Abstract
A major challenge in eukaryotic cells is the proper distribution of nuclear-encoded proteins to the correct organelles. For a subset of mitochondrial proteins, a signal sequence at the N terminus (matrix-targeting sequence [MTS]) is recognized by protein complexes to ensure their proper translocation into the organelle. However, the early steps of mitochondrial protein targeting remain undeciphered. The cytosolic chaperone nascent polypeptide–associated complex (NAC), which in yeast is represented as the two different heterodimers αβ-NAC and αβ′-NAC, has been proposed to be involved during the early steps of mitochondrial protein targeting. We have previously described that the mitochondrial outer membrane protein Sam37 interacts with αβ′-NAC and together promote the import of specific mitochondrial precursor proteins. In this work, we aimed to detect the region in the MTS of mitochondrial precursors relevant for their recognition by αβ′-NAC during their sorting to the mitochondria. We used targeting signals of different mitochondrial proteins (αβ′-NAC-dependent Oxa1 and αβ′-NAC-independent Mdm38) and fused them to GFP to study their intracellular localization by biochemical and microscopy methods, and in addition followed their import kinetics in vivo. Our results reveal the presence of a positively charged amino acid cluster in the MTS of select mitochondrial precursors, such as Oxa1 and Fum1, which are crucial for their recognition by αβ′-NAC. Furthermore, we explored the presence of this cluster at the N terminus of the mitochondrial proteome and propose a set of precursors whose proper localization depends on both αβ′-NAC and Sam37.
Keywords: mitochondria, protein import, chaperone, mitochondrial transport, Saccharomyces cerevisiae
Abbreviations: ER, endoplasmic reticulum; HA, hemagglutinin; hsp, heat shock protein; MTS, matrix-targeting sequence; NAC, nascent polypeptide–associated complex; PK, proteinase K; RBD, ribosomal-binding domain; RCF, relative centrifugal force; TOM, translocase of the outer membrane; YP, yeast peptone
Mitochondria are one of the most important organelles in eukaryotic cells because of the vital functions that occur within their membranes. As a result of their endosymbiotic origin (1, 2), the mitochondrial proteome is encoded in both the nuclear and the mitochondrial genomes (3). During evolution, most mitochondrial genes have been transferred to the nucleus (4). Hence, one of the most challenging events during the biogenesis of nuclear-encoded mitochondrial proteins is their proper sorting to the mitochondrial surface to achieve their transport inside the organelle (5, 6).
It is widely accepted that, after being translated by cytosolic ribosomes, mitochondrial proteins are post-translationally directed to the translocase of the outer membrane (TOM) complex, the entrance gate at the mitochondria. Nevertheless, substantial evidence supports that some elements act cotranslationally to ensure the fidelity and specificity of mitochondrial import. Such evidence involves (i) the visualization of cytosolic ribosomes and polysomes on the mitochondrial outer membrane (7, 8, 9, 10); (ii) the localization of some mRNAs encoding mitochondrial proteins at the mitochondrial surface (11, 12, 13); and (iii) the inability to import some mitochondrial proteins, such as fumarase (Fum1), in the well-established in vitro import assays (14).
A factor involved in the cotranslational recognition of mitochondrial proteins is the cytosolic chaperone nascent polypeptide–associated complex (NAC) (15, 16). This heterodimeric complex is constitutively bound to the ribosomal exit tunnel ready to escort nascent polypeptides preventing their misfolding (17, 18, 19, 20). NAC prevents resident proteins in the endoplasmic reticulum (ER) from being wrongly targeted to mitochondria and vice versa (21, 22, 23, 24, 25), supporting the role of NAC as a pivotal factor that maintains proper targeting and shows the intricate relationship between protein transport to mitochondria and the ER. Recently, NAC has been related to the correct import of selected mitochondrial proteins and to interact with mitochondrial outer membrane proteins such as Om14 and Sam37 (26, 27). The relevance of NAC during protein import, an essential process, is further supported by the lethal phenotype obtained after deletion of the genes encoding its subunits in Mus musculus, Drosophila melanogaster, and Caenorhabtidits elegans (28, 29, 30). In contrast, NAC is not essential in the yeast Saccharomyces cerevisiae where there are two versions of the NAC (αβ-NAC and αβ′-NAC) as the result of a duplication of the gene encoding the β-NAC subunit. It has been described that each one of the yeast NACs can bind to ribosomes translating different sets of mRNAs and that the αβ′-NAC is present in ribosomes specifically translating mRNAs that encode mitochondrial proteins (16). In particular, the simultaneous deletion of αβ′-NAC and Sam37, a protein located in the outer mitochondrial membrane, impairs the import of several proteins, such as Oxa1, Sod2, and Mmf1 (27). In the present study, we describe the relevance of a positively charged patch of amino acids at the N terminus of a subset of mitochondrial proteins that mediates their early recognition by αβ′-NAC and Sam37 during their transport to the mitochondria.
Results
Different matrix-targeting sequences are differentially affected by the absence of the αβ′-NAC and Sam37
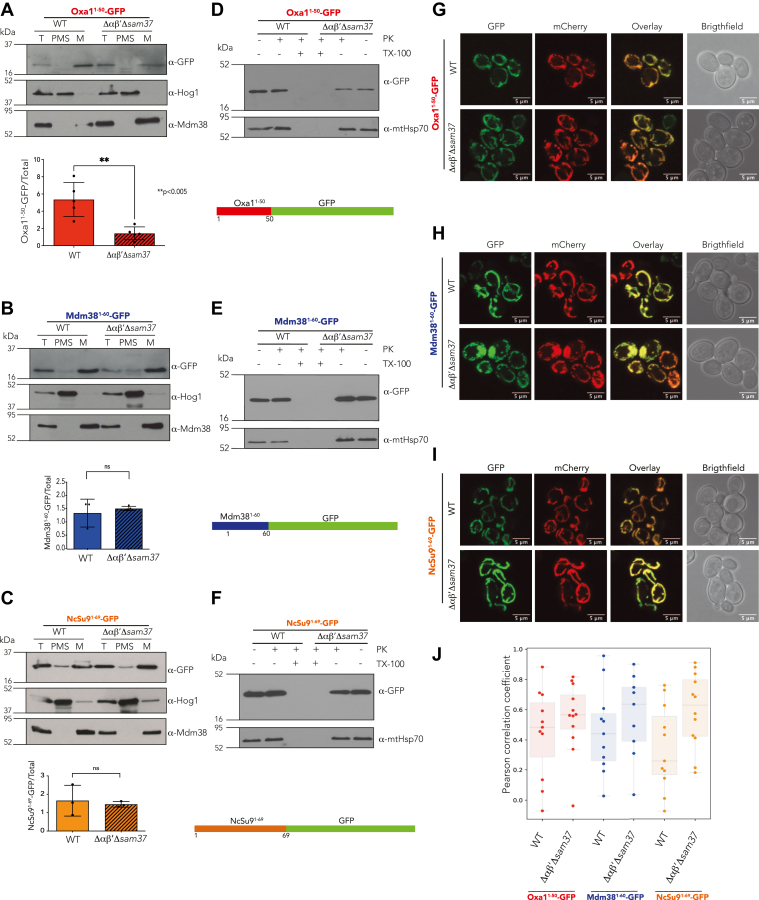
We previously described that the αβ′-NAC (composed of the proteins Egd2 and Btt1) interacts physically and functionally with Sam37 at the mitochondrial outer membrane, affecting the presence of selected proteins within the organelle (27). To identify the features found in the region recognized by αβ′-NAC on nascent polypeptides, we designed chimeric proteins fusing different matrix-targeting sequences (MTSs) (31, 32) at the N terminus of the GFP and analyzed their mitochondrial localization in a mutant lacking αβ′-NAC and Sam37 (Δαβ′Δsam37). The rationale is that, during translation, the MTS is the first part of the protein exposed to the cytosol and, thereby, to αβ′-NAC, which associates with the ribosomal exit tunnel. The MTSs of inner membrane proteins Oxa1 (Oxa11–50-GFP) and Mdm38 (Mdm381–60-GFP) were chosen since we previously showed that they were oppositely affected in a mutant lacking both αβ′-NAC and Sam37 (Δαβ′Δsam37) (27). As a control, we used the widely studied MTS of subunit 9 of the ATPase from Neurospora crassa (NcSu91–69-GFP).
These engineered MTS-GFP variants were expressed in WT and Δαβ′Δsam37 cells, and their localization was determined by cellular fractionation. While Oxa11–50-GFP is mitochondrially localized, its steady-state levels are significantly decreased in the absence of αβ′-NAC and Sam37 (Fig. 1A). In contrast, Mdm381–60-GFP and NcSu91–69-GFP can be localized in the mitochondrial fraction in WT and Δαβ′Δsam37 cells, and their steady-state levels appeared unaffected (Fig. 1, B and C). To confirm that the chimeric proteins were internalized into the organelle, purified mitochondria were treated with proteinase K (PK). The three proteins were protected from the PK treatment, indicating that all chimeric proteins are inside mitochondria (Fig. 1, D–F). Furthermore, confocal microscopy revealed that the yellow color that is observed in the superposition of the three chimeric proteins colocalize with a mitochondrially targeted mCherry (mt-mCherry) confirming that the three chimeric proteins are mitochondrially localized (Fig. 1, G–I). The colocalization was evaluated using spatial correlation analysis by pixel intensity using the Pearson’s correlation coefficient, finding only statistically significant differences (Student's t test, p < 0.05) between WT and Δαβ′Δsam37 strains expressing NcSu91–69-GFP (Fig. 1J). In this approach, no accumulation of the proteins in the cytosol or any other cellular compartment was visible.
Figure 1.
The Oxa1 N terminus 50 amino acids are relevant for αβ′-NAC recognition. Mitochondria were purified from a WT strain or a mutant lacking αβ′-NAC and SAM37 (Δαβ′Δsam37), transformed with plasmids containing the indicated constructs, and the presence of the chimeric protein within the organelle or in the postmitochondrial supernatant (PMS) was analyzed by Western blotting (left panels). Endogenous Mdm38 was used as mitochondrial marker and Hog1 as cytosolic marker. The signals were quantified, and the amount of the corresponding chimera found in mitochondria was compared with the total fraction (T) (left panels, bottom). To further explore if the chimeric protein was internalized into mitochondria, the samples were treated with proteinase K (PK) and 0.5% Triton X-100 (Tx), where indicated (middle panel). The cultures were also analyzed by confocal microscopy after labeling mitochondria with the fluorescent protein mt-mCherry (right panels). A, D, and G, the first 50 amino acids of Oxa1 fused to GFP. B, E, and H, the first 60 amino acids of Mdm38 fused to GFP. C, F, and I, the first 69 amino acids of Neurospora crassa ATPase 9 fused to GFP. J, colocalization analysis was evaluated using spatial correlation analysis calculating the Pearson's correlation coefficient. ∗∗p < 0.005. NAC, nascent polypeptide–associated complex.
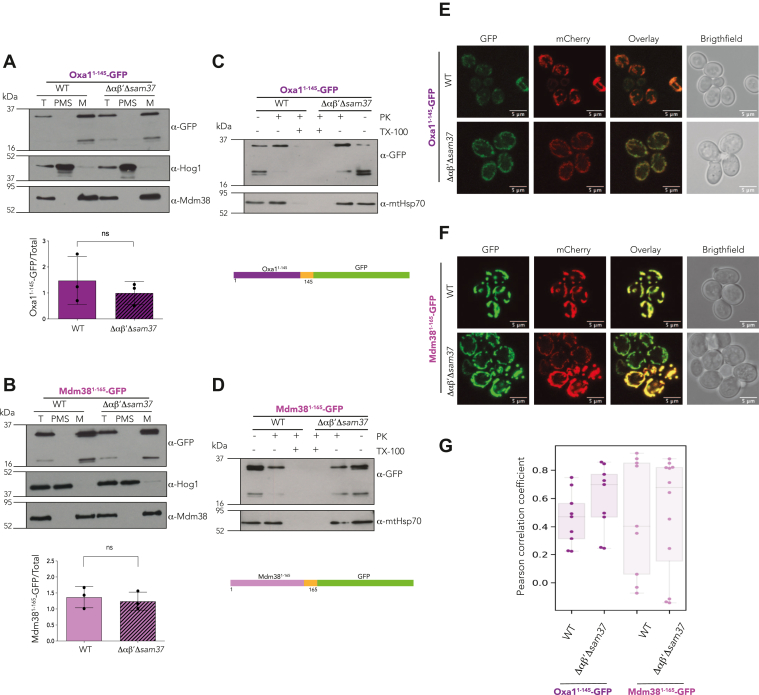
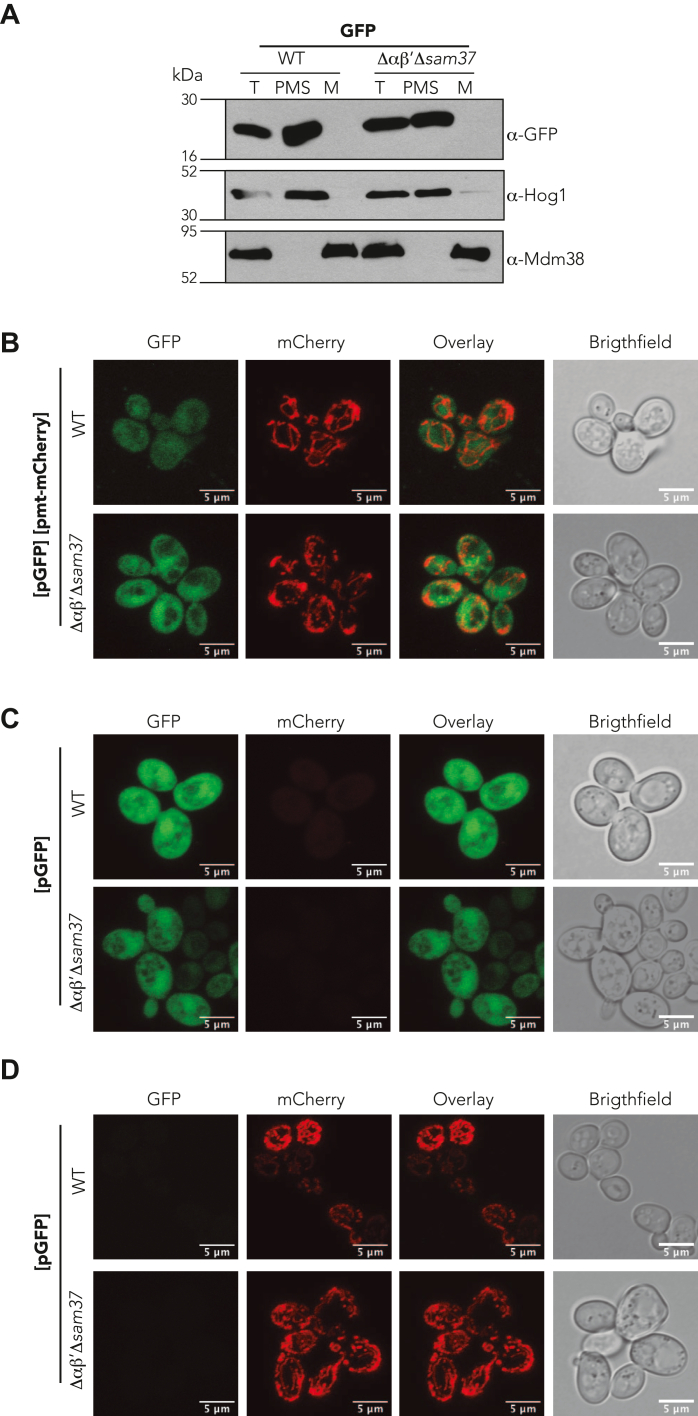
Previous reports indicate that hydrophobic domains, such as those found in transmembrane stretches, are important during the biogenesis of inner membrane proteins. Moreover, they could be wrongly directed to the ER under certain conditions (21, 33, 34, 35). To explore if the first hydrophobic segment of Oxa1 and Mdm38 could modulate their mitochondrial localization in the absence of αβ′-NAC and Sam37, two additional chimeras were analyzed under similar conditions as described previously: Oxa11–145-GFP and Mdm381–165-GFP, which includes the MTS and the first transmembrane stretch of each protein (Fig. 2). As observed for Oxa11–50-GFP, Oxa11–145-GFP was correctly localized into the mitochondria, and even though not significant, a trend of decreased protein levels was observed in mitochondria of the Δαβ′Δsam37 mutant (Fig. 2A). Similarly to Mdm381–60-GFP, Mdm381–165-GFP levels remain unaffected in the absence of αβ′-NAC and Sam37 (Fig. 2B). GFP alone, that is, without an MTS, was found to be cytosolically located independently on the presence or the absence of αβ′-NAC and Sam37 (Fig. 3). Overall, our results suggest that the MTS of Oxa1 contains relevant information for its αβ′-NAC-mediated recognition and import into the mitochondria.
Figure 2.
The Oxa1 first hydrophobic transmembrane segment does not increase αβ′-NAC dependence during mitochondrial import. Mitochondria were purified from a WT strain or a mutant lacking αβ′-NAC and SAM37 (Δαβ′Δsam37), transformed with plasmids containing the indicated constructs, and the presence of the chimeric protein within the organelle or in the postmitochondrial supernatant (PMS) was analyzed by Western blotting (left panels). Endogenous Mdm38 was used as mitochondrial marker and Hog1 as cytosolic marker. The signals were quantified, and the amount of the corresponding chimera found in mitochondria was compared with the total fraction (T) (left panels, bottom). To further explore if the chimeric protein was internalized within mitochondria, the samples were treated with proteinase K (PK) and 0.5% Triton X-100 (Tx), where indicated (middle panel). The cultures were also analyzed by confocal microscopy where mitochondria were labeled with the fluorescent protein mCherry that was targeted to the organelles by the means of the NcSu9 MTS (right panels). A, C, and E, import of the first 145 amino acids of Oxa1 fused to GFP. B, D, and F, import of the first 165 amino acids of Mdm38 fused to GFP. G, colocalization analysis was evaluated using spatial correlation analysis calculating the Pearson's correlation coefficient. MTS, matrix-targeting sequence; NAC, nascent polypeptide–associated complex.
Figure 3.
GFP without a signal sequence is localized in the cytosol.A, mitochondria were purified from a WT strain or a mutant lacking αβ′-NAC and SAM37 (Δαβ′Δsam37), transformed with plasmid encoding a GFP lacking an MTS, the presence of the protein within the organelle or in the postmitochondrial supernatant (PMS) was analyzed by Western blotting. Endogenous Mdm38 was used as mitochondrial marker and Hog1 as cytosolic marker. B–D, the proper localization of GFP lacking an MTS and of the mt-mCherry was evaluated by confocal microscopy in both strains studied WT and Δαβ′Δsam37. MTS, matrix-targeting sequence; NAC, nascent polypeptide–associated complex.
αβ′-NAC and Sam37 are important during the early stages of mitochondrial protein import
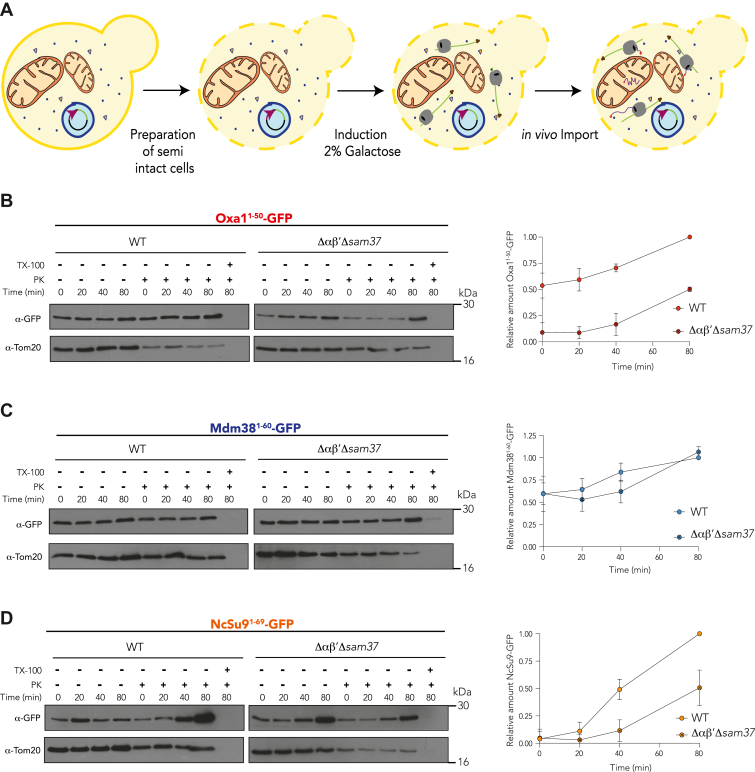
To further delineate whether αβ′-NAC is crucial for the targeting of Oxa1, the import of the MTS-GFP chimeric proteins was evaluated in vivo using semipermeabilized cells (36). Our goal in these experiments was to employ a system that allowed us to evaluate the import kinetics of specific proteins using the cytosol from desired yeast mutant strains, an essential incorporation to assess the relevance of ribosomal chaperones during the early steps of mitochondrial protein import. To this end, the MTS-GFP constructs were cloned under the control of the inducible GAL1 o GAL10 promoters and expressed in a WT strain or a Δαβ′Δsam37 mutant. The presence of each protein was followed in a time course for 20, 40, and 80 min after adding galactose to the cell suspension (Fig. 4A). To determine if the newly synthesized proteins had been internalized into mitochondria, the reactions were also subjected to PK treatment.
Figure 4.
In vivo import guided by the Oxa1 first 50 amino acids is delayed in αβ′-NAC and Sam37 absence.A, whole cells of the indicated strains, transformed with plasmids encoding the chimeric Oxa11–50, Mdm381–60, or NcSu9 fused to GFP under an inducible promoter GAL1 or GAL10, were grown in rich media with raffinose and permeabilized with zymolyase to produce semi-intact cells, transferred to import buffer before adding 2% galactose to induce the expression of the chimeric proteins, and follow the import of the corresponding proteins at 20, 40, and 80 min. Tom20 was used as a control for the digestion of the proteins exposed or contained within the cytosol. Samples were treated with proteinase K (PK) and 3% Triton X-100 (Tx), where indicated. Samples were analyzed by Western blot, and the signals were quantified: the 80 min sample obtained in the WT strain was considered as 1.0 as it represents the maximum amount of imported protein, and the 0, 20, 40, and 80 min samples were analyzed as a fraction of that reference sample. B, import kinetics of the first 50 amino acids of Oxa1 fused to GFP. C, import kinetics of the first 60 amino acids of Mdm38 fused to GFP. D, import kinetics of the first 69 amino acids of Neurospora crassa ATPase 9 fused to GFP. NAC, nascent polypeptide–associated complex.
The in vivo Oxa11–50-GFP import was strongly affected in the absence of αβ′-NAC and Sam37 (Fig. 4B). In contrast, the import of Mdm381–60-GFP was unaffected (Fig. 4C). This result is in line with the hypothesis that αβ′-NAC recognizes the MTS of Oxa1 during translation and modulates its mitochondrial import from early stages of this process. Surprisingly, NcSu91–69-GFP was also affected in these experiments, indicating that while its steady-state levels are not affected in the Δαβ′Δsam37 mutant, the early stages of its import are affected by the lack of αβ′-NAC and Sam37 (Fig. 4D).
The first 10 amino acids in the MTS of Oxa1 are crucial for the recognition by αβ′-NAC
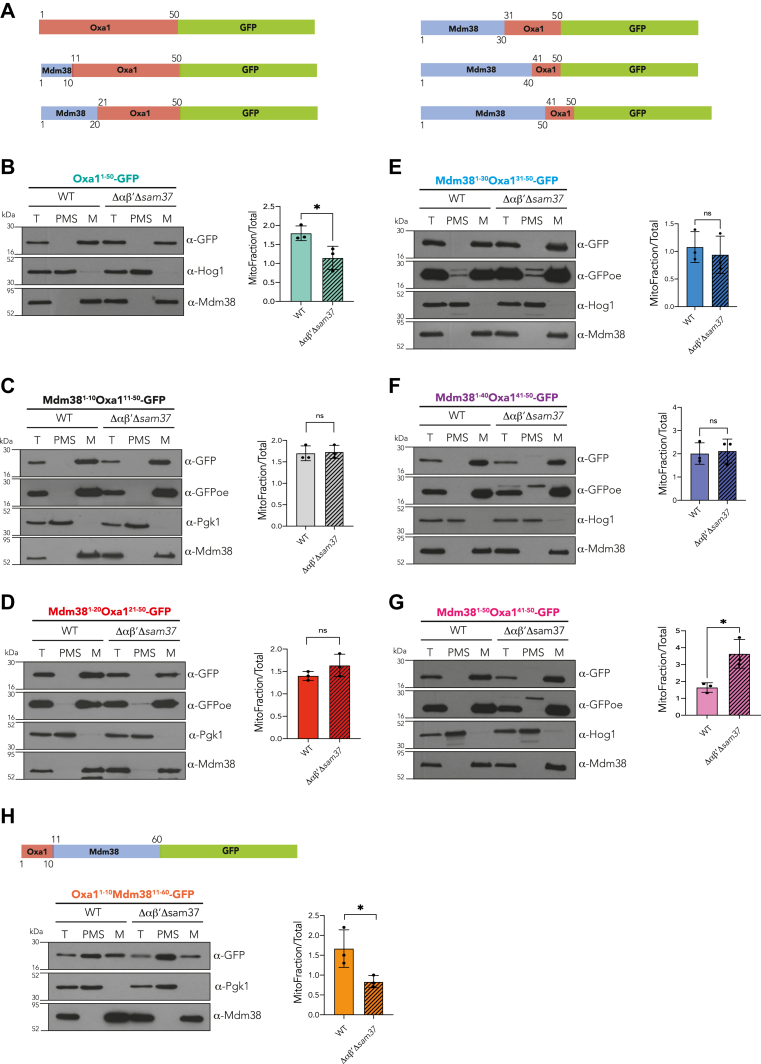
To further dissect the region recognized by αβ′-NAC in the MTS of Oxa1, a new series of chimeric proteins were designed by fusing the MTS of Oxa1 with the one of Mdm38 in sequential fragments of 10 amino acids (Fig. 5A), aiming to detect the region that renders the MTS of Oxa1 insensitive to the presence of αβ′-NAC and Sam37. Mitochondrial localization of the chimeric proteins was analyzed by cellular fractionation (Fig. 5, B–G). Remarkably, the replacement of the first 10 amino acids of Oxa1 by those of Mdm38 (Mdm381–10Oxa111–50-GFP) was sufficient to restore the levels of GFP within the mitochondria in the Δαβ′Δsam37 mutant (Fig. 5C). This result suggests that the information required for the recognition by αβ′-NAC is located within the first 10 amino acids of the MTS of Oxa1. In the following studied chimeras, the mitochondrial amount of GFP was similar in both the WT and Δαβ′Δsam37 strains (Fig. 5, D–G). It is noteworthy that we were able to visualize a higher molecular weight protein for the first time in our study, presumably the precursor version of the proteins (Fig. 5, E–G). To confirm the relevance of the N terminus of Oxa1 for protein import, we constructed an additional chimera by fusing its first 10 amino acids with the MTS of Mdm38 (Oxa11–10Mdm3811–60-GFP). The result is striking: the steady-state levels of the chimera are reduced in the mitochondrial fraction of the mutant strain Δαβ′Δsam37 (Fig. 5H) in the same manner as observed when the complete MTS of Oxa1 was used (compare with Figs. 1A and 5B).
Figure 5.
The Oxa1 first 10 amino acids contain relevant information for αβ′-NAC recognition.A, schematic representation of the chimeric proteins used. Red boxes represent fractions of the Oxa1 MTS, blue boxes represent fractions of the Mdm38 MTS, and green boxes represent the sequence corresponding to GFP. Numbers indicate the amino acid position in the protein according to the Saccharomyces cerevisiae genome annotation. B–H, mitochondria were purified from a WT strain or the Δαβ′Δsam37 mutant, transformed with plasmids encoding the indicated constructs, and the presence of the chimeric protein within the organelle or in the postmitochondrial supernatant (PMS) was analyzed by Western blotting. Endogenous Mdm38 was used as mitochondrial marker, and Hog1 or Pgk1 was used as cytosolic marker. The signals were quantified, and the amount of the corresponding chimera found in mitochondria was compared with the total fraction (T). Asterisks represent p < 0.05. MTS, matrix-targeting sequence; NAC, nascent polypeptide–associated complex; ns, nonsignificant; OE, overexposure.
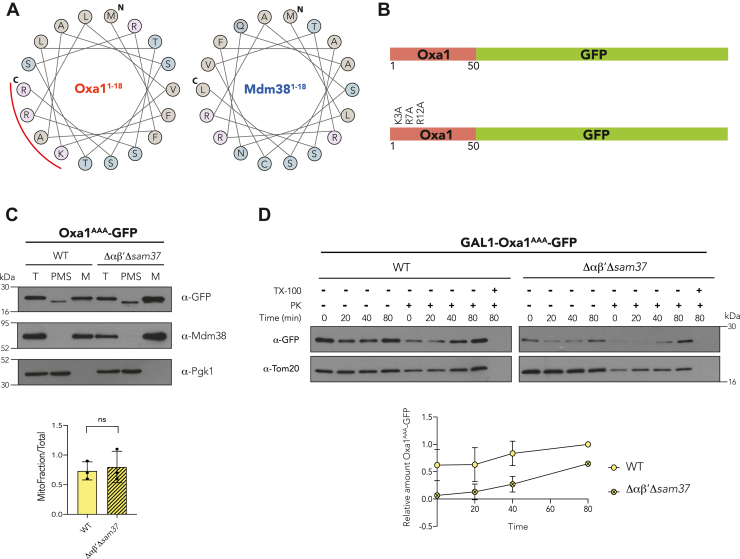
Both Oxa1 and Mdm38 contain a classic MTS with similar values of hydrophobicity (0.355 versus 0.356 as calculated with a standard hydrophobicity scale); however, they exhibit different net charges (4 versus 3) and hydrophobic moments (μH) (0.291 versus 0.203). The hydrophobic moment represents the amphiphilicity of a helix (37), that is, if positively charged amino acids are exposed on one face of the MTS α-helix while hydrophobic residues cluster on the opposite face. Hence, we analyzed the first 18 amino acids of the two MTSs within the α-helix (Fig. 6A). It was evident that the higher hydrophobic moment of the Oxa1-MTS is due to a cluster of positively charged amino acids at one side of the α-helix that is not present in the Mdm38-MTS. To determine if this positive cluster was responsible for the αβ′-NAC recognition, three of those positive amino acids (K3, R7, and R12, Fig. 6B) were replaced by alanine residues, and the mitochondrial localization and import kinetics of the corresponding precursors were analyzed. The steady-state levels of Oxa1AAA-GFP were similar both in WT and Δαβ′Δsam37 strains (Fig. 6C). In contrast, when the import kinetics in semi-intact cells was analyzed, there was a delay in the mitochondrial internalization of the Oxa1AAA-GFP construct in Δαβ′Δsam37 background, similar to the scenario with the Oxa11–50-GFP construct (Fig. 6D).
Figure 6.
The Oxa1 first 18 amino acids harbor a cluster of positive amino acids important for αβ′-NAC recognition.A, helical wheel diagram of the first 18 amino acids of Oxa1 and Mdm38: N indicates the first methionine and C indicates the last amino acid of the analyzed sequence; the red curved line shows a positive amino acid cluster found in the Oxa1 MTS. B, schematic representation of the Oxa11–50-GFP construct (up) and the location of the amino acid residues that were changed for alanines (K3A, R7A, and R12A). C, mitochondria were purified from a WT strain or a mutant lacking αβ′-NAC and SAM37 (Δαβ′Δsam37), transformed with the Oxa1AAA-GFP construct plasmid, and the presence of the chimeric protein within the organelle or in the postmitochondrial supernatant (PMS) was analyzed by Western blotting. Endogenous Mdm38 was used as mitochondrial marker and Pgk1 as cytosolic marker. The signals were quantified, and the amount of the corresponding chimera found in mitochondria was compared with the total fraction (T). D, import kinetics of Oxa1AAA-GFP followed in semipermeabilized cells of a WT or a Δαβ′Δsam37 as for Figure 4. MTS, matrix-targeting sequence; NAC, nascent polypeptide–associated complex.
To investigate how general is the presence of a positively charged amino acid stretch within the mitochondrial proteome, we took each mitochondrial protein reported (31, 38, 39) and performed peep-wheel analyses of their first 18 N-terminal amino acids to observe the distribution of the charged amino acids, mean hydrophobicity (H), hydrophobic moment (μH), and net charge (Table S1). To complement our analysis, we included the association of mRNAs to the mitochondrial outer membrane (11, 12, 13) and if precursors are translated by ribosomes localized to the mitochondrial periphery (10). From these data, we defined a subset of mitochondrial proteins that we propose require αβ′-NAC and Sam37 for their correct and efficient import into the organelle (Table 1 and Table S1).
Table 1.
Proteins that require mechanisms acting at early stages of protein synthesis for proper targeting to mitochondria
| Gene | Submitochondrial localization | MTS |
|---|---|---|
| ATP2 | IMM | X |
| ATP22 | IMM | X |
| CCP1 | Intermembrane space | X |
| CIR2 | IMM | X |
| COR1 | IMM/Matrix | X |
| COX10 | IMM | X |
| CYB2 | Intermembrane space | X |
| DLD2 | IMM | X |
| FMP40 | IMM | |
| FUM1 | Matrix | X |
| GCV1 | Matrix | X |
| ILV1 | IMM | |
| KGD2 | Matrix | X |
| MCR1 | Intermembrane space | X |
| OXA1 | IMM | X |
| TIM50 | IMM | X |
| TIM54 | IMM | |
| YER077C | IMM |
Abbreviation: IMM, inner mitochondrial membrane.
Selected mitochondrial proteins with a high probability to follow an import mediate by αβ′-NAC and Sam37.
The import of the mitochondrial protein Fum1 depends on αβ′-NAC and Sam37
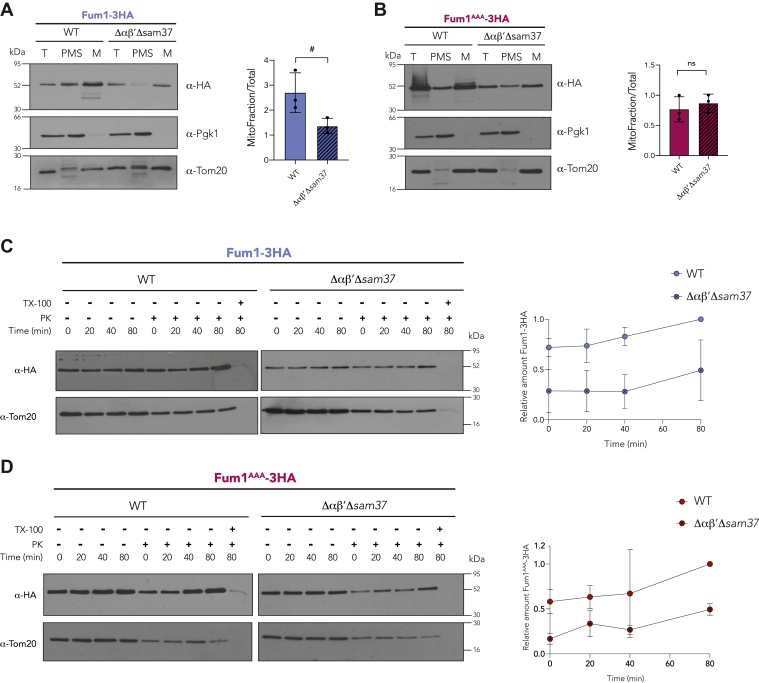
To validate the information gathered in Table 1, we decided to study the case of fumarase (Fum1), a protein previously shown to depend on a coupled protein synthesis and import process (14, 40). To this end, we cloned the gene FUM1 fused with a C-terminal 3× hemagglutinin tag (3XHA) under the regulation of the GAL1 promoter (Fum1-3HA). In addition, positively charged amino acids R3, K10, and K14 at its N terminus were changed for alanines (Fum1AAA-3HA). We then determined the localization of each Fum1 version by cellular fractionation (Fig. 7, A and B). A similar behavior is observed as for Oxa11–50-GFP; the steady-state levels of Fum1-3HA are reduced in the Δαβ′Δsam37 strain (Fig. 7A). However, the difference is lost when the positive amino acids are absent at its N terminus (Fum1AAA-3HA, Fig. 7B), confirming the importance of these residues at the N terminus of some mitochondrial proteins during the first steps of mitochondrial import. In addition, we analyzed the import kinetics in vivo of both Fum1-3HA and Fum1AAA-3HA. The import of these proteins is delayed in the mutant lacking αβ′-NAC and Sam37 (Fig. 7, C and D), similarly to what was observed with the Oxa1 chimeras.
Figure 7.
The mitochondrial protein Fum1 follows an import mediated by αβ′-NAC and Sam37.A and B, mitochondria were purified from a WT strain or a mutant lacking αβ′-NAC and SAM37 (Δαβ′Δsam37), transformed with a plasmid harboring either Fum1-3HA or Fum1AAA-3HA constructs; the presence of the protein within the organelle (M) or in the postmitochondrial supernatant (PMS) was analyzed by Western blotting. Tom20 was used as mitochondrial marker and Pgk1 as cytosolic marker. The signals were quantified, and the amount of the corresponding chimera found in mitochondria was compared with the total fraction (T), #p = 0.05. C and D, import kinetics of Fum1-3HA and Fum1AAA-3HA followed in semipermeabilized cells of a WT or a Δαβ′Δsam37 as for Figure 4. NAC, nascent polypeptide–associated complex.
The ribosome-binding domain of β′-NAC subunit plays an important role mediating mitochondrial protein import
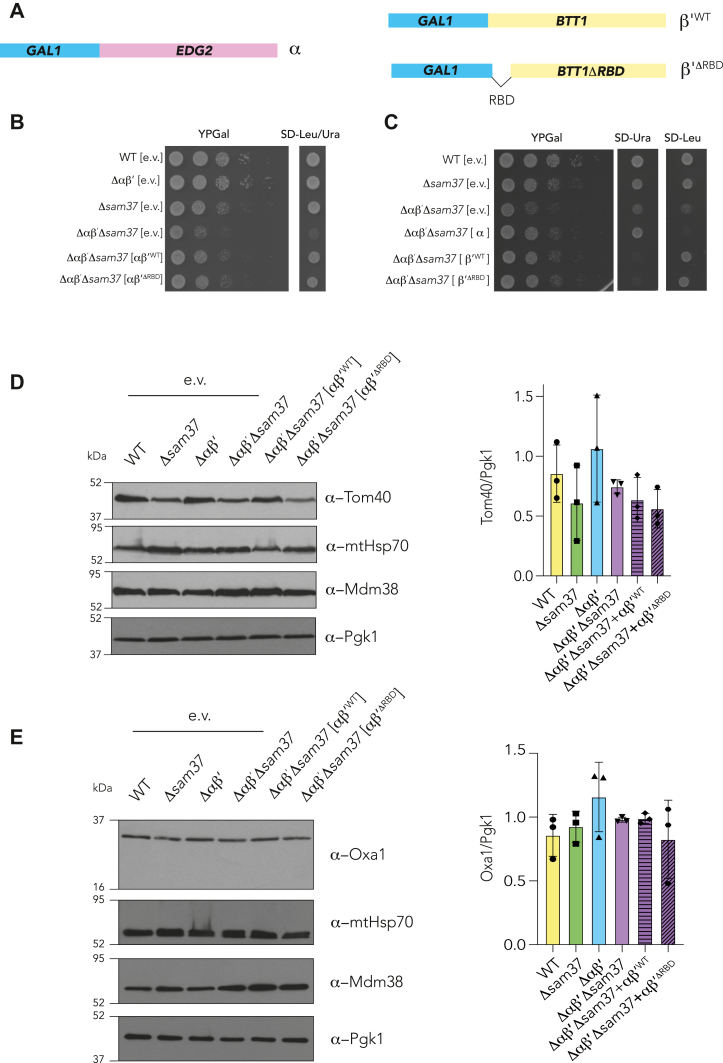
The NAC has been extensively studied as a ribosomal chaperone, able to bind the ribosome at the exit tunnel and, thereby, contacting the nascent proteins in a cotranslational manner (20, 21, 41). This binding depends on the first 11 amino acids of the β-NAC subunit of the corresponding heterodimer, the so-called ribosomal-binding domain (RBD) (19, 42, 43). To identify if the association to the ribosome is a requirement for the function of αβ′-NAC during early steps of mitochondrial import, a WT version of β′-NAC or a mutant lacking the RBD (β′ΔRBD-NAC) was cloned under the regulation of the inducible GAL1 promoter and expressed alone or simultaneously with the α-NAC subunit in the Δαβ′Δsam37 strain (Fig. 8A). As indicated before, the absence of αβ′-NAC and Sam37 triggers a negative growth phenotype characteristic of their genetic interaction (27). The coexpression of the α-NAC and β′-NAC subunits restores the observed growth phenotype, independently of the presence or the absence of the RBD in β′-NAC (Fig. 8B). Furthermore, the expression of each subunit independently also restored the growth phenotype (Fig. 8C). It is important to mention that in these strains, the EGD1 gene (encoding β-NAC) is still present. Therefore, the observed growth phenotype can be explained by the formation of heterodimers (αβ-, ββ′WT-, or ββ′ΔRBD) that could functionally compensate for the absence of αβ′-NAC and restore the growth phenotype. To further address this point, the steady-state levels of Oxa1 and Tom40, mitochondrial proteins previously reported to be affected by the absence of αβ′-NAC and Sam37, were evaluated (27). While αβ′WT-NAC was able to restore the steady-state levels of Tom40, in the strain carrying αβ′ΔRBD-NAC, the levels of this protein remain affected (Fig. 8D). However, it is interesting to notice that the effect is subtle for Oxa1 (Fig. 8E), indicating that either the ribosome binding is not crucial for its import; or that until certain extent, β-NAC keeps β′-NAC ribosomally bound because of the presence of heterodimers.
Figure 8.
The ribosome-binding domain of β′-NAC is important during the mitochondrial protein import.A, schematic representation of NAC subunits regulated by the GAL promoter used in these experiments. β′ΔRBD represents the construct where amino acids 2 to 11 of β′-NAC have been deleted. B and C, indicated strains were transformed with combinations of plasmids expressing either αβ′WT-NAC, αβ′ΔRBD-NAC, or the individual subunits, under the regulation of the GAL promoter. Strains were grown to logarithmic phase, and serial 1:10 dilutions were spotted on rich media with galactose as carbon source. Plates were incubated at 30 °C. Synthetic media were used to confirm either both plasmids (SD-Leu/Ura) or individual ones (SD-Ura or SD-Leu) were present. D and E, whole cell extracts (25 μg of protein) of the indicated strains harboring plasmids expressing either αβ′WT-NAC or αβ′ΔRBD-NAC were grown in SRaf-Leu/Ura until an absorbance of 1 at 600 nm followed by galactose induction for 3 h. Protein extracts were separated by electrophoresis on denaturing SDS-PAGE. Samples were transferred to nitrocellulose membranes and decorated with the specific antibodies as indicated. Cytosolic Pgk1 was analyzed as loading control. e.v., empty vector; NAC, nascent polypeptide–associated complex; RBD, ribosomal-binding domain.
Discussion
Most of the mitochondrial proteome is encoded in the nuclear genome. Hence, cells have developed precise mechanisms to identify and target substrates to their final subcellular destination and preventing protein mislocalization. It has been widely demonstrated that mitochondrial proteins can be recognized post-translationally by cytosolic chaperones and escorted to the mitochondrial surface where they are handed over to the import machineries. However, it is increasingly accepted that the participation of additional elements mediates and ensures a cotranslational transport of proteins. In this study, we address the function of the ribosomal chaperone αβ′-NAC during the early steps of mitochondrial protein transport and propose a relevant feature within the presequences crucial for substrate identification by this chaperone.
We chose the mitochondrial inner membrane protein Oxa1 as a model protein because its steady-state levels are affected by the simultaneous absence of αβ′-NAC and Sam37 (27). Furthermore, the OXA1 mRNA has been found in the proximity of mitochondrial outer membrane (11, 13, 44), and its translation is performed by cytosolic ribosomes associated to the periphery of mitochondria (10). While the Oxa1 import has been successfully achieved using in vitro systems (32), our data suggest that the import of Oxa1 is mediated by αβ′-NAC. Contrary to Oxa1, Mdm38 was selected as an example of the mitochondrial protein whose biogenesis is unaffected in cells lacking αβ′-NAC and Sam37.
First, we took the initial 50 amino acids of Oxa1, corresponding to its MTS, and fused them to a GFP, which yielded a reporter where the only relevant information for its targeting was located at the N terminus. We found that, in the absence of αβ′-NAC and Sam37, the steady-state levels of the chimeric reporter in the mitochondria are decreased (Fig. 1A). In contrast, the steady-state levels of Mdm38 or NcSu9, guided by their stronger MTSs, seemed to be unaffected in the same mutant (Fig. 1, D and G). The analyses of colocalization showed no apparent differences for the proteins visualized in our study in the strains analyzed (Figs. 1J and 2G). The exception being NcSu91–69-GFP expressed in WT and Δαβ′Δsam37, where it was possible to observe a statistically significant difference (Student's t test, p < 0.05). At this point, we do not have a possible explanation for this result since NcSu91–69 is a well-established substrate for mitochondrial protein import in yeast. In addition, it is worth mentioning that the Oxa1 chimeras were observed with a higher laser intensity compared with the Mdm38 and NcSu9 chimeras. Therefore, further and more sophisticated analyses are required to reach the resolution needed to identify those differences invisible to the human eye.
In addition, the import kinetics in permeabilized cells of Oxa11–50-GFP was severely affected in a Δαβ′Δsam37 mutant (Fig. 4B). These results show that even though the import of Oxa11–50-GFP occurs in the absence of αβ′-NAC and Sam37, its kinetics of import are severely affected. It is possible that in this scenario, Oxa11–50-GFP may either be wrongly retained in the cytosol or mistargeted to the ER, as it has been previously shown in other organisms (23, 33, 35), or degraded by the proteasome.
Our results agree with previous observations on the import of the entire Oxa1 protein (27), strongly suggesting that the information for the early recognition of Oxa1 by αβ′-NAC is contained at its MTS. Furthermore, we were able to limit the first 10 amino acids of the Oxa1 MTS as crucial for the recognition by αβ′-NAC (Fig. 5). This result is in line with previous data showing that the β subunit of NAC interacts with the first 10 amino acids in nascent peptides (20).
Remarkably, from all the experimental conditions tested, this was the first time that we observed the presence of a higher molecular weight band that could be the precursor form of the studied GFP reporters (Fig. 5). One explanation for this could be that, within Oxa1-MTS, there may be a signal for degradation when the protein is retained in the cytosol, and therefore, in the absence of αβ′-NAC, this signal is exposed, and the protein is rapidly degraded. Such a protein biogenesis control mechanism would be relevant in conditions where the mitochondrial function becomes compromised (45, 46, 47) and the membrane potential is affected, which otherwise would drive hydrophobic precursor accumulation in the cytosol.
To dissect the physicochemical characteristics in the MTS responsible for its recognition by αβ′-NAC, we analyzed the amino acid composition and the physicochemical characteristics of Oxa1 and Mdm38 targeting sequences (Fig. 6). We found a cluster of positively charged amino acids in Oxa1-MTS α-helix that contributes to increase the MTS hydrophobic moment. Such residues have a clear role in the import of Oxa11–50-GFP since the steady-state levels of Oxa1AAA-GFP are restored to those of the WT levels (Fig. 6C). Despite this, the mutations do not seem to restore the import kinetics of the protein (Fig. 6D). While these results seem contradictory, it is possible that decreasing the amphiphilicity in the MTS of Oxa11–50-GFP increases its accessibility for other cytosolic chaperones such as heat shock proteins Hsp70s and/or Hsp90s, creating a continuous but slower flow of post-translationally imported Oxa1, which ultimately would restore its basal level in the mitochondria. αβ′-NAC, thus may act by finely tuning a faster cotranslational import of mitochondrial proteins. We further tested the relevance of positive residues in the MTS of Fum1, a matrix mitochondrial protein that has been proposed to follow a cotranslational mode of import (14, 40) and contains several cotranslationally acting features as those analyzed in Table S1. Certainly, Fum1 has a similar import behavior as that seen for Oxa1 (Fig. 7) confirming the relevance of this positive cluster of amino acids at its N terminus and suggesting that αβ′-NAC, with the help of Sam37, guides its proper import as well. However, the role of other signals such as those present in the mRNA sequence that may contribute to the import process cannot be discarded.
It is known that any obstacles during mitochondrial protein targeting, particularly for Oxa1, can result in its mistargeting to other organelles such as the ER. In this scenario, different outcomes can occur depending on the ER factor that receives the precursor in the ER: (i) the protein can be relocated to mitochondria guided by Djp1 through the ER surface retrieval pathway (33); (ii) if the incoming protein reaches the membrane, Spf1 dislocates it to the cytosol, where it can be degraded by the proteasome (48); or (iii) it could be degraded directly at the ER by Ema19 (49). In particular, the ER surface retrieval pathway could well explain the observed delay in mitochondrial internalization and processing of the MTS in our experiments (Figs. 4B and 7C).
The absence of a precursor version of Oxa11–50-GFP in our initial experiments, either when analyzing whole or permeabilized cells, strongly suggests that the protein degradation systems remove any precursor retained in the cytosol. A precursor was only observed in the cytosolic fraction of the Δαβ′Δsam37 mutant when chimeric MTSs of Oxa1 and Mdm38 were constructed, which could imply that a degradation signal is located within amino acids 1 to 30. Analysis of the MTS sequence of Oxa1 reveals a Lysine (K3), which could be a potential substrate for ubiquitination and targeting for degradation by the proteasome (49).
It has been already pointed out that there are different kinds of MTSs (50), and our results agree with this notion. Oxa1 seems to possess an MTS that is dependent on early recognition at the exit ribosomal tunnel by αβ’-NAC, which is supported by the altered steady-state levels and the in vivo import of Oxa11–50-GFP in the Δαβ′Δsam37 mutant. Interestingly, the in vivo import of Su9-GFP was affected in the same mutant (Fig. 4D). Nevertheless, its steady-state levels were the same both in the WT and in the mutant strains (Fig. 1, G–I). These data indicate that some mitochondrial proteins may still arrive to mitochondria even when the speed of the process occurs at the lower rate. A similar trend was observed for Oxa1AAA-GFP (Fig. 6, C and D). Other mitochondrial proteins, such as Mdm38, are completely independent of αβ′-NAC and plausibly rely on cytosolic chaperones such as Hsp70.
The yeast mitochondrial proteome is composed of nearly 1000 proteins, and, after our in silico analysis, we propose that approximately 200 of them possess a signal relevant for early recognition by αβ′-NAC at the ribosomal exit tunnel (Table S1). Most of these proteins reside in the inner membrane or the mitochondrial matrix. Overall, the N terminus hydrophobicity is very similar, as well as their hydrophobic moment, while their net charge oscillates between two and four. These features align well with our experimental results since proteins residing in the matrix and the inner membrane exhibit MTSs that must also be recognized by the import machineries at the inner membrane to be properly distributed to its final compartment, similar to Oxa1. Therefore, it is not surprising that they have both a higher hydrophobic moment and a positive net charge, given the intrinsic characteristics found in such MTSs (51). Some studies suggest that the mitochondrial proteins that require elements acting at a cotranslational level, such as the mRNA targeting to the mitochondrial surface, are those that originated from the endosymbiont from where mitochondria evolved (52). It is possible that the appearance of the MTSs as targeting elements, in combination with other elements acting at a cotranslational level such as signals in the mRNA, occurred to enhance the specificity during targeting and distribution of proteins in early eukaryotic cells.
In early studies regarding the function of the NAC, it was proposed that NAC is important to prevent ER proteins from being wrongly directed to other compartments such as the mitochondria (21, 22, 23, 24, 25). Recent investigations demonstrated that NAC also assists the early recognition and transport of mitochondrial proteins (26, 27). Our results indicate that the RBD of the β′-NAC subunit is instrumental in conducting precursor proteins toward mitochondria (Fig. 8). For these reasons, NAC may act as a quality control factor for newly synthesized proteins and/or a regulator of their import rate. The fact that the α-NAC subunit possesses a ubiquitination domain (53) reinforces this hypothesis; however, more experiments are required to test this notion.
Most of our results involve the function of Sam37 simultaneously with αβ′-NAC since previous work from our group (27) demonstrated that the phenotype of the strain lacking EGD2 and BTT1 (Δαβ′) is similar to the WT strain, both in growth and steady-state levels of mitochondrial proteins. Sam37 was initially described as a possible receptor for mitochondrial proteins, whose function is shared to that of Tom70 (54). Sam37 was afterward described as a subunit of the SAM complex, which assembles β-barrels in the outer membrane (55), and subsequently reported as the subunit, along Tom22, that transiently binds the TOM and the SAM complexes (56, 57). It may be possible that Sam37 has an additional role as a receptor for mitochondrial proteins escorted by αβ′-NAC and hands them to the TOM complex. A nonexcluding possibility could be that αβ′-NAC recognizes the N terminus signal present in mitochondrial proteins, whereas Sam37 functions as the receptor for αβ′-NAC allowing the ribosome to localize in the proximity of the outer membrane. While the level of Tom40 decreases in a sam37-null mutant, we have previously demonstrated that such a lower Tom40 level is enough to support the import of proteins to the organelle (27). Importantly, Tom40 seems to be an additional substrate of the import route mediated by αβ′-NAC (Fig. 8D). Similar to the data obtained for the MTS of Oxa1, it may be possible that this route ensures no exposure of β-barrels to the cytosol, which would be otherwise detrimental for the cell. This effect is dependent on the association of αβ′-NAC to the ribosomes, since a β′ΔRBD-NAC mutant is unable to restore the steady-state levels of Tom40 (Fig. 8). In contrast, it seems that the ribosome binding of β′-NAC is not as crucial for Oxa1, possibly explained by heterodimers formed with β-NAC, or because substrate recognition through the cluster of positive amino acids does not depend on the ribosomal location of αβ′-NAC. It is important to recall that the 3′-UTR of the OXA1 mRNA ensures a localized translation on the mitochondrial periphery, hence boosting the internalization of the encoded protein. It is also possible that the structural and submitochondrial location differences between Tom40 and Oxa1 define the relevance of αβ′-NAC for their biogenesis. This intricated relationship between αβ′-NAC and Sam37 can explain the severe phenotype of the mutant strain lacking both components and the importance to perform all the experiments in this background. In addition, the contrasting results found for different substrates reveal the necessity to further investigate the functions of the αβ-NAC and αβ′-NAC and their relationship with other mitochondrial import components.
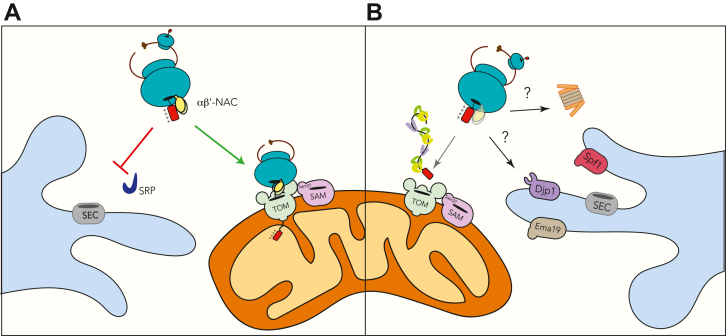
Based on our results and the available data in the literature, we propose that αβ′-NAC recognizes a patch of positive amino acids present in the MTS of a subset of mitochondrial proteins, and, in doing so, αβ′-NAC directs the translational machinery to the mitochondrial periphery (Fig. 9). The binding of αβ′-NAC to the ribosome avoids recognition of nascent polypeptides by other chaperones such as signal recognition particle and ensures a continuous and efficient flow of mitochondrial proteins to the organelle. It is well known that the import of proteins to the mitochondria is a robust process and, therefore, in the absence of αβ′-NAC, a protein harboring an MTS with a high hydrophobic moment and a cluster of positive residues could still reach the mitochondria with the help of other chaperones belonging to the HSP family, even at a slower rate. When protein import cannot be achieved, degradation systems come in place to eliminate nonimported substrates. Finally, failure to eliminate mitochondrial substrates or their mistargeting will initiate cellular stress aimed to restore protein import and cellular homeostasis.
Figure 9.
Final model.A, the αβ′-NAC mediates some mitochondrial protein import because of the recognition of a positive amino acid cluster present at the N terminus of precursors that are being actively translated. In addition, αβ′-NAC prevents recognition by other factors such as SRP that would lead to mistargeting to the endoplasmic reticulum. B, in the absence of αβ′-NAC, mitochondrial precursors are still able to arrive to mitochondria with the help of other chaperones such as Hsp70s and Hsp90s or other signals present in the mRNA. They can also be mistargeted to the endoplasmic reticulum, where some factors like Ema19, Djp1, or Spf1 may redirect the precursors to the mitochondria or toward its degradation. Hsp, heat shock protein; NAC, nascent polypeptide–associated complex; SRP, signal recognition particle.
Experimental procedures
Yeast strains and growth conditions
Strains were grown on standard laboratory conditions at 25 or 30 °C using rich yeast peptone (YP) media (yeast extract [1% w/v], peptone [2% w/v], and carbon source [2% v/v]); synthetic minimum media (yeast nitrogen base without amino acids and ammonium sulfate [0.17% w/v], (NH4)2SO4 [0.5% w/v], carbon source [2% v/v], adenine [0.002% v/v], uracil [0.002% v/v], l-tryptophan [0.002% v/v], l-histidine [0.002% v/v], l-leucine [0.01% v/v], l-lysine [0.003% v/v], and l-arginine [0.002% v/v]); or synthetic complete media (yeast nitrogen base without amino acids and ammonium sulfate [0.17% w/v], monosodic glutamic acid [0.1% w/v], drop out amino acid mix [0.2% w/v], and carbon source [2% v/v]). Glucose, galactose, glycerol, or raffinose was used as carbon sources. Drop out amino acid mix is adenine (3 g), uracil (2 g), inositol (2 g), p-aminobenzoic acid (0.2 g), l-alanine (2 g), l-arginine (2 g), l-aspartic acid (2 g), l-asparagine (2 g), l-glutamic acid (2 g), l-glutamine (2 g), l-glycine (2 g), l-histidine (2 g), l-isoleucine (2 g), l-leucine (10 g), l-lysine (2 g), l-methionine (2 g), l-phenylalanine (2 g), l-proline (2 g), l-serine (2 g), l-threonine (2 g), l-tryptophan (2 g), l-tyrosine (2 g), l-valine (2 g), and l-cysteine (2 g). The entire open reading frames of EGD2, BTT1, and SAM37 were deleted by homologous recombination with the resistance modules KanMX4 (pFa6A, (58)), nat1 (pAG25, (59)), or hph (pAG32, (59)). The following are the genotypes of the resulting strains: BY4741 (MATa hisΔ1, leu2Δ0, met15Δ0, ura3Δ0), Δsam37 (MATa hisΔ1, leu2Δ0, met15Δ0, ura3Δ0, sam37Δ0::KanMX4), Δαβ’ (MATa hisΔ1, leu2Δ0, met15Δ0, ura3Δ0, btt1Δ0::KanMX4, egd2Δ0::Hph), and Δαβ′Δsam37 (MATa hisΔ1, leu2Δ0, met15Δ0, ura3Δ0, sam37Δ0::KanMX4, egd2Δ0::Hph, btt1Δ0::Nat1).
Plasmids
All the plasmids were constructed using standard molecular biology techniques. pVT100u (60) was used as template for PCR amplification of GFP and of the MTS signal of NcSu9; yeast genomic DNA was used as template for amplifications of promoters, terminators, and the coding sequences of MTS signals and N terminus regions of Oxa1 and Mdm38. All PCRs were performed using Phusion High Fidelity DNA polymerase (New England Biolabs). The fragments were digested with the appropriate restriction enzymes (New England Biolabs) followed by ligation with T4 DNA ligase (New England Biolabs) into the vector pRS315 (61). pYES (Thermo Fisher Scientific) was used as template for amplification of GAL1 promoter using Phusion High Fidelity DNA polymerase (New England Biolabs), the fragment was digested with the appropriate restriction enzymes followed by ligation with T4 DNA ligase (New England Biolabs) into the vectors pOxa11–50-GFP and pMdm381–60-GFP replacing their endogenous promoters. The plasmids pMdm381–10Oxa111–50-GFP, pMdm381–20Oxa121–50-GFP, pMdm381–30Oxa131–50-GFP, pMdm381–40Oxa141–50-GFP, and pMdm381–50Oxa141–50-GFP were constructed by fusion PCR; a pair of primers were designed for amplification of the first region (promoter and Mdm381-X); and another pair of primers were designed for amplification of the second region (Oxa1X-50, GFP, and terminator) with a homology region of 15 bp. The fragment was digested with the appropriate restriction enzymes followed by ligation by T4 DNA ligase into the pRS315 vector. The plasmid pOxa11–10Mdm3811–60-GFP was constructed by fusion PCR; a pair of primers were designed for amplification of the first region (promoter and Oxa11–10); and an additional pair of primers were designed for amplification of the second region (Mdm3810–60, GFP, and terminator) with a homology region of 15 bp. The fragment was digested with the appropriate restriction enzymes followed by ligation by T4 DNA ligase into the pRS315 vector (61). FUM1 gene was amplified from genomic DNA, a second PCR was performed to add the 3HA tag at its 3′-end, and the fragments were digested with restriction enzymes followed by ligation with T4 DNA ligase into pRS316 vector (61). Single mutations on the plasmids pOxa1KRR/AAA-GFP and pFum1AAA-3HA were achieved by PCR using specific primers that included the mutations. Plasmid pBS34 (Yeast Resource Center, University of Washington; https://yeastrc.org/yeastrc/pages/pBS34.html) was used as template for amplification of mCherry, and the fragment was digested with restriction enzymes and inserted into the pYX122-mtGFP vector (60). EGD2 and BTT1 genes were amplified from genomic DNA, second PCR was performed to add the 3HA tag at their 3′-end, and the fragments were digested with restriction enzymes followed by ligation with T4 DNA ligase into pRS316 vector (61), where the GAL1 promoter had been previously cloned as described previously. To coexpress both genes, GAL1-Egd2-3HA was subcloned into pRS315. To delete the sequence corresponding to the first 11 amino acids of Btt1, a primer was designed to amplify the BTT1 coding sequence starting at nucleotide 33 (corresponding to amino acid 11) adding an AUG codon. All plasmids were verified by Sanger sequencing (Unidad de Biología Molecular, Instituto de Fisiología Celular, UNAM). Primers and plasmids used in this work are listed in the Supporting information section (Tables S2 and S3).
Whole protein cell extracts
Cells contained in an absorbance of 1.0 at 600 nm from a culture in exponential phase were harvested by centrifugation at 8600 relative centrifugal force (RCF) for 2 min at room temperature. After washing with distilled water, the cell pellet was resuspended in 250 μl of Tris–HCl 50 mM (pH 8.0) and lysed by adding 50 μl of a solution containing NaOH 1.85 N, β-mercaptoethanol 7.4%, and PMSF 20 mM. Proteins were precipitated by the addition of 12% trichloroacetic acid, washed with cold acetone, and resuspended in sample buffer (62).
Yeast growth test by serial dilutions
Cells contained in 0.5 ml of a culture in exponential phase with an absorbance of 1.0 at 600 nm were harvested by centrifugation at 8600 RCF and washed with distilled water before diluting them in a 1:10 ratio five sequential times. Drops of 3.5 μl were spotted onto appropriated fresh solid media as indicated.
Cell fractionation
Crude mitochondrial fractions were prepared essentially as described (63). Briefly, yeast strains were grown on synthetic medium with galactose as carbon source to an absorbance between 1.0 and 2.0 at 600 nm. Cells were pelleted and washed with distilled water. The cell pellet was resuspended and incubated in MP1 buffer (Tris 100 mm, DTT 10 mm, pH 11) for 10 min at 30 °C. Cells were pelleted and resuspended in MP2 buffer (sorbitol 1.2 M, phosphate buffer 20 mM, zymolyase 20T 3 mg/g wet weight [MP Biomedicals]) and incubated for 1 h with gentle shaking. The resulting spheroplasts were then pelleted and resuspended in homogenization buffer (0.6 M sorbitol, 10 mM Tris [pH 7.4], 1 mM EDTA, 0.2% [w/v] fatty acid–free bovine serum albumin, 1 mM PMSF, and broken with 30 strokes applied on a glass–glass Dounce homogenizer. Cell debris and nuclei were cleared by a first centrifugation at 3900 RCF for 5 min. Supernatant was then centrifuged at 16,000 RCF for 10 min. The mitochondrial pellet was resuspended in SH buffer (0.6 M sorbitol, 20 mM Hepes, pH 7.4). Mitochondrial (M) and postmitochondrial supernatant fractions were stored at −80 °C. The protein concentration in each fraction was determined using a modified Lowry method for membrane proteins (64).
PK assay
Crude mitochondria (5 μg) were diluted into 500 ml of SH buffer (sorbitol 0.6 M, Hepes 20 mM, pH 7.4) before adding PK (final concentration of 50 μg/ml) and, if indicated, Triton X-100 (final concentration of 1.5%). Samples were incubated on ice for 20 min before adding 2 mM of PMSF (Phenylmethylsulfonyl fluoride). Samples were recovered by centrifugation at 12,000 RCF for 10 min at 4 °C, washed with SH–KCl–PMSF buffer (sorbitol 0.6 M, Hepes 20 mM [pH 7.4], KCl 80 mM, and 2 mM PMSF), and resuspended in sample buffer (62).
Import assays in permeabilized cells
Permeabilized cells were prepared essentially as described previously (36). Briefly, strains were grown in complete synthetic media without uracil and using raffinose as carbon source (SRaf-Ura or SRaf-His) until they reached an absorbance of 1 at 600 nm. About 100 ml of each culture was harvested by centrifugation at 700 RCF for 7 min at room temperature. The cell pellet was resuspended in 25 ml of SP1 buffer (Tris 100 mM [pH 11] and DTT 10 mM) and incubated 10 min at 30 °C with gentle shaking. Cells were recovered by centrifugation at 1000 RCF for 5 min at room temperature and resuspended in 6 ml of SP2 buffer (sorbitol 0.6 M, 1× YP, glucose 0.2%, phosphate buffer 50 mM [pH 7.4], zymolyase 3–5 mg/g wet weight) and incubated 30 min at 30 °C with gentle shaking. Spheroplasts were harvested by centrifugation at 1000 RCF for 5 min at room temperature and carefully resuspended in 40 ml of SP3 buffer (sorbitol 0.7 M, YP 1×, and glucose 1%) and incubated for 20 min at 30 °C with gentle shaking, recovered by centrifugation at 1000 RCF for 5 min at 4 °C, and washed twice 20 ml of permeabilization buffer chilled on ice (Hepes [pH 6.8] 20 mM, potassium acetate 150 mM, magnesium acetate 2 mM, and sorbitol 0.4 M). The resultant pellet was resuspended in 1 ml of permeabilization buffer supplemented with 0.5 mM EGTA, aliquoted, frozen slowly by liquid nitrogen steam, and stored at −80 °C.
Import reactions were performed using an absorbance of at 600 nm of permeabilized cells in 100 μl of adjust B88 buffer (Hepes [pH 6.8] 20 mM, sorbitol 100 mM, potassium acetate 150 mM, magnesium acetate 8 mM, ATP 2 mM, NADH 2 mM, creatine phosphate 5 mM, creatine phosphatase 100 μg/ml, and galactose 2%). Samples were incubated at 30 °C for the indicated times before stopping the reaction by the addition of AVO (1.25 μM antimycin A, 80 μM valinomycin, and 0.65 mM oligomycin). Next, the samples were treated with or without PK to a final concentration of 750 μg/ml, and samples were treated as described previously. Samples were finally resuspended in 20 ml in sample buffer (62) and heated at 95 °C for 2 min.
Confocal microscope analysis
Strains were grown on complete synthetic media lacking leucine and/or histidine and using galactose as carbon source (SGal-Leu/His) at 25 °C until the cell suspension reached an absorbance of 1.0 at 600 nm. Cells contained in 1 ml of the cell suspension were recovered by centrifugation at 8600 RCF for 3 min at room temperature, washed with sterile distilled water, and resuspended in 100 μl of sterile distillated water. About 7 μl of the cell suspension was spotted on an agarose cushion (65) previously mounted on a microscope slide and gently covered with a coverslip. Samples were then analyzed with a confocal ZEISS LSM 800 microscope (Unidad de Imagenología, Instituto de Fisiología Celular, UNAM).
Miscellaneous
Protein samples were resolved by SDS-PAGE using 17.5% acrylamide/0.2% bis–acrylamide gels and transferred onto 0.2 μm nitrocellulose membranes (Amersham GE Healthcare).
For immunoblotting, primary antibodies were incubated overnight at 4 °C, washed once with Tris–HCl 50 mM (pH 7.5), NaCl 150 mM, and Tween 0.1% during 10 min and twice with Tris–HCl 50 mM (pH 7.5) and NaCl 150 mM during 10 min each. All the washing steps were performed at room temperature. Secondary antibodies were incubated at least 1 h and washed as described previously. Because of the close molecular weight between Fum1-3HA and Pgk1, these membranes were stripped with acid stripping buffer (glycine–HCl 0.1 M [pH 2.1], Tween-20 0.5% w/v, β-Mercaptoethanol 0.1 M; (66)) between the two immunodetections. Membranes were developed by chemiluminescence using Immobilon Western Chemiluminescent Horseradish Peroxidase Substrate (Millipore) and exposing them into films Biomax XAR Carestream (Kodak). The images were analyzed using ImageJ software (Wayne Rasband at the National Institutes of Health).
The following were the antibodies used in this study: α-GFP (1:2000 dilution; catalog no.: SC-9996; Santa Cruz Biotechnology, Inc), α-Hog1 (1:2000 dilution; catalog no.: SC-9079; Santa Cruz Biotechnology, Inc), α-Pgk1 (1:5000 dilution; catalog no.: 459250; Thermo Fisher Scientific), α-Tom20 (1:1000 dilution; Walter Neupert’s Lab), α-Ssc1 (1:5000 dilution; Walter Neupert’s Lab), α-Mdm38 (1:5000 dilution; Walter Neupert’s Lab), α-Tom40 (1:20,000 dilution; Walter Neupert’s Lab), α-Oxa1 (1:1000 dilution; Johannes M. Hermann’s Lab), α-HA (1:5000 dilution; catalog no.: 12013819001; MilliporeSigma), rabbit α-IgG (1:5000 dilution; catalog no.: 31461; Invitrogen), and mouse α-IgG (1:10,000 dilution; catalog no.: 715035150; Jackson ImmunoResearch Laboratories, Inc).
Unless otherwise specified, all experiments were performed at least three times. After quantification, Western blot data were analyzed using GraphPad Prism (GraphPad Software, Inc) with a Student's t test carried out. ∗∗p < 0.005, ∗p < 0.05, and #p = 0.05.
Colocalization analysis
The analysis of colocalization was evaluated using spatial correlation analysis by pixel intensity using ad hoc Phyton scripts with the scikit-image, numpy, pandas, and matplotlib libraries. Briefly, each image was processed through a gaussian blur, followed by a local equalization and background subtraction, then the most relevant pixels were selected by the intersection of the Otsu threshold masks of the colocalization channels. The Pearson's correlation coefficient was applied to the intersection pixels.
In silico analyses
The N terminus of each mitochondrial protein was analyzed by drawing a helical wheel diagram (pepwheel at EMBOSS explorer, https://www.bioinformatics.nl/cgi-bin/emboss/pepwheel), and the hydrophobicity, hydrophobic moment, and net charge were calculated using the HELIQUEST web server (67).
Data availability
All the data described in the article are located within the article and/or its supporting information.
Supporting information
This article contains supporting information (10, 11, 12, 13, 60, 61).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We extend our thank to Dr Benedikt Westermann for the kind gift of pVT100u-mtGFP, pYX122-mtGFP, and pYX223-mtGFP plasmids, to Walter Neupert and Johannes M. Herrmann for kindly providing the antibodies used in this study, to Janina Laborenz for her help with the import assays in permeabilized cells, and to Guadalupe Códiz Huerta and Laura Ongay Larios from Unidad de Biología Molecular (IFC, UNAM) for the technical support. We also thank all the staff at Instituto de Fisiología Celular whose work throughout the last months, despite the difficult circumstances, made it possible to complete this article. We specially thank Dr Imelda López-Villaseñor and Dr Diego González-Halphen for their helpful discussions throughout this project and critically reading this article.
Author contributions
M. C. A.-M., J. C. P.-R., and S. F. conceptualization; M. C. A.-M., A. E. M.-M., J. C. P.-R., A. C. P.-H., R. R.-H., and S. F. methodology; M. C. A.-M., J. C. P.-R., A. C. P.-H., and R. R.-H. formal analysis; M. C. A.-M. and A. E. M.-M. investigation; A. C. P.-H. and S. F. resources; A. C. P.-H. data curation; M. C. A.-M. and S. F. writing–original draft; M. C. A.-M., A. E. M.-M., J. C. P.-R., A. C. P.-H., R. R.-H., and S. F. writing–review & editing; M. C. A.-M., J. C. P.-R., and S. F. visualization; S. F. supervision; S. F. project administration; S. F. funding acquisition.
Funding and additional information
This work was supported by the Consejo Nacional de Ciencia y Tecnología, Mexico (CB 237344 and CF 58550; to S. F.) and the Dirección General de Asuntos del Personal Académico de la Universidad Nacional Autnoma de Mxico (DGAPA-UNAM; IN207518 and IN208921; to S. F.). M. C. A.-M. is a doctoral student from the Programa de Doctorado en Ciencias Bioquímicas, Universidad Nacional Autónoma de México, and was supported by fellowships/scholarships from Consejo Nacional de Ciencia y Tecnología, Mexico (464718) and from Ministerio de Ciencia, Tecnología e Innovación de Colombia (Convocatoria 885, Doctorados en el exterior). J. C. P.-R. was sponsored by the UC-MEXUS postdoctoral fellowship 2020.
Edited by Phyllis Hanson
Footnotes
Present address for José Carlos Ponce-Rojas: OmniVis Inc, South San Francisco, CA, USA.
Supporting information
References
- 1.Eme L., Spang A., Lombard J., Stairs C.W., Ettema T.J.G. Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 2017;15:711–723. doi: 10.1038/nrmicro.2017.133. [DOI] [PubMed] [Google Scholar]
- 2.Sagan L. On the origin of mitosing cells. 1967. J. NIH Res. 1993;5:65–72. [PubMed] [Google Scholar]
- 3.Chacinska A., Koehler C.M., Milenkovic D., Lithgow T., Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karakaidos P., Rampias T. Mitonuclear interactions in the maintenance of mitochondrial integrity. Life (Basel) 2020;10:173. doi: 10.3390/life10090173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avendano-Monsalve M.C., Ponce-Rojas J.C., Funes S. From cytosol to mitochondria: the beginning of a protein journey. Biol. Chem. 2020;401:645–661. doi: 10.1515/hsz-2020-0110. [DOI] [PubMed] [Google Scholar]
- 6.Bykov Y.S., Rapaport D., Herrmann J.M., Schuldiner M. Cytosolic events in the biogenesis of mitochondrial proteins. Trends Biochem. Sci. 2020;45:650–667. doi: 10.1016/j.tibs.2020.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Kellems R.E., Allison V.F., Butow R.A. Cytoplasmic type 80 S ribosomes associated with yeast mitochondria. II. Evidence for the association of cytoplasmic ribosomes with the outer mitochondrial membrane in situ. J. Biol. Chem. 1974;249:3297–3303. [PubMed] [Google Scholar]
- 8.Kellems R.E., Butow R.A. Cytoplasmic-type 80 S ribosomes associated with yeast mitochondria. I. Evidence for ribosome binding sites on yeast mitochondria. J. Biol. Chem. 1972;247:8043–8050. [PubMed] [Google Scholar]
- 9.Gold V.A., Chroscicki P., Bragoszewski P., Chacinska A. Visualization of cytosolic ribosomes on the surface of mitochondria by electron cryo-tomography. EMBO Rep. 2017;18:1786–1800. doi: 10.15252/embr.201744261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams C.C., Jan C.H., Weissman J.S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadir N., Haim-Vilmovsky L., Kraut-Cohen J., Gerst J.E. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA. 2011;17:1551–1565. doi: 10.1261/rna.2621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marc P., Margeot A., Devaux F., Blugeon C., Corral-Debrinski M., Jacq C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia M., Darzacq X., Delaveau T., Jourdren L., Singer R.H., Jacq C. Mitochondria-associated yeast mRNAs and the biogenesis of molecular complexes. Mol. Biol. Cell. 2007;18:362–368. doi: 10.1091/mbc.E06-09-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knox C., Sass E., Neupert W., Pines O. Import into mitochondria, folding and retrograde movement of fumarase in yeast. J. Biol. Chem. 1998;273:25587–25593. doi: 10.1074/jbc.273.40.25587. [DOI] [PubMed] [Google Scholar]
- 15.George R., Walsh P., Beddoe T., Lithgow T. The nascent polypeptide-associated complex (NAC) promotes interaction of ribosomes with the mitochondrial surface in vivo. FEBS Lett. 2002;516:213–216. doi: 10.1016/s0014-5793(02)02528-0. [DOI] [PubMed] [Google Scholar]
- 16.del Alamo M., Hogan D.J., Pechmann S., Albanese V., Brown P.O., Frydman J. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.George R., Beddoe T., Landl K., Lithgow T. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2296–2301. doi: 10.1073/pnas.95.5.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S., Sakai H., Wiedmann M. NAC covers ribosome-associated nascent chains thereby forming a protective environment for regions of nascent chains just emerging from the peptidyl transferase center. J. Cell Biol. 1995;130:519–528. doi: 10.1083/jcb.130.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wegrzyn R.D., Hofmann D., Merz F., Nikolay R., Rauch T., Graf C., et al. A conserved motif is prerequisite for the interaction of NAC with ribosomal protein L23 and nascent chains. J. Biol. Chem. 2006;281:2847–2857. doi: 10.1074/jbc.M511420200. [DOI] [PubMed] [Google Scholar]
- 20.Gamerdinger M., Kobayashi K., Wallisch A., Kreft S.G., Sailer C., Schlomer R., et al. Early scanning of nascent polypeptides inside the ribosomal tunnel by NAC. Mol. Cell. 2019;75:996–1006.e8. doi: 10.1016/j.molcel.2019.06.030. [DOI] [PubMed] [Google Scholar]
- 21.Wiedmann B., Sakai H., Davis T.A., Wiedmann M. A protein complex required for signal-sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- 22.Moller I., Jung M., Beatrix B., Levy R., Kreibich G., Zimmermann R., et al. A general mechanism for regulation of access to the translocon: competition for a membrane attachment site on ribosomes. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13425–13430. doi: 10.1073/pnas.95.23.13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gamerdinger M., Hanebuth M.A., Frickey T., Deuerling E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science. 2015;348:201–207. doi: 10.1126/science.aaa5335. [DOI] [PubMed] [Google Scholar]
- 24.Powers T., Walter P. The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr. Biol. 1996;6:331–338. doi: 10.1016/s0960-9822(02)00484-0. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Berndt U., Golz H., Tais A., Oellerer S., Wolfle T., et al. NAC functions as a modulator of SRP during the early steps of protein targeting to the endoplasmic reticulum. Mol. Biol. Cell. 2012;23:3027–3040. doi: 10.1091/mbc.E12-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lesnik C., Cohen Y., Atir-Lande A., Schuldiner M., Arava Y. OM14 is a mitochondrial receptor for cytosolic ribosomes that supports co-translational import into mitochondria. Nat. Commun. 2014;5:5711. doi: 10.1038/ncomms6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponce-Rojas J.C., Avendano-Monsalve M.C., Yanez-Falcon A.R., Jaimes-Miranda F., Garay E., Torres-Quiroz F., et al. αβ’-NAC cooperates with Sam37 to mediate early stages of mitochondrial protein import. FEBS J. 2017;284:814–830. doi: 10.1111/febs.14024. [DOI] [PubMed] [Google Scholar]
- 28.Deng J.M., Behringer R.R. An insertional mutation in the BTF3 transcription factor gene leads to an early postimplantation lethality in mice. Transgenic Res. 1995;4:264–269. doi: 10.1007/BF01969120. [DOI] [PubMed] [Google Scholar]
- 29.Markesich D.C., Gajewski K.M., Nazimiec M.E., Beckingham K. Bicaudal encodes the Drosophila beta NAC homolog, a component of the ribosomal translational machinery. Development. 2000;127:559–572. doi: 10.1242/dev.127.3.559. [DOI] [PubMed] [Google Scholar]
- 30.Bloss T.A., Witze E.S., Rothman J.H. Suppression of CED-3-independent apoptosis by mitochondrial betaNAC in Caenorhabditis elegans. Nature. 2003;424:1066–1071. doi: 10.1038/nature01920. [DOI] [PubMed] [Google Scholar]
- 31.Vogtle F.N., Wortelkamp S., Zahedi R.P., Becker D., Leidhold C., Gevaert K., et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 32.Herrmann J.M., Neupert W., Stuart R.A. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 1997;16:2217–2226. doi: 10.1093/emboj/16.9.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen K.G., Aviram N., Laborenz J., Bibi C., Meyer M., Spang A., et al. An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science. 2018;361:1118–1122. doi: 10.1126/science.aar8174. [DOI] [PubMed] [Google Scholar]
- 34.Xiao T., Shakya V.P., Hughes A.L. ER targeting of non-imported mitochondrial carrier proteins is dependent on the GET pathway. Life Sci. Alliance. 2021;4 doi: 10.26508/lsa.202000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shakya V.P., Barbeau W.A., Xiao T., Knutson C.S., Schuler M.H., Hughes A.L. A nuclear-based quality control pathway for non-imported mitochondrial proteins. Elife. 2021;10 doi: 10.7554/eLife.61230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laborenz J., Hansen K., Prescianotto-Baschong C., Spang A., Herrmann J.M. In vitro import experiments with semi-intact cells suggest a role of the Sec61 paralog Ssh1 in mitochondrial biogenesis. Biol. Chem. 2019;400:1229–1240. doi: 10.1515/hsz-2019-0196. [DOI] [PubMed] [Google Scholar]
- 37.Eisenberg D., Weiss R.M., Terwilliger T.C. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982;299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 38.Morgenstern M., Stiller S.B., Lubbert P., Peikert C.D., Dannenmaier S., Drepper F., et al. Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 2017;19:2836–2852. doi: 10.1016/j.celrep.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogtle F.N., Burkhart J.M., Gonczarowska-Jorge H., Kucukkose C., Taskin A.A., Kopczynski D., et al. Landscape of submitochondrial protein distribution. Nat. Commun. 2017;8:290. doi: 10.1038/s41467-017-00359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yogev O., Karniely S., Pines O. Translation-coupled translocation of yeast fumarase into mitochondria in vivo. J. Biol. Chem. 2007;282:29222–29229. doi: 10.1074/jbc.M704201200. [DOI] [PubMed] [Google Scholar]
- 41.Hsieh H.H., Lee J.H., Chandrasekar S., Shan S.O. A ribosome-associated chaperone enables substrate triage in a cotranslational protein targeting complex. Nat. Commun. 2020;11:5840. doi: 10.1038/s41467-020-19548-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franke J., Reimann B., Hartmann E., Kohlerl M., Wiedmann B. Evidence for a nuclear passage of nascent polypeptide-associated complex subunits in yeast. J. Cell Sci. 2001;114:2641–2648. doi: 10.1242/jcs.114.14.2641. [DOI] [PubMed] [Google Scholar]
- 43.Pech M., Spreter T., Beckmann R., Beatrix B. Dual binding mode of the nascent polypeptide-associated complex reveals a novel universal adapter site on the ribosome. J. Biol. Chem. 2010;285:19679–19687. doi: 10.1074/jbc.M109.092536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zabezhinsky D., Slobodin B., Rapaport D., Gerst J.E. An essential role for COPI in mRNA localization to mitochondria and mitochondrial function. Cell Rep. 2016;15:540–549. doi: 10.1016/j.celrep.2016.03.053. [DOI] [PubMed] [Google Scholar]
- 45.Weidberg H., Amon A. MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science. 2018;360 doi: 10.1126/science.aan4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boos F., Kramer L., Groh C., Jung F., Haberkant P., Stein F., et al. Mitochondrial protein-induced stress triggers a global adaptive transcriptional programme. Nat. Cell Biol. 2019;21:442–451. doi: 10.1038/s41556-019-0294-5. [DOI] [PubMed] [Google Scholar]
- 47.Wrobel L., Topf U., Bragoszewski P., Wiese S., Sztolsztener M.E., Oeljeklaus S., et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature. 2015;524:485–488. doi: 10.1038/nature14951. [DOI] [PubMed] [Google Scholar]
- 48.McKenna M.J., Sim S.I., Ordureau A., Wei L., Harper J.W., Shao S., et al. The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. Science. 2020;369 doi: 10.1126/science.abc5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laborenz J., Bykov Y.S., Knoringer K., Raschle M., Filker S., Prescianotto-Baschong C., et al. The ER protein Ema19 facilitates the degradation of nonimported mitochondrial precursor proteins. Mol. Biol. Cell. 2021;32:664–674. doi: 10.1091/mbc.E20-11-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rolland S.G., Schneid S., Schwarz M., Rackles E., Fischer C., Haeussler S., et al. Compromised mitochondrial protein import acts as a signal for UPR(mt) Cell Rep. 2019;28:1659–1669.e5. doi: 10.1016/j.celrep.2019.07.049. [DOI] [PubMed] [Google Scholar]
- 51.von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986;5:1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sylvestre J., Vialette S., Corral Debrinski M., Jacq C. Long mRNAs coding for yeast mitochondrial proteins of prokaryotic origin preferentially localize to the vicinity of mitochondria. Genome Biol. 2003;4:R44. doi: 10.1186/gb-2003-4-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ott A.K., Locher L., Koch M., Deuerling E. Functional dissection of the nascent polypeptide-associated complex in Saccharomyces cerevisiae. PLoS One. 2015;10 doi: 10.1371/journal.pone.0143457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gratzer S., Lithgow T., Bauer R.E., Lamping E., Paltauf F., Kohlwein S.D., et al. Mas37p, a novel receptor subunit for protein import into mitochondria. J. Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiedemann N., Kozjak V., Chacinska A., Schonfisch B., Rospert S., Ryan M.T., et al. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- 56.Qiu J., Wenz L.S., Zerbes R.M., Oeljeklaus S., Bohnert M., Stroud D.A., et al. Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell. 2013;154:596–608. doi: 10.1016/j.cell.2013.06.033. [DOI] [PubMed] [Google Scholar]
- 57.Wenz L.S., Ellenrieder L., Qiu J., Bohnert M., Zufall N., van der Laan M., et al. Sam37 is crucial for formation of the mitochondrial TOM-SAM supercomplex, thereby promoting β-barrel biogenesis. J. Cell Biol. 2015;210:1047–1054. doi: 10.1083/jcb.201504119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bahler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., 3rd, Steever A.B., et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 59.Goldstein A.L., McCusker J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 60.Westermann B., Neupert W. Mitochondria-targeted green fluorescent proteins: Convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 2000;16:1421–1427. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 61.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 63.Herrmann J.M., Fölsch H., Neupert W., Stuart R.A. In: Cell Biology: A Laboratory Handbook. Celis D.E., editor. Academic Press; San Diego: 1994. Isolation of yeast mitochondria and study of mitochondrial protein translation; pp. 538–544. [Google Scholar]
- 64.Markwell M.A., Haas S.M., Bieber L.L., Tolbert N.E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 65.Pemberton L.F. Preparation of yeast cells for live-cell imaging and indirect immunofluorescence. Methods Mol. Biol. 2014;1205:79–90. doi: 10.1007/978-1-4939-1363-3_6. [DOI] [PubMed] [Google Scholar]
- 66.Litovchick L. Stripping of the immunoblot for reprobing. Cold Spring Harb. Protoc. 2020;2020 doi: 10.1101/pdb.prot098491. [DOI] [PubMed] [Google Scholar]
- 67.Gautier R., Douguet D., Antonny B., Drin G. HELIQUEST: A web server to screen sequences with specific alpha-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data described in the article are located within the article and/or its supporting information.