microtubules and eventually work synergistically with compound 9f.
Fig. 3.
Immunofluorescence images of BM4-5 Feo cells after incubation with daclatsvir (125 pM) and compound 9f (25 nM) and the combination group respectively after (A)24 h (B) 48h. Cells were fixed and immunostained for nuclei (blue), microtubule (green) and HCV NS5A (red). Photos were captured by confocal SP8 microscope (630 magnification). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Collectively, our findings suggested that compound 9f, as a microtubule inhibitor, could be a promising agent which deserves further attention and exploration for the development of anti-HCV therapies. This suggests that other clinically used microtubule inhibitors could also be investigated for their potential antiviral activities, which may be more readily applicable in the clinic if they were effective because of the advantage of repurposed drugs. Additionally, the combination of daclatasvir with compound 9f presents a synergistic approach to decrease HCV production. Further characterization of the mechanism at the molecular level would help better understand how these compounds work synergistically. It is also worth examining the effect of this combination strategy on drug resistant HCV strains. As microtubules play a role in many viruses, including HIV and many flaviviruses such as Dengue virus, Zika virus, Yellow fever virus, and West-Nile virus, the combinations of microtubule inhibitors with clinically used drugs may offer an additional strategy for blocking infection and managing resistance to current DAA regimens.
Author contributions
Conceptualization, H.Z. and J.A.; methodology, H.Z., X.Z., L.H., X.F., M.K. and Y.X.; software, H.Z.; validation, H.Z.; formal analysis, H.Z.; investigation, H.Z.; resources, X.F. and H.Z.; data curation, H.Z.; writing – original draft preparation, H.Z.; writing – review and editing, Z.H., R.S. and J.A.; visualization, H.Z.; supervision, Z.H., R.S. and J.A.; project administration, Z.H.. All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgements
We thank Dr. Aleem Siddiqui (University of California, San Diego) who provided BM4-5 Feo subgenomic replicon cells.
Contributor Information
Jing An, Email: jan@health.ucsd.edu.
Robert T. Schooley, Email: rschooley@health.ucsd.edu.
Ziwei Huang, Email: zhuang@health.ucsd.edu.
Data availability
Data will be made available on request.
References
- 1.Choo Q.-L., Kuo G., Weiner A.J., Overby L.R., Bradley D.W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Taherkhani R., Farshadpour F. Global elimination of hepatitis C virus infection: progresses and the remaining challenges. World J. Hepatol. 2017;9:1239–1252. doi: 10.4254/wjh.v9.i33.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvaruso V., Petta S., Craxi A. Is global elimination of HCV realistic? Liver Int. 2018;38(Suppl 1):40–46. doi: 10.1111/liv.13668. [DOI] [PubMed] [Google Scholar]
- 4.Lahser F.C., Bystol K., Curry S., McMonagle P., Xia E., Ingravallo P., Chase R., Liu R., Black T., Hazuda D., Howe A.Y., Asante-Appiah E. The combination of Grazoprevir, a hepatitis C virus (HCV) NS3/4A protease inhibitor, and Elbasvir, an HCV NS5A inhibitor, demonstrates a high genetic barrier to resistance in HCV genotype 1a replicons. Antimicrob. Agents Chemother. 2016;60:2954–2964. doi: 10.1128/AAC.00051-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyles D.L., Kaihara K.A., Schooley R.T. Synergy of a hepatitis C virus (HCV) NS4A antagonist in combination with HCV protease and polymerase inhibitors. Antimicrob. Agents Chemother. 2008;52:1862–1864. doi: 10.1128/AAC.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lobato C.M.O., Codes L., Silva G.F., Souza A.F.M., Coelho H.S.M., Pedroso M.L.A., Parise E.R., Lima L., Borba L.A., Evangelista A.S., Rezende R.E.F., Cheinquer H., Kuniyoshi A.S.O., Aires R.S., Quintela E.H.D., Mendes L.S.C., Nascimento F.C.V., Medeiros Filho J.E.M., Ferraz M., Abdala E., Bittencourt P.L., t H.C.V. Members of the Brazilian Real-Life Study about, H.C.V.t. Members of the Brazilian Real-Life Study about, Direct antiviral therapy for treatment of hepatitis C: a real-world study from Brazil. Ann. Hepatol. 2019;18:849–854. doi: 10.1016/j.aohep.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Bacon B.R., Gordon S.C., Lawitz E., Marcellin P., Vierling J.M., Zeuzem S., Poordad F., Goodman Z.D., Sings H.L., Boparai N., Burroughs M., Brass C.A., Albrecht J.K., Esteban R., Investigators H.R.- Boceprevir for previously treated chronic HCV genotype 1 infection. N. Engl. J. Med. 2011;364:1207–1217. doi: 10.1056/NEJMoa1009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherman K.E., Flamm S.L., Afdhal N.H., Nelson D.R., Sulkowski M.S., Everson G.T., Fried M.W., Adler M., Reesink H.W., Martin M., Sankoh A.J., Adda N., Kauffman R.S., George S., Wright C.I., Poordad F., I.S. Team Response-guided telaprevir combination treatment for hepatitis C virus infection. N. Engl. J. Med. 2011;365:1014–1024. doi: 10.1056/NEJMoa1014463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeuzem S., Andreone P., Pol S., Lawitz E., Diago M., Roberts S., Focaccia R., Younossi Z., Foster G.R., Horban A., Ferenci P., Nevens F., al e. Telaprevir for retreatment of HCV infection. N. Engl. J. Med. 2011;364:2417–2428. doi: 10.1056/NEJMoa1013086. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi N., Okanoue T., Tsubouchi H., Toyota J., Chayama K., Kumada H. Efficacy and safety of telaprevir, a new protease inhibitor, for difficult-to-treat patients with genotype 1 chronic hepatitis C. J. Viral Hepat. 2012;19:e134–142. doi: 10.1111/j.1365-2893.2011.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartenschlager R., Lohmann V., Penin F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat. Rev. Microbiol. 2013;11:482–496. doi: 10.1038/nrmicro3046. [DOI] [PubMed] [Google Scholar]
- 12.Keating G.M. Daclatasvir: A Review in Chronic Hepatitis C, Drugs. 2016;76:1381–1391. doi: 10.1007/s40265-016-0632-x. [DOI] [PubMed] [Google Scholar]
- 13.Kim B.K., Ahn S.H. The remaining challenges of HCV treatment in the direct-acting antivirals era. J. Gastroenterol. Hepatol. 2019;34:1891–1892. doi: 10.1111/jgh.14916. [DOI] [PubMed] [Google Scholar]
- 14.Hayes C.N., Imamura M., Tanaka J.A.-O., Chayama K.A.-O. Liver International; 2021. Road to Elimination of HCV: Clinical Challenges in HCV Management. LID - 10.1111/liv; p. 15150. [doi] [DOI] [PubMed] [Google Scholar]
- 15.Saito Y., Imamura M., Uchida T., Osawa M., Teraoka Y., Fujino H., Nakahara T., Ono A., Murakami E., Kawaoka T., Miki D., Tsuge M., Serikawa M., Aikata H., Abe-Chayama H., Hayes C.N., Chayama K. Ribavirin induces hepatitis C virus genome mutations in chronic hepatitis patients who failed to respond to prior daclatasvir plus asunaprevir therapy. J. Med. Virol. 2020;92:210–218. doi: 10.1002/jmv.25602. [DOI] [PubMed] [Google Scholar]
- 16.Aguiar B.F., Campos G.R.F., Rodrigues J.P.V., Marques N.N., Molina B.F., Bittar C., Souza F.F., Martinelli A.L.C., Rahal P., Pereira L.R.L. Baseline resistance associated substitutions in HCV genotype 1 infected cohort treated with Simeprevir, Daclatasvir and Sofosbuvir in Brazil. Clin. Res. Hepatol. Gastroenterol. 2020;44:329–339. doi: 10.1016/j.clinre.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 17.Zakaria M.K., Carletti T., Marcello A. Cellular targets for the treatment of flavivirus infections. Front. Cell. Infect. Microbiol. 2018;8:398. doi: 10.3389/fcimb.2018.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaufmann S.H.E., Dorhoi A., Hotchkiss R.S., Bartenschlager R. Host-directed therapies for bacterial and viral infections, Nature reviews. Drug Discov. 2018;17:35–56. doi: 10.1038/nrd.2017.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan M.A., Wilson L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer. 2004;4:253–265. doi: 10.1038/nr1317. [DOI] [PubMed] [Google Scholar]
- 20.Gg G., Ta C. Microtubules and signal transduction. Curr. Opin. Cell Biol. 1999;11:81–94. doi: 10.1016/s0955-0674(99)80010-6. [DOI] [PubMed] [Google Scholar]
- 21.Honore S., Pasquier E., Braguer D. Understanding microtubule dynamics for improved cancer therapy. Cell. Mol. Life Sci. 2005;62:3039–3056. doi: 10.1007/s00018-005-5330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dohner K., Sodeik B. The role of the cytoskeleton during viral infection. Curr. Top. Microbiol. Immunol. 2005;285:67–108. doi: 10.1007/3-540-26764-6_3. [DOI] [PubMed] [Google Scholar]
- 23.Radtke K., Dohner K., Sodeik B. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell Microbiol. 2006;8:387–400. doi: 10.1111/j.1462-5822.2005.00679.x. [DOI] [PubMed] [Google Scholar]
- 24.Miranda-Saksena M., Denes C.E., Diefenbach R.J., Cunningham A.L. Infection and transport of herpes simplex virus type 1 in neurons: role of the cytoskeleton. Viruses. 2018;10 doi: 10.3390/v10020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roohvand F., Maillard P., Lavergne J.P., Boulant S., Walic M., Andreo U., Goueslain L., Helle F., Mallet A., McLauchlan J., Budkowska A. Initiation of hepatitis C virus infection requires the dynamic microtubule network: role of the viral nucleocapsid protein. J. Biol. Chem. 2009;284:13778–13791. doi: 10.1074/jbc.M807873200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spichtin H.P., Gudat F., Schmid M., Plrovino M., Altorfer J., Blanchi L. Microtubular aggregates in human chronic non-A, non-B hepatitis with bridging hepatic necrosis and multinucleated hepatocytic giant cells. Liver. 1982;2:355–360. doi: 10.1111/j.1600-0676.1982.tb00834.x. [DOI] [PubMed] [Google Scholar]
- 27.Gudat F., Eder G., Eder C., Bianchi L., Stöcklin E., Kre G., Dürmüller U., Spichtin H.P. Experimental non-A, non-B hepatitis in chimpanzees: light, electron and immune microscopical observations. Liver. 1983;3:110–121. doi: 10.1111/j.1600-0676.1983.tb00857.x. [DOI] [PubMed] [Google Scholar]
- 28.Schaff Z., Eder G., Eder C., Lapis K. Intracytoplasmic crystalline inclusions in the hepatocytes of humans and chimpanzees. Ultrastruct. Pathol. 2009;14:303–309. doi: 10.3109/01913129009032245. [DOI] [PubMed] [Google Scholar]
- 29.Akil A., Song P., Peng J., Gondeau C., Samuel D., Gassama-Diagne A. PIAS1 regulates hepatitis C virus-induced lipid droplet accumulation by controlling septin 9 and microtubule filament assembly. Pathogens. 2021;10 doi: 10.3390/pathogens10101327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong S.S., Ng M.L. Involvement of microtubules in Kunjin virus replication. Brief report. Arch. Virol. 1987;97:115–121. doi: 10.1007/BF01310739. [DOI] [PubMed] [Google Scholar]
- 31.Ng M.L., Pedersen J.S., Toh B.H., Westaway E.G. Immunofluorescent sites in Vero cells infected with the flavivirus Kunjin. Arch. Virol. 1983;78:177–190. doi: 10.1007/BF01311313. [DOI] [PubMed] [Google Scholar]
- 32.Bost A.G., Venable D., Liu L., Heinz B.A. Cytoskeletal requirements for hepatitis C virus (HCV) RNA synthesis in the HCV replicon cell culture system. J. Virol. 2003;77:4401–4408. doi: 10.1128/jvi.77.7.4401-4408.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoder A., Guo J., Yu D., Cui Z., Zhang X.E., Wu Y. Effects of microtubule modulators on HIV-1 infection of transformed and resting CD4 T cells. J. Virol. 2011;85:3020–3024. doi: 10.1128/JVI.02462-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naghavi M.H. HIV-1 capsid exploitation of the host microtubule cytoskeleton during early infection. Retrovirology. 2021;18:19. doi: 10.1186/s12977-021-00563-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H., Fang X., Meng Q., Mao Y., Xu Y., Fan T., An J., Huang Z. Design, synthesis and characterization of potent microtubule inhibitors with dual anti-proliferative and anti-angiogenic activities. Eur. J. Med. Chem. 2018;157:380–396. doi: 10.1016/j.ejmech.2018.07.043. [DOI] [PubMed] [Google Scholar]
- 36.Syed G.H., Siddiqui A. Effects of hypolipidemic agent nordihydroguaiaretic acid on lipid droplets and hepatitis C virus. Hepatology. 2011;54:1936–1946. doi: 10.1002/hep.24619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Krausslich H.G., Mizokami M., Bartenschlager R., Liang T.J. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong J., Gastaminza P., Chung J., Stamataki Z., Isogawa M., Cheng G., McKeating J.A., Chisari F.V. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 2006;80:11082–11093. doi: 10.1128/JVI.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D.R., Wieland S.F., Uprichard S.L., Wakita T., Chisari F.V. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wyles D.L., Kaihara K.A., Vaida F., Schooley R.T. Synergy of small molecular inhibitors of hepatitis C virus replication directed at multiple viral targets. J. Virol. 2007;81:3005–3008. doi: 10.1128/JVI.02083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Veroli G.Y., Fornari C., Wang D., Mollard S., Bramhall J.L., Richards F.M., Jodrell D.I. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics. 2016;32:2866–2868. doi: 10.1093/bioinformatics/btw230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greco W.R., Bravo G., Parsons J. The search for synergy: a critical review from a response surface perspective. Pharmacol. Rev. 1995;47:337–385. [PubMed] [Google Scholar]
- 43.Yeh P.J., Hegreness M.J., Aiden A.P., Kishony R. Drug interactions and the evolution of antibiotic resistance. Nat. Rev. Microbiol. 2009;7:460–466. doi: 10.1038/nrmicro2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang J., Wennerberg K., Aittokallio T. What is synergy? The Saariselka agreement revisited. Front. Pharmacol. 2015;6:181. doi: 10.3389/fphar.2015.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chou T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 46.Chou T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.