Highlights
-
•
mRNA 3′ UTRs facilitate an intricate network of interactions between miRNA-AGO and RBPs.
-
•
The mode of interaction between miRNA-AGO and RBPs on target mRNAs determine the outcome of gene regulation in cancer.
-
•
RBP-RBP interactions and their cumulative binding patterns on mRNA 3′ UTRs influence miRNA-AGO recognition of targets for repression.
-
•
Transcriptome wide approaches exploring AGO-RBP interactions in primary tumors is paramount to have a comprehensive understanding of mRNA metabolism in cancer.
Keywords: miRNAs, Argonaute protein, RBPs, mRNAs, miRISC, Cancer
Abstract
MicroRNAs (miRNAs) and RNA-binding proteins (RBPs) are important regulators of mRNA translation and stability in eukaryotes. While miRNAs can only bind their target mRNAs in association with Argonaute proteins (AGOs), RBPs directly bind their targets either as single entities or in complex with other RBPs to control mRNA metabolism. miRNA binding in 3′ untranslated regions (3′ UTRs) of mRNAs facilitates an intricate network of interactions between miRNA-AGO and RBPs, thus determining the fate of overlapping targets. Here, we review the current knowledge on the interplay between miRNA-AGO and multiple RBPs in different cellular contexts, the rules underlying their synergism and antagonism on target mRNAs, as well as highlight the implications of these regulatory modules in cancer initiation and progression.
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs that repress gene expression by base-pairing with complementary sites, typically found in 3′ untranslated regions (3′ UTRs) of target mRNAs [1]. miRNAs are predicted to target over 50% of all human protein-coding genes post-transcriptionally, enabling their regulatory roles in physiological and pathological processes [2]. Global dysregulation of miRNA expression is a discernible feature in cancer [3], and miRNAs can either promote or impede tumor development depending on the presence of their targets and the cellular context [4]. Alterations in miRNA-mediated gene regulation are implicated in key processes of tumorigenesis, such as apoptosis, angiogenesis, migration, and invasion [5].
MiRNAs are short double-stranded RNA molecules of 21-23 nucleotides (nt) in length. Long hairpin transcripts, called primary miRNAs (pri-miRNAs), are processed by the Drosha-DGCR8 complex in the nucleus yielding precursor miRNAs (pre-miRNAs). The pre-miRNAs are then processed by the Dicer complex in the cytoplasm, producing mature miRNAs [6]. Fully processed mature miRNAs consist of a guide strand and a passenger strand which are loaded onto Argonaute (AGO) proteins, the essential components of miRNA-induced silencing complexes (miRISCs). Among the four AGO proteins (AGO1-4) found in mammals, AGO2 is the most studied and has endonuclease activity. Briefly, mature miRNAs are loaded onto AGO, the passenger strand is released, and the guide strand directs the AGO complex to complementary sites on mRNAs. Once the miRNA-mRNA interaction is initiated, AGO recruits TNRC6 and CCR4-NOT complexes, to facilitate translational repression and/or de-adenylation followed by degradation of targeted mRNAs [7]. However, multiple local and global determinants modulate target binding and silencing efficacy, including target abundance, number and accessibility of miRNA binding sites, and binding of RBPs on the 3′ UTR of targets [8].
RBPs are essential players in mRNA metabolism which regulate splicing, transport, translation, and degradation [9]. Interestingly, about half of the RBPs identified thus far bind mRNAs and manifest their function by regulating the fate of target mRNAs. RBPs together with mRNAs form ribonucleoprotein complexes (mRNPs) and the composition of mRNPs is dynamic and depends on the specifics of mRNA metabolism. Altered expression, localization, and post-translational modification of RBPs contribute to tumorigenesis, by increasing the expression of oncogenes or decreasing the expression of tumor suppressor genes [10,11].
RBPs bind RNAs through RNA binding domains (RBDs) which include the K- homology domain (KH), RNA recognition motif (RRM), Zinc finger domain (ZNF), Pumilio homology domain (PUM), cold shock domain (CSD), double stranded RNA binding domain (dsRBD) and others [10]. They recognize common mRNA features such as the 3′ poly(A) tail and the sequence motifs or secondary structures present in mRNA 3′ UTRs [9]. The 3′ UTRs of mammalian mRNAs can be as long as 10 kilobases and are bound by different miRNAs and RBPs [12]. Genome-wide analysis of mRNAs reveals that nearly half of the mammalian genes express isoforms varying in 3′ UTR length as a result of alternative polyadenylation (APA) [13]. For several genes, mRNA isoforms with shorter 3′ UTRs are expressed at higher levels in transformed cells than in non-transformed cells [14]. Variations in 3′ UTR lengths determine the degree of association of RBPs and the miRISCs with potential consequences for tumorigenesis [10,15,16].
In this review, we will discuss various RBPs pertinent to their involvement in the function of miRNA-AGO complexes on specific target mRNAs and the outcome of gene silencing (Tables 1 and 2, Figs. 1 and 2) with a focus on recent advances and new insights into malignant transformation.
Table 1.
Examples of antagonistic interactions between miRNA-AGO and RBPs
| RBP | miRNA | Type of Cancer/Cells | Target mRNA(s) | Effect | Refs. |
|---|---|---|---|---|---|
| HuR | miR-16 | Colorectal | COX-2 | Promotion of tumorigenesis | [17] |
| miR-331-3p | Prostate | ERRB2 | Resistance to therapy | [18] | |
| miR-122 | Multiple cancer lines | CAT-1 | Control of cellular stress | [19] | |
| miR-548c-3p | Cervical cancer cells | TOP2A | Control of cell cycle | [20] | |
| miR-494 | Cervical cancer cells | Nucleolin | Control of cell proliferation and survival | [21] | |
| miR-200b | Macrophage | VEGF-A | Angiogenesis | [22] | |
| miR-25-3p | Human 293 cells | BTG2, CDK16 | ND | [23] | |
| circAGO2 | Gastric | EIF4EBP3, MAP4K1, SLC2A4 | ND | [24] | |
| Pumilio | miR-30, miR-25, miR-17 | Human 293 cells | FNIP1, TOB1, VLDLR | ND | [25] |
| hnRNPL | miR-297, miR-299 | Myeloid cells | VEGF-A | Hypoxia | [26] |
| hnRNPE2 | miR-328 | Leukemia | CEPBA | Promotion of oncogenic signalling | [27] |
| hnRNPK | miR-149-3p, miR-193b-5p | Cervical cancer cells | PLK1 | Inhibition of apoptosis Drug resistance |
[28] |
| IGF2BP1 | miR-183 | Colorectal | BTRC | Inhibition of apoptosis | [29] |
| miR-340 | Melanoma | MITF | Cell survival and invasion | [30] | |
| Multiple miRNAs | Ovarian | SIRT1 | Promotion of tumor growth and metastasis | [31] | |
| Let-7 | Ovarian | IGF2BP1, HMGA2, LIN28B | Promotion of tumor cell growth, self-renewal and migration | [32] | |
| IGF2BP2 | Let-7 | Glioblastoma | CCND1, PEG10, HMGA1/2, IGF2BP3 | Preservation of cancer stem cells | [33] |
| miR-195 | Colorectal cancer | RAF-1 | Promotion of cell proliferation and survival | [34] | |
| IGF2BP3 | Let-7 | Multiple | HMGA2 | Progression of tumors | [35] |
| Rbm38 | miR-150, miR-206 | Breast cancer | c-Myb, Cx43, p21 | Cellular stress and cell cycle control | [36] |
| FMRP | miR-328 | Neuronal cells | MAZ | ND | [37] |
| DND1 | miR-221, miR-372 | Germ cell tumors | p27, LATS-1 | suppression of tumorigenesis | [38] |
| miR-21 | Squamous cell carcinoma | MSH2 | suppression of tumorigenesis | [39] | |
| FAM120A | ND | Embryonic stem cells | Poly(G) rich | ND | [40] |
| CSDE1 | miR-129-5p | Melanoma cells | PMEPA1 | Promotion of tumorigenesis | [41] |
Table 2.
Examples of co-operative interaction between miRNA-AGO and RBPs
| RBP | miRNA | Type of Cancer/Cells | Target mRNA(s) | Effect | Refs. |
|---|---|---|---|---|---|
| HuR | Let-7a | Cervical cancer cells | c-Myc | Promotion of tumorigenesis | [42] |
| miR-26 | Primary neurons | Rgs4 | ND | [43] | |
| miR-19a/b-3p, miR-130b-3p, miR-17 |
HEK293 | MSMO1 | ND | [23] | |
| Pumilio | miR-221/222 | Glioblastoma | p27Kip1 | Cell cycle control | [44] |
| miR-502, miR-125b |
Bladder | E2F3 | Inhibition of tumorigenesis | [45] | |
| hnRNPI | miR-101 | Prostate and Lung cancer cells | MCL1 | Induction of apoptosis | [46] |
| hnRNPD | Let-7 | Cervical cancer cells and fibroblasts | PDP2, POLR2D |
ND | [47,48] |
| miR-196a | Cervical cancer and Leukemia cells | HOXB8, IFI16 |
ND | [49] | |
| TTP | miR-16 | HEK293, S2 | TNF, COX2 |
ND | [50] |
| IGF2BP3 | miR-9, miR-128 | Pancreatic cancer cells | ZFP36L1, DCBLD2, CLND1 |
Promotion of invasion | [51] |
| Rbm38 | miR-203 | Breast and colorectal cancer cells | p63 | ND | [52] |
| FUS | miR-200c | HEK293 | ZEB1 | ND | [53] |
| CFIm25 | miR-95 and miR-124 | Hepatocellular carcinoma | CCT5, UBA5 |
Inhibition of cell proliferation | [54] |
| CPEB1/2 | miR-580 | Breast cancer | TWIST-1 | Inhibition of EMT | [55] |
| SFPQ | Let-7 | HEK293, RAW 264.7 | LIN28A | ND | [56] |
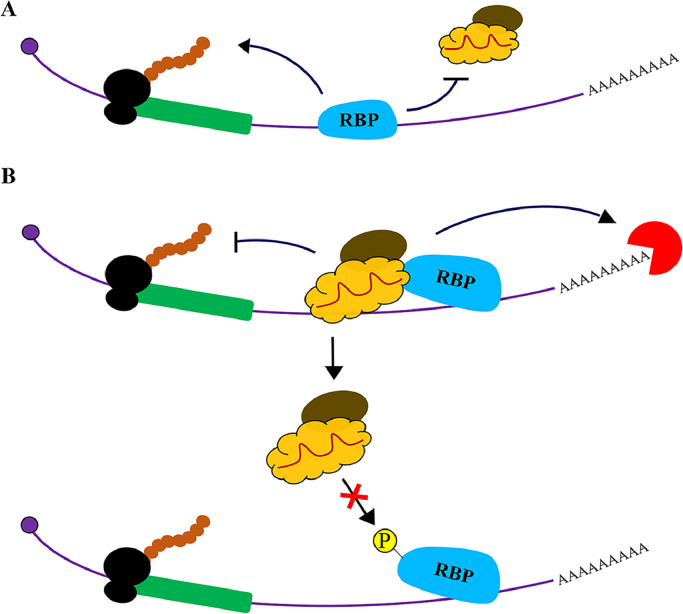
Fig. 1.
Models of antagonistic miRNA-AGO: RBP interactions on target mRNAs. (A) The RBP competes with miRNA-AGO to bind the mRNA 3′ UTR and antagonizes miRNA-guided target repression. (B) The interaction between an RBP and miRNA-AGO is disrupted by the phosphorylation of the RBP, therefore hindering the repression of miRNA-targeted mRNAs. RBPs are presented in Blue, AGO in Yellow, TNRC6 in Brown, Exonucleases in Red, and Ribosomes in Black while the Coding region of the mRNA in Green.
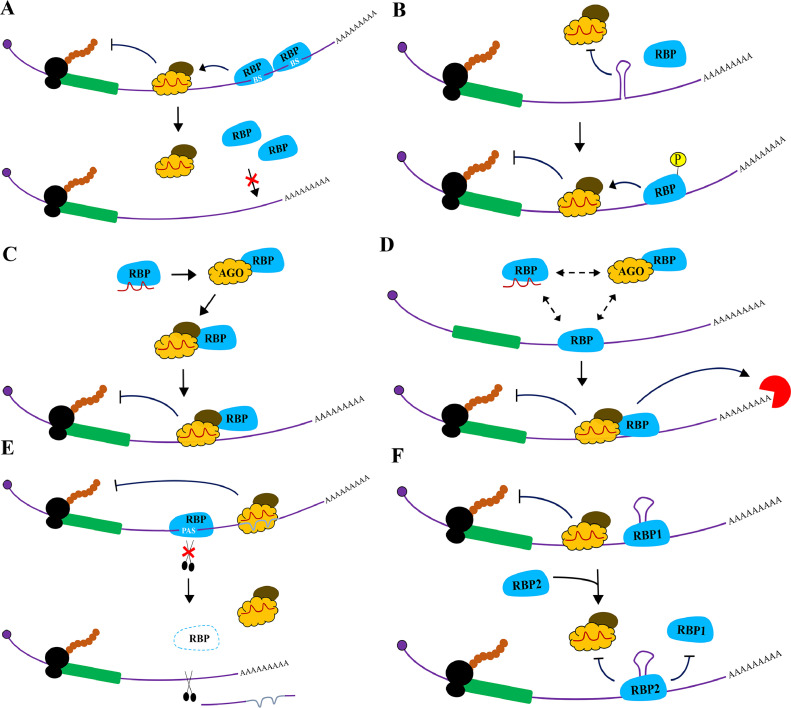
Fig. 2.
Models of co-operative miRNA-AGO: RBP interactions on target mRNAs. (A) The RBP binds multiple sites on the target mRNA and facilitates the interaction between miRNA-AGO and complementary miRNA sequences in the 3′ UTR, thereby cooperating in gene silencing. However, shortening of 3′ UTR results in the loss of RBP binding sites (BS), thus affecting miRNA-AGO association with the target mRNA and its regulation. (B) Structures in the 3′ UTR impede the binding of miRNA-AGO to target sites. Phosphorylated RBPs resolve these secondary structures, thereby allowing the binding of miRNA-AGO to miRNA complementary sites in mRNA 3′ UTRs towards their repression. (C) The RBP assists in the loading of miRNAs into AGO and promotes miRNA-AGO binding to target sites in 3′ UTR for gene silencing. (D) The RBP can simultaneously bind both miRNA and its targets, and its interaction with AGO facilitates the formation of miRISC on respective mRNA 3′ UTRs, thus enabling miRNA-guided decay of targets. (E) The RBP protects Poly(A) Signals (PAS) from cleavage, and loss of its expression leads to deletion of complementary sites for miRNA-AGO binding to the mRNA, resulting in loss of miRNA function. (F) Different RBPs (RBP1 and RBP2) have varied binding affinities towards secondary structures in the 3′ UTR of mRNAs. Depending on their nature of interaction with AGO, they either facilitate the binding of miRNA-AGO to the miRNA complementary sites, leading to target repression or form a stable complex surrounding the secondary structures in 3′ UTR of target mRNAs. Such an arrangement limits the access of miRNA-AGO complex to the target sites embedded in the secondary structures, thus negatively affecting gene silencing. RBPs are presented in Blue, AGO in Yellow, TNRC6 in Brown, Exonucleases in Red, and Ribosomes in Black while the Coding region of the mRNA in Green.
Interplay between miRNA-AGO and RBPs on target mRNAs
miRNAs control the translation and abundance of their target mRNAs even when there is no significant change in miRNA expression levels. Among the potential targets of a specific miRNA, only a subset of mRNAs are subjected to miRNA regulation. These phenomena imply regulatory mechanisms downstream of miRNA biogenesis that influences miRNA activity. An expanding number of studies report the interplay between miRNA-AGO and RBPs on target 3′ UTRs in determining the outcome of miRNA-guided gene regulation.
HuR
Human antigen R (HuR) is a ubiquitously expressed protein that binds AU-rich element (ARE) containing transcripts and controls their stability [57]. Subcellular localization of HuR to the cytoplasm is critical for its role in mRNA metabolism [58]. HuR is overexpressed in numerous cancers, including ovarian, breast, lung, colorectal and pancreatic. Besides its enhanced expression, HuR regulates miRNA-targeted mRNAs involved in cell proliferation, metastasis, invasion, and angiogenesis [59]. For instance, in human liver cells, under stress, HuR binds the cationic amino acid transporter 1 (CAT1) mRNA and inhibits the recruitment of miR-122/AGO complex to target sites in the 3′ UTR, thus relieving CAT1 mRNA repression [19]. Similarly, in colorectal cancer, HuR binds AREs in target mRNAs and hinders miR-16 guided repression of COX2 to promote tumorigenesis of intestinal tumors [17]. In prostate cancer, HuR binds U-rich elements in the 3′ UTR of ERRB-2 and antagonizes miR-331-3p function [18]. Likewise, HuR enhances the translation of DNA topoisomerase 2-alpha (TOP2A) and Nucleolin by competing with miR-548c-3p and miR-494 in cervical cancer lines [[20], [21]]. In macrophages, HuR and miR-200b compete to regulate the expression of Vascular Endothelial Growth Factor-A (VEGF-A) and angiogenesis [22]. Furthermore, HuR interaction with the intronic circular RNA, circAGO2 facilitates HuR binding to target 3′ UTRs of EIF4EBP3, MAP4K1 and SLC2A4 mRNAs associated with gastric cancer progression [24]. In contrast, HuR binding of targets can also promote miRNA-AGO association in let-7-mediated repression of MYC in cervical cancer cells [42]. In neurons, HuR and miR-26/AGO2 synergistically act on Rgs4 mRNA, involved in the regulation of tumorigenic capacities [43]. Additionally, recent transcriptome wide analysis by Li et al., uncovered higher order regulatory modules of interaction between miRNA-AGO and HuR on specific mRNA 3′ UTRs [23]. The study demonstrated that 10% of AGO2 binding sites overlap with 18% of HuR sites, suggesting AGO2 and HuR association has evolved to frequently co-occur towards a combinatorial regulation of mRNAs. Individual characterization of high-fidelity targets revealed that HuR antagonizes miRNA-guided repression of BTG2 and CDK16 but cooperates with miRNA-AGO on MSMO1 3′ UTR [23]. While BTG2 acts as a tumor suppressor [60], the roles of CDK16 and MSMO1 need to be determined in terms of cancer development. Collectively, the interactions between HuR and miRNA-AGO on different mRNAs indicate that the overall contribution of these modules to mRNA control is substantial in malignant transformation.
Pumilio
Pumilio (PUM) proteins function as repressive factors through mRNA destabilization and translational inhibition. They contain PUF RNA-binding domains that recognize the motif, UGUANAUA, located primarily in the 3′ UTR of target mRNAs [61]. Abnormal expression of PUM proteins (PUM1 and PUM2) is associated with several types of cancers such as bladder, breast and ovarian [62]. The effects of PUM proteins on mRNA metabolism are case-specific and involve interaction with additional factors and changes in 3′ UTR secondary structures upon PUM binding [63]. For instance, phosphorylation of PUM facilitates its binding to 3′ UTR of p27Kip1 mRNA, which induces a local conformational rearrangement, making the miR-221/222 complementary sites accessible for AGO binding and thus enabling miRNA-induced repression of p27Kip1 [44]. Interestingly, p27Kip1 is a crucial cell cycle inhibitor and acts as a tumor suppressor in glioblastoma [64]. In addition, PUM shares co-operative interactions with AGO2 on E2F3 mRNA, an oncogene that regulates cell proliferation and apoptosis. PUM1 and PUM2 proteins downregulate E2F3 expression in association with miR-503 and miR-125b in bladder carcinoma [45]. Of note, several cancer cells escape PUM-mediated regulation by shortening the 3′ UTR of E2F3 mRNA, eliminating PUF RNA-binding motifs and thus disabling PUM-mediated repression [25]. Recent transcriptomic studies uncovered that miRNA binding sites are enriched in the vicinity of PUM sites, and co-occurrence of PUM along with miRNA sites in stem loops or sites of low accessibility correlates with repression of the mRNA [25]. Furthermore, Sternburg et al., revealed an overall predominance of co-operative interactions between PUM and AGO2 in altering gene expression [25]. Individual validation for a subset of targets confirmed an antagonistic co-regulation of FNIP1, TOB1 and VLDLR, which shows that Pumilio, a normally repressive factor, can take on a stabilizing role in a context-dependent manner. Interestingly, VLDLR is abnormally expressed in multiple cancers, namely gallbladder, gastric and breast and has critical roles in tumor development [[65], [66], [67]]. TOB1 acts as a tumor suppressor in multiple cancers, and FNIP1 is a binding partner of FLCN, a tumor suppressor in kidney cancer [68]. Despite these relationships, whether AGO2-PUM interactions on these targets contribute to the pathogenesis of each type of cancer remains an open question.
hnRNPs
Heterogeneous nuclear ribonucleoproteins (hnRNPs) are sequence specific regulators of mRNA splicing but also control mRNA stabilization and translation [69]. The altered expression of hnRNPs in multiple cancer types suggest an association with tumorigenesis [70]. hnRNPs are predominantly present in the nucleus during steady state; however, their localization to cell cytoplasm alters the translational output of target mRNAs [[69], [70]]. For instance, during hypoxia, hnRNPL translocates to the cytoplasm, competes with the miRNAs, miR-297 and miR-299 and binds to CA-rich elements (CAREs) in the 3′ UTR of VEGF-A to hinder its repression [26]. Similarly, hnRNPE2 was shown to bind C-rich regions in the 5′ UTR of CCAAT/enhancer binding protein-α (CEPBA) mRNA, a key regulator of myeloid differentiation. Here, the C-rich mature form of miR-328 competes with CEPBA for binding to hnRNPE2 in a RISC-independent manner [27]. Likewise, hnRNPK influences binding of PLK1 mRNA to AGO2 and competes for C-rich motifs on 3′ UTR with miR-149-3p and miR-193b-5p, regulating PLK1 expression and associated apoptosis and drug resistance [28]. In contrast, hnRNPI enhances miR-101-guided AGO2 interaction with MCL1 mRNA, thereby regulating miR-101-induced apoptosis and cell survival [46]. Apart from hnRNP interactions with either the miRNA or the target mRNA, hnRNPD facilitates the binding of let-7b to AGO2 and therefore enhance let-7b/AGO2 interaction with target mRNAs towards let-7b mediated repression of PDP2 and POLR2D [[47], [48]]. In addition, Wu et al. reported that hnRNPD and AGO2 binding is co-operative for HOXB8 and IFI16 mRNAs [49]. Therefore, it is likely that direct hnRNPD-AGO2 association facilitates their co-binding to the mRNA. Alternatively, hnRNPD binding to HOXB8 mRNA may expose the miR-196a binding site, much like Pumilio that exposes miR-221/222 sites in p27 mRNA by altering local RNA structures [44,49]. Altogether, these studies show that hnRNP proteins, through their expression in specific contexts and interactions with miRNA-AGO, alter the metabolic outcomes of target mRNAs in tumors.
Tristetraprolin
Tristetraprolin (TTP) is an ARE-binding protein that mediates mRNA decay and translational repression by interacting with their 3′ UTRs [71]. TTP targets Tumor necrosis factor (TNF), cyclooxygenase 2 (COX2), and MYC, thereby regulating apoptosis and proliferation [[72], [73]]. TTP is downregulated in several tumors such as glioma, colon, gastric and liver cancers, and low TTP mRNA levels correlate with poor prognosis [74]. Several studies reported interactions between TTP, miRISC components, and the mRNA degradation machinery [75]. For example, TTP can induce mRNA decay by decapping, which requires binding to an ARE in the 3′ UTR and interaction with the decapping complex. Additionally, TTP interaction with AGO2 enables miR-16-mediated repression of TNF and COX2 in human cells [50]. Besides, TTP dissociation from AGO2 increases ARE-mRNA stabilization [76]. These data suggest multiple and yet poorly understood mechanisms, where TTP and miRNAs promote mRNA degradation with implications in pathological phenomena.
IGF2BPs
The oncofetal IGF2 mRNA binding proteins (IGF2BPs) control mRNA transport, translation, and turnover during development and in cancer [77]. IGF2BP isoforms 1 and 3 enhance the viability, migration, invasion, and metastatic potential of tumors in vitro and in vivo [[31], [51]]. IGF2BP1 controls the degradation of BTRC mRNA, which encodes the ubiquitin ligase βTrCP1, by disrupting the association of miR-183 and AGO2 with a target site in the BTRC coding region [29]. IGF2BP1 also targets the 3′ UTR of a microphthalmia-induced transcription factor (MITF) isoform that is predominantly expressed in melanoma and prevents miR-340-mediated repression [30]. In addition, IGF2BP1 recruits mRNAs encoding IGF2BP1, HMGA2 and LIN28B into mRNPs devoid of let-7/AGO2, thereby protecting them from miRNA-guided silencing. It also interferes with miRNA-mediated decay of SIRT1 mRNA in ovarian cancer cells to promote tumor cell growth, self-renewal, and migration [31,32]. Likewise, in glioblastoma, IGF2BP2 interacts with AGO2 in an RNA-dependent fashion and binds to let-7 target sites. Such activity impairs miRNA-guided repression of targets, namely CCND1, PEG10, HMGA1/2 and IGF2BP3; responsible for glioblastoma stem cell maintenance [33]. In addition, IGF2BP2 promotes cell proliferation and survival in colorectal cancer by interfering with miR-195 driven RAF-1 degradation [34]. Similarly, IGF2BP3 protects and upregulates HMGA2 expression by opposing the interaction between let-7/AGO2 and HMGA2 mRNA [35]. In contrast, IGF2BP3 promotes the association of AGO2 with invasion-associated transcripts, namely ZFP36L1, DCBLD2, and CLDN1 in pancreatic ductal adenocarcinoma [51]. Furthermore, Muller S et al., 2018 discovered that IGF2BP binding sites are enriched ∼40 nucleotides upstream of miRNA target sites which overlap with AGO2-binding, suggesting broader regulation [31]. Collectively, these reports demonstrate that all three IGF2BPs promote tumorigenesis by modulating the miRNA-AGO directed degradation of oncogene-encoding mRNAs in cancer cells.
Rbm38
The RNA-binding protein Rbm38 is a target of the p53 family. Rbm38 regulates gene expression underlying several cellular processes, including cell growth and differentiation [78]. Rbm38 regulates the stability of its targets depending on the nucleotide composition of binding sites. For instance, it reduces p63 α/β mRNA levels by binding to AU/U- rich elements but enhances p63γ mRNA levels by association with GU-rich elements [[78], [79]]. In breast cancer, following DNA damage, p53 induces the expression of Rbm38 which in turn limits the accessibility of target sites of the miRNAs, miR-150 and miR-206 on 3′ UTRs of c-MYB, CX43, and p21 mRNA to regulate cellular stress and cell cycle [36]. Further, Zhang Y et al., 2019 showed that Rbm38 binds U-rich elements in the 3′ UTR of p63 and recruits miRNA-AGO complex by interacting with AGO2. However, the Ser-195 phosphorylation of Rbm38 abrogates its association with AGO2, thereby hindering the binding of miR-203/AGO2 complex to p63α mRNA for degradation. [52]. Of note, p63 over-expression is associated with malignant conditions, such as squamous carcinomas of the head, neck, and skin [[80], [81]]. Since tumors with mutated p53 exhibit changes in Rbm38 expression, it is likely that the controlling action of Rbm38 over miRNA-AGO function on target mRNAs contributes to p53 activity in tumor progression.
FUS
FUS is a DNA/RNA-binding protein with a conserved RNA recognition motif (RRM) [82]. FUS is involved in multiple cellular processes, including transcription, pre-mRNA splicing, and APA [83]. FUS expression is inversely correlated with prostate tumor grade, and patients with high levels of FUS have longer survival rates and are less likely to have bone metastases [84]. Recently, Zhang T et al., 2018 showed that FUS interacts with AGO2 in an RNA-dependent manner [53]. FUS binds to miR-200c and is required for the optimal association between miR-200c and AGO2. The impairment of FUS's interaction with the 3′ UTR of target mRNA ZEB1 is sufficient to reduce miRNA silencing. Interestingly, ZEB1 expression is associated with many aggressive tumors, including breast, lung, colorectal, liver and glioblastoma [[85], [86]]. ZEB1 plays a crucial role in tumor dissemination, metastasis, and therapy resistance [87]. These reports add to the growing number of studies demonstrating the involvement of RBPs in a three-way interaction with AGO, miRNA and the mRNA [88].
FMRP
The Fragile X Mental Retardation Protein (FMRP) is an RNA binding protein that binds ∼4% of mRNAs in the brain and affects their translation [89]. FMRP binds its target mRNAs in the coding region as well as in the 3′ UTR, depending on cellular identity [90]. FMRP contains two RNA binding domains, protein binding K-homology domains KH1 and KH2 [91], as well as an arginine-glycine-glycine (RGG) box that binds G-Quadruplex RNA structures (rG4s) in vitro [92]. FMRP exhibits a bifunctional role in translational regulation of bound mRNAs through its interaction with the RNA helicase MOV10 [93]. Both FMRP and MOV10 directly interact with AGO and rG4s in mRNA 3′ UTRs. MOV10 binding to FMRP increases the ability of FMRP to bind rG4s in vitro, forming a stable FMRP-MOV10 complex. This co-operation between FMRP and MOV10 inhibits AGO binding to miR-328 target sites embedded in rG4 on Myc-associated zinc finger protein (MAZ) mRNA, preventing its translational repression [37]. MAZ is abnormally expressed in multiple tumors including breast, prostate, and liver. It promotes cancer cell proliferation, migration, and invasion [[94], [95]]. However, considering the ambiguity over the occurrence of rGs in the in vivo context of mRNAs, it is yet to be determined how FMRP interaction with MOV10 pertinent to rG4 resolution and the miRNA-AGO function contributes to cancer development.
DND1
Dead end 1 (DND1) is expressed in germ cells and binds uridine-rich regions (URRs) in the 3′ UTRs of germline-specific genes. DND1 either sequesters mRNAs or physically displaces miRISC to alleviate miRNA-mediated suppression [38]. DND1 alleviates miR-372-mediated repression of large tumor suppressor 2 (LATS2) and miR-221 and miR-222-mediated repression of p27 by competing with these miRNAs to bind their targets in germ cell tumors [38]. Similarly, in squamous cell carcinoma, DND1 impairs miR-21 action on its target MSH2, thus suppressing tumorigenesis in skin [96]. In contrast, DND1 recruits the CCR4-NOT deadenylase complex to destabilize and repress RNAs associated with apoptosis and inflammation [[39], [97]]. Together, these studies indicate that DND1 forms a network of post-transcriptional regulation for the maintenance of stemness and these mechanisms could be relevant in tumors with acquired multipotency.
FAM120A
FAM120A is a putative RBP and a component of Purα-containing mRNP complexes through direct binding of IGF-II mRNA [98]. It acts as a scaffold for activation of several intracellular kinases, including Src, FAK, and PI3K, in response to oxidative stress, and IL13 receptor α2 signalling in cancer cells [[99], [100]]. Recently, FAM120A was shown to interact with AGO2 in mESCs. FAM120A binds homopolymeric tracts in 3′ UTRs of mRNAs with poly(G) sequences, and its targets overlap with more than one third of mRNAs bound by AGO2 [40]. Further, FAM120A-bound targets are less sensitive to AGO2-mediated target degradation, suggesting FAM120A attenuates AGO2-mediated gene silencing. While the specific mechanism by which FAM120A binding counteracts miRNA-guided mRNA degradation is unclear, it is likely independent of miRNA-AGO2 binding to its targets, as the binding sites of FAM120A and AGO2 do not directly overlap. Also, in light of FAM120A association with other RBPs, including IGF2BP1, IGF2BP3 and HuR, which cooperate and compete with miRNA-AGO to bind target mRNAs, it is possible that the regulatory effects of FAM120A can only be observed in a combinatorial module with the other regulatory RBPs.
CSDE1
Cold Shock Domain containing protein E1 (CSDE1), also known as UNR (upstream of N-Ras), is a member of the evolutionarily conserved CSD containing protein family [101]. CSDE1 plays an important role in a wide range of biological processes such as cell cycle, apoptosis, and differentiation [[102], [103]]. CSDE1 acts as an oncogene in melanoma, glioma, colorectal and breast cancers while performing tumor suppressor functions in pheochromocytoma, paraganglioma, oral squamous cell carcinoma and in keratinocytes [[104], [105]]. Recently, we reported that CSDE1 interacts with AGO2 within miRISC and particularly, in melanoma cells, this association is facilitated by target mRNAs [[41], [106]]. CSDE1 and AGO2 share a total of 93 targets, including Prostate Transmembrane Protein, Androgen Induced 1 (PMEPA1), a protein that enhances tumorigenic properties in breast, lung cancers and glioblastoma whilst displaying anti-tumoral roles in prostate cancer [[107], [108], [109]]. CSDE1 competes with miR-129-5p/AGO2 to bind PMEPA1 mRNA and promote its expression at protein levels. Further, the loss of CSDE1 N-terminal cold shock domain (CSD1), which is required for CSDE1 RNA-binding, enhances AGO2 association with PMEPA1 mRNA leading to its repression [41]. This suggests that target recognition and occupancy by CSDE1 determines the degree of recruitment of miRNA-AGO2 complex to specific mRNAs involved in the development of tumorigenic phenotypes.
NUDT21/CFIm25
CFIm25 is a subunit of the heterodimer CFIm, a major factor that governs APA [110]. CFIm25 is necessary and sufficient to bind sequences specific for the poly(A)-site upstream component UGUA [111]. CFIm25 can function as a tumor suppressor and an oncogene in glioblastoma and chronic myelocytic leukemia, respectively [110,112]. CFIm25 interacts with AGO2 and co-localizes to discrete cytoplasmic loci called P-bodies in liver tumors [113]. Functional studies showed that CFIm25 promoted APA at distal sites, resulting in elongated mRNA 3′ UTRs, which enhanced AGO2 binding to the targets of miR-95 and miR-124, notably CCT5 and UBA5 [54]. In addition, the loss of CFIm25 shortened the 3′ UTRs of oncogenes in Hepatocellular Carcinoma (HCC) cells. The shorter 3′ UTRs contained fewer miRNA binding sites, enabling the oncogenes to evade miRNA regulation, and become overexpressed in HCC, leading to unregulated cancer cell proliferation.
CPEB
The Cytoplasmic Polyadenylation Element Binding protein (CPEB) family includes 4 members (CPEB1-4), which regulate translation of their target mRNAs by binding to Cytoplasmic Polyadenylation Elements (CPE) in the 3′ UTR [114]. CPEB proteins can repress or activate translation of target mRNAs by shortening or elongating the poly-A tail. CPEB1 acts as a tumor suppressor in mammary epithelial cells and controls Epithelial-to-Mesenchymal Transition (EMT) and metastatic phenotypes [115]. Similarly, CPEB2 acts as a tumor-suppressor by binding to Hypoxia Inducible Factor 1 Subunit Alpha (HIF1α) mRNA and suppressing its translation under normoxic conditions but releasing it to allow translation under hypoxic conditions [116]. HIF1α, under hypoxic conditions, stimulates genes promoting angiogenesis, migration, metastasis, and therapeutic resistance [117]. Twist-related protein 1 (TWIST1) is a known HIF1α target and is highly relevant in EMT and metastasis formation in head and neck squamous cell carcinoma and non-small cell lung cancer [[55], [117]]. Nairismagi ML et al., 2012 identified miR-580, CPEB1 and CPEB2 as negative regulators of TWIST1 in an in vitro model of breast cancer and demonstrated co-operative effects between the CPEB and miR-580 sites [55]. Also, CPEB2 is unable to bind the shorter form of TWIST1 mRNA, preferentially expressed in metastatic cells which correlates with high TWIST1 expression, indicating CPEB-miR-580 axis regulates TWIST1 during tumor pathogenesis. These findings add to the growing number of studies demonstrating that cancer cells use 3′ UTR shortening as a strategy to control the interplay between the miRNA-AGO and RBPs on mRNAs with oncogenic or tumor suppressor functions.
SFPQ
Splicing factor proline- and glutamine-rich protein (SFPQ) is a ubiquitous and abundant nuclear RBP involved in splicing, RNA transport, and apoptosis [118], [119], [120]. SFPQ is involved in multiple cancer types, including renal cell carcinoma, colorectal, liver, lung, and breast cancers [121]. Recently, Bottini S et al., 2017. showed that SFPQ interacts with AGO2 in an RNA-dependent manner [56]. SFPQ associates to nucleoplasmic AGO2/miRISC through its interacting partners, Paraspeckle component 1 (PSPC1) and Non-POU domain-containing octamer-binding protein (NONO). SFPQ preferentially binds long 3′ UTRs that harbour multiple copies of SFPQ-binding motifs, CUGU and UGUA and regulate the accessibility of miRNA-target sites. Here, SFPQ forms long aggregates on target 3′ UTRs in the nucleus to modulate their folding for proper positioning/recruitment of miRNA-AGOs to select binding sites, namely on LIN28A 3′UTR. These results highlight the importance of nuclear miRNA targeting and the sequence features of mRNA 3′ UTRs for subcellular allocation of post-transcriptional miRNA regulation. However, it is yet to be determined how PSPC1, NONO are involved in these mechanisms and the overall contribution of SFPQ-PSCP1-NONO complex to nuclear miRNA-AGO function in tumor pathogenesis.
Summary
miRNA function is often deregulated in cancer, and these variations are rarely caused by miRNA gene amplification or disruption. RBPs and their interacting partners mostly determine the changes observed in miRNA activity associated with tumorigenesis. The fact that 3′ UTRs of mRNAs frequently contain multiple evolutionary conserved binding sites for both miRNAs and RBPs suggests that the interplay between RBPs and miRNAs is a crucial component of gene regulation. Although the list of miRNA-AGO: RBP interactions presented here keeps growing (Table 1–2), our comprehension of this functional relationship is relatively recent and still in its nascent stage.
Studies thus far provided us a glimpse of intricate associations between miRNA-AGO and RBPs on target mRNAs. However, it is yet to be determined, what's the net effect of miRNA-AGO: RBP interactions in the development of tumors and whether miRNA-AGO: RBP pairs are specific to the cancer type or share a pattern of target repression between different cancers and their cellular phenotypes. Since the earlier studies strongly suggest that the effects of interaction are dependent on the nature of target mRNAs, whether oncogenic or tumor suppressor, it would be intriguing for further studies to focus on the repertoire of target mRNAs bound by the miRISC in the absence or the over expression of each RBP and evaluate how the changes in mRNA binding by both miRNA-AGO and RBP manifest into tumorigenic capacities of cancer cells in vivo.
Challenges
For the past few years, considerable advances have been made in dissecting miRNA-AGO: RBP interactions underlying the tumorigenic properties of different cancers. However, there remain challenges in defining the contribution of these associations to the development of tumors. For example, an RBP can have context dependent opposing effects on miRNA-AGO function in the repression of specific mRNAs. Also, a single miRNA can target multiple mRNAs, and RBP association with a particular miRISC varies depending on the extent of its binding to the repertoire of target mRNAs with oncogenic and/or tumor suppressor functions. Therefore, to determine the accuracy/mode of influence exerted by an RBP on miRNA-AGO function, at first, we need to know the number of miRNA binding sites along with their position relative to RBP binding and the contribution of each site to the silencing of target mRNAs under endogenous conditions. To acquire such information, we could make use of improved high throughput techniques such as iCLiP, which produces miRNA-target chimeras. These hybrid sequences will help identify and confer specificity to miRNA-target interactions in vivo and establish the link between a given miRNA and its target mRNA [122]. Additionally, another daunting task now faced by the scientific community studying AGO-RBP contribution to cancer is the comprehension of non-canonical interactions of AGO proteins in gene regulation. For instance, in quiescent cells, AGO2 promotes translation of specific ARE mRNAs in association with Fragile-X-Mental-Retardation syndrome Related protein 1a (FXR1a), p97 and PARN [[123], [124]]. In Pancreatic Ductal Adenocarcinoma (PDAC) cells, AGO2 interacts with oncogenic KRAS, which affects the unwinding of miRNA duplexes and thus the binding of mature miRNA-AGO complex to target mRNAs in order to promote KRAS-mediated oncogenesis [[125], [126]]. These complex layers of bi-directional regulation associated with AGO-RBPs often make it impossible to neatly classify their involvement in the control of gene expression into oncogenic or tumor-suppressive.
Outlook
Despite the challenges surrounding the context dependent function of miRNA-AGO and RBPs, studies over the last few years measuring RNA-RNA and RNA-protein interactions, as well as mRNA secondary structures, enabled us to connect networks of post-transcriptional regulation involving miRNAs, RBPs and decipher their relevance for cancer initiation and progression [[9], [10],122] (Fig. 1, Fig. 2). And yet, the data generated so far is mostly from cell lines such as HEK or HeLa, of which may not reflect the real time scenarios that occur during the stages of malignant transformation in vivo. Thus, we need to revisit the already known -as well as explore new AGO-RBP interactions on specific mRNAs using transcriptome wide approaches in primary tumor cells along the stages of cancer progression to have a comprehensive understanding of their regulation. Alternatively, we could make use of animal cancer models that are physiologically relevant to gain the insights into AGO-RBP interactions on miRNA-targeted mRNAs with pathological consequences. Furthermore, in light of studies emerging to support the notion that multiple RBPs coordinately act on the miRNA-AGO complex, we could explore RBP-RBP interactions and their individual and cumulative binding patterns to define their combinatorial effects over miRNA-AGO recognition of targets towards gene silencing. We believe that a thorough knowledge of the interplay among RBPs, miRNAs and other mRNA-interacting molecules is paramount to understanding the complexity of post-transcriptional gene control surrounding aberrant mRNA metabolism in cancers. This exciting playground of RBPs and miRNA-AGOs still holds secrets that, when uncovered, will reveal networks with potential therapeutic benefits.
Funding
This work was supported by the Dean of Science Startup funds from Memorial University of Newfoundland (MUN), St. John's, NL, Canada.
CRediT authorship contribution statement
Pavan Kumar Kakumani: Conceptualization, Investigation, Visualization, Methodology, Data curation, Formal analysis, Validation, Resources, Supervision, Funding acquisition, Project administration, Writing – original draft.
Conflict of Interest
The author(s) declare that no conflict of interests exists.
Acknowledgements
I sincerely thank Dr. Martin Simard and Dr. Fatima Gebauer for their helpful comments and suggestions. I am thankful to Grace Christopher for assistance with compilation and formatting of references.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101434.
Appendix. Supplementary materials
References
- 1.Bartel D.P. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Downing J.R., Jacks T., Horvitz H.R., Golub T.R. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 4.Farazi T.A., Spitzer J.I., Morozov P., Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olson P., Lu J., Zhang H., Shai A., Chun M.G., Wang Y., Libutti S.K., Nakakura E.K., Golub T.R., Hanahan D. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Treiber T., Treiber N., Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 7.Gebert L.F.R., MacRae I.J. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quévillon Huberdeau M., Simard M.J. A guide to microRNA-mediated gene silencing. FEBS J. 2019;286:642–652. doi: 10.1111/febs.14666. [DOI] [PubMed] [Google Scholar]
- 9.Hentze M.W., Castello A., Schwarzl T., Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 10.Pereira B., Billaud M., Almeida R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer. 2017;3:506–528. doi: 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Neelamraju Y., Gonzalez-Perez A., Bhat-Nakshatri P., Nakshatri H., Janga S.C. Mutational landscape of RNA-binding proteins in human cancers. RNA Biol. 2018;15:115–129. doi: 10.1080/15476286.2017.1391436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang P., Coller H. Functional interactions between microRNAs and RNA binding proteins. Microrna. 2012;1:70–79. doi: 10.2174/2211536611201010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H., Lee J.Y., Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayr C., Bartel D.P. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam J.W., Rissland O.S., Koppstein D., Abreu-Goodger C., Jan C.H., Agarwal V., Yildirim M.A., Rodriguez A., Bartel D.P. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol Cell. 2014;53:1031–1043. doi: 10.1016/j.molcel.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Kouwenhove M., Kedde M., Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 17.Young L.E., Moore A.E., Sokol L., Meisner-Kober N., Dixon D.A. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol Cancer Res. 2012;10:167–180. doi: 10.1158/1541-7786.MCR-11-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epis M.R., Barker A., Giles K.M., Beveridge D.J., Leedman P.J. The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331-3p in prostate cancer cells. J Biol Chem. 2011;286:41442–41454. doi: 10.1074/jbc.M111.301481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 20.Srikantan S., Abdelmohsen K., Lee E.K., Tominaga K., Subaran S.S., Kuwano Y., Kulshrestha R., Panchakshari R., Kim H.H., Yang X., Martindale J.L., Marasa B.S., Kim M.M., Wersto R.P., Indig F.E., Chowdhury D., Gorospe M. Translational control of TOP2A influences doxorubicin efficacy. Mol Cell Biol. 2011;31:3790–3801. doi: 10.1128/MCB.05639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tominaga K., Srikantan S., Lee E.K., Subaran S.S., Martindale J.L., Abdelmohsen K., Gorospe M. Competitive regulation of nucleolin expression by HuR and miR-494. Mol Cell Biol. 2011;31:4219–4231. doi: 10.1128/MCB.05955-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang S.H., Lu Y.C., Li X., Hsieh W.Y., Xiong Y., Ghosh M., Evans T., Elemento O., Hla T. Antagonistic function of the RNA-binding protein HuR and miR-200b in post-transcriptional regulation of vascular endothelial growth factor-A expression and angiogenesis. J Biol Chem. 2013;288:4908–4921. doi: 10.1074/jbc.M112.423871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Estep J.A., Karginov F.V. Transcriptome-wide Identification and Validation of Interactions between the miRNA Machinery and HuR on mRNA Targets. J Mol Biol. 2018;430:285–296. doi: 10.1016/j.jmb.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y., Yang F., Fang E., Xiao W., Mei H., Li H., Li D., Song H., Wang J., Hong M., Wang X., Huang K., Zheng L., Tong Q. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sternburg E.L., Estep J.A., Nguyen D.K., Li Y., Karginov F.V. Antagonistic and cooperative AGO2-PUM interactions in regulating mRNAs. Sci Rep. 2018;8:15316. doi: 10.1038/s41598-018-33596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jafarifar F., Yao P., Eswarappa S.M., Fox P.L. Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J. 2011;30:1324–1334. doi: 10.1038/emboj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eiring A.M., Harb J.G., Neviani P., Garton C., Oaks J.J., Spizzo R., Liu S., Schwind S., Santhanam R., Hickey C.J., Becker H., Chandler J.C., Andino R., Cortes J., Hokland P., Huettner C.S., Bhatia R., Roy D.C., Liebhaber S.A., Caligiuri M.A., Marcucci G., Garzon R., Croce C.M., Calin G.A., Perrotti D. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin C.H., Lee H., Kim H.R., Choi K.H., Joung J.G., Kim H.H. Regulation of PLK1 through competition between hnRNPK, miR-149-3p and miR-193b-5p. Cell Death Differ. 2017;24:1861–1871. doi: 10.1038/cdd.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elcheva I., Goswami S., Noubissi F.K., Spiegelman V.S. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol Cell. 2009;35:240–246. doi: 10.1016/j.molcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goswami S., Tarapore R.S., Teslaa J.J., Grinblat Y., Setaluri V., Spiegelman V.S. MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor mRNA is inhibited by the coding region determinant-binding protein. J Biol Chem. 2010;285:20532–20540. doi: 10.1074/jbc.M110.109298. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Müller S., Bley N., Glaß M., Busch B., Rousseau V., Misiak D., Fuchs T., Lederer M., Hüttelmaier S. IGF2BP1 enhances an aggressive tumor cell phenotype by impairing miRNA-directed downregulation of oncogenic factors. Nucleic Acids Res. 2018;46:6285–6303. doi: 10.1093/nar/gky229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Busch B., Bley N., Müller S., Glaß M., Misiak D., Lederer M., Vetter M., Strauß H.G., Thomssen C., Hüttelmaier S. The oncogenic triangle of HMGA2, LIN28B and IGF2BP1 antagonizes tumor-suppressive actions of the let-7 family. Nucleic Acids Res. 2016;44:3845–3864. doi: 10.1093/nar/gkw099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Degrauwe N., Schlumpf T.B., Janiszewska M., Martin P., Cauderay A., Provero P., Riggi N., Suvà M.L., Paro R., Stamenkovic I. The RNA Binding Protein IMP2 Preserves Glioblastoma Stem Cells by Preventing let-7 Target Gene Silencing. Cell Rep. 2016;15:1634–1647. doi: 10.1016/j.celrep.2016.04.086. [DOI] [PubMed] [Google Scholar]
- 34.Ye S., Song W., Xu X., Zhao X., Yang L. IGF2BP2 promotes colorectal cancer cell proliferation and survival through interfering with RAF-1 degradation by miR-195. FEBS Lett. 2016;590:1641–1650. doi: 10.1002/1873-3468.12205. [DOI] [PubMed] [Google Scholar]
- 35.Jønson L., Christiansen J., Hansen T.V.O., Vikeså J., Yamamoto Y., Nielsen F.C. IMP3 RNP safe houses prevent miRNA-directed HMGA2 mRNA decay in cancer and development. Cell Rep. 2014;7:539–551. doi: 10.1016/j.celrep.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Léveillé N., Elkon R., Davalos V., Manoharan V., Hollingworth D., Oude Vrielink J., le Sage C., Melo C.A., Horlings H.M., Wesseling J., Ule J., Esteller M., Ramos A., Agami R. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat Commun. 2011;2:513. doi: 10.1038/ncomms1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenny P.J., Kim M., Skariah G., Nielsen J., Lannom M.C., Ceman S. The FMRP-MOV10 complex: a translational regulatory switch modulated by G-Quadruplexes. Nucleic Acids Res. 2020;48:862–878. doi: 10.1093/nar/gkz1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kedde M., Strasser M.J., Boldajipour B., Oude Vrielink J.A., Slanchev K., le Sage C., Nagel R., Voorhoeve P.M., van Duijse J., Ørom U.A., Lund A.H., Perrakis A., Raz E., Agami R. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 39.Yamaji M., Jishage M., Meyer C., Suryawanshi H., Der E., Garzia A., Morozov P., Manickavel S., McFarland H.L., Roeder R.G., Hafner M., Tuschl T. DND1 maintains germline stem cells via recruitment of the CCR4-NOT complex to target mRNAs. Nature. 2017;543:568–572. doi: 10.1038/nature21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelly T.J., Suzuki H.I., Zamudio J.R., Suzuki M., Sharp P.A. Sequestration of microRNA-mediated target repression by the Ago2-associated RNA-binding protein FAM120A. RNA. 2019;25:1291–1297. doi: 10.1261/rna.071621.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kakumani P.K., Guitart T., Houle F., Harvey L.M., Goyer B., Germain L., Gebauer F., Simard M.J. CSDE1 attenuates microRNA-mediated silencing of PMEPA1 in melanoma. Oncogene. 2021;40:3231–3244. doi: 10.1038/s41388-021-01767-9. [DOI] [PubMed] [Google Scholar]
- 42.Kim H.H., Kuwano Y., Srikantan S., Lee E.K., Martindale J.L., Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ehses J., Fernández-Moya S.M., Schröger L., Kiebler M.A. Synergistic regulation of Rgs4 mRNA by HuR and miR-26/RISC in neurons. RNA Biol. 2021;18:988–998. doi: 10.1080/15476286.2020.1795409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kedde M., van Kouwenhove M., Zwart W., Oude Vrielink J.A., Elkon R., Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 45.Miles W.O., Tschöp K., Herr A., Ji J.Y., Dyson N.J. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012;26:356–368. doi: 10.1101/gad.182568.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui J., Placzek W.J. PTBP1 enhances miR-101-guided AGO2 targeting to MCL1 and promotes miR-101-induced apoptosis. Cell Death Dis. 2018;9:552. doi: 10.1038/s41419-018-0551-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon J.H., Jo M.H., White E.J., De S., Hafner M., Zucconi B.E., Abdelmohsen K., Martindale J.L., Yang X., Wood W.H., Shin Y.M., Song J.J., Tuschl T., Becker K.G., Wilson G.M., Hohng S., Gorospe M. AUF1 promotes let-7b loading on Argonaute 2. Genes Dev. 2015;29:1599–1604. doi: 10.1101/gad.263749.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min K.W., Jo M.H., Shin S., Davila S., Zealy R.W., Kang S.I., Lloyd L.T., Hohng S., Yoon J.H. AUF1 facilitates microRNA-mediated gene silencing. Nucleic Acids Res. 2017;45:6064–6073. doi: 10.1093/nar/gkx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu X., Chesoni S., Rondeau G., Tempesta C., Patel R., Charles S., Daginawala N., Zucconi B.E., Kishor A., Xu G., Shi Y., Li M.L., Irizarry-Barreto P., Welsh J., Wilson G.M., Brewer G. Combinatorial mRNA binding by AUF1 and Argonaute 2 controls decay of selected target mRNAs. Nucleic Acids Res. 2013;41:2644–2658. doi: 10.1093/nar/gks1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 51.Ennajdaoui H., Howard J.M., Sterne-Weiler T., Jahanbani F., Coyne D.J., Uren P.J., Dargyte M., Katzman S., Draper J.M., Wallace A., Cazarez O., Burns S.C., Qiao M., Hinck L., Smith A.D., Toloue M.M., Blencowe B.J., Penalva L.O., Sanford J.R. IGF2BP3 Modulates the Interaction of Invasion-Associated Transcripts with RISC. Cell Rep. 2016;15:1876–1883. doi: 10.1016/j.celrep.2016.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y., Feng X., Sun W., Zhang J., Chen X. Serine 195 phosphorylation in the RNA-binding protein Rbm38 increases p63 expression by modulating Rbm38′s interaction with the Ago2-miR203 complex. J Biol Chem. 2019;294:2449–2459. doi: 10.1074/jbc.RA118.005779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang T., Wu Y.C., Mullane P., Ji Y.J., Liu H., He L., Arora A., Hwang H.Y., Alessi A.F., Niaki A.G., Periz G., Guo L., Wang H., Elkayam E., Joshua-Tor L., Myong S., Kim J.K., Shorter J., Ong S.E., Leung A.K.L., Wang J. FUS Regulates Activity of MicroRNA-Mediated Gene Silencing. Mol Cell, 2018;69:787–801. doi: 10.1016/j.molcel.2018.02.001. e788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun M., Ding J., Li D., Yang G., Cheng Z., Zhu Q. NUDT21 regulates 3′-UTR length and microRNA-mediated gene silencing in hepatocellular carcinoma. Cancer Lett. 2017;410:158–168. doi: 10.1016/j.canlet.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 55.Nairismägi M.L., Vislovukh A., Meng Q., Kratassiouk G., Beldiman C., Petretich M., Groisman R., Füchtbauer E.M., Harel-Bellan A., Groisman I. Translational control of TWIST1 expression in MCF-10A cell lines recapitulating breast cancer progression. Oncogene. 2012;31:4960–4966. doi: 10.1038/onc.2011.650. [DOI] [PubMed] [Google Scholar]
- 56.Bottini S., Hamouda-Tekaya N., Mategot R., Zaragosi L.E., Audebert S., Pisano S., Grandjean V., Mauduit C., Benahmed M., Barbry P., Repetto E., Trabucchi M. Post-transcriptional gene silencing mediated by microRNAs is controlled by nucleoplasmic Sfpq. Nat Commun. 2017;8:1189. doi: 10.1038/s41467-017-01126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ripin N., Boudet J., Duszczyk M.M., Hinniger A., Faller M., Krepl M., Gadi A., Schneider R.J., Šponer J., Meisner-Kober N.C., Allain F.H. Molecular basis for AU-rich element recognition and dimerization by the HuR C-terminal RRM. Proc Natl Acad Sci U S A, 2019;116:2935–2944. doi: 10.1073/pnas.1808696116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grammatikakis I., Abdelmohsen K., Gorospe M. Posttranslational control of HuR function. Wiley Interdiscip Rev RNA. 2017;8 doi: 10.1002/wrna.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdelmohsen K., Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsui K.H., Chiang K.C., Lin Y.H., Chang K.S., Feng T.H., Juang H.H. BTG2 is a tumor suppressor gene upregulated by p53 and PTEN in human bladder carcinoma cells. Cancer Med. 2018;7:184–195. doi: 10.1002/cam4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bohn J.A., Van Etten J.L., Schagat T.L., Bowman B.M., McEachin R.C., Freddolino P.L., Goldstrohm A.C. Identification of diverse target RNAs that are functionally regulated by human Pumilio proteins. Nucleic Acids Res. 2018;46:362–386. doi: 10.1093/nar/gkx1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smialek M.J., Ilaslan E., Sajek M.P., Jaruzelska J. Role of PUM RNA-Binding Proteins in Cancer. Cancers (Basel) 2021;13 doi: 10.3390/cancers13010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldstrohm A.C., Hall T.M.T., McKenney K.M. Post-transcriptional Regulatory Functions of Mammalian Pumilio Proteins. Trends Genet. 2018;34:972–990. doi: 10.1016/j.tig.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morris-Hanon O., Furmento V.A., Rodríguez-Varela M.S., Mucci S., Fernandez-Espinosa D.D., Romorini L., Sevlever G.E., Scassa M.E., Videla-Richardson G.A. The Cell Cycle Inhibitors p21 Cip1 and p27 Kip1 Control Proliferation but Enhance DNA Damage Resistance of Glioma Stem Cells. Neoplasia. 2017;19:519–529. doi: 10.1016/j.neo.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou H., Guo W., Zhao Y., Wang Y., Zha R., Ding J., Liang L., Yang G., Chen Z., Ma B., Yin B. MicroRNA-135a acts as a putative tumor suppressor by directly targeting very low density lipoprotein receptor in human gallbladder cancer. Cancer Sci. 2014;105:956–965. doi: 10.1111/cas.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takada H., Imoto I., Tsuda H., Nakanishi Y., Sakakura C., Mitsufuji S., Hirohashi S., Inazawa J. Genomic loss and epigenetic silencing of very-low-density lipoprotein receptor involved in gastric carcinogenesis. Oncogene. 2006;25:6554–6562. doi: 10.1038/sj.onc.1209657. [DOI] [PubMed] [Google Scholar]
- 67.Webb D.J., Nguyen D.H., Sankovic M., Gonias S.L. The very low density lipoprotein receptor regulates urokinase receptor catabolism and breast cancer cell motility in vitro. J Biol Chem. 1999;274:7412–7420. doi: 10.1074/jbc.274.11.7412. [DOI] [PubMed] [Google Scholar]
- 68.Lee H.S., Kundu J., Kim R.N., Shin Y.K. Transducer of ERBB2.1 (TOB1) as a Tumor Suppressor: A Mechanistic Perspective. Int J Mol Sci. 2015;16:29815–29828. doi: 10.3390/ijms161226203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaudhury A., Chander P., Howe P.H. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1′s multifunctional regulatory roles. RNA. 2010;16:1449–1462. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geuens T., Bouhy D., Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu R., Olsen M.T., Webb K., Bennett E.J., Lykke-Andersen J. Recruitment of the 4EHP-GYF2 cap-binding complex to tetraproline motifs of tristetraprolin promotes repression and degradation of mRNAs with AU-rich elements. RNA. 2016;22:373–382. doi: 10.1261/rna.054833.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carballo E., Lai W.S., Blackshear P.J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 73.Marderosian M., Sharma A., Funk A.P., Vartanian R., Masri J., Jo O.D., Gera J.F. Tristetraprolin regulates Cyclin D1 and c-Myc mRNA stability in response to rapamycin in an Akt-dependent manner via p38 MAPK signaling. Oncogene. 2006;25:6277–6290. doi: 10.1038/sj.onc.1209645. [DOI] [PubMed] [Google Scholar]
- 74.Brennan S.E., Kuwano Y., Alkharouf N., Blackshear P.J., Gorospe M., Wilson G.M. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009;69:5168–5176. doi: 10.1158/0008-5472.CAN-08-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.von Roretz C., Gallouzi I.E. Decoding ARE-mediated decay: is microRNA part of the equation? J Cell Biol. 2008;181:189–194. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qi M.Y., Song J.W., Zhang Z., Huang S., Jing Q. P38 activation induces the dissociation of tristetraprolin from Argonaute 2 to increase ARE-mRNA stabilization. Mol Biol Cell. 2018;29:988–1002. doi: 10.1091/mbc.E17-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lederer M., Bley N., Schleifer C., Hüttelmaier S. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol. 2014;29:3–12. doi: 10.1016/j.semcancer.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 78.Jiang Y., Xu E., Zhang J., Chen M., Flores E., Chen X. The Rbm38-p63 feedback loop is critical for tumor suppression and longevity. Oncogene. 2018;37:2863–2872. doi: 10.1038/s41388-018-0176-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan W., Zhang Y., Chen X. TAp63γ and ΔNp63γ are regulated by RBM38 via mRNA stability and have an opposing function in growth suppression. Oncotarget. 2017;8:78327–78339. doi: 10.18632/oncotarget.18463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Denaro N., Lo Nigro C., Natoli G., Russi E.G., Adamo V., Merlano M.C. The Role of p53 and MDM2 in Head and Neck Cancer. ISRN Otolaryngol. 2011;2011 doi: 10.5402/2011/931813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benjamin C.L., Ananthaswamy H.N. p53 and the pathogenesis of skin cancer. Toxicol Appl Pharmacol. 2007;224:241–248. doi: 10.1016/j.taap.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang X., Schwartz J.C., Cech T.R. Nucleic acid-binding specificity of human FUS protein. Nucleic Acids Res. 2015;43:7535–7543. doi: 10.1093/nar/gkv679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Masuda A., Takeda J., Okuno T., Okamoto T., Ohkawara B., Ito M., Ishigaki S., Sobue G., Ohno K. Position-specific binding of FUS to nascent RNA regulates mRNA length. Genes Dev. 2015;29:1045–1057. doi: 10.1101/gad.255737.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brooke G.N., Culley R.L., Dart D.A., Mann D.J., Gaughan L., McCracken S.R., Robson C.N., Spencer-Dene B., Gamble S.C., Powell S.M., Wait R., Waxman J., Walker M.M., Bevan C.L. FUS/TLS is a novel mediator of androgen-dependent cell-cycle progression and prostate cancer growth. Cancer Res. 2011;71:914–924. doi: 10.1158/0008-5472.CAN-10-0874. [DOI] [PubMed] [Google Scholar]
- 85.Sreekumar R., Emaduddin M., Al-Saihati H., Moutasim K., Chan J., Spampinato M., Bhome R., Yuen H.M., Mescoli C., Vitale A., Cillo U., Rugge M., Primrose J., Hilal M.A., Thirdborough S., Tulchinsky E., Thomas G., Mirnezami A., Sayan A.E. Protein kinase C inhibitors override ZEB1-induced chemoresistance in HCC. Cell Death Dis. 2019;10:703. doi: 10.1038/s41419-019-1885-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Siebzehnrubl F.A., Silver D.J., Tugertimur B., Deleyrolle L.P., Siebzehnrubl D., Sarkisian M.R., Devers K.G., Yachnis A.T., Kupper M.D., Neal D., Nabilsi N.H., Kladde M.P., Suslov O., Brabletz S., Brabletz T., Reynolds B.A., Steindler D.A. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med. 2013;5:1196–1212. doi: 10.1002/emmm.201302827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sánchez-Tilló E., Siles L., de Barrios O., Cuatrecasas M., Vaquero E.C., Castells A., Postigo A. Expanding roles of ZEB factors in tumorigenesis and tumor progression. Am J Cancer Res. 2011;1:897–912. [PMC free article] [PubMed] [Google Scholar]
- 88.Herzog V.A., Ameres S.L. Approaching the Golden Fleece a Molecule at a Time: Biophysical Insights into Argonaute-Instructed Nucleic Acid Interactions. Mol Cell. 2015;59:4–7. doi: 10.1016/j.molcel.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 89.Ashley C.T., Wilkinson K.D., Reines D., Warren S.T. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 90.Ascano M., Mukherjee N., Bandaru P., Miller J.B., Nusbaum J.D., Corcoran D.L., Langlois C., Munschauer M., Dewell S., Hafner M., Williams Z., Ohler U., Tuschl T. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Darnell J.C., Fraser C.E., Mostovetsky O., Stefani G., Jones T.A., Eddy S.R., Darnell R.B. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vasilyev N., Polonskaia A., Darnell J.C., Darnell R.B., Patel D.J., Serganov A. Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP. Proc Natl Acad Sci U S A. 2015;112:E5391–E5400. doi: 10.1073/pnas.1515737112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kenny P., Ceman S. RNA Secondary Structure Modulates FMRP's Bi-Functional Role in the MicroRNA Pathway. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17060985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smits M., Wurdinger T., van het Hof B., Drexhage J.A., Geerts D., Wesseling P., Noske D.P., Vandertop W.P., de Vries H.E., Reijerkerk A. Myc-associated zinc finger protein (MAZ) is regulated by miR-125b and mediates VEGF-induced angiogenesis in glioblastoma. FASEB J. 2012;26:2639–2647. doi: 10.1096/fj.11-202820. [DOI] [PubMed] [Google Scholar]
- 95.Zhu X., Luo W., Liang W., Tang F., Bei C., Ren Y., Qin L., Tan C., Zhang Y., Tan S. Over expression and clinical significance of MYC-associated zinc finger protein in pancreatic carcinoma. Onco Targets Ther. 2016;9:7493–7501. doi: 10.2147/OTT.S124118. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 96.Bhandari A., Gordon W., Dizon D., Hopkin A.S., Gordon E., Yu Z., Andersen B. The Grainyhead transcription factor Grhl3/Get1 suppresses miR-21 expression and tumorigenesis in skin: modulation of the miR-21 target MSH2 by RNA-binding protein DND1. Oncogene. 2013;32:1497–1507. doi: 10.1038/onc.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Suzuki E., Umezawa K., Bonavida B. Rituximab inhibits the constitutively activated PI3K-Akt pathway in B-NHL cell lines: involvement in chemosensitization to drug-induced apoptosis. Oncogene. 2016;35:5576. doi: 10.1038/onc.2016.207. [DOI] [PubMed] [Google Scholar]
- 98.Kobayashi Y., Suzuki K., Kobayashi H., Ohashi S., Koike K., Macchi P., Kiebler M., Anzai K. C9orf10 protein, a novel protein component of Puralpha-containing mRNA-protein particles (Puralpha-mRNPs): characterization of developmental and regional expressions in the mouse brain. J Histochem Cytochem. 2008;56:723–731. doi: 10.1369/jhc.2008.950733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tanaka M., Sasaki K., Kamata R., Hoshino Y., Yanagihara K., Sakai R. A novel RNA-binding protein, Ossa/C9orf10, regulates activity of Src kinases to protect cells from oxidative stress-induced apoptosis. Mol Cell Biol. 2009;29:402–413. doi: 10.1128/MCB.01035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bartolomé R.A., García-Palmero I., Torres S., López-Lucendo M., Balyasnikova I.V., Casal J.I. IL13 Receptor α2 Signaling Requires a Scaffold Protein, FAM120A, to Activate the FAK and PI3K Pathways in Colon Cancer Metastasis. Cancer Res. 2015;75:2434–2444. doi: 10.1158/0008-5472.CAN-14-3650. [DOI] [PubMed] [Google Scholar]
- 101.Mihailovich M., Militti C., Gabaldón T., Gebauer F. Eukaryotic cold shock domain proteins: highly versatile regulators of gene expression. Bioessays. 2010;32:109–118. doi: 10.1002/bies.200900122. [DOI] [PubMed] [Google Scholar]
- 102.Avolio R., Inglés-Ferrándiz M., Ciocia A., Coll O., Bonnin S., Guitart T., Ribó A., Gebauer F. Coordinated post-transcriptional control of oncogene-induced senescence by UNR/CSDE1. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2021.110211. [DOI] [PubMed] [Google Scholar]
- 103.Ju Lee H., Bartsch D., Xiao C., Guerrero S., Ahuja G., Schindler C., Moresco J.J., Yates J.R., Gebauer F., Bazzi H., Dieterich C., Kurian L., Vilchez D. A post-transcriptional program coordinated by CSDE1 prevents intrinsic neural differentiation of human embryonic stem cells. Nat Commun. 2017;8:1456. doi: 10.1038/s41467-017-01744-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guo A.X., Cui J.J., Wang L.Y., Yin J.Y. The role of CSDE1 in translational reprogramming and human diseases. Cell Commun Signal. 2020;18:14. doi: 10.1186/s12964-019-0496-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wurth L., Papasaikas P., Olmeda D., Bley N., Calvo G.T., Guerrero S., Cerezo-Wallis D., Martinez-Useros J., García-Fernández M., Hüttelmaier S., Soengas M.S., Gebauer F. UNR/CSDE1 Drives a Post-transcriptional Program to Promote Melanoma Invasion and Metastasis. Cancer Cell. 2016;30:694–707. doi: 10.1016/j.ccell.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 106.Kakumani P.K., Harvey L.M., Houle F., Guitart T., Gebauer F., Simard M.J. CSDE1 controls gene expression through the miRNA-mediated decay machinery. Life Sci Alliance. 2020;3 doi: 10.26508/lsa.201900632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fournier P.G., Juárez P., Jiang G., Clines G.A., Niewolna M., Kim H.S., Walton H.W., Peng X.H., Liu Y., Mohammad K.S., Wells C.D., Chirgwin J.M., Guise T.A. The TGF-β Signaling Regulator PMEPA1 Suppresses Prostate Cancer Metastases to Bone. Cancer Cell. 2015;27:809–821. doi: 10.1016/j.ccell.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Amalia R., Abdelaziz M., Puteri M.U., Hwang J., Anwar F., Watanabe Y., Kato M. TMEPAI/PMEPA1 inhibits Wnt signaling by regulating β-catenin stability and nuclear accumulation in triple negative breast cancer cells. Cell Signal. 2019;59:24–33. doi: 10.1016/j.cellsig.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 109.Ji J., Ding K., Luo T., Xu R., Zhang X., Huang B., Chen A., Zhang D., Miletic H., Bjerkvig R., Thorsen F., Wang J., Li X. PMEPA1 isoform a drives progression of glioblastoma by promoting protein degradation of the Hippo pathway kinase LATS1. Oncogene. 2020;39:1125–1139. doi: 10.1038/s41388-019-1050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Masamha C.P., Xia Z., Yang J., Albrecht T.R., Li M., Shyu A.B., Li W., Wagner E.J. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412–416. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang Q., Gilmartin G.M., Doublié S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proc Natl Acad Sci U S A. 2010;107:10062–10067. doi: 10.1073/pnas.1000848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang L., Zhang W. Knockdown of NUDT21 inhibits proliferation and promotes apoptosis of human K562 leukemia cells through ERK pathway. Cancer Manag Res. 2018;10:4311–4323. doi: 10.2147/CMAR.S173496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang H., Sheng C., Yin Y., Wen S., Yang G., Cheng Z., Zhu Q. PABPC1 interacts with AGO2 and is responsible for the microRNA mediated gene silencing in high grade hepatocellular carcinoma. Cancer Lett. 2015;367:49–57. doi: 10.1016/j.canlet.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 114.Fernández-Miranda G., Méndez R. The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res Rev. 2012;11:460–472. doi: 10.1016/j.arr.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 115.Nagaoka K., Fujii K., Zhang H., Usuda K., Watanabe G., Ivshina M., Richter J.D. CPEB1 mediates epithelial-to-mesenchyme transition and breast cancer metastasis. Oncogene. 2016;35:2893–2901. doi: 10.1038/onc.2015.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hägele S., Kühn U., Böning M., Katschinski D.M. Cytoplasmic polyadenylation-element-binding protein (CPEB)1 and 2 bind to the HIF-1alpha mRNA 3′-UTR and modulate HIF-1alpha protein expression. Biochem J. 2009;417:235–246. doi: 10.1042/BJ20081353. [DOI] [PubMed] [Google Scholar]
- 117.Ajduković J. HIF-1–a big chapter in the cancer tale. Exp Oncol. 2016;38:9–12. [PubMed] [Google Scholar]
- 118.Yarosh C.A., Iacona J.R., Lutz C.S., Lynch K.W. PSF: nuclear busy-body or nuclear facilitator? Wiley Interdiscip Rev RNA. 2015;6:351–367. doi: 10.1002/wrna.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tsukahara T., Matsuda Y., Haniu H. PSF knockdown enhances apoptosis via downregulation of LC3B in human colon cancer cells. Biomed Res Int. 2013;2013 doi: 10.1155/2013/204973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lim Y.W., James D., Huang J., Lee M. The Emerging Role of the RNA-Binding Protein SFPQ in Neuronal Function and Neurodegeneration. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21197151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bi O., Anene C.A., Nsengimana J., Shelton M., Roberts W., Newton-Bishop J., Boyne J.R. SFPQ promotes an oncogenic transcriptomic state in melanoma. Oncogene. 2021;40:5192–5203. doi: 10.1038/s41388-021-01912-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sternburg E.L., Karginov F.V. Global Approaches in Studying RNA-Binding Protein Interaction Networks. Trends Biochem Sci. 2020;45:593–603. doi: 10.1016/j.tibs.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 123.Bukhari S.I., Vasudevan S. FXR1a-associated microRNP: A driver of specialized non-canonical translation in quiescent conditions. RNA Biol. 2017;14:137–145. doi: 10.1080/15476286.2016.1265197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bukhari S.I.A., Truesdell S.S., Lee S., Kollu S., Classon A., Boukhali M., Jain E., Mortensen R.D., Yanagiya A., Sadreyev R.I., Haas W., Vasudevan S. A Specialized Mechanism of Translation Mediated by FXR1a-Associated MicroRNP in Cellular Quiescence. Mol Cell. 2016;61:760–773. doi: 10.1016/j.molcel.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Waninger J.J., Beyett T.S., Gadkari V.V., Siebenaler R.F., Kenum C., Shankar S., Ruotolo B.T., Chinnaiyan A.M., Tesmer J.J.G. Biochemical characterization of the interaction between KRAS and Argonaute 2. Biochem Biophys Rep. 2022;29 doi: 10.1016/j.bbrep.2021.101191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tien J.C., Chugh S., Goodrum A.E., Cheng Y., Mannan R., Zhang Y., Wang L., Dommeti V.L., Wang X., Xu A., Hon J., Kenum C., Su F., Wang R., Cao X., Shankar S., Chinnaiyan A.M. AGO2 promotes tumor progression in KRAS-driven mouse models of non-small cell lung cancer. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2026104118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.