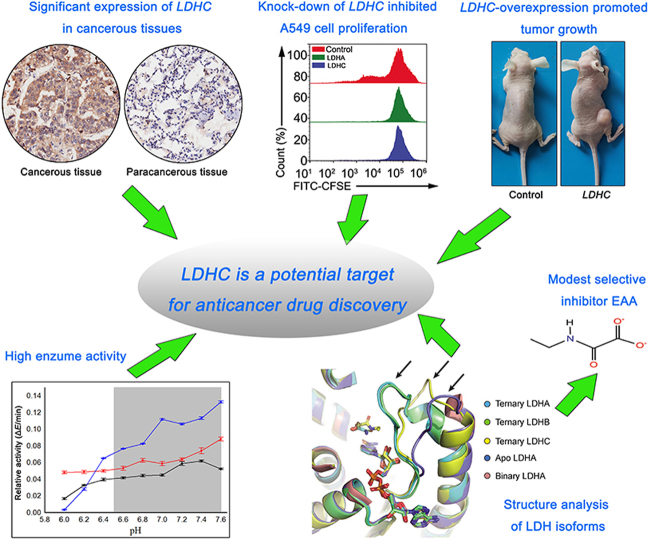
Abstract
One of the distinct hallmarks of cancer cells is aerobic glycolysis (Warburg effect). Lactate dehydrogenase A (LDHA) is thought to play a key role in aerobic glycolysis and has been extensively studied, while lactate dehydrogenase C (LDHC), an isoform of LDHA, has received much less attention. Here we showed that human LDHC was significantly expressed in lung cancer tissues, overexpression of Ldhc in mice could promote tumor growth, and knock-down of LDHC could inhibit the proliferation of lung cancer A549 cells. We solved the first crystal structure of human LDHC4 and found that the active-site loop of LDHC4 adopted a distinct conformation compared to LDHA4 and lactate dehydrogenase B4 (LDHB4). Moreover, we found that (ethylamino) (oxo)acetic acid shows about 10 times selective inhibition against LDHC4 over LDHA4 and LDHB4. Our studies suggest that LDHC4 is a potential target for anticancer drug discovery and (ethylamino) (oxo)acetic acid provides a good start to develop lead compounds for selective drugs targeting LDHC4.
KEY WORDS: LDHC, Anticancer target, Warburg effect, LDHC4 structure, (Ethylamino) (oxo)acetic acid, LDH isoforms, Lung cancer, Tissue microarray
Graphical abstract
Tissue chip, overexpression, knock-down, and enzyme experiments indicate that LDHC is a potential target for cancer therapy. Structural data will facilitate LDHC selective inhibitor design in the future.
1. Introduction
Lactate dehydrogenase (LDH), a tetrameric enzyme composed of four subunits, is an enzyme that catalyzes the interconversion of pyruvate and lactate with concomitant interconversion of nicotinamide adenine dinucleotide (NADH, reduced form) and NAD+ (oxidized form)1. The two most common subunits of LDH are the LDHA (or LDHM) and LDHB (or LDHH) proteins, encoded by LDHA and LDHB genes, respectively. The two subunits can form five tetramers (isoforms): LDHA4, LDHA3B1, LDHA2B2, LDHA1B3, and LDHB4. These five isoforms show different tissue distribution. For example, LDHA4, which is the major isoform in skeletal muscle where oxygen deficiency happens frequently and glycolysis is required to satisfy metabolic needs, kinetically favors the conversion of pyruvate to lactate. LDHB4, which is the main isoform in heart muscle that is dependent upon aerobic metabolism pathways, is thought to convert lactate to pyruvate that is further oxidized2, 3, 4. The third subunit is LDHC encoded by the LDHC gene, which forms the isoform LDHC4 that shows a specific distribution in male5 and female6 germ cells. LDHC4, representing more than 80% of the total LDH activities of spermatozoa7, is thought to convert pyruvate to lactate in the pathway of aerobic glycolysis which is thought to be one of the two major sources of adenosine triphosphate (ATP) for sperm motility and capacitation8. The most distinctive feature of LDHC4 is the ability to catalyze the conversion of 2-oxo acids with longer carbon chain into 2-hydroxy acids, such as 2-oxobutanoate into 2-hydroxybutanoate9.
Among the LDH subunits, LDHA is the most studied one because it plays a key role in the Warburg effect of cancer cells. Cancer cells tend to take up glucose and convert it to lactate through glycolysis pathway for obtaining energy, even in aerobic conditions, which is known as the Warburg effect or aerobic glycolysis10. The metabolism of one molecule of glucose to two molecules of pyruvate through glycolysis has a net yield of two molecules of ATP and two molecules of NADH. For the continuance of glycolysis, the NADH produced in the process needs to be oxidized into NAD+. LDHA is thought to catalyze the key step of converting pyruvate into lactate with concomitant oxidizing NADH into NAD+11,12. So, LDHA has been treated as a potential target for cancer therapy and received extensive studies13, 14, 15. Actually, studies showed that LDHA is important for tumor initiation, maintenance, and progression16, 17, 18. Some inhibitors against LDHA have already been identified and further proceeded into preclinical studies18, 19, 20, 21, 22.
Compared with LDHA, LDHC received much less attention and only several papers published in the area of cancer research. In 2002, Koslowski et al.23 reported for the first time the expression of LDHC at mRNA level in a variety of cancer cells. From then on, LDHC was classified as a cancer/testis antigen gene. The high frequencies of expression of LDHC have been found in lung cancer, melanoma, and breast cancer23,24. Due to the high frequency of ectopic expression in cancers, LDHC is suggested to be a potential biomarker for diagnosis and prognosis of hepatocellular carcinoma and breast cancer25,26. Besides the aforementioned studies on LDHC expression level, Thomas et al.27 reported that LDHC-derived peptides-primed T cells exhibited cytolytic activity against HLA-A∗0201 breast cancer cell lines with endogenous LDHC expression. Naik and Decock28 reported that knockdown of LDHC expression in breast cancer cells increased DNA damage and apoptosis. These studies highly suggested that LDHC, similar as LDHA, may also plays a role in cancer.
Here we show that LDHC is significantly expressed in lung cancer tissues, knock-down of LDHC can inhibit the proliferation of lung cancer A549 cell, and overexpression of Ldhc in mice can promote tumor growth. We also solved the crystal structure of human LDHC4 which remains absent previously, hoping to help identify its largely unknown biological function, unlike LDHA and LDHB. It was found that the active-site loop of LDHC4 adopted a much more open conformation, which might be the selective reason why the detected (ethylamino) (oxo)acetic acid showed about 10-fold inhibitory effect against LDHC4 over LDHA4 and LDHB4. Our studies suggest that LDHC is a potential target for anticancer drug discovery and selective inhibitors could be developed for targeting LDHC4, to some degree, without disturbing the activity of LDHA and LDHB.
2. Materials and methods
2.1. Tissue microarrays
The tissue samples from 93 patients with lung cancer (including 84 cases of adenocarcinoma, 8 cases of bronchoalveolar carcinoma, 1 case of mucoepidermoid carcinoma) were obtained from tissue specimen bank of Shanghai Super Biological Biotech Co., Ltd. (Shanghai, China). The 93 patients (50 males and 43 females) aged 30–84 years old with a median of 62 years. All the patients were pathologically diagnosed with lung cancer and without pre-surgery, chemotherapy, and radiotherapy treatment. Each study specimen was provided with cancer tissue and adjacent-carcinoma tissue that was 1.5 cm distanced from cancer. No distant metastases occurred in all patients. We followed ethical standards and respected welfare of animals. The work was approved by the Ethics Committee of Shanghai Super Biological Biotech Co., and the informed consent forms from all the patients were retained by tissue specimen bank of Shanghai Super Biological Biotech Co.
The tissue chip (Lot. XT16-022; matrix code: HLugA180Su02) with 180 array points (1.5 mm diameter of each point) was prepared using the aforementioned samples including the tumor tissues (93 array points) and corresponding para-cancerous tissues (87 array points) from the 93 cases.
Immunohistochemistry was performed according to the two-step EnVision method29. Briefly, the slices were dewaxed and incubated with 0.3% H2O2 at room temperature for 20 min to eliminate the activity of endogenous peroxidase, and washed three times with PBS for 3 min each. They were repaired by microwave for 20 min, followed by incubation at 4 °C overnight with monoclonal antibody against human LDHC (Cat. #ab52747, Abcam). Subsequently, they were further washed and incubated at 37 °C for 30 min with EnVision secondary antibody (Cat. #GK500705, Dako), then washed and incubated with DAB to develop the color. The tissue chip was scanned and converted to images and further analyzed by Olympus cell sense software. The images were assessed by two experienced pathologists to determine whether the samples are LDHC positive or negative without knowledge of the clinical data separately.
2.2. RNA interference
Commercial siRNAs from ThermoFisher Scientific were used in this study. To knockdown the expression of LDHA, the validated siRNA s350 (Cat. #4390824, sense strand: CCAGCGUAACGUGAACAUATT; antisense strand: UAUGUUCACGUUACGCUGGAC) was used following the manufacturer's instructions. To knockdown the expression of LDHC, the siRNA s8129 (Cat. #4392420, sense strand: GUUAAGGGAUUAUAUGGAATT; antisense strand: UUCCAUAUAAUCCCUUAACCA) was used. Transfection using the negative control siRNA (Cat. #4390843, ThermoFisher Scientific) was also parallelly used.
A549 cells were maintained in Dulbecco's modified Eagle's medium (Cytiva) supplemented with 10% fetal bovine serum (ThermoFisher Scientific) containing 100 U/mL penicillin and streptomycin at 37 °C in a humidified atmosphere with 5% CO2.
For cell transfection and qRT-PCR, A549 cells were plated in 6-well plates with antibiotic free growth medium at a density of 1 × 106 cells per well for 24 h and transfected with the siRNAs using Lipofectamine 2000 according to the manufacturer's instructions. The growth mediums were replaced with fresh ones after 6 h from transfection. The cells were incubated for another 48 h and harvested. Total RNAs were isolated from the harvested A549 cells by Trizol reagents (ThermoFisher Scientific) according to the manufacturer's instructions and were reversely transcribed with a cDNA Synthesis Kit (Cat. #6110A, TaKaRa). The resulting cDNAs were used for qPCR analysis. The qPCR was performed using SYBR Green Kit (Cat. #740703, TaKaRa) and gene-specific primers as shown in Supporting Information Table S1. β-Actin was set as internal control.
For cell viability assay, MTT assay was used. A549 cells were seeded in 96-well plates at a density of 2000 cells/well, incubated for 24 h, and transfected with the siRNAs. The growth mediums were replaced with fresh ones 6 h after transfection. The cells were then incubated for 24, 48, 72 and 96 h, respectively. Then 20 μL MTT regent (5 mg/mL) was added to each well, and incubated for another 4 h until purple precipitate was visible. The medium was discarded, and 150 μL DMSO was added and oscillated for 10 min. The absorbance of each well was measured at 570 nm in a multimode plate reader (PerkinElmer).
For flow cytometric analysis, A549 cells were seeded in 6-well plates in antibiotic free growth medium at a density of 5 × 105 cells per well for 24 h and transfected with the siRNAs. The growth mediums were replaced with fresh ones 6 h after transfection. The cells were incubated for another 12 h, and digested with trypsin. After adjusted to 5.0 × 105/mL, the cells were incubated with 10 μmol/L CFSE at 37 °C for 30 min, after which 5 times the volume of pre-cooled medium were added to stop staining. After sitting on ice for 5 min and washed 3 times with fresh medium by centrifugation, the washed cells were plated in 12-well plates at a density of 2.0 × 105 cells per well, incubated for 24, 48, 72, and 96 h, respectively. The incubated cells were further digested with trypsin, washed with PBS, and resuspended in 200 μL PBS for flow cytometric analysis30.
2.3. LDHC overexpression mice tumor model
The LDHC gene with His-tag was constructed into the pMJL1 lentiviral vector (GenePharma, Shanghai, China). The Lentiviruses were packaged in 293T cells using the lentiviral vector pMJL1, the plasmid pMD2.G, pMDLg-pRRE and pRSV-Rev. The Lentiviruses were isolated and purified for transfection. The purified lentiviruses were used to transfect A549 cells and the stable cell lines overexpressing LDHC were screened using puromycin dihydrochloride of 2.0 μg/mL.
The cryopreserved A549 cells overexpressing LDHC and the control A549 cells were resuscitated in 100 μL serum-free medium, and inoculated subcutaneously in BALB/c-nu nude mice at 5 × 106 cell per mouse. When the subcutaneous tumor volumes reached about 50 mm3, the mice were numbered, and the length and width of the tumors were measured every three days till the 45th day from inoculation. The tumor volumes were calculated using Eq. (1):
| (1) |
and the tumor growth curve was drawn by using the volume data against the days. On the 45th day, the mice were euthanized, and the subcutaneous tumors were taken out, photographed, weighed, and stored at −80 °C.
In order to determine whether or not the introduced LDHC gene was overexpressed in tumors of mice, detection of the His6-tag was performed by Western blot because fusion of His6-tag was introduced to LDHC gene. About 10 mg of each tumor sample was sonicated in 200 μL RIPA lysis buffer and centrifuged. The protein concentrations of the supernatants were determined using the BCA kit (KeyGEN BioTECH, Nanjing, China) and equal amount of protein from each sample was loaded onto 12.5% polyacrylamide gel for electrophoresis. Subsequently, the proteins were transferred to a 0.22 μm polyvinylidene fluoride (PVDF) membrane. The membrane was blocked for 2 h using 5% skimmed milk, washed with TBST buffer for 3 times, and then incubated with the mouse-origin His-Tag primary antibody (Cat. #T0009, Affinity Biosciences, Changzhou, China) overnight at 4 °C. After washed and incubated with the goat anti-mouse secondary antibody (Cat. #5500, ZENBIO) for 1 h at 37 °C, the bands were visualized by detecting chemiluminescence.
2.4. Purification and characterization of LDH isoforms
The three genes encoding human LDHA, LDHB, and LDHC subunits were synthesized with codon usage adapted for expression in E. coli by the Tsingke Biotechnology Co., Ltd. (Beijing, China). The three synthesized genes were inserted into vector pET28a using restriction sites NcoI and XhoI. Each of the constructed vectors was theoretically ensured to express recombinant LDH with a His6-tag at the C-terminal and no additional amino acids added at the N-terminal.
To increase the solubility of LDHC, L56E and I332Q mutations were introduced by site-directed mutation31. The double-mutation resulted in significantly higher solubility of LDHC. The process of purification of LDHC-mutant is the same as that of LDHA and LDHB.
The recombinant plasmids pET28a-LDHA, pET28a-LDHB, pET28a-LDHC and pET28a-LDHC-mutant were transformed into E. coli BL21 (DE3) strain (Tiangen Biotech, Beijing, China). A single colony was picked and grown at 37 °C overnight in 20 mL of 2TY medium containing 50 μg/mL kanamycin. A 5 mL aliquot of the culture was introduced into 4 × 1000 mL of 2TY medium and grown at 37 °C to an optical density (OD600) of ∼0.8. Protein expression was induced with 0.2 mmol/L isopropyl-β-d-thiogalactoside and cells were further cultured for 16 h at 16 °C. Cells were harvested by centrifugation at 4 °C. The cell pellets of LDHA, LDHB, and LDHC-mutant were suspended in 60 mL lysis buffer (50 mmol/L Tris-Cl pH 7.5, 500 mmol/L NaCl, 10 mmol/L imidazole, 10% glycerol), while the cell pellet of LDHC was suspended in lysis buffer with a different pH value (50 mmol/L Tris-Cl pH 8.0, 500 mmol/L NaCl, 10 mmol/L imidazole, 10% glycerol). The cell pellets were disrupted by sonication. The cell debris was removed by centrifuged at 35,000×g for 60 min at 4 °C and the supernatant was filtered using 0.45 μm filter and stored at 4 °C for protein purification.
The procedures of purification of LDHA, LDHB, and LDHC-mutant are the same. The resultant supernatant was collected and applied to precharged Ni Sepharose column (Cat. #17531901, Cytiva) pre-equilibrated with buffer A (20 mmol/L Tris-Cl pH 7.5, 500 mmol/L NaCl, 10 mmol/L imidazole, 10% glycerol). The column was washed with buffer A supplemented with 10% of buffer B (20 mmol/L Tris-Cl pH7.5, 500 mmol/L NaCl, 500 mmol/L imidazole, 10% glycerol). The target proteins were eluted with buffer A mixed with 40% buffer B, and concentrated using a 30 kDa cut-off concentrator (Millipore). Proteins were further purified using HiLoad 160/600 Superdex 200 size exclusion chromatography column (Cat. #28989335, Cytiva) pre-equilibrated with 20 mmol/L Tris-Cl pH 7.5, 300 mmol/L NaCl, and 10% glycerol. Fractions containing the target protein were pooled, concentrated to 10 mg/mL and stored at −80 °C.
The purification of LDHC is partly different from the above-mentioned procedure mainly in pH value of buffer and size exclusion chromatography. The supernatant of LDHC was applied to precharged Ni Sepharose column pre-equilibrated with buffer A (20 mmol/L Tris-Cl pH8.0, 500 mmol/L NaCl, 10 mmol/L imidazole, 10% glycerol). The target proteins were eluted with buffer A mixed with 40% buffer B (20 mmol/L Tris-Cl pH8.0, 500 mmol/L NaCl, 500 mmol/L imidazole, 10% glycerol), concentrated, and loaded onto HiLoad 160/600 Superdex 200 size exclusion chromatography column pre-equilibrated with 20 mmol/L Tris-Cl pH8.0, 300 mmol/L NaCl, and 10% glycerol. Fractions containing the target protein were pooled, concentrated and loaded onto Superdex 200 increase 10/300 GL column pre-equilibrated with 20 mmol/L Tris–HCl pH 8.0, 300 mmol/L NaCl, and 10% glycerol. Fractions containing the LDH activities were pooled, concentrated, and stored at −80 °C for enzyme assay.
The LDHs activities were determined by recording the absorbance changes at 340 nm produced by NADH oxidation. The standard reaction buffer contained 100 mmol/L Tris–HCl (pH 7.5), 0.15 mmol/L NADH and 1.0 mmol/L pyruvate. LDH was then added to the mixture to start the reaction. In order to make the reaction velocity in an appropriate range that can be recorded by spectrophotometer, the amounts of enzymes (0.15 μg/mL) added were set to provide a ΔE340 of 0.060–0.070 per min in a 1 cm light path. All assays were performed at 25 °C unless otherwise stated. One unit of enzyme activity was defined as the amount of enzyme that converted 1 μmol of substrate to product in 1 min under standards conditions.
The standard assay described above was modified for kinetic analysis by varying the specific parameters tested. To determine the Michaelis constant (Km) of LDHs for pyruvate, pyruvate concentrations ranged 0.005–0.4 mmol/L were used for LDHA, 0.005–0.8 mmol/L for LDHB, and 0.005–0.25 mmol/L for LDHC with a constant concentration of 0.15 mmol/L NADH. To determine Km of LDHs for NADH, NADH concentrations ranged 0.005–0.4 mmol/L were used for LDHA, 0.005–0.36 mmol/L for LDHB, and 0.002–0.12 mmol/L for LDHC with a constant concentration of 1 mmol/L pyruvate. The Km and Vmax were calculated by nonlinear-regression fitting to the Michaelis–Menten equation using the software Origin Pro 9.1 (Origin Lab), and the results are expressed as mean ± SD. Every experiment was repeated at least three times with similar results.
To determine the inhibition constant (Ki) of (ethylamino) (oxo)acetic acid, reaction buffer of 50 mmol/L PBS (pH7.4) was used with 0.2 mmol/L pyruvate, 0.15 mmol/L NADH, 1.2 μg/mL enzyme, and varied concentration of inhibitor. The concentration of inhibitor ranged 0–40 mmol/L. The reactions were started by adding the pyruvate at 25 °C. Ki was calculated by linear regression fitting to Eq. (2):
| (2) |
where V0 is the velocity without inhibitor, Vobs is the velocities under different concentrations of inhibitor, [I] is inhibitor concentration, [S] is 0.2 mmol/L, Km was calculated in advance with the same buffer without inhibitor at 25 °C.
To determine the optimal pH of the LDH isoforms, reaction buffers of 100 mmol/L glycine-HCl (pH 3.84–6.28) and glycine-NaOH (pH 6.63–12.46) were used with 0.15 mmol/L NADH, 1.0 mmol/L pyruvate, and 0.78 μg/mL enzyme. In order to be comparable among LDH isoforms, the equal amounts of enzymes were used. NADH auto-oxidation in acidic pH buffers were also counted in by omitting the enzyme from the standard reaction mixture and subtracting it from the ΔE340 obtained when the enzyme was present. The optimal pH is defined as the pH at which the maximum activity (ΔE/min) was reached.
Thermal stabilities of LDH isoforms were assayed by incubating the enzyme solutions in a thermocycler at temperatures ranging from 10 °C to 70 °C for 10 min, respectively. The enzymes were taken out from thermocycler and put in ice bath for 5 min. The residue activities of the enzymes were determined at 25 °C with reaction mixture containing 100 mmol/L Tris–HCl (pH7.5), 0.15 mmol/L NADH and 1.0 mmol/L pyruvate and expressed as relative activity (ΔE340/min). The optimal temperature for each isoform is defined as the temperature at which the maximum residue (ΔE/min) was remained.
2.5. Crystallization, data collection and structure determination
The LDHC–NAD+–inhibitor ternary complex was prepared by mixing LDHC (8 mg/mL), NAD+ and (ethylamino) (oxo)acetic acid in a 1:10:10 molar ratio. The crystals of the ternary complex were grown using the hanging-drop vapour diffusion method at 20 °C, in a buffer consisting of 0.1 mol/L NaCl, 0.1 mol/L HEPES pH 7.5, and 1.6 mol/L ammonium sulfate.
Crystals were flash frozen in liquid nitrogen after briefly soaking in crystallization solution containing 2.5 mol/L ammonium sulfate as cryoprotectant. Diffraction data were collected on beamline BL19U1 of National Facility for Protein Science Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility32. The data collected were processed by the HKL-3000 program suite33. Details of data collection and statistics are summarized in Supporting Information Table S2. Structures were determined by molecular replacement using the crystal structure of human LDHA (PDB ID 4L4R) as the model. Structure refinement and model building were performed with PHENIX34 and Coot35. All models were validated with MolProbity36. All structure figures were prepared with PyMOL (www.pymol.org).
2.6. Effects of EAA on A549 cells
To detect the effects on proliferation, A549 cells were seeded in 96-well plates at a density of 2000 cells per well, cultured for 24 h, and EAA was added at a final concentration of 5 mmol/L. The cells were added 20 μL MTT reagent (5 mg/mL) per well after incubated for 24, 48, and 72 h, respectively. The cells were then incubated for another 4 h until purple precipitate was visible. The medium was discarded, after which 150 μL DMSO was added. The products were oscillated for 10 min, and the absorbance of each well was measured at 570 nm in a multimode plate reader.
To detect the effects on apoptosis, A549 cells were seeded in 6-well plates in antibiotic-free growth medium at a density of 5 × 105 cells/well for 24 h and EAA was added at a final concentration of 5 mmol/L. The cells were collected after incubation for 24, 48, and 72 h, respectively, after which the cells were collected and stained with Annexin A5 apoptosis detection kit (Cat. #640930, BioLegend) according to the manufacturer's protocol. Subsequently, apoptotic cells were analyzed using flow cytometry.
2.7. Statistical analysis
All the statistical calculations were performed using SPSS 19.0 software. A P value < 0.05 or less was considered statistically significant. Results are expressed as mean ± standard deviation (SD). The expression of LDHC in lung cancer and adjacent cancer tissues were compared with chi-square test. Other differences between two or three groups were compared with Student's t-test.
3. Results
3.1. Up-regulation of LDHC in patients with lung cancer
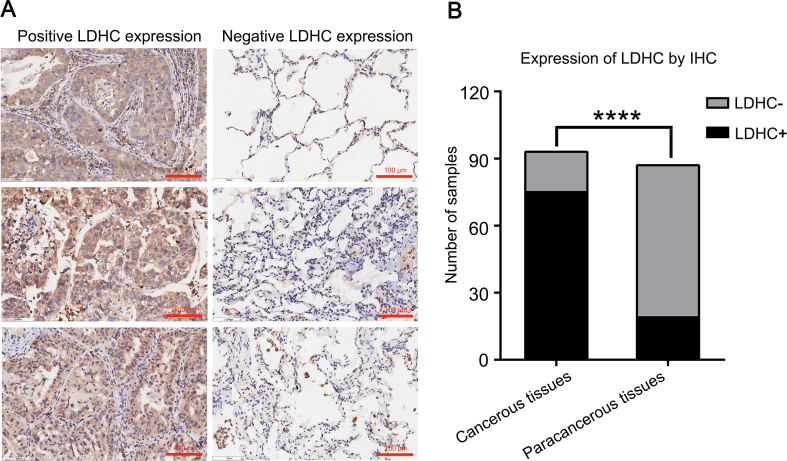
To detect the expression of LDHC in cancer tissues, a tissue microarray with 180 array points of cancerous and paracancerous tissues from 93 patients with lung cancer was used. The results show that there are 75 positive samples and 18 negative samples from cancerous tissues, while 19 positive samples and 68 negative samples from paracancerous tissues. The LDHC positive ratio of 80.6% in cancerous tissues vs. 21.8% in paracancerous tissues indicates that the expression of LDHC is significantly higher in lung cancerous tissues than that in paracancerous tissues (Fig. 1).
Figure 1.
High expression of LDHC in patients with lung cancer. (A) The selected positive expressions vs. negative expressions under magnification of 200 times, the brown color indicates the positive staining. (B) Statistical analysis of the expression of LDHC in cancerous and paracancerous tissues of the 93 patients. There were 75 positive samples and 18 negative samples from cancerous tissues, while 19 positive samples and 68 negative samples from paracancerous tissues. The rates of positive LDHC expression are 80.6% and 21.8% in cancerous and paracancerous samples, respectively, which indicates that the expression of LDHC is significantly higher in lung cancerous tissues than that in paracancerous tissues (χ2 = 18.710, ∗∗∗∗P < 0.001).
3.2. Overexpression of LDHC promotes tumor growth in mice
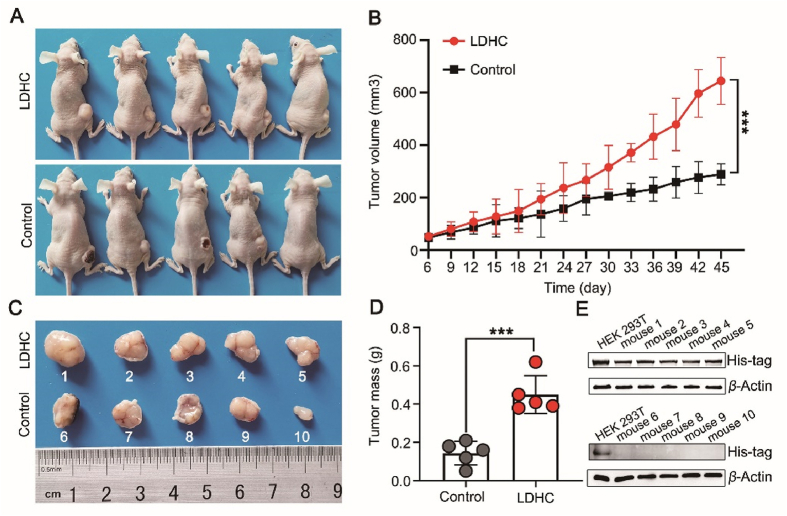
To study the biological role of LDHC overexpression in cancer tumorigenesis, we constructed a stable cell line of A549 overexpressing LDHC and inoculated it subcutaneously in nude mice (Fig. 2A). The volume and mass of the tumors overexpressing LDHC are significantly higher than that of the control tumors on the 45th day from inoculation (Fig. 2B–D), indicating that overexpression of LDHC promoted the A549 tumor growth in nude mice. The presence of the LDHC fused with a His6-tag in tumors were identified by Western blot (Fig. 2E).
Figure 2.
Overexpression of LDHC promotes tumor growth in mice. (A) The nude mice inoculated with A549 cells overexpressing LDHC (upper) and with control A549 cells (lower), the picture was taken on the 45th day after inoculation. (B) Tumor growth curves of the LDHC-overexpression group (red) and the control group (black). The volumes of the tumors overexpressing LDHC are significantly higher than that of the control tumors (n = 5, data are mean ± SD, ∗∗∗P < 0.001). (C) The picture of the tumors from the LDHC-overexpression group and the control group with a ruler to show the diameters of the tumors. (D) Statistical analysis of tumor mass. The masses of the tumors overexpressing LDHC are significantly higher than that of the control tumors (n = 5, data are mean ± SD, ∗∗∗P < 0.001). (E) Detection of His6-tag of the recombinant LDHC in tumors by Western blot. Mice 1 to 5 were inoculated with the LDHC-overexpression A549 cells, while mice 6 to 10 were inoculated with the control A549 cells. HEK 293 cells were used as the positive control.
3.3. Knock-down of LDHC expression inhibits viability and proliferation of A549 cells
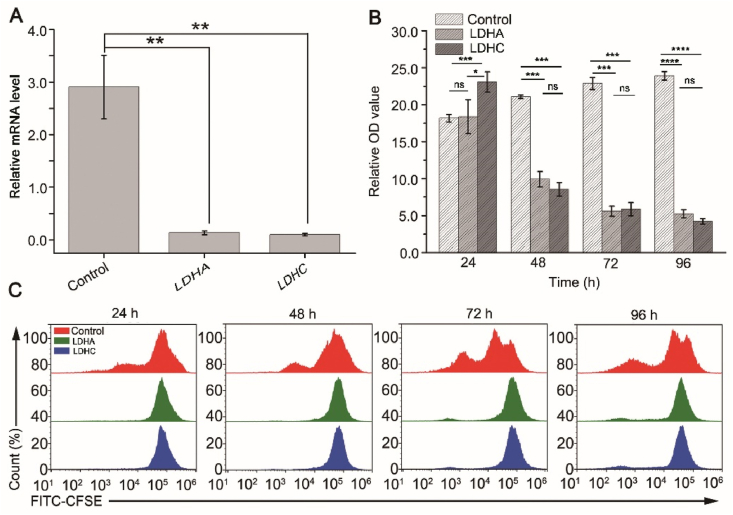
To evaluate the down-regulation effects of LDHC, we identified two siRNAs (s350 for LDHA and s8129 for LDHC) that significantly repressed the expression of LDHA and LDHC in A549 cells by real-time PCR (Fig. 3A). The knock-down of LDHA significantly inhibited the viability and proliferation of A549 cells as indicated by MTT (Fig. 3B) and flow cytometry (Fig. 3C). The inhibition of viability was significant on the 48th hour after transfection and lasted for at least 96 h, while the inhibition of proliferation was significant on the 24th hour after transfection and lasted for at least 96 h. These results are consistent with previous reports for LDHA14,37. It is intriguing that the knock-down of LDHC of A549 cells showed very similar results as that of LDHA (Fig. 3B and C), indicating that LDHC may also play a key role in the metabolism of A549 cells.
Figure 3.
Knock-down of LDHC inhibits the proliferation and growth of A549 cells. (A) Detection of the expressions of LDHA and LDHC by quantitative PCR. The expressions of LDHA and LDHC of the knock-down groups are significantly lower than that of the control groups (n = 3, data are mean ± SD, ∗∗P < 0.01). (B) Detection of cell viability by MTT assay. Data are mean ± SD, n = 3 (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001; ns, no significant difference). (C) Detection of cell proliferation by flow cytometry.
3.4. Comparison of the activities of human LDH isoforms
To further characterize the properties of LDHC, we then compared the activities of different human LDH isoforms. LDHA and LDHB were easily purified from crude extracts with high yields, while the yield of the recombinant LDHC was very low due to its low solubility. The three purified recombinant LDH proteins were all active tetramers (Supporting Information Fig. S1, hereafter referred to as LDHA4, LDHB4, and LDHC4) that displayed a single band on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), with an apparent subunit molecular weight of ∼35 kDa, consistent with the molecular weights calculated from the amino acid sequences (Supporting Information Fig. S2).
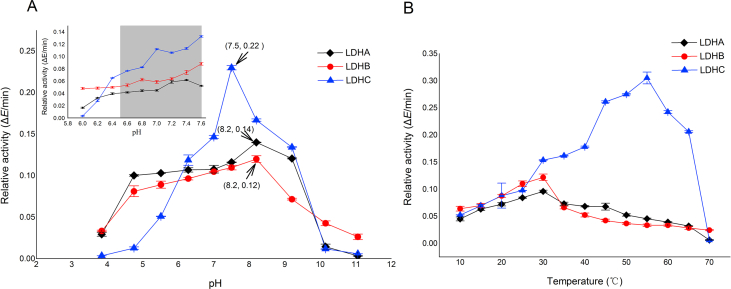
Besides the low solubility of LDHC4 compared with that of LDHA4 and LDHB4, the optimum pH of 7.5 for LDHC4 was significantly different from 8.2 for the other two isoforms. The optimum pH determination showed that LDHC4 has a pH value of 7.5 compared to 8.2 of both LDHA4 and LDHB4. LDHC4 also had the highest activities at the pH range of 6.4–7.6 (Fig. 4A) among the three LDH isoforms. The enzyme kinetic analysis showed that the LDHC4 had the highest values of Kcat/Km both for pyruvate and NADH among the three isoforms (Table 1). The Kcat/Km of LDHC4 for NADH was 2.4-fold higher than that of LDHA4 and 5.9-fold higher than that of LDHB4. The Kcat/Km of LDHC4 for pyruvate was about 3.3-fold higher than that of LDHA4 and 2.1-fold higher than that of LDHB4, which indicate that LDHC4 had the highest catalyzing efficiency among the three isoforms.
Figure 4.
Properties of LDH isoforms. (A) Optimal pH of the LDH isoforms. Glycine-HCl and glycine-NaOH buffers were used to determine the optimal pH. The result shows that LDHC4 has an optimum pH value of 7.5 compared to 8.2 of both LDHA4 and LDHB4. LDHC4 also had the highest activities at the pH range 6.4–7.6 among the three LDH isoforms. (B) Thermal stability of LDH isoforms. The highest activity of LDHC4 achieved at 55 °C, while the highest activities of LDHA4 and LDHB4 were both achieved at 30 °C, indicating that LDHC4 is much more thermally stable than LDHA4 and LDHB4. The catalytic activity was measured in 100 mmol/L Tris-Cl, pH 7.5. The LDH proteins concentration used in each reaction were all 0.78 μg/mL. Each spot represents an average value of triplicate experiments (n = 3, data are mean ± SD).
Table 1.
Kinetic parameters of LDH isozyme (100 mmol/L Tris-Cl, pH7.4).
| Kinetic parameter | LDHA4 |
LDHB4 |
LDHC4 |
|||
|---|---|---|---|---|---|---|
| Pyruvate | NADH | Pyruvate | NADH | Pyruvate | NADH | |
| Vmax (μmol/L∙s) | 0.61 ± 0.03 | 0.65 ± 0.01 | 0.75 ± 0.02 | 0.77 ± 0.04 | 1.09 ± 0.04 | 0.29 ± 0.01 |
| Km (μmol/L) | 53.91 ± 7.84 | 16.38 ± 1.43 | 42.83 ± 3.51 | 48.39 ± 6.78 | 28.89 ± 4.19 | 3.05 ± 0.01 |
| Kcat (1/s) | 153.46 ± 7.55 | 163.52 ± 2.52 | 188.45 ± 5.03 | 193.48 ± 10.05 | 272.00 ± 9.98 | 72.37 ± 2.50 |
| Kcat/Km (L/μmol∙s) | 2.85 | 9.98 | 4.40 | 4.00 | 9.42 | 23.73 |
Besides the canonical substrate pyruvate (Supporting Information Fig. S3A), we also evaluated the activities of the three human LDH isoforms against substrate analogues with longer carbon chains and found that LDHC had a higher activity than the other two isoforms (Fig. S3B). This suggests that substrates with a longer carbon chain exhibited specificity to LDHC.
Thermal stability assay showed that LDHC4 was much more thermally stable than LDHA4 and LDHB4 (Fig. 4B). The activity of LDHC4 increased along with the increasing temperature, and the highest activity of LDHC4 achieved at 55 °C, while the highest activities of LDHA4 and LDHB4 were both achieved at 30 °C.
3.5. EAA shows the modest selective inhibition against LDHC4 over LDHA4 and LDHB4
It has been reported that oxamate is a non-selective inhibitor of mouse LDH, while N-substituted oxamic acids are selective inhibitors of mouse LDHC438. Since substrates with a long chain exhibited specificity to LDHC (Fig. S3B), here we evaluated the inhibition specificity of a long-chain N-substituted oxamic acid, (ethylamino) (oxo)acetic acid (EAA), against human LDH isoforms (Fig. S3A). We determined the inhibition constants (Ki) of EAA against human LDH isoforms, which were 6.79, 4.89, and 0.53 mmol/L for LDHA4, LDHB4, and LDHC4, respectively. (Ethylamino) (oxo)acetic acid shows about 10 times selective inhibition against LDHC4 over LDHA4 and LDHB4 base on the Ki values. The kinetic analysis of human LDHC inhibition by EAA suggested that EAA exhibited a mixed inhibition pattern against LDHC4 (Supporting Information Fig. S4). We also tested the effects of EAA on the growth of lung cancer cell A549, and found that 5 mmol/L EAA significantly inhibited the cell growth rate, while had no effects on the cell apoptosis (Supporting Information Fig. S5).
3.6. Crystal structure of human LDHC4
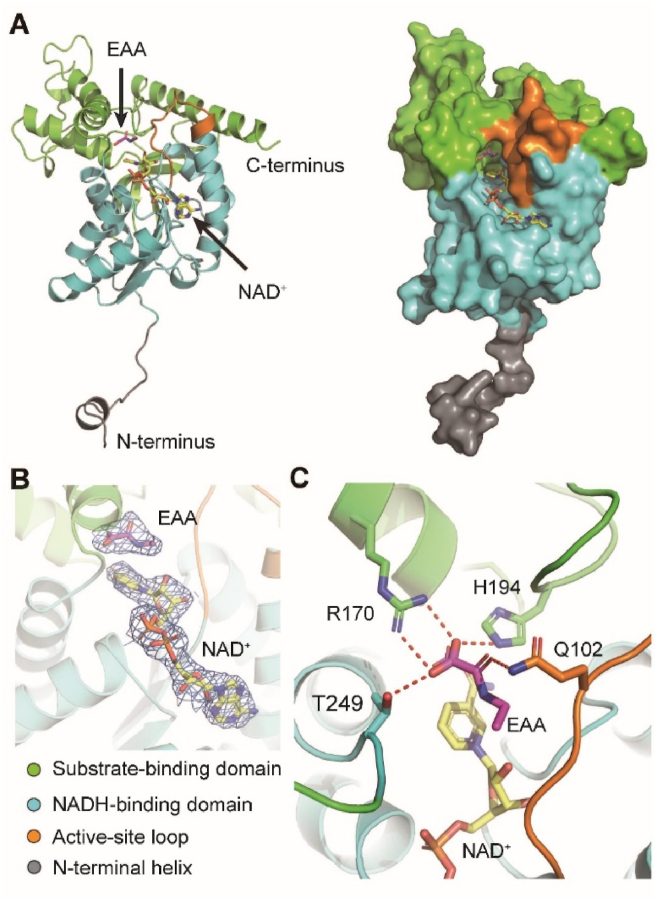
To elucidate the structural basis of LDHC and how the inhibitor EAA binds to LDHC, we used X-ray crystallography to solve the complex structure of LDHC and EAA. We introduced two mutations (L56E and I332Q) to human LDHC to increase its solubility and facilitate crystallization. We have solved the crystal structure of human LDHC4 complexed with NAD+ and the inhibitor EAA at 3.0 Å resolution (Fig. 5A). The clear density map enabled us to determine the position and orientation of the cofactor NAD+ and the small-molecule inhibitor EAA unambiguously (Fig. 5B). The inhibitor EAA made hydrogen bonds with Q102 from active-site loop, R170 and H194 from substrate-binding domain, and T249 from NADH-binding domain (Fig. 5C).
Figure 5.
Structure of human LDHC complexed with NAD+ and (ethylamino) (oxo)acetic acid (EAA). (A) A protomer of human LDHC is shown as cartoon and surface, respectively. The cofactor NAD+ (yellow) and inhibitor EAA (purple) are shown as sticks. (B) The Fo-Fc omit maps of NAD+ and EAA are colored blue and contoured at 3 σ. (C) Interactions between EAA and LDHC. Residues that make hydrogen bonds with EAA are shown as sticks and labeled. Hydrogen bonds are drawn as red dashed lines.
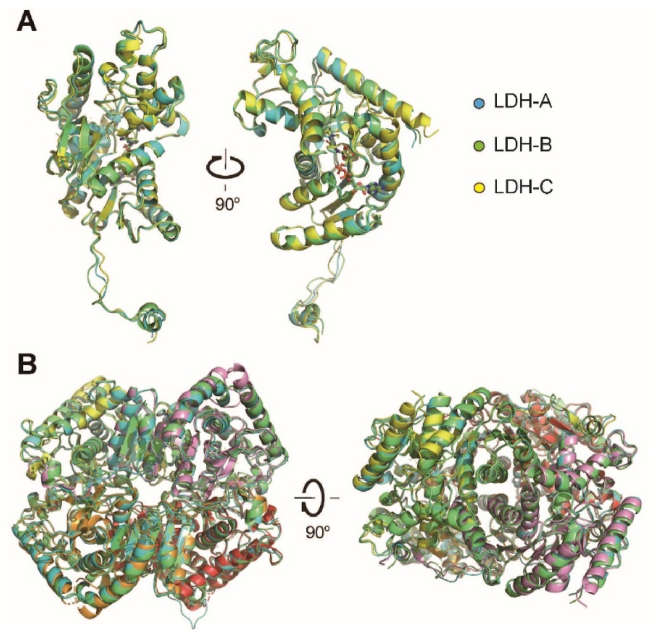
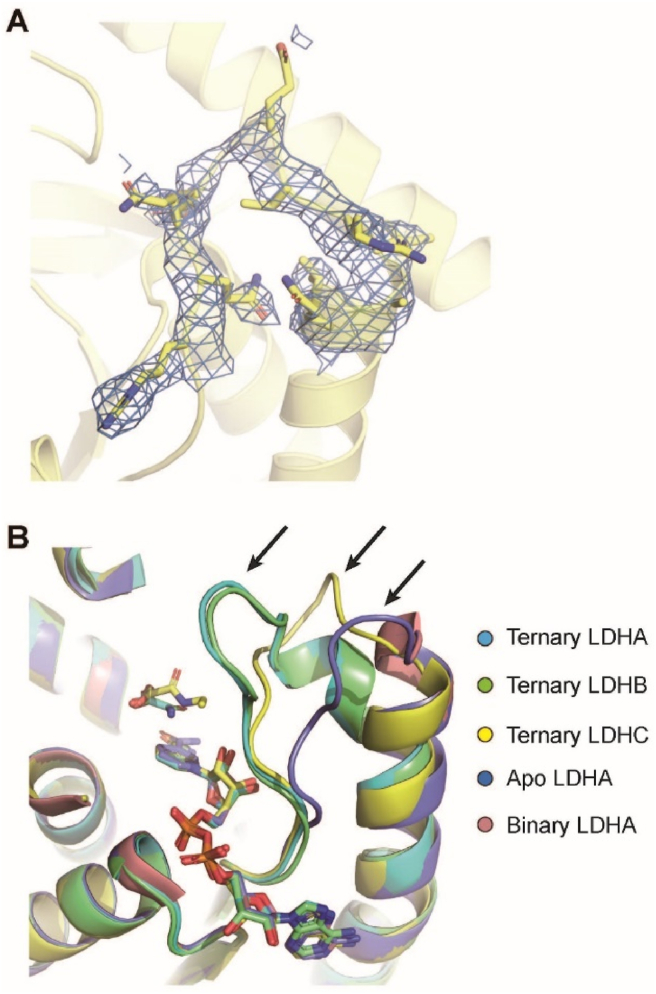
In human, LDHC shares 75.4% and 69.7% protein sequence identity with LDHA and LDHB, respectively (Supporting Information Fig. S6). Although the two mutations (L56E and I332Q) are away from the active site of LDHC, they seemed to change the kinetic properties of LDHC4 to some extent (Supporting Information Fig. S7), which may be due to the changes of LDHC surface electrostatic potentials. The two mutations are unlikely to affect LDHC structure, since the determined crystal structure of LDHC with mutations adopted a very similar fold and tetramer conformation as LDHA and LDHB (Fig. 6). Comparison of the ternary complex structures of human LDHA4, LDHB4 and LDHC4 showed that the root-mean-square deviation over 270 Cα atoms is less than 0.5 Å for each pairwise comparison of the three isoforms. The binding sites for cofactor and inhibitor are very similar for all the three isoforms, and the cofactors and inhibitors in all the three crystal structures could be well superimposed (Fig. 7). The major conformational differences occurred at the active-site loop (Fig. 7). The active-site loops in the ternary complex of LDHA and LDHB shared an identical closed conformation2, while that of apo or NADH-bound binary complex LDHA adopted an open conformation39. However, in the ternary complex structure of LDHC, the active-site loop adopted a distinct conformation (Fig. 7).
Figure 6.
Comparison of crystal structures of human LDHA, LDHB and LDHC. The crystal structures of the three human LDH isoforms are compared. LDHA (PDB No. 1I10), LDHB (PDB No. 1I0Z) and LDHC are colored in cyan, green and yellow, respectively. (A) Superpositions of the protomers of three LDH isoforms. The cofactors and substrate-like inhibitors are shown as sticks. (B) Superpositions of the tetramers of three LDH isoforms. For clarity, the cofactors and substrate-like inhibitors are not shown. To underline the tetrameric architecture, the four subunits of LDHC are colored differently.
Figure 7.
Comparison of the active-site loops of human LDH isoforms. (A) The 2Fo-Fc map of LDHC active-site loop is colored blue and contoured at 1 σ. (B) The crystal structures of ternary complex of human LDHA (PDB No. 1I10), LDHB (PDB No. 1I0Z) and LDHC, apo LDHA (PDB No. 4L4R) and binary complex of LDHA (PDB No. 4L4S) are superimposed. The cofactors and substrate-like inhibitors are shown as sticks. The active-site loops are indicated by arrows.
4. Discussion
LDHC is expressed in a variety of cancer cells. Furthermore, like the role of LDHA in the aerobic glycolysis of cancer cells, LDHC catalyzes the key step of aerobic glycolysis in spermatozoa, which is thought to be one of the major ATP sources of spermatozoa motility8. Here, we set out to identify if LDHC is a potential target for cancer therapy.
Analysis of LDHC expression in cancerous and paracancerous tissues from lung cancer patients suggested that the LDHC was indeed upregulated in lung cancer tissues (Fig. 1). Then, we further studied the effects of overexpression or know-down of LDHC on lung cancer. Our results indicated that the dysregulation of LDHC in lung cancer was associated with the tumor proliferation (Figure 2, Figure 3). Accordingly, LDHC is a promising candidate diagnostic marker and therapeutic target for cancers. Furthermore, in normal condition, LDHC is characteristically confined to express in male testicular germ cells but not in adult somatic tissues, implying it is a potential target with selective therapeutic benefit.
From the point of view of cancer energy metabolism, the most significant features of LDHC4 while compared with LDHA4 and LDHB4 are the highest catalytic efficiency and the highest activity at pH range of 6.4–7.6 (Fig. 4). Generation of NAD+ is the key step for the continuance of glycolysis, and the high catalyzing efficiency of LDHC4 would be more favorable to the continuance of glycolysis than LDHA4 and LDHB4. Studies reported that some tumor cells have acidic extracellular microenvironments with pH values ranging 6.5–6.9, and slightly alkaline intracellular microenvironments with pH values ranging 7.2–7.540, 41, 42, 43. The relatively high catalytic activity of LDHC at this pH range would favor Warburg effect. The highest thermal stability of LDHC4 among the isoforms is also an interesting feature. LDHC4 homologs from other species also show better thermal stability than their isoforms44, 45, 46, which indicates the conservation of LDHC during evolution. However, there were no reports of relationship of thermal stability to the cellular function of LDHC4.
From the point of view of protein structure, the active-site loop of LDHC4 adopted a distinct conformation compared with that of LDHA4 and LDHB4 (Fig. 7), which will facilitate the design of specific inhibitors. We found that the inhibitor EAA owned a ∼10-fold higher selection against human LDHC4 over LDHA4 and LDHB4, which made a basis for the development of selective inhibitors targeting human LDHC.
5. Conclusions
In this study, we found that LDHC was up-regulated in lung cancer tissues, and overexpression of LDHC in mice promoted tumor growth. Furthermore, the knock-down of LDHC inhibited the proliferation of lung cancer A549 cell. Our results suggest that LDHC is a potential target for anticancer drug discovery. We also showed that the inhibitor EAA exhibited preferred inhibition against LDHC, which provided evidence for the feasibility of developing selective inhibitors targeting human LDHC. We determined the structure of human LDHC to elucidate the interactions between LDHC and EAA for the first time, and revealed an active-site loop with a distinct conformation compared to LDHA and LDHB, providing the structural basis to facilitate the structure-based rational drug design for human LDHC.
Acknowledgments
Funds were provided by National Natural Science Foundation of China (81602653) and the Fundamental Research Funds for the Central Universities, Southwest Minzu University (2020NYBPY13, China). We thank the staff from BL19U1 beamline of National Facility for Protein Science in Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility for assistance during data collection. The atomic coordinates and structure factors of human LDHC complexed with NAD+ and (ethylamino) (oxo)acetic acid have been deposited in the Protein Data Bank under the accession code 7EPM.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.12.002.
Contributor Information
Qinghua He, Email: demeatry@gmail.com.
Qiang Chen, Email: qiang_chen@scu.edu.cn.
Qinglian Zhang, Email: qlzhang80@163.com.
Author contributions
Qinghua He, Qiang Chen, and Qinglian Zhang designed the research, and wrote the manuscript. Hong Tan, Huali Wang, Jinhu Ma, and Hui Deng carried out the experiments and analyzed the data with input from Qiang Chen, Qinghua He, and Qinglian Zhang.
Conflicts of interest
The authors declare no conflict of interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Everse J., Kaplan N.O. Lactate dehydrogenases: structure and function. Adv Enzymol Relat Area Mol Biol. 1973;37:61–133. doi: 10.1002/9780470122822.ch2. [DOI] [PubMed] [Google Scholar]
- 2.Read J.A., Winter V.J., Eszes C.M., Sessions R.B., Brady R.L. Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase. Proteins. 2001;43:175–185. doi: 10.1002/1097-0134(20010501)43:2<175::aid-prot1029>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan N.O., Everse J., Admiraal J. Significance of substrate inhibition of dehydrogenases. Ann N Y Acad Sci. 1968;151:400–412. doi: 10.1111/j.1749-6632.1968.tb11903.x. [DOI] [PubMed] [Google Scholar]
- 4.Eventoff W., Rossmann M.G., Taylor S.S., Torff H.J., Meyer H., Keil W., et al. Structural adaptations of lactate dehydrogenase isozymes. Proc Natl Acad Sci U S A. 1977;74:2677–2681. doi: 10.1073/pnas.74.7.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberg E., Eddy E.M., Duan C., Odet F. LDHC: the ultimate testis-specific gene. J Androl. 2010;31:86–94. doi: 10.2164/jandrol.109.008367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coonrod S., Vitale A., Duan C., Bristol-Gould S., Herr J., Goldberg E. Testis-specific lactate dehydrogenase (LDH-C4; Ldh3) in murine oocytes and preimplantation embryos. J Androl. 2006;27:502–509. doi: 10.2164/jandrol.05185. [DOI] [PubMed] [Google Scholar]
- 7.Zinkham W.H., Blanco A., Clowry L.J., Jr. An unusual isozyme of lactate dehydrogenase in mature testes: localization, ontogeny, and kinetic properties. Ann N Y Acad Sci. 1964;121:571–588. doi: 10.1111/j.1749-6632.1964.tb14227.x. [DOI] [PubMed] [Google Scholar]
- 8.Odet F., Gabel S.A., Williams J., London R.E., Goldberg E., Eddy E.M. Lactate dehydrogenase C and energy metabolism in mouse sperm. Biol Reprod. 2011;85:556–564. doi: 10.1095/biolreprod.111.091546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanco A., Burgos C., Gerez de Burgos N.M., Montamat E.E. Properties of the testicular lactate dehydrogenase isoenzyme. Biochem J. 1976;153:165–172. doi: 10.1042/bj1530165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatenby R.A., Gillies R.J. Why do cancers have high aerobic glycolysis?. Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 11.Tseng P.L., Chen C.W., Hu K.H., Cheng H.C., Lin Y.H., Tsai W.H., et al. The decrease of glycolytic enzyme hexokinase 1 accelerates tumor malignancy via deregulating energy metabolism but sensitizes cancer cells to 2-deoxyglucose inhibition. Oncotarget. 2018;9:18949–18969. doi: 10.18632/oncotarget.24855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolappan S., Shen D.L., Mosi R., Sun J., McEachern E.J., Vocadlo D.J., et al. Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms. Acta Crystallogr D Biol Crystallogr. 2015;71:185–195. doi: 10.1107/S1399004714024791. [DOI] [PubMed] [Google Scholar]
- 13.Arseneault R., Chien A., Newington J.T., Rappon T., Harris R., Cumming R.C. Attenuation of LDHA expression in cancer cells leads to redox-dependent alterations in cytoskeletal structure and cell migration. Cancer Lett. 2013;338:255–266. doi: 10.1016/j.canlet.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 14.Sheng S.L., Liu J.J., Dai Y.H., Sun X.G., Xiong X.P., Huang G. Knockdown of lactate dehydrogenase A suppresses tumor growth and metastasis of human hepatocellular carcinoma. FEBS J. 2012;279:3898–3910. doi: 10.1111/j.1742-4658.2012.08748.x. [DOI] [PubMed] [Google Scholar]
- 15.Kim H.S., Lee H.E., Yang H.K., Kim W.H. High lactate dehydrogenase 5 expression correlates with high tumoral and stromal vascular endothelial growth factor expression in gastric cancer. Pathobiology. 2014;81:78–85. doi: 10.1159/000357017. [DOI] [PubMed] [Google Scholar]
- 16.Fantin V.R., St-Pierre J., Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 17.Xie H., Valera V.A., Merino M.J., Amato A.M., Signoretti S., Linehan W.M., et al. LDH-A inhibition, a therapeutic strategy for treatment of hereditary leiomyomatosis and renal cell cancer. Mol Cancer Therapeut. 2009;8:626–635. doi: 10.1158/1535-7163.MCT-08-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le A., Cooper C.R., Gouw A.M., Dinavahi R., Maitra A., Deck L.M., et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granchi C., Roy S., Giacomelli C., Macchia M., Tuccinardi T., Martinelli A., et al. Discovery of N-hydroxyindole-based inhibitors of human lactate dehydrogenase isoform A (LDH-A) as starvation agents against cancer cells. J Med Chem. 2011;54:1599–1612. doi: 10.1021/jm101007q. [DOI] [PubMed] [Google Scholar]
- 20.Koukourakis M., Tsolou A., Pouliliou S., Lamprou I., Papadopoulou M., Ilemosoglou M., et al. Blocking LDHA glycolytic pathway sensitizes glioblastoma cells to radiation and temozolomide. Biochem Biophys Res Commun. 2017;491:932–938. doi: 10.1016/j.bbrc.2017.07.138. [DOI] [PubMed] [Google Scholar]
- 21.Flack M.R., Pyle R.G., Mullen N.M., Lorenzo B., Wu Y.W., Knazek R.A., et al. Oral gossypol in the treatment of metastatic adrenal cancer. J Clin Endocrinol Metab. 1993;76:1019–1024. doi: 10.1210/jcem.76.4.8473376. [DOI] [PubMed] [Google Scholar]
- 22.Deiab S., Mazzio E., Messeha S., Mack N., Soliman K.F. High-throughput screening to identify plant derived human LDH-A inhibitors. Eur J Med Plants. 2013;3:603–615. doi: 10.9734/ejmp/2013/5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koslowski M., Türeci O., Bell C., Krause P., Lehr H.A., Brunner J., et al. Multiple splice variants of lactate dehydrogenase C selectively expressed in human cancer. Cancer Res. 2002;62:6750–6755. [PubMed] [Google Scholar]
- 24.Yen C.C., Liang S.C., Jong Y.J., Chen Y.J., Lin C.H., Chen Y.M., et al. Chromosomal aberrations of malignant pleural effusions of lung adenocarcinoma: different cytogenetic changes are correlated with genders and smoking habits. Lung Cancer. 2007;57:292–301. doi: 10.1016/j.lungcan.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Cui Z., Chen Y., Hu M., Lin Y., Zhang S., Kong L., et al. Diagnostic and prognostic value of the cancer-testis antigen lactate dehydrogenase C4 in breast cancer. Clin Chim Acta. 2020;503:203–209. doi: 10.1016/j.cca.2019.11.032. [DOI] [PubMed] [Google Scholar]
- 26.Cui Z., Li Y., Gao Y., Kong L., Lin Y., Chen Y. Cancer-testis antigen lactate dehydrogenase C4 in hepatocellular carcinoma: a promising biomarker for early diagnosis, efficacy evaluation and prognosis prediction. Aging. 2020;12:19455–19467. doi: 10.18632/aging.103879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas R., Shaath H., Naik A., Toor S.M., Elkord E., Decock J. Identification of two HLA-A∗0201 immunogenic epitopes of lactate dehydrogenase C (LDHC): potential novel targets for cancer immunotherapy. Cancer Immunol Immunother. 2020;69:449–463. doi: 10.1007/s00262-020-02480-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naik A., Decock J. Targeting of lactate dehydrogenase C dysregulates the cell cycle and sensitizes breast cancer cells to DNA damage response targeted therapy. Mol Oncol. 2021 doi: 10.1002/1878-0261.13024. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong L., Du W., Cui Z., Wang L., Yang Z., Zhang H., et al. Expression of lactate dehydrogenase C in MDA-MB-231 cells and its role in tumor invasion and migration. Mol Med Rep. 2016;13:3533–3538. doi: 10.3892/mmr.2016.4963. [DOI] [PubMed] [Google Scholar]
- 30.Moore N., Houghton J., Lyle S. Slow-cycling therapy-resistant cancer cells. Stem Cell Dev. 2012;21:1822–1830. doi: 10.1089/scd.2011.0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H., Naismith J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91–101. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W.Z., Tang J.C., Wang S.S., Wang Z.J., Qin W.M., He J.H. The protein complex crystallography beamline (BL19U1) at the Shanghai Synchrotron Radiation Facility. Nucl Sci Tech. 2019;30:170–181. [Google Scholar]
- 33.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. HKL-3000: the integration of data reduction and structure solution-from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 34.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao F., Zhao T., Zhong C., Zhu J., Zhao H. LDHA is necessary for the tumorigenicity of esophageal squamous cell carcinoma. Tumour Biol. 2013;34:25–31. doi: 10.1007/s13277-012-0506-0. [DOI] [PubMed] [Google Scholar]
- 38.Rodríguez-Páez L., Chena-Taboada M.A., Cabrera-Hernández A., Cordero-Martínez J., Wong C. Oxamic acid analogues as LDH-C4-specific competitive inhibitors. J Enzym Inhib Med Chem. 2011;26:579–586. doi: 10.3109/14756366.2011.566221. [DOI] [PubMed] [Google Scholar]
- 39.Dempster S., Harper S., Moses J.E., Dreveny I. Structural characterization of the apo form and NADH binary complex of human lactate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2014;70:1484–1490. doi: 10.1107/S1399004714005422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estrella V., Chen T., Lloyd M., Wojtkowiak J., Cornnell H.H., Ibrahim-Hashim A., et al. Acidity generated by the tumor microenvironment drives local invasion. Cancer Res. 2013;73:1524–1535. doi: 10.1158/0008-5472.CAN-12-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mishra D., Banerjee D. Lactate dehydrogenases as metabolic links between tumor and stroma in the tumor microenvironment. Cancers. 2019;11:750–771. doi: 10.3390/cancers11060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gillies R.J., Raghunand N., Karczmar G.S., Bhujwalla Z.M. MRI of the tumor microenvironment. J Magn Reson Imag. 2002;16:430–450. doi: 10.1002/jmri.10181. [DOI] [PubMed] [Google Scholar]
- 43.Persi E., Duran-Frigola M., Damaghi M., Roush W.R., Aloy P., Cleveland J.L., et al. Systems analysis of intracellular pH vulnerabilities for cancer therapy. Nat Commun. 2018;9:2997–3008. doi: 10.1038/s41467-018-05261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee C.Y., Yuan J.H., Goldberg E. Lactate dehydrogenase isozymes from mouse. Methods Enzymol. 1982;89:351–358. doi: 10.1016/s0076-6879(82)89063-0. [DOI] [PubMed] [Google Scholar]
- 45.Schatz L., Segal H.L. Reduction of alpha-ketoglutarate by homogeneous lactic dehydrogenase X of testicular tissue. J Biol Chem. 1969;244:4393–4397. [PubMed] [Google Scholar]
- 46.Zhang Q., Yang M., Zhao Y., Zhang S., He Q., Meng X., et al. Cloning and characterization of lactate dehydrogenase C4 from pika ochotona curzoniae. Mol Biol (Mosk) 2014;48:124–132. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.