Abstract
Gametogenesis, including spermatogenesis and oogenesis, are unique differentiation processes involving extraordinarily complex and precise regulatory mechanisms that require the interactions of multiple cell types, hormones, paracrine factors, genes and epigenetic regulators, and extensive chromatin 3D structure re-organization. In recent years, the development of 3D genome technology represented by Hi-C, enabled mapping of the 3D re-organization of chromosomes during zygogenesis at an unprecedented resolution. The 3D remodeling is achieved by folding chromatin into loops, topologically associating domains (TADs), and compartments (A and B), which ultimately affect transcriptional activity. In this review, we summarize the research progresses and findings on chromatin 3D structure changes during spermatogenesis and oogenesis.
Keywords: Spermatogenesis, Oogenesis, Chromatin 3D structure, Hi-C, TADs, Loops, Compartments
1. Introduction
Infertility is a growing health and social problem. Meiosis errors are very common in humans, and the frequency of chromosomal abnormalities is at least an order of magnitude higher than in other animals. The incidence of chromosomal abnormalities is about 6% in stillbirths, and 60% in spontaneous abortions[1]. Most chromosomal abnormalities are fatal, disappear early in embryonic development and manifest as infertility or spontaneous abortion. Detailed studies of spermatogenesis and oogenesis have led to better understanding and treatment of infertility.
Spermatogenesis and oogenesis refers to the differentiation and development process from germline stem cells to mature sperm and ova, including dozens of finely regulated developmental stages[2]. This process involves an important transformation of cell morphology and function, including DNA double strand breaking (DSB) and repair, synaptonemal complex (SC) formation, homologous chromosome separation, sperm acrosome and flagella formation and a series of landmark events. These events are often accompanied by dramatic changes in chromatin structure. Over the past 100 years, scientists have largely relied on the development of microscopy to explore chromatin spatial structure. As early as 1879, German biologist W. Fleming discovered that when cells divide, the scattered chromatin in the nucleus folds into highly condensed rods (later named chromosomes in 1888)[3]. Since then, chromosomal morphological changes have been widely reported. During the leptotene stage of spermatogenesis, telomeres combine with the nuclear membrane and assemble into bouquets. The bouquet stage was first observed in Helix Pomatia in 1885, and has since been observed occasionally in all meiotic organisms[4]. Synaptonemal complex (SC) was first independently observed in a number of mammals and an invertebrate by Fawcett and Moses in 1956 through a series of slides under electron microscopy[5], [6]. In 1977, Moses successfully observed the synaptic complex using a light microscope with the help of silver impregnation methods[7]. Under electron microscopy, SC is a ladder-like structure formed between homologous chromosomes[8]. There are lateral elements about 40 nm on both sides with high electron density, and a bright intermediate space about 100 nm wide in the middle, and a central element about 30 nm wide in the middle[9] (Fig. 1a). Between the lateral component and the central component, there are 7–10 nm SC fibers arranged horizontally, making the SC look like a ladder[10]. Morphologically, SC is formed in zygotene stage, matured in pachytene stage, exists for several days and disappears in diplotene stage. The formation of SC is related to DNA synthesis, and the addition of DNA synthesis inhibitors in leptotene stage can inhibit the formation of SC. In 1980, the Sperm Chromatin Structure Assay (SCSA) was first proposed by Evenson et al., and has been widely used to detect sperm DNA damage, evaluate male fertility and predict live birth rate via natural conception [11], [12] or in vitro fertilization and intracytoplasmic sperm injection (IVF/ICSI) and embryo transfer[13], [14]. The principle of the SCSA test is to treat sperm with pH 1.2 buffer to partially denature nuclear DNA at sites with single and double strand breaks, stain with acridine orange that distinguishes between native ds DNA (green fluorescence) and ss DNA (red fluorescence) and measure by flow cytometry. In the same year, Solari used microspreading technique to isolate the chromosomes of human pachytene spermatocytes, and accurately measured the length of each SC. In recent years, with the development of sequencing-based technologies, such as Hi-C (High-throughput whole-genome chromatin conformation capture), large new progress has been made in the study of dynamic changes of chromatin 3D structure during gametogenesis. In this review, we summarize the research progresses and findings on chromatin 3D structure changes during spermatogenesis and oogenesis.
Fig. 1.
Illustration of synaptonemal complex structure (a) and an overview of mitotic division of mouse spermatogonia (b).
2. Biological stages of spermatogenesis and oogenesis
With the help of microscopy and other imaging-based techniques, scientists have gradually broken down spermatogenesis into dozens of distinct stages. The spermatogonial cell is originally a single (As) spermatogonia, located in the basement membrane between the wall of the spermatogenic tubule and the sertoli cells. These As cells undergo some unscheduled, incomplete cytokinesis mitosis and form spermatogonial cells that are attached to each other in pairs (A paired, Ap) or chains (A aligned, Aal). Aal differentiated A1 spermatogonial cells were isolated in spermatogenic tubules and gradually entered a highly programmed process. A1 spermatogonial cells undergo 5 more mitosis to produce A2, A3, A4, intermediate (In), and finally B spermatogonial cells. After the last mitosis, B type spermatogonial cells differentiated into primary spermatogonial cells and entered the meiosis stage[15] (Fig. 1b). According to cytological characteristics, meiosis I can be substaged into leptonema (DNA replication is over, DSBs occur and recombination start), zygonema (DSBs repair and homologs synapsis initiation), pachynema (complete synapsis of homologs and DSBs repair is over) and subsequently diplonema (desynapsis and crossover) followed by homologous chromosome separation and sister chromatid separation in late meiosis I and II, respectively, resulting in haploid round spermatids (Fig. 2). Round spermatids undergo further extensive and complex cytological changes, including acrosome and flagellum formation, chromatin remodeling, and removal of residues, and eventually mature spermatozoa[15], [16]. During late spermatogenesis, the whole genome of haploid sperm cells gradually undergo a drastic chromatin remodeling, in which 85% of histone bound to DNA is replaced by protamine, a basic nuclear protein rich in arginine [17]. As a result, gene transcription stops, the genome becomes highly folded and the sperm nucleus becomes extremely compact, a process designed to ensure the safe, accurate and nondestructive transmission of DNA information from sperm to offspring.
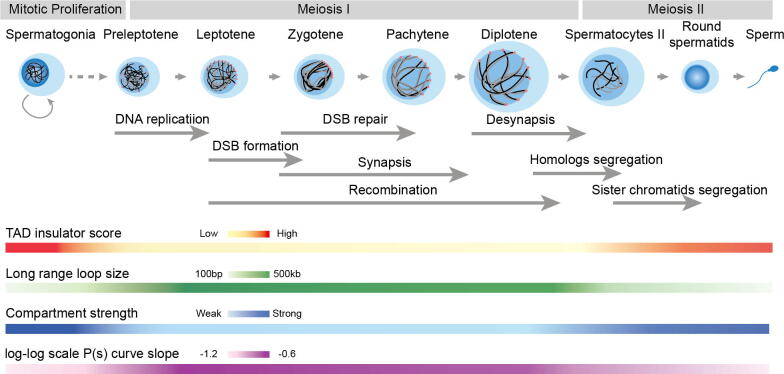
Fig. 2.
Overview of high-order chromosomal structure dynamics during mammalian spermatogenesis. The gray arrow in the middle marks the hallmark events during spermatogenesis. The colored bands at the bottom indicate the intensity of 3D chromatin structural features. TAD and compartment strengths are weakened or even disappeared during meiosis I and recovered right after the entry of meiosis II. (It should be noted that TAD insulator score color bar does not apply to humans, which, unlike those in mice, human sperm cells are reported not to have TADs). Long range loop arrays (>500 kb) are gradually formed during meiosis I, with the slope of log-log scale P (s) curve rising from −1.2 to −0.6.
Unlike spermatogenesis, which takes place continuously throughout lifetime in male individuals, the complete process of oogenesis only takes place for limited times in a female mammal's lifetime. The initiation of oogenesis takes place before birth. Primordial germ cells (PGC) are differentiated from the proximal epiblast and then migrate into the fetal gonad. Colonized PGCs in the female fetal gonad are called oogonia[18]. After vast number of times of mitosis, oogenia differentiate into oocytes and are committed into the prophase of meiosis I which, like that in spermatogenesis, contains preleptotene, leptotene, zygotene, pachytene, and diplotene[19], [20]. Oocytes get arrested in the stage of diplotene as they are surrounded by a layer of pre-granulosa cells to form primordial follicles. The follicles are then stored in the ovary after birth until the individual grows into adolescence. During that time, a small number of the oocytes gradually increase in size and acquire the competence of development through complex communications with the surrounding somatic cells in the follicles, like granulosa and theca-cells. Meanwhile, the follicles grow through primary, secondary and finally into the antral stage. The developed oocytes in antral follicles got a large nucleus termed germinal vesicle (GV) [21], [22], and were thus called GV oocytes or fully-grown oocytes (FGO). Once simulated by hormones, GV oocytes will resume the meiosis I process, and then got arrested again in the metaphase in meiosis II (MII), waiting for fertilization.
3. Chromatin 3D structure remodeling in spermatogenesis
Advances in sequencing based chromosomal conformation capture techniques, such as Hi-C, have enabled the global characterization of three-dimensional chromatin structures with higher resolution[23]. A/B compartmentalization and topologically associating domains (TADs) are two important hierarchical structural feature of chromatin organization at multi-megabase and hundreds of kilobases scale respectively[23], [24]. Histones in sperm are replaced by protamine, chromatin is highly compressed, and transcription is completely terminated[25]. The 3D chromatin structure changes from mature sperm to zygote have attracted extensive attention in different species. Previous work in mice and rhesus monkey have shown strong similarities between sperm and somatic cells. Compartment A/B and TADs are present among sperms, fibroblasts and mouse embryonic stem cells. At the same time, many enhancers and super-enhancers common in mature somatic cells are already present in sperms[26], [27], [28], [29]. Unlike mammals, zebrafish sperm chromatin is packaged by histones. 3D structure of chromatin in zebrafish sperm is largely different from somatic cells, lacks TADs and compartment A/B, instead displays aperiodic self-associating “hinge-like” chromosome domains of ∼150 kb that repeat every 1–2 Mbs. This “hinge” domain is highly correlated with H3K27ac modification[30]. Unlike mice and rhesus monkeys, human sperm cells have a strong compartment, but do not contain TADs and they do not express chromatin regulator CTCF. Sperm chromatin showed strong interactions at longer distances (>15 Mb), indicating dense packing[31].
During meiosis, chromatin 3D structure undergoes dramatic dynamic changes with the occurrence of landmark events such as homologous recombination of chromosomes. In yeast, Muller et al. find that the interactions between chromatin telomere and centromere regions of early-zygotene and early-pachynema cells change dynamically during meiosis. They found that, chromatins are arranged into arrays of Rec8-delimited loops of various sizes[32]. In recent years, several research groups have studied more different stages of spermatogenesis in mice and rhesus monkey. Alavattam et al. compared the 3D genomic structure changes of pachytene spermatocytes, round spermatids, mature spermatozoa and ESCs of C57BL/6J mice[33]. They reported the deterioration of compartment and TADs in pachytene spermatocytes. The power law of intra-chromosome interaction probability between spermatozoa and ESCs are P (s) – S−1. While, pachynema and round spermatids follow a power law of P (s) – S−0.61 within 3 Mb of the genome, and the interaction frequency decreases sharply at longer distances[33]. Histone modifications can affect the 3D structure of chromatin. For example, H3K27ac marked enhancers can form loops with other enhancers or promoters to promote distant gene expression. They observed that H3K27ac, H3K4me3, and H3K27me3 are enriched at the pachynema and round spermatids TAD boundary. In addition, they found strong X-shaped inter-chromosomal interactions between telomeres during meiosis, which is consistent with the bouquet phase[33]. Meanwhile, Patel et al. found a loss of TAD, but retained physically separated compartment A/B in the hybrid offspring of C57BL/6J and CAST/EiJ F1 mice[34]. Their data support the meiotic chromosome loop array model, in which zygonema and pachynema chromosomes were organized into a compact loop array and followed the internal power ratio of P (s) – S−0.5, inferring that loops average 0.8–1.0 Mb in zygonema and expand to 1.5–2.0 Mb in pachynema. They observed a transcriptionally active long distance (1–10 MB) central interaction in pachytene spermatocytes[34]. Vara et al. 's Hi-C results show that compartments A/B disappear from spermatogonia to leptotene, zygotene, pachytene and diplotene stage, and reestablish in round spermatids[35]. In P/D (mixture of pachytene and B diplotene), 80.7% of RAD21L peaks and 83.3% of REC8 peaks locate in promoter regions and nearly 80% of cohesin and CTCF peaks locate in A compartment. Genes with cohesin peaks in their promoter region have significantly higher expression than genes without peaks[35]. Luo et al. again demonstrate the disappearance of TADs and loop and the weakening of compartment in pachytene spermatocytes and round spermatids, while spermatocyte and As spermatogonia have similar compartment and TADs. In addition, they found no significant difference in chromatin accessibility between As spermatogonia and pachytene spermatocytes[36]. Chromatin accessibility declined significantly until mature spermatozoa. Wu et al. recently conducted a more detailed study of chromosomal 3D structural changes during sperm development. They accurately synchronized and isolated Sertoli, spermatogonia, preleptotene, leptotene, zygotene, pachytene, diplotene and meiosis II cells[37]. In general, the average contact probability P(s) is used to show the chromatin interaction frequency as a function of chromatin distance. Here, they inferred the size of chromosome loops size for all stages by inspecting the inflection point of the derivatives of log P(s) versus genomic distances curve. The size of meiotic chromatin loops increased from ∼500 kb in leptotene to ∼700 kb in zygotene stage, ∼1.4 Mb in pachytene and ∼1.6 Mb in diplotene. In addition, Compartment A and compartment B consist of shorter and longer chromatin loops (560 kb and 730 kb in leptotene, and 800 kb and 1.05 Mb in zygotene) respectively[37]. The force-transmitting LINC complex promotes alignment of different chromosomal ends up to 20% of the length of the chromosome, and this effect increases gradually from preleptotene to diplotene[37]. Similar to mice, in rhesus monkey, instead of the typical compartments and TADs, pachytene chromosomes contain smaller domains detected by local principal component analysis, named refined compartments, which are highly correlated with transcriptional level of the corresponding sequences within the compartments. Transcription inhibitor treatment has no effect on this refined A/B compartments. They found that TADs recovered in KO mice for Sycp2, a core component of the synaptic complex, or Top6bl, a topoisomerase that regulates DSB formation during spermatogenesis, suggesting that the synaptic complex may restricts conventional TADs and promotes the formation of refined A/B compartments[29].
4. 3D genomic and epigenome features on DSB hotspots and sex chromosomes during spermatogenesis
Meiosis recombination is initiated by DSB induced by SPO11[38]. In most mammals, histone methyltransferase PRDM9 guides the targeting of SPO11, and ATM kinases control the number of meiosis DSBs[39]. DSB repair occurs in zygotene and pachytene. RAD51 and DMC1 bind to chromatin during these periods, helping to relocate homologous chromosomes and form single-ended invasion exchange intermediates[40]. DSBs modification is characterized by the accumulation of specific chromatin modifications (γH2AX) and DDR factors (53BP1 and MDC1)[41]. It has been found that TADs are the basic functional unit of DSB repair in artificially induced DSB, and contributes to the correct establishment of γH2AX-53BP1 chromatin domain, which contains one-sided cohesin-mediated loop extrusion on both sides of the DSB[42]. DSB in sperm also corresponds to some characteristics of 3D chromatin structure. First, DSB and crossover are more likely to occur in compartment A[34]. The DSB hotspots are pre-opened in type A spermatogonia[36]; Using DMC1 single-stranded DNA ChIP data, they classified DSB into either crossover-favored or crossed-disfavored DSB hotspots (CO-DSBs and NCO-DSBs). In pachytene spermatocytes, chromatin interaction patterns change significantly in DSB regions, especially those associated with CO-DSBs, during homologous pairing and coarrangement in prophase I of meiosis. During preleptotene and Leptotene periods, CO-DSBs (but not NCO-DSBs) exhibits a transient, TAD-boundary-like pattern[37]. This pattern is also consistent with the loop extrusion model.
Unlike autosomes, homologous recombination occurs only in a short terminal region between XY sex chromosomes. During pachynema, the transcription of sex chromosomes stops, namely meiotic sex chromosome inactivation (MSCI)[43]. At this point, XY chromatin is reconstituted into heterochromatin, forming a specific and distinctly distinct region within the nucleus of mammalian spermatocytes, known as the XY body, or sex body. In recent years, Hi-C technology has led to a clearer understanding of the unique dynamic organization of 3D structure in sex chromosomes during meiosis I. In spermatogonia, mature sperm and fibroblasts, sex chromosomes are similar to autochromosomes and have strong compartment and TADs. But during the Meiosis I, and especially during pachytene, it almost disappears. During zygonema, the X chromosome behaves like the autosomes, showing strong interchromosomal interactions. By pachynema, X chromosome displays a 3D organization distinct from the transcriptionally active autosomes, completely losing the X-shaped interchromosomal contact that had always been observed in autosomes. There is no transcriptionally active distal interaction hub in the X chromosome, and X chromosome shows much stronger distal interactions beyond 10 Mb than autosomes[29], [33], [34], [37].
5. Chromosomal morphodynamics during oogenesis
Comparing to spermatocytes, the rate of oocytes to be aneuploid is higher due to the lack of cell cycle checkpoint mechanisms, and aging contributes much to the risk of maternal aneuploidy [44], [45]. Therefore, the chromosomal behavior during oogenesis have long been noticed. Due to the limited number of growing oocytes, it is harder to get abundant cells for 3D genome sequencing. Rather, image-based methods are more suitable in such case. The synapsis and recombination of homologs have been visualized by light and electron microscope[46], [47], [48], and in combination with genetic studies, some of the molecular mechanisms have been uncovered. Like spermatocytes, the chromatin in meiosis I (MI) oocytes becomes linearly-arranged loop arrays anchored upon meiotic chromosome axis, which is composed of a cohesin core and synaptonemal complex (SC). There are more than one type of SCs in oocytes, and some of the subunits of SC, including REC8, RAD21L, STAG3 and SMC1β, are specific in meiotic process [44]. Inactivation of SMC1β in oocytes will cause the shortening of chromosome axis and the increase of loop size, suggesting the crucial roles of SC complex components to regulate the genome structure[49].
Of notice, the structure changes of chromatin inside GV are unique in oocytes and are studied by many researches on various species[21]. Two remarkable configurations are described. One is called non-surrounded nucleolus (NSN), where the chromosomes are loosely distributed inside the whole nucleus. The other is called surrounded nucleolus (SN). In this type of GV oocyte, the chromosomes are condensed and re-arranged to surround the nucleolus-like body (a oocyte nuclear organelle derived from nucleolus[50]), thus forming a ring-shaped structure, which was named karyosphere by Blackman in early 1900s[51]. This structure is conserved among many species, though they are given different names in the earlier studies[50]. Both NSN and SN oocytes can proceed to the next stage and complete meiosis I, but SN oocytes are suggested to be more mature than NSN oocytes, with higher successful rate of finishing meiosis II and fertilization[52], [53], [54]. Consistently, the gene transcription is mostly active in NSN oocytes and silent in SN oocytes, but factors essential for embryo development are up-regulated in SN oocytes[55], probably making it more suitable for development. In aged mice, the typical SN and NSN configurations are partially replaced by other irregular patterns, such as clumped inside the nuclear[56]. That might be a cause of the declining of fertilization with age.
In healthy individuals, GV oocytes are gradually switching from NSN to SN state. This transition of chromosomal structure is associated with a lot of molecular behavior and functions. Centromere-specific immunochemical imaging[57], as well as a FISH study on satellite repeats[58] show that the centromeric heterochromatin was re-distributed to the close region of nucleolus-like body during NSN-SN switch. That might be led by the binding between the rDNA inside the nucleolus-like body and major satellite on the chromatin. Histone modifications also take parts in NSN-SN transition. H3K9me3 and H4K20me3 were found by FISH method to ocondensate on pericentromere chromatins and contribute to the condensation of heterochromatin and the formation of nucleolus-like body[58] (Fig. 3).
Fig. 3.
Illustration of the nuclear organization and related epigenome marks inside the non-surrounded nucleolus (NSN) and surrounded nucleolus (SN) germinal vesicles.
Cell-cell communication can also promote NSN-SN transition. Oocytes communicate with nearby somatic cells through gap junction. When stimulated by sex hormones, the gap junctions between oocyte and cumulus cells will promote NSN-SN transition[59], while interrupting the communication by knocking out related proteins will obstruct the condensation of chromatins and gene silencing[60]. The cAMP mechanism was found to be related to the regulation of cell communication and chromatin structure regulation[61], [62]. Both oocytes and cumulus cells produce cAMP, and gap junction helps to balance cAMP level[63]. Inversely, cAMP level will also regulate the throughput of gap junction by inactivating the MAPK protein, which will also influence other sex hormones like FSH, and stimulate the formation of karyosphere[64].
6. Sequencing-based 3D genome mapping during oogenesis
In the past few years, the optimization of Hi-C technique, either lowering the input of sequencing[27], [65], or enhancing the resolution on single cell level[66], [67], along with the respective algorithm updates, promote the study of chromatin structure in different stages of oogenesis on the whole genome. Currently, the sequencing-based chromosomal structure analysis was mostly focused upon the GV and MII stages[27], [66], [68], [69], and a recent work of Du et al.[65] systematically mapped six major time points during the whole process of oogenesis, from progenitor germ cell (PGC) to fully grown oocytes. Those researches give rise to our knowledge about 3D genome structure dynamics during that process.
At the beginning of oogenesis, the genome structure of PGC is similar to mammalian somatic cells, with some local difference. When comparing to mouse ESC cells, the long range (>50 M) contact probability of PGC is increased gradually in both male and female embryo. This feature was lost when the PGCs are differentiated into oocytes and arrested at diplotene. The long-range contacts gradually disappear comparing to somatic cells on post-natal day 7 to day 14, when the oocytes are arrested at diplotene and start growing in size. On the contrary, the contact between 1 and 10 Mb of distance are increased at that stage, suggesting an increase of high-order structure at shorter range. This pattern is conserved in the fully-grown GV oocytes[65]. When separating the two configurations of GV oocytes, the SN type have relatively higher long-range interactions and lower cell-cell diversity comparing to NSN type[66], consistent with the fact that the chromatins are more condensed and stiff in SN oocytes. When the oocyte is released from diplotene and come to the metaphase of meiosis II, the short-range (1–10 M) interaction become stronger than all the previous stages and the power-law exponent of P(s) curve is about −0.5[27], similar to the metaphase in mitosis[70] (Fig. 4).
Fig. 4.
Overview of high-order chromosomal structure dynamics during mammalian oogenesis. The gray arrow in the middle marks the hallmark chromosomal behaviors during oogenesis. The colored bands at the bottom indicate the intensity of 3D chromatin structural features. TADs and compartment strengths are weakened during the whole process, and almost totally disappeared at the metaphase of meiosis II, while the PAD structures are gradually established as the oocytes growing in size inside the follicles and reach a peak of strength in both NSN and SN GV oocytes before fading away at GVBD. The slope of log-log scale P(s) curve in meiosis II oocytes is increased to about −0.5 within 3Mbp.
The high-order genome structures show very special patterns during oogenesis. Both the strength and distribution of A/B compartments on PGCs are similar to mESC. Same similarity was shown on TADs, supporting that PGCs are still somewhat like somatic stem cells in biological morphology and functions[65]. In oocytes, the compartments and TAD boundaries begin to melt. Fully grown oocytes at diplotene arrest have strongly weakened the compartmentalization and TAD boundary[65], which was similar with spermatocytes at mid-prophase of meiosis I[71], [72]. Single-cell analysis found that the TADs identified in contact map are not truly from the physically isolated DNA clutches, but rather an enrichment of interaction tendency among cell population[66]. SN type have even weaker TAD and compartmentalization comparing to NSN[66]. WAPL-mediated release of SCC1 take some part in the melting of high-order structure in meiosis. SCC1 was thought to be replaced by REC8 for its function of forming linear loop arrays on chromosomes in meiosis[73], but Silva, at al. [68] demonstrated that the binding of SCC1 on chromatin may play some alternative roles in oogenesis, like the formation of some other high-order structures. Knocking out SCC1 leads to further elimination of some loops and TADs in GV oocytes, while knocking out WAPL will increase the time of SCC1 binding and cause a little increase of compartment. SCC1 can also regulate loop extrusion and, if not released at proper time, form small loops inside the REC8-formed loop arrays in the GV oocytes[68]. Given that WAPL knock-out or hyper-phosphorylation [74] also causes the accumulation of other SC subunits, like SMC3 and SMC1β, thus forming a 'vermicelli ' structure, the mechanism of WAPL regulation on cohesin and genome structure might be more complex in oocytes. Chiasmata number on genome also increase after WAPL deletion[68], suggesting a higher risk of segregation error might relate to the abnormal pattern of high-order structure. The above studies suggest that the dynamic of high-order genome structures in GV probably contributes much to the biological functions specific for oocyte development.
As canonical compartment and TAD are strongly weakened, a special local compartment-like pattern initiates in diplotene-arrest stage and becomes clearly established in fully-grown GV oocytes. This high-order structure is marked by H3K27me3, a repressive mark catalyzed the polycomb complex, and was thus named polycomb-associated domains (PADs) by Du et al[65]. PADs are very commonly found in GV genome and their distributions are neither associated with GC level nor transcriptional region, unlike canonical compartments. The emergence of PADs coincident with the dynamics of other high-order structures. Those TADs whose boundaries are overlapped with PAD boundaries will be strengthened in GV oocytes, while others will be weakened or eliminated. Re-analysis of single cell Hi-C found PADs in both NSN and SN oocytes, suggesting that the whole genome-wide chromatin condensation and transcriptional silence do not take much part in the local high-order structure[65], [66]. Consistently, artificially silencing the transcription of growing oocytes did not block the emergence of PADs[65]. The exact forming mechanism and biological function of PADs are still a mystery. Nevertheless, polycomb is certain to play crucial roles in the formation of PADs. Especially, PRC1 is important in PAD formation, enhancing PAD-PAD interaction and repressing gene expressions inside PAD regions[65]. PAD structure is found disappeared when oocytes are released from diplotene arrest and surprisingly reconstructed in the maternal genome of two-cell stage in early embryo. And another polycomb protein, PRC2, does not contribute much on the formation of PADs, but is crucial for the re-construction of PADs in 2-cell embryos[65].
The later part of meiosis II, during which sister chromatids segregate in oocytes, happens abruptly almost at the same time as fertilization, making it harder to get pure haploid ova for the study on genome structure. Alternatively, the genome structure of oocytes arrested in MII mid-term stage was often studied, and compared with genome structures of sperm and zygotes in the field of early development[27], [69]. The MII oocytes almost eliminated all high-order structures despite the linear loop arrays arranged on chromosomal axis for the proper segregation of sister chromatids. TAD insulation score is the lowest in MII oocytes comparing with sperms and early embryos[27], suggesting the chromosomes are loosely constructed inside the nucleus in maternal gamates while tightly compacted in paternal gamates. Current proofs indicate that the maternal genome structures in early embryo are arranged de novo instead of directly inherited from oocytes. However, there might be some epigenome markers reserved in MII oocytes, which can record the structure of maternal genome for re-construction in early development, like histone modifications. H3K27me3 level is reserved during MII stage, and its distribution is strongly enriched in the loci that forms allele-specific domains in the early embryos [69]. The reconstruction of PADs may also be located by the H3K27me3 distribution[65]. In addition, there are broad peaks of H3K4me3 in MII oocytes, which cover the genome regions mostly outside PADs[65]. Those regions are hypo-methylated and untranscribed, forming genome domains that are called partially methylated domains (PMDs)[21], [75], which might contribute to the regulation on gene expression inside the domains. H3K4me3 modified genome regions are also reported to become enriched in A compartment in early embryo[27]. However, there is no further evidence that H3K4me3 is involved in genome construction during early development.
In the follicle, the cell-cell interaction between oocytes and adjacent somatic cells is crucial to promote oogenesis. A very recent study by Li, et al.[76] investigated the 3D genome structure dynamics of chicken granulosa cells across 10 stages of oogenesis. Their results show minor changes of compartmentalization and relatively stable TAD distribution during the process, and the genome regions with switched configuration are always related to hormone activity and signaling processes crucial for follicle maturation. The respective study on mammals is still missing, and we suggest that the comprehensive study of transcriptome, 3D genome and epigenome on both oocytes and adjacent somatic cells might be interesting and provide more insights on the complex process of oogenesis.
7. Summary
In conclusion, current studies drew a unique map of genome structure organization and dynamics during the process of spermatogenesis and oogenesis. At different stages, the chromosomal structures of germ cells would change, performing various functions like regulating global or local gene expression, ensuring the recombination, or preparing for the genome reconstruction in the coming early embryo development. Some structures in germ cells are like somatic cells, either interphase or mitotic metaphase, while others are special during gamate production, like the karyosphere structure and PADs patterns in oocytes. Further studies are still needed for their mechanism and functions in both 3D genome and epigenome fields. Nevertheless, due to the imbalanced time scale on different stages of gamate production, some crucial time points during the whole dynamic processes, like the detailed mitotic-to-meiotic differentiation step from PGCs, are still missing for 3D genome mapping. Other obstacles in technology, like the synchronization and sorting of meiosis I prophase oocytes in fetal ovary, also wait for future advances to overcome. Cell-cell communication might be another interesting direction for studying the regulation of germ cell 3D genome structures. Also, the changes of the 3D genome structure with age will be essential to decipher the age-related fertility competence declining and birth defects[44], empowering human beings to tackle the increasingly urgent reproductive crisis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by grants from China Ministry of Science and Technology (2020YFA0804000), National Natural Science Foundation of China (92049302, 32088101), and Shanghai Municipal Science and Technology Major Project (2017SHZDZX01) to J.D.J.H.
References
- 1.Martin R.H. Meiotic errors in human oogenesis and spermatogenesis. Reproductive Biomedicine Online. 2008;16:523–531. doi: 10.1016/s1472-6483(10)60459-2. [DOI] [PubMed] [Google Scholar]
- 2.Clermont Y. Kinetics of Spermatogenesis in Mammals – Seminiferous Epithelium Cycle and Spermatogonial Renewal. Physiological Reviews, (1972), 52: 198-&. [DOI] [PubMed]
- 3.Paweletz N. Walther Flemming: pioneer of mitosis research (Reprinted from Nature Reviews Molecular Cell Biology, vol 2, pg 72–75, 2001) Nat Rev Mol Cell Biol. 2001:S32–S35. doi: 10.1038/35048077. [DOI] [PubMed] [Google Scholar]
- 4.Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Biol. 2001;2:621–627. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- 5.Fawcett D W. The Fine Structure of Chromosomes in the Meiotic Prophase of Vertebrate Spermatocytes. Journal of Biophysical and Biochemical Cytology, (1956), 2: 403-&. [DOI] [PMC free article] [PubMed]
- 6.Moses M.J. Chromosomal Structures in Crayfish Spermatocytes. Journal of Biophysical and Biochemical Cytology. 1956;2:215-+. doi: 10.1083/jcb.2.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moses M.J., Russell L.B., Cacheiro N.L.A. Mouse Chromosome Translocations – Visualization and Analysis by Electron-Microscopy of Synaptonemal Complex. Science. 1977;196:892–894. doi: 10.1126/science.870966. [DOI] [PubMed] [Google Scholar]
- 8.Moses M J. Structure and Function of Synaptonemal Complex. Genetics, (1969), 61: 41-&. [PubMed]
- 9.Bojko M. Synaptic Adjustment of Inversion Loops in Neurospora-Crassa. Genetics. 1990;124:593–598. doi: 10.1093/genetics/124.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heyting C. Synaptonemal complexes: Structure and function. Curr Opin Cell Biol. 1996;8:389–396. doi: 10.1016/s0955-0674(96)80015-9. [DOI] [PubMed] [Google Scholar]
- 11.Evenson D.P., Darzynkiewicz Z., Melamed M.R. Relation of mammalian sperm chromatin heterogeneity to fertility. Science. 1980;210:1131–1133. doi: 10.1126/science.7444440. [DOI] [PubMed] [Google Scholar]
- 12.Evenson D.P., Jost L.K., Marshall D., et al. Utility of the sperm chromatin structure assay as a diagnostic and prognostic tool in the human fertility clinic. Hum Reprod. 1999;14:1039–1049. doi: 10.1093/humrep/14.4.1039. [DOI] [PubMed] [Google Scholar]
- 13.Bungum M., Bungum L., Giwercman A. Sperm chromatin structure assay (SCSA): a tool in diagnosis and treatment of infertility. Asian journal of andrology. 2011;13:69–75. doi: 10.1038/aja.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evenson D.P., Djira G., Kasperson K., et al. Relationships between the age of 25,445 men attending infertility clinics and sperm chromatin structure assay (SCSA®) defined sperm DNA and chromatin integrity. Fertil Steril. 2020;114:311–320. doi: 10.1016/j.fertnstert.2020.03.028. [DOI] [PubMed] [Google Scholar]
- 15.Lin Z., Tong M.H. m(6)A mRNA modification regulates mammalian spermatogenesis. Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms. 2019;1862:403–411. doi: 10.1016/j.bbagrm.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 16.Hunter N. Meiotic Recombination: The Essence of Heredity. Cold Spring Harbor Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon G.H., Aiken J.M., Jankowski J.M., et al. In: Chromosomal Proteins and Gene Expression. Reeck G.R., Goodwin G.H., Puigdomènech P., editors. Springer; US, Boston, MA: 1985. Organization and Evolution of the Protamine Genes of Salmonid Fishes; pp. 287–314. [Google Scholar]
- 18.Oktem O., Urman B. Understanding follicle growth in vivo. Hum Reprod. 2010;25:2944–2954. doi: 10.1093/humrep/deq275. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Pepling M E. Regulation of Meiotic Prophase One in Mammalian Oocytes. Frontiers in Cell and Developmental Biology, (2021), 9: 667306-667306. [DOI] [PMC free article] [PubMed]
- 20.Borum K. Oogenesis in the mouse: A study of the meiotic prophase. Exp Cell Res. 1961;24:495–507. doi: 10.1016/0014-4827(61)90449-9. [DOI] [PubMed] [Google Scholar]
- 21.Bogolyubova I., Bogolyubov D. Heterochromatin Morphodynamics in Late Oogenesis and Early Embryogenesis of Mammals. Cells. 2020;9:1497. doi: 10.3390/cells9061497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mihajlovic A.I., Fitzharris G. Segregating Chromosomes in the Mammalian Oocyte. Curr Biol. 2018;28:R895–R907. doi: 10.1016/j.cub.2018.06.057. [DOI] [PubMed] [Google Scholar]
- 23.Lieberman-Aiden E., Van Berkum N.L., Williams L., et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixon J.R., Selvaraj S., Yue F., et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammoud S.S., Nix D.A., Zhang H.Y., et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–U447. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Battulin N., Fishman V.S., Mazur A.M., et al. Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach. Genome Biol. 2015;16:77. doi: 10.1186/s13059-015-0642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ke Y., Xu Y., Chen X., et al. 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell. 2017;170(367–381):e320. doi: 10.1016/j.cell.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 28.Du Z., Zheng H., Huang B., et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature. 2017;547:232–235. doi: 10.1038/nature23263. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., Wang H., Zhang Y., et al. Reprogramming of Meiotic Chromatin Architecture during Spermatogenesis. Mol Cell. 2019;73(547–561):e546. doi: 10.1016/j.molcel.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 30.Wike C.L., Guo Y., Tan M., et al. Chromatin architecture transitions from zebrafish sperm through early embryogenesis. Genome Res. 2021;31:981–994. doi: 10.1101/gr.269860.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X., Ke Y., Wu K., et al. Key role for CTCF in establishing chromatin structure in human embryos. Nature. 2019;576:306–310. doi: 10.1038/s41586-019-1812-0. [DOI] [PubMed] [Google Scholar]
- 32.Muller H., Scolari V.F., Agier N., et al. Characterizing meiotic chromosomes' structure and pairing using a designer sequence optimized for Hi-C. Mol Syst Biol. 2018;14:e8293. doi: 10.15252/msb.20188293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alavattam K.G., Maezawa S., Sakashita A., et al. Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development. Nat Struct Mol Biol. 2019;26:175–184. doi: 10.1038/s41594-019-0189-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel L., Kang R., Rosenberg S.C., et al. Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase. Nat Struct Mol Biol. 2019;26:164–174. doi: 10.1038/s41594-019-0187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vara C., Paytuvi-Gallart A., Cuartero Y., et al. Three-Dimensional Genomic Structure and Cohesin Occupancy Correlate with Transcriptional Activity during Spermatogenesis. Cell Rep. 2019;28(352–367):e359. doi: 10.1016/j.celrep.2019.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo Z., Wang X., Jiang H., et al. Reorganized 3D Genome Structures Support Transcriptional Regulation in Mouse Spermatogenesis. iScience. 2020;23 doi: 10.1016/j.isci.2020.101034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuo W., Chen G., Gao Z., et al. Stage-resolved Hi-C analyses reveal meiotic chromosome organizational features influencing homolog alignment. Nat Commun. 2021;12:5827. doi: 10.1038/s41467-021-26033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romanienko P.J., Camerini-Otero R.D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 39.Lange J., Yamada S., Tischfield S.E., et al. The Landscape of Mouse Meiotic Double-Strand Break Formation, Processing, and Repair. Cell. 2016;167:695–708.e616. doi: 10.1016/j.cell.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinohara M., Gasior S.L., Bishop D.K., et al. Tid1/Rdh54 promotes colocalization of Rad51 and Dmc1 during meiotic recombination. PNAS. 2000;97:10814–10819. doi: 10.1073/pnas.97.20.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stucki M., Jackson S.P. gamma H2AX and MDC1: Anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair. 2006;5:534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 42.Arnould C., Rocher V., Finoux A.L., et al. Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature. 2021;590:660–665. doi: 10.1038/s41586-021-03193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner J.M.A. Meiotic sex chromosome inactivation Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- 44.Lee J. Is age-related increase of chromosome segregation errors in mammalian oocytes caused by cohesin deterioration? Reproductive medicine and biology. 2019;19:32–41. doi: 10.1002/rmb2.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herbert M., Kalleas D., Cooney D., et al. Meiosis and maternal aging: insights from aneuploid oocytes and trisomy births. Cold Spring Harbor Perspect Biol. 2015;7:a017970–a. doi: 10.1101/cshperspect.a017970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Speed R.M. The prophase stages in human foetal oocytes studied by light and electron microscopy. Hum Genet. 1985;69:69–75. doi: 10.1007/BF00295532. [DOI] [PubMed] [Google Scholar]
- 47.Bojko M. Human meiosis VIII. Chromosome pairing and formation of the synaptonemal complex in oocytes. Carlsberg Research. Communications. 1983;48:457. [Google Scholar]
- 48.Moses M J, Poorman P A. Synapsis, synaptic adjustment and DNA synthesis in mouse oocytes. In: M.D. Bennett, A. Gropp, U. Wolf (Eds.) Chromosomes Today: Volume 8 Proceedings of the Eighth International Chromosome Conference held in Lübeck, West Germnay. 21–24 September 1983. Springer Netherlands, Dordrecht. 1984. 90-103.
- 49.Bolcun-Filas E., Schimenti J.C. In: International Review of Cell and Molecular Biology. Jeon K.W., editor. Academic Press; 2012. Chapter Five – Genetics of Meiosis and Recombination in Mice; pp. 179–227. [DOI] [PubMed] [Google Scholar]
- 50.Bogolyubov D.S. In: International Review of Cell and Molecular Biology. Galluzzi L., editor. Academic Press; 2018. Chapter One – Karyosphere (Karyosome): A Peculiar Structure of the Oocyte Nucleus; pp. 1–48. [DOI] [PubMed] [Google Scholar]
- 51.Blackman M.W. The Spermatogenesis of the myriapods.—II. On the chromatin in the spermatocytes of scolopendra heros. The Biological Bulletin. 1903;5:187–217. [Google Scholar]
- 52.Zuccotti M., Giorgi Rossi P., Martinez A., et al. Meiotic and Developmental Competence of Mouse Antral Oocytes1. Biol Reprod. 1998;58:700–704. doi: 10.1095/biolreprod58.3.700. [DOI] [PubMed] [Google Scholar]
- 53.Schramm R.D., Tennier M.T., Boatman D.E., et al. Chromatin configurations and meiotic competence of oocytes are related to follicular diameter in nonstimulated rhesus monkeys. Biol Reprod. 1993;48:349–356. doi: 10.1095/biolreprod48.2.349. [DOI] [PubMed] [Google Scholar]
- 54.Hinrichs K. The equine oocyte: Factors affecting meiotic and developmental competence. Mol Reprod Dev. 2010;77:651–661. doi: 10.1002/mrd.21186. [DOI] [PubMed] [Google Scholar]
- 55.Ma J.-Y., Li M., Luo Y.-B., et al. Maternal factors required for oocyte developmental competence in mice: Transcriptome analysis of non-surrounded nucleolus (NSN) and surrounded nucleolus (SN) oocytes. Cell cycle (Georgetown, Tex) 2013;12:1928–1938. doi: 10.4161/cc.24991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manosalva I., González A. Aging changes the chromatin configuration and histone methylation of mouse oocytes at germinal vesicle stage. Theriogenology. 2010;74:1539–1547. doi: 10.1016/j.theriogenology.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 57.Longo F., Garagna S., Merico V., et al. Nuclear localization of NORs and centromeres in mouse oocytes during folliculogenesis. Mol Reprod Dev. 2003;66:279–290. doi: 10.1002/mrd.10354. [DOI] [PubMed] [Google Scholar]
- 58.Bonnet-Garnier A., Feuerstein P., Chebrout M., et al. Genome organization and epigenetic marks in mouse germinal vesicle oocytes. Int J Dev Biol. 2012;56:877–887. doi: 10.1387/ijdb.120149ab. [DOI] [PubMed] [Google Scholar]
- 59.De La Fuente R., Eppig J.J. Transcriptional Activity of the Mouse Oocyte Genome: Companion Granulosa Cells Modulate Transcription and Chromatin Remodeling. Dev Biol. 2001;229:224–236. doi: 10.1006/dbio.2000.9947. [DOI] [PubMed] [Google Scholar]
- 60.Carabatsos M.J., Sellitto C., Goodenough D.A., et al. Oocyte-Granulosa Cell Heterologous Gap Junctions Are Required for the Coordination of Nuclear and Cytoplasmic Meiotic Competence. Dev Biol. 2000;226:167–179. doi: 10.1006/dbio.2000.9863. [DOI] [PubMed] [Google Scholar]
- 61.Luciano A.M., Franciosi F., Dieci C., et al. Changes in large-scale chromatin structure and function during oogenesis: a journey in company with follicular cells. Anim Reprod Sci. 2014;149:3–10. doi: 10.1016/j.anireprosci.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 62.Luciano A.M., Franciosi F., Modina S.C., et al. Gap junction-mediated communications regulate chromatin remodeling during bovine oocyte growth and differentiation through cAMP-dependent mechanism(s) Biol Reprod. 2011;85:1252–1259. doi: 10.1095/biolreprod.111.092858. [DOI] [PubMed] [Google Scholar]
- 63.Anderson E., Albertini D.F. Gap junctions between the oocyte and companion follicle cells in the mammalian ovary. The Journal of cell biology. 1976;71:680–686. doi: 10.1083/jcb.71.2.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun M.J., Zhu S., Li Y.W., et al. An essential role for the intra-oocyte MAPK activity in the NSN-to-SN transition of germinal vesicle chromatin configuration in porcine oocytes. Sci Rep. 2016;6:23555. doi: 10.1038/srep23555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du Z., Zheng H., Kawamura Y.K., et al. Polycomb Group Proteins Regulate Chromatin Architecture in Mouse Oocytes and Early Embryos. Mol Cell. 2020;77(825–839):e827. doi: 10.1016/j.molcel.2019.11.011. [DOI] [PubMed] [Google Scholar]
- 66.Flyamer I.M., Gassler J., Imakaev M., et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544:110–114. doi: 10.1038/nature21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gassler J., Flyamer I.M., Tachibana K. Single-nucleus Hi-C of mammalian oocytes and zygotes. Methods Cell Biol. 2018;144:389–407. doi: 10.1016/bs.mcb.2018.03.032. [DOI] [PubMed] [Google Scholar]
- 68.Silva M.C.C., Powell S., Ladstatter S., et al. Wapl releases Scc1-cohesin and regulates chromosome structure and segregation in mouse oocytes. The Journal of cell biology. 2020;219 doi: 10.1083/jcb.201906100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Collombet S., Ranisavljevic N., Nagano T., et al. Parental-to-embryo switch of chromosome organization in early embryogenesis. Nature. 2020;580:142–146. doi: 10.1038/s41586-020-2125-z. [DOI] [PubMed] [Google Scholar]
- 70.Gibcus J.H., Samejima K., Goloborodko A., et al. A pathway for mitotic chromosome formation. Science. 2018;359:eaao6135. doi: 10.1126/science.aao6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y., Wang H., Zhang Y., et al. Reprogramming of Meiotic Chromatin Architecture during Spermatogenesis. Mol Cell. 2019;73:547–561.e546. doi: 10.1016/j.molcel.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 72.Alavattam K.G., Maezawa S., Sakashita A., et al. Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development. Nat Struct Mol Biol. 2019;26:175-+. doi: 10.1038/s41594-019-0189-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tedeschi A., Wutz G., Huet S., et al. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature. 2013;501:564-+. doi: 10.1038/nature12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brieño-Enríquez M.A., Moak S.L., Toledo M., et al. Cohesin Removal along the Chromosome Arms during the First Meiotic Division Depends on a NEK1-PP1γ-WAPL Axis in the Mouse. Cell Reports. 2016;17:977–986. doi: 10.1016/j.celrep.2016.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dahl J.A., Jung I., Aanes H., et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537:548–552. doi: 10.1038/nature19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li D, Ning C, Zhang J, et al. Dynamic transcriptome and chromatin architecture in granulosa cells during chicken folliculogenesis. Nature Communications. (2022). 13. 131-131. [DOI] [PMC free article] [PubMed]