Abstract
Head and neck squamous cell carcinoma (HNSCC) is one of the most common human cancers; however, its outcome of pharmacotherapy is always very limited. Herein, we performed a batch query in the connectivity map (cMap) based on bioinformatics, queried out 35 compounds with therapeutic potential, and screened out parbendazole as a most promising compound, which had an excellent inhibitory effect on the proliferation of HNSCC cell lines. In addition, tubulin was identified as a primary target of parbendazole, and the direct binding between them was further verified. Parbendazole was further proved as an effective tubulin polymerization inhibitor, which can block the cell cycle, cause apoptosis and prevent cell migration, and it exhibited reasonable therapeutic effect and low toxicity in the in vivo and in vitro anti-tumor evaluation. Our study repositioned an anthelmintic parbendazole to treat HNSCC, which revealed a therapeutic utility and provided a new treatment option for human cancers.
Keywords: HNSCC, cMap, Bioinformatics, Parbendazole, Drug repositioning, Anti-tumor
Graphical abstract
In this work, with the help of cMap, anthelmintic parbendazole was repositioned as an anti-head and -neck squamous cell carcinoma, and its mechanism of action was studied.

1. Introduction
Head and neck squamous cell carcinoma (HNSCC) is an epithelial malignancy originating from the mucosal epithelium in the mouth, pharynx, and larynx as the most common malignant tumor that arises in the head and neck. Considering that more than 600,000 new cases are diagnosed yearly with less than 50% five-year survival rate, HNSCC has become the sixth most common human cancer worldwide. Clinic treatments of HNSCC mainly rely on surgical resection, chemoradiotherapy, and targeted therapy. In general, the HNSCC patients who receive the surgical resection and local radiation should undergo the systemic conservative treatment, in which the combination therapy with cetuximab, cisplatin, and 5-fluorouracil (5-FU) combination therapy is commonly used1, 2, 3. These chemotherapeutic or targeted drugs are often used as radiosensitizers and have very restricted direct effects against HNSCC. In addition to these routine treatments, the promising immune checkpoint blockade-based immunotherapy is also being developed, and two PD1 inhibitors, pembrolizumab and nivolumab, have been approved by US Food and Drug Administration (FDA). However, due to the high frequency of local recurrence or distant metastasis and considerable heterogeneity of HNSCC, the therapeutic outcome of those above-mentioned treatments is very limited4, 5, 6, 7. Fast discovery of novel and potent compounds that can fight against HNSCC efficiently will contribute much to improve the treatment and prognosis of HNSCC patients.
Given the numerous advantages in hit rate, development costs and research timelines, drug repositioning represents a considerable strategy in drug discovery and development8. Nowadays, as a rapid and convenient method to analyze gene expression and the corresponding interventions, bioinformatics is increasingly becoming popular in repositioning an “old” drug to its new applications9. The connectivity map (cMap), developed by Lamb et al.10 in 2006, is a large public database containing drugs and genes signatures, within which genes, drugs and diseases are connected. Subramanian et al.11,12 fully expanded cMap (L1000) in the dimensions of small molecule compounds (from 164 drugs to 19,811), genetic disturbance (knocking down and overexpressing 5075 genes), and cell lines (nine cell lines), and developed connectivity score to evaluate the relationship between query and disturbance. Querying one or a group of test gene signatures through the cMap database will result in a “connectivity score”. For small-molecule drugs, a positive connectivity score indicates that the corresponding compound in the cMap database induced the expression of the query signature; on the contrary, a negative connectivity score indicates that the corresponding compound inhibits the expression, which implies that the compound has therapeutic potential. So far, cMap has made good progress in the field of drug repositioning. For example, new small molecule compounds for the treatment of osteoporosis13,14, diabetes, muscle atrophy15,16, inflammatory bowel disease17, ischemic stroke18 and obesity19 have also been discovered through cMap. It should not be overlooked that cMap plays a positive role in cancer treatment. Some new compounds have been found to be used to treat gastric cancer20, hepatoblastoma21, multiple myeloma22, papillary renal cell carcinoma23, and other cancers24,25.
In this study, according to the differential gene expression data of HNSCC and normal tissue samples from Gene Expression Omnibus (GEO)26, the Cancer Genome Atlas (TCGA)27 and Genotype-Tissue Expression (GTEx)28,29, we screened out the differential expression genes (DEGs) between tumor tissues and adjacent normal tissues, searched for their potential therapeutic compounds in cMap, and analyzed the mechanisms of action using bioinformatics [Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes (KEGG)]. Through a preliminary screening of the queried compounds at the cellular level, we determined the anthelmintic drug parbendazole as a further research object, and identified tubulin as its target, studied the mechanism and the therapeutic effect on tumors, which suggests that parbendazole has the potential to become a therapeutic molecule with low toxicity for HNSCC.
2. Results
2.1. Screening of DEGs in HNSCC, cMap query and bioinformatics enrichment analysis
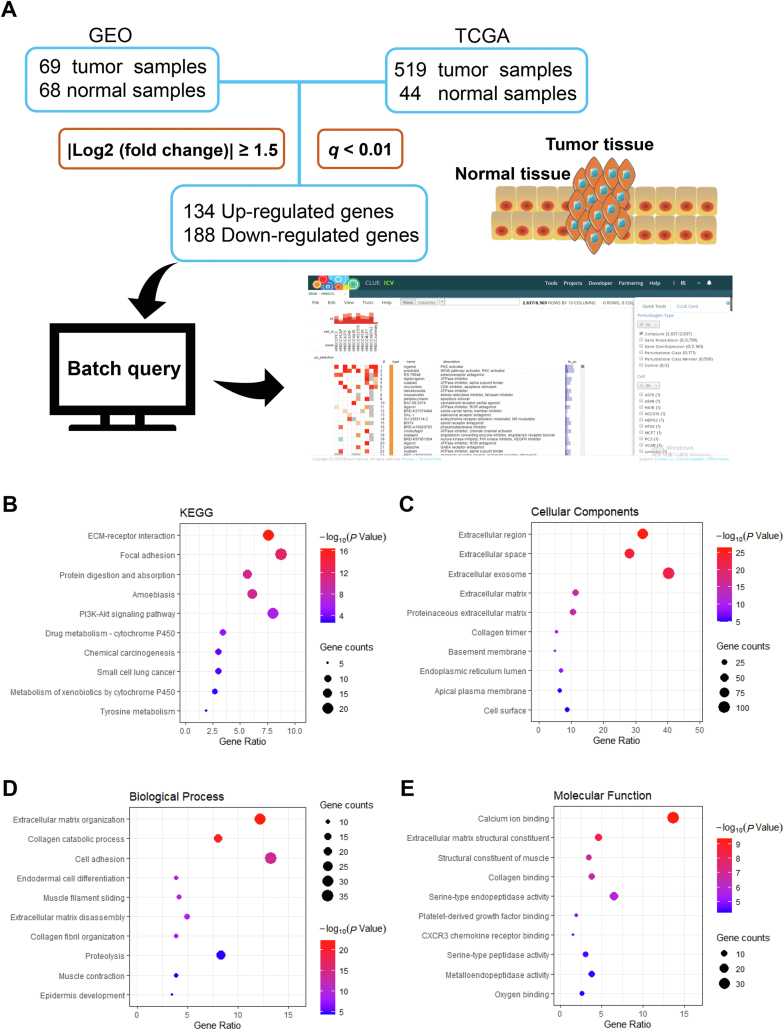
To achieve the drug repositioning screening through cMap, we investigated the DEGs between HNSCC and adjacent normal tissues. Four gene expression datasets (GSE6631, GSE13397, GSE58911 and GSE107591) were selected from the GEO database for GEO2R analysis, and totally 341 up-regulated genes and 543 down-regulated genes were collected. Also, GEPIA was utilized to compare the gene expression profiles of 519 tumor tissue samples and 44 normal tissue samples in TCGA and GTEx, in which 482 up-regulated genes and 329 down-regulated genes were involved. Through the further GO and KEGG enrichment analysis, we found that these DEGs mainly enriched in focal adhesion pathway and ECM-receptor interaction pathway (Fig. 1B–E), which are strongly correlated to cellular activities, including adhesion, migration, differentiation, proliferation, and apoptosis30,31. The intersected 134 up-regulated genes and 188 down-regulated genes (Supporting Information Table S1) from the above analyses were transferred as the signature to make a query in cMap to gain a list of small molecules that hold the potentials to treat HNSCC (Supporting Information Table S2).
Figure 1.
Screening of DEGs in HNSCC, cMap query and bioinformatics enrichment analysis. (A) is the usual procedure of cMap query; (B)‒(E) are KEGG and GO (CC, BP and MF) analysis of DEGs. The size of the dot represents the number of genes, and the color represents the P-value.
2.2. Parbendazole exhibits good inhibition on the proliferation of HNSCC cell lines (HN6, Fadu and CAL-27)
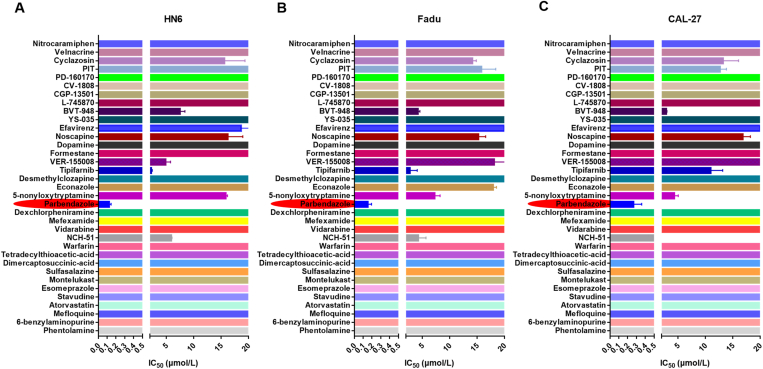
After obtaining the list of “old” drug molecules, we conducted a cell-based preliminary screening to verify their actual potency in inhibiting HNSCC cells. Three HNSCC cell lines, including HN6, Fadu, and CAL-27, were treated with these compounds for 72 h, respectively, followed by determining the cell viabilities using CCK8. As shown in Fig. 2, nine compounds from the list exhibited relatively low IC50 ranges, implying their potentials for the treatment of HNSCC (detailed data was shown in Supporting Information Table S3). Among them, the parbendazole is the most eye-catching owing to its highly excellent IC50 values on these three cell lines, which are 126, 153, and 270 nmol/L to HN6, Fadu, and CAL-27 cell lines, respectively. As we know, parbendazole belongs to the benzimidazole-based anthelmintics with a broad-spectrum activity32. Inspired by the fantastic performance of parbendazole on anti-HNSCC, we next investigated whether this type of benzimidazole derivatives have similar effects. Parbendazole and its 9 analogs were assessed in another series of 72 h cell viability assays. We found that the majority of these parbendazole analogs showed very similar in vitro anticancer activity with parbendazole, indicating that the active benzimidazole scaffold is amenable to various modification lines (Supporting Information Fig. S1A‒S1C). Compared to the HNSCC cell lines, the non-cancerous cell line, IMR90, was very tolerable for the treatment with parbendazole, demonstrating its high safety to normal tissues cell lines (Fig. S1D). In addition, parbendazole exhibited a broad-spectrum activity against various cancer types (Supporting Information Table S4). The chemical structure can be found in Fig. 3E.
Figure 2.
Summary of IC50 values of 35 small molecule compounds on three HNSCC cell lines (HN6, Fadu and CAL-27). (A)IC50 of compounds inhibiting the proliferation of HN6 cells; (B) IC50 of compounds inhibiting the proliferation of Fadu cells; (C) IC50 of compounds inhibiting the proliferation of CAL-27 cells. Cells were seeded in 96-well plate and treated with a series of concentrations of compounds for 72 h. Cell viability was evaluated by the CCK8 method. Data are represented as mean ± SEM (n = 3).
Figure 3.
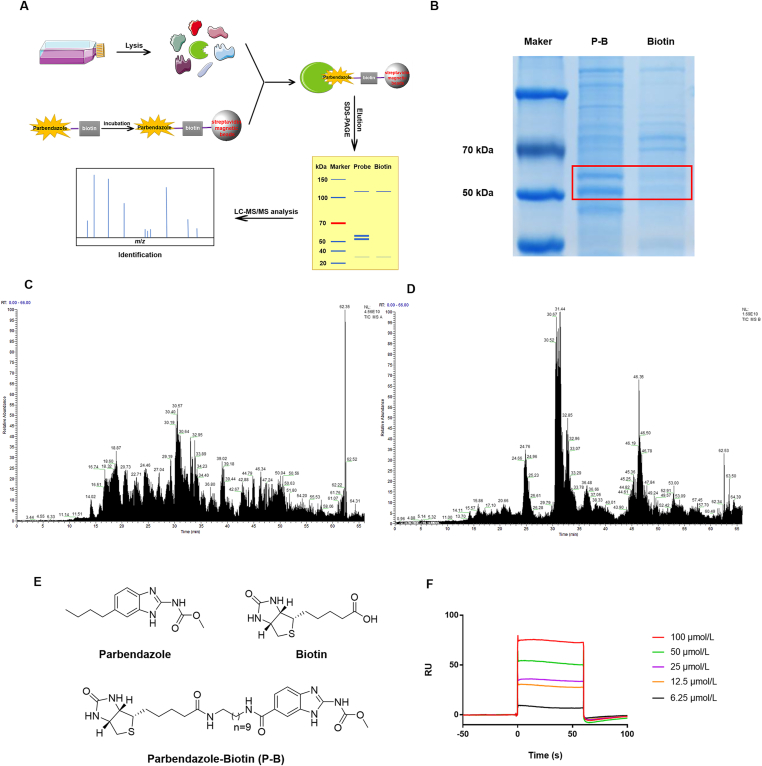
Identification of tubulin as a direct interacting protein of parbendazole. (A) is the process of protein capture and LC‒MS/MS analysis; (B) shows the differential bands. The lysates of HN6 cells were incubated with streptavidin magnetic bead-immobilized probe P–B or biotin in advance for 2 h at room temperature. Differential bands are separated by SDS-PAGE from proteomics, stained with Coomassie brilliant blue. (C) and (D) are total ion current chromatogram obtained by LC‒MS/MS of 55 kDa bands (C) and 52 kDa bands (D); (E) is chemical structures of parbendazole, biotin and parbendazole-biotin (P-B); (F) shows interactions of tubulin-coupled CM5 chip with parbendazole at different concentrations (6.25, 12.5, 25, 50 and 100 μmol/L).
2.3. Tubulin is the direct protein target of parbendazole
To confirm the direct target of parbendazole, we synthesized parbendazole-biotin (P-B) and performed a pull-down assay, accompanied by biotin as a negative control (Fig. 3C). The detailed synthetic route and steps can be found in Supporting Information Scheme S1. Based on the compound-centric chemical proteomics (CCCP) approach33,34 (Fig. 3A), HN6 cells lysates were incubated with streptavidin magnetic beads, which were preincubated with probe or biotin, respectively. Subsequently, the captured protein was eluted and separated by SDS-PAGE, followed by staining by Coomassie brilliant blue. Two clear bands at about 52 and 55 kDa were captured by P-B, not biotin alone (Fig. 3B). The LC‒MS/MS analysis and database search exhibited that the two bands match tubulin, with the highest scores. According to the scores, the top five of the two samples are listed respectively (Supporting Information Table S5). Furthermore, we verified this combination through an SPR assay (Fig. 3D). In short, we can basically confirm that tubulin is the target of parbendazole.
2.4. Parbendazole binds to tubulin at the colchicine binding site
Studies have shown that colchicine releases enhanced fluorescence when it binds to tubulin, and the competition between the compound and colchicine can reduce the fluorescence intensity of the colchicine–tubulin complex35,36. To study the binding site of parbendazole on tubulin, a competitive binding assay was performed. In detail, 5 μmol/L parbendazole and colchicine were incubated with tubulin together, combretastatin A-4 (CA-4) was used as a positive control, and DMSO was a negative control. It is shown that both parbendazole and CA-4 can reduce the fluorescence intensity of the system by competing with colchicine for the binding site (Fig. 4A). Therefore, the colchicine binding site can be considered as the direct binding site of parbendazole.
Figure 4.
Parbendazole binds to tubulin at the colchicine binding site. (A) shows the RFI after the compound and colchicine competitively bind to tubulin. Parbendazole or CA-4 was incubated with tubulin and colchicine at 37 °C for 30 min, and fluorescent intensity was measured. The fluorescence of colchicine and tubulin was set to 1 for normalization. RFI means relative fluorescent intensity. Data is represented as mean ± SEM (n = 3, ∗∗∗∗P < 0.0001); (B) and (C) are the final figures of ITC assay of CA-4 (B) and parbendazole (C). Tubulin (20 μmol/L) was in the sample and small molecule (100 μmol/L) was in the syringe. The fitting model was one set of sites; (D) shows small molecule could improve the thermal stability of tubulin in PBS. The temperature-programmed program was executed in the LC480, and the fluorescent signals were collected. The first derivative of the fluorescence emission was plotted as a function of temperature (−dF/dT). The inflection point of the curve is Tm.
In addition, isothermal titration calorimetry (ITC) assay was carried out based on measurement of the sum of all exothermic and endothermic heat generated in the interaction. As shown in Fig. 4B and C, the KD values of parbendazole and CA-4 are 3.12 and 1.17 μmol/L, respectively. In conclusion, the binding between parbendazole and tubulin is proved and calculated.
The application of environmentally sensitive dyes in protein folding is common37,38. In this section, SYPRO orange dye (λex 470 nm/λem 570 nm) was used to characterize the state of the protein during the heating process in a thermal shift assay. Compared with DMSO, the addition of parbendazole or CA-4 in PBS increased the melting temperature (Tm) of tubulin by 2.5 and 3.5 °C, respectively. These results indicate that parbendazole could increase the thermal stability of tubulin, reflecting the binding between parbendazole and tubulin (Fig. 4D).
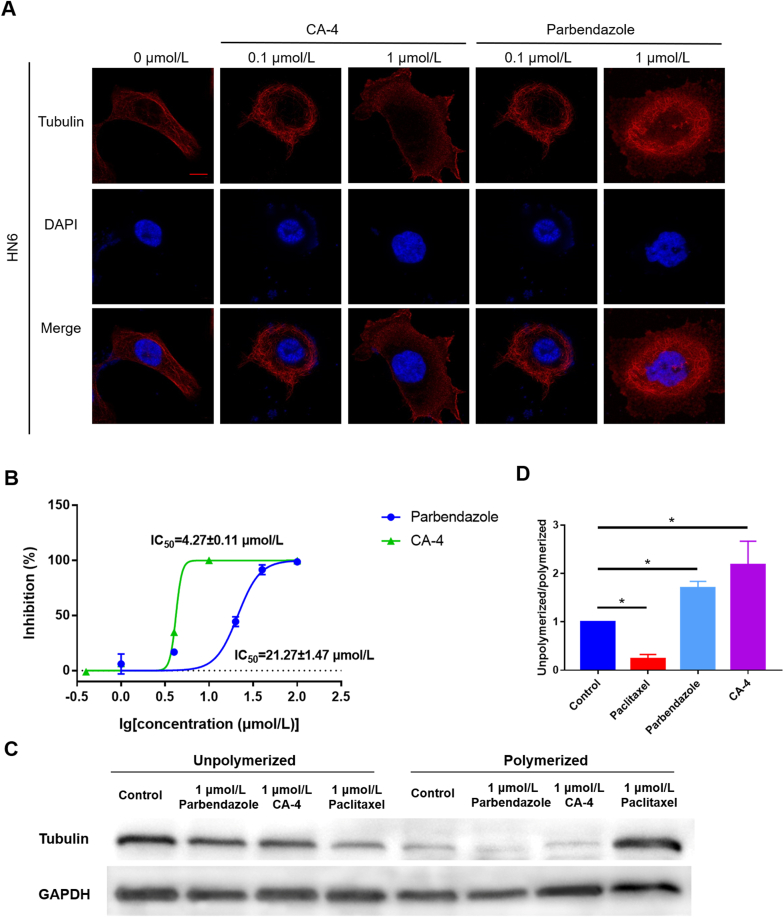
2.5. Parbendazole inhibits tubulin polymerization
So far, there are two types of drugs that target tubulin. One type inhibits the polymerization of unpolymerized tubulin, such as colchicine and vinblastine, while the other inhibits the depolymerization of polymerized tubulin, such as paclitaxel39. In order to clarify the effect of parbendazole on tubulin, we conducted a tubulin polymerization assay, and found that parbendazole inhibited tubulin polymerization just as CA-4 did (Fig. 5B). Subsequently, Western blotting was performed to evaluate the ratio of unpolymerized and polymerized tubulin in parbendazole-treated cells (Fig. 5C and D). The results showed that the ratios of unpolymerized tubulin to polymerized tubulin in parbendazole group, CA-4 group, paclitaxel group, and control group were significantly different (∗P < 0.05). More intuitively, the immunofluorescence assay allows us to clearly observe the effect of parbendazole and CA-4 on tubulin; that is, the regular microtubule structure in the cell was destroyed. After treatment with parbendazole or CA-4 for 24 h, cells lost their regular microtubule network structure compared with the control group (Fig. 5A).
Figure 5.
Parbendazole inhibits tubulin polymerization. (A) is the result of immunofluorescence assay which displays the microtubule structure of cells. Red (Alexa Fluor® 555) was used to mark tubulin, and blue (DAPI) was used to mark cell nuclei. Images were obtained by Laser Confocal Microscope (LSM780). The scale bar was 20 μm; (B) shows compounds inhibited tubulin polymerization in vitro. The spontaneous polymerization of tubulin in the absence of the compound was used as a negative control; (C) is the result of Western blotting of the amount of unpolymerizated and polymerizated tubulin; (D) shows unpolymerizated/polymerizated tubulin ratio. Data is represented as mean ± SEM (n = 3, ∗P < 0.05).
2.6. Parbendazole induces apoptosis, arrests the cell cycle and prevents cell migration
To investigate whether the inhibition ability of parbendazole on cell proliferation was related to its induction of cell apoptosis, we used flow cytometry to detect the effect of parbendazole on cell apoptosis. After parbendazole or CA-4 treatment for 24 h, the apoptotic ratio of HN6 cells increased compared with the control group (Fig. 6A). As the concentration increased, the proportion of apoptotic cells was more remarkable, and there was a significant difference between the control group and treated groups (Fig. 6B and C).
Figure 6.
Parbendazole induces apoptosis, arrests the cell cycle and prevents cell migration. (A) is the result of cell apoptosis by flow cytometry. HN6 cells were treated with or without compounds (0.1 or 1 μmol/L) for 24 h. Apoptotic cells were stained by Annexin-FITC. Representative dot plots of flow cytometry are shown; (B), (C) show the percentage of apoptotic cells of groups treated by CA-4 and parbendazole. Data is represented as mean ± SEM (n = 3, ∗∗P < 0.01, ∗∗∗P < 0.001); (D) is representative DNA distribution histogram of HN6 cells. HN6 cells were treated with or without compounds (0.1 or 1 μmol/L) for 24 h, the DNA content of cells was detected by PI staining and flow cytometry. The purple represents cells at the G1 phase (2N). The blue and red represent cells at (G2/M phase 4N and 8N); (E) is representative cell migration assay picture for HN6 cells treated with or without parbendazole or CA-4. HN6 cells were treated with or without compounds (0.1 or 1 μmol/L) for 24 h, and the pictures were captured at 0, 8 and 24 h under a Zeiss Axio Observer A1 fluorescence microscope (brightfield, objective lens: 10×).
In view of the irreplaceability of tubulin in cell division, and the disordered microtubule structure may block the cell cycle, we used flow cytometry to analyze the effect of the compound on the cell cycle of HN6 cells. Flow cytometry analysis showed that parbendazole could induce cell apoptosis and also block the cell cycle in the G2/M phase (4N). After treatment with 0.1 μmol/L parbendazole for 24 h, cells in the G2/M phase (4N) increased, and cells treated with CA-4 appeared 8-ploid (8N, G2/M phase); after treatment with 1 μmol/L compounds for 24 h, 2N disappeared, accompanied by an increase in 4N and 8N in both parbendazole and CA-4 groups (Fig. 6D).
Considering that cell migration plays an essential role in the tumor pathological process, the effect of parbendazole on HN6 cell migration was evaluated by cell migration assay (Fig. 6E). Cells were cultured in a normal growth medium containing compounds, and scratch healing was observed. The scratches gradually healed over time. In the control group, the scratches were almost completely healed after 24 h. In the test groups, with the increase of compound concentration, the degree of scratch healing was decreased.
2.7. Parbendazole is potent to inhibit HNSCC tumor growth in vivo
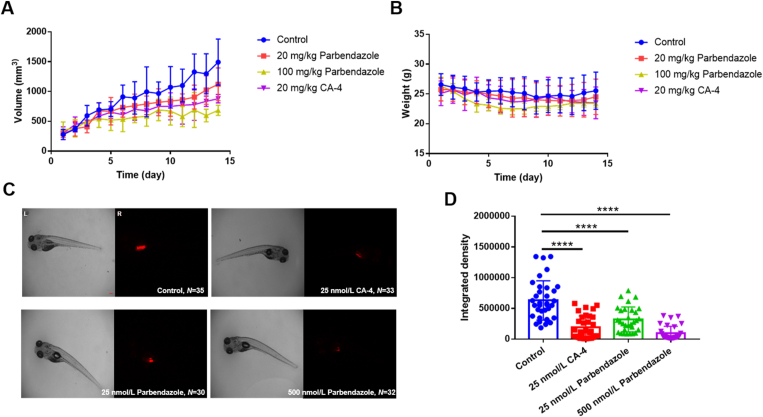
HN6 xenograft model was used to evaluate parbendazole anti-tumor efficacy. In this study, parbendazole showed dose-dependent tumor growth inhibition without altering the bodyweight of mice (Fig. 7A and B), indicating its excellent in vivo anti-tumor efficacy and low toxicity. Furthermore, we further conducted another evaluation of in vivo anticancer activity by using a zebrafish bearing HN6 xenograft and observed a good therapeutic effect of parbendazole as well (Fig. 7C and D). HN6 cells were stained with CM-Dil, which would emit intense red fluorescence, the fluorescence intensity can be calculated, and the therapeutic effect can be evaluated. After treatment with parbendazole, the fluorescence in the zebrafish was significantly weaker than that of the control group, indicating the reduction of tumor cells. Interestingly, parbendazole disclosed the same obvious curative effect at the nanomolar level as CA-4.
Figure 7.
Parbendazole shows good anti-tumor activity in vivo. (A) is the tumor volume of mice; (B) is the body weight of mice. The xenograft tumor mice were randomly divided into four groups (n = 5 for each group) and given different doses of CA-4, parbendazole or solvent, once a day by intragastric administration. The tumor volumes were measured by an electronic caliper, and the body weight of mice were measured by an electronic balance. Results are expressed as mean ± SD; (C) indicates the therapeutic effect of papendazole on zebrafish bearing xenografts. The zebrafish bearing xenografts were randomly divided into four groups (n ≥ 30 for each group) and were administered different doses of CA-4, parbendazole, or solvent alone. Images were obtained by an OLYMPUS SZX16 microscope, and the fluorescent integrated density was measured by Image J software. The scale bar is 100 μmol/L; (D) is the fluorescent integrated density of each group of zebrafish. Graphs were draw by GraphPad Prism 7.00. Results are expressed as mean ± SD (∗∗∗∗P < 0.0001).
2.8. Parbendazole is at the level of practical non-toxicity
Whether a compound can be used as a drug depends not only on its therapeutic efficacy but also on its low toxicity. Generally, the more active the compound, the more toxic it is, and researchers need to balance the relationship between the two. The acute toxicity of colchicine and its analogs to humans is one of their limitations40; therefore, in this study, we tested the acute toxicity of parbendazole to determine whether it has the potential to become a drug. SPF Kunming mice were given different doses of parbendazole by gavage, and their weights and survival were recorded. It can be seen from the survival curve, and weight change that the highest dose of 10 g/kg is more toxic to mice, and mice were given other doses survive normally (Fig. 8). The LD50 is greater than 5 g/kg, which is at the level of practical non-toxicity, indicating the safety of parbendazole. In addition, after human embryonic lung fibroblasts (IMR-90) were treated with parbendazole for 72 h, the cell viability was evaluated by the CCK8 method, and the IC50 value was greater than 20 μmol/L, presenting its safety against normal human cells (Fig. S1D).
Figure 8.
Parbendazole is at the level of practical non-toxicity. (A) is the survival curve change of mice, all the mice in the first nine groups survived, and their survival curves overlapped. (B) is the survival weight change of mice. Mice were randomly grouped and given the drug (0, 0.1, 0.316, 1, 3.16, and 10 g/kg) by gavage. M means male, and F means female. The design and implementation of the experiment are carried out following the rules of the Korbor method described in GB 15193.3-2014.
2.9. Parbendazole inhibits the expression of tumor microenvironment-related ECM-receptor interaction pathway genes (verification of DEGs)
To verify whether the DEGs we screened out is consistent with the actual and whether parbendazole could reverse these changes, we performed a qPCR assay to detect the gene expression at the mRNA level. According to the KEGG analysis, the number of DEGs enriched by the ECM-receptor interaction pathway and focal adhesion pathway was the largest, so we initially selected these genes, which are up-regulated in HNSCC for verification. Overall, after treated with parbendazole for 24 h, 25 genes were down-regulated (P < 0.05) (Supporting Information Fig. S2A), the expression of 4 genes indicated no significant difference (Fig. S2B), and 2 genes were up-regulated (Fig. S2C) in expression. Thus, the DEGs are credible and reasonable, and parbendazole has been proven to reverse these differences, especially in the expression of the ECM-receptor interaction pathway and focal adhesion pathway, indicating that parbendazole can treat cancers by inhibiting the expression of genes related to the tumor microenvironment.
2.10. Parbendazole and cisplatin have synergistic or additive effects
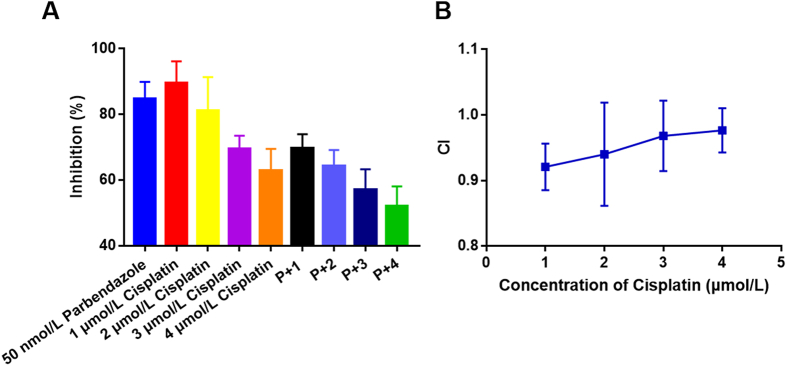
Cisplatin is a commonly used drug for chemotherapy of HNSCC in the clinic, and a cell proliferation assay was performed to evaluate the effect of cisplatin in combination with parbendazole by combination indexes (CIs). At a certain range of concentrations (50 nmol/L parbendazole, 1, 2, 3 or 4 μmol/L cisplatin), CI < 1 (Fig. 9B), indicating that the two drugs have a synergistic or additive effect. In practical applications, their combined use can reduce the dose of drugs used alone.
Figure 9.
Parbendazole and cisplatin have synergistic or additive effects. Cell viability was tested by the CCK8 method after treated with parbendazole or cisplatin alone or their combination (50 nmol/L parbendazole, 1, 2, 3 or 4 μmol/L cisplatin) for 72 h. Data are represented as mean ± SEM, n = 3. . CI > 1, antagonism. CI = 1, additive effect. CI < 1, synergy. P + n means 50 nmol/L parbendazole + n μmol/L cisplatin (n = 1, 2, 3, 4).
3. Discussion
HNSCC is one of the most common human cancers, and its treatment remains a dilemma41,42. In recent years, cMap based on bioinformatics has become an effective means of drug repositioning8,10. In this study, we performed a batch query in cMap, queried out 35 compounds with therapeutic potential, screened out parbendazole as a most promising drug from them, and demonstrated that it had an excellent inhibitory effect on the proliferation of HNSCC cell lines.
We verified the credibility of cMap query based on DEGs at the mRNA level. According to the GO and KEGG analysis of DEGs, we selected 31 genes enriched in ECM-receptor interaction43 and focal adhesion, which were up-regulated compared with normal tissues for verification. Parbendazole significantly reduced the expression of up-regulated genes (P < 0.05), which was consistent with the design of cMap, and indicated that the down-regulation of tumor microenvironment-related genes may also be one of the mechanisms of parbendazole.
In pull-down assay, we identified tubulin as a primary target of parbendazole, and further verified the direct binding between tubulin and parbendazole through SPR, ITC and TSA methods. The results proved that parbendazole binding to the colchicine site is an effective tubulin polymerization inhibitor, which can block the cell cycle, cause apoptosis and inhibit cell migration. Compared with normal cells, a distinguishing feature of tumor cells is the ability to proliferate indefinitely and rapidly, and tubulin is irreplaceable in the process of cell division. In the in vivo anti-tumor evaluation, parbendazole still exhibits good efficacy on xenograft tumor models in mice and zebrafish.
Generally, acute toxicity is one of the disadvantages of tubulin polymerization inhibitors acting on colchicine sites. Fortunately, our results suggest that parbendazole can overcome this shortcoming, its LD50 > 5 g/kg, at a non-toxic level. Moreover, parbendazole shows low toxicity to normal cell lines (IMR-90), and the low toxicity is one of the significant advantages of being a candidate.
Drug repositioning refers to the development of new indications for existing drugs or chemical entities. It can shorten the time to market and reduce investment risks. Some successful cases such as the repositioning of sildenafil44 and thalidomide45 have attracted the attention of many researchers and companies as well as many anti-tumor drugs such as itraconazole46, nelfinavir47, etc. cMap conducts drug screening based on the genetic differences between tumors and normal tissues, providing great help for drug repositioning.
Our study based on the cMap that connects diseases, genes and drugs, established a general process for drug repositioning from the screening of DEGs, cMap query, potential drug identification and verification, mechanism research, pharmacological, and toxicological evaluation. Herein, with the help of cMap, an anthelmintics parbendazole, was repositioned to treat HNSCC, showing sound therapeutic effects in in vivo and in vitro studies and low toxicity as well.
4. Conclusions
HNSCC is one of the cancer types that seriously endanger human health. Our study found that parbendazole can be used to treat HNSCC based on connections genes, diseases and drugs in cMap. In vivo and in vitro studies have confirmed the reasonable efficacy and low toxicity of parbendazole. Our research also provides a general research idea and workflow for drug repositioning using cMap to promote drug development.
5. Experimental
5.1. Reagents
5.1.1. Antibodies
GAPDH (Affinity, Cat# AF7021), α-tubulin antibody (Cell Signaling Technology, Cat# 2144), Goat anti-Rabbit lgG-HRP antibody (Abmart, Cat# M21002), Anti-Rabbit IgG (H + L), F(ab')2 Fragment (Alexa Fluor® 555 Conjugate) (Cell Signaling Technology, Cat# 4413).
5.1.2. Chemicals
Phentolamine (Santa Cruz, Cat# sc-279964), YS-035 (Santa Cruz, Cat# sc-203722), CGP-13501 (Santa Cruz, Cat# sc-203542), CGP-13501 (Santa Cruz, Cat# sc-203542), BVT-948 (Santa Cruz, Cat# sc-203536), 6-benzylaminopurine (HEOWNS, Cat# B-21640), mefloquine (HEOWNS, Cat# S81484), atorvastatin (9ding Chem, Cat# B5H43), stavudine (9ding Chem, Cat# A-NB665), CV-1808 (Santa Cruz, Cat# sc-203556), esomeprazole (Hwrk Chem, Cat# E881773), montelukast (Bidepharm, Cat# BD223092), sulfasalazine (HEOWNS, Cat# sc-77000), parbendazole (Hwrk Chem, Cat# P882528), dimercaptosuccinic-acid (Yuanye Biology, Cat# S45699), L-745870 (Abcam, Cat# ab120598), 5-nonyloxytryptamine (Santa Cruz, Cat# sc-203480), tetradecylthioacetic-acid (9ding Chem, Cat# PJ717), econazole (Bidepharm, Cat# BD41492), PD-160170 (Santa Cruz, Cat# sc-204836), desmethylclozapine (Macklin, Cat# N878220), warfarin (Santa Cruz, Cat# sc-205888), PIT (Santa Cruz, Cat# sc-203665), tipifarnib (Bidepharm, Cat# BD226736), VER-155008 (Bidepharm, Cat# BD233406), formestane (9ding Chem, Cat# A-MZ236), NCH-51 (Yuanye Biology, Cat# S86720), vidarabine (Dibo BIological Technology, Cat# K100553), cyclazosin (Santa Cruz, Cat# sc-239599), ispinesib (Bidepharm, Cat# BD164168), dopamine (9ding Chem, Cat# L-BB471), noscapine (Macklin, Cat# N873311), efavirenz (Yuanye Biology, Cat# S62062), dexchlorpheniramine (Topscience, Cat# T0785), velnacrine (Bidepharm, Cat# BD156385), nocodazole (Bidepharm, Cat# BD145296), oxibendazole (Bidepharm, Cat# BD137812), fenbendazole (Macklin, Cat# F810011), flubendazole (Bidepharm, Cat# BD130449), albendazole (Bidepharm, Cat# BD34058), albendazole sulfoxide (Bidepharm, Cat# BD14062), mebendazole (Bidepharm, Cat# BD102436), carbendazim (Bidepharm, Cat# BD17433), methyl [5(6)-chlorobenzimidazole-2-yl] carbamate (Bidepharm, Cat# BD139235), SYPRO Orange (Sigma–Aldrich, Cat# S5692), combretastatin A-4 (Sigma–Aldrich, Cat# C7744), colchicine (Hwrk Chem, Cat# HWBD110348), streptavidin magnetic beads (Beyotime, Cat# P2151), Series S Sensor Chip CM5 (GE Health Care, Cat# 10274087).
5.1.3. Critical commercial assays
CM-Dil (BestBio, Cat# BB-441924), total protein extraction kit (BestBio, Cat# BB-31301), Annexin V-FITC Apoptosis Detection Kit (Solarbio, Cat# CA1020), DNA Content Quantitation Assay (Cell Cycle) (Solarbio, Cat# CA1510), BCA Protein Assay Kit (Beyotime, Cat# P0012), Tubulin Polymerization Assay Kit (Cytoskeleton, Cat# BK011P), RNAeaseTM kit (Beyotime, Cat# R0026), HiScript III All-in-one RT SuperMix Perfect for qPCR kit (Vazyme, Cat# R333), FastStart Essential DNA Green Master (Roche, Cat# 06402712001).
5.1.4. Cell lines
HN6 (Shandong Ent Hospital, N/A), Fadu (Shandong Ent Hospital, N/A), CAL-27 (Shandong Ent Hospital, N/A), IMR-90 (iCell Bioscience, iCell-h260). HNSCC cell lines CAL-27, Fadu and HN6 were obtained from Shandong Ent Hospital. Human embryonic lung fibroblast cell line (IMR-90) was purchased from iCell Bioscience. HNSCC cells were cultured in DMEM/F12 medium and human embryonic lung fibroblast cells were cultured in MEM medium, both supplemented with 10% fetal bovine serum (FBS) and 5% Penicillin-Streptomycin liquid at 37 °C in an atmosphere of 5% CO2 and 100% humidity.
5.1.5. Software and algorithms
GraphPad Prism 7.00 (GraphPad Software Inc., https://www.graphpad.com/scientific-software/prism/), MicroCal PEAQ-ITC Analysis Software (Malvern Panalytical Ltd., https://www.malvernpanalytical.com.cn/), 2.0.3 BIA evaluation software (GE Healthcare, https://www.gehealthcare.com/), LC480 software (Roche, http://www.roche-diagnostics.cn/).
5.2. Animals
Mouse: nude (SiPeiFu, SCXK 2019-0010), Mouse: SPF Kunming (SiPeiFu, SCXK 2019-0010), zebrafish (Institute of Hydrobiology, Chinese Academy of Sciences, Z-005). All animal care and experiments were in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals; and approved by Ethics Committee of Cheeloo College of Medicine Shandong University (Approval No. 18032, Jinan, China). The zebrafish were cultured under the conditions of 14 h light and 10 h dark photoperiod and 28 °C.
5.3. Method details
5.3.1. Screening of DEGs in HNSCC, bioinformatics enrichment analysis and cMap query
In this study, GEO2R was used to compare the gene expression profiles of the tumor tissues and adjacent normal tissues of patients with HNSCC of GSE6631, GSE13397, GSE58911 and GSE107591 gene expression datasets in GEO, and the DEGs were combined. Subsequently, GEPIA was used to compare the gene expression profiles of 519 tumor tissue samples and 44 normal tissue samples in TCGA and GTEx. The threshold of DEGs is |log2 (fold change) | ≥ 1.5 and q < 0.01. Finally, the intersected DEGs of the two datasets was taken, so that 134 up-regulated genes and 188 down-regulated genes were obtained (Table S1). GO and KEGG pathway enrichment analysis for DEGs were performed in the Database for Annotation, Visualization, and Integrated Discovery (DAVID) 6.8. At the same time, we submitted the DEGs as a signature to cMap for query and determined the 35 small molecule compounds to be screened in preliminary experiments based on the connectivity scores (Table S2).
5.3.2. Cell proliferation/survival assay
CCK8 method was used to study the antiproliferative activities of compounds against tumor and normal cell lines. Cells were seeded in 96-well transparent plates at 5000 per well and cultured (5% CO2, 37 °C) for 12 h to adhere. Then, 100 μL of a series of compounds solutions prepared in serum-free medium were added to the wells, and the cells were cultured for another 72 h. Next, cell viability was evaluated by CCK8 method. The absorbance values were recorded by a BMG CLARIOstar microplate reader at 450 nm, and the IC50 values were calculated though the GraphPad Prism 7.00.
5.3.3. Xenograft tumor assay on mice
Four-week-old male nude mice were obtained from SiPeiFu (Beijing) Biotechnology Co., Ltd. After 4 days of adaptive feeding, the animals were injected with tumor cells (HN6, 10 million each) subcutaneously and allowed to grow to a suitable size. Mice were divided into experimental and control groups randomly and each group contains five mice. Parbendazole powder was formulated into a suspension with 0.5% sodium carboxymethyl cellulose solution and administered by oral gavage once a day. The size of the tumor was measured by an electronic caliper, calculated according to the following formula: length × width2/2. All procedures were carried out in accordance with the laboratory animal care and use guidelines of laboratory animal research institutions.
5.3.4. Xenograft tumor assay on zebrafish
The zebrafish were cultured under the conditions of 14 h light and 10 h dark photoperiod and 28 °C. In the evening, put an adult male zebrafish and an adult female zebrafish into the mating tank, separated by a partition, and placed in the incubator. Pull out the partition in the early morning of the next day, collect the fertilized eggs 4 h later, and culture them in the zebrafish embryo culture medium (5 mmol/L NaCl, 0.17 mmol/L KCl, 10 mmol/L HEPES, 0.33 mmol/L MgSO4, 0.33 mmol/L CaCl2, pH 7.8) containing phenylthiourea (3 mg/100 mL) for 96 h. The culture medium needs to be replaced every day to remove shed egg membranes and dead embryos. Normally developed zebrafish embryos were selected and then microinjected on 0.5% sodium carboxymethylcellulose with CM-Dil stained HN6 cells, 200 count per fish (HN6 cells were collected, washed twice with PBS, resuspended in serum-free medium containing CM-Dil staining solution, incubated at 37 °C for 20 min, centrifuged to remove the supernatant, washed twice with DPBS, and resuspended in 2% polyvinylpyrrolidone in DPBS). Zebrafish were randomly divided into experimental and control groups. After 24 h, the culture medium was replaced with a culture medium containing parbendazole. After 48 h, images were obtained by using an OLYMPUS SZX16 microscope (4 ×) equipped with an AxioCam MRm CCD camera and a light source for RFP excitation (532 nm), and the fluorescence integrated density was calculated with Image J.
5.3.5. Acute toxicity test (Korbor method)
SPF Kunming mice were obtained from SiPeiFu (Beijing) Biotechnology Co., Ltd. After adaptive feeding for one week, the mice were randomly divided into twelve groups (control, 0.1, 0.316, 1, 3.16, and 10 g/kg, with 12 mice in each dose, half male and female). The male and female are reared separately. Fasting for 6 h before the test, drinking freely, and fasting for 2 h after the test. Parbendazole powder was formulated into a suspension with 0.5% sodium carboxymethyl cellulose solution and administered by oral gavage. The scheduled dose is administered twice within 24 h, with an interval of 6 h. Two weeks after the administration, the survival of the mice was observed, the weight was recorded, and the LD50 was calculated as Eq. (1):
| (1) |
where Xi and Xi+1 represented the dose logarithm of the adjacent two groups, Pi+1-Pi represented the death percentage of the adjacent two groups of animals. This test was performed in accordance with the National Standards of the Republic of China (GB 15193.3-2014).
5.3.6. Cell apoptosis assay
HN6 cells were seeded in 6-well plates at a density of 1.5 × 105 cells per well. Cells were treated with 0.1, 1 μmol/L parbendazole or CA-4 for 24 h and collected, washed with 1 mL PBS and 1 mL binding buffer. Then, they were resuspended in binding buffer, mixed 100 μL with 5 μL Annexin-FITC, and incubated for 10 min at room temperature in the dark. The staining result was analyzed by flow cytometry.
5.3.7. Cell cycle assay
HN6 cells were seeded in 6-well plates at a density of 1.5 × 105 cells per well. Cells were treated with 0.1, 1 μmol/L parbendazole or CA-4 for 24 h and then collected, washed twice with PBS, resuspended in 300 μL of pre-cooled PBS, slowly dripped with 750 μL of absolute ethanol, and fixed overnight. Next, the cells were washed 3 times with cold PBS, resuspended in 100 μL RNase A solution, and placed in a 37 °C water bath for 30 min. Continue to add 400 μL PI staining solution and incubate at 4 °C in the dark for 30 min. The staining result was analyzed by flow cytometry.
5.3.8. Western blotting assay
HN6 cells were seeded in 6-well plates at a density of 1.5 × 105 cells per well. After being treated with 0.1 μmol/L, 1 μmol/L parbendazole, CA-4 or 0.1 μmol/L paclitaxel for 24 h, they were collected and washed twice with cold PBS. 200 μL of hypotonic cell lysate (1 mmol/L MgCl2, 2 mmol/L EGTA, 1 mmol/L sodium metavanadate, 20 mmol/L Tris, 0.5% NP-40) was added, protease and phosphatase inhibitors were added at the same time. The cells were lysed at 4 °C for 20 min and scraped off by cell scraping. Then, they were transferred to tubes, centrifuged at 14,000×g for 10 min at 4 °C. The supernatant obtained contains free unpolymerized tubulin. The precipitated protein was extracted by a total protein extraction kit (Cat# BB-31301, BestBio), which was the total protein containing polymerized tubulin. The protein concentration was determined with the BCA kit (Cat# P0012, Beyotime). Western blotting was performed using α-tubulin antibody (Cat# 2144, Cell Signaling Technology, 1:1000 dilution). Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel, then transferred to a PVDF membrane by BioRad transfer system and sealed for 1 h at room temperature. The membranes were incubated with primary antibody overnight at 4 °C, then washed three times with TBST, and incubated with secondary antibody (Cat# AF7021, Affinity) for 1 h at room temperature. Subsequently, membranes were washed three times with TBST and imagined via chemiluminescent gel imaging system (ChemiDoc XRS+, BIO RAD, USA). Chemiluminescence intensity was quantified by ImageJ software.
5.3.9. Cell migration assay
HN6 cells were seeded in a 12-well plate at a density of 1 × 106 cells per well. 12 h later, the scratches were created by a sterile pipette tip. Then the wells were washed 3 times by PBS to remove cell debris and serum-free medium containing compounds was added. After the cells were treated with 0.1, 1 μmol/L parbendazole or CA-4 for 24 h, scratch healing was observed and photographed via a Zeiss Axio Observer A1 fluorescence microscope (objective lens: 10×).
5.3.10. Tubulin polymerization assay
This test was performed using the tubulin polymerization kit in accordance with the manufacturer's protocol (BK011P, Cytoskeleton, Inc.). Tubulin, compound and GTP were mixed into the well at 37 °C. As the microtubes polymerize, the reporter group was incorporated into the microtubes, and the fluorescence gradually increased. The fluorescent signal was recorded by a BMG CLARIOstar microplate reader every minute within 1 h. The IC50 of the compound inhibiting the polymerization of microtubules was calculated by the rate of tubulin polymerization.
5.3.11. Immunofluorescence assay
HN6 cells were seeded in laser confocal small dishes at 40,000 per dish, and treated with 0.1, 1 μmol/L parbendazole or CA-4 for 24 h. Then, the medium was discarded, and the cells were then covered with 1 mL 4% fixative at room temperature for 15 min. Then the fixative was discarded, and the plate was washed three times with PBS. Subsequently, the specimen was blocked in blocking buffer for 60 min. After that, α-tubulin antibody (Cat# 2144, Cell Signaling Technology, 1:25 dilution) was added and incubated overnight at 4 °C before the blocking buffer was discarded. After washing three times with PBS, Anti-Rabbit IgG (H + L), F(ab')2 Fragment (Alexa Fluor® 555 Conjugate) (Cat# 4413, Cell Signaling Technology, 1:500 dilution) was added, and the specimen was incubated at room temperature for 1–2 h in the dark. DAPI (10 μg/mL) was used to stain the nucleus. The cells were photographed by Laser Confocal Microscope (LSM780).
5.3.12. qPCR assay
HN6 cells were seeded in a 6-well plate at a density of 1.5 × 105 cells per well, treated with 200 nmol/L parbendazole for 24 h and then collected. RNA was extracted using RNAeaseTM kit (Cat# R0026, Beyotime), and the reverse transcription reaction was completed using HiScript III All-in-one RT SuperMix Perfect for qPCR kit (Cat# R333, Vazyme), fluorescence quantitative analysis was performed on a Quantitative Real-time PCR (LC96, Roche) with FastStart Essential DNA Green Master (Cat# 06402712001, Roche). GAPDH and RPLPO were defined as internal reference. The primer sequence can be found in supplemental information (Table S6).
5.3.13. Pull-down assay and LC‒MS/MS analysis
HN6 cells were lysed and total protein was collected by protein extraction kit. Streptavidin magnetic beads (Cat #P2151, Beyotime) were incubated with probe (50 μmol/L) or biotin (50 μmol/L) for 1 h at room temperature, and then washed three times with binding buffer (PBS, pH 7.4, 0.05% Tween-20, 0.05% BSA). Then, magnetic beads coupled with probe or biotin were incubated with cell lysates for 2 h at room temperature to catch target protein. Finally, the magnetic beads were collected and heated in the SDS-PAGE (1×) loading buffer for 3 min at 95 °C to collected binding protein, then separated by SDS-PAGE and visualized by Coomassie blue staining. Differential bands were cut down and analyzed by mass spectrometry. The synthesis of probe can be found in Scheme S1. The gels were decolorized and then digested by trypsin. The processed samples were analyzed by LC‒MS/MS to obtain the raw file of the original mass spectra, which were analyzed by the software MaxQuant (1.6.2.10) to match the data and obtain the identification results.
5.3.14. SPR assay
First, the tubulin (100 μg/mL) dissolved in a sodium acetate solution at pH 4.0 was coupled on Series S Sensor Chip CM5 (Cat# 10274087, GE Healthcare) by an amino coupling kit. Binding analysis of parbendazole was carried out in HBS-P running buffer (0.01 mol/L HEPES pH 7.4, 0.15 mol/L NaCl, 0.0005% Surfactant P20). The binding time is 60 s, the dissociation time is 60 s, and the flow rate is 50 μL/min. The tests were performed on BIAcore T200, and the data were analysed using the 2.0.3 BIA evaluation software.
5.3.15. Thermal shift assay (TSA)
Tubulin solution and compound solution dissolved in PBS (pH 7.4) in 96-well plate was incubated for 15 min at 4 °C, then SYPRO Orange (Cat# S5692, Sigma–Aldrich) were added and the plate was covered with a sheet of optically clear adhesive and centrifuged at 800×g for 2 min. The final volume was 50 μL and the final concentrations of protein, compounds and SYPRO Orange are 5 μmol/L, 100 μmol/L and 5× , respectively. Thermal scanning (30–85 °C) was performed using a real-time PCR instrument (LC480, Roche). Fluorescence intensity was measured 100 times per °C. The melting curve and melting temperature (Tm) are calculated by LC480 software.
5.3.16. Isothermal titration calorimetry (ITC) assay
Experiments were performed on the MicroCal PEAQ-ITC (Malvern Panalytical Ltd.) at 25 °C in a running buffer (80 mmol/L PIPES, pH 6.9, 0.5 mmol/L EGTA, 2 mmol/L MgCl2). Tubulin (20 μmol/L) was in sample and small molecules (100 μmol/L) was in syringe. The data was analyzed by MicroCal PEAQ-ITC Analysis Software.
5.3.17. Competitive binding assay
Tubulin, colchicine and parbendazole diluted by tubulin buffer (80 mmol/L PIPES, pH 6.9, 0.5 mmol/L EGTA, 2 mmol/L MgCl2) complemented with 1 mmol/L GTP were added to the 96-well full blackboard, and the final concentrations of tubulin, colchicine and parbendazole were all 5 μmol/L. Then, the mixture was incubated for 30 min at 37 °C and the fluorescence intensity was measured at excitation wavelength of 380 nm and emission wavelength of 435 nm. CA-4 was positive control and tubulin buffer complemented with 1 mmol/L GTP was blank. After subtracting the blank from the original fluorescence value, the fluorescence values of colchicine and tubulin were set to 1 for normalization.
5.4. Quantification and statistical analysis
Experiments were repeated at least three times, unless otherwise stated in the legend. Data is represented as mean ± SD or SEM. P-values < 0.05 were considered significant. Significant differences between groups were determined by unpaired one or two-tailed Student t-test. All statistical analyses employed were performed using GraphPad Prism 7.00.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81673393 and 81874308), the Taishan Scholar Program at Shandong Province and the Shandong Natural Science Foundation (ZR2018ZC0233, China). The author would like to thank the Translational Medicine Core Facility of Shandong University for instrument availability that supported this work.
Author contributions
Dong Liang designed experiments, performed experiments, analyzed data, and wrote the paper. Chen Yu and Zhao Ma performed experiments and wrote the paper. Xingye Yang, Zhenzhen Li, Xiaojun Qin and Xuhui Dong performed experiments and analyzed data. Lupei Du and Minyong Li designed and supervised experiments and provided conceptual advice.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2021.12.005.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lau A., Yang W.F., Li K.Y., Su Y.X. Systemic therapy in recurrent or metastatic head and neck squamous cell carcinoma—a systematic review and meta-analysis. Crit Rev Oncol Hematol. 2020;153:102984. doi: 10.1016/j.critrevonc.2020.102984. [DOI] [PubMed] [Google Scholar]
- 2.Blasco M.A., Svider P.F., Raza S.N., Jacobs J.R., Folbe A.J., Saraf P., et al. Systemic therapy for head and neck squamous cell carcinoma: historical perspectives and recent breakthroughs. Laryngoscope. 2017;127:2565–2569. doi: 10.1002/lary.26629. [DOI] [PubMed] [Google Scholar]
- 3.Sacco A.G., Cohen E.E. Current treatment options for recurrent or metastatic head and neck squamous cell carcinoma. J Clin Oncol. 2015;33:3305–3313. doi: 10.1200/JCO.2015.62.0963. [DOI] [PubMed] [Google Scholar]
- 4.Ling D.C., Bakkenist C.J., Ferris R.L., Clump D.A. Role of immunotherapy in head and neck cancer. Semin Radiat Oncol. 2018;28:12–16. doi: 10.1016/j.semradonc.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Loganathan S.K., Schleicher K., Malik A., Quevedo R., Langille E., Teng K., et al. Rare driver mutations in head and neck squamous cell carcinomas converge on NOTCH signaling. Science. 2020;367:1264–1269. doi: 10.1126/science.aax0902. [DOI] [PubMed] [Google Scholar]
- 6.Marur S., Forastiere A.A. Head and neck cancer: changing epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2008;83:489–501. doi: 10.4065/83.4.489. [DOI] [PubMed] [Google Scholar]
- 7.Leemans C.R., Snijders P.J.F., Brakenhoff R.H. The molecular landscape of head and neck cancer. Nat Rev Cancer. 2018;18:269–282. doi: 10.1038/nrc.2018.11. [DOI] [PubMed] [Google Scholar]
- 8.Gns H.S., Gr S., Murahari M., Krishnamurthy M. An update on drug repurposing: re-written saga of the drug's fate. Biomed Pharmacother. 2019;110:700–716. doi: 10.1016/j.biopha.2018.11.127. [DOI] [PubMed] [Google Scholar]
- 9.Zerbini L.F., Bhasin M.K., de Vasconcellos J.F., Paccez J.D., Gu X., Kung A.L., et al. Computational repositioning and preclinical validation of pentamidine for renal cell cancer. Mol Cancer Therapeut. 2014;13:1929–1941. doi: 10.1158/1535-7163.MCT-13-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 11.Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao S.J., Zhu X.C., Deng H., Zhou W.P., Yang W.Y., Yuan L.K., et al. Gene expression profiling coupled with connectivity map database mining reveals potential therapeutic drugs for hirschsprung disease. J Pediatr Surg. 2018;53:1716–1721. doi: 10.1016/j.jpedsurg.2018.02.060. [DOI] [PubMed] [Google Scholar]
- 13.Brum A.M., van de Peppel J., van der Leije C.S., Schreuders-Koedam M., Eijken M., van der Eerden B.C., et al. Connectivity map-based discovery of parbendazole reveals targetable human osteogenic pathway. Proc Natl Acad Sci U S A. 2015;112:12711–12716. doi: 10.1073/pnas.1501597112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brum A.M., van de Peppel J., Nguyen L., Aliev A., Schreuders-Koedam M., Gajadien T., et al. Using the connectivity map to discover compounds influencing human osteoblast differentiation. J Cell Physiol. 2018;233:4895–4906. doi: 10.1002/jcp.26298. [DOI] [PubMed] [Google Scholar]
- 15.Farooq F., Balabanian S., Liu X., Holcik M., MacKenzie A. P38 mitogen-activated protein kinase stabilizes SMN mRNA through RNA binding protein HuR. Hum Mol Genet. 2009;18:4035–4045. doi: 10.1093/hmg/ddp352. [DOI] [PubMed] [Google Scholar]
- 16.Dyle M.C., Ebert S.M., Cook D.P., Kunkel S.D., Fox D.K., Bongers K.S., et al. Systems-based discovery of tomatidine as a natural small molecule inhibitor of skeletal muscle atrophy. J Biol Chem. 2014;289:14913–14924. doi: 10.1074/jbc.M114.556241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudley J.T., Sirota M., Shenoy M., Pai R.K., Roedder S., Chiang A.P., et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3:96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo S.J., Li H.Q., Mo Z.H., Lei J.J., Zhu L.J., Huang Y.X., et al. Connectivity map identifies luteolin as a treatment option of ischemic stroke by inhibiting MMP9 and activation of the PI3K/Akt signaling pathway. Exp Mol Med. 2019;51:1–11. doi: 10.1038/s12276-019-0229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J., Lee J., Salazar Hernandez M.A., Mazitschek R., Ozcan U. Treatment of obesity with celastrol. Cell. 2015;161:999–1011. doi: 10.1016/j.cell.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L., Kang W.Q., Lu X.L., Ma S.Y., Dong L., Zou B.C. Weighted gene co-expression network analysis and connectivity map identifies lovastatin as a treatment option of gastric cancer by inhibiting HDAC2. Gene. 2019;681:15–25. doi: 10.1016/j.gene.2018.09.040. [DOI] [PubMed] [Google Scholar]
- 21.Beck A., Eberherr C., Hagemann M., Cairo S., Haberle B., Vokuhl C., et al. Connectivity map identifies HDAC inhibition as a treatment option of high-risk hepatoblastoma. Cancer Biol Ther. 2016;17:1168–1176. doi: 10.1080/15384047.2016.1235664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu F.X., He Y.C., Zhang J.Y., Wang H.F., Zhong C., Wang X.T. Using prognosis-related gene expression signature and connectivity map for personalized drug repositioning in multiple myeloma. Med Sci Mon Int Med J Exp Clin Res. 2019;25:3247–3255. doi: 10.12659/MSM.913970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pang J.S., Li Z.K., Lin P., Wang X.D., Chen G., Yan H.B., et al. The underlying molecular mechanism and potential drugs for treatment in papillary renal cell carcinoma: a study based on TCGA and cmap datasets. Oncol Rep. 2019;41:2089–2102. doi: 10.3892/or.2019.7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S.E., Xian B., Guix M., Olivares M.G., Parker J., Chung C.H., et al. Transforming growth factor beta engages TACE and ErbB3 to activate phosphatidylinositol-3 kinase/Akt in ErbB2-overexpressing breast cancer and desensitizes cells to trastuzumab. Mol Cell Biol. 2008;28:5605–5620. doi: 10.1128/MCB.00787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenbluth J.M., Mays D.J., Pino M.F., Tang L.J., Pietenpol J.A. A gene signature-based approach identifies mTOR as a regulator of p73. Mol Cell Biol. 2008;28:5951–5964. doi: 10.1128/MCB.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patra B.G., Maroufy V., Soltanalizadeh B., Deng N., Zheng W.J., Roberts K., et al. A content-based literature recommendation system for datasets to improve data reusability―a case study on gene expression omnibus (GEO) datasets. J Biomed Inf. 2020;104:103399. doi: 10.1016/j.jbi.2020.103399. [DOI] [PubMed] [Google Scholar]
- 27.Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R.M., Ozenberger B.A., Ellrott K., et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium G.T. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cox T.R. The matrix in cancer. Nat Rev Cancer. 2021;21:217–238. doi: 10.1038/s41568-020-00329-7. [DOI] [PubMed] [Google Scholar]
- 31.Machackova T., Vychytilova-Faltejskova P., Souckova K., Trachtova K., Brchnelova D., Svoboda M., et al. MiR-215-5p reduces liver metastasis in an experimental model of colorectal cancer through regulation of ECM-receptor interactions and focal adhesion. Cancers. 2020;12:3518. doi: 10.3390/cancers12123518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nath J., Paul R., Ghosh S.K., Paul J., Singha B., Debnath N. Drug repurposing and relabeling for cancer therapy: emerging benzimidazole antihelminthics with potent anticancer effects. Life Sci. 2020;258:118189. doi: 10.1016/j.lfs.2020.118189. [DOI] [PubMed] [Google Scholar]
- 33.Dai L., Li Z., Chen D., Jia L., Guo J., Zhao T., et al. Target identification and validation of natural products with label-free methodology: a critical review from 2005 to 2020. Pharmacol Ther. 2020;216:107690. doi: 10.1016/j.pharmthera.2020.107690. [DOI] [PubMed] [Google Scholar]
- 34.Chen X., Wang Y., Ma N., Tian J., Shao Y., Zhu B., et al. Target identification of natural medicine with chemical proteomics approach: probe synthesis, target fishing and protein identification. Signal Transduct Target Ther. 2020;5:72. doi: 10.1038/s41392-020-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamal A., Shaik A.B., Polepalli S., Kumar G.B., Reddy V.S., Mahesh R., et al. Synthesis of arylpyrazole linked benzimidazole conjugates as potential microtubule disruptors. Bioorg Med Chem. 2015;23:1082–1095. doi: 10.1016/j.bmc.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Bhattacharyya B., Howard R., Maity S.N., Brossi A., Sharma P.N., Wolff J. B ring regulation of colchicine binding kinetics and fluorescence. Proc Natl Acad Sci U S A. 1986;83:2052–2055. doi: 10.1073/pnas.83.7.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu D., Zhao H., Jin M., Zhu H., Shan B., Geng J., et al. Modulating TRADD to restore cellular homeostasis and inhibit apoptosis. Nature. 2020;587:133–138. doi: 10.1038/s41586-020-2757-z. [DOI] [PubMed] [Google Scholar]
- 38.Huynh K., Partch C.L. Analysis of protein stability and ligand interactions by thermal shift assay. Curr Protein Pept Sci. 2015;79:28. doi: 10.1002/0471140864.ps2809s79. 9.1-9.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinmetz M.O., Prota A.E. Microtubule-targeting agents: strategies to hijack the cytoskeleton. Trends Cell Biol. 2018;28:776–792. doi: 10.1016/j.tcb.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Lu Y., Chen J.J., Xiao M., Li W., Miller D.D. An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm Res-Dordr. 2012;29:2943–2971. doi: 10.1007/s11095-012-0828-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 42.Ferlay J., Colombet M., Soerjomataram I., Parkin D.M., Pineros M., Znaor A., et al. Cancer statistics for the year 2020: an overview. Int J Cancer. 2021;149:778–789. doi: 10.1002/ijc.33588. [DOI] [PubMed] [Google Scholar]
- 43.Wang J., Zhang Q., Li S., Chen Z., Tan J., Yao J., et al. Low molecular weight fucoidan alleviates diabetic nephropathy by binding fibronectin and inhibiting ECM-receptor interaction in human renal mesangial cells. Int J Biol Macromol. 2020;150:304–314. doi: 10.1016/j.ijbiomac.2020.02.087. [DOI] [PubMed] [Google Scholar]
- 44.Boolell M., Allen M.J., Ballard S.A., Gepi-Attee S., Muirhead G.J., Naylor A.M., et al. Sildenafil: an orally active type 5 cyclic GMP-specific phosphodiesterase inhibitor for the treatment of penile erectile dysfunction. Int J Impot Res. 1996;8:47–52. [PubMed] [Google Scholar]
- 45.Majumdar S., Lamothe B., Aggarwal B.B. Thalidomide suppresses NF-kappa B activation induced by TNF and H2O2, but not that activated by ceramide, lipopolysaccharides, or phorbol ester. J Immunol. 2002;168:2644–2651. doi: 10.4049/jimmunol.168.6.2644. [DOI] [PubMed] [Google Scholar]
- 46.Rudin C.M., Brahmer J.R., Juergens R.A., Hann C.L., Ettinger D.S., Sebree R., et al. Phase 2 study of pemetrexed and itraconazole as second-line therapy for metastatic nonsquamous non-small-cell lung cancer. J Thorac Oncol. 2013;8:619–623. doi: 10.1097/JTO.0b013e31828c3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shim J.S., Rao R., Beebe K., Neckers L., Han I., Nahta R., et al. Selective inhibition of HER2-positive breast cancer cells by the HIV protease inhibitor nelfinavir. J Natl Cancer Inst. 2012;104:1576–1590. doi: 10.1093/jnci/djs396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.