Highlight
-
•
The 2,6-Dihydroxyacetophenone (DHAP), 4-Hydroxyphenethyl alcohol (4-HPEA), and 4-Hydroxybenzaldehyde (HBA) in pickled radish showed a good affinity for xanthine oxidase (XOD) in the molecular docking results.
-
•
DHAP, 4-HPEA and HBA inhibit in vitro XOD enzymatic activity by affecting secondary structure and hydrophobic groups, IC50 were: 1.24 ± 0.02 mM, 24.52 ± 0.8 mM, and 2.67 ± 0.9 µM, respectively.
-
•
DHAP, 4-HPEA and HBA reduce xanthine-induced high uric acid levels in BRL 3A cells by inhibiting XOD enzyme activity (p < 0.05).
Chemical compounds studied in this article: 2,6-Dihydroxyacetophenone (DHAP, PubChem CID: 69687); 4-Hydroxyphenethyl alcohol (4-HPEA, PubChem CID: 10393); 4-Hydroxybenzaldehyde (HBA, PubChem CID: 126); 5-Hydroxymethylfurfural (5-HMF, PubChem CID: 237332); α-Linolenic (PubChem CID: 5280934); Methyl linoleate (PubChem CID: 5284421); 1-Monopalmitin (PubChem CID: 14900); Chaenomic acid A (PubChem CID: 102339344); β-Sitosterol (PubChem CID: 222284); Daucosterol (PubChem CID: 5742590); Allopurinol (AP, PubChem CID: 135401907); Genistein (PubChem CID: 5280961); Kaempferol (PubChem CID: 5280863); Quercetin (PubChem CID: 5280343); Resveratrol (PubChem CID: 445154); Luteolin (PubChem CID: 5280445); Piceatannol (PubChem CID: 667639); Carvacrol (PubChem CID: 10364); Isohapontigenin (PubChem CID: 5318650); p-coumaric acid (PubChem CID: 637542); Catechol (PubChem CID: 289)
Keywords: Pickled radish, Phenols, Xanthine oxidase, Molecular simulation, BRL 3A cell
Abstract
Pickled radish is a general source of natural bioactive compounds that include phenols. Here, we used molecular docking, fluorescence quenching, circular dichroism spectroscopy and molecular dynamics simulations to identify potential inhibitors against xanthine oxidase from a library of pickled radish compounds. The most effective compounds were selected for validation through in vitro experiments including enzyme activity inhibition tests, and cell-based assays. Molecular docking results revealed that 2,6-Dihydroxyacetophenone, 4-Hydroxyphenethyl alcohol, and 4-Hydroxybenzaldehyde exhibited significant effects on xanthine oxidase inhibition. Three phenols have varying degrees of inhibition on xanthine oxidase, which is driven by hydrophobic interactions and hydrogen bonds and affects the secondary structure and hydrophobic homeostasis of xanthine oxidase. The stability of xanthine oxidase inhibition by three phenols was analyzed by molecular dynamics simulation. Finally, cellular experiments confirmed that three phenols reduced uric acid levels by inhibiting the xanthine oxidase enzyme activity of BRL 3A cells.
1. Introduction
Xanthine oxidase (XOD), a key rate-limiting enzyme of purine metabolism in human body, it is responsible for catalyzing hypoxanthine to xanthine, which is further oxidized to uric acid (UA) (Cao, Pauff, & Hille, 2014). Uric acid oxidase in non-primate mammals can effectively oxidize UA to allantoin and CO2. However, UA is the final product of human purine metabolism due to the lack of uric acid oxidase in the body (So, Dumusc, & Nasi, 2018). The disorder of purine metabolism or the obstructions of UA excretion led to the abnormally elevated levels of serum UA, which is the characteristic of hyperuricemia (HUA). Furthermore, abnormal serum UA level is associated with multiple diseases, such as hypertension, hyperlipidemia, cardiovascular disease and chronic kidney disease (Sharaf El Din, Salem, & Abdulazim, 2017). Currently, the main treatment for HUA in clinical practice is to reduce UA generation and promote UA excretion as a supplement (Mackenzie et al., 2020). Hence, inhibition of the activity of XOD could be used as an effective strategy to reduce UA level.
As a homodimer with a molecular mass of approximately 290 kDa, each subunit of XOD has independent catalysis. The active center of XOD consists of molybdopterin (Mo-Pt), flavin adenine dinucleotide (FAD), and two iron sulfur centers (Fe2S2) (Enroth, Eger, Okamoto, Nishino, Nishino, & Pai, 2000). XOD is widely distributed in mammals, mostly in the liver and intestines (Pacher, Nivorozhkin, & Szabo, 2006). Although allopurinol is effective in reducing UA production in the body, long-term consumption can lead to side effects such as rashes, chronic kidney damage and cardiovascular disease (Badve et al., 2020). Therefore, the development of safe and efficient natural XOD inhibitors has promising prospect.
Pickled radish, as a popular pickled food all over the world, has the characteristics of unique flavor, long shelf life and rich nutritional value (Kim, Zheng, & Shin, 2008). Radishes undergo complex biochemical changes after the unique curing process, resulting in changes in their microbial composition, nutrient composition and product texture (Cheigh & Park, 1994). In our previous research, a series of natural compounds containing phenols were identified from pickled radishes (Li, Huang, Deng, Li, Su, Liu, et al., 2020). At present, due to the limitations of cost, experimental conditions, and manpower, it is often difficult to screen out effective inhibitors according to specific target proteins. Computer-aided drug design (such as molecular docking) has emerged in this context as an efficient way to screen potential small molecules from a large number of compounds (Rester, 2008). In recent years, the effective screening of target small molecule compounds by molecular docking has been reported frequently. Liu et al used Autodock 4.2 to screen for potential inhibitors of the key target enzymes acetylcholinesterase (AchE) and Butyrylcholinesterase (BuchE) in Alzheimer's disease (AD). The results revealed Ajmalicine, a prominent natural alkaloid, showing promising inhibitory potential against AChE and BuChE (Liu et al., 2020). Cao et al used the molecular docking method to identify curcumin as an effective inhibitor of α-glucosidase and dipeptidyl-peptidase 4 (DPP-4) from 22 food source compounds and verified its inhibitory activity in vitro and in vivo (Cao et al., 2021). Virtual screening of potential functional small molecule compounds using molecular docking is increasingly being widely used in drug design and natural product development.
In this study, molecular docking was used to screen potential XOD inhibitors from a collection of 10 bioactive compounds from pickled radish. After the promising compounds were selected, circular dichroism, fluorescence quenching and molecular simulation were carried out to clarify the inhibition mechanism between XOD and the phenols. The effects of the phenols on XOD were further conducted in enzyme activity inhibition tests and xanthine-induced high-UA BRL 3A cells model. The present study demonstrated a viable way for screening the potential target compounds of XOD inhibition from a series of bioactive compounds, which also provided a theoretical basis for the nutritional value of pickled radish.
2. Materials and methods
2.1. Materials
MEM and antibiotics (streptomycin and penicillin) were from HyClone, USA. Fetal bovine serum (FBS) was from Gemini Bio-products, USA. 3-(4, 5-dimethylthiazol-2yl)-2, 5-diphenyltetrazolium bromide (MTT) and xanthine oxidase (XOD, from bovine milk) were purchased from Sigma-Aldrich Chemicals Co. (St. Louis, MO, USA). 2,6-dihydroxyacetophenone (DHAP, 99%), 4-Hydroxyphenethyl alcohol (4-HPEA, 98%), and 4-Hydroxybenzaldehyde (HBA, 98%) were purchased from Macklin Biochemical Co., Ltd (Shanghai, China). Xanthine and allopurinol were purchased from Shanghai Macklin Biochemical Co., Ltd. Dimethyl sulfoxide (DMSO) was obtained from Xilong Scientific Co., Ltd (Guangdong, China). All reagents were analytical grade. The deionized water was obtained from the Milli-Q Reagent Water System of Milliore Co. (Milford, MA).
2.2. Virtual screening using molecular docking
The most favorable binding small molecules were screened by molecular docking, the top three compounds with the best binding were used for subsequent studies (Li & Li, 2010). Using XOD as target, AutoDock 4.2 was used for virtual screening of previously identified pickled radish compounds and using PYMOL further investigates the probable binding interactions of compounds with XOD. The 3D structures of all compounds were obtained from NCBI. Complex crystal structure of XOD and salicylic acid (PDB ID: 3BDJ) from RCSB protein data bank (http://www.rcsb.org/pdb) Download it. The 3D structure of the compounds was optimized by using Sybyl-X 2.0 according to the principle of minimizing the energy of the Tripos force field. The 3D structure was manually removed water and added polar hydrogen atoms according to the inhibitor and protein, and each atom of the protein was assigned Kollman charges. The grid box containing the active center is defined as 40 Å * 40 Å * 40 Å and the grid spacing is 0.375 Å. Docking parameters adopt Lamarkian genetic algorithm (LGA), while the other parameters adopt default values.
2.3. In vitro XOD activity assay
In vitro XOD activity assay method modified from previous study (Yan, Zhang, Hu, & Ma, 2013). The XOD inhibitory activity by screened compounds (DHAP, 4-HPEA, and HBA) was examined spectrophotometrically at 290 nm by the Multimode microplate reader (SYNERGY, Biotek, USA). Different concentration of three phenols were prepared using DMSO, including DHAP solution (0.1, 0.25, 0.5, 1, 1.5, 2 mM), 4-HPEA solution (10, 20, 40, 60, 80, 100 mM), and HBA solution (1, 2, 4, 6, 8, 10 μM). Add 25 µL sample solution, 25 µL XOD (0.1 U/mL), and 100 µL phosphate buffer sodium (PBS, pH = 7.5) to 96-well plate for uniform oscillation. After incubation at 37℃ for 10 min, 50 µL xanthine was added for 30 s, and the absorbance of the obtained solution was immediately determined at 290 nm. In blank group, PBS was used instead of sample and xanthine. In the complete response group, PBS was used instead of the sample. The XOD enzyme was replaced by PBS in the sample control group. Allopurinol was used as a positive control. The inhibition rate of the sample on XOD activity was calculated according to the following formula:
In the formula, C1 is the zero-setting group; C2 was the complete response group.C3 was the sample reaction group (positive group); C4 was the sample control group.
2.4. CD measurements
CD measurements using previously reported and slightly modified method (Li et al., 2018). The CD spectra of XOD in the absence and presence of three phenols were recorded from 260 nm to 190 nm by a CD spectrometer (Applied Photo Physics Ltd., Surrey, UK) under nitrogen atmosphere. The concentration of XOD was kept at 0.1 U mL−1, and phenols of different concentrations were prepared and mixed with XOD, respectively. Three successive spectral scans were carried out, and the mean value was taken to present. 0.2 M PBS (pH = 7.5) was used as the zero-setting solvent.
2.5. Fluorescence titration
The quenching of XOD endogenous fluorescence by DHAP, 4-HPEA, and HBA was detected, and methodologies refer to previous report (Li, Zhao, Su, Lin, & Wang, 2016). In the reaction system of 0.2 M PBS (pH = 7.5), 0.1 U·mL−1 XOD working solution was mixed with different concentrations of DHAP, 4-HPEA and HBA, respectively, and incubated at 25℃ for 5 min. The multi-mode enzyme plate analyzer (Synergy/H1, Biotek, USA) was used for detection. The excitation wavelength was set at 280 nm and the emission wavelength was 300 ∼ 500 nm, the width of the slit was set to 2 nm.
2.6. Molecular dynamics simulation
Complexes were committed to 120 ns of all-atom MD simulation using the GROMACS ver. 2019.3 package followed by subsequent analysis. Amber99sb-ildn Force Field is used to process XOD. Moreover, the ligands restrained electrostatic potential (RESP) was calculated by Multiwfn and parameterized by General Amber Force Field (GAFF) (Wang et al., 2006, Wang et al., 2004). Acubic box with dimension of 12 Å layer was defined and the complexes were solvated using the TIP3P water model. Kt counterions were added to the system until charge neutralization and further Na and Cl ions were added. The long-range electrostatic interactions were computed by means of the Particle Mesh Ewald (PME) method (Darden, York, & Pedersen, 1993), Use 1.0 Å spacing in periodic boundary conditions to set the grid. Subjected to a double minimization step to preprocessing this system: minimization of water and ions constraining the solute; global minimization without any restriction. Before MD simulation, both NVT and NPT were balanced with a 0.1 ns ensemble. The 120 ns trajectory was analyzed by MD simulation, and the integration time step is 2 fs.
2.7. Cell culture and cell viability assay
Methods refer to the study by Ma et al. (Ma, Lian, & Wang, 2019). BRL 3A (Procell CL-0036) were kindly provided by Procell Life Science&Technology Co., Ltd. The complete growth medium of BRL 3A cells were MEM containing 10% FBS and 1% antibiotics (streptomycin and penicillin), then cells were grown at 37 °C in 5% CO2. Around 1 × 104 cells per well were seeded in a 96-well plate for overnight. After treated with phenols for 48 h, 15 μL MTT (5 mg/mL) was added to the cells for 4 h at 37℃. The supernatant was then removed and 150 μL DMSO was added to solubilize the formazan products with 10 min continuous shaking before OD490 nm was taken. The cell viability was determined, and DMSO served as the control. The calculation formula of cell survival rate is as follows:
In the formula, OD0 represents the background value, OD1 is the light absorption value of the blank control, and OD2 is the light absorption value of the sample treatment group.
2.8. Optimization of inductive agent, concentration, and incubation time
BRL 3A cells at the exponential growth stage were digested with 0.25% trypsin, and resuspended with complete medium, inoculated into 12-well plates at a density of 5 × 105. After 16 h, the medium was discarded, and complete medium containing 1% inducer (adenosine, hypoxanthine, and xanthine) was added at different concentrations. After 48 h of incubation, the cell supernatant was collected and analyzed for uric acid test kit (Nanjing Jiancheng).
BRL 3A cells in the exponential growth phase were digested with 0.25% trypsin, resuspended in complete medium and seeded in 12-well plates at a density of 5 × 105. The culture medium was discarded after 16 h of normal culture. At a ratio of 1%, different concentrations of xanthine were dissolved in complete medium and added to the well plate. After incubating for 12 h, 24 h, and 48 h, the cell supernatant was collected for UA detection.
2.9. Determination of UA levels and XOD enzyme activity in BRL 3A cells
The test method is modified with reference to Li et al (Li, Liu, Xie, Liu, & Zou, 2020). BRL 3A cells in the logarithmic growth phase were resuspended with complete growth medium and seeded in 12-well plates at a density of 5 × 105 cells per well. After a 16 h inoculation period, discarded the supernatant of each well and washed twice with PBS (pH = 7.5). The cells were treated with 1 mM xanthine and three phenols at various concentrations. Set allopurinol as a positive control. After 48 h, the cell supernatant was collected, and the UA level of the treated cells was quantitatively detected using the uric acid test kit (Nanjing Jiancheng). At the same time, the cells were collected, and the cells were treated with a cell lysate containing 1 mM Protease Inhibitor (PMSF) and used for the determination of protein concentration. XOD assay kit (Nanjing Jiancheng) was used to determine the XOD enzyme activity in the cells, following the manufacturer's agreement.
2.10. Statistical analysis
Experimental data of this study were expressed as mean ± standard deviation (SD). Analysis of ANOVA and Duncan's multiple comparison tests were used to determine significance. p < 0.05 means a significant difference, p < 0.01 and p < 0.001 means a highly significant difference.
3. Results
3.1. Molecular docking and visual analysis
By predicting the optimal conformation of the ligand-binding site to the receptor, Ligands with good docking pose were selected. The output filter results were shown in Table 1. The interaction between the three phenols- 2,6-Dihydroxyacetophenone (DHAP), 4-Hydroxyphenethyl alcohol (4-HPEA), and 4-Hydroxybenzaldehyde (HBA) and XOD was further elucidated, and the molecular structure was visualized by molecular docking model Fig. 1(A-D). The active cavity of Xanthine oxidase (XOD) is a multi-channel, and three phenols can enter the active cavity containing molybdopterin effectively, and three phenols were anchored in the active cavity containing molybdopterin in the same conformational posture. The –OH on the benzene ring of DHAP directly forms hydrogen bonds with bond lengths of 1.9 Å, 2.4 Å and 2.3 Å with GLU-802, ARG-880 and THR-1010 respectively, and the O atom on the ketone group forms a hydrogen bond with VAL-1011 with a bond length of 2.4 Å. The –OH directly connected to the benzene ring of 4-HPEA forms hydrogen bonds with bond lengths of 2.1 Å and 2.2 Å with ARG-880 and THR-1010. The hydrogen formed between the other hydroxyl group and ALA-1079 and GLU-1261, and the hydrogen-bond length is 1.9 Å and 2.1 Å. The O atom on the hydroxyl group of HBA forms a hydrogen bond with ARG-880, and the bond length is 2.5 Å; the H atom on the hydroxyl group forms a hydrogen bond with THR-1010, and the bond length is 2.2 Å. The O atom on the aldehyde group of HBA and ALA-1079 formed a hydrogen bond with a bond length of 2.0 Å. In the hydrophobic cavity of XOD, there is π-π conjugation between PHE-914, PHE-1009, and ALA-1078 and the benzene rings of the three phenols Fig. 1(E-G). In addition, we summarized ten reported natural phenols with XOD inhibitory activity and docked them with XOD to verify the stability of this molecular docking mode. The results were shown in Table S1. Compared with other phenols, flavonoids used in this study were more dominant in binding with XOD, and the binding energy of other phenols was also in line with expectations.
Table 1.
Virtual screening results of 10 compounds and allopurinol in pickled radish with XOD as target.
| Name | Structure | Molecules Weight | Binding_Energy (Kcal/mol) |
|---|---|---|---|
| DHAP |  |
151.14 | −6.63 |
| 4-HPEA |  |
138.16 | −6.03 |
| HBA |  |
126.11 | −5.63 |
| 5-HMF |  |
126.11 | −4.86 |
| α-Linolenic | 278.43 | −4.19 | |
| Methyl linoleate | 294.47 | −0.53 | |
| 1-Monopalmitin | 330.50 | >0 | |
| Chaenomic Acid A | 344.40 | >0 | |
| β-Sitosterol |  |
428.73 | >0 |
| Daucosterol |  |
576.85 | >0 |
| Allopurinol |  |
136.11 | −4.91 |
Fig. 1.
The conformations of the three phenolic compounds are superimposed in the active pocket of XOD (A). Binding mode of DHAP (B), 4-HPEA (C) and HBA (D) in the active site of XOD (PDB ID: 3BDJ) and the visualization of interaction between XOD and DHAP (E), 4-HPEA(F), and HBA (G).
3.2. XOD inhibitory assay in vitro
Allopurinol as the positive control, the inhibitory rates of different concentrations of phenols on XOD in vitro were shown in Fig. 2(A). With the increase of the concentration of the three phenols, the inhibition rate increased significantly (p < 0.05). When allopurinol (positive control group) concentration was 0.01 mM, the inhibition rate of XOD was 80.06 ± 0.6%, which was significantly higher than the Dimethyl sulfoxide (DMSO) group (control group) (p < 0.001). The concentration of DHAP, 4-HPEA, and HBA reached 2 mM, 80 mM, and 0.006 mM respectively, and the inhibition rate of XOD was close to the positive control allopurinol. The half inhibiting concentration (IC50) of DHAP, 4-HPEA, and HBA against XOD were calculated as 1.24 ± 0.02 mM, 24.52 ± 0.8 mM, and 2.67 ± 0.9 µM, respectively. The results showed that three phenols had significant inhibitory effects on XOD in vitro, and the order of the inhibitory strength of the three phenols on XOD: HBA> DHAP> 4-HPEA.
Fig. 2.
(A) Inhibiting effect of phenols on pickled radish in vitro. Allopurinol was used in the positive control. DMSO was used as a solvent control. Error bars represent SD (n = 4). *** p < 0.001, (ai) Indicates the difference between sample groups (p < 0.05). (B) CD spectra of XOD in the absence and presence of DHAP, 4-HPEA and HBA. (C) Fluorescence spectroscopic analysis and the Stern-Volmer curves of XOD after radish compounds at treatment at different concentrations. indicate that the inhibitor concentration increases from small to large.
3.3. CD spectrum detection
To elucidate the effect of three phenols on the secondary structure of XOD, CD spectroscopy was used to analyze. As shown in Fig. 2(B), there were two negative bands at 208 nm and 220 nm in the CD spectrum, indicating that three phenols changed the secondary structure of XOD and affect the secondary structure of the protein by adding α-helix. Meanwhile, negative absorption occurs at 218 nm, which was the characteristic peak of antiparallel β-folding. It could be seen that three phenols can increase the number of α-helix and β-folding of XOD, increase the rigid structure of a protein, lead to the formation of binding sites difficult to reduce the catalytic activity of XOD.
3.4. Fluorescence quenching experiments
Fluorescence spectroscopic analysis can be used to elucidate the interactions between small and large molecular proteins. To understand the effect of three phenols on the hydrophobic microenvironment of XOD, fluorescence spectroscopy was used to further explore the mechanism of protein quenching by the inhibitors. Tryptophan (Try) is the main endogenous fluorescence source of XOD, which is sensitive to the changes of microenvironment. With the increase of compound concentration, the fluorescence intensity of XOD decreased, and the emission peak did not appear obvious shift. These results indicate that phenols interact with XOD, quench the intrinsic fluorescence of XOD, and change the hydrophobic microenvironment.
In Fig. 2(C), the phenols were inversely correlated with XOD expression based on the Stern-Volmer curves of XOD. The quenching constant was decreased with the temperature rising, which indicated the quenching reaction between the three compounds and XOD was of a single type. An obvious linear relationship between the three compounds at different temperatures (298 K, 304 K, and 310 K). It suggested that the fluorescence quenching mechanism of DHAP, 4-HPEA and HBA for XOD were a single type. Hence, it could be inferred that the compound and XOD combine to form a stable complex in the process of fluorescence quenching, and the fluorescence quenching type of XOD for the three were static quenching.
3.5. Molecular dynamics analysis
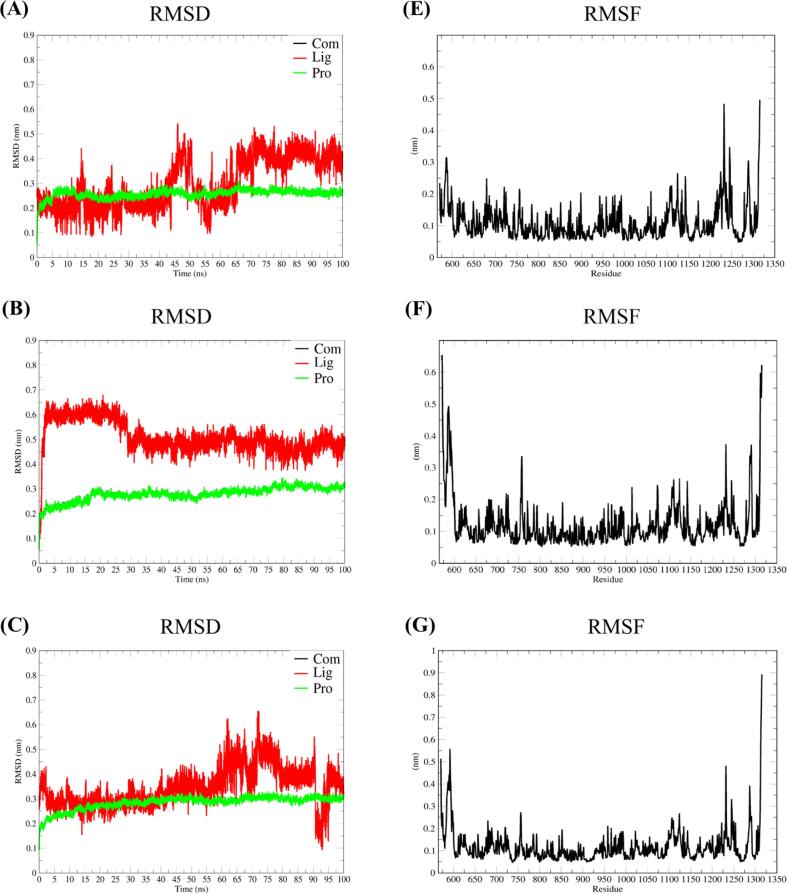
The stability of three phenols with XOD complexes was further studied by molecular dynamics simulation. RMSD was used to evaluate the stability of each complexes system. As shown in Fig. 3(A–C), all the complexes’ systems were in a stable state after 10 ns, and the RMSD value was stable at 0.2–0.3 nm. Among them, the small molecule HBA has the smallest RMSD value. It shows that the combination of HBA and protein was more stable. This might be the reason why HBA has the best inhibition of XOD. RMSF analysis was performed according to the kinetic trajectories of the complexes. After the three phenols were combined with XOD, the RMSF curves of the three compounds fluctuated in the same way. The Fig. 3(E–F) shows that the three complexes have three identical obvious fluctuations, which were roughly concentrated at amino acid residues 571 to 600, amino acid residues 1225 to 1250, and amino acid residues 1275 to 1330. The three regions with large fluctuations were all located at the entrance of the active pocket, and the regions composed of these amino acid residues were all located in the Loop region and flexible side chain of the protein. In addition, the amino acid residues (648 ∼ 1015) located in the active pocket region, the RMSF fluctuated at 0.1 nm, indicate that the complexes have good stability.
Fig. 3.
Root mean square deviations (RMSD) and Root mean square fluctuations (RMSF) calculation of three complexes. (A) and (D) DHAP-XOD complex, (B) and (E) 4-HPEA-XOD complex, (C) and (F) HBA-XOD complex.
3.6. Optimization of the induction conditions of HUA cell model and the UA-lowering effect of three phenols on BRL 3A cells
In order to establish a suitable hyperuricemia cell model, using adenosine, hypoxanthine and xanthine as candidate inductive agent, the level of UA produced by the cells was measured at different times. Results as shown in Fig. 4, Fig. 1 mM xanthine stimulated BRL 3A cells to produce higher level of UA than other inducers. And the UA produced after 48 h incubation with 1 mM xanthine and cells was higher than other induction time.
Fig. 4.
Determination the content of uric acid produced by BRL 3A cells with different inducers and induction time. * p < 0.05, ** p < 0.01.
The effects of three phenols lowering UA were further performed in BRL 3A cells. MTT assay was used to detect the effects of three phenols on the survival rate of BRL 3A cells. Fig. 5 showed that the survival rate of BRL 3A cells decreased with the increase of the working concentration and culture time of the three phenols. In particular, when the concentration of DHAP, 4-HPEA, and HBA was 200 μM, 800 μM, and 200 μM, respectively, the cell survival rate decreased significantly (48 h) (p < 0.05). Hence, this dose was set as the high-concentration for subsequent experiments. A high UA cell model was established by inducing BRL 3A cells with xanthine, and the level of UA in the model group was significantly higher than that in the normal group (p < 0.001). When allopurinol 10 μM (positive control group) was added, the UA content was significantly decreased compared with the model group (p < 0.05), reaching the level of UA in the normal group. After that, we observed that the UA level of BRL 3A cells was significantly revising after treatment with three phenols and xanthine, except for the low dose of HBA (p < 0.05). The effects of three phenols on the XOD enzyme activity of BRL 3A cells were further tested, and it was observed that the addition of three phenols could effectively reduce the XOD enzyme activity of BRL 3A cells (p < 0.05). Except for the 4-HPEA group, the ability to inhibit XOD is positively correlated with the concentration of phenols, as shown in Fig. 5.
Fig. 5.
Effect of three phenols on the survival rate of BRL 3A cells and evaluation of their uric acid lowering efficacy in vitro. Error bars represent SD (n = 6). *** p < 0.001, ** p < 0.01. (a-i) indicates the difference between sample groups (p < 0.05).
4. Discussion
In recent years, the number of patients with hyperuricemia and gout has been increasing year by year, which has become a major metabolic disease worldwide (Dehlin, Jacobsson, & Roddy, 2020). However, the current clinical drugs for the treatment of hyperuricemia were often associated with a certain degree of side effects, which greatly limit their use (Li, et al., 2016). Therefore, special attention has been paid to functional foods that can reduce uric acid (UA). A growing number of natural phenols have been shown to have UA-lowering effects. Considering the important role of xanthine oxidase (XOD) in UA metabolism, phenolic compounds have become popular for research on XOD inhibitors due to their healthy and efficient properties.
In this study, ten natural compounds in pickled radish were docked with XOD, and three phenols include 2,6-Dihydroxyacetophenone (DHAP), 4-Hydroxyphenethyl alcohol (4-HPEA), and 4-Hydroxybenzaldehyde (HBA), which were more dominant in binding to XOD, were successfully screened. Especially, 5-Hydroxymethylfurfural (5-HMF) has been reported to have XOD inhibitory activity (Li, Zhao, Su, Lin, & Wang, 2016). Meanwhile, 5-Hydroxymethylfurfural (5-HMF), which has previously been shown to have XOD inhibitory activity, has shown good binding energy (Li, Zhao, Su, Lin, & Wang, 2016). In addition, ten phenols that have been reported to have XOD inhibitory activities were summarized in this study, and their binding energy to XOD was evaluated using this molecular docking mode (Table S1). The results were expected.
By observing the binding modes between the three phenols and XOD, it was found that there were multiple hydrogen bonds between ligand and receptor. ARG-880 and THR-1010 located in the active XOD cavity are more likely to form hydrogen bonds with hydroxyl groups directly attached to the benzene ring. Therefore, it can be considered that this is one of the shreds of evidence of effective binding between the three phenols and XOD. In the docking results, it can be observed that three phenols have similar binding effects with XOD. The –OH on the benzene ring of phenols was close to the entrance of the hydrophobic pocket, while the other side (–CH2CH2OH, –CHO) was closer to the inside of the active pocket. This result was consistent with the previously reported interaction between phenols and XOD (Ou, Lin, Zhao, & Xie, 2020). In the active pocket of XOD, the amino acid residues GLU-802 and ARG-880 located inside the hydrophobic pocket were generally considered to be the key amino acids when XOD catalyzes the action of substrates (Okamoto, Matsumoto, Hille, Eger, Pai, & Nishino, 2004). Hydrogen bonds play a key role in ligand–protein interactions. In our results, the amino acid residues GLU-802 and ARG-880 formed hydrogen bonds with the target small molecule. In addition, there is a π-π interaction between the benzene ring of the phenols and the amino acid residues PHE-914 and PHE-1009 of XOD. Since the amino acid residues PHE-914 and PHE-1009 regulate the recognition and entry of small molecules, this binding mode facilitates the stable binding of the compound in the pocket, thereby blocking the entry of purine substrates (Pauff, 2008).
Three phenols, DHAP, 4-HPEA and HBA had varying degrees inhibitory effect on XOD in vitro, and IC50 values were 1.24 ± 0.02 mM, 24.52 ± 0.8 mM and 2.67 ± 0.9 µM, respectively. By comparing the structural pairs of the three phenols and their inhibition relationship to XOD, it can be obtained that the presence of –CHO on the benzene ring of HBA contributes more to the XOD inhibitory activity than -C O- and –CH2CH2OH. Therefore, this may be the key to the difference in the ability of the three phenols to inhibit XOD.
In general, the ultraviolet region (<250 nm) can be used to reveal the characteristics of the secondary structure of proteins (α-helix, β-folding, random coil, etc.) (Gao et al., 2010, Kamat and Seetharamappa, 2004). Due to the anchoring of three phenols, the number of α-helix and β-fold in the secondary structure of XOD was increased, which leads to the reduction of flexible structure, the formation of binding sites was difficult. Hence, the chance of substrate binding to the active site of XOD is reduced, resulting in a decrease of UA production.
Fluorescence quenching reaction usually depends on the reaction temperature or the life of the excited state, so the quenching reaction could be distinguished as static quenching or dynamic quenching. Static quenching is caused by the combination of fluorescence molecule and quenching agent to form a stable complex (Acharya, Sanguansri, & Augustin, 2013). While the increase of the reaction system temperature will lead to the decrease of the stability of the composite during static quenching (Chen, Jiang, Chen, & Chen, 2008). Among the endogenous fluorescence sources of XOD, tryptophan (Trp) is the most sensitive to the changes in the microenvironment. The XOD has two strong fluorescence emission peaks at 328 nm and 398 nm at a certain excitation wavelength, indicating that the tryptophan residues in XOD are located in the hydrophobic region of the active site (Qi, Zhang, Liao, Ou-Yang, Liu, & Yang, 2008). It was found that three phenols quenched the endogenous fluorescence of XOD, and destroy the hydrophobic homeostasis of XOD. Further, the quenching of XOD endogenous fluorescence by the three phenols were all dominated by static quenching. These results confirmed our conjecture and are consistent with the previously confirmed effect of phenols on XOD (Yan, Zhang, Hu, & Ma, 2013).
The stability of phenols binding to XOD can be observed by molecular dynamics simulation. All three complexes showed reasonable stability in this system. Among them, we observed that the RMSD value of HBA was better than that of the other two phenols, which might be one of the reasons for the strongest inhibition of XOD activity of HBA. In addition, the key amino acids with large fluctuations were evenly distributed around the active pocket. These areas where the RMSF value fluctuates greatly have Loop regions and long flexible side chains. They were in frequent contact with solvents, so they have greater flexibility (Pan et al., 2021).
Li et al. indicated that hydrophobic interactions play a key role in the inhibition of the XOD enzymatic response (Li, et al., 2018). In the present study, the molecular docking study showed that the binding of the three phenols to XOD was driven by hydrophobic interactions and hydrogen bonding. Fluorescence spectroscopy, circular dichroism (CD) and molecular dynamics simulations were powerful approaches for characterizing the hydrophobic homeostasis of proteins, secondary structure changes and the internal stability of systems (Pan et al., 2021, Tang and Zhao, 2019). Our fluorescence quenching assays and CD results confirmed the effect of three phenols on the endogenous fluorescence source (hydrophobic amino acid residues) and secondary structure of XOD. In order to verify the stability of the inhibitor-protein complex, we performed molecular dynamics simulations, which also further proved that our molecular docking results were reliable. This is consistent with previous studies (Pan, et al., 2021). Furthermore, one study has demonstrated that phenols inhibited the production of superoxide anion generation (O2–) when combined with the molybdopterin-containing active pocket of XOD (Zhang, Zhang, Liao, & Gong, 2017). Meanwhile, in another study, it was reported that XOD inhibitors reduced superoxide anion generation (O2–) and alleviated hepatic lipid peroxidation in rats (Fang, Seki, Qin, Bharate, Iyer, & Maeda, 2010). Therefore, there could be a possible association between the XOD inhibition and lipid peroxidation suppression, and additional research were required.
In cell experiments, compared with 4-HPEA, DHAP and HBA cause damage to BRL 3A cells at a lower dose. Therefore, we chose a relatively safe dose for BRL 3A cells. We took BRL 3A cells as the modeling object and successfully established a hyperuricemia cell model after 48 h of inducing BRL 3A cells with xanthine. Subsequently, we used this model to successfully evaluate the UA lowering ability of three phenols at the cellular level. The results showed that except for the low-dose HBA group, the other experimental groups effectively reduced the level of UA. This result is consistent with the previously reported hypouricemic effect of phenols on BRL 3A cells (Li, Liu, Xie, Liu, & Zou, 2020). Although the positive control group treated with allopurinol was the best to reduce the UA level of BRL 3A cells, considering the potential toxicity of allopurinol, we believed that the UA lowering effect of the three phenols in cells was as expected.
As a preliminary study, there were still some limitations in the present study. For one thing, this study did not explore the absorption and bioavailability of the three phenols in the human body. In the next place, is there coordinate repression of three phenols on XOD? In addition, some in vivo experiments are needed for verification.
5. Conclusion
In conclusion, through virtual screening of 10 compounds previously identified in pickled radish, three phenols with good docking pose to xanthine oxidase (XOD) were screened out, namely 2,6-Dihydroxyacetophenone (DHAP), 4-Hydroxyphenethyl alcohol (4-HPEA), and 4-Hydroxybenzaldehyde (HBA). Then, three pickled radish phenols were taken as the object to study the inhibition mechanism of XOD by multispectral methods and molecular simulation. It speculated that hydrophobic forces and hydrogen bonds mainly drove the interaction of three phenols with XOD. The anti-hyperuricemia effects of three phenols were confirmed further by XOD activity in vitro assay. Established an ideal hyperuricemia cell model and tested the effect of three phenols on improving hyperuricemia at the cellular level. The improvement of hyperuricemia by diet has always been a concern. This study provided base information that explains the inhibitory effect of salted radish phenols on XOD activity, which is used further as theoretical support for the XOD inhibitors' development.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by the Major Applied Agricultural Technology Innovation Projects of Shandong Province (grant numbers: SD2019ZZ009), the Ministry of Science and Technology (grant numbers: MOST 108-2221-E-005-044), Natural Science Foundation of Fujian Province (grant numbers: 2021 J01133236), and the opening project of Fujian Provincial Engineering Technology Research Center of Marine Functional Food (grant numbers: No. 2900/Z820235).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2022.100343.
Contributor Information
Lingyu Zhang, Email: zhanglingyu@jmu.edu.cn.
Jian Li, Email: lijian2013@jmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Acharya D.P., Sanguansri L., Augustin M.A. Binding of resveratrol with sodium caseinate in aqueous solutions. Food Chemistry. 2013;141(2):1050–1054. doi: 10.1016/j.foodchem.2013.03.037. [DOI] [PubMed] [Google Scholar]
- Badve S.V., Pascoe E.M., Tiku A., Boudville N., Brown F.G., Cass A.…Investigators C.-F.-S. Effects of Allopurinol on the Progression of Chronic Kidney Disease. New England Journal of Medicine. 2020;382(26):2504–2513. doi: 10.1056/NEJMoa1915833. [DOI] [PubMed] [Google Scholar]
- Cao H., Pauff J.M., Hille R. X-ray crystal structure of a xanthine oxidase complex with the flavonoid inhibitor quercetin. Journal of Natural Products. 2014;77(7):1693–1699. doi: 10.1021/np500320g. [DOI] [PubMed] [Google Scholar]
- Cao W., Chen X., Chin Y., Zheng J., Lim P.E., Xue C., Tang Q. Identification of curcumin as a potential alpha-glucosidase and dipeptidyl-peptidase 4 inhibitor: Molecular docking study, in vitro and in vivo biological evaluation. Journal of Food Biochemistry. 2021;e13686 doi: 10.1111/jfbc.13686. [DOI] [PubMed] [Google Scholar]
- Cheigh H.S., Park K.Y. Biochemical, microbiological, and nutritional aspects of kimchi (Korean fermented vegetable products) Critical Reviews in Food Science and Nutrition. 1994;34(2):175–203. doi: 10.1080/10408399409527656. [DOI] [PubMed] [Google Scholar]
- Chen J., Jiang X.Y., Chen X.Q., Chen Y. Effect of temperature on the metronidazole–BSA interaction: Multi-spectroscopic method. Journal of Molecular Structure. 2008;876(1–3):121–126. [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: AnN⋅log(N) method for Ewald sums in large systems. The Journal of Chemical Physics. 1993;98(12):10089–10092. [Google Scholar]
- Dehlin M., Jacobsson L., Roddy E. Global epidemiology of gout: Prevalence, incidence, treatment patterns and risk factors. Nature reviews. Rheumatology. 2020;16(7):380–390. doi: 10.1038/s41584-020-0441-1. [DOI] [PubMed] [Google Scholar]
- Enroth C., Eger B.T., Okamoto K., Nishino T., Nishino T., Pai E.F. Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: Structure-based mechanism of conversion. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(20):10723–10728. doi: 10.1073/pnas.97.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Seki T., Qin H., Bharate G.Y., Iyer A.K., Maeda H. Tissue protective effect of xanthine oxidase inhibitor, polymer conjugate of (styrene-maleic acid copolymer) and (4-amino-6-hydroxypyrazolo[3,4-d]pyrimidine), on hepatic ischemia-reperfusion injury. Experimental Biology and Medicine (Maywood) 2010;235(4):487–496. doi: 10.1258/ebm.2009.009304. [DOI] [PubMed] [Google Scholar]
- Gao W., Li N., Chen Y., Xu Y., Lin Y., Yin Y., Hu Z. Study of interaction between syringin and human serum albumin by multi-spectroscopic method and atomic force microscopy. Journal of Molecular Structure. 2010;983(1–3):133–140. [Google Scholar]
- Kamat B.P., Seetharamappa J. In vitro study on the interaction of mechanism of tricyclic compounds with bovine serum albumin. Journal of Pharmaceutical and Biomedical Analysis. 2004;35(3):655–664. doi: 10.1016/j.jpba.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Zheng Z.B., Shin D.H. Growth inhibitory effects of kimchi (Korean traditional fermented vegetable product) against Bacillus cereus, Listeria monocytogenes, and Staphylococcus aureus. Journal of Food Protection. 2008;71(2):325–332. doi: 10.4315/0362-028x-71.2.325. [DOI] [PubMed] [Google Scholar]
- Li F., Liu Y., Xie Y., Liu Z., Zou G. Epigallocatechin gallate reduces uric acid levels by regulating xanthine oxidase activity and uric acid excretion in vitro and in vivo. Annals of Palliative Medicine. 2020;9(2):331–338. doi: 10.21037/apm.2019.11.28. [DOI] [PubMed] [Google Scholar]
- Li H., Li C. Multiple ligand simultaneous docking: Orchestrated dancing of ligands in binding sites of protein. Journal of Computational Chemistry. 2010;31(10):2014–2022. doi: 10.1002/jcc.21486. [DOI] [PubMed] [Google Scholar]
- Li H., Zhao M., Su G., Lin L., Wang Y. Effect of Soy Sauce on Serum Uric Acid Levels in Hyperuricemic Rats and Identification of Flazin as a Potent Xanthine Oxidase Inhibitor. Journal of Agriculture and Food Chemistry. 2016;64(23):4725–4734. doi: 10.1021/acs.jafc.6b01094. [DOI] [PubMed] [Google Scholar]
- Li J., Huang S.Y., Deng Q., Li G., Su G., Liu J., David Wang H.M. Extraction and characterization of phenolic compounds with antioxidant and antimicrobial activities from pickled radish. Food and Chemical Toxicology. 2020;136 doi: 10.1016/j.fct.2019.111050. [DOI] [PubMed] [Google Scholar]
- Li S., Yang H., Guo Y., Wei F., Yang X., Li D.…Wang Y. Comparative efficacy and safety of urate-lowering therapy for the treatment of hyperuricemia: A systematic review and network meta-analysis. Scientific Reports. 2016;6:33082. doi: 10.1038/srep33082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Kang X., Li Q., Shi C., Lian Y., Yuan E.…Ren J. Anti-hyperuricemic peptides derived from bonito hydrolysates based on in vivo hyperuricemic model and in vitro xanthine oxidase inhibitory activity. Peptides. 2018;107:45–53. doi: 10.1016/j.peptides.2018.08.001. [DOI] [PubMed] [Google Scholar]
- Liu S., Dang M., Lei Y., Ahmad S.S., Khalid M., Kamal M.A., Chen L. Ajmalicine and its Analogues Against AChE and BuChE for the Management of Alzheimer's Disease: An In-silico Study. Current Pharmaceutical Design. 2020;26(37):4808–4814. doi: 10.2174/1381612826666200407161842. [DOI] [PubMed] [Google Scholar]
- Ma D., Lian F., Wang X. PLCG2 promotes hepatocyte proliferation in vitro via NF-kappaB and ERK pathway by targeting bcl2, myc and ccnd1. Artificial Cells, Nanomedicine, and Biotechnology. 2019;47(1):3786–3792. doi: 10.1080/21691401.2019.1669616. [DOI] [PubMed] [Google Scholar]
- Mackenzie I.S., Ford I., Nuki G., Hallas J., Hawkey C.J., Webster J.…Waller T. Long-term cardiovascular safety of febuxostat compared with allopurinol in patients with gout (FAST): A multicentre, prospective, randomised, open-label, non-inferiority trial. The Lancet. 2020;396(10264):1745–1757. doi: 10.1016/S0140-6736(20)32234-0. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Matsumoto K., Hille R., Eger B.T., Pai E.F., Nishino T. The crystal structure of xanthine oxidoreductase during catalysis: Implications for reaction mechanism and enzyme inhibition. Proceedings of the National academy of Sciences of the United States of America. 2004;101(21):7931–7936. doi: 10.1073/pnas.0400973101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou R., Lin L., Zhao M., Xie Z. Action mechanisms and interaction of two key xanthine oxidase inhibitors in galangal: Combination of in vitro and in silico molecular docking studies. International Journal of Biological Macromolecules. 2020;162:1526–1535. doi: 10.1016/j.ijbiomac.2020.07.297. [DOI] [PubMed] [Google Scholar]
- Pacher P., Nivorozhkin A., Szabo C. Therapeutic effects of xanthine oxidase inhibitors: Renaissance half a century after the discovery of allopurinol. Pharmacological reviews. 2006;58(1):87–114. doi: 10.1124/pr.58.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Lu Z., Li C., Qi R., Chang H., Han L., Han W. Molecular Dockings and Molecular Dynamics Simulations Reveal the Potency of Different Inhibitors against Xanthine Oxidase. ACS Omega. 2021;6(17):11639–11649. doi: 10.1021/acsomega.1c00968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauff, J. M. (2008). Structure-Function Studies of Xanthine Oxidoreductase. Xanthine Oxidase.
- Qi Z.D., Zhang Y., Liao F.L., Ou-Yang Y.W., Liu Y., Yang X. Probing the binding of morin to human serum albumin by optical spectroscopy. Journal of Pharmaceutical and Biomedical Analysis. 2008;46(4):699–706. doi: 10.1016/j.jpba.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Rester U. From virtuality to reality - Virtual screening in lead discovery and lead optimization: A medicinal chemistry perspective. Current Opinion in Drug Discovery & Development. 2008;11(4):559–568. [PubMed] [Google Scholar]
- Sharaf El Din U.A.A., Salem M.M., Abdulazim D.O. Uric acid in the pathogenesis of metabolic, renal, and cardiovascular diseases: A review. Journal of Advanced Research. 2017;8(5):537–548. doi: 10.1016/j.jare.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So A., Dumusc A., Nasi S. The role of IL-1 in gout: From bench to bedside. Rheumatology (Oxford) 2018;57(suppl_1) doi: 10.1093/rheumatology/kex449. i12-i19. [DOI] [PubMed] [Google Scholar]
- Tang H., Zhao D. Investigation of the interaction between salvianolic acid C and xanthine oxidase: Insights from experimental studies merging with molecular docking methods. Bioorganic Chemistry. 2019;88 doi: 10.1016/j.bioorg.2019.102981. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang W., Kollman P.A., Case D.A. Automatic atom type and bond type perception in molecular mechanical calculations. Journal of Molecular Graphics and Modelling. 2006;25(2):247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. Journal of Computational Chemistry. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Yan J., Zhang G., Hu Y., Ma Y. Effect of luteolin on xanthine oxidase: Inhibition kinetics and interaction mechanism merging with docking simulation. Food Chemistry. 2013;141(4):3766–3773. doi: 10.1016/j.foodchem.2013.06.092. [DOI] [PubMed] [Google Scholar]
- Zhang C., Zhang G., Liao Y., Gong D. Myricetin inhibits the generation of superoxide anion by reduced form of xanthine oxidase. Food Chemistry. 2017;221:1569–1577. doi: 10.1016/j.foodchem.2016.10.136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.