Abstract
The potential medicinal value of Ma bamboo (Dendrocalamus latiflorus), one of the most popular and economically important bamboo species in China, has been underestimated. In the present study, we found that D. latiflorus leaf extract (DLE) reduced fasting blood glucose levels, body weight, and low-density lipoprotein cholesterol with low liver toxicity in db/db mice. In addition, gene expression profiling was performed and pathway enrichment analysis showed that DLE affected metabolic pathways. Importantly, DLE activated the AKT signaling pathway and reduced glucose production by downregulating glucose-6-phosphatase (G6PC) and phosphoenolpyruvate carboxykinase 1 (PCK1) expression. Moreover, network pharmacology analysis identified rutin as an active component in DLE through targeting insulin growth factor 1 receptor (IGF1R), an upstream signaling transducer of AKT. Due to its hypoglycemic effects and low toxicity, DLE may be considered an adjuvant treatment option for type 2 diabetes patients.
Key words: Dendrocalamus latiflorus leaf extract, Type 2 diabetes mellitus, FOXO1, AKT, Rutin, IGF1R, Gene expression profiling, Network pharmacology
Abbreviations: DLE, Dendrocalamus latiflorus leaf extract; G6PC, glucose 6 phosphatase; PCK1, phosphoenolpyruvate carboxykinase 1; PKB/AKT, protein kinase B; IGF1R, insulin growth factor 1 receptor; T2DM, type 2 diabetes mellitus; PIN, protein interaction network; FOXO1, forkhead box O1
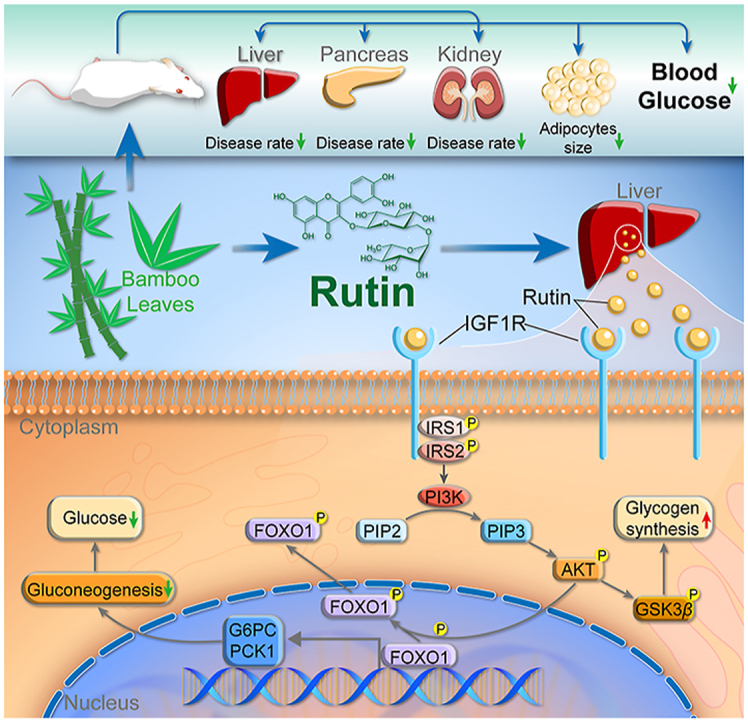
Graphical abstract
Ma bamboo extract and its active component rutin exert hypoglycemic activities by inhibiting hepatic gluconeogenesis through activating AKT/FOXO signaling pathway in an IGF1R dependent manner.
1. Introduction
Humans have struggled with diabetes for more than 20 years. Currently, diabetes is still a major threat to public health in most parts of the world. More than 425 million people are living with diabetes, which leads to 727 billion dollars in healthcare costs worldwide1. Importantly, China is facing the largest diabetes epidemic in the world. According to a recent epidemiology study that involved 98,658 Chinese adults, the overall prevalence of diabetes and prediabetes was estimated to be 11.6% and 50.1%, respectively2. Notably, type 2 diabetes mellitus (T2DM) is the major subtype and accounts for 90% of cases of diabetes3.
T2DM is characterized by insulin resistance (in the early stage) and insulin deficiency (in the late stage). Insulin resistance, also called impaired glucose tolerance, refers to the fact that insulin cannot function properly, which leads to an increased demand for insulin. Different organs exhibit different insulin resistance phenotypes. In fat and muscle tissue, insulin resistance manifests as a reduced ability to absorb glucose, whereas in the liver, insulin resistance is characterized by excess glucose production4.
Clinical antidiabetic drugs are divided into several types based on different targets, including sulfonylureas, biguanides, thiazolidinediones, glucagon-like peptide 1 analogs, α-glucosidase inhibitors, dipeptidyl peptidase IV inhibitors, and sodium/glucose cotransporter 2 inhibitors5. Unfortunately, none of these antidiabetic drugs are free of adverse effects, including hypoglycemia, lactic acidosis, weight gain, cardiovascular disease and so on6,7. Moreover, as a complex systemic metabolic disease, the glycemic control of T2DM patients cannot be sustained using a single drug that affects only a single target8. Therefore, there is an urgent need for novel antidiabetic drugs with lower toxicities and/or multiple targets. Interestingly, natural medicinal plants and plant-derived drugs make up for the shortcomings of monomeric drugs. These medicinal plants and plant-derived drugs, such as Coptidis Rhizoma and its active component berberine, have gained particular interest in recent years as antidiabetic agents with low toxicity9.
Bamboo is a large perennial grass that is widely distributed from tropical to subtropical zones. In ancient China, different parts of bamboo were used for medicinal purposes to treat metabolic syndromes such as diabetes and hyperlipidemia. In addition, bamboo was used to treat inflammatory diseases caused by bacterial infection10. Modern medicine indicates that bamboo extracts play important roles in detoxification and anti-inflammation11. Ma bamboo (Dendrocalamus latiflorus) is one of the most popular and economically important bamboo species in China12. However, the potential medicinal value of Ma bamboo has been underestimated, and its pharmaceutical activities have not yet been systematically studied.
The current study aimed to discover the antidiabetic potential of Ma bamboo and to identify its active components and working mechanism. Leptin receptor-deficient diabetic (db/db) mice were used to study the hypoglycemic effects of D. latiflorus leaf extract (DLE). Transcriptome analysis was performed to reveal DLE-regulated genes and signaling pathways from a systematic perspective. Finally, a protein interaction network (PIN) was constructed to identify the potential targets and predict the active components of DLE.
2. Methods and materials
2.1. Materials
D. latiflorus leaves were purchased from Maoshennongye Co., Ltd. (Shandong, China). HepG2 cell line was obtained from the National Infrastructure of Cell Line Resource, Peking Union Medical College (Beijing, China). Fetal bovine serum (FBS) Uruguay was purchased from Biowest (Nuaillé, France). Dulbecco's modified Eagle's medium (DMEM) with high glucose, liquid and other cell culture reagents were purchased from HyClone (Logan, UT, USA). AKT antibody, phospho-AKT (Ser473) (D9E) XP rabbit mAb, FOXO1 (C29H4) rabbit mAb, phospho-FOXO1 (Ser256) antibody, GSK-3β (D5C5Z) XP rabbit mAb, phospho-GSK-3β (Ser9) (D85E12) XP rabbit mAb, and GAPDH (14C10) rabbit mAb were purchased from Cell Signaling Technology (Beverly, MA, USA). The insulin growth factor 1 receptor (IGF1R) inhibitor GSK1904529A was purchased from Selleck Chemicals (Houston, TX, USA).
2.2. Preparation of DLE
A total of 100 g of leaves were milled and extracted with 150 mL of 90% ethanol in a distillation bottle. Solvents were maintained at 80 °C for 2 h for extraction. The raw extract was concentrated at 45 °C for 2 h. Finally, the concentrate was lyophilized at −45 °C and 80 Pa for 24 h to obtain the dry powder. The DLE powder was dissolved in saline for animal study and in dimethyl sulfoxide (DMSO) for in vitro experiments to ensure consistent preparation of DLE and rutin.
2.3. Animal study
Eight-week-old male C57BL/6J and T2DM db/db mice were purchased and housed at the animal facility of the Model Animal Research Center of Nanjing University (the program conforms with the AAALAC International Standard). Mouse experiments were conducted with the approval of the Institutional Animal Care and Use Committee (IACUC) of GemPharmatech Co. Ltd. [Nanjing, China; Permit Number: SYXK (Jiangsu) 2018-0027; Animal Protocol No: GPTAP009]. The mice were randomly divided into 4 groups of 8 animals based on equal body weight and fasting blood glucose (FBG) levels (mice were fasted for 6 h). The C57BL/6J mice (normal group) were treated with saline. The T2DM db/db mice were treated with saline, DLE (200 mg/kg), or pioglitazone (PGZ, 20 mg/kg) for 12 weeks, and were named model group, DLE group, and PGZ group, respectively. All mice received oral administration of the indicated treatments once a day. Considering the extraction rate of D. latiflorus leaves (26.5%), the intragastric dose of 200 mg/kg of DLE equals to 755 mg/kg of D. latiflorus leaves. After dose conversion between humans and mice, this is equivalent to 5 g D. latiflorus leaves taken by a 60 kg human every day, which is a reasonable amount for daily consumption.
2.3.1. Body weight, food/water intake, and plasma biochemical assay
Body weight (g) and food/water intake were recorded once a week. The mice were treated with the same method for the normal group, model group, and DLE group, and the body weight differences were evaluated between the model group (db/db mice treated with saline) and the DLE group (db/db mice treated with DLE). FBG (6 h) was measured once per month using a blood glucose meter V1.3.0 1455 (Bayer, Germany). At the endpoint of the experiment (drug treatment for 12 weeks), mouse plasma samples were collected, centrifuged for 10 min (Centrifuge 5810R, Eppendorf, Hamburg, Germany, 3000 rpm) and stored at −80 °C until further analysis. High-density lipoprotein cholesterol (HDL-c), low-density lipoprotein cholesterol (LDL-c), triglyceride (TG), total cholesterol (TC), aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels were measured using a Hitachi 7020 Automatic Analyzer (Hitachi, Japan). Plasma insulin concentration was determined using a Rat/Mouse Insulin ELISA Kit (Millipore, Burlington, MA, USA).
2.3.2. Insulin tolerance test (ITT)
The ITT was performed one week before the endpoint of the experiment. Fasted mice (6 h) were orally challenged with insulin (0.6 U/kg). Blood glucose was measured at 0, 15, 30, 60, 90, and 120 min after insulin administration.
2.3.3. Histological observation
At the endpoint, the mice were sacrificed, and their organs were collected. Liver, fat, pancreas, and kidney tissue sections (5 μm thickness) were fixed and frozen, followed by hematoxylin–eosin (HE) staining for histological observation or oil red O (ORO) staining for lipid droplet evaluation. In each HE stained fat tissue picture, ten adipocytes were randomly selected for ImageJ quantification. The mean adipocyte size of the normal group, model group, and DLE group was calculated by GraphPad Prism (version 8.0, GraphPad Software, San Diego, CA, USA).
2.3.4. RNA-seq analysis of mouse livers
Liver tissues of three randomly selected mice from the normal, model, PGZ, and DLE groups were preserved after sacrifice and ground into powder in liquid nitrogen for the following RNA-seq analysis. Total RNA was extracted by TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) and purified by a TURBO DNA-free Kit (Invitrogen) according to the manufacturer's instructions. RNA gel electrophoresis and an Agilent 2100 Bioanalyzer System (Agilent Technologies, Santa Clara, CA, USA) were used to evaluate the quality of the extracted RNA. Quality-controlled and purified RNA was used for cDNA library construction. The amplification and sequencing of the cDNA library were performed by a NovaSeq 6000 Sequencing System (Illumina, San Diego, CA, USA) with the PE150 strategy. Quality control was performed on the FASTQ files using FastQC software (v0.11.5). StringTie (version 1.3.3b) was used to align the transcript sequences obtained by RNA-seq to the Mus musculus GRCm38 reference genome. The differential expression analysis was performed by DESeq2 (version 1.16.1, R language package), and the differentially expressed genes (DEGs) were identified with a fold change >1.5 and P-value < 0.05. Heatmap generation, cluster analysis, principal component analysis (PCA), and t-distributed stochastic neighbor embedding (tSNE) were performed by R (version 3.5.2). Pearson correlation coefficients and Euclidean distance were selected as the default parameters during gene/sample clustering.
2.4. RNA-mediated oligonucleotide annealing, selection, and ligation with next-generation sequencing (RASL-seq) analysis of DLE-treated HepG2 cells
HepG2 cells were treated with DMSO or DLE for 24 h, and gene expression profiling was performed by a modified RNA-seq procedure based on an RASL strategy13,14. A total of 3265 genes that play important roles in glucose metabolism, lipid metabolism, inflammation, and cancer progression were selected for detection, and a pair of probes was designed for each gene. The RASL procedure was automatically implemented by the Bravo Automated Liquid Handling Platform and the Agilent BenchBot Robot (Agilent Technologies). In the RNA annealing step, target mRNAs were bound with gene probes and biotinylated oligo-dTs (Sangon Biotech, Shanghai, China), and Sera-Mag Magnetic Streptavidin Coated Particles (General Electric, Boston, MA) were used to capture the biotinylated oligo-dTs. In the selection and ligation step, each pair of gene probes was ligated by T4 DNA ligase (NEB, Ipswich, MA). The unligated gene probes were eluted and the ligated gene probes were tagged with bar codes by PCR. The PCR products were pooled, purified, and sequenced by HiSeq X Ten (Illumina), and the data were analyzed in the same manner as the RNA-seq data. DLE regulated dose dependent genes were defined using IsoGene (R package, version 1.0.24)15.
2.5. Gene Ontology (GO) and pathway analysis
DEG functions were annotated by GO analysis, and DEG-enriched biological signaling pathways were identified by Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. GO and KEGG analyses were performed using the DAVID Bioinformatics Resources (http://david.ncifcrf.gov, NIH).
2.6. Cell viability assay
Cell viability was measured using a Cell Counting Kit-8 (Dojindo Molecular Technologies, Kumamon, Japan). Cells were seeded in 96-well plates and treated with various concentrations of DLE for 24 h. Each well was incubated with 100 μL of 10% Cell Counting Kit-8 working solution for 2 h at 37 °C. The absorbance at 450 nm was measured using a SpectroMax Spectrophotometer (SpectroMax Solutions Ltd., London, UK).
2.7. Glucose production assay
The glucose production assay was performed as previously described16. Briefly, HepG2 cells were treated with DMSO, the indicated concentrations of DLE, or rutin for 24 h. Cells were rinsed twice with PBS and incubated with glucose production buffer with glucose-free DMEM, pH 7.4, containing 20 mmol/L sodium lactate (Sigma, Saint Louis, MO, USA), 0.1 mmol/L pCPT-cAMP (Sigma), and 2 mmol/L sodium pyruvate without phenol red (Gibco, Brooklyn, NY, USA) for 3 h. At the end of incubation, 100 μL of the medium was collected to measure the glucose concentration with a Glucose Oxidase Method Kit (Applygen Technologies, Beijing, China) per the manufacturer's instructions. Protein concentration was determined and used for normalization.
2.8. Construction of the reporter gene system and luciferase assays
The promoter regions of human glucose-6-phosphatase (G6PC) (−1200 to +128) and phosphoenolpyruvate carboxykinase 1 (PCK1) (−1500 to 0) were amplified from genomic DNA extracted from HepG2 cells. The PCR products were cloned into the pGL3-enhancer vector. Plasmids containing the G6PC and PCK1 promoters were designated pGL3-G6PC and pGL3-PCK1, respectively. HepG2 cells were cotransfected with 1000 ng of the indicated luciferase plasmids (pGL3-G6PC or pGL3-PCK1) and 100 ng of pRL-TK Renilla plasmid using the jetPEI (Polyplus, New York, NY, USA). After 24 h, the cell medium was replaced with medium supplemented with DMSO, 200 μg/mL DLE or 50 μmol/L rutin. The luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Renilla activity was used to normalize the transfection efficiency.
2.9. RNA isolation, cDNA synthesis, and quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Total RNA was isolated by TRIzol Reagent (Invitrogen) according to the manufacturer's instructions. A total of 1 μg RNA was reverse transcribed into cDNA by a High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific, Rochester, NY, USA). Quantitative real-time PCR was conducted by incorporating KAPA SYBR FAST qPCR Master Mix (2 ×) Kit (Kapa Biosystems, Woburn, MA, USA) using a CFX Opus 96 Real-Time PCR System (BIO-RAD, Hercules, CA, USA), and relative mRNA levels were determined by normalizing to the level of GAPDH. The relative gene expression data were analyzed using the ΔΔCT method17.
2.10. Western blot
Cells and animal tissues were lysed with lysis buffer, and cell debris was removed by centrifugation at 28,340 × g for 15 min at 4 °C. Equal amounts of protein from each sample (30 μg for cells and 60 μg for animal tissues) were subjected to SDS-PAGE and transferred onto 0.2 μm PVDF membranes (Millipore). After being blocked with 7% skim milk, the membranes were incubated with primary antibodies at 4 °C overnight and then with horseradish peroxidase-conjugated secondary antibodies for 1 h. After each antibody incubation, the membranes were washed 3 times with TBST. Finally, the membranes were developed with ECL Prime Western Blotting System (General Electric), and the results were recorded on X-ray film.
2.11. Small interfering RNA (siRNA) and transient transfection
HepG2 cells were transfected with siRNAs using jetPRIME (Polyplus) after reaching 50% confluence according to the manufacturer's protocol. The FOXO1 (forkhead box O 1)-specific siRNA and control scrambled siRNA were synthesized by GenePharma (Shanghai, China). The sequence of FOXO1-specific siRNA was as follows: 5′-CCAUGGACAACAACAGUAATT-3′. The sequence of control scrambled siRNA was 5′-UUCUCCGAACGUGUCACGUTT-3′.
2.12. Isolation of primary mouse hepatocytes
Hepatocytes were isolated from mice by two-step collagenase perfusion. Briefly, after placing a catheter into the portal vein, the inferior vena cava was cut and the liver was perfused at 5–7 mL/min with a prewarmed perfusion solution (D-Hanks with 5 mmol/L ethylene glycol tetraacetic acid) for 5 min. Then, perfusion was performed with prewarmed digestion solution (D-Hanks containing 0.3 mg/mL collagenase IV and 5 mmol/L CaCl2) for 10 min. After dissociation, cells were filtered through a 70 μm filter. Hepatocytes were further separated and purified by centrifugation at low speed (50 g, 5 min, 4 °C).
2.13. Protein interaction network analysis of DLE
First, a total of 530 DEGs with |fold change| > 2 were selected by RASL-seq (DLE 200 μg/mL) as seed proteins to construct PINs. Next, the protein–protein interactions (PPIs) of the seed proteins with a confidence score >0.99 were obtained from the String database (http://string-db.org/, ELIXIR, UK). Finally, PPIs were visualized by Cytoscape 3.6.0 (https://cytoscape.org, NRNB, La Jolla, CA), and the largest connected subgraph was selected as the PIN of DLE18. Functional modules of the PIN were identified by the fast agglomerate algorithm based on the edge clustering coefficients (FAG-EC)19. GO enrichment analysis was used to analyze the molecular functions of the identified functional modules by the BinGO method.
2.14. Molecular docking of insulin growth factor 1 receptor (IGF1R) agonists
The molecular docking process was performed as follows: first, the crystal structure and binding pockets (sites A and B) of IGF1R were obtained from the Protein Data Bank (https://www.rcsb.org, Rutgers, NJ, USA) and the literature20, respectively, and a total of 14 chemical components in DLE were identified based on literature mining21,22. Next, low-energy three-dimensional conformations of the 14 candidate components were minimized within the CHARMm force field for molecular docking of IGF1R. Finally, the LibDock algorithm was utilized to identify potential IGF1R agonists in DLE using Discovery Studio (https://discoverystudios.com, Discovery Inc., LA)23.
2.15. Statistical analysis
The results of the in vitro study are expressed as the mean ± standard deviation (SD), followed by Student's t-test or linear regression analysis. The results of the in vivo study are expressed as the mean ± standard error of the mean (SEM), followed by nonparametric Kolmogorov–Smirnov test. Kolmogorov–Smirnov test and linear regression analysis were performed by GraphPad Prism. A P-value <0.05 was considered statistically significant.
3. Results
3.1. DLE improved glucose and lipid metabolism-related parameters in db/db mice
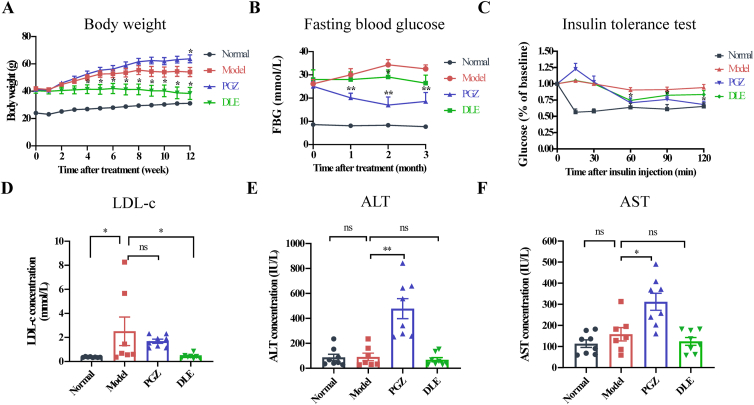
As one of the key clinical symptoms in db/db diabetic mice, body weight increases rapidly over time and is a major cause of insulin resistance. After 4 weeks of administration, DLE treatment resulted in a reduction in body weight until the end of the experiment (Fig. 1A). However, PGZ, a well-studied hypoglycemic drug that was used as a positive control, further increased the body weight of the mice, which is an undesirable side effect. Moreover, the food and water intake in the DLE group was reduced compared with those in the model group (Supporting Information Fig. S1A and S1B).
Figure 1.
DLE improved glucose and lipid metabolism-related parameters in db/db mice. (A) The body weight and (B) FBG level (mice were fasted for 6 h) of C57BL/6J mice treated with saline (normal group, black) and db/db mice treated with saline (model group, red), DLE (DLE group, 200 mg/kg, green), or PGZ (PGZ group, 20 mg/kg, blue). (C) Insulin tolerance test. Mice were fasted for 6 h, followed by intraperitoneal injection of insulin (0.6 U/kg). Mouse blood glucose was detected every 30 min for 2 h. (D) LDL-c concentration, (E) ALT concentration, and (F) AST concentration in mouse plasma samples at the endpoint (drug treatment for 12 weeks). The food intake, water intake, FBG increment, TC, HDL-c, and TG in each group are shown in Fig. S1. The data are shown as the mean ± SEM (n = 7 for model group, n = 8 for normal, DLE, and PGZ group). Statistical analyses were conducted using Kolmogorov–Smirnov test. ∗P < 0.05, ∗∗P < 0.01 vs. the model group, ns means not significant. DLE, Dendrocalamus latiflorus leaf extract; FBG, fasting blood glucose; PGZ, pioglitazone; LDL-c, low-density lipoprotein cholesterol; ALT, alanine aminotransferase; AST, aspartate aminotransferase.
FBG was measured once a month, and the results indicated that PGZ lowered FBG within one month, while DLE showed hypoglycemic effects after two months of administration and maintained the FBG in a relatively stable state (Fig. 1B). Considering the baseline FBG was different among the groups, the FBG increment was analyzed in each group. The FBG increment were defined as the current blood glucose level minus the baseline blood glucose level. After 2 months of treatment, the average FBG increment was 1.31 in the DLE group and 8.12 in the model group (Fig. S1C). The ITT was performed to evaluate insulin resistance in each group. Mice were fasted for 6 h, and glucose concentration was measured at 0, 15, 30, 60, 90, and 120 min after insulin administration (0.6 U/kg). At 60 min after insulin loading, glucose levels were decreased by 9.58%, 29.2%, 24.84%, and 36.18% in the model, PGZ, DLE, and normal groups, respectively (Fig. 1C). Compared with the model group, DLE lowered glucose levels at 60 min after insulin injection, indicating that DLE increases mouse insulin sensitivity to some extent.
At the endpoint (after 12 weeks of drug administration), the animals were sacrificed, and blood plasma was extracted for biochemical assays. Lipid metabolism parameters, including LDL-c, HDL-c, TG, and TC, were analyzed. DLE reduced LDL-c concentrations compared with those in the model group, while PGZ did not (Fig. 1D). There were no differences in TC, HDL-c, or TG between the model and DLE groups (Fig. S1D–S1F). To estimate drug-induced liver toxicity, we measured the concentrations of ALT and AST in each group. The results show that DLE did not alter ALT or AST levels, while PGZ exhibited liver toxicity with induced ALT and AST (Fig. 1E and F). In summary, DLE ameliorated dysregulated glucose and lipid metabolism in db/db mice without obvious liver toxicity.
3.2. DLE improved the histopathological changes in db/db mice
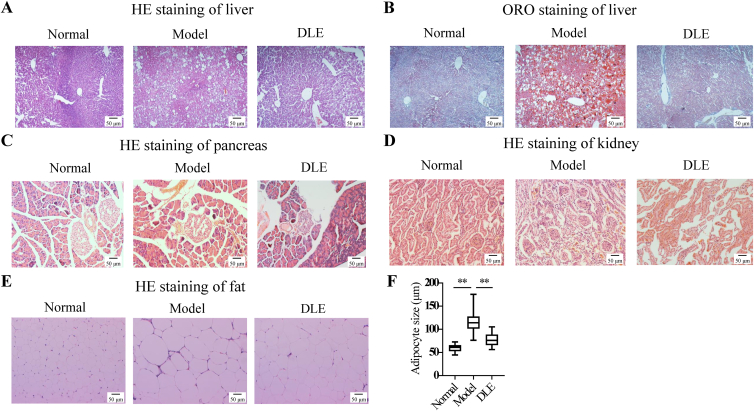
Different mouse tissues were sectioned for histopathological observation. HE staining of liver sections in the model group indicated that the liver tissue from diabetic mice was swelled, denatured, and vacuolated, and these effects were alleviated and rescued in the DLE group (Fig. 2A). ORO staining of liver sections demonstrated that the number and size of lipid droplets in DLE-treated mice were decreased compared with those in the model group (Fig. 2B).
Figure 2.
DLE improved the histopathological changes in db/db mice. At the endpoint, the mice were sacrificed, and different tissues were sectioned for HE staining or ORO staining for histopathological observation. HE staining of (A) liver tissue, (C) pancreatic tissue, (D) kidney tissue, and (E) fat tissue, and (B) ORO staining of liver tissue are shown. The complete panel was presented in Fig. S2. (F) The mean adipocyte size of the normal group, model group, and DLE group. The sizes of ten randomly selected adipocytes stained by HE were quantified by ImageJ. The data are shown as the mean ± SD (n = 3). Statistical analyses were conducted using unpaired Student's t-tests. ∗∗P < 0.01 vs. the model group. HE, hematoxylin–eosin; ORO, oil red O.
Moreover, HE staining of the pancreatic islets showed that DLE treatment rescued the disorganized structures and vascular degeneration compared with the model group (Fig. 2C). Diabetic nephropathy is one of the main complications of diabetes during the late stage. In the present study, we found that the glomerulus in the model group was enlarged compared with that in the normal group, indicating the occurrence of diabetic nephropathy. However, the glomerulus size was reduced in the DLE group (Fig. 2D). As obesity is a common symptom in T2DM patients, adipose tissues were analyzed by HE staining. The model group showed enlarged adipocytes compared with those in the normal group, while DLE reduced adipocyte size (Fig. 2E). The quantitative results show that DLE reduced adipocyte size by 30.27% (Fig. 2F). The complete panel of HE staining and ORO staining of three animals in each group is shown in Supporting Information Fig. S2A–S2E. These results suggest that DLE is effective in improving T2DM-related histopathological changes in the liver, pancreas, kidney, and fat.
3.3. DLE affected glucose and lipid metabolic pathways and partly reversed diabetes-induced transcriptional changes in vivo
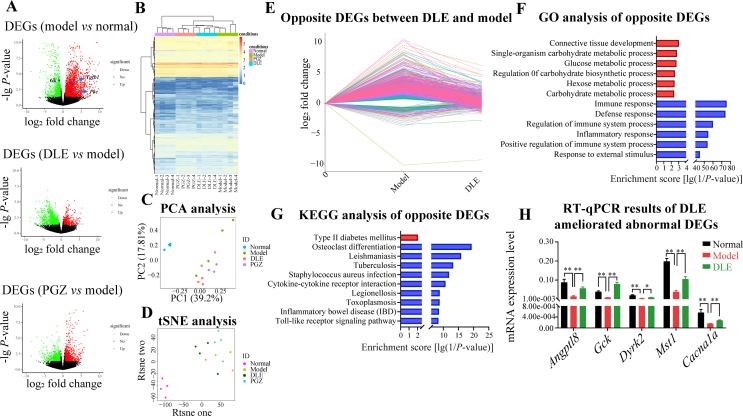
The liver plays an important role in maintaining the balance of glucose metabolism and accounts for ∼90% of endogenous glucose production. Clinical antidiabetic drugs that target the liver include metformin, which works primarily by reducing hepatic glucose production through inhibiting gluconeogenesis24. To further explore the molecular mechanisms underlying the hypoglycemic activities of DLE, the liver tissues of three randomly selected mice from the normal, model, PGZ, and DLE groups were used for RNA-seq analysis. There were 3708 DEGs in the model group versus the normal group. Among these DEGs, 2533 were upregulated (expressed in red), and 1175 were downregulated (expressed in green, Fig. 3A). Importantly, many abnormally expressed gene markers in T2DM patients, including glucokinase (GK)25, protein kinase C (PKC)26, and transforming growth factor beta 1 (TGFB1)27 (Fig. 3A), were identified as DEGs (model group versus normal group) in the present study. There were 1743 DEGs identified in the DLE group versus the model group, with 496 upregulated and 1247 downregulated (Fig. 3A).
Figure 3.
DLE affected glucose and lipid metabolic pathways and partly reversed diabetes-induced transcriptional changes in vivo. (A) Volcano plots of DEGs among different comparisons (upregulation in red and downregulation in green). Each comparison of DEGs was determined by comparing model vs. normal, DLE vs. model, and PGZ vs. model using fold change >1.5 and P-value < 0.05 as the cut-off. (B) Hierarchical clustering, (C) PCA, and (D) tSNE analysis of all groups was performed using the normalized RNA-seq read counts that corresponded to all DEG unions as the input. (E) Line chart of DLE improved 1004 DEGs in db/db mice (opposite DEGs between DLE and model group). (F) The GO annotated biological processes enriched by the DLE improved 1004 DEGs. Among them, 101 were upregulated (red) and 903 were downregulated (blue). (G) Enriched KEGG pathways in the 101 DLE-upregulated genes (red) and 903 DLE-downregulated genes (blue). (H) RT-qPCR of representative genes involved in glucose metabolism pathways (n ≥ 3). The data are shown as the mean ± SD. Statistical analyses were conducted using unpaired Student's t-tests. ∗P < 0.05, ∗∗P < 0.01 as indicated in the figure. DEG, differentially expressed genes; PCA, principal component analysis; tSNE, t-distributed stochastic neighbor embedding; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; RT-qPCR, quantitative reverse transcription polymerase chain reaction.
The normalized RNA-seq read counts that correspond to the DEG union of the model, PGZ, and DLE groups were clustered with the normal group. The cluster analysis results show that each group could be distinguished (Fig. 3B). The model group was the furthest from the normal group while the PGZ and DLE group reversed the gene expression pattern to the normal group to some degree. The PCA and tSNE analysis results were consistent with the cluster results (Fig. 3C and D), that the normal group was far away from other groups and the PGZ and DLE group was somehow deviated from the model group. Importantly, in each analysis, the DLE cluster was closely associated with the PGZ cluster, which clustered in parallel, suggesting DLE was similar to PGZ in gene regulation compared with the model and normal groups (Fig. 3B–D). Next, GO and KEGG analyses were performed on DEGs in DLE group to uncover DLE-mediated signaling pathways. We found biological processes involved in carbonhydrate metabolism were enriched by DLE (Supporting Information Fig. S3A) and lipid metabolic pathways such as fatty acid biosynthesis, fatty acid elongation, and fatty acid metabolism were highly enriched by DLE (Fig. S3B). These results indicate that DLE regulates glucose and lipid metabolism.
A shift in the transcriptome was observed in the model group compared with the normal group. Therefore, it was important to identify genes associated with DLE-mediated improvements that were abnormal in the model group. A total of 1004 opposite DEGs were found between the DLE/model and model/normal comparisons. Among them, 903 were upregulated in the model group but reduced by DLE. Another 101 genes were downregulated in the model group but induced by DLE (Fig. 3E).
Next, the 1004 opposite DEGs were used for functional annotation and pathway enrichment determination by GO and KEGG analysis. GO analysis showed that four glycometabolism-related pathways, including glucose metabolic process, regulation of carbohydrate biosynthetic process, hexose metabolic process, and carbohydrate metabolic process, were enriched in the DLE-upregulated DEGs (Fig. 3F). KEGG analysis indicated that the type 2 diabetes mellitus signaling pathway was enriched in DLE-upregulated DEGs (Fig. 3G). These results validate that DLE reversed the abnormal expression of diabetes-related genes at the transcriptional level. On the other hand, the enrichment results of DLE-downregulated DEGs suggest that DLE was involved in immunity and inflammation regulation. Immune- and inflammation-related signaling pathways, including immune response, defense response, regulation of immune system process, inflammatory response, inflammatory bowel disease, and Toll-like receptor signaling pathway, were enriched in DLE-downregulated DEGs (Fig. 3F and G). DLE not only regulated glucose and lipid metabolic genes but also played roles in anti-inflammatory processes in db/db mice.
Furthermore, the DLE-modulated genes involved in glucose and lipid metabolic pathways, including the lipid regulator gene Angptl8, glycolysis gene Gck, and signaling transduction genes Dyrk2, Mst1, and Cacna1a, were selected for RT-qPCR validation. Notably, compared with expression of these genes in the normal group, expression of these genes was downregulated in the model group but was rescued in the DLE group (Fig. 3H).
According to the transcriptome analysis in vivo, pathways including type II diabetes mellitus were enriched by the genes that were altered in the model group but were reversed by DLE. Those pathways indicated DLE's potential in treating diabetes. Therefore, we switched to an in vitro study of DLE to explore its underlying mechanism.
3.4. Transcriptome analysis of DLE in vitro revealed that DLE affected gluconeogenesis via the protein kinase B (PKB/AKT)/FOXO1 signaling pathway
To further explore the molecular mechanism underlying the hypoglycemic functions of DLE on hepatic cells, HepG2 cell line was selected for subsequent in vitro studies. The cell viability assay was performed to detect the cytotoxicity of DLE, followed by RASL-seq and pharmacological studies. HepG2 cells were treated with the indicated concentrations of DLE (12.5–400 μg/mL) for 24 h, and the results indicate that DLE had little cytotoxicity within the analyzed concentration range (Supporting Information Fig. S4A). HepG2 cells were treated with DMSO or 100, 200, and 300 μg/mL DLE in triplicate for 24 h, followed by RASL-seq-based gene profiling. To determine the effects of DLE on gene regulation in vitro, a total of 3265 probes targeting genes related to glucose metabolism, lipid metabolism, inflammation, and cancer progression were selected for RASL-seq analysis.
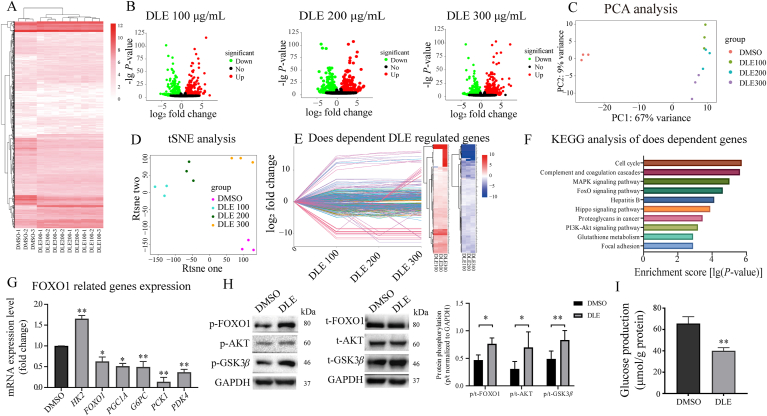
The normalized RNA-seq read counts corresponding to the DEG unions of all groups (DLE vs. DMSO) were used for clustering analysis. The clustering results indicated that all DLE groups were far from the DMSO group branch and DEGs were differentially regulated by different DLE concentrations, as each concentration group was distinguished from the others (Fig. 4A). DEGs that were affected by each DLE concentration were visualized by volcano plot with the upregulated DEGs shown in red and downregulated DEGs shown in green (Fig. 4B). A total of 770 (396 upregulated and 374 downregulated), 722 (373 upregulated and 349 downregulated) and 712 (363 upregulated and 349 downregulated) DEGs were identified in the DLE 100, 200, and 300 μg/mL treatment groups, respectively.
Figure 4.
Transcriptome analysis of DLE in vitro revealed that DLE affected gluconeogenesis via the AKT/FOXO1 signaling pathway. (A) Hierarchical clustering, (C) PCA, and (D) tSNE analysis of all groups, using the normalized read counts of RNA-mediated oligonucleotide annealing, selection, and ligation with next-generation sequencing that corresponded to all DEG unions as the input. (B) The volcano plots of DEGs induced by the indicated concentrations of DLE (upregulation in red and downregulation in green). (E) Line chart and heatmap of dose-dependent DLE regulated genes (upregulated in red and downregulated in blue). (F) Enriched KEGG pathways in DLE regulated dose responsive genes. (G) RT-qPCR results of 200 μg/mL DLE-regulated FOXO1-related genes (n = 3). (H) Western blot analysis of the protein expression and phosphorylation levels of FOXO1 (n = 3), AKT (n = 4), and GSK3β (n = 3). The results were quantified by ImageJ software, the data were quantified by ImageJ. (I) The inhibitory effects of DLE on the glucose production in HepG2 cells treated with 200 μg/mL DLE for 24 h (n = 3). The data are shown as the mean ± SD. Statistical analyses were conducted using paired Student's t-tests. ∗P < 0.05, ∗∗P < 0.01 vs. the DMSO-treated group. PKB/AKT, protein kinase B; FOXO1, forkhead box O1; GSK3β, glycogen synthase kinase 3 β; DMSO, dimethyl sulfoxide.
PCA and tSNE analyses were performed using the same data as was used for the clustering analysis. The results show that DLE-treated samples were obviously distinguished from the DMSO group, and the genes were differentially regulated among the different DLE concentrations, which was consistent with the cluster analysis results (Fig. 4C and D).
Next, we explored the potential targets of DLE whose expression was regulated by DLE in a dose-dependent manner. We found 476 dose-dependent DLE-regulated genes (210 upregulated and 266 downregulated, Fig. 4E) using an R package IsoGene. Moreover, KEGG analysis was performed to investigate the function of 476 dose–response genes. As shown in Fig. 4F, the cell cycle, complement and coagulation cascades, mitogen-activated protein kinase (MAPK) signaling pathway, FOXO signaling pathway, and phosphatidylinositol 3-kinase (PI3K)–AKT signaling pathway were enriched. To validate the reliability of the RASL-seq findings, six DEGs involved in the top 4 enriched pathways were selected as representative genes and validated by RT-qPCR. The MAPK signaling pathway gene EGFR and the FOXO signaling pathway gene SKP2, the cell cycle genes TGFB1 and TFDP1, and the complement and coagulation cascade genes C5AR1 and FGA were analyzed. The RT-qPCR results agreed well with the RASL-seq results (Fig. S4B).
Although the type II diabetes mellitus was not enriched directly by the DEGs of DLE in the HepG2 cell line as the in vivo study, pathways related to glucose metabolism, such as the MAPK signaling pathway, FOXO signaling pathway, and PI3K–AKT signaling pathway were enriched. The FOXO signaling pathway plays important roles in gluconeogenesis, oxidative stress resistance, the cell cycle, DNA damage repair, and anti-inflammation28,29. Interestingly, the PI3K–AKT signaling pathway, which is the upstream of the FOXO signaling pathway and plays a critical role in glucose metabolism, was also enriched, suggesting that DLE may affect glucose metabolism by mediating the FOXO signaling pathway through AKT activation.
To confirm this hypothesis, glucose metabolism genes, including HK2, FOXO1, PGC1A, G6PC, PCK1, and PDK4, were selected from the FOXO signaling pathway for RT-qPCR validation. After DLE treatment of HepG2 cells for 24 h, the glycolysis gene HK2 was upregulated, while the gluconeogenesis genes, including FOXO1, PGC1A, G6PC, and PCK1, and the citric acid cycle-related gene PDK4 were all downregulated, indicating gluconeogenesis process was inhibited by DLE (Fig. 4G). Similar results were shown that Pgc1a, G6pc, Pck1, and Pdk4 were all downregulated in mouse primary hepatocytes treated with DLE (Supporting Information Fig. S5A). Gluconeogenesis accounts for ∼50% of hepatic glucose production during overnight fasting in humans and is subtly regulated at the transcriptional level. Key genes involved in gluconeogenesis include G6PC and PCK1, both of which encode the rate-limiting enzymes for gluconeogenesis. It was widely reported that the first-line antidiabetic drug metformin and the hypoglycemic hormone insulin reduced hepatic glucose production by inhibiting G6PC and PCK1 expression30,31.
Importantly, insulin works through AKT-induced phosphorylation of FOXO1, which is the key protein in the FOXO signaling pathway. Phosphorylation of FOXO1 leads to nuclear translocation, which induces downregulation of G6PC and PCK1 expression and eventually affects glucose production32. Therefore, we explored whether FOXO1 and its upstream AKT were activated by DLE at the protein level. The Western blot results showed that phosphorylation of both AKT and FOXO1 was increased by DLE treatment (Fig. 4H). Phosphorylation of glycogen synthase kinase 3 β (GSK3β), another protein downstream of AKT, that is involved in glycogen production, was also increased (Fig. 4H). To verify whether FOXO1 is indispensable during DLE regulation, we knocked down FOXO1 by siRNA in the HepG2 cells to observe the expression changes of G6PC and PCK1 with or without DLE treatment. FOXO1 was knocked down by approximately 75% in HepG2 cells (Fig. S5B). As expected, G6PC and PCK1 were reduced by DLE in the scrambled siRNA-transfected HepG2 cells, and this reduction was abolished in the FOXO1-specific siRNA-transfected cells (Fig. S5B). To validate the function of DLE at the phenotype level, a glucose production assay was performed. The results show that DLE treatment for 24 h reduced hepatic glucose production in HepG2 cells (Fig. 4I). These results demonstrate that DLE inhibits glucose production by inhibiting G6PC and PCK1 expression via AKT-mediated phosphorylation of FOXO1.
3.5. Network pharmacology analysis indicated that DLE activated AKT through targeting IGF1R and rutin was a potential IGF1R agonist in DLE
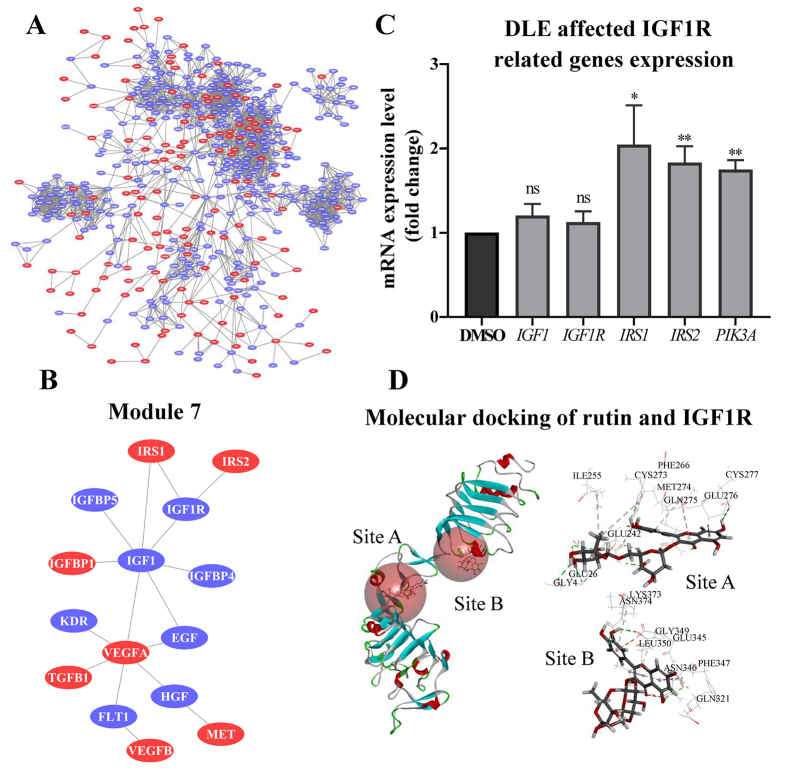
Exploring factors that are upstream of AKT is challenging because AKT is activated by a variety of pathways. Recently, network pharmacology has attracted the attention of researchers as a method to identify molecular pathways and targets underlying drug action. In the present study, PIN and molecular docking, which are both network pharmacology methods, were used to identify DLE targets. A total of 530 DEGs of DLE with |fold change| > 2 were selected from the RASL-seq results (DLE 200 μg/mL) as seed proteins for PPI analysis. After PPIs visualization, the largest connected sub-graph was obtained as the PIN of DLE, which consisted of 148 seed proteins (plotted in red) and 359 interacting proteins searched from String (plotted in blue) (Fig. 5A). The pharmacological activities and key nodes were identified from the PIN by the FAG-EC method, from which 9 modules were obtained. In addition, functional enrichment analysis was performed by the BinGO method to annotate the biological functions of each module, and the results are shown in Supporting Information Table S1.
Figure 5.
Rutin was identified as an active component in DLE and was predicted as a potential IGF1R agonist. (A) The DLE-regulated protein interaction network. DLE affected 148 proteins, shown in red, while 359 interacting proteins searched from String are shown in blue. (B) Illustration of module 7 of the protein interaction network (other modules were presented in Fig. S6). Module 7 was involved in the insulin receptor binding pathway. (C) RT-qPCR validation of DLE-mediated IGF1R-related genes (n = 3). The data are shown as the mean ± SD. Statistical analyses were conducted using paired Student's t-tests. ∗P < 0.05, ∗∗P < 0.01 vs. the DMSO-treated group, ns means not significant. (D) Molecular docking results of rutin and IGF1R using the LibDock algorithm. IGF1R, insulin growth factor 1 receptor.
Among the 9 modules, module 7 was involved in the insulin receptor binding pathway (Fig. 5B and Table S1, the specific content of the other 8 modules is shown in Supporting Information Fig. S6) which could explain the observed DLE-induced AKT activation. Therefore, the direct target of DLE may be present in module 7. Module 7 consists of 15 proteins, including insulin receptor substrates 1 and 2 (IRS1 and IRS2), which are critical proteins involved in the initiation of insulin signaling transduction and eventually leads to AKT activation. Importantly, IGF1R is the closest protein connected with IRS1 and IRS2 and located on cell membrane, which indicates that IGF1R is the potential direct target of DLE that leads to AKT activation (Fig. 5B). As a member of the receptor tyrosine kinase family that binds with IGF1, IGF1R shares a similar structure with the insulin receptor33. Binding of insulin to IGF1R initiates insulin-related cascade activation34. Thus, DLE may interact with IGF1R to regulate downstream effector genes. RT-qPCR results show that the expression of IGF1R downstream genes, such as IRS1, IRS2, and PIK3A, was indeed all upregulated by DLE, while the expression of IGF1R or its ligand IGF1 was not affected (Fig. 5C).
Next, molecular docking was performed to identify the potential bioactive components in DLE that bind IGF1R. According to the study by Young-Lai Cho, the cysteine-rich domains (site A and site B) of IGF1R are protein pocket binding sites for IGF1R agonists, whose binding leads to insulin signaling pathway activation20. After literature mining, 14 DLE constituents were identified and analyzed for molecular docking with IGF1R at the pockets of sites A and B (Table 1). The docking results show that rutin had the highest binding score for site A and ranked 4th in binding score for site B (Table 1, the IGF1R agonist Rg5 was selected as positive control). The binding modes of rutin with site A/B are shown in Fig. 5D. Interestingly, rutin shares similar IGF1R-interacting amino acids as the IGF1R agonist Rg5, including Glu26, Glu242, Gln275, Gly4, Ile255, Phe266, and Glu276 at site A and Asn346, Gln321, and Glu345 at site B. Importantly, the predicted binding site Phe266 was previously validated, which objectively confirmed the reliability of the present prediction20. In summary, DLE activates the insulin signaling pathway, which is probably achieved by targeting IGF1R. The component rutin in DLE is a potential IGF1R agonist.
Table 1.
Molecular docking results of Dendrocalamus latiflorus leaf extract components and IGF1R.
| Site A | Libscore | Site B | Libscore |
|---|---|---|---|
| Rutin | 155.51 | Kaempferol-3-O-rutinoside | 130.66 |
| Kaempferol-3-O-rutinoside | 147.47 | Ginsenoside Rg5 | 125.78 |
| Ginsenoside Rg5 | 144.55 | Vitexin | 115.92 |
| Tricin-7-O-glucopyranoside | 131.35 | Rutin | 115.86 |
| Tricin-4-O-glucopyranoside | 129.36 | Tricin-7-O-glucopyranoside | 113.45 |
| Apigenin 7-O-beta-d-glucoside | 121.51 | Lignan | 111.93 |
| Isovitexin | 120.90 | Luteolin | 104.36 |
| Lignan | 119.95 | 7-Methoxy-tricin | 103.81 |
| Vitexin | 110.49 | Apigenin | 102.37 |
| 7-Methoxy-tricin | 102.11 | Apigenin 7-O-beta-d-glucoside | 99.74 |
| Luteolin | 98.07 | Tricin | 99.43 |
| Tricin | 93.81 | Tricin-4-O-glucopyranoside | 94.90 |
| Apigenin | 91.18 | Isovitexin | 73.46 |
| Loliolide | 64.06 | p-Hydroxybenzaldehyde | 63.39 |
| p-Hydroxybenzaldehyde | 51.62 | Loliolide | 0.00 |
Molecular docking was performed to analyze the docking potential of site A or site B of IGF1R with different components in Dendrocalamus latiflorus leaf extract.
3.6. Rutin was a hypoglycemic component of DLE working through IGF1R
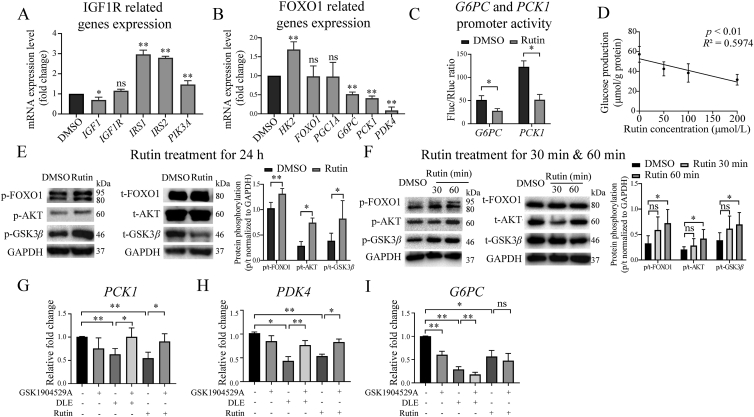
A cell viability assay was performed to detect the cytotoxicity of rutin. HepG2 cells were treated with the indicated concentrations of rutin (12.5–400 μmol/L) for 24 h, and the results indicate that rutin has little cytotoxicity within the tested concentration range (Supporting Information Fig. S7A). Next, RT-qPCR was performed with the same DLE-regulating gene panel to validate whether rutin affects the IGF1R signaling pathway (Fig. 6A) and FOXO signaling pathway (Fig. 6B). The results indicate that rutin and DLE had similar effects on IGF1R and FOXO signaling pathway-related genes, that it up-regulated IRS1, IRS2, PIK3A and HK2, while down-regulated G6PC, PCK1 and PDK4. Importantly, the gluconeogenesis genes G6PC and PCK1 were downregulated by rutin treatment, and we hypothesized that rutin regulated the expression of G6PC and PCK1 at the transcriptional level.
Figure 6.
Rutin was a hypoglycemic component of DLE working through IGF1R. RT-qPCR validation of rutin-mediated (A) IGF1R and (B) FOXO1-related genes in HepG2 cells treated with 50 μmol/L rutin for 24 h (n = 3). (C) The inhibitory effects of rutin on the transcriptional activity of the G6PC and PCK1 promoters (n = 3). (D) Dose responsive inhibitory effects of rutin on glucose production. HepG2 cells were treated with indicated concentrations of rutin for 24 h, followed by glucose production analysis. Data were analyzed using linear regression (n = 3). (E) Western blot showing FOXO1, AKT, and GSK3β expression in HepG2 cells treated with rutin for 24 h (n ≥ 3), (F) 30 or 60 min (n = 5), the data were quantified by ImageJ. (G–I) RT-qPCR results of FOXO1 downstream gene PCK1 (n = 4), G6PC (n = 4), and PDK4 (n = 3) in HepG2 cells. Cells were treated by DLE or rutin with or without IGF1R inhibitor GSK1904529A for 24 h, respectively. All the data are shown as the mean ± SD. Statistical analyses were conducted using paired Student's t-tests. ∗P < 0.05, ∗∗P < 0.01 vs. the DMSO group, ns means not significant. PCK1, phosphoenolpyruvate carboxykinase 1; PDK4, pyruvate dehydrogenase lipoamide kinase isozyme 4; G6PC, glucose-6-phosphatase.
The endogenous promoters of G6PC and PCK1 were cloned into the PGL3 vector for the luciferase reporter assay. After treatment with rutin for 24 h, the promoter activity of G6PC and PCK1 was repressed (Fig. 6C). As expected, the glucose production assay indicated that rutin inhibited glucose production dose-dependently (Fig. 6D).
Next, Western blot was performed to detect the effect of rutin on the phosphorylation of AKT, FOXO1, and GSK3β. The phosphorylation of the three proteins was upregulated (Fig. 6E) by 50 μmol/L rutin for 24 h in HepG2 cells. Based on the hypothesis that rutin activates the AKT signaling pathway through direct binding with IGF1R, we treated HepG2 cells with 50 μmol/L rutin for a relatively short time (30 and 60 min). As shown in Fig. 6F, rutin activated AKT and its downstream targets FOXO1 and GSK3β within 1 h. These results suggest that rutin is a hypoglycemic component of DLE and activates the AKT/FOXO1 signaling pathway probably through binding with IGF1R.
To further verify if DLE and rutin exert their functional activities through IGF1R, the IGF1R inhibitor GSK1904529A was used. Representative genes downstream of FOXO1 were examined, including gluconeogenesis genes PCK1 and G6PC, and the citric acid cycle-related gene PDK4. The IGF1R inhibitor did not affect the expression of IGF1R itself (Fig. S7B). The expression of PCK1 (Fig. 6G) and PDK4 (Fig. 6H) was reduced by DLE and rutin treatment, and this reduction was rescued by GSK1904529A. Unexpected, the reduction of FOXO1 downstream gene G6PC by DLE or rutin was not rescued by GSK1904529A. This is probably because G6PC was inhibited by GSK1904529A alone (Fig. 6I). The reason as why GSK1904529A functions differently on G6PC and PCK1 is not yet clear. Previous studies have shown that although G6PC and PCK1 are regulated by overlapping sets of transcription factors such as FOXO1, they also have unique regulatory elements on their promoters35, which may explain their different changes in response to GSK1904529A. Taken together, the results indicate that DLE and rutin regulated FOXO1 signaling pathway in an IGF1R dependent manner.
4. Discussion
Considering that China is facing the largest diabetes epidemic in the world, it is important to develop novel medicinal plant-based antidiabetic drugs with low toxicity and/or high efficacy. Interestingly, bamboo has gained interest due to its anti-inflammatory and antioxidative properties. According to a recent study, Ma bamboo shoot fiber reduced body weight, lowered blood glucose levels, and improved insulin resistance in high-fat diet-induced diabetic model mice36. In the present study, we found that Ma bamboo leaves also played critical roles in reducing body weight, lowering FBG levels, and improving insulin resistance in gene defect-induced diabetic mice. Importantly, we compared the hypolipidemic potential and hepatic toxicities of DLE with those of the antidiabetic drug PGZ, which is a former popular product for lowering blood glucose but was delisted due to excessive side effects. DLE but not PGZ ameliorated abnormal lipid metabolism by decreasing LDL-c (Fig. 1D). Moreover, compared with the normal group, the DLE group exhibited no hepatorenal toxicity, while the PGZ group did exhibit hepatorenal toxicity. As a natural product with hypoglycemic/hypolipidemic effects and low toxicity, DLE is a potential adjuvant treatment for type 2 diabetes.
Another insightful finding was that DLE achieved a hypoglycemic effect through the AKT/FOXO signaling pathway. As nuclear receptors, the FOXO family is mainly involved in energy regulation. FOXO1, a member of the FOXO family that is primarily expressed in hepatic cells, negatively regulates adipogenesis37 and hepatic glucose production29. Importantly, the downstream FOXO1 proteins G6PC and PCK1 are rate-limiting enzymes in gluconeogenesis. Previous studies have validated that hepatic glucose production is reduced by activating AKT, which leads to the phosphorylation and repression of FOXO1, followed by the transcriptional inhibition of G6PC and PCK138. Our study suggests that DLE reduced hepatic glucose production by inhibiting G6PC and PCK1 expression via AKT-mediated FOXO1 phosphorylation.
Identifying targets of herbal extracts is challenging because herbal substances usually contain multiple chemical components, which affect hundreds or thousands of genes. We used a network pharmacology method and found that IGF1R is an important target that is involved in DLE-mediated AKT activation. As a growth factor receptor, the main function of IGF1R is to bind with IGF1 and mediate cell proliferation, survival, differentiation, and transformation partially through activating AKT39. Interestingly, IGF1R shares high structural homology with insulin receptor (84% similarity)40. Therefore, it interacts with multiple insulin signaling pathway related factors, including IRS1 and IRS2, which are key proteins involved in signal transduction and eventually lead to AKT activation41. Although IGF1R is involved in energy metabolism regulation, there are no antidiabetic drugs on the market that target IGF1R. In our study, we demonstrated that IGF1R is the most likely target of DLE-mediated hypoglycemic effects.
Natural plants are a treasure trove for drug discovery. Metformin, an antidiabetic drug with first-line treatment qualification, and artemisinin, an antimalarial drug whose discovery led to a Nobel Prize, were identified from natural plants. A prior study demonstrated the hypoglycemic effect of Ma bamboo shoot fiber; however, the active compounds remained unclear36. As a well-studied molecule, rutin exhibits pharmacological effects including antioxidant, anticancer, organ protection, antiallergy, and anti-inflammatory effects42,43. However, little is known about its hypoglycemic effects. According to a report by Hsu et al.44 in 2014, rutin showed antidiabetic effects by increasing glucose transport efficiency through AKT mediated glucose transporter activation. Interestingly, in the present study, we found that rutin exerted antidiabetic effects by AKT mediated suppression of hepatic gluconeogenesis. Specifically, rutin inhibited the transcriptional activity of FOXO1 by activating AKT, followed by downregulating G6PC and PCK1 expression, which led to reduced glucose production. Importantly, the mechanism by which rutin activates AKT has not been fully investigated in the previous studies. In the present study, we proposed that rutin binds to IGF1R based on molecular docking which in turn triggers AKT activation. We need to point out that rutin showed antidiabetic effects in vitro at a concentration of 50 μmol/L and we are not sure about the concentration of ruin after DLE was absorbed into blood. Different studies have reported that rutin can be detected in the plasma after oral administration of herbal extracts. However, the blood concentrations of rutin vary a lot among different studies45,46. More work is necessary to evaluate the blood concentrations of rutin after oral administration of DLE at a concentration of 200 mg/kg used in our study. The bioavailability of rutin is low due to its hydrophilic nature. However, the development of delivery systems for rutin or transformation towards highly soluble derivatives by enzymatic or chemical tools will hold the potential for its clinical application47.
In the meanwhile, our studies have some limitations. In the present study, we focused on exploring the hypoglycemic efficacy of DLE in diabetic model mice. C57BL/6J mice can be treated with PGZ and DLE to detect the effects of DLE in normal mice in future work. We noticed that DLE treatment affected food intake and body weight gain in the current study. More work is needed to investigate how DLE regulates food intake and to test if it works on metabolic hormone secretion such as glucagon-like peptide 1.
5. Conclusion
In summary, we validated the antidiabetic effects and uncovered the molecular mechanisms of DLE in vivo and in vitro. Importantly, we found that IGF1R was a potential target of DLE and that rutin was an active component that represented the hypoglycemic functions of DLE. Due to its hypoglycemic effects and low toxicity, DLE may be considered an adjuvant treatment option for patients with T2DM, and clinical studies can be performed in the future.
Acknowledgments
This study was supported by Beijing Talents Foundation (No. 2017000021223ZK30, China), Consulting Research Project of the Chinese Academy of Engineering (No. 2020-XZ-23, China), and Beijing Lab Foundation (China). The authors are grateful to Zhi Zhang, Yujun Chang, Qing Zhou, Ke Zhang and Haohao Deng for useful advices.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.11.017.
Contributor Information
Lan Xie, Email: xielan@tsinghua.edu.cn.
Jing Cheng, Email: jcheng@tsinghua.edu.cn.
Author contributions
Kun Luo, Lan Xie and Jing Cheng designed research. Kun Luo, Wenting Huang, Chengmei Ma, Di Yan and Zhiyu Ning conducted research. Xiaoling Zhang, Liansheng Qiao and Honglei Dang analyzed data. Dong Wang and Hongyan Guo audited the experiment procedures. Kun Luo, Wenting Huang, Lan Xie and Jing Cheng wrote and/or revised the manuscript and had primary responsibility for final content.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cho N.H., Shaw J.E., Karuranga S., Huang Y., da Rocha Fernandes J.D., Ohlrogge A.W., et al. IDF diabetes atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract. 2018;138:271–281. doi: 10.1016/j.diabres.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 2.Xu Y., Wang L., He J., Bi Y., Li M., Wang T., et al. Prevalence and control of diabetes in Chinese adults. JAMA. 2013;310:948–959. doi: 10.1001/jama.2013.168118. [DOI] [PubMed] [Google Scholar]
- 3.Chen L., Magliano D.J., Zimmet P.Z. The worldwide epidemiology of type 2 diabetes mellitus—present and future perspectives. Nat Rev Endocrinol. 2012;8:228–236. doi: 10.1038/nrendo.2011.183. [DOI] [PubMed] [Google Scholar]
- 4.Gastaldelli A., Cusi K., Pettiti M., Hardies J., Miyazaki Y., Berria R., et al. Relationship between hepatic/visceral fat and hepatic insulin resistance in nondiabetic and type 2 diabetic subjects. Gastroenterology. 2007;133:496–506. doi: 10.1053/j.gastro.2007.04.068. [DOI] [PubMed] [Google Scholar]
- 5.Zhou K., Pedersen H.K., Dawed A.Y., Pearson E.R. Pharmacogenomics in diabetes mellitus: insights into drug action and drug discovery. Nat Rev Endocrinol. 2016;12:337. doi: 10.1038/nrendo.2016.51. [DOI] [PubMed] [Google Scholar]
- 6.Philippe J., Raccah D. Treating type 2 diabetes: how safe are current therapeutic agents?. Int J Clin Pract. 2009;63:321–332. doi: 10.1111/j.1742-1241.2008.01980.x. [DOI] [PubMed] [Google Scholar]
- 7.Inzucchi S.E., Bergenstal R.M., Buse J.B., Diamant M., Ferrannini E., Nauck M., et al. Management of hyperglycemia in type 2 diabetes, 2015: a patient-centered approach: update to a position statement of the american diabetes association and the european association for the study of diabetes. Diabetes Care. 2015;38:140–149. doi: 10.2337/dc14-2441. [DOI] [PubMed] [Google Scholar]
- 8.Wright E. Jr, Stonehouse A., Cuddihy R. In support of an early polypharmacy approach to the treatment of type 2 diabetes. Diabetes Obes Metabol. 2010;12:929–940. doi: 10.1111/j.1463-1326.2010.01255.x. [DOI] [PubMed] [Google Scholar]
- 9.Pang B., Yu X.T., Zhou Q., Zhao T.Y., Wang H., Gu C.J., et al. Effect of Rhizoma coptidis (huang lian) on treating diabetes mellitus. Evid Based Complement Alternat Med. 2015;2015:921416. doi: 10.1155/2015/921416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singhal P., Bal L.M., Satya S., Sudhakar P., Naik S.N. Bamboo shoots: a novel source of nutrition and medicine. Crit Rev Food Sci Nutr. 2013;53:517–534. doi: 10.1080/10408398.2010.531488. [DOI] [PubMed] [Google Scholar]
- 11.Panee J. Potential medicinal application and toxicity evaluation of extracts from bamboo plants. J Med Plants Res. 2015;9:681–692. doi: 10.5897/jmpr2014.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Z.M., Li C.L., Peng Z.H. Generation and analysis of expressed sequence tags from a normalized cDNA library of young leaf from Ma bamboo (Dendrocalamus latiflorus Munro) Plant Cell Rep. 2011;30:2045–2057. doi: 10.1007/s00299-011-1112-0. [DOI] [PubMed] [Google Scholar]
- 13.Li H., Zhou H., Wang D., Qiu J., Zhou Y., Li X., et al. Versatile pathway-centric approach based on high-throughput sequencing to anticancer drug discovery. Proc Natl Acad Sci U S A. 2012;109:4609–4614. doi: 10.1073/pnas.1200305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Qiu J., Fu X.D. RASL-seq for massively parallel and quantitative analysis of gene expression. Curr Protoc Mol Biol. 2012;4:4131–4139. doi: 10.1002/0471142727.mb0413s98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pramana S., Lin D., Haldermans P., Shkedy Z., Verbeke T., Göhlmann H., et al. IsoGene: an R package for analyzing dose-response studies in microarray experiments. R J. 2010;2:5–12. [Google Scholar]
- 16.Waltner-Law M.E., Wang X.L., Law B.K., Hall R.K., Nawano M., Granner D.K. Epigallocatechin gallate, a constituent of green tea, represses hepatic glucose production. J Biol Chem. 2002;277:34933–34940. doi: 10.1074/jbc.M204672200. [DOI] [PubMed] [Google Scholar]
- 17.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Shao Y., Qiao L., Wu L., Sun X., Zhu D., Yang G., et al. Structure identification and anti-cancer pharmacological prediction of triterpenes from Ganoderma lucidum. Molecules. 2016;21:678. doi: 10.3390/molecules21050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M., Wang J., Chen J. International conference on bioMedical engineering and informatics; 2008 may 27-30. IEEE; Sanya, China: 2008. A fast agglomerate algorithm for mining functional modules in protein interaction networks; pp. 3–7. [Google Scholar]
- 20.Cho Y.L., Hur S.M., Kim J.Y., Kim J.H., Lee D.K., Choe J., et al. Specific activation of insulin-like growth factor-1 receptor by ginsenoside Rg5 promotes angiogenesis and vasorelaxation. J Biol Chem. 2015;290:467–477. doi: 10.1074/jbc.M114.603142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang H.G., Zheng W.D., Chen Z.D. Component study on flavonoids from leaves of Dendrocalamus latiflorus. Chin Agric Sci Bull. 2005;49:114–118. [Google Scholar]
- 22.Wang S., Yue Y., Tang F., Sun J., Wei Q., Yu J. Chemical constituents from the leaves of Dendrocalamus latiflorus. Sci Silvae Sin. 2013;49:135–140. [Google Scholar]
- 23.Qiao L.S., He Y.S., Huo X.Q., Chen Y.K., Chen X., Zhang Y.L., et al. Construction and evaluation of merged pharmacophore based on peroxisome proliferator receptor-alpha agonists. Chin J Chem Phys. 2016;29:508. [Google Scholar]
- 24.Madiraju A.K., Erion D.M., Rahimi Y., Zhang X.M., Braddock D.T., Albright R.A., et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature. 2014;510:542–546. doi: 10.1038/nature13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haeusler R.A., Camastra S., Astiarraga B., Nannipieri M., Anselmino M., Ferrannini E. Decreased expression of hepatic glucokinase in type 2 diabetes. Mol Metab. 2015;4:222–226. doi: 10.1016/j.molmet.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Considine R.V., Nyce M.R., Allen L.E., Morales L.M., Triester S., Serrano J., et al. Protein kinase C is increased in the liver of humans and rats with non-insulin-dependent diabetes mellitus: an alteration not due to hyperglycemia. J Clin Invest. 1995;95:2938–2944. doi: 10.1172/JCI118001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takamura T., Sakurai M., Ota T., Ando H., Honda M., Kaneko S. Genes for systemic vascular complications are differentially expressed in the livers of type 2 diabetic patients. Diabetologia. 2004;47:638–647. doi: 10.1007/s00125-004-1366-y. [DOI] [PubMed] [Google Scholar]
- 28.Nakae J., Oki M., Cao Y. The FoxO transcription factors and metabolic regulation. FEBS Lett. 2008;582:54–67. doi: 10.1016/j.febslet.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 29.Haeusler R.A., Kaestner K.H., Accili D. FoxOs function synergistically to promote glucose production. J Biol Chem. 2010;285:35245–35248. doi: 10.1074/jbc.C110.175851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao J., Meng S., Chang E., Beckwith Fickas K., Xiong L., Cole R.N., et al. Low concentrations of metformin suppress glucose production in hepatocytes through AMP-activated protein kinase (AMPK) J Biol Chem. 2014;289:20435–20446. doi: 10.1074/jbc.M114.567271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puigserver P., Rhee J., Donovan J., Walkey C.J., Yoon J.C., Oriente F., et al. Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1α interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 32.Lin H.V., Accili D. Hormonal regulation of hepatic glucose production in health and disease. Cell Metabol. 2011;14:9–19. doi: 10.1016/j.cmet.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dupont J., LeRoith D. Insulin and insulin-like growth factor I receptors: similarities and differences in signal transduction. Horm Res. 2001;55:22–26. doi: 10.1159/000063469. [DOI] [PubMed] [Google Scholar]
- 34.Denley A., Cosgrove L.J., Booker W., Wallace J.C., Forbes B.E. Molecular interactions of the IGF system. Cytokine Growth Factor Rev. 2005;16:421–439. doi: 10.1016/j.cytogfr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Wang L.H., Liu Q.M., Kitamoto T., Hou J.J., Qin J., Accili D. Identification of insulin-responsive transcription factors that regulate glucose production by hepatocytes. Diabetes. 2019;68:1156–1167. doi: 10.2337/db18-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X., Fu B., Guo J., Ji K., Xu Y., Dahab M.M., et al. Bamboo shoot fiber improves insulin sensitivity in high-fat diet-fed mice. J Funct Foods. 2018;49:510–517. [Google Scholar]
- 37.Nakae J., Kitamura T., Kitamura Y., Biggs W.H., Arden K.C., Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell. 2003;4:119–129. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- 38.Nakae J., Kitamura T., Silver D.L., Accili D. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J Clin Invest. 2001;108:1359–1367. doi: 10.1172/JCI12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hakuno F., Takahashi S.I. 40 years of IGF1: IGF1 receptor signaling pathways. J Mol Endocrinol. 2018;61:69–86. doi: 10.1530/JME-17-0311. [DOI] [PubMed] [Google Scholar]
- 40.Werner H., Weinstein D., Bentov I. Similarities and differences between insulin and IGF-I: structures, receptors, and signalling pathways. Arch Physiol Biochem. 2008;114:17–22. doi: 10.1080/13813450801900694. [DOI] [PubMed] [Google Scholar]
- 41.Meyts P.D., Whittaker J. Structural biology of insulin and IGF1 receptors: implications for drug design. Nat Rev Drug Discov. 2002;1:769–783. doi: 10.1038/nrd917. [DOI] [PubMed] [Google Scholar]
- 42.Sharma S., Ali A., Ali J., Sahni J.K., Baboota S. Rutin: therapeutic potential and recent advances in drug delivery. Expet Opin Invest Drugs. 2013;22:1063–1079. doi: 10.1517/13543784.2013.805744. [DOI] [PubMed] [Google Scholar]
- 43.Ganeshpurkar A., Saluja A.K. The pharmacological potential of rutin. Saudi Pharmaceut J. 2017;25:149–164. doi: 10.1016/j.jsps.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu C.Y., Shih H.Y., Chia Y.C., Lee C.H., Ashida H., Lai Y.K., et al. Rutin potentiates insulin receptor kinase to enhance insulin-dependent glucose transporter 4 translocation. Mol Nutr Food Res. 2014;58:1168–1176. doi: 10.1002/mnfr.201300691. [DOI] [PubMed] [Google Scholar]
- 45.Nugrahaningsih W.H., Zahroh F., Lisdiana, Ari Y., Ely R. The effect of Cassava leaves extract on pharmacokinetics profile of rutin plasma. Sains Malays. 2019;48:1707–1712. [Google Scholar]
- 46.Smirnova L.A., Suchkov E.A., Kuznetsov K.A., Ryabukha A.F., Perfilova V.N., Popova T.A., et al. Quantitative determination of rutin in biological samples of rats upon administration of Primula veris L. herbal dense extract. Pharm Chem J. 2021;54:1235–1238. [Google Scholar]
- 47.Beatriz G., Thelmo A., Chau L., Moreira M.T., Lema J.M., Gemma E. Rutin: a review on extraction, identification and purification methods, biological activities and approaches to enhance its bioavailability. Trends Food Sci Technol. 2017;67:220–235. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.