Summary
Hydrogen sulfide (H2S) and downstream reactive sulfur species (RSS), including organic persulfides, protect bacterial cells against diverse oxidative stressors. Specialized dithiol-based transcriptional repressors sense persulfides directly to control cellular H2S/RSS and avoid toxicity. Here, we present a protocol to quantify the kinetics of chemical reactivity of cysteines in two bacterial persulfide sensors toward cysteine persulfide and glutathione persulfide, with a LC-ESI-MS analysis that results in a kinetic model. This protocol has potential applications to other cysteine-containing proteins and oxidants.
For complete details on the use and execution of this protocol, please refer to Fakhoury et al. (2021) and Capdevila et al. (2021).
Subject areas: Protein Biochemistry, Mass Spectrometry
Graphical abstract
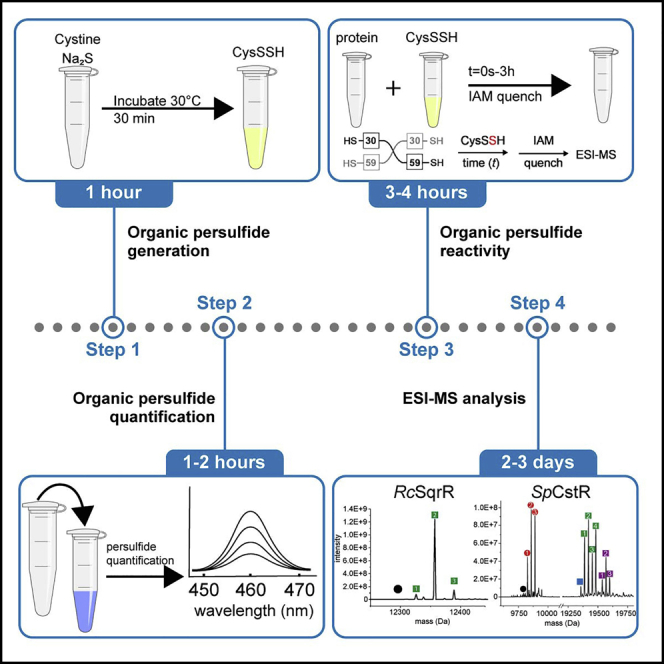
Highlights
-
•
Persulfide generation from reduction with sulfide of oxidized cysteine and glutathione
-
•
Cysteine reactivity profiling strategy for proteins of interest that respond to RSS
-
•
LC-ESI-MS analysis to study product distribution and kinetics of cysteines with RSS
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Hydrogen sulfide (H2S) and downstream reactive sulfur species (RSS), including organic persulfides, protect bacterial cells against diverse oxidative stressors. Specialized dithiol-based transcriptional repressors sense persulfides directly to control cellular H2S/RSS and avoid toxicity. Here, we present a protocol to quantify the kinetics of chemical reactivity of cysteines in two bacterial persulfide sensors toward cysteine persulfide and glutathione persulfide, with a LC-ESI-MS analysis that results in a kinetic model. This protocol has potential applications to other cysteine-containing proteins and oxidants.
Before you begin
The protocol described below outlines a mass spectrometry (MS)-based kinetic profiling scheme that is used to elucidate the mechanism of chemical reactivity of in situ-generated CysSSH and GSSH with two bacterial persulfide sensors, SqrR (sulfide-responsive transcriptional repressor) from Rhodobacter capsulatus (Capdevila et al., 2021; Shimizu et al., 2017) and CstR (CsoR-like sulfurtransferase repressor) (Luebke et al., 2014) from Streptococcus pneumoniae (Fakhoury et al., 2021). These two proteins are representative of two classes of kinetic profiles obtained with thiol persulfides: a simple time course of formation of a major product, with little to no accumulation of intermediates or side products, and a far more complex time dependence characterized by many species, including several highly populated intermediates, respectively. This protocol can be used for other proteins that form disulfide crosslinks that may or may not proceed through persulfidated intermediates and other persulfide sulfur donors, including naturally occurring thiol persulfides, coenzyme A persulfide and bacillithiol persulfide, and as well as synthetic persulfide sulfur donors.
Generation of the organic thiol persulfide
Timing: 1–2 h
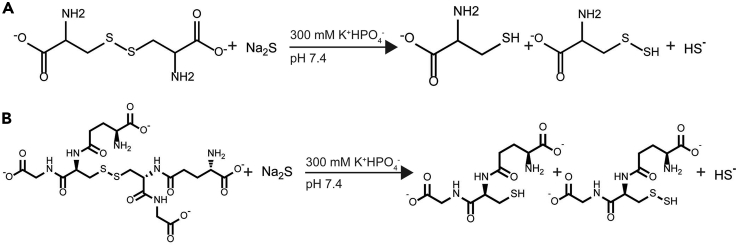
The initial steps of the protocol are to generate the persulfide of interest, in this case cysteine persulfide (Figure 1A) and glutathione persulfide (Figure 1B). These steps can also be applied to other oxidants, such as oxidized bacillithiol, oxidized CoA and oxidized homocysteine, to generate their respective persulfide.
-
1.Weigh out cystine (or glutathione disulfide) in a 1.5 mL screwcap tube.
-
a.Due to the low solubility of cystine, dissolve in less than 3% v/v HCl to ensure that all the cystine is dissolved.
-
b.Fill to the desired volume with ddH2O to obtain a final concentration of 50 mM.
-
a.
-
2.Prepare 250 mM of Na2S.
-
a.Na2S is kept in an argon-filled glovebox (≤1 ppm O2) to avoid oxidation.
-
b.First weigh an empty, capped 1.5 mL screwcap microfuge tube.
-
c.Next, bring the tube into the glovebox and add a small amount of Na2S into the capped 1.5 mL tube.
-
i.To obtain approximately 250 mM Na2S, 19.5 mg of Na2S is dissolved in 1 mL of 300 mM sodium phosphate, pH 7.4. also stored in the anaerobic glovebox.
-
i.
-
d.Remove the capped 1.5 mL tube with the Na2S and obtain a final weight of the tube with Na2S.
-
e.Return the 1.5 mL tube to the glovebox and add the appropriate amount of buffer needed to obtain 250 mM Na2S.
-
a.
-
3.Mix 50 mM cystine and 250 mM Na2S in a 1:1 volume ratio.
-
a.For a 1 mL sample, mix 500 μL of 50 mM cystine and 500 μL of 250 mM Na2S.
-
b.Once mixed, the solution will turn slightly yellow.
-
a.
-
4.Incubate this reaction mixture at 30°C for 30 min outside the glovebox.
-
a.This mixture was placed in a screwcap tube to avoid potential contact with the air and incubated in a water bath set to 30°C.
-
a.
-
5.
After 30 min, return the reaction mixture to the glovebox to begin a cold cyanolysis assay to determine the persulfide concentration.
CRITICAL: First, when dissolving cystine in HCl, ensure that less than 3% v/v of HCl should be used. If too much HCl is added, the color of the solution following addition of cystine to Na2S will be white and unusable. The expected color of the solution is yellow. If a white solution is obtained, discard the solution.
Note: To make GSSH instead of CysSSH, generally lower concentrations are used for similar incubation times (30 mM glutathione disulfide in step 1 and 150 mM Na2S in step 2). Step 1a is not necessary when preparing GSSH.
Figure 1.
Persulfide generation reaction
(A and B) Reaction scheme for generating cysteine persulfide (A) and glutathione persulfide (B) by reduction of the corresponding disulfide with excess sulfide salt under anaerobic conditions.
Cold cyanolysis assay to determine the persulfide concentration
Timing: 1–2 h
This section of the protocol describes steps to calculate the calculated concentration of the generated persulfide. The concentration of the generated persulfide is determined spectrophotometrically through an Fe-thiolate bond with an absorbance at 460 nm. Once the concentration of the persulfide is determined, the organic thiol persulfide (CysSSH or GSSH) should be used immediately in reactions with a protein of interest.
-
6.Prepare 25 μL KSCN standards that will be used to generate a standard curve and determine the persulfide concentration.
-
a.Freshly prepare a 1 M stock of KSCN.
-
b.Prepare serial dilutions from the 1 M stock to a total volume of 100 μL for each dilution.
[KSCN] (mM) Volume of KSCN from previous tube (μL) Milli-Q H2O (μL) Prepare Fresh 1 M KSCN 100∗ 10 90 10∗ 10 90 4 40 60 2 50 50 1 50 50 0.5 50 50 0 0 25 ∗Prepared only to minimize the error in the serial dilution, do not proceed with these tubes for the rest of the assay. -
c.Remove 25 μL from the 4 mM, 2 mM, 1 mM, and 0.5 mM tubes and transfer to separate microfuge tubes.
-
a.
-
7.
Make 5× and 10× dilutions of the freshly prepared CysSSH or GSSH solution inside the glovebox to 25 μL total, brought to volume with milli-Q H2O and remove from the glovebox.
-
8.To each 25 μL aliquot for both the standards and CysSSH or GSSH, add the following reagents in this order:
-
a.20 μL of 1 M ammonium hydroxide.
-
b.180 μL of milli-Q H2O.
-
c.25 μL of 0.5 M KCN, freshly prepared, and incubate at room temperature (22°C–26°C) for 45 min.
-
d.5 μL of 37% formaldehyde.
-
e.50 μL of Goldstein reagent.
-
a.
-
9.Once the Goldstein reagent is added, measure the absorbance of each sample at 460 nm (Figure 2A).
-
a.Use the 0 mM KSCN standard as the blank and measure the absorbance for the diluted standards first before measuring the absorbance of the 5× and 10× diluted cysteine persulfide solutions.
-
a.
-
10.
After obtaining the absorbance measurements, construct a standard curve that plots the known concentrations of the standard solutions against their A460 values (Figure 2B).
-
11.Calculate the concentration of both diluted CysSSH or GSSH samples from the standard curve to determine the stock concentration.
-
a.Average stock concentrations typically range from 15–23 mM CysSSH and 9–13 mM GSSH using this procedure.
-
a.
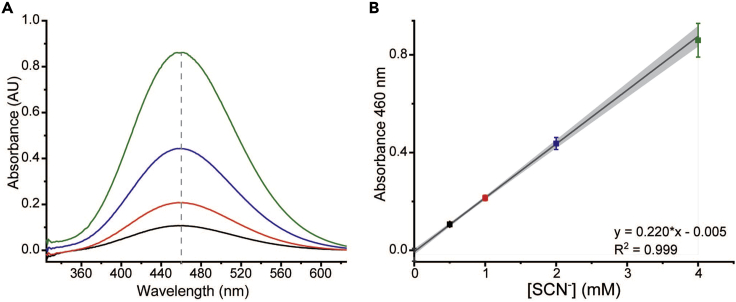
Figure 2.
Cold cyanolysis standard curve for persulfide quantification
(A) Representative UV-Vis spectra from the cold cyanolysis standard curve (1 mM SCN-, black; 2 mM SCN-, red; 2 mM SCN- in, blue; 4 mM SCN- in, green). The absorbance at 460 nm is indicated by a dashed gray line. Black line corresponds to the 0.5 mM standard, red line corresponds to the 1 mM standard, blue line corresponds to the 2 mM standard and the green line represents the 4 mM standard.
(B) Triplicate dataset of the standard curve at 460 nm fitted to a linear equation. The concentrations and colors from panel A match the square points plotted in panel B.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| L-Cystine | Sigma-Aldrich | Cat#C8755 |
| L-Glutathione oxidized | Sigma Aldrich | Cat#G4376 |
| Sodium sulfide | Sigma-Aldrich | Cat#407410 |
| Sodium phosphate Monobasic | Macron Chemicals | Cat#7892-04 |
| Sodium phosphate Dibasic | Fisher Scientific | Cat#S374 |
| Potassium thiocyanate | Sigma-Aldrich | Cat#P3011 |
| Ammonium hydroxide | EMD Chemicals | Cat#AX1303 |
| Potassium cyanide | Fluka Analytical | Cat#60180 |
| Formaldehyde | Macron Chemicals | Cat#5016-02 |
| Ferric nitrate nonahydrate | Sigma-Aldrich | Cat#216828 |
| Nitric acid | Fluka Analytical | Cat#84385 |
| EDTA | Alfa Aesar | Cat#A10713 |
| Iodoacetamide | Sigma-Aldrich | Cat#I6125 |
| Software and algorithms | ||
| MassLynx | Waters | https://www.waters.com/nextgen/us/en.html |
| DynaFit | BioKin | http://www.biokin.com/dynafit/ |
| Other | ||
| Anaerobic glovebox | VAC | https://usedglovebox.com/ |
| Temperature controlled centrifuge for 2 mL tubes under anaerobic conditions | Eppendorf | Eppendorf® Centrifuge 5425 R |
| UV-Vis spectrometer | HP | HP 8452A UV-Vis Diode Array Spectrophotometer with Multicell |
| HPLC-ESI-MS instrument, for example Waters/Micromass LCT Classic time-of-flight (TOF, Milford, MA) mass spectrometer | Waters | https://www.waters.com/nextgen/us/en.html |
| Agilent BioBasic C4 reversed-phase column (5 μm particle size, 300 Å pore size) | Agilent | https://www.agilent.com/ |
Materials and equipment
Persulfide reactivity buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| sodium phosphate pH 7.4 (300 mM) | 150 mM | 25 mL |
| EDTA (0.5 M) | 2 mM | 200 μL |
| ddH2O | n/a | 24.8 mL |
| Total | n/a | 50 mL |
Stored anaerobically at room temperature, (22°C–26°C).
Step-by-step method details
Organic thiol persulfide reactivity experiments
Timing: 3–4 h
The organic thiol persulfide (CysSSH or GSSH) stock solution prepared above should be used immediately in reactions with a protein of interest. This step is carried out entirely in an anaerobic glovebox to prevent the reaction of protein-derived cysteines with molecular oxygen. The same protocol is used for SqrR and CstR as outlined below.
-
1.Transport an aliquot of protein of interest into the glovebox.
-
a.Buffer exchange the protein into the persulfide reactivity buffer detailed above, stored at 22°C–26°C for two months, using a PD MiniTrap G-25 spun-column (GE, Boston, Massachusetts). This is essentially a gel filtration column with low resolution that can be used for buffer exchange. Alternatively, Amicon filters can be used (Amicon filter protocol).
-
i.The protocol for exchanging the protein buffer into the persulfide reactivity buffer is provided by the manufacturer and followed precisely. The column is first equilibrated in the persulfide reactivity buffer, then the protein sample in the original buffer is loaded, and finally, the protein is eluted with persulfide reactivity buffer, and the salts are retained in the column.If a reducing agent, TCEP or dithiothreitol, is still present after buffer exchange, the desired reaction products will not form with the addition of the persulfide. It is therefore useful to have a positive control that can be used to evaluate the integrity of the reactant, prior to starting the reaction.
-
i.
-
b.Set up 13 microfuge tubes (39 total tubes if the experiment is performed in triplicate, which is recommended to reach significant conclusions) to accommodate reactions run for specific time durations ranging from 0 to 3 h. These can be set up while the protein is exchanged into the intended buffer.
-
c.In a typical experiment, timepoints were taken at these specified times; 0 s, 30 s, 1 min, 5 min, 10 min, 15 min, 30 min, 45 min, 1 h, 1.5 h, 2 h, 2.5 h, 3 h. However, these can be varied depending on the rates of reaction.Note: For an initial trial, it would be best to start with a simple end point assay, with a sample taken at a single (long) timepoint, 3 h timepoint for example, and subjected to MS-based analysis (see below).
-
a.
-
2.Prepare a stock solution of Iodoacetamide (IAM) at a concentration where when used as a quenching solution, it would result in a 900-fold molar excess over protein thiolates. IAM caps any unreacted cysteine thiolates and persulfides that form during the course of the reaction.
-
a.Ensure that the reduced protein can be fully capped before starting the reaction by mixing the protein of interest with IAM and analyzing through LC-ESI-MS. With the two persulfide sensors discussed here, the cysteines are readily capped, but this may not be true for other proteins. The molar excess of IAM can also be optimized empirically with the reduced protein of interest.
-
a.
-
3.
Add 50 μL of the 900-fold molar excess IAM stock into each labeled microfuge tube.
-
4.Calculate the volume of protein, persulfide solution and buffer needed for the reaction and mix the protein and buffer in a separate microfuge tube.
-
a.The final concentration of the protein used is typically 60 μM.
-
b.The final concentration of persulfide corresponds to a 20-fold molar excess over the protein, or 1.2 mM.
-
c.Before adding persulfide to the reaction mixture, remove 50 μL unreacted protein and add to the first tube containing 50 μL of IAM, which will be labeled as the 0 s timepoint.
-
i.If done in triplicate, make sure to adjust volumes to ensure sufficient sample for three sets of timepoints. For example, if 13 points are run in triplicate, a total volume of 2000 mL of reaction mixture will be used.
-
i.
-
a.
-
5.
Add the calculated volume of persulfide to start the experiment. Start the timer with the addition of persulfide to the reaction mixture.
-
6.At each specified timepoint, remove 50 μL from the reaction mixture and add to a microfuge tube containing 50 μL of IAM to quench the reaction.
-
a.Repeat this step for each timepoint.
-
b.Clean-up step(s) can be added after quenching the reaction to minimize any reaction of IAM with other residues beyond Cys.
-
a.
-
7.Once all timepoints are collected, all quenched samples can be removed from the glovebox.
-
a.All cysteines in the protein have either reacted with CysSSH or GSSH to generate a product and/or have been capped with IAM, preventing any oxidation with molecular oxygen.
-
a.
Clean-up step for the preparation of persulfide reactions for LC-ESI-MS analysis
Timing: 1–2 h
This step allows for the removal of excess persulfides and IAM to prepare samples for ESI-MS injection and downstream analysis. This clean-up step should be performed right after the quenching occurs to prevent additional undesired modifications with IAM. This step can be removed if no additional peaks appear in the chromatogram.
-
8.Obtain a 10 kDa mini-concentrator (MilliporeSigma, Burlington, Massachusetts) for each sample obtained from the reactivity experiments described above.
-
a.Since the molecular weights of reduced CstR and SqrR are approximately 40 kDa (tetramer) and 24 kDa (dimer) respectively, 10 kDa mini-concentrators were used. Other molecular weight cut-off filters can be used depending on the native molecular weight of the protein of interest.
-
b.Make sure to label each mini-concentrator identically to each timepoint used.
-
a.
-
9.Add 400 μL of persulfide reactivity buffer to each tube containing the 100 μL mixture of IAM and oxidized protein.
-
a.The persulfide reactivity buffer used in this clean-up step is the same as the persulfide reactivity buffer used in the reaction mixture.
-
b.Any excess buffer within the glovebox can be transported out with the reacted protein to be used in the clean-up steps.
-
a.
-
10.
Remove all the solution (500 μL) from each timepoint sample and add to the labeled 10 kDa mini-concentrator corresponding to the same timepoint.
-
11.
Place all tubes into a tabletop microcentrifuge and spin samples for 10 min at 15,871 × g.
-
12.
Next, remove the flow-through and add an additional 400 μL of buffer to all samples and repeat. Repeat this step at least two more times to ensure removal of all unreacted cysteine persulfide and IAM.
-
13.5 μL of each sample was aliquoted into a screwcap vial (Waters, Milford, Massachusetts) with 45 μL of persulfide reactivity buffer and analyzed by LC-ESI-MS as described below. Samples are stable at this point and can be stored at –80°C prior to analysis if needed.
-
a.The mass spectrometer used is a Waters/Micromass LCT Classic time-of-flight (TOF, Milford, MA) mass spectrometer with a CapLC inlet following chromatography on a 50 μm Agilent BioBasic C4 reversed-phase column (5 μm particle size, 300 Å pore size).
-
b.5 μL of each sample is injected and subjected to a 10 min linear gradient from Solvent A (5% acetonitrile, 95% water, 0.1% formic acid) to Solvent B (95% acetonitrile, 5% water, 0.1% formic acid) with the elution monitored at 215 nm. Data were acquired and analyzed using MassLynx Software (Waters, Milford, MA).
-
a.
Pause point: There are places to pause briefly after determining the concentration of the persulfide and after cleaning up all samples with buffer, prior to the mass spectrometry.
Quantification and statistical analysis of LC-ESI-MS data
Timing: 2–3 days
This section outlines the steps that we routinely take to convert the raw “intact protein” LC-ESI-MS chromatograms into progress curves that present the fractional population of all significant features as a function of reaction time t.
-
14.
Chromatograms are processed individually by calculating peak intensities for those features that are characterized by an intensity that is ≥20% that of the most intense feature in each chromatogram.
-
15.
As the peak intensities will depend on the mass of covalently linked unit, a factor that takes into account the differential ionization efficiencies of the monomeric species vs. oligomers need to be estimated to calculate the relative concentrations in solution from the individual peak intensities. e.g., a disulfide bond linking two Cys from opposite protomers would give rise to a dimeric peak of relative less intensity compared to the same concentration of reduced monomer.
Note: This factor can be determined empirically by measuring the peak intensities associated with the monomeric vs dimeric species. For example, it can be calculated as the ratio between the intensity of the reduced and IAM-capped monomer and the intensity fully oxidized dimeric species, e.g., obtained with excess hydrogen peroxide or with the potent oxidant tetrathionate. This factor was found to be 4 for the CstR systems we have studied in detail and was found to be independent of the CstR species (data not shown).
-
16.
The adjusted peak intensities are summed, and fractional peak intensities obtained and averaged for individual features (defined on a basis of mass, ±1 Da) from triplicate experiments with standard deviations of the mean and then plotted as function of reaction time, t.
Note: The multiplicative factor is introduced to account for the different ionization efficiencies of each covalent oligomeric species. The total signal should remain constant among all time points and no significant deviations in signal over time should be detected.
-
17.
For CstRs, which harbor a cysteine pair across the protomer interface on each end of the dimeric unit, we binned individual features into one of five (minimal) species that share an underlying common structural feature but are otherwise heterogeneous with respect to the number of sulfur additions or mixed di- and trisulfide linkages to the organic thiol.
These five species are (1) underivatized, IAM-capped monomer (starting material); (2) derivatized monomer, typically characterized by per- and polysulfide chains on one or both cysteines; (3) closed/open dimeric species, in which one side of the dimer is crosslinked across the protomer interface, where on the other side, are not disulfide crosslinked, but chemically modified; (4) di/di species, corresponding to disulfide crosslinks on both sides of the dimer; and (5) closed/closed species, which contain more complex tri- and tetrasulfide crosslinked species on both sides of dimer.
Note: How one bins MS features is completely operationally defined by the user. Our goal was to use a kinetic model(s) to analyze the progress curves for individual species globally, thus motivating the use of a minimal number of “species”, or more accurately a reduced representation “meta-species node” approach. This reduced representation approach readily permits comparisons between different proteins or a mutant vs. a wild-type protein, while also minimizing the possibility of “over-fitting” the data.
Note: Although we could detect no crosslinked tetrameric species in these reactions, this should be carefully evaluated. All data are analyzed with the simplifying assumption that the individual dimer units react independently of one another within the tetramer.
-
18.
The resulting progress curves are then globally analyzed (no weighting function was used) by applying the simplest possible kinetic model, e.g., a linear model, unless otherwise stated, using DynaFit (Kuzmic, 1996). Schematic representations of these fitting models are shown in each case. Data were fitted using a pseudo-first order reaction, justified by the ≈20-fold molar excess of oxidant to protomer or ≈10-fold molar excess of oxidant to Cys residues.
Expected outcomes
This protocol will yield a series of LC-ESI-MS chromatograms that define the relative concentrations of macromolecular species that are formed as function of reaction time (t) with a persulfide donor, in this case, cysteine and glutathione persulfide.
The protocol is effectively oxidant-agnostic, and in unpublished data we have used this protocol to obtain kinetic profiles with other persulfide donors, and indeed other oxidants (cf. Dillon et al., 2021). Reactions carried out with persulfide donors leverage the nucleophilicity of both the thiolate and the thiol persulfide anion, so that both can be capped with the electrophile, IAM (Capdevila et al., 2021; Fakhoury et al., 2021).
With each family of reactive sulfur species dependent regulators (namely, SqrR and CstR), dramatically different outcomes were observed when reacted with in situ generated organic persulfides (Figures 3A and 3B).
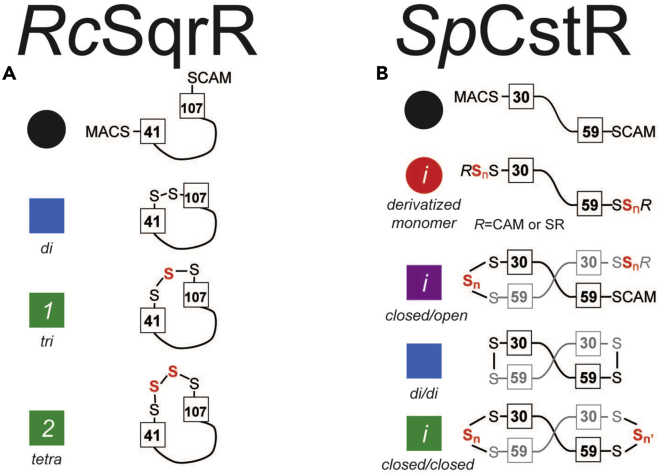
Figure 3.
Reactants, intermediates, and final products observed from the reaction with persulfides
(A) Representation of the different species identified in for the mass spectrometry data from SqrR reactivity experiment with glutathione persulfide, where the cysteine residues, C41 and C107, are boxed and the rest of the protein is depicted by a black line. The main species observed are reduced (black), di (blue), tri (green 1), and tetra (green 2).
(B) Representation of the different species identified in for the mass spectrometry data from CstR reactivity experiment with cysteine persulfide, where the cysteine residues, C30 and C59, are boxed and the first protomer is depicted by a black line and the second protomer, which forms the intermolecular crosslink, is depicted by a gray line. The main species observed are reduced (black), derivatized monomer (red), closed/open (purple), di/di (blue) and closed/closed (green).
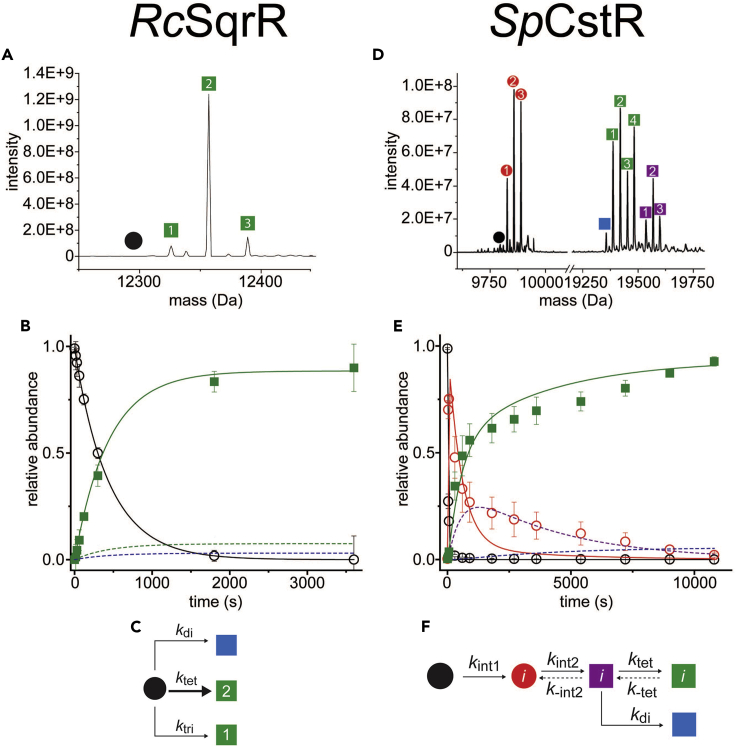
In case of RcSqrR, reduced protein capped with IAM is observed at the initial timepoint without glutathione persulfide (GSSH), generated, in situ, in a way that is identical to the preparation of CysSSH. Addition of GSSH yields a single major feature with a mass shift of +62 Da, consistent with an intraprotomeric tetrasulfide linkage, observed nearly exclusively at all timepoints (Figures 3A, 4A, and 4B). The formation of the tetrasulfide bridge is kinetically well-modeled using a pseudo-first-order rate equation. The two minor species observed, di- and trisulfide species, are modeled as parallel reactions derived from reduced protein rather than on-pathway intermediates (Figure 4C) (Capdevila et al., 2021). RcSqrR is representative of a simple outcome of reactivity toward a persulfide donor (Figures 3A and 4A).
Figure 4.
Mass spectrometry results and mechanism
(A) Representative regions of the ESI-MS data obtained for a reaction of GSSH with RcSqrR. The symbols correspond to the reduced and IAM-capped (black) and tetrasulfide (green) monomeric species. No crosslinked dimer species are observed here since the cysteines that form the tetrasulfide linkage are derived from the same protomer (Capdevila et al., 2021).
(B) Relative abundance of the indicated species as a function of pulse time t fitted to the kinetic mechanism in panel C, represented by the continuous and dashed lines. Datapoints for the dashed lines corresponding to disulfide (red) and trisulfide (green) were omitted to improve the clarity.
(C) A minimal kinetic model with respect to the protein concentration without an intermediate step. Major species formed (tetrasulfide) is represented with a bold arrow. The two minor species (disulfide and trisulfide) were fitted to parallel reactions from the same starting material.
(D) Representative regions of the ESI-MS data obtained for a 10 min reaction of CysSSH with SpCstR. In each case, the monomer region is shown at left, and the crosslinked dimer region is shown at right. The symbols correspond to the following: capped monomer (black), polysulfidated monomer (red), closed/open (purple), di/di (blue), and closed/closed (green).
(E) Global analysis of the kinetic profiles for SpCstR using the generalized kinetic scheme and fitted to the minimal bifurcated model shown in panel F, represented by continuous and dashed lines. Datapoints for the dashed lines corresponding to closed/open (purple) and di/di (blue) were omitted to improve the clarity.
(F) Generalized kinetic scheme used to analyze the reaction profiles obtained with persulfide donors. Continuous lines represent the processes that were determined in all profiles, whereas dashed lines represent the processes that could only be determined for some profiles (Fakhoury et al., 2021).
In contrast, with SpCstR, far more MS features are observed during the course of a reaction. Reactivity of SpCstR with CysSSH yields a complex distribution of crosslinked products, where nearly quantitative formation of a more symmetric closed/closed product (green) is observed at longer timepoints (Figures 3B, 4D, and 4E). The interprotomer dithiol persulfide sensing site in CstR forms a variety of crosslinked products upon persulfide treatment in a way that depends on the incubation time. At early timepoints, we first observe a heterogeneous mixture of polysulfidated monomers (red) formed from the reduced monomer. This is followed by the formation of a “closed/open” dimeric intermediate (purple) which slowly gives way to formation of a heterogeneous mixture of closed/closed species (green) characterized by tri- and tetrasulfide linkages on either side of the dimer (Figure 4E). Interestingly, the symmetric disulfide (di/di) species (blue) does not accumulate to any significant degree in these reactions (Figure 4E). This finding, coupled with the appearance and subsequent disappearance of the asymmetric closed/open species, is consistent with an asymmetry of reactivity within each dimer unit in the tetramer (Fakhoury et al., 2021). A kinetic model consistent with these reactivity data is shown in Figure 4F.
A critical point to emphasize is that these kinetic models are by design, minimal models that consider the fewest number of reaction intermediates that globally capture the observed precursor-product relationships found in the data. These intermediates, which themselves are heterogeneous, are numbered to signify distinct species with mass differences of +32 Da (the addition of one S atom) and binned and considered as a single representative intermediate. Note also that the mass spectrometer cannot resolve structural isomers in these MS1 spectra, characterized here by distinct adductions on each cysteine but which sum to the total mass, providing further justification for grouping like species. This “reduced representation” of the data is done to constrain the kinetic modeling to the fewest number of kinetically resolvable steps so as to capture and quantify major features of these progress curves.
Limitations
A major limitation of the current protocol is the organic persulfide preparation itself, which is effectively an in situ generated, anaerobic, equilibrium mixture of the disulfide starting material, the component thiol and persulfide, and residual HS–. One can show clearly that the disulfide, the thiol and the HS– (provided this is free of polysulfide impurities) do not react with the reduced protein in control experiments carried out under the same conditions (Shimizu et al., 2017). However, as persulfide is pulled out from this in situ mixture, contaminating thiol and HS– may impact the species that are formed in these kinetic traces. These thiols and HS– can for example react with protein thiols to form protein derived persulfides (which can be monitored). Note however, this mixture of RSS mirrors what is likely observed in cells.
Another major limitation is the stability of persulfide that is generated. The generated persulfide product lasts approximately 48 h at room temperature (22°C–26°C) inside a glovebox. Cold cyanolysis was performed on each day after the initial reaction and the concentration of persulfide decreased between each experiment. The yellow color that is observed becomes less yellow as the persulfide donor degrades. Generating a freshly prepared stock solution of persulfide for each experiment is best practice to ensure maximal persulfide starting material.
Troubleshooting
Problem 1
White precipitation is observed with the formation of cysteine persulfide. (step 1).
Potential solution
-
•
Use less HCl to dissolve cystine.
Problem 2
Concentration of persulfide is low during cold cyanolysis measurement. (step 11).
Potential solution
-
•
Remake cystine and Na2S stocks to ensure stocks are freshly prepared.
-
•
Preform the reaction again to generate the organic persulfide using the newly made stocks.
Problem 3
Yield of reduced protein capped with IAM is low. (step 2).
Potential solution
-
•
Increase concentration of IAM relative to the protein concentration.
-
•
Ensure that the protein of interest isn’t already oxidized because IAM will not react with crosslinked cysteines.
Problem 4
Reactivity of protein of interest with cysteine persulfide is not observed. (step 7).
Potential solution
-
•
Confirm that the generated persulfide has not aggregated.
-
•
Preform the reaction again to generate the organic persulfide using the newly made stocks.
Problem 5
Observation of low protein signal on LC-ESI-MS instrument. (step 14).
Potential solution
-
•
Increase ratio of protein to buffer and rerun the samples.
-
•
Make sure that protein concentration has not decreased due to precipitation which will result in lower signal.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, David P. Giedroc (giedroc@indiana.edu).
Materials availability
This study did not generate new, unique reagents.
Acknowledgments
We thank members of the Giedroc and Capdevila laboratories for helpful comments on the manuscript. We gratefully acknowledge Dr. J. Trinidad for his assistance in the Laboratory for Biological Mass Spectrometry at IU-Bloomington; US National Institutes of Health (R35 GM118157 to D.P.G.); Bunge & Born, Argentina; Williams Foundations; and MinCyT Argentina (PICT 2019-00011 to D.A.C.). D.A.C. is a staff member from CONICET, Argentina. Funding for open access charge: US National Institutes of Health (R35 GM118157 to D.P.G.).
Author contributions
J.N.F. and D.A.C. wrote the manuscript as well as developed the experimental methods and performed the experiments. D.P.G. wrote the manuscript and supervised the project.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Joseph N. Fakhoury, Email: jfakhour@iu.edu.
Daiana A. Capdevila, Email: dcapdevila@leloir.org.ar.
David P. Giedroc, Email: giedroc@indiana.edu.
Data and code availability
Raw LC-ESI-MS datasets are available from the lead contact upon request. Processed LC-ESI-MS datasets are provided in Fakhoury et al. (2021) and Capdevila et al. (2021).
References
- Capdevila D.A., Walsh B.J.C., Zhang Y., Dietrich C., Gonzalez-Gutierrez G., Giedroc D.P. Structural basis for persulfide-sensing specificity in a transcriptional regulator. Nat. Chem. Biol. 2021;17:65–70. doi: 10.1038/s41589-020-00671-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon K.M., Morrison H.A., Powell C.R., Carrazzone R.J., Ringel-Scaia V.M., Winckler E.W., Mcalister Council-Troche R., Allen I.C., Matson J.B. Targeted delivery of persulfides to the gut: effects on the microbiome. Angew. Chem. Int. Ed. 2021;60:6061–6067. doi: 10.1002/ange.202014052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhoury J.N., Zhang Y., Edmonds K.A., Bringas M., Luebke J.L., Gonzalez-Gutierrez G., Capdevila D.A., Giedroc D.P. Functional asymmetry and chemical reactivity of CsoR family persulfide sensors. Nucleic Acids Res. 2021;49:12556–12576. doi: 10.1093/nar/gkab1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- Luebke J.L., Shen J., Bruce K.E., Kehl-Fie T.E., Peng H., Skaar E.P., Giedroc D.P. The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol. Microbiol. 2014;94:1343–1360. doi: 10.1111/mmi.12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Shen J., Fang M., Zhang Y., Hori K., Trinidad J.C., Bauer C.E., Giedroc D.P., Masuda S. Sulfide-responsive transcriptional repressor SqrR functions as a master regulator of sulfide-dependent photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 2017;114:2355–2360. doi: 10.1073/pnas.1614133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw LC-ESI-MS datasets are available from the lead contact upon request. Processed LC-ESI-MS datasets are provided in Fakhoury et al. (2021) and Capdevila et al. (2021).