Summary
RNA-binding proteins (RBPs) are multifunctional proteins that shuttle between the nucleus and the cytoplasm where they assemble with target RNAs to form multi-molecular complexes. Here, we describe a protocol to selectively identify RNAs associated with RBPs of interest in the cytoplasmic and nuclear compartments of adult Drosophila brain cells. Cytoplasmic and nuclear fractions are differentially collected and used for immunoprecipitation-based purification of GFP-tagged RBPs. This protocol can be applied to samples expressing ectopic or endogenous tagged RBPs.
Subject areas: Cell separation/fractionation, Gene Expression, Model Organisms, Molecular Biology, Protein Biochemistry
Graphical abstract

Highlights
-
•
Protocol to identify RNAs associated with RBPs in adult Drosophila brain
-
•
Fractionation of Drosophila head lysates into cytoplasmic and nuclear extracts
-
•
Optimized RNA-IP protocol from cytoplasmic and nuclear fractions using GFP-trap beads
-
•
Provides RNA samples that can be used for deep-sequencing and/or qRT-PCR
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
RNA-binding proteins (RBPs) are multifunctional proteins that shuttle between the nucleus and the cytoplasm where they assemble with target RNAs to form multi-molecular complexes. Here, we describe a protocol to selectively identify RNAs associated with RBPs of interest in the cytoplasmic and nuclear compartments of adult Drosophila brain cells. Cytoplasmic and nuclear fractions are differentially collected and used for immunoprecipitation-based purification of GFP-tagged RBPs. This protocol can be applied to samples expressing ectopic or endogenous tagged RBPs.
Before you begin
The protocol below describes how to immunoprecipitate GFP-tagged RNA-binding proteins (RBPs) and their associated RNAs from cytoplasmic and nuclear head fractions of adult Drosophila. We have used this protocol starting from flies expressing tagged RBPs specifically in adult neurons, using the Gal4/UAS system (Brand and Perrimon, 1993). Such experiments require generating fly line(s) expressing the GFP-tagged protein(s) of interest under the control of UAS sequences, and crossing this line(s) with flies expressing both Gal4 in neurons and the temperature sensitive Gal4 inhibitor Gal80ts (McGuire et al., 2004). Flies expressing sole GFP under the control of UAS sequences can be used as a reference. In these conditions, three weeks are needed to obtain a progeny raised at restrictive temperature (18°C) and an additional week to age the flies at permissive temperature (29°C) (Figure 1, green boxes). Notably, RNA-immunoprecipitation (RIP) can also be performed on lines expressing GFP-tagged RBPs from their endogenous locus. These lines may already be available in collections of stocks generated through protein-trap (Kelso et al., 2004; Lowe et al., 2014; Morin et al., 2001) or recombination-mediated cassette exchange (Nagarkar-Jaiswal et al., 2015a, 2015b) approaches. Alternatively, they can be generated using the CRISPR-Cas9 methodology (Kina et al., 2019). It is possible to adapt this protocol to the immunoprecipitation of proteins fused to tags adapted to biochemical approaches (Flag, V5…), although fly lines generated with these tags may be more difficult to obtain and will need to be generated on purpose.
Figure 1.
Overall schematic workflow of the protocol
For each main stage (box), the expected duration and the corresponding steps are indicated.
Sieves, funnels, mortars, brushes and Dounce Tissue Grinders should be washed with RNase-free distilled water and cleared of RNase traces using RNase ZAP at least one day before the experiment. They should then be wrapped in aluminum foils and stored at −20°C.
To minimize RNase contamination during the experiment, it is recommended to clean the workspace and pipettes with RNase ZAP before starting, and to use filter tips and RNAse-free tubes throughout the procedure.
Refrigerated centrifuges should be used and set to 4°C before starting.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat Anti-mouse IRDye 800 (dilution 1:10,000) | Thermo Fisher Scientific/Invitrogen | Cat#SA5-10156; RRID: AB_2556736 |
| Goat Anti-rabbit AF680 (dilution 1:10,000) | Thermo Fisher Scientific/Invitrogen | Cat#A21076; RRID: AB_2535736 |
| Mouse monoclonal Anti-Lamin Dm0 (dilution 1:2,000) | DSHB | Cat#ADL 67.10; RRID: AB_528336 |
| Mouse monoclonal Anti-Lamin Dm0 (dilution 1:2,000) | DSHB | Cat#ADL 84.12; RRID: AB_528338 |
| Mouse monoclonal Anti-Tubulin (dilution 1:5,000) | Merck/Sigma-Aldrich | Cat#T9026; RRID: AB_477593 |
| Rabbit polyclonal Anti-GFP (dilution 1:2,500) | Torrey Pines Biolabs | Cat#TP401; RRID: AB_10013661 |
| Chemicals, peptides, and recombinant proteins | ||
| Bromophenol blue | Euromedex | Cat#EM-11470; CAS: 115-39-9 |
| CaCl2 (Calcium Chloride) | Merck/Sigma-Aldrich | Cat#C1016-500G; CAS: 10043-52-4 |
| CAPS | Merck/Sigma-Aldrich | Cat#C2632; CAS: 1135-40-6 |
| CHAPS | Euromedex | Cat#1083-B; CAS: 75621-03-3 |
| Chloroform | Merck | Cat#1024451000; CAS: 67-66-3 |
| CompleteTM, EDTA-free Protease Inhibitor Cocktail | Merck | Cat#11873580001 |
| CompleteTM mini, EDTA-free Protease Inhibitor Cocktail | Merck | Cat#11836170001 |
| Binding Control Agarose Beads | Chromotek | Cat#Bab-20 |
| DEPC (Diethyl Pyrocarbonate) | Merck/Sigma-Aldrich | Cat#D5758; CAS: 1609-47-8 |
| DNaseI AmbionTM (RNase-free) | Thermo Fisher Scientific | Cat#AM2222 |
| DTT (1,4 dithiothreitol) | Euromedex | Cat#EU0006-E; CAS: 3483-12-3 |
| EDTA | Euromedex | Cat#EU0007-B; CAS: 6381-92-6 |
| Ethanol Absolute ≥ 99,9% | VWR Chemicals BDH | Cat#EM1.00983.2500; CAS: 64-17-5 |
| GFP-Trap Agarose Beads | Chromotek | Cat#Gta-20 |
| Glycerol High purity | Euromedex | Cat#EU3550; CAS56-81-5 |
| GlycoBlueTM Coprecipitant (15 mg/mL) | Thermo Fisher Scientific | Cat#AM9515 |
| HEPES | Euromedex | Cat#10-110-C; CAS: 7365-45-9 |
| IGEPAL® CA-630 (NP40) | Merck/Sigma-Aldrich | Cat#I3021; CAS: 9002-93-1 |
| KCl (Potassium Chloride) | Merck | Cat#1049361000; CAS: 7447-40-7 |
| Methanol | Merck/Sigma-Aldrich | Cat#322415; CAS: 67-56-1 |
| MgCl2 (Magnesium Chloride) | Euromedex | Cat#2189-C; CAS: 7791-18-6 |
| NaCl (Sodium Chloride) | Merck | Cat#1064041000; CAS: 7647-14-5 |
| 2-propanol (Isopropanol) | Merck | Cat#1096341000; CAS: 67-63-0 |
| PBS 1× (no calcium, no magnesium) | Thermo Fisher Scientific | Cat#14190136 |
| Proteinase K | Thermo Fisher Scientific/Invitrogen | Cat#AM2546 |
| Skimmed milk powder | Dutscher | Cat#711160 |
| RNase free Water | Merck/Sigma-Aldrich | Cat#95284; CAS: 7732-18-5 |
| RNaseOUTTM Recombinant Ribonuclease Inhibitor | Thermo Fisher Scientific/Invitrogen | Cat#10777-019 |
| SDS | Euromedex | Cat#BI-SB0485; CAS: 151-21-3 |
| Sucrose | Merck/Sigma-Aldrich | Cat#S0389-500G; CAS: 57-50-1 |
| TRI Reagent® | Merck/Sigma-Aldrich | Cat#93289 |
| Tris HCl | Euromedex | Cat#EU0011; CAS: 1185-53-1 |
| Tris MBG (Tris Molecular Biology Grade) | Euromedex | Cat#200923-A; CAS: 77-86-1 |
| Tween®20 | Euromedex | Cat#2001-B; CAS: 9005-64-5 |
| Urea Ultra-Pure | Euromedex | Cat#EU0014-B; CAS: 57-13-6 |
| Critical commercial assays | ||
| Agilent RNA 6000 Pico Kit | Agilent Technologies | Cat#5067-1513 |
| QubitTM RNA HS Kit | Thermo Fisher Scientific | Cat#Q32852 |
| Experimental models: Organisms/strains | ||
| Drosophila: elav-Gal4 (C155) | Bloomington Drosophila Stock Center | BDSC #458 |
| Drosophila: tub-Gal80ts; TM2/TM6 | Bloomington Drosophila Stock Center | BDSC #7019 |
| Other | ||
| Agilent 2100 bioanalyzer | Agilent Technologies | Cat#G2939BA |
| Benchtop Liquid Nitrogen Containers | Fisher Scientific | Cat#11-670-4B |
| BrandTechTM Polypropylene Funnels | Fisher Scientific | Cat#12624930 |
| MμltiFlex Flat tips, 200 μL, 17 mm OD | Sorenson BioScience | Cat#17310 |
| FalconTM, 50 mL conical centrifuge tubes | Merck | Cat#10788561 |
| Falcon TM, 15 mL conical centrifuge tubes | Merck | Cat#10136120 |
| 3D shaker GyroMiniTM | Labnet International | Cat#S0500 |
| HaldenwangerTM Porcelain Pestles, 115 mm | Fisher Scientific | Cat#10405011 |
| Immobilon-P PVDF membrane | Merck/ Millipore | Cat#IPVH00010 |
| InvitrogenTM NuPAGETM 4%–12%, Bis-Tris,1.5 mm, Mini Protein Gel, 10-well | Fisher Scientific/Invitrogen | Cat#12020166 |
| InvitrogenTM NuPAGETM 4%–12%, MOPS SDS Running Buffer (20×) | Fisher Scientific/Invitrogen | Cat#11589156 |
| Life Sciences kimbleTM KontesTM Dounce Tissue Grinders, 15 mL | Fisher Scientific | Cat#10145564 |
| Minicellule XCell SureLockTM | Thermo Fisher Scientific | Cat#EI0001 |
| Odyssey® DLx imaging System | LI-COR Biosciences | N/A |
| Porcelaines AvignonTM Porcelain Mortar, 250 mL | Fisher Scientific | Cat#11832851 |
| Qubit 4 Fluorometer | Thermo Fisher Scientific | Cat#Q33226 |
| RNAse-free 1.5 mL microtube, TREFF CapLock | Sorenson BioScience | Cat#11510 |
| RNAse-free 1.5 mL low binding microtube | Sorenson BioScience | Cat#39640T |
| RNase ZapTM | Merck/Sigma-Aldrich | Cat#R2020-250ML |
| Refrigerated centrifuge | Eppendorf | Cat#5415R or 5418R |
| Refrigerated centrifuge | Beckman Coulter | Cat#Allegra 25R |
| RETSCH Stainless-Steel Sieves, diameter 100 mm, height 40 mm, pore size 400 μm | Fisher Scientific | Cat#10594862 |
| RETSCH Stainless-Steel Sieves, diameter 100 mm, height 40 mm, pore size 630 μm | Fisher Scientific | Cat#10034891 |
| Sterile syringe filter, porosity 0.20 μm, ClearLine® | Sarstedt | Cat#1826001 |
| ThermoMixer® C | Eppendorf | Cat#5382000015 |
| Western blot filter paper | Thermo Fisher Scientific | Cat#84783 |
Materials and equipment
Note: Stock solutions required to prepare working buffers are filtered (0.2 μm) and can be stored at room temperature for several months (with the exception of the sucrose solution).
Note: Distilled water treated with 0.1% diethyl pyrocarbonate (DEPC) was used to prepare all solutions. DEPC treatment was performed overnight (∼12–16 h) and the solutions were autoclaved.
Alternatives: Commercial RNase-free water can be used instead of DEPC water.
CRITICAL: DEPC is a hazardous product (CAS: 1609-47-8; GHS07) that should be manipulated under a fume hood, with lab coat, eye shields and gloves.
Note: Lysis, Sucrose, high salt and urea buffers should be prepared freshly, in RNAse-free conditions, using 50 mL and 15 mL Falcon tubes. They are kept on ice during the experiment.
Lysis buffer
| Reagent | Final concentration | Amount/Volume |
|---|---|---|
| HEPES, pH 8 (0.5 M) | 20 mM | 2 mL |
| KCl (1 M) | 125 mM | 6.250 mL |
| MgCl2 (1 M) | 4 mM | 200 μL |
| NP40 | 0.05% | 25 μL |
| DEPC water | n/a | 41.525 mL |
| Total | n/a | 50 mL |
Note: Immediately before use, dissolve one Complete EDTA-free protease inhibitor tablet (final concentration 1×) in 50 mL lysis buffer. Transfer 30 mL to another tube and add 150 μL RNaseOUT (dilution 1:200, final concentration 0.2 U/μL) and 30 μL DTT (dilution 1:1,000, final concentration 1 mM). Prepare freshly before use.
Sucrose buffer
| Reagent | Final concentration | Amount/Volume |
|---|---|---|
| Tris, pH 7.65 (1 M) | 20 mM | 400 μL |
| NaCl (5 M) | 60 mM | 240 μL |
| KCl (1 M) | 15 mM | 300 μL |
| Sucrose (1 M) | 0.34 M | 6.8 mL |
| DEPC water | n/a | 12.260 mL |
| Total | n/a | 20 mL |
Note: Immediately before use, dissolve two complete mini EDTA-free protease inhibitor tablets (final concentration 1 ×). Add 100 μL RNaseOUT (dilution 1:200, final concentration 0.2 U/μL) and 20 μL DTT (dilution 1:1,000, final concentration 1 mM). Prepare freshly before use.
High Salt buffer
| Reagent | Final concentration | Amount/Volume |
|---|---|---|
| Tris, pH 7.65 (1 M) | 20 mM | 100 μL |
| EDTA (0.5 M) | 0.2 mM | 2 μL |
| Glycerol (80%) | 25% | 1.56 mL |
| NaCl (5 M) | 900 mM | 900 μL |
| MgCl2 (1 M) | 1.5 mM | 7.5 μL |
| DEPC water | n/a | 2.43 mL |
| Total | n/a | 5 mL |
Note: Immediately before use, dissolve ½ complete mini EDTA-free protease inhibitor tablet (final concentration 1 ×), 25 μL RNaseOUT (dilution 1:200, final concentration 0.2 U/μL) and 5 μL DTT (dilution 1:1,000, final concentration 1 mM). Prepare freshly before use.
Urea buffer
| Reagent | Final concentration | Amount/Volume |
|---|---|---|
| Urea | 9 M | 0.54 g |
| Tris HCl, pH 8 (0.1 M) | 50 mM | 500 μL |
| CHAPS (20%) | 1% | 50 μL |
| DEPC water | n/a | up to 1 mL |
| Total | n/a | 1 mL |
Note: Prepare freshly before use.
CaCl2 buffer
| Reagent | Final concentration | Amount/Volume |
|---|---|---|
| CaCl2 | 1 M | 1.110 g |
| DEPC water | n/a | up to 10 mL |
| Total | n/a | 10 mL |
Note: This buffer is filtered (0.2 μm) and can be stored at room temperature (22°C–25°C) for several months.
EDTA buffer
| Reagent | Final concentration | Amount/Volume |
|---|---|---|
| EDTA | 0.5 M | 1.861 g |
| DEPC water | n/a | up to 10 mL |
| Total | n/a | 10 mL |
Note: This buffer is filtered (0.2 μm) and can be stored at room temperature (22°C–25°C) for several months.
5× Laemmli buffer
| Reagent | Final concentration | Amount/Volume |
|---|---|---|
| Tris HCl pH6.8 (1 M) | 312.5 mM | 6.25 mL |
| SDS | 10% | 2 g |
| Glycerol (100%) | 50% | 10 mL |
| β-Mercaptoethanol (100%) | 2.5% | 500 μL |
| Bromophenol Blue (0.25%) | 0.025% | 2 mL |
| Water | n/a | 1.25 mL |
| Total | n/a | 20 mL |
Note: Make aliquots (1 mL) and store at −20°C for up to a year.
Blocking buffer
| Reagent | Final concentration | Amount/Volume |
|---|---|---|
| Skimmed milk powder | 5% | 2.5 g |
| PBS (1×) | 1 × | up to 50 mL |
| Total | n/a | 50 mL |
Note: This buffer can be stored at 4°C for several days.
Transfer buffer
| Reagent | Final concentration | Amount/Volume |
|---|---|---|
| CAPS (10×) | 1 × | 100 mL |
| Methanol (100%) | 10% | 100 mL |
| Water | n/a | 800 mL |
| Total | n/a | 1 L |
Note: This buffer can be stored at 4°C for several months.
PBS-Tween
| Reagent | Final concentration | Amount/Volume |
|---|---|---|
| PBS (1×) | 1 × | 2 L |
| Tween (100%) | 0,1% | 2 mL |
| Total | n/a | 2 L |
Note: This buffer can be stored at room temperature for several months.
Step-by-step method details
The main steps of the procedure are displayed in Figure 1, in blue boxes.
Fly head collection
Timing: 45 min
In this step, heads from 5–7 day-old adult male and female flies are dissociated from the rest of the bodies and selectively collected.
Note: This protocol is modified from a previous publication (Tian et al., 2013).
-
1.Collect and freeze adult flies.
-
a.Transfer 5–7 day-old anesthetized flies into 50 mL Falcon tubes.
-
b.Snap freeze flies by immediately plunging the tubes into liquid nitrogen.
-
a.
Pause point: Flies can be stored at −80°C at this stage.
Note: 10–15 mL adult flies can be recovered from about 20 fly bottles.
Note: We recommend to not collect more than 25 mL of adult flies per 50 mL Falcon tube for optimal separation of fly heads in step 2.
-
2.Dissociate heads from fly carcasses.
-
a.Vortex the tubes for 15 s and shake vigorously to separate the heads, legs, and wings from the bodies.
-
b.Repeat this step twice or thrice.
-
a.
-
3.Selectively collect fly heads.
-
a.Assemble the sieves pre-chilled at −20°C together, such that the sieve with the bigger mesh size (630 μm) lies on top of the sieve with the smaller mesh size (400 μm) (Figures 2A and 2B).
-
b.Transfer the frozen fly material to the assembled sieves and sift by vigorous shaking. Fly bodies are retained on the top sieve, while heads are retained on the bottom one (Figure 2C). Smaller elements such as legs or wings are not retained and pass through both sieves.
-
c.Transfer the head fraction to 1.5 mL Eppendorf tubes using a funnel pre-chilled at −20°C and a brush. Prepare aliquot of 1 mL heads.
-
d.Store the collected head fractions at −80°C.
-
a.
Note: For sieves of a 100 mm diameter, we recommend to not use more than 60–80 mL of adult flies in step 3.
Note: About 1 mL of fly heads can be recovered from 30 mL adult flies.
CRITICAL: Step 2 must be performed in a cold room (4°C), and step 3 on powdered dry ice.
Figure 2.
Material used to collect fly heads
(A and B) Sieves of different mesh sizes (630 μm and 400 μm) are stacked such that the sieve with the bigger mesh size lies on top of the one with the smaller mesh size.
(C) After sifting, fly carcasses are found in the upper sieve (right) while fly heads are found in the lower one (left).
Preparation of head lysates
Timing: 30 min
In this step, fly heads are crushed and lysed.
- 4.
Note: Although starting from 1 mL should work for most proteins, the amount of fly heads to be used can be optimized depending on the expression level and pattern of the tagged RBP.
-
5.Prepare the head lysate.
-
a.Transfer the powder to a pre-chilled 15 mL Dounce homogenizer using a brush.
-
b.Add 8.5 mL of ice-cold Lysis Buffer (+ RNaseOUT + protease inhibitors + DTT).
- c.
-
a.
Note: We recommend using the smaller Dounce pestle for homogenization of the head lysate.
CRITICAL: These steps should be performed on ice, in a cold room.
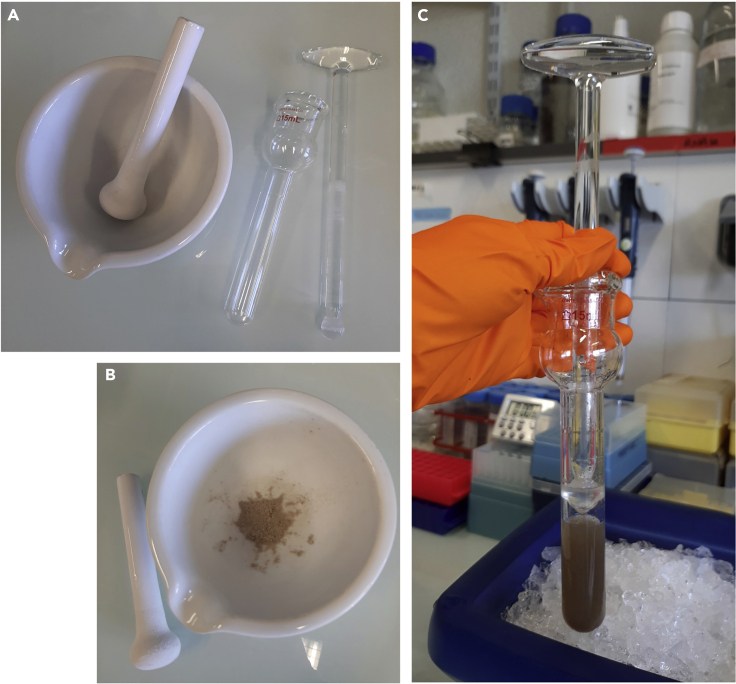
Figure 3.
Material used to prepare head lysates
(A) Porcelain mortar and pestle (left) and glass Dounce Tissue Grinder and pestle (right).
(B) Fly head powder obtained after grinding heads into liquid nitrogen-cooled mortar and pestle.
(C) Fly head lysate obtained after homogenization with the glass Dounce Tissue Grinder.
Preparation of cytoplasmic and nuclear fractions
Timing: 2–3 h
In this step, head lysates are fractionated into cytoplasmic and nuclear fractions and nuclear proteins are extracted (Figure 4).
-
6.
Transfer the homogenate to a 15 mL Falcon tube.
-
7.
Centrifuge at 100 × g for 5 min at 4°C to eliminate most of the cuticular and cellular debris.
-
8.
Transfer the supernatant to a new 15 mL Falcon tube. Save two aliquots: one of 100 μL for RNA extraction and one of 50 μL for protein extraction. These aliquots represent input fractions.
-
9.
Separate cytoplasmic and nuclear fractions by centrifuging at 900 × g for 10 min at 4°C.
-
10.Clarify and collect the cytoplasmic fraction.
-
a.Transfer the supernatant (cytoplasmic fraction) into a new 15 mL Falcon tube.
-
b.Centrifuge at 16,000 × g for 20 min at 4°C.
-
c.Repeat steps 10.a and 10.b.
-
d.Discard the remaining pelleted debris and transfer the supernatant to a 15 mL Falcon tube. Save two aliquots: one of 100 μL for RNA extraction and one of 50 μL for protein extraction. These aliquots represent cytoplasmic input fractions.
-
a.
-
11.Extract and collect the soluble nuclear fraction.
-
a.Wash the pellet recovered after step 9 in 1 mL of Sucrose Buffer (+ RNaseOUT + protease inhibitors + DTT). Centrifuge at 900 × g for 10 min at 4°C. Discard the supernatant.
-
b.Break the nuclear membrane.
-
i.Resuspend the pellet in 2 mL of Sucrose Buffer (+ RNaseOUT + protease inhibitors + DTT) supplemented with 800 μL of High Salt Buffer (+ RNaseOUT + protease inhibitors + DTT).
-
ii.Homogenize by pipetting up and down and incubate for 30 min on ice. Invert the tube every 5 min.
-
i.
-
c.Digest DNA.
-
i.Add 4.6 mL of Sucrose Buffer (+ RNaseOUT + protease inhibitors + DTT) supplemented with 7.5 μL of CaCl2 1 M to reach a final concentration of 150 mM NaCl and 1 mM CaCl2.
-
ii.Add 75 μL of DNaseI, invert the tube several times and incubate for 15 min at 37°C. Agitate every 5 min.
-
iii.Stop the reaction by adding 60 μL of EDTA 0.5 M (4 mM final concentration) and invert the tube 5 times.Note: This step is particularly important for chromatin-associated RNA-binding proteins.
-
i.
-
d.Collect the soluble nuclear fraction.
-
i.Centrifuge at 16,000 × g for 20 min at 4°C.
-
ii.Transfer the supernatant (soluble nuclear fraction) into a 15 mL Falcon tube on ice. Save two aliquots: one of 100 μL for RNA extraction and one of 50 μL for protein extraction. These represent soluble nuclear input fractions.Note: If desired, an aliquot corresponding to the nuclear insoluble fraction can be prepared by transferring 50 μL of the nuclear fraction collected in step 11.c.iii to a 1.5 mL Eppendorf tube, centrifuging it at 16,000 × g for 20 min at 4°C and resuspending the pellet into 50 μL of urea buffer.Note: To save time, we recommend performing steps 10.b and 10.c in parallel to steps 11.b.ii and 11.d.i.
 CRITICAL: Steps 6–11 should be performed on ice (except 11.c.ii).
CRITICAL: Steps 6–11 should be performed on ice (except 11.c.ii).
-
i.
-
a.
Figure 4.
The main steps of lysate preparation and fractionation
The images were taken after the indicated centrifugation steps.
Immunoprecipitation of GFP-tagged RNA-binding proteins
Timing: 4 h
In this step, GFP-tagged RNA-binding proteins are immuno-precipitated from the cytoplasmic and nuclear fractions using GFP-Trap beads.
-
12.Pre-clear the cytoplasmic and soluble nuclear fractions.
-
a.Prepare control agarose beads.
-
i.Transfer the beads to a 15 mL Falcon tube.Note: 120 μL agarose beads are used per fraction. Calculate the total amount needed (which depends on the number of conditions analyzed in parallel) and pipet the corresponding volume with a pre-cut tip.
-
ii.Add 500 μL of Lysis Buffer (per 120 μL of beads) and gently tap the tube.
-
iii.Centrifuge at 400 × g for 2 min at 4°C and discard the supernatant.
-
iv.Repeat two times steps 12.a.ii and 12.a.iii.
-
v.Add 500 μL of Lysis Buffer per 120 μL of beads, pipet up and down several times and split into independent 15 mL Falcon tubes. The number of Falcon tubes depends on the number of fractions prepared.
-
vi.Centrifuge at 400 × g for 2 min at 4°C and discard the supernatant.Note: Use MμltiFlex flat tips to avoid pipetting the beads.
-
i.
-
b.Pre-clear cytoplasmic and nuclear fractions with agarose beads.
-
i.Transfer each nuclear and cytoplasmic fraction to one of the Falcon Tubes prepared in step 12.a.v.
-
ii.Resuspend the beads by gently tapping the tubes.
-
iii.Agitate on a nutator for 30 min at 4°C.
-
iv.Centrifuge at 400 × g for 2 min at 4°C.
-
i.
-
a.
-
13.Immunoprecipitate GFP-tagged proteins.
-
a.Prepare agarose beads coupled to anti-GFP nanobody/VHH (GFP-Trap beads) as described in step 12.a.Note: we recommend performing step 13.a in parallel to step 12.a to spare time. Washed GFP-Trap beads can be stored on ice during step 12.b.
-
b.Immunoprecipitate GFP-tagged proteins from cytoplasmic and nuclear fractions.
-
i.Transfer each of the pre-cleared supernatant recovered in step 12.b to one of the individual Falcon Tubes prepared in step 13.a.
-
ii.Resuspend the beads by gently tapping the tubes.
-
iii.Agitate on a nutator for 1 h 30 min at 4°C.
-
iv.Centrifuge at 400 × g for 2 min at 4°C and discard the supernatant.Note: If desired, save two aliquots of the supernatant: one of 100 μL for RNA extraction and one of 50 μL for protein extraction. These represent unbound fractions.
-
i.
-
a.
-
14.Wash and elute the bound fractions.
-
a.Add 500 μL of Lysis Buffer (+ RNaseOUT + protease inhibitors + DTT) and resuspend the beads by gently tapping the tubes.
-
b.Centrifuge at 400 × g for 2 min at 4°C and discard the supernatant.
-
c.Repeat steps 14.a and 14.b three times.
-
d.Repeat step 14.a and transfer the resuspended bead solution to a 1.5 mL low binding Eppendorf tube. Save an aliquot of 20 μL for protein extraction. This represents the bound fraction.
-
e.Repeat step 14.b and discard the supernatant.
-
f.Resuspend the beads in 100 μL of Lysis Buffer (+ RNaseOUT + protease inhibitors + DTT).
-
g.To elute the bound RNAs, digest proteins by adding 1.5 μL of Proteinase K solution and incubating for 30 min at 55°C in a Thermomixer under agitation. Gently tap the tubes 3–4 times during the incubation.
-
h.Collect the supernatant in a 1.5 mL low binding Eppendorf tube.
-
a.
CRITICAL: Steps 12–14.f should be performed on ice.
RNA extraction and purification
Timing: 1 day
In this step, total RNA is extracted from cytoplasmic and nuclear bound fractions through TRI Reagent extraction.
CRITICAL: RNA should be extracted immediately after sample collection to preserve integrity.
CRITICAL: Both TRI Reagent and chloroform are hazardous solutions, always work under a fume hood, with gloves and lab coat, when using them.
-
15.Extract RNA.
-
a.Add 600 μL of TRI Reagent in each tube and mix by pipetting up and down.
-
b.Incubate for 5 min at room temperature.Note: Tri Reagent should be in at least 4-fold excess.
 Pause point: Samples can be stored at −80°C.
Pause point: Samples can be stored at −80°C. -
c.Add 150 μL of chloroform.
-
d.Shake the tubes vigorously for 15 s.
-
e.Incubate for 10 min at room temperature.
-
f.Centrifuge at 12,000 × g for 10–15 min at 4°C.Note: Different phases are obtained after centrifugation: a lower pink TRI Reagent-chloroform phase containing proteins, an interphase containing DNA and a colorless upper aqueous phase containing RNA.
-
g.Tilt the tube, pipet the aqueous phase and transfer into a 1.5 mL low binding Eppendorf tube.
 CRITICAL: Avoid transferring any of the interphase or organic phase into the pipette when pipetting the aqueous phase.
CRITICAL: Avoid transferring any of the interphase or organic phase into the pipette when pipetting the aqueous phase.
-
a.
-
16.Precipitate RNA.
-
a.Add 1.5 μL of GlycoBlue.
-
b.Add an equivalent volume (about 500 μL) of isopropanol and vortex for 5–10 s.
-
c.Precipitate overnight (∼12–16 h) at −20°C.
-
d.Centrifuge at 12,000 × g for 25 min at 4°C.
-
a.
Note: GlycoBlue corresponds to a blue dye covalently linked to glycogen. While glycogen serves as a nucleic acid co-precipitant, the attached dye increases the visibility of the pellet.
-
17.Wash the RNA pellet.
-
a.Add 600 μL of 70% ethanol.
-
b.Centrifuge at 12,000 × g for 5 min at 4°C, remove and discard the supernatant without touching the pellet.
-
c.Centrifuge briefly to remove potential remaining ethanol.Note: Remove as much ethanol as possible at this step.
-
d.Let the pellet dry for 5–10 min at room temperature.
 CRITICAL: Do not let the RNA pellet dry for too long, otherwise re-solubilization (step 18) will be difficult.
CRITICAL: Do not let the RNA pellet dry for too long, otherwise re-solubilization (step 18) will be difficult.
-
a.
-
18.
Resuspend the RNA pellet. Add 20 μL of RNase free-water and pipet up and down until dissolution of the pellet. Save 2–3 μL for quality check (see step 20).
Pause point: Samples can be stored at −80°C.
Validation of cell fractionation and RNA integrity
Timing: 2 days
In this step, the purity of the cytoplasmic and nuclear fractions is estimated and the integrity of recovered RNA validated.
-
19.Assess the quality of the lysate fractionation by western-blot.
-
a.Sample and gel preparation.
-
i.Use a pre-casted gel.
-
ii.Add 5× Laemmli buffer to the aliquots collected in steps 8, 10.d, 11.d.ii, 13.b.iv and 14.d and boil the samples for 5 min at 95°C.Alternatives: Prepare a polyacrylamide gel with a percentage adapted to the molecular weight of protein(s) of interest.
-
i.
-
b.Gel loading and running.
-
i.Mount the gel in the electrophoresis chamber and fill the chamber with running buffer.
-
ii.Load samples on the gel (35 μL), including a molecular weight marker.
-
iii.Run the gel for 1 h 30 min at 110 V.
-
i.
-
c.Protein transfer.
-
i.Cut a gel-sized PVDF membrane and activate it with methanol for 1 min.
-
ii.Prepare the transfer sandwich in the gel holder cassette by positioning the gel and the membrane between foam pads and filter papers.
-
iii.Place the cassette in the electrophoresis chamber and fill the chamber with transfer buffer.
-
iv.Transfer overnight (∼12–16 h) at 40 mA at 4°C.
-
i.
-
d.Immunoblotting.
-
i.Rinse the membrane with PBS.
-
ii.Block the membrane with blocking buffer for 1 h at room temperature under agitation.
-
iii.Incubate the membrane with primary antibodies diluted in blocking buffer overnight (∼12–16 h), at 4°C, under agitation.Note: We used a combination of two anti-Lamin antibodies recognizing different epitopes to label the nuclear fraction (dilution 1:2,000 each), anti-Tubulin antibodies to label the cytoplasmic fraction (dilution 1:5,000) and anti-GFP antibodies to label RNA-binding proteins of interest (dilution 1:2,500). References are listed in the key resources table.
-
iv.Wash the membrane three times with PBS-Tween for 10 min at room temperature under agitation.
-
v.Incubate the membrane with fluorescent secondary antibodies diluted in blocking buffer for 2 h at room temperature, under agitation.Note: Secondary antibodies excited at different wavelengths can be combined to simultaneously detect two or more proteins.Note: Protect from light to avoid exciting the antibodies with ambient light.
-
vi.Wash the membrane three times 10 min with PBS-Tween at room temperature under agitation.
-
vii.Image using a fluorescence Odyssey imaging system.
-
i.
-
a.
-
20.Assess RNA quality.
-
a.Measure the concentration of RNA samples using the QuBit RNA high sensitivity assay, according to manufacturer’s indications (https://www.thermofisher.com/document-connect/document-connect.html?url=https%253A%252F%252Fassets.thermofisher.com%252FTFS-Assets%252FLSG%252Fmanuals%252FQubit_RNA_HS_Assay_UG.pdf).Note: Given the low concentration of bound fractions, we recommend performing high sensitivity measurements, as regular spectrophotometers tend to overestimate the concentration of poorly-concentrated samples.
-
b.For fractions with a concentration higher than 1 ng/μL, dilute the RNA samples with commercial RNAse-free water to reach a concentration of 1 ng/μL.
-
c.Determine the integrity of RNA samples with an Agilent bioanalyzer. Use the Agilent RNA 6000 Pico Kit according to the manufacturer’s instructions (https://www.agilent.com/cs/library/usermanuals/public/G2938-90046_RNA600Pico_KG_EN.pdf).
-
a.
Expected outcomes
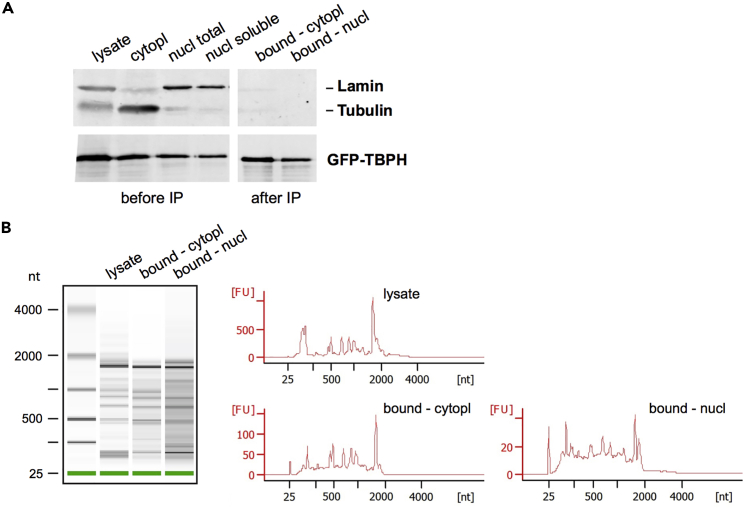
This protocol describes a method to selectively purify the nuclear and cytoplasmic complexes formed by GFP-tagged RBPs expressed in Drosophila brain, and to extract associated RNA. In contrast to classical RIP protocols (e.g., Wessels et al., 2016), it thus allows for the selective recovery and comparison of the populations of mRNAs bound in the nuclear and cytoplasmic compartments. A successful fractionation of Drosophila head lysates should produce a cytoplasmic lysate enriched in Tubulin and depleted of nuclear Lamin and a nuclear lysate enriched in Lamin and depleted of Tubulin (Figure 5A; troubleshooting problem 3). GFP-RBP(s) of interest should be detectable in the cytoplasmic and nuclear input fractions used for immunoprecipitation and should be enriched in the respective bound fractions (Figure 5A; troubleshooting problem 4). 50–100 ng of total RNA are typically recovered from the bound fractions. Although these fractions are typically depleted of rRNA, a 18S pic is still frequently detected (Figure 5B). RIN values provided by the bioanalyzer should however not be used to evaluate the integrity of RNA fractions, as Drosophila 28S rRNA is known to be cleaved into two smaller products migrating close to the 18S pic (Winnebeck et al., 2010). Rather, integrity should be evaluated based on the presence of a significant amount of RNA molecules with a size higher than about 200 bp (Figure 5B; troubleshooting problem 5).
Figure 5.
Validation of the purity of nuclear and cytoplasmic fractions and analysis of RNA integrity
(A) Western blots performed on the input and bound nuclear and cytoplasmic fractions. Lamin and Tubulin were used as markers of the nuclear and cytoplasmic fractions, respectively (upper panel). Note that Tubulin is depleted from the nuclear fraction while Lamin is depleted from the cytoplasmic one. The amount of GFP-TBPH, an RBP found both in the nucleus and in the cytoplasm, is shown for input and bound fractions (lower panel).
(B) Examples of bioanalyzer electrophoresis profiles obtained for initial head lysate and bound cytoplasmic and nuclear fractions. Note that no clear 28S pic is observed in Drosophila samples, as the 28S rRNA is cleaved into two smaller fragments.
RNA extracted from the bound fractions can be used to prepare libraries for deep-sequencing analyses and/or for RT-qPCR experiments with candidate transcripts. Different strategies can be used to identify RNAs enriched in the bound fractions: comparison of the RNA content of the input and bound fractions, or comparison of the RNA content of the bound fractions recovered after immunoprecipitation of GFP-RBP(s) and sole GFP. We used the latter method to better estimate the fraction of RNAs associating non-specifically with beads.
Limitations
In this protocol, RBPs and their associated RNAs are purified through high affinity capture of GFP-fusion proteins. Adding a 27 kDa GFP tag may not be neutral and could impact the localization, binding properties and/or function of the tagged-RBPs. The subcellular distribution of GFP-RBPs, as well as their capacity to rescue the mutant phenotypes, should thus be assessed before performing RNA-immunoprecipitation. The respective properties of N-terminal and C-terminal GFP-fusions could be compared during the validation process.
In contrast to methods developed to identify RNAs directly bound by RBPs of interest (e.g., CLIP and its variants (Lee and Ule, 2018)), this protocol identifies RNAs found in complex with the tagged-RBPs of interest. RNAs associating either through direct interaction, or through indirect interaction, are thus recovered. Furthermore, the identity of associated mRNA species, but not the sequence mediating binding, is recovered.
This protocol has been optimized to recover RBPs from the nucleus and their associated RNAs. As extraction of nuclear proteins is performed using a high salt buffer, weak RBP-RNA interactions may however be disrupted under such stringent conditions. In addition, some of the nuclear RBP-RNA interactions may be dependent on DNA and will be lost upon DNase treatment.
Troubleshooting
Problem 1
When ground, fly heads agglomerate and form a paste instead of fine powder.
Potential solution
Avoid formation of moisture in the Eppendorf tubes used for collection of heads by immediately closing the tubes and transferring them at −80°C (step 3). Transfer the heads in the mortar immediately after cooling both the pestle and the mortar with liquid nitrogen and grind the material as fast as possible (step 4).
Problem 2
A clot of fly head material is forming at the bottom of the Dounce homogenizer.
Potential solution
Avoid using the Dounce homogenizer right after transferring it from −20°C to 4°C, but rather wait for a few minutes before transferring the fly head material (step 5). If a clot has formed, it is still possible to break it into small fragments using a plastic pipette and to resuspend it.
Problem 3
The nuclear fraction is not pure.
Potential solution
If the nuclear fraction is contaminated with cytoplasmic proteins, one or two additional washes can be performed before extraction of the nuclear fraction (step 11.a).
Problem 4
The proteins of interest are not detected on the Western-Blot.
Potential solution
Low level of detected proteins may be due to weak initial expression of the tagged RBP. For endogenously expressed proteins, increase the amount of fly heads used to prepare the lysate (step 4). For Gal4/UAS-expressed proteins, use a Gal4 line with higher expression.
It is also possible that the proteins get degraded when preparing the extracts. This can be prevented by ensuring that all buffers are freshly supplemented with protease inhibitors and by keeping the samples on ice or at 4°C to avoid proteolysis.
If proteins are detectable in the extracts, but not in the bound fraction, ensure that the proper amount of beads was pipetted initially (a pre-cut tip should be used to pipet the bead slurry) and that beads were not pipetted out during washes (MμltiFlex flat tips should be used). Also ensure that GFP-trap beads do not settle during incubation with the lysates to favor optimal capture of GFP-tagged proteins (step 13.b).
Problem 5
RNA degradation.
Potential solution
To better identify at which step RNA gets degraded, it is recommended to compare the bioanalyzer profiles of the input, unbound and bound fractions. If RNA is already degraded in the input fractions, this means RNase contamination is already high in the initial lysate. Special care should then be taken to collect fly heads in RNase-free conditions (step 2), using RNase Zap-treated material. Gloves should be worn. If RNA is degraded in the unbound and bound fractions, care should be taken to maintain the samples at 4°C throughout the immunoprecipitation and washing steps (steps 12–14). If RNA is degraded exclusively in the bound fraction, ensure that the proteinase K solution used to elute the RNA is RNase-free (step 14.g) and extract RNA immediately after elution.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Florence Besse, PhD (florence.besse@univ-cotedazur.fr).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This study was supported by the CNRS, by the JPND project “Fly-SMALS,” and by the Agence Nationale pour la Recherche (ANR-18-CE13-0020-02 grant). We thank L. Palin for excellent technical assistance and members of the Besse group for discussion and advice. We are grateful to the Bloomington Drosophila Stock Center and the Developmental Studies Hybridoma Bank for reagents.
Author contributions
F.B. and M.H. conceived and designed the protocol. M.H. performed the laboratory experiments. F.B., M.H., and L.B. prepared and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101415.
Supplemental information
Data and code availability
This study did not generate/analyze datasets/code.
References
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Kelso R.J., Buszczak M., Quiñones A.T., Castiblanco C., Mazzalupo S., Cooley L. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 2004;32:D418–D420. doi: 10.1093/nar/gkh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kina H., Yoshitani T., Hanyu-Nakamura K., Nakamura A. Rapid and efficient generation of GFP-knocked-in Drosophila by the CRISPR-Cas9-mediated genome editing. Dev. Growth Differ. 2019;61:265–275. doi: 10.1111/dgd.12607. [DOI] [PubMed] [Google Scholar]
- Lee F.C.Y., Ule J. Advances in CLIP technologies for Studies of protein-RNA interactions. Mol. Cel. 2018;69:354–369. doi: 10.1016/j.molcel.2018.01.005. [DOI] [PubMed] [Google Scholar]
- Lowe N., Rees J.S., Roote J., Ryder E., Armean I.M., Johnson G., Drummond E., Spriggs H., Drummond J., Magbanua J.P., et al. Analysis of the expression patterns, subcellular localisations and interaction partners of Drosophila proteins using a pigP protein trap library. Development. 2014;141:3994–4005. doi: 10.1242/dev.111054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire S.E., Mao Z., Davis R.L. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- Morin X., Daneman R., Zavortink M., Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkar-Jaiswal S., DeLuca S.Z., Lee P.T., Lin W.W., Pan H., Zuo Z., Lv J., Spradling A.C., Bellen H.J. A genetic toolkit for tagging intronic MiMIC containing genes. eLife. 2015;4:e08469. doi: 10.7554/elife.08469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkar-Jaiswal S., Lee P.T., Campbell M.E., Chen K., Anguiano-Zarate S., Cantu Gutierrez M., Busby T., Lin W.W., He Y., Schulze K.L., et al. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. eLife. 2015;4:e05338. doi: 10.7554/elife.05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Zhu M., Li L., Wu C. Identifying protein-protein interaction in Drosophila adult heads by Tandem Affinity Purification (TAP) J. Vis. Exp. 2013:50968. doi: 10.3791/50968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels H.H., Hirsekorn A., Ohler U., Mukherjee N. Identifying RBP targets with RIP-seq. Methods Mol. Biol. 2016;1358:141–152. doi: 10.1007/978-1-4939-3067-8_9. [DOI] [PubMed] [Google Scholar]
- Winnebeck E.C., Millar C.D., Warman G.R. Why does insect RNA look degraded? J. Insect Sci. 2010;10:1–7. doi: 10.1673/031.010.14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze datasets/code.