Abstract
N6-Methyladenosine (m6A) is the most abundant internal modification in eukaryotic mRNA, playing critical role in various bioprocesses. Like other epigenetic modifications, m6A modification can be catalyzed by the methyltransferase complex and erased dynamically to maintain cells homeostasis. Up to now, only two m6A demethylases have been reported, fat mass and obesity-associated protein (FTO) and alkylation protein AlkB homolog 5 (ALKBH5), involving in a wide range of mRNA biological progress, including mRNA shearing, export, metabolism and stability. Furthermore, they participate in many significantly biological signaling pathway, and contribute to the progress and development of cancer along with other diseases. In this review, we focus on the studies about structure, inhibitors development and biological function of FTO and ALKBH5.
KEY WORDS: FTO, ALKBH5, RNA demethylation, Diseases, Inhibitors, Screening
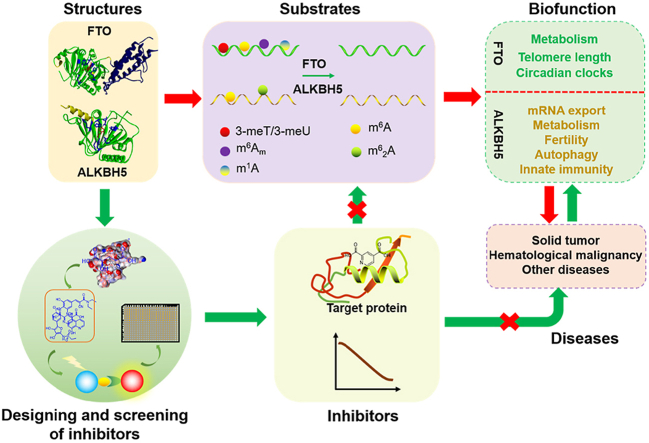
Graphical abstract
This review systematically describes the chemical and biological functions of RNA demethylases FTO and ALKBH5, from their crystal structure to inhibitors development and screening, demethylation mechanism and substrates to biological functions and roles in diseases.
1. Introduction
Among more than 100 structurally distinct posttranscriptional modifications of RNA in higher eukaryotic organisms, m6A is the most abundant modification in eukaryotic mRNA, which was first discovered in the 1970s1. Recently, the development of new techniques, like methylated RNA immunoprecipitation sequencing (MeRIP-seq), made the m6A research progress rapidly2. With the discovery of m6A methyltransferases, readers and demethylases, m6A modification was considered reversibly. Up to now, the m6A methyltransferases complex that constitutes of methyltransferase like 3 (METTL3, also known as MTA70), methyltransferase Like 14 (METTL14), pre-mRNA-splicing regulator WTAP, protein virilizer homolog (VIRMA) and RNA-binding protein 15 (RBM15) has been discovered to write the RNA m6A modification3, 4, 5. METTL3 was firstly identified as a writer for m6A, working with other catalytic component METTL14 and the accessory WTAP to achieve methylation function3,4,6.
In addition to methyltransferases, m6A readers that can recognize m6A substrates to regulate RNA translation and stability by binding to m6A, including YTH domain-containing family protein 1/2/3 (YTHDF1/2/3), YTH domain-containing protein 1/2 (YTHDC1/2), insulin-like growth factor-binding protein 1/2/3 (IGF2BP1/2/3), have been identified7, 8, 9. YTHDF1 can interact with translation machinery and then accelerate their loading on the ribosome to promote translation10. Differently, YTHDF2 is involved in regulating the stability of cytoplasmic mRNAs and promoting the translational initiation of transcripts11. As a partner of YTHDF1 and YTHDF2, YTHDF3 mainly impacts on cytoplasmic metabolism and translation of methylated mRNAs2,12. Besides, YTHDC1 is a nuclear m6A reader and recognizes transcribed pre-mRNA and promotes the recruitment of other YTHDC1-associated splicing factors7. YTHDC2, selectively binding m6A at its consensus motif, not only enhances the translation efficiency of its target genes but also decreases their mRNA abundance13. IGF2BP1/2/3, members of conserved family of single-stranded RBPs (RNA binding proteins), show preferential recognition for m6A of mRNAs and increase the stability of mRNA in an m6A-dependent manner9.
As another kind of crucial proteins, discovery of RNA demethylases confirms that m6A is a reversible modification. In 2011, FTO was first identified to exert demethylase activity to m6A14. After that, ALKBH5, another member of AlkB family, was identified as the second RNA m6A demethylase in 2013, playing key roles in RNA splicing, export, stability, metabolism along with FTO by regulating RNA m6A level in eukaryote15. In recent years, more and more studies have revealed functions of FTO and ALKBH5 in physiological process. Meanwhile, a large number of inhibitors targeting them have also been discovered. As the biological roles and inhibitors of FTO and ALKBH5 have not been well reviewed, we summarized the research about structure, inhibitors, biological function in diseases development, as well as screening methods for FTO and ALKBH5 in this review, providing a relatively comprehensive perspective for in-depth understanding of m6A demethylases.
2. Structure of FTO and ALKBH5
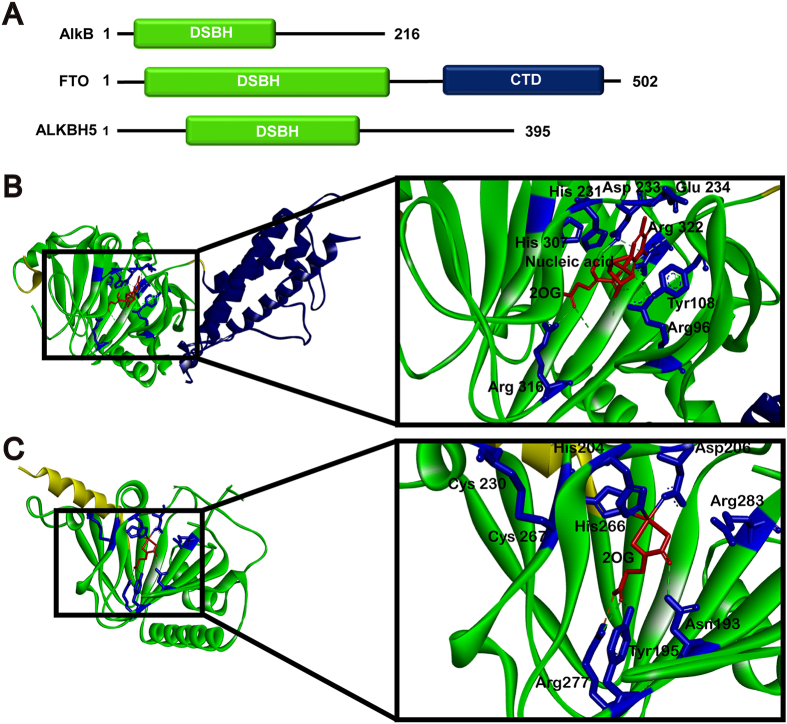
Both FTO and ALKBH5 belong to the nonheme Fe(II)-2-oxoglutarate (2OG)-dependent dioxygenase AlkB family proteins that repair N-alkylated nucleobases by oxidative demethylation16. AlkB family contains nine AlkB homologs, ALKBH1‒8 and FTO. In structure, they all contain a conserved double-stranded β-helix (DSBH) domain to mediate demethylase activity and some extra functional domains17. Undoubtedly, FTO and ALKBH5 both have a core catalytic domain containing DSBH domain for demethylation of RNA and DNA (Fig. 1A).
Figure 1.
Structures of FTO and ALKBH5. (A) DSBH domain in AlkB family. (B) and (C) Crystal structure of FTO (PDB: 3LFM) and ALKBH5 (PDB: 4NRO). The DSBH active region is shown in green, and the other domain is shown in yellow and mazarine. In the larger version, the ligand group is highlighted with red region, the side chains of active sites are shown in blue (stick), the interaction of ligand and protein is shown with dotted line.
2.1. Crystal structure of FTO
In 2010, the crystal structure of human FTO in complex with 3-methylthymine (3-meT) was first determined by Han et al.18. It contains 505 residues which can be divided into N-terminal domain (NTD, residues 32–326) and C-terminal domain (CTD, residues 327–498) (Fig. 1A). The catalytic core of NTD is mainly composed of a distorted DSBH construction, called the jelly-roll motif for RNA demethylase activity. In this domain, two α-helices and one long loop linking β5 and β6 coordinately form the two sides of this jelly-roll motif. The highly conserved residues His231, Asp233 and His307 chelate to Fe2+, while Arg316 and Arg322 form salt bridges with carboxylic acid of 2OG (Fig. 1B). For the binding of substrate, the conserved residues Tyr108 and His231 provide appropriate binding position. Beyond that, the carbonyl oxygen of RNA m6A is stabilized by forming hydrogen-bond with Arg96 and Glu234 residues, which also defines its substrate specificity. The unique structure of FTO, the β1‒2 loop, which enables FTO to recognize double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) or single-stranded RNA (ssRNA). The CTD of FTO is formed a three-helix bundle with α7, α8 and α10. In function, the CTD region can interact with NTD containing helix bundle to destabilize the conformation of the NTD. Thereby, the CTD region also plays a significant role in activating the demethylation activity18.
2.2. Crystal structure of ALKBH5
After the discovery of ALKBH5 working as an RNA m6A demethylase in 2013 by Zheng et al.15, crystal structures of ALKBH5 from human and zebrafish have been solved by many groups in following years19,20. ALKBH5 has 395 amino acids in full length (Fig. 1A). As a 2OG oxygenase, the catalytic core of ALKBH5 also contains a DSBH domain, that consists of 11 β strands (β1‒β11) and 5 α helices (α1‒α5). Like other AlkB family members, ALKBH5 binds to 2OG and metal ion in a conserved manner. The metal ion is coordinated by His204, Asp206 and His266 in CTD of ALKBH5, while binding of 2OG is well stabilized by electrostatic and hydrogen-bond with Asn193, Tyr195, Arg277 and Arg283 in the ALKBH5 active (Fig. 1C). In substrate specificity, different from other AlkB family members, ALKBH5 harbors a unique disulfide bond between Cys230 and Cys267 that confers ALKBH5 to specifically recognize single-stranded nucleic acids21. These residues mentioned above may be the key sites for the designing of selective inhibitors.
Though both FTO and ALKBH5 have the DSBH domain and are Fe(II) and 2OG-dependent dioxygenases, there are also some differences. In structure, FTO has the CTD region to destabilize the conformation of the NTD, while ALKBH5 does not. Besides, the 2OG binding site in FTO comes from DSBH β6, while the 2OG binding site in ALKBH5 is derived from DSBH β120. In substrate recognization, FTO can recognize dsDNA and ssDNA or ssRNA through its β1‒2 loop, while the loop between β7 and β8 of ALKBH5 links to β10 strand via a disulfide bond formed between Cys230 and Cys267, which can exclude the binding of dsDNA. These differences in structure may be the key sites for the designing of selective inhibitors22.
3. Development of FTO and ALKBH5 inhibitors
Based on the crystal structure of these m6A demethylases, diverse small molecule inhibitors have been designed and discovered, along with their mechanism and biological function have been revealed. Among them, FTO inhibitors mainly consists of substrate competitive inhibitors, 2OG-derivatives or Fe(II) chelating agents and other inhibitors with different mechanism. For ALKBH5 inhibitors, only several non-selective compounds have been characterized and most of them are 2OG oxygenase inhibitors.
3.1. Substrate competitive inhibitors of FTO
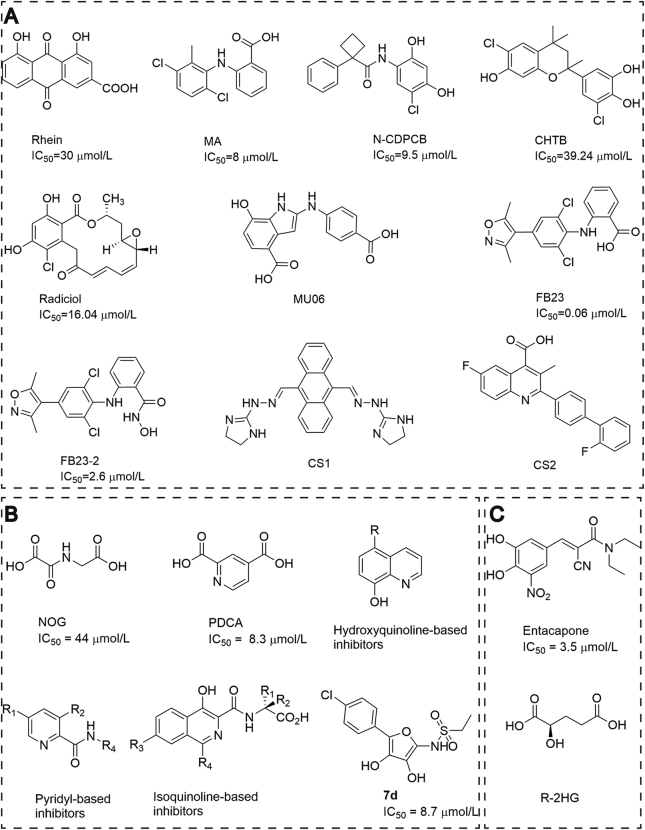
3.1.1. Rhein
The first FTO inhibitor is rhein, a natural product identified by Chen et al.23 in 2012 (Fig. 2A). Unlike most of the reported 2OG-derivatives based inhibitors, it is neither a 2OG analogue nor chelator of iron. Kinetics studies showed that rhein inhibited FTO by competitively binding the catalytic domain against ssRNA substrate. The negative-charged carboxyl of rhein interacts with the extensive positive-charged region around Arg316 of FTO. Besides, Arg316 also forms several hydrogen bonds with rhein, contributing to the important role of Arg316 in mediating an interacting between rhein and FTO. The IC50 value of rhein against FTO is around 30 μmol/L, which was measured by high performance liquid chromatography (HPLC) assay. Not just at the molecular level, rhein can also increase mRNA m6A levels in cells in a concentration-dependent manner23.
Figure 2.
Structure and activity of FTO inhibitors. (A) Substrate competitive inhibitors of FTO; (B) 2OG-derivatives or Fe(II) chelating agent based inhibitors of FTO; (C) Other mechanism inhibitors of FTO.
3.1.2. Meclofenamic acid (MA)
In 2015, Huang and others24 identified MA, an anti-inflammatory drug, as a highly selective FTO inhibitor with IC50 value of 8 μmol/L using drug repurposing strategy (Fig. 2A). MA is neither a chemical mimic of 2OG nor an iron chelator and it is likely to bind FTO competitively instead of its substrate m6A-containing ssDNA. One chlorine atom in MA directly contacts the complex guanidinium group in Arg96 so that to form a hydrogen bond with the carbonyl oxygen of RNA m6A. Of note, the first loop in the FTO nucleotide recognition lid (NRL), β3 and β4 provides hydrophobic interactions with MA, while ALKBH5 lacks β-hairpin motif of NRL, which provides extra interactions between MA and FTO, thus resulting in leakage between ALKBH5 and MA. As this site is crucial for FTO selective inhibitor, MA could not inhibit the conversion of m1A repairment in dsDNA mediated by alpha-ketoglutarate-dependent dioxygenase AlkB homolog 2 (ALKBH2), or in ssDNA by alpha-ketoglutarate-dependent dioxygenase AlkB homolog 3 (ALKBH3). Extendedly, the ethyl ester form of MA can also increase the level of m6A modification in HeLa cell mRNA and suppress self-renewal and tumorigenesis of glioblastoma stem cells24. In addition, Rhein and MA were reported to either prevent or reverse tyrosine kinase inhibitor resistance25.
3.1.3. N-(5-chloro-2,4 dihydroxyphenyl)-1-phenylcyclobutanecarboxamide (N-CDPCB)
N-CDPCB was a new FTO inhibitor with IC50 of 4.95 μmol/L (Fig. 2A). The crystal structure of FTO in complex with N-CDPCB shows N-CDPCB binds with FTO at a new site, but not 2OG-binding sites, where N-CDPCB is sandwiched between an antiparallel β-sheet and the L1 loop of FTO. This site is not conserved in other mammalian AlkB members, implying that it may be an FTO specific inhibitor, and can be considered as a basis for designing selective inhibitors. On the other hand, the structure of N-CDPCB bound to FTO is nearly identical with 3meT-bound to FTO, which indicates N-CDPCB as a substrate competitive inhibitor of FTO. Surprisingly, though their chemical structures are completely different, the binding sites of N-CDPCB and MA in FTO overlap partially26.
3.1.4. 4-chloro-6-(6′-chloro-7′-hydroxy-2′,4′,4′-trimethyl-chroman-2′-yl) benzene-1,3-diol (CHTB)
CHTB was identified as a new FTO inhibitor in 2016 with IC50 of 39.24 μmol/L (Fig. 2A). Similar to N-CDPCB, CHTB binds to FTO at a novel site, which is sandwiched between an antiparallel-sheet and extended C-terminal of the long loop. And CHTB overlaps completely with the methylated strand in the dsDNA bound ALKBH2, which indicates that CHTB may act as a competitive inhibitor of FTO. Moreover, residues that can interact with CHTB are not conserved among AlkB members, suggesting that CHTB may be an FTO specific inhibitor27.
3.1.5. Radicicol
Chang's group28 identified another natural compound radicicol as an effective FTO inhibitor, which showed dose-dependent inhibition against FTO m6A demethylation activity with IC50 value of 16.04 μmol/L (Fig. 2A). By analyzing the crystal structure of radicicol‒FTO complex, the result showed that radicicol partially overlapped the binding sites of MA and CHTB in FTO. Moreover, radicicol adopted an L-shaped conformation and the 4-Cl-1,3-diol group of radicicol bound to the same site of FTO with N-CDPCB, suggesting the 4-Cl-1,3-diol group is important when interacting with FTO28.
3.1.6. Derivatives of rhein and MA
Considering structure and activity of rhein and MA, N-phenyl-1H-indol-2-amine was found as a new scaffold as FTO inhibitor by applying scaffold hopping approach29. From their rigid and flexible docking, MU06 showed high binding affinities to FTO and was docked into the active cavity of FTO by forming H-bond interactions with Arg96, Asp233, and His231 residues of FTO (Fig. 2A). Meanwhile, FTO protein also showed stable interaction with MU06 in simulation of their binding mode29.
3.1.7. FB23 and FB23-2
Based on the structure of MA, FB23 and FB23-2 were designed, synthesized and identified as FTO inhibitors by Huang et al.30 (Fig. 2A). Both compounds directly bound to FTO and selectively inhibited its m6A demethylase activity and FB23 was significantly more potent than MA in inhibiting activity with IC50 of 0.06 μmol/L. Co-crystal structure of FB23 in complex with FTO showed FB23 occupied the whole substrate-binding pocket of FTO. Similar to the selectivity of MA to FTO, the phenyl ring and carboxyl acid substituent of FB23 formed hydrophobic interaction with FTO. Therefore, FB23 could avoid nonspecific binding to other AlkB members. Besides, hydrogen bond was formed directly between the carboxyl group in FB23 and Ser229 residue of FTO. And additional hydrogen bond between nitrogen or oxygen in FB23 and Glu234 in FTO may allow FB23 to show increased inhibitory activity against FTO, compared to MA30. Meanwhile, FB23-2, the derivative of FB23, whose cell permeability and activity as well as selectivity were better than FB23, can increase RNA methylation in acute myelogenous leukemia (AML) cells and dramatically suppress proliferation and promote the differentiation and apoptosis of AML cell line as well as primary AML cells in vitro (Fig. 2A)30.
3.1.8. CS1 and CS2
Recently, Chen et al.31 found two efficient FTO inhibitors CS1 and CS2 by structure based virtual screening of National Cancer Institute Developmental Therapeutics Program (NCI DTP) library with IC50 at nanomolar range in AML cells (Fig. 2A). CS1 and CS2 could selectively bind to and occupy the catalytic pocket of FTO, thereby blocking m6A-modified substrate from entering into FTO's catalytic pocket to inhibit FTO's demethylase activity. Excitingly, CS1 and CS2 also showed much higher efficacy in inhibiting AML differentiation and the signaling pathways of FTO as well as decreased leukocyte immunoglobulin-like receptor subfamily B 4 (LILRB4) expression and sensitized AML cells to T cell cytotoxicity and overcame immune evasion31.
Thus, due to the distinct substrate binding of FTO and ALKBH5, substrate competitive inhibitors are selective, which provides skeleton and designing ideas of selective inhibitors.
3.2. 2OG-derivatives and Fe(II) chelating agent
Considering that FTO is a 2OG-dependent demethylase, both cyclic and acyclic 2OG analogues were shown to inhibit FTO without selectivity. For instance, NOG (IC50 = 44 μmol/L) and PDCA (IC50 = 8.3 μmol/L), occupied 2OG binding pocket of FTO by forming electrostatic and hydrogen bond (Fig. 2B) and chelated with the metal in a bidentate manner. Other compounds, such as hydroxyquinoline-, pyridyl-, and isoquinoline-based compounds were also identified as FTO inhibitors. All the inhibitors bound across both co-substrate and primary substrate binding sites. These results gave an important path to the development of more effective and selective FTO inhibitors (Fig. 2B)32.
To design and synthesize novel antiepileptogenic compounds, Zheng et al.33 described a new class of ascorbic acid analogues, which could inhibit the 2-oxoglutarate-dependent hydroxylase and induce erythropoietin production in the central nervous system (CSN). One of the compounds, 7d showed FTO inhibitory activity with IC50 of 8.7 μmol/L (Fig. 2B). A crystal structure analysis of 7d binding to FTO shows that the dihydroxy furan moiety of 7d is involved in Fe(II) chelating sites, while both sulfonamide oxygens form hydrogen bonds with Arg322 and Arg96, respectively. The oxygen of the furan ring is proposed to form a hydrogen bond with Asn20533.
3.3. Other mechanism inhibitors of FTO
Other than above inhibitors, Peng et al.34 identified entacapone as an FTO inhibitor with IC50 of 3.5 μmol/L (Fig. 2C), which is structurally different from other known FTO inhibitors. Entacapone can occupy both binding sites of cofactor and substrate in FTO. The meta-position hydroxyl group of the nitrocatechol ring could bind to Arg322 and Tyr106 of FTO by forming hydrogen bounds, and the nitrile group can chelate with metal ion. Besides, carbonyl group of entacapone interacts with Asn205 by forming a hydrogen bond and the diethyl-propanamide tail is deeply stretched into the cofactor binding domain34. In 2018, R-2HG, an endogenous oncometabolite, was reported to suppress FTO enzymatic activity competitively, thereby increasing global m6A modification in R-2HG sensitive leukemia cells and leading to the suppression of relevant pathways by leading to decreased MYC mRNA stability, down-regulation of MYC signaling, and then inhibiting cancer cell proliferation (Fig. 2C)35.
3.4. ALKBH5 inhibitors
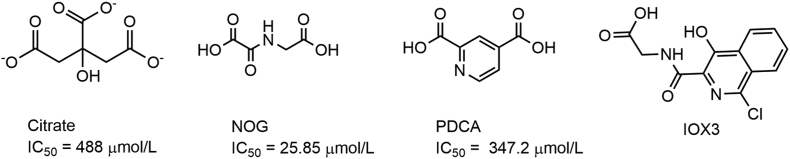
Xu et al.22 firstly identified citrate as a modest ALKHB5 inhibitor (IC50 = 488 μmol/L) and co-crystal structure of citrate in complex with ALKBH5 showed that citrate excluded both metal ion and 2OG in ALKBH5. The C-5-carboxylate of citrate molecule formed hydrogen bond with Lys132 and Asn193 side chains as well as the C-1 and C-6 carboxylates with Ser217 and Arg283, and the citrate C-3 carbonyl oxygen with His266, respectively22. Meanwhile, Aik et al.32 reported that citrate can also act as a modest FTO inhibitor. But with a different manner, the citrate only replaces 2OG and the metal ion site is not occupied32, which indicates that citrate has no selectivity for ALKBH5 and FTO. Aik and others20,36 also reported that 2OG oxygenase inhibitors NOG, PDCA and HIF PHD inhibitor IOX3 can weakly inhibit ALKBH5 activity by competing 2OG (Fig. 3). Up to now, only several compounds have been characterized as ALKBH5 inhibitors, and none of them is selective.
Figure 3.
Structures and activity of ALKBH5 inhibitors.
To solve these current problems, firstly, the design of compounds is critical. Computer aided design and evaluation according to the structure of ALKBH5 to search active skeleton structure, or modifying FTO inhibitors according to the structural differences between FTO and ALKBH5 can be the effective means of design. Meanwhile, combined with virtual screening, establishment of stable and efficient high-throughput screening methods (we discussed below) to expand the screening of compound library may help to discover and identify potential lead compounds. To improve the selectivity of compounds, though both FTO and ALKBH5 have the DSBH domain and are Fe(II) and 2OG-dependent dioxygenase, the sites where they bind to the substrate are quite different. The loop between β7 and β8 of ALKBH5 linked to β10 strand via a disulfide bond formed between Cys230 and Cys267, which can exclude the binding of dsDNA. These above mentioned residues may be the key sites for the designing of selective inhibitors. In addition, Arg130 and Tyr139 play essential roles in substrate recognition of ALKBH5, which may also as an important site for the designing of selective inhibitors22.
4. Demethylation mechanism of FTO and ALKBH5 against m6A
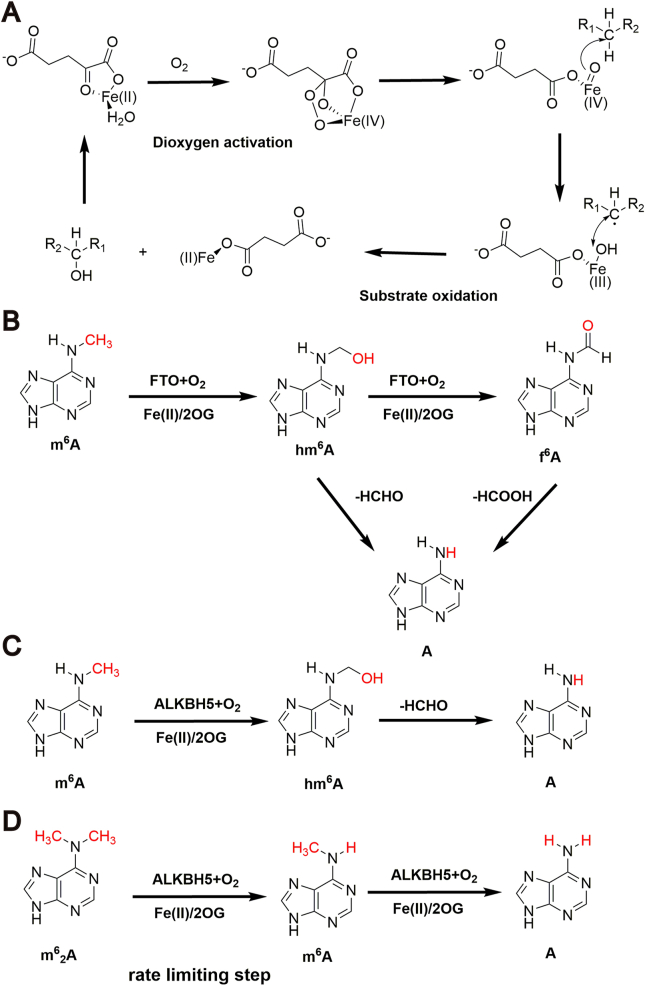
Generally, two oxidation reaction stages are involved in the demethylation mechanism of Fe(II) and 2OG dependent oxygenase: dioxygen activation and substrate oxidation. At the first stage, Fe(II) and 2OG may each contribute two electrons to activate a dioxygen molecule. The active dioxygen molecule first enters bridged peroxo and then into the Fe(IV)-oxo intermediate. In the second substrate oxidation stage, the inert C‒H bond of the RNA or other substrate is oxidized to hydroxy by the highly active Fe (IV)-oxo species, then one molecule of formaldehyde is removed from this intermediate product to obtain final demethylation product. Meanwhile, Fe(IV) is reduced back into Fe(II) to complete all catalytic cycle with 2OG being reduced to succinate. Because of the instability of the C‒N bond, N-methylated substrates undergo hydrolytic deformylation and trigger direct demethylation37 (Fig. 4A).
Figure 4.
Demethylation mechanism of Fe(II) and 2OG dependent oxygenase. (A) The demethylation mechanism of general oxygenases. Two stages of oxidation reaction are involved: dioxygen activation and substrate oxidation. (B) Demethylation mechanism of FTO. FTO can oxidize m6A to short-lived hm6A and f6A in a stepwise manner, and then formaldehyde or formic acid is removed from hm6A and f6A to obtain adenine. (C) Demethylation mechanism of ALKBH5 to m6A. m6A can be oxidized by ALKBH5 to hydroxymethyl-A intermediates and then formaldehyde is removed from hydroxymethyl-A to obtain adenine. (D) Demethylation mechanism of ALKBH5 to m62A. The first demethylation step is rate limiting step. The demethylation site is highlighted with red.
While in FTO-mediated demethylation process, FTO can oxidize m6A to short-lived and previously unknown intermediates, N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (f6A), in a stepwise manner. By removing formaldehyde and formic acid group respectively, both hm6A and f6A can be transformed into the shared product adenine (Fig. 4B)37.
For ALKBH5, the N-methylated bond of m6A or other substrate is oxidized to hydroxymethyl-A intermediates and then one molecule of formaldehyde is removed from these intermediates to final demethylation product with concomitant production of succinate, formaldehyde, and carbon dioxide (Fig. 4C)37. While for demethylation of the m62A, the reaction is more complex and the demethylation by ALKBH5 is fast while the first demethylation step is rate limiting step (Fig. 4D)38.
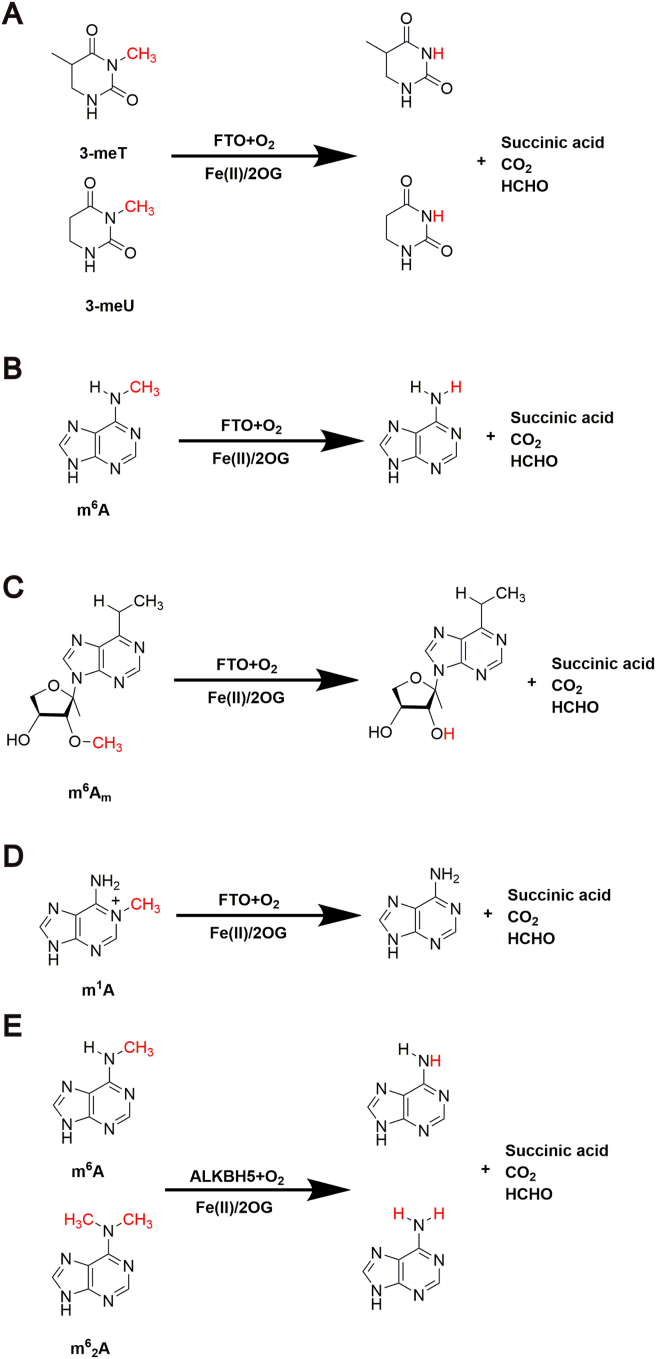
4.1. FTO demethylates 3-meT in ssDNA and 3-methyluracil (3-meU) in ssRNA
Demethylation of FTO for nuclear acid was first discovered by Thomas Gerken and others39 in 2007. FTO shares similar sequence with other Fe(II) and 2OG dependent oxygenase and mouse FTO catalyzes the demethylation of 3-meT in ssDNA (Fig. 5A)39. Soon after that, Jia et al.40 demonstrated that recombinant human FTO protein can completely demethylate the methylation of 3-meU in ssRNA in 2008 (Fig. 5A)40. Meanwhile, different affinity of FTO to various substrates reveal that both human and mouse FTO proteins preferentially repair 3-meT in ssDNA over others. In addition, FTO performs neglectable weak affinity to 3-meT in dsDNA40. This is the first study for the biochemical activity of FTO and provides basics for investigating the functional role of FTO.
Figure 5.
The structures and demethylation site of FTO and ALKBH5 substrates. (A–D) The structures and demethylation site of FTO in 3-meT/U (A), m6A (B), m6Am (C), m1A (D). (E) The structures and demethylation site of ALKBH5 in m6A and m62A. The demethylation site is highlighted with red.
4.2. FTO demethylates m6A in mRNA
m6A residues in mRNA was reported as another substrate of FTO in vitro and in vivo in 201114. m6A in ssDNA and ssRNA can be completely converted to adenosine upon FTO treatment in an Fe(II) and 2OG dependent manner (Fig. 5B)14. This discovery of the FTO-mediated demethylation of m6A in mRNA further shows the regulatory function of FTO on reversible chemical modification of RNA and also makes FTO the first characterized m6A demethylase.
4.3. FTO demethylates m6Am in mRNA and snRNA
In mRNA biogenesis, an N7-methylguanosine (m7G) cap with a triphosphate linker always exists in the 5′ end of mRNAs. 2′-O-Methyladenosine, if following closely with the m7G cap, can be further methylated at the N6-position to form N6,2′-O-dimethyladenosine (m6Am) (Fig. 5C). Recently, FTO was revealed to target methyladenine at transcription start sites and the m6Am of mRNA was another substrate of FTO41. More than that, Mauer et al.41 indicated that FTO preferentially demethylates m6Am rather than m6A and controlled the balance between m6Am and Am.
Not only mRNA, m6Am modifications also occur in small nuclear RNAs (snRNAs). Recent research has shown that FTO also demethylates spliceosomal snRNA m6Am to Am and determined relative level of m6Am and Am, which provides a direct link between FTO activity and the snRNA splicing machinery42.
4.4. FTO demethylates m1A in tRNA
Utilized enhanced crosslinking and immunoprecipitation sequence (eCLIP-seq), tRNA was discovered to be enriched in FTO-bound RNA species and FTO decreased total m1A level in tRNA with no selectivity in nuclear or cytoplasmic tRNA m1A (Fig. 5D). The proximity of FTO to tRNA m1A is due to its similar structure to tRNA m5C methyltransferase NOP2/Sun RNA methyltransferase 6 (NSUN6). As m1A in tRNAs is easily recognized and delivered to activate translational polysomes, FTO-induced demethylation of tRNA m1A has negative effect on translation43.
4.5. ALKBH5 demethylates m6A in ssRNA and ssDNA
For ALKBH5, it is stood out as it completely demethylated m6A in the ssRNA/DNA substrates (Fig. 5E). Similar to FTO, ALKBH5 also demethylates its substrate in an iron center dependent manner, which indicates the physiologically function of ALKBH5 in RNA biological progress15. This study identified ALKBH5 as the second RNA demethylase, indicating the significant function of ALKBH5 to erase m6A modification and also the critical role in fundamental biological processes.
4.6. ALKBH5 demethylates m62A in rRNA
Ribosomal RNA (rRNA) contains special m6A modification, the double methylated species m62A, in which the two methyl groups are both situated at the exocyclic of N6-purine heteroatom (Fig. 5E). Ensfelder et al.38 reported that m62A as well as m6A in rRNA can be demethylated by ALKBH5. Because each step of the demethylation reaction requires 2OG in the active site, demethylation of ALKBH5 on mono-methylated m6A is much more efficient compared to m62A38.
The demethylation mechanism of FTO and ALKBH5 in different methylation substrates reveals the biochemical activity of FTO and ALKBH5, providing solid support for the discovery of more biological functions and therapeutic target of related diseases.
5. Biological function of FTO and ALKBH5
m6A in eukaryotic mRNA influences splicing, export, stability, metabolism of mRNA. So m6A demethylases, FTO and ALKBH5, can be involved in many physiological functions in human beings, including eukaryotic metabolism, telomere length, circadian clocks, spermiogenesis, autophagy and so on.
5.1. Crucial role of FTO in metabolic related function
FTO is widely expressed in fetal and adult tissues, especially in brain and affects obesity susceptibility. From childhood to old age, FTO gene variation is highly associated with their body mass index (BMI) and obesity44. Loss of FTO in mice leads to frequently postnatal death and an obvious decrease in adipose tissue and lean body mass as well as increasing energy expenditure but uncoupling invariable mitochondrial, thyroid function and glucose metabolism45.
Skeletal muscle can also produce and expend energy in human body thus participating in the metabolism control of the whole body. Wang et al.46 provided the evidence that FTO downregulation suppresses mitochondria biogenesis and energy production by positively regulating mammalian target of rapamycin-peroxisome proliferator-activated receptor-γ coactivator-1α (mTOR-PGC-1α) pathway and makes a large contribution for skeletal muscle differentiation depending on its m6A demethylation activity46.
In various heart failure models, RNA m6A is overexpressed compared to normal hearts. Deficiency of FTO decreases cardiomyocyte contractile function through accelerating Ca2+ decay and increasing sarcomere shortening during cardiac remodeling and repair. Consistently, overexpression of FTO in myocardial enhanced the cardiac function at ischemia site by selectively demethylating RNA m6A in cardiac contractile transcripts hypermethylation, and then increasing RNA stability47.
In brief, FTO can directly affect the energy metabolism of adipose tissue and muscle tissue through its m6A demethylation activity, thus involving in obesity, mitochondrial metabolism and cardiomyocyte contractile function.
5.2. Role of FTO in telomere length and circadian clocks
By affecting retinoblastoma-like protein 2 (RBL2) expression or interacting with global energy sensors, such as mTOR, AMP activated protein kinase (AMPK) and uncoupling protein 2 (UCP2), FTO is involved in the telomere length (TL) regulation48,49. Furthermore, FTO deficiency can lead to alter response to light and a prolonged circadian period which is resulted from FTO mediated decreasing expression of core clock genes cryptochrome1 and 2 (CRY1 and CRY2). Otherwise, FTO is involved in circadian transcription-translation negative-feedback loop via decreasing circadian locomoter output cycles protein kaput-aryl hydrocarbon receptor nuclear translocator-like protein 1 (CLOCK-BMAL1) transactivation activity50. The involvement of FTO in TL and circadian rhythm modulation indirectly explains the regulatory role of FTO in metabolism and may be solid evidence for FTO broad function.
5.3. Effect of ALKBH5 in mRNA export and metabolism
ALKBH5 is located in nuclear speckles and colocalized well with mRNA-processing factors15, regulating the translocation of mRNA from nuclear to cytoplasm by affecting the function of serine/arginine-rich splicing factor 1 (ASF/SF2) via its demethylation activity. Besides, ASF/SF2 acts as a bridge for the interaction of mRNA export factor TAP‒p15 complex and mRNA cargo to facilitate mRNA export. On RNA metabolism, ALKBH5 can decrease the synthesis rate of nascent RNA and maintain the global RNA stability51.
5.4. Effect of ALKBH5 in spermiogenesis process, pregnancy and recurrent miscarriage
ALKBH5 mRNA is highest expressed in testis and ALKBH5 deficiency strongly results in spermatogenic maturation arrest and failed to finish the spermiogenesis process, thereby inducing apoptosis of pachytene and metaphase-stage spermatocytes15. ALKBH5-mediated m6A erasure which is essential for longer 3′-UTR mRNAs splicing and production in the spermatocytes and round spermatids, makes ALKBH5 extremely important in spermiogenesis52. Moreover, ALKBH5 can decrease the stability of cysteine-rich angiogenic inducer 61 (CYR61) mRNA, which is a matricellular protein regulating embryogenesis cell survival, proliferation, differentiation, migration, adhesion and synthesis of extracellular matrix (ECM), thus controlling trophoblast invasion at the maternal‒fetal interface in recurrent miscarriage (RM)53. So, ALKBH5 is mainly involved in spermatogenesis and embryogenesis, which is one of the research directions of related function and diseases. All these functions also show ALKBH5 indeed exert an additional layer of biological regulation on multifaceted life processes through m6A demethylation.
5.5. Effect of ALKBH5 in autophagy and antiviral innate responses
In autophagy process, ALKBH5 can reverse H/R-mediated m6A modification of transcription factor EB (TFEB) nascent transcript mRNA, thereby enhancing autophagic flux and inhibiting the apoptosis of H/R-treated cardiomyocytes. In turn, TFEB induces ALKBH5 expression via binding to ALKBH5 promoter with its conserved E-box elements in cardiomyocytes54. In addition, ALKBH5 can inhibit autophagy by interacting with HuR and activating receptor protein-tyrosine kinase‒phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform-RAC-alpha serine/threonine-protein kinase‒serine/threonine-protein kinase mTOR (EGFR‒PIK3CA‒AKT‒mTOR) signaling pathway to enhance the stability of B-cell lymphoma 2 (BCL-2) mRNA and promote interaction between BCL-2 and beclin1 in epithelial ovarian cancer55.
ALKBH5 also participates in the inhibition of antiviral innate responses through removing m6A modification and entrapping selected antiviral transcripts exportation from the nucleus by DEAD box polypeptide 46 (DDX46) recruitment56. These broad roles indicate significant status of mRNA m6A modification, and also provide more strategies for the treatment of many related diseases. Certainly, due to the widespread presence of m6A modification, more biological functions and target genes of ALKBH5 still need to be explored and revealed.
5.6. Functions comparation of FTO and ALKBH5
On the whole, both FTO and ALKBH5 are involved in RNA metabolism by removing mRNA m6A modification and accordingly regulating translation of target genes. While because of their different tissue distribution characteristics and target genes, FTO mainly performs metabolic related function while ALKBH5 mainly affects spermatogenesis. FTO is directly involved in metabolism of adipose tissue and muscle tissue thus increasing energy expenditure ingestion and energy production as well as cardiomyocyte contractile function57, and it indirectly interacts with global energy sensors mTOR, AMPK and UCP2 to regulate TL and mediate circadian clocks through CRY1, CRY2 and CLOCK-BMAL148,50. But ALKBH5 is expressed in testis highest and regulates spermatogenesis through 3′-UTR mRNA splicing and decreasing mRNA m6A level of spermatogenesis genes. Through removing m6A modification of related genes, ALKBH5 is widely involved in embryonic development, autophagy and antiviral innate responses55. Moreover, due to the widespread presence of m6A modification in the body, the function of FTO and ALKBH5 will be further expanded except metabolism or there may be different selective regulatory mechanisms in different tissues and genes. These broad functions of FTO and ALKBH5 indicate prominent role of m6A in extensive biological processes, suggesting that FTO and ALKBH5 may be involved in many diseases and as potential therapeutic targets.
6. Role of FTO and ALKBH5 in diseases
FTO and ALKBH5 are involved in various human diseases, especially in cancers by regulating related genes transcription and translation. Meanwhile, the molecular mechanism studies of FTO and ALKBH5 in diseases provide scientific support for feasibility of therapy targeting these targets.
6.1. Role of FTO in cancers
FTO is overexpressed in breast cancer tissues compared to normal breast tissues, particularly in HER2-overexpressed breast cancer58. Because of its close association with energy metabolism, overexpression of FTO promotes glycolysis in breast cancer cells and FTO inhibition can significantly decrease the expression of PI3 kinase (PI3K), p-PI3K, AKT and p-AKT, following by inhibiting breast cancer cell energy metabolism, which can also be mediated by estrogen in estrogen receptor (ER) positive patients59. Through demethylating BCL2 and adenovirus E1B 19-kDa-interacting protein 3 (BNIP3) 3′-UTR mRNA m6A modification and inducing its degradation, FTO promotes carcinogenesis and tumor growth in breast cancer60.
mRNA m6A modification plays a crucial role in self-renewal and tumorigenesis of glioblastoma stem cell (GSC)24. Inhibition of FTO can suppress tumor progression and prolong lifespan of GSC-grafted mice by regulating mRNA m6A enrichment and related gene expression24, which suggests that FTO can be a potential target in glioblastoma.
FTO is overexpressed and correlated with endometrial cancer risk in non-Hispanic white women and estrogen induces FTO nuclear accumulation, thereby enhancing endometrial cancer cell proliferation via increasing the phosphorylation of mTOR61. While the latest report by Gaudet et al.62 showed that FTO was not associated with endometrial cancer risk. Therefore, the relationship between FTO and endometrial cancer still needs further research to find a clearer regulatory mechanism. In another gynecological tumor, cervical squamous cell carcinoma (CSCC), FTO can induce the chemo-radiotherapy resistance in patients by increasing β-catenin expression and in turn strengthening activity of excision repair cross-complementation group 1 (ERCC1) through reducing β-catenin m6A levels in its mRNA transcripts63.
FTO expression is also upregulated in both protein and mRNA levels in gastric cancer (GC) tissues, closely relating to low differentiation, lymph node metastasis, TNM stage as well as poor prognosis in GC64. The consistent roles were uncovered that FTO induces the proliferation, migration, and invasion ability of lung cancer cells by decreasing the m6A level and increasing mRNA stability of ubiquitin carboxyl-terminal hydrolase 7 (USP7) and myeloid zinc finger protein 1 (MZF1)65. Thus FTO may be as a critical molecular marker in GC and lung cancer diagnosis and prognosis, while discoveries of detailed mechanism underlying FTO activity and FTO targets are still necessary.
Not only in solid tumors, FTO also is an oncogene in AML, especially in some certain subtypes with fms-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD), t (15; 17)/protein PML-retinoic acid receptor alpha (PML-RARA), nucleophosmin (NPM1) mutations, or myeloid-lymphoid leukemia (MLL) rearrangements. FTO can inhibit all-trans-retinoic acid (ATRA)-induced AML cell differentiation through reducing m6A levels in target genes mRNA transcripts66, as well as decreasing the sensitivity of leukemic cells to R-2-hydroxyglutarate (R-2HG)35. The functional significance of FTO in leukemogenesis and drug resistance provide a promising therapeutic strategy to treat leukemia with FTO inhibitors and others' drug combination (Table 1).
Table 1.
The role of FTO in cancers and other diseases.
| Cancer type | Expression | Regulated pathway or target | Ref. |
|---|---|---|---|
| Breast cancer | High | PI3K/AKT | 59 |
| BNIP3 | 60 | ||
| Glioblastoma | High | mRNA m6A enrichment | 24 |
| Endometrial cancer | High | mTOR | 61 |
| Cervical squamous cell carcinoma | High | β-Catenin; ERCC1 | 63 |
| GC | High | Low differentiation, lymph node metastasis and TNM stage | 64 |
| Lung cancer | High | USP7 | 65 |
| Acute myeloid leukemia | High | Leukemic oncogene-mediated cell transformation and leukemogenesis | 66 |
| Type 2 diabetes/cardiovascular disease/polycystic ovary syndrome | Minor allele A of FTO variant rs9939609 | 67 | |
| Alzheimer's disease | High | Tau/TSC1/mTOR | 68 |
| Kidney diseases | High | α-SMA | 70 |
| Premature ovarian insufficiency | Low | High RNA m6A modification | 71 |
6.2. Effect of FTO in other disease
Besides cancers, FTO is considered as an important factor for the development of age-related and metabolic disease, such as type 2 diabetes mellitus, cardiovascular disease, and dementia67. The minor allele A of FTO variant rs9939609 was reported to be associated with increased risk of type 2 diabetes mellitus and cardiovascular disease along with polycystic ovary syndrome (PCOS). Because of the commonly association among diabetes, obesity and Alzheimer's disease (AD), Li68 revealed that FTO was involved in the insulin defects-associated AD with activating the phosphorylation of Tau by increasing the mRNA level of TSC complex subunit 1 (TSC1). Knockout FTO in the neurons could also decrease the cognitive disorder in AD mice68. FTO deficiency can also improve metabolic syndrome, glucose tolerance and reduced ectopic fat in the liver69, which indicates that FTO can be used as a regulatory target for metabolic diseases. Similarly, FTO was overexpressed after ureteral obstruction and renal fibrosis. In FTO deficient kidney, global gene transcriptions amplitude is reduced after ureteral obstruction renal tubular cells produce decreasing α-smooth muscle actin (SMA) when treated with transforming growth factor beta (TGF-β). These results revealed significant role of FTO in obstructive nephropathy and had potential therapeutic implications70. While in premature ovarian insufficiency (POI) disease, FTO mRNA and protein expression levels are significantly lower in POI patients and RNA m6A modification is higher. Thus, FTO may be as a novel potential biomarker of POI diagnosis71 (Table 1).
6.3. Distinct roles of ALKBH5 in cancers
As a crucial direct target of hypoxia-inducible factor 1α (HIF-1α), ALKBH5 had significant effect in regulating cellular response to hypoxia, which was a common characteristic of various cancers and was connected with chemo-radio resistance, invasiveness, and angiogenesis72. Unlike FTO through metabolic and adipose pathways, ALKBH5 regulates the capacity of tumor initiation in breast cancer cells by demethylating 3′-UTR m6A of pluripotency factor nanog homeobox (NANOG) under hypoxia. Exposure to hypoxia stimulated HIF-1α- and hypoxia-inducible factor 2α (HIF-2α)-dependent expression of ALKBH5, which increased NANOG level and breast cancer stem cell phenotype73. In addition, ALKBH5 also combines with zinc finger protein 217 (ZNF217) and then induces ZNF217-dependent m6A inhibition of NANOG and Krueppel-like factor 4 (KLF4) mRNAs to promote breast cancer progress74. Similar to FTO, ALKBH5 is overexpressed in glioblastoma stem-like cells and contributes to tumorgenicity. The difference lies in the clear mechanism that ALKBH5 could demethylate Forkhead box protein M1 (FOXM1) nascent transcripts, thus resulting in increased FOXM1 expression75. Based on the demethylation of ALKBH5 in FOXM1 and NANOG, DEAD-Box helicase 3 (DDX3), a potential factor in chemoresistance and recurrence and involving in numerous aspects of RNA metabolism and translation, can decrease ALKBH5-mediated m6A methylation and suppress expression of FOXM1 and NANOG to inhibit cisplatin resistance in oral squamous cell carcinoma (OSCC)76. Through downregulating FOXM1 mRNA m6A modification and promoting FOXM1 expression as well as targeting metallopeptidase inhibitor 3 (TIMP3) 3′-UTR and repressing TIMP3 transcript stability, ALKBH5 can induce proliferation and invasion of lung adenocarcinoma cells77,78. Overexpressed ALKBH5 in epithelial ovarian cancer inhibits autophagy as well as promotes the proliferation and invasion of the epithelial ovarian cancer cells in vitro and in vivo through the EGFR‒PIK3CA‒AKT‒mTOR signaling pathway by interacting with HuR or stabilizing BCL-2 mRNA and promoting its interaction with beclin155.
Except by acting on mRNA, ALKBH5 was characterized to bind to lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) and affect its m6A level and then promote invasion and metastasis of GC79. Otherwise, low-m6A mediated by ALKBH5 can activate oncogenic Wnt/phosphoinositide 3-kinase‒RAC-alpha serine/threonine-protein kinase (Wnt/PI3K‒AKT) signaling in GC, showing ALKBH5 can be used to predict adverse clinicopathological features of GC80. Similarly, by interacting with another lncRNA growth arrest specific 5-antisense RNA 1 (GAS5-AS1), ALKBH5 enhances tumor suppressor GAS5 stability by decreasing GAS5 m6A modification in YTHDF2 dependent manner, thus promoting cervical cancer distant metastasis, lymphatic metastasis and poor prognosis81. In osteosarcoma, ALKBH5 is a critical factor to cause tumor by decreasing the m6A of lncRNA PVT1 to participate the growth and chemosensitivity82. ALKBH5 is overexpressed and essential for the development and maintenance of AML as well as self-renewal of leukemia stem/initiating cells through affecting mRNA stability of transforming acidic coiled-coil containing protein 3 (TACC3), which shows close correlation with poor prognosis in patients83 (Table 2). These studies provide insights into the crucial roles of ALKBH5 in various cancers.
Table 2.
The role of ALKBH5 in cancers.
| Cancer type | Expression | Regulated pathway or target | Ref. |
|---|---|---|---|
| Breast cancer | High | 3′-UTR m6A of NANOG | 73 |
| KLF4 | 74 | ||
| Glioblastoma | High | FOXM1 | 75 |
| OSCC | High | DDX3 | 76 |
| Lung adenocarcinoma cells | High | FOXM1 | 77 |
| NSCLC | TIMP3 | 78 | |
| Epithelial ovarian cancer | High | EGFR-PIK3CA-AKT-mTOR | 55 |
| BCL-2 | |||
| Gastric cancer | High | lncRNA NEAT1 | 79 |
| Wnt/PI3K-Akt | 80 | ||
| Cervical cancer | High | GAS5 | 81 |
| Osteosarcoma | High | lncRNA PVT1 | 82 |
| AML | High | TACC3 | 83 |
| Pancreatic Cancer cells | Low | KCNK15-AS1 | 84 |
| Colon cancer | Low | 85 | |
| Hepatocellular carcinoma | Low | LYPD1 | 86 |
Not only considered as an oncogene, ALKBH5 also shows opposite roles in pancreatic cancer, colon cancer and hepatocellular carcinoma84, 85, 86. In pancreatic cancer, ALKBH5 can demethylate potassium two pore domain channel subfamily K member 15‒antisense RNA 1 (KCNK15‒AS1) and downregulate KCNK15‒AS1-mediated cell motility to inhibit metastasis ability of pancreatic cancer cells84. Similarly, ALKBH5 expression is reduced in human colon cancer tissues, showing distinctly negative correlation with distant metastasis and cancer stage identified by American Joint Committee86. According to the latest research, ALKBH5 suppresses malignancy of hepatocellular carcinoma by erasing the m6A modification of LY6/PLAUR domain containing 1 (LYPD1) and leads to its post-transcriptional inhibition. Moreover, ALKBH5 is down-regulated in hepatocellular carcinoma and is closely related to the survival outcome of hepatocellular carcinoma patients (Table 2)86. These discoveries provide a novel understanding of the function of ALKBH5 in cancer and the opposite effects show the tissue function specificity of ALKBH5.
Thus, both FTO and ALKBH5 display enhanced roles on various cancers development, which can be novel therapeutic targets for cancers treatment. Moreover, these studies also suggest the urgency of the discovery of selective inhibitors of ALKBH5.
7. Inhibitor screening methods of FTO and ALKBH5
7.1. Polyacrylamide gel electrophoresis (PAGE)-based assay
PAGE-based assay is the earliest method that was used to quantify the activity of m6A demethylase. Nucleic acid substrates of FTO or ALKBH5 are annealed to the complementary strand for endonucleases digestion and the digested samples can be checked by non-reducing PAGE. As endonucleases can't recognize the methylated substrate, the PAGE can distinguish methylated and unmethylated nucleic acids and the results reveal activity of FTO or ALKBH5 and inhibition effect of compounds against demethylation activity of FTO or ALKBH5. This method is low cost, but also low throughput with complicated process and inexact quantification. So, other detection methods are always needed to verify the results40.
7.2. Liquid chromatography-tandem mass spectrometry (LC‒MS/MS)
As a classical separation and quantification method, HPLC plays an important role in identifying m6A demethylases inhibitors. Depending on adsorption difference of stationary phase to various material in mobile phase, m6A can be separated from adenine in specific nucleotide sequence. After quantify the m6A demethylation assay results, compounds which can inhibit FTO or ALKBH5 demethylation activity will be verified30. In addition, combination of LC and mass spectrometry is also used to discover m6A demethylases inhibitors. After separating by HPLC, the nucleosides are subsequently analyzed by online mass spectrometry, making the m6A demethylation assay precisely performed14,33. Nevertheless, these methods are low throughput and high cost for compounds screening. They can be used to identify the inhibitors effect but are not suitable for early compounds screening.
7.3. Fluorescence polarization (FP) assay
FP, an assay based on the light polarization, was used to screen FTO inhibitors by Huang et al87. In their assay, the ssDNA substrate containing m6A was labeled with carboxyfluorescein (FAM). They detected the fluorescence polarization to reflect the substrate binding to FTO, thereby indicating the inhibitory activity of compound to the interaction between FTO and substrate. This method can be conducted in a multiple-well plate, so it is applicable for high throughput screening. However, this method may miss the compound competing coenzyme and is easily influenced by many factors. So, the result also needs to be verified and confirmed by other methods87.
7.4. Formaldehyde dehydrogenase (FDH)-coupled assay
In our previous research, we reported a FDH coupled assay to detect ALKBH5 activity88. This assay quantifies the formaldehyde produced by ALKBH5 demethylation on m6A. The formaldehyde is oxidized by FDH to formic acid, along with the reduction its cofactor from nicotinamide adenine dinucleotide (NAD) to NADH with fluorescence. The fluorescence intensity can be quantified by microplate reader (λex = 360 nm, λem = 460 nm), reflecting the inhibitory activity of compounds against ALKBH5. This method can be used for high throughput ALKBH5 inhibitors screening with low cost. While the compound with self-fluorescence or quenching property can obstruct the assay88.
7.5. Alphascreen assay
With the development of biotechnology, antibody-based assays such as time-resolved fluorescence resonance energy transfer (TR-FRET) and alphascreen have been used to analyze biomolecules interaction89,90. In our previous report, we used alphascreen assay to establish a new assay for ALKBH5 inhibitor screening. In alphascreen beads-based technology, photoactive donor beads can convert ambient oxygen to excited singlet oxygen at 680 nm. If the acceptor bead is within approximately 200 nm distance from donor beads, the energy will be transferred to the acceptor beads, resulting in light production at 570 nm. Based on this, m6A antibody and biotinated-ALKBH5 substrate are captured by accepter beads and donor beads, respectively. If ALKBH5 demethylates m6A, the donor beads would not be in proximity of the acceptor beads, and the singlet oxygen energy would not be transferred, thus no signal is produced at 570 nm. The ALKBH5 activity and inhibitory activity of inhibitors would be indicated by the light intensity. This antibody-based assay performs excellent in selectivity and sensitivity and can be applied to high throughput screening. While its cost may be higher because of biotin labeled substrate and antibody requirement88.
The above methods represent screening methods based on different principles, PAGE-based, mass spectrometry-based, light polarization based, coupled enzyme-based, and antibody-based. The different characteristics of these methods can be complemented and verified with each other, providing effective tools for the discovery of FTO and ALKBH5 inhibitors.
8. Conclusions
mRNA plays a crucial role in carrying genetic information and guiding protein synthesis in eukaryotic. m6A was discovered as the most abundant internal modification in eukaryotic mRNA. Like other epigenetic modifications, the discovery of FTO and ALKBH5 suggests that the m6A modification is a reversible and dynamic process. Using various crystallographic structural analysis, their substrate binding sites and demethylation mechanism have been revealed clearly in recent years. Though both FTO and ALKBH5 have the similar demethylation mechanism, there also are some differences in structure and substrates recognition, thus resulting in differences in substrate species and functions. FTO can recognize dsDNA and ssDNA or ssRNA and mainly affect the metabolic related functions and diseases, while ALKBH5 can only recognize single stranded nucleic acid because of its special disulfide bond structure. And their structural characteristics also become an important basis for the designing of inhibitors. As these two enzymes are considered as novel drug targets for numerous diseases, especially for tumors and diabetes, the discovery of small molecular inhibitors offered new treatment strategy. Presently, there have been various inhibitors for FTO with different mechanisms. Among substrate competitive inhibitors, due to the distinct substrate binding of FTO from ALKBH5, many inhibitors are selective, which provides skeleton and designing ideas of many selective inhibitors. As to 2OG-derivatives and Fe(II) chelating agent, both cyclic and acyclic 2OG analogues were shown to have FTO inhibitory activity without selectivity. There are also some other mechanism inhibitors of FTO that expends the structural types of FTO inhibitors. However, only several compounds have been characterized as ALKBH5 inhibitors up to now, and there are no outstanding and selective inhibitors. To ameliorate the problem, designing of compounds and screening of compound library are key strategies, and differential structural characterization between FTO and ALKBH5 as well as the binding principle of discovered inhibitors can provide ideas for designing or modification of ALKBH5 inhibitors. In this review, we discussed the various screening methods based on different principles, and expected to provide more solutions for ALKBH5 inhibitor discovery.
In functions, because m6A is the most abundant modification in eukaryotic mRNA, FTO and ALKBH5 have broad functions, covering RNA metabolism, adipose tissue and muscle tissue metabolism, TL and circadian clocks as well as spermiogenesis and autophagy et al. These functions also determine the important role of FTO and ALKBH5 in many diseases, particularly in cancers, although their roles in various disease are not consistent, which needs more in-depth and sophisticated research. Certainly, there may also be other substrates catalyzed by FTO and ALKBH5, such as other methylated bases in RNA or DNA, as well as other m6A modified downstream genes. Their demethylation mechanism and biological functions need further excavation. With the aid of more advanced sequencing technologies and disease models, deeper understanding of m6A and new strategies to the treatment of related diseases will be revealed.
Acknowledgments
This work was supported by the National Key Research Program (No. 2018YFE0195100); the National Natural Science Foundation of China (No. 82020108030, No. U21A20416 and No. 82103997); Science and Technology Innovation Talents of Henan Provincial Education Department (No. 19IRTSTHN001, China); Basic and Frontier Technology Research Project of Henan Province (No. 212102310313, China). Basic Research of the Key Project of the High Education from the Education Department of Henan Province (No. 22ZX008, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences
Contributor Information
Yi-Chao Zheng, Email: yichaozheng@zzu.edu.cn.
Hong-Min Liu, Email: liuhm@zzu.edu.cn.
Author contributions
Dandan Shen wrote the manuscript. Bo Wang, Ya Gao, and Lijuan Zhao contributed to critical revision of the manuscript. Yaping Bi, Jinge Zhang, Ning Wang, Huiqin Kang, Jingru Pang, Ying Liu, Luping Pang, Zhe-Sheng Chen contributed to minor revision of the manuscript. Hong-Min Liu and Yi-Chao Zheng revised and finalized the manuscript.
Conflicts of interest
The authors declare no competing financial interests.
References
- 1.Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 3.Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 7.Xu C., Liu K., Ahmed H., Loppnau P., Schapira M., Min J. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J Biol Chem. 2015;290:24902–24913. doi: 10.1074/jbc.M115.680389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu T., Roundtree I.A., Wang P., Wang X., Wang L., Sun C., et al. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014;24:1493–1496. doi: 10.1038/cr.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., et al. N6-Methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu P.J., Zhu Y., Ma H., Guo Y., Shi X., Liu Y., et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fedeles B.I., Singh V., Delaney J.C., Li D., Essigmann J.M. The AlkB family of Fe(II)/α-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J Biol Chem. 2015;290:20734–20742. doi: 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsujikawa K., Koike K., Kitae K., Shinkawa A., Arima H., Suzuki T., et al. Expression and sub-cellular localization of human ABH family molecules. J Cell Mol Med. 2007;11:1105–1116. doi: 10.1111/j.1582-4934.2007.00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Z., Niu T., Chang J., Lei X., Zhao M., Wang Q., et al. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010;464:1205–1209. doi: 10.1038/nature08921. [DOI] [PubMed] [Google Scholar]
- 19.Chen W., Zhang L., Zheng G., Fu Y., Ji Q., Liu F., et al. Crystal structure of the RNA demethylase ALKBH5 from zebrafish. FEBS Lett. 2014;588:892–898. doi: 10.1016/j.febslet.2014.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aik W., Scotti J.S., Choi H., Gong L., Demetriades M., Schofield C.J., et al. Structure of human RNA N⁶-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 2014;42:4741–4754. doi: 10.1093/nar/gku085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng C., Liu Y., Wang G., Deng Z., Zhang Q., Wu W., et al. Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J Biol Chem. 2014;289:11571–11583. doi: 10.1074/jbc.M113.546168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu C., Liu K., Tempel W., Demetriades M., Aik W., Schofield C.J., et al. Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation. J Biol Chem. 2014;289:17299–17311. doi: 10.1074/jbc.M114.550350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen B., Ye F., Yu L., Jia G., Huang X., Zhang X., et al. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc. 2012;134:17963–17971. doi: 10.1021/ja3064149. [DOI] [PubMed] [Google Scholar]
- 24.Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., et al. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan F., Al-Kali A., Zhang Z., Liu J., Pang J., Zhao N., et al. A dynamic N6-methyladenosine methylome regulates intrinsic and acquired resistance to tyrosine kinase inhibitors. Cell Res. 2018;28:1062–1076. doi: 10.1038/s41422-018-0097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He W., Zhou B., Liu W., Zhang M., Shen Z., Han Z., et al. Identification of a novel small-molecule binding site of the fat mass and obesity associated protein (FTO) J Med Chem. 2015;58:7341–7348. doi: 10.1021/acs.jmedchem.5b00702. [DOI] [PubMed] [Google Scholar]
- 27.Qiao Y., Zhou B., Zhang M., Liu W., Han Z., Song C., et al. A novel inhibitor of the obesity-related protein FTO. Biochem. 2016;55:1516–1522. doi: 10.1021/acs.biochem.6b00023. [DOI] [PubMed] [Google Scholar]
- 28.Wang R., Han Z., Liu B., Zhou B., Wang N., Jiang Q., et al. Identification of natural compound radicicol as a potent FTO inhibitor. Mol Pharmaceut. 2018;15:4092–4098. doi: 10.1021/acs.molpharmaceut.8b00522. [DOI] [PubMed] [Google Scholar]
- 29.Padariya M., Kalathiya U. Structure-based design and evaluation of novel N-phenyl-1H-indol-2-amine derivatives for fat mass and obesity-associated (FTO) protein inhibition. Comput Biol Chem. 2016;64:414–425. doi: 10.1016/j.compbiolchem.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y., Su R., Sheng Y., Dong L., Dong Z., Xu H., et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35:677–691.e10. doi: 10.1016/j.ccell.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su R., Dong L., Li Y., Gao M., Han L., Wunderlich M., et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. 2020;38:79–96.e11. doi: 10.1016/j.ccell.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aik W., Demetriades M., Hamdan M.K.K., Bagg E.A.L., Yeoh K.K., Lejeune C., et al. Structural basis for inhibition of the fat mass and obesity associated protein (FTO) J Med Chem. 2013;56:3680–3688. doi: 10.1021/jm400193d. [DOI] [PubMed] [Google Scholar]
- 33.Zheng G., Cox T., Tribbey L., Wang G.Z., Iacoban P., Booher M.E., et al. Synthesis of a FTO inhibitor with anticonvulsant activity. ACS Chem Neurosci. 2014;5:658–665. doi: 10.1021/cn500042t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng S., Xiao W., Ju D., Sun B., Hou N., Liu Q., et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci Transl Med. 2019;11:eaau7116. doi: 10.1126/scitranslmed.aau7116. [DOI] [PubMed] [Google Scholar]
- 35.Su R., Dong L., Li C., Nachtergaele S., Wunderlich M., Qing Y., et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 2018;172:90–105.e23. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan S., Zhang J., Zhang D., Wei D. Cu(OTf)2-catalyzed intramolecular radical cascade reactions for the diversity-oriented synthesis of quinoline-annulated polyheterocyclic frameworks. Org Lett. 2021;23:1445–1450. doi: 10.1021/acs.orglett.1c00129. [DOI] [PubMed] [Google Scholar]
- 37.Shen L., Song C.-X., He C., Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ensfelder T.T., Kurz M.Q. ALKBH5-induced demethylation of mono- and dimethylated adenosine. ChemComm. 2018;54:8591–8593. doi: 10.1039/c8cc03980a. [DOI] [PubMed] [Google Scholar]
- 39.Gerken T., Girard C.A., Tung Y.C., Webby C.J., Saudek V., Hewitson K.S., et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jia G., Yang C.G., Yang S., Jian X., Yi C., Zhou Z., et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mauer J., Sindelar M., Despic V., Guez T., Hawley B.R., Vasseur J.J., et al. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat Chem Biol. 2019;15:340–347. doi: 10.1038/s41589-019-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei J., Liu F., Lu Z., Fei Q., Ai Y., He P.C., et al. Differential m6A, m6A(m), and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell. 2018;71:973–985.e5. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stratigopoulos G., Padilla S.L., LeDuc C.A., Watson E., Hattersley A.T., McCarthy M.I., et al. Regulation of Fto/Ftm gene expression in mice and humans. Am J Physiol-Reg I. 2008;294:R1185–R1196. doi: 10.1152/ajpregu.00839.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia L., Xi Q., Wang H., Zhang Z., Liu H., Cheng Y., et al. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem Bioph Res Co. 2017;488:425–431. doi: 10.1016/j.bbrc.2017.05.074. [DOI] [PubMed] [Google Scholar]
- 47.Mathiyalagan P., Adamiak M., Mayourian J., Sassi Y., Liang Y., Agarwal N., et al. FTO-dependent N6-methyladenosine regulates cardiac function during remodeling and repair. Circulation. 2019;139:518–532. doi: 10.1161/CIRCULATIONAHA.118.033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cota D., Proulx K., Smith K.A.B., Kozma S.C., Thomas G., Woods S.C., et al. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- 49.Andrews Z.B., Liu Z.W., Walllingford N., Erion D.M., Borok E., Friedman J.M., et al. UCP2 mediates Ghrelin's action on NPY/AgRP neurons by lowering free radicals. Nature. 2008;454:846–851. doi: 10.1038/nature07181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang C.Y., Shie S.S., Hsieh I.C., Tsai M.L., Wen M.S. FTO modulates circadian rhythms and inhibits the CLOCK-BMAL1-induced transcription. Biochem Bioph Res Co. 2015;464:826–832. doi: 10.1016/j.bbrc.2015.07.046. [DOI] [PubMed] [Google Scholar]
- 51.Dauksaite V., Akusjärvi G. Human splicing factor ASF/SF2 encodes for a repressor domain required for its inhibitory activity on pre-mRNA splicing. J Biol Chem. 2002;277:12579–12586. doi: 10.1074/jbc.M107867200. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y., Tang C., Yu T., Zhang R., Zheng H., Yan W. MicroRNAs control mRNA fate by compartmentalization based on 3′ UTR length in male germ cells. Genome Biol. 2017;18:105. doi: 10.1186/s13059-017-1243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X.C., Jin F., Wang B.Y., Yin X.J., Hong W., Tian F.J. The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics. 2019;9:3853–3865. doi: 10.7150/thno.31868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song H., Feng X., Zhang H., Luo Y., Huang J., Lin M., et al. METTL3 and ALKBH5 oppositely regulate m6A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019;15:1419–1437. doi: 10.1080/15548627.2019.1586246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu H., Gan X., Jiang X., Diao S., Wu H., Hu J. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J Exp Clin Cancer Res. 2019;38:163. doi: 10.1186/s13046-019-1159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng Q., Hou J., Zhou Y., Li Z., Cao X. The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus. Nat Immunol. 2017;18:1094–1103. doi: 10.1038/ni.3830. [DOI] [PubMed] [Google Scholar]
- 57.Church C., Moir L., McMurray F., Girard C., Banks G.T., Teboul L., et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. 2010;42:1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan A., Dang Y., Chen G., Mo Z. Overexpression of the fat mass and obesity associated gene (FTO) in breast cancer and its clinical implications. Int J Clin Exp Pathol. 2015;8:13405–13410. [PMC free article] [PubMed] [Google Scholar]
- 59.Gholamalizadeh M., Jarrahi A.M., Akbari M.E., Bourbour F., Mokhtari Z., Salahshoornezhad S., et al. Association between FTO gene polymorphisms and breast cancer: the role of estrogen. J Clin Endocrinol Metab. 2020;15:115–121. doi: 10.1080/17446651.2020.1730176. [DOI] [PubMed] [Google Scholar]
- 60.Niu Y., Lin Z., Wan A., Chen H., Liang H., Sun L., et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18:46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lurie G., Gaudet M.M., Spurdle A.B., Carney M.E., Wilkens L.R., Yang H.P., et al. The obesity-associated polymorphisms FTO rs9939609 and MC4R rs17782313 and endometrial cancer risk in non-Hispanic white women. PLoS One. 2011;6:e16756. doi: 10.1371/journal.pone.0016756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaudet M.M., Yang H.P., Bosquet J.G., Healey C.S., Ahmed S., Dunning A.M., et al. No association between FTO or HHEX and endometrial cancer risk. Cancer Epidem Biomar. 2010;19:2106–2109. doi: 10.1158/1055-9965.EPI-10-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou S., Bai Z.L., Xia D., Zhao Z.J., Zhao R., Wang Y.Y., et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog. 2018;57:590–597. doi: 10.1002/mc.22782. [DOI] [PubMed] [Google Scholar]
- 64.Li Y., Zheng D., Wang F., Xu Y., Yu H., Zhang H. Expression of demethylase genes, FTO and ALKBH1, is associated with prognosis of gastric cancer. Dig Dis Sci. 2019;64:1503–1513. doi: 10.1007/s10620-018-5452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J., Ren D., Du Z., Wang H., Zhang H., Jin Y. m6A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Bioph Res Co. 2018;502:456–464. doi: 10.1016/j.bbrc.2018.05.175. [DOI] [PubMed] [Google Scholar]
- 66.Li Z., Weng H., Su R., Weng X., Zuo Z., Li C., et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sabarneh A., Ereqat S., Cauchi S., AbuShamma O., Abdelhafez M., Ibrahim M., et al. Common FTO rs9939609 variant and risk of type 2 diabetes in Palestine. BMC Med Genet. 2018;19:156. doi: 10.1186/s12881-018-0668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H., Ren Y., Mao K., Hua F., Yang Y., Wei N., et al. FTO is involved in Alzheimer's disease by targeting TSC1-mTOR-Tau signaling. Biochem Bioph Res Co. 2018;498:234–239. doi: 10.1016/j.bbrc.2018.02.201. [DOI] [PubMed] [Google Scholar]
- 69.Ikels K., Kuschel S., Fischer J., Kaisers W., Eberhard D., Rüther U. FTO is a relevant factor for the development of the metabolic syndrome in mice. PLoS One. 2014;9 doi: 10.1371/journal.pone.0105349. e105349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang C.Y., Shie S.S., Tsai M.L., Yang C.H., Hung K.C., Wang C.C., et al. FTO modulates fibrogenic responses in obstructive nephropathy. Sci Rep. 2016;6:18874. doi: 10.1038/srep18874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ding C., Zou Q., Ding J., Ling M., Wang W., Li H., et al. Increased N6-methyladenosine causes infertility is associated with FTO expression. J Cell Physiol. 2018;233:7055–7066. doi: 10.1002/jcp.26507. [DOI] [PubMed] [Google Scholar]
- 72.Manoochehri Khoshinani H., Afshar S., Najafi R. Hypoxia: a double-edged sword in cancer therapy. Cancer Invest. 2016;34:536–545. doi: 10.1080/07357907.2016.1245317. [DOI] [PubMed] [Google Scholar]
- 73.Zhang C., Samanta D., Lu H., Bullen J.W., Zhang H., Chen I., et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m⁶A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113:E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang C., Zhi W.I., Lu H., Samanta D., Chen I., Gabrielson E., et al. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7:64527–64542. doi: 10.18632/oncotarget.11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang S., Zhao B.S., Zhou A., Lin K., Zheng S., Lu Z., et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shriwas O., Priyadarshini M., Samal S.K., Rath R., Panda S., Das Majumdar S.K., et al. DDX3 modulates cisplatin resistance in OSCC through ALKBH5-mediated m6A-demethylation of FOXM1 and NANOG. Apoptosis. 2020;25:233–246. doi: 10.1007/s10495-020-01591-8. [DOI] [PubMed] [Google Scholar]
- 77.Chao Y., Shang J., Ji W. ALKBH5-m6A-FOXM1 signaling axis promotes proliferation and invasion of lung adenocarcinoma cells under intermittent hypoxia. Biochem Bioph Res Co. 2020;521:499–506. doi: 10.1016/j.bbrc.2019.10.145. [DOI] [PubMed] [Google Scholar]
- 78.Zhu Z., Qian Q., Zhao X., Ma L., Chen P. N6-Methyladenosine ALKBH5 promotes non-small cell lung cancer progress by regulating TIMP3 stability. Gene. 2020;731:144348. doi: 10.1016/j.gene.2020.144348. [DOI] [PubMed] [Google Scholar]
- 79.Zhang J., Guo S., Piao H.Y., Wang Y., Wu Y., Meng X.Y., et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75:379–389. doi: 10.1007/s13105-019-00690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang J.Q., Wang B., Lei Z.N., Teng Q.X., Li J.Y., Zhang W., et al. Derivative of 5-cyano-6-phenylpyrimidin antagonizes ABCB1- and ABCG2-mediated multidrug resistance. Eur J Pharmacol. 2019;863:172611. doi: 10.1016/j.ejphar.2019.172611. [DOI] [PubMed] [Google Scholar]
- 81.Wang X., Zhang J., Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019;11:4909–4921. [PMC free article] [PubMed] [Google Scholar]
- 82.Chen S., Zhou L., Wang Y. ALKBH5-mediated m6A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020;20:34. doi: 10.1186/s12935-020-1105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shen C., Sheng Y., Zhu A.C., Robinson S., Jiang X., Dong L., et al. RNA Demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell. 2020;27:64–80.e9. doi: 10.1016/j.stem.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He Y., Hu H., Wang Y., Yuan H., Lu Z., Wu P., et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem. 2018;48:838–846. doi: 10.1159/000491915. [DOI] [PubMed] [Google Scholar]
- 85.Yang P., Wang Q., Liu A., Zhu J., Feng J. ALKBH5 holds prognostic values and inhibits the metastasis of colon cancer. Pathol Oncol Res. 2020;26:1615–1623. doi: 10.1007/s12253-019-00737-7. [DOI] [PubMed] [Google Scholar]
- 86.Chen Y., Zhao Y., Chen J., Peng C., Zhang Y., Tong R., et al. ALKBH5 suppresses malignancy of hepatocellular carcinoma via m6A-guided epigenetic inhibition of LYPD1. Mol Cancer. 2020;19:123. doi: 10.1186/s12943-020-01239-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang Y., Yan J., Li Q., Li J., Gong S., Zhou H., et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373–384. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shen D.D., Suo F.Z., Song Q.M., Chang J., Zhang T., Hong J.J., et al. Development of formaldehyde dehydrogenase-coupled assay and antibody-based assays for ALKBH5 activity evaluation. J Pharm Biomed Anal. 2019;162:9–15. doi: 10.1016/j.jpba.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 89.Yasgar A., Jadhav A., Simeonov A., Coussens N.P. AlphaScreen-Based Assays: ultra-high-throughput screening for small-molecule inhibitors of challenging enzymes and protein‒protein interactions. Methods Mol Biol. 2016;1439:77–98. doi: 10.1007/978-1-4939-3673-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Degorce F., Card A., Soh S., Trinquet E., Knapik G.P., Xie B. HTRF: a technology tailored for drug discovery―a review of theoretical aspects and recent applications. Curr Chem Genom. 2009;3:22–32. doi: 10.2174/1875397300903010022. [DOI] [PMC free article] [PubMed] [Google Scholar]