Abstract
Obesity is increasingly prevalent globally, searching for therapeutic agents acting on adipose tissue is of great importance. Equisetin (EQST), a meroterpenoid isolated from a marine sponge-derived fungus, has been reported to display antibacterial and antiviral activities. Here, we revealed that EQST displayed anti-obesity effects acting on adipose tissue through inhibiting adipogenesis in vitro and attenuating HFD-induced obesity in mice, doing so without affecting food intake, blood pressure or heart rate. We demonstrated that EQST inhibited the enzyme activity of 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1), a therapeutic target of obesity in adipose tissue. Anti-obesity properties of EQST were all offset by applying excessive 11β-HSD1's substrates and 11β-HSD1 inhibition through knockdown in vitro or 11β-HSD1 knockout in vivo. In the 11β-HSD1 bypass model constructed by adding excess 11β-HSD1 products, EQST's anti-obesity effects disappeared. Furthermore, EQST directly bond to 11β-HSD1 protein and presented remarkable better intensity on 11β-HSD1 inhibition and better efficacy on anti-obesity than known 11β-HSD1 inhibitor. Therefore, EQST can be developed into anti-obesity candidate compound, and this study may provide more clues for developing higher effective 11β-HSD1 inhibitors.
KEY WORDS: EQST, Obesity, Adipocyte, Lipid accumulation, Preadipocyte differentiation, Adipose tissue, 11β-HSD1, 11β-HSD1 inhibitor
Graphical abstract
Equisetin suppresses lipid accumulation in adipose tissue to curb diet-induced obesity through inhibiting 11β-HSD1 activity.
1. Introduction
Given the known central roles of adipose tissue in obesity, adipose-targeted therapies for obesity are gaining substantial interest1. On the one hand, proteins including peroxisome proliferator-activated receptor gamma (PPARγ), fatty acid-binding protein (FABP4), fatty acid synthase (FAS), and acetyl-CoA carboxylase (ACC) are now understood to function in adipocyte formation2, in inhibiting adipocyte differentiation, and in fat accumulation, and targeted inhibition of the expression and/or activity of these factors has been considered as a potential therapeutic approach for treating obesity3,4. On the other hand, promotion of energy expenditure, such as browning in white adipose tissue, has been associated with health benefits for body weight regulation and obesity phenotypes5. Therefore, it is of great importance to search for pharmacological molecules capable of regulating lipid metabolism and promoting energy expenditure for combating obesity and the associated diseases5, 6, 7, 8. However, few anti-obesity products act on adipose tissue directly, so candidate adipose-tissue-targeted anti-obesity agents would be highly welcomed.
11β-hydroxysteroid dehydrogenase type I (11β-HSD1) is an attractive and promising pharmaceutical target for the treatment of obesity and the related diseases9 due to its roles in adipogenesis10,11 and energy expenditure12,13. 11β-HSD1 converts biologically inactive glucocorticoids (GCs) to their active form14, which contribute to enhanced adipogenic differentiation by upregulating adipogenic transcription factors such as PPARγ, FABP4, and various C/EBP family members (e.g., C/EBPα, β, and δ)11. Suppression of 11β-HSD1 expression15 or activity16 attenuates the differentiation of adipocytes. Beyond its known roles in adipocyte formation17, 11β-HSD1 also inhibits white adipose tissue browning18,19, suggesting the potential inhibitory roles of 11β-HSD1 in energy expenditure.
Recent years have seen efforts to develop natural products into 11β-HSD1 inhibitors for treating obesity and/or type 2 diabetes mellitus20,21. Glycyrrhetinic acid22, carbenoxolone23, emodin24 and other small molecular compounds from natural sources have been developed as 11β-HSD1 inhibitors, but all of them stopped due to poor 11β-HSD1 selectivity and(or) poor anti-obesity efficacy25, 26, 27, given that no trials of these inhibitors have met primary endpoints for treating obesity, it is clear that the translation of initial findings to clinical impact has proved challenging so far28.
In this study, we demonstrated that equisetin (EQST), a natural product29, 30, 31,97 obtained from the marine Fusarium equisetin strain NRRL 553732, is a 11β-HSD1 inhibitor and that confers potent anti-obesity activity in vitro and in vivo by accumulating in adipocyte tissue and through binding to and inhibiting the activity of 11β-HSD1. Our results support the further development of EQST as a drug candidate for the treatment of obesity.
2. Materials and methods
2.1. Materials
EQST was isolated and characterized as described in a previous study31. The normal diet (“chow”, 10% kcal fat, D12450B) and high-fat diet (HFD, 60% kcal fat, D12492) were purchased from Research Diets, Inc. (New Brunswick, NJ, USA). Antibodies against fatty acid-binding protein 4 (FABP4), uncoupling protein1 (UCP1), peroxisome proliferator-activated receptor gamma coactivator (PPARγ) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were purchased from Cell Signaling Technology (Beverly, MA, USA). Orlistat and BVT.2733 were purchased from MedChemExpress (Monmouth Junction, NJ, USA). PF-915275 was purchased from Santa Cruz (Santa Cruz, CA, USA), 3-isobutyl-1-methylxanthine (IBMX), dexamethasone, insulin and transferrin were purchased from MERK (Darmstadt, Germany).
2.2. Animal experiments
All animal experiments were approved by the Medical Ethics Committee of Peking Union Medical College, following the National Institutes of Health (NIH) regulations for the care and use of animals in research. 8-Week-old male C57BL/6J mice were purchased from Vital River Laboratories Co., Ltd. (Beijing, China). Animals were randomly assigned to each group (n = 8) and fed with normal diet (chow) or high-fat diet (HFD). EQST powder were solved with anhydrous alcohol at a concentration of 1 mg/mL and 1% (v/v) Tween-80, and then diluted with distilled water for daily oral gavage. A matched volume of distilled water was given to the control group. Whole-body energy expenditure and respiratory exchange ratio (RER) were monitored by a comprehensive lab animal monitoring system (CLAMS; Columbus, OA, USA). Body-weight was monitored once a week. At the end of the experiment, animals were fasted for 12 h before collecting blood samples, and then culled by cervical dislocation. Livers, epididymal fats, retroperitoneal fats, and subcutaneous fats were collected and weighed separately, some were fixed in 4% (v/v) paraformaldehyde; the rest were snap-frozen using liquid nitrogen for later analyses. Blood samples were stored at 4 °C for measurement of serum levels of total cholesterol (TC), triglyceride (TG), low density lipoprotein-cholesterol (LDL-C), high density lipoprotein-cholesterol (HDL-C), leptin, and adiponectin using assay kits (Jian Cheng Biotechnology Company, Nanjing, China).
2.3. Preparation for 11β-HSD1 knockout mice
The global 11β-HSD1 knockout mouse model (in a C57BL/6N genetic background) was generated by deleting the genomic DNA fragment from exon 3 of the 11β-HSD1 locus using a CRISPR-Cas9 mediated genome editing system (Cyagen Biosciences Inc., Wuhan, China). The founders were genotyped by PCR, followed by DNA sequencing analysis. Animals were housed at 22–24 °C on a 12 h light, 12 h dark cycle, and allowed free access to HFD and drinking water supplemented with corticosterone (100 μg/mL) to establish a 11β-HSD1-bypassed DIO model. PCR primers for the 11β-HSD1 mutation was characterized by PCR with the primers 5′- TTCCATGGAGATACTGATAGCCGA-3′ (forward) and 5′-TGGGGTAACTGGGGTTTAAATGAA-3′ (reverse), yielding a 567-base pair (bp) fragment for the mutant allele, or a 617-bp fragment for the WT allele.
2.4. 3T3-L1 cell culture and adipocyte differentiation
3T3-L1 preadipocytes were obtained from Peking Union Medical College; human primary adipocytes (HPA) were purchased from ScienCell Ltd. (Carlsbad, CA, USA). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with high glucose (Corning, Corning, NY, USA) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco, Grand Island, NY, USA), penicillin (100 U/mL) and streptomycin (100 mg/mL) (Thermo Scientific, Waltham, MA, USA) in a 100% humidified incubator with 5% CO2 at 37 °C. Culture medium was changed every 2 days. For adipocyte differentiation, over-confluent 3T3-L1 preadipocytes were treated with differentiation induction medium (growth medium containing 0.5 mmol/L IBMX, 1 μmol/L dexamethasone and 10 μg/mL insulin) during Days 0–4. On Day 4, the medium was replaced with maintenance medium (growth medium containing 10 μg/mL insulin, and 50 nmol/L transferrin) for another 4 days.
For the continuous treatment during the differentiation process, EQST, BVT.2733, or cordycepin were added to the differentiation induction medium, and the final concentration was maintained at 10 μmol/L. For mature adipocytes, 3T3-L1-derived mature adipocytes were exposed to EQST or BVT.2733 (final concentration was maintained at 10 μmol/L) for 24 h.
2.5. Cytotoxicity assay
3T3-L1 preadipocytes were seeded in a 96-well plate with 5 × 103 cells per well, and treated with the following conditions: fresh culture medium alone (control), fresh culture medium with different concentrations (0.1, 1, and 10 μmol/L) of EQST for 24 h. Cell viability was assessed by a cell counting kit-8 (CCK-8; Dojindo Molecular Technologies, Gaithersburg, MD, USA) according to manufacturer's instructions. Briefly, after treatment, the CCK-8 solution was added to the culture medium and incubated at 37 °C for 1 h. The absorbance was read at 450 nm with a microplate reader (Thermo, Waltham, MA, USA). Cell viability was calculated by the following Eq. (1):
| Cell viability (%)=(Experimental group absorbance value/Control group absorbance value) × 100 | (1) |
2.6. Construction for 11β-HSD1 knockdown 3T3-L1 preadipocyte cell line
11β-HSD1-knockdown preadipocyte cells were prepared by lentiviral transduction. A small guide RNA (sgRNA) against 11β-HSD1 mRNA (5′-GGAAGAGCACCAGGATCGGG-3′) was inserted into the lentiviral expression vector pHS-ACR-LW720 (and 5′-GGTATTGACTGCCAGGTCGG-3′ was inserted into vector pHS-ACR-LW721), which contains an enhanced green fluorescent protein (EGFP) gene as a reporter (activated by the CMV promoter). Recombinant lentiviral plasmid was produced by co-transduction of HEK293T with reconstructive vector and packaging vector using Lipofectamine 2000 transfection reagent (Thermo Scientific, Waltham, Massachusetts, USA). Lentiviral particles were harvested after 2 days, centrifuged to get rid of cell debris, and filtered through 0.45 μm filters. For lentivirus transduction, 3T3-L1 cells were seeded at 5 × 103 cells per well into 6-well plates. When grown to the density of 30%–40% confluence, cells were transfected with lentivirus containing sgRNA or control lentivirus Neg control sgRNA at a multiplicity of infection (MOI) of 10. After 72 h infection, cells were harvested, and transfection efficiency was evaluated by GFP-positive cells. The 11β-HSD1 knock down 3T3-L1 preadipocytes were harvested and cultured as above.
2.7. Construction of exogenous 11β-HSD1 expressed HepG2 cells
HepG2 cells were purchased from American Type Culture Collection (ATCC) (Rockville, MD, USA). HepG2 cells were plated into a 6-well plate with 3 × 105 cells per well one day before plasmid transfection. Cells were transfected with a 11β-HSD1-expressing plasmid using Lipofectamine 2000 transfection reagent (Thermo Scientific, Waltham, MA, USA) according to the manufacturer's protocol. Briefly, the consensus coding sequence of Mus musculus 11β-HSD1 (CCDS15635.1) was amplified by PCR and cloned into the pEGFP-N1 vector between XhoI and BamHI sites to obtain the recombinant expression plasmid. Primers for PCR were as follows: Primer F: 5′-CCGCTCGAGATGGCAGTTATGAAAAATTACCT-3′; Primer R: 5′-CGGGATCCCGGTTACTTACAAACATGTCCTTATTATA-3′. After 5 h, the me-dium was replaced with culture medium [DMEM supplemented with 10% (v/v) FBS, 100 U/mL penicillin and 100 mg/mL streptomycin] supplemented with or without EQST (final concentration was maintained at 10 μmol/L). 11β-HSD1 activity assays were performed 48 h post transfection.
2.8. Histological analysis
Adipose tissues were fixed in 4% paraformaldehyde, and then embedded in paraffin wax. The embedded samples were cut into 4-μm thick slides and mounted on glass slides. The slides were stained with hematoxylin and eosin (H&E).
2.9. Oil red O staining
Cells were washed with PBS, and then fixed with 4% paraformaldehyde for 1 h at room temperature, followed by staining with a mixture of 0.5% oil red O dye (resolved in isopropanol and ddH2O at a 3:2 ratio) for 20 min. Cells were washed three times with PBS, the images were captured under a microscope. The oil red O dye was resolved with DMSO and subjected for absorbance measurement at 358 nm.
2.10. Intracellular triglyceride (TG) quantification
Cells were scraped and re-suspended in 1% TritonX-100 and heated to 80–100 °C for 5 min to solubilize the intracellular triglycerides. The triglyceride content was measured using a triglyceride quantification assay kit (BioVision, Inc., Milpitas, CA, USA) according to the manufacturer's instructions.
2.11. Excessive glucocorticoids (GCs) treatment
3T3-L1 preadipocytes were seeded into 6-well culture plates and induced into mature adipocytes as above. For the “excessive” 11-dehydrocorticosterone treatment, 3T3-L1-derived mature adipocytes were exposed to 40 nmol/L 11-dehydrocorticosterone and/or 10 μmol/L EQST for 24 h. For the excessive corticosterone treatment, 3T3-L1-derived mature adipocytes were exposed to 40 nmol/L corticosterone and/or 10 μmol/L EQST for 24 h.
2.12. 11β-HSD1 and 11β-HSD2 enzyme activity assays
The enzyme activity of 11β-HSD1 in cells was evaluated according to its capability to convert cortisone to cortisol, and the enzyme activity of 11β-HSD2 in cells was evaluated according to its capability to convert corticosterone to 11-dehydrocorti-costerone33,34. 3T3-L1 preadipocytes were seeded into 6-well culture plates and induced into mature adipocyte as above. 3T3-L1-derived mature adipocyte were exposed to 160 nmol/L cortisone or 160 nmol/L corticosterone for 24 h. The reaction mixtures were collected and analyzed for cortisol levels using an ELISA kit (R&D Systems, Minneapolis, MN, USA), according to the manufacturer's instructions.
2.13. Quantitative real time-PCR analysis
Total RNA was isolated from cells using TRIzol Reagent (Ambion, Austin, TX, USA) and was reverse-transcribed into cDNA using EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). Gene transcript levels of Pparg, Ucp-1, Fabp4, Fas, Acc-1, Srebp1, Perilipin, Hsl, Lpl was quantified using the TransStart Top Green qPCR SuperMix (TransGen Biotech, Beijing, China) according to the manufacturer's protocol. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was used as housekeeping control. Primers used for quantitative PCR are listed in Supporting Information Table S1.
2.14. Western blotting
Homogenized adipose tissue and differentiated adipocytes were lysed using tissue lysis buffer reagent (CWBIO, Jiangsu, China) and RIPA buffer (strong) (CWBIO, Jiangsu, China). Protein concentrations were determined using a BCA protein assay kit (CWBIO, Jiangsu, China). The total proteins separated by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA USA), then immunoblotted with antibodies against FABP4, UCP1 (both from Abcam, Cambridge, UK), GAPDH, and PPARγ (from Cell Signaling Technology, Beverly, MA, USA). The protein signals were visualized with enhanced chemiluminescence reagent (Millipore, Billerica, MA, USA) according to the manufacturer's protocol, with quantification using Image J 4.1 software (NIH, Bethesda, MD, USA).
2.15. Multiple reaction monitoring (MRM)
MRM analysis was performed on a 6495 Triple-Quadrupole Mass Spectrometer coupled with an Agilent 1260 Infinity HPLC system (Agilent, Santa Clara, CA, USA). Twenty micrograms of tryptic-digested peptides from each sample were loaded onto a reverse-phase analytical column (Agilent ZORBAX SB-C18, 3.5 μm, 15 cm in length, 500 μm inner diameter). The aforementioned mass spectrometer was used to measure m/z values and signal intensities for peptides sequentially eluted from the analytical column. MRM-MS analysis was performed in positive ion mode, with the ion spray capillary voltage and nozzle voltage set at 3000 and 1200 V, respectively. All raw MRM-MS data were processed using Skyline platform. All integrated peaks were manually inspected to confirm correct peak detection and accurate integration. The MRM acquisition method was initially built with eight to fourteen precursor/fragment ion pairs (transitions) for each light and heavy peptide. Reverse response curves for examining linearity were generated from MRM-MS data from each serially diluted heavy peptide spiked into 25 μg of tryptic-digested sample. Eight concentration points (approximately 1.56–200 fmol/μL) were used, and a tryptic-digested pooled sample (without heavy peptides added) was used as a blank to estimate background levels. Triplicate MRM-MS analyses were performed for each group.
2.16. Reverse docking
The two-dimensional molecular structure of EQST (Fig. 1A) was downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov, PubChem CID: 54684703), and minimized in Discovery Studio ver. 4.5. The RCSB Protein Data Bank (https://www.rcsb.org) is used to retrieve the released three-dimensional structures of human proteins. DS 4.5 has been used to hydrogenate proteins, remove water and the ligand molecules. AutoDockTools ver. 1.5.6 was used to convert EQST and protein molecules into “pdbqt” format, and finally vina was been used for molecular docking. Generally, if the affinity value is less than −6 kcal/mol, the small molecule can be considered as a potential ligand of the receptor35. The receptors with a docking score less than −9 kcal/mol are considered as the potential target proteins of EQST. Obesity-related proteins with a relevance score higher than 10 were retrieved from the GeneCards database (https://www.genecards.org/) by searching the term of “obesity”. The two sets of the proteins were subjected to Venn analysis using the Venny 2.1.0 online tool to identify candidate obesity-related targets of EQST.
Figure 1.
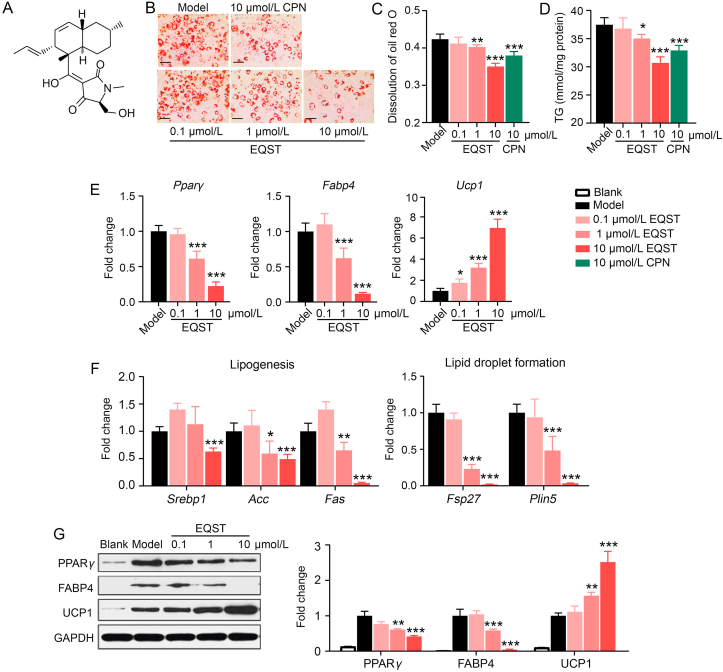
EQST inhibited terminal differentiation of preadipocytes and promoted UCP1 expression. (A) Chemical structure of Equisetin (EQST). (B) Representative Oil Red O-stained images (scar bar = 100 μm, n = 3). (C) Quantitative analysis of dissolved oil red O at OD 358 nm, n = 3. (D) Triglycerides (TG) content, n = 3. (E) qPCR analysis of Pparg, Fabp4 and Ucp1 expression normalized by Gapdh. (F) qPCR analysis of lipogenesis genes (Srebp1, Acc, Fas) and lipid droplet formation genes (Fsp27, Plin5) normalized by Gapdh, n = 3. (G) Immunoblotting against the PPARγ, FABP4 and UCP1 proteins, n = 3. All data are presented as mean ± SD of at least three independent experiments. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.005 vs. Model group. P value was assessed by two-tailed Student's t-test. CPN: Cordycepin group.
2.17. Molecular docking simulation of EQST to 11β-HSD1
The molecular docking simulation was conducted using Molecular Operating Environment (MOE; Chemical Computing Group Inc., Montreal, Canada) v2018.01. The 2D structure of EQST was drawn in ChemBioDraw 2014 and was converted to its 3D conformations through energy minimization. The 3D structure of the protein 11β-HSD1 was retrieved from the RCSB Protein Data Bank (https://www.rcsb.org/; PDB code 3D5Q). The protein structure was prepared by using the QuickPrep module. The protonation states and the orientations of the hydrogens were optimized by LigX at a pH 7 at 300 K. The docking process was performed using an AMBER10/EHT force field, along with an internal dielectric constant of 1 and an external dielectric constant of 80, as well as an implicit solvation model of reaction field (R-field). The Triangle Matcher algorithm was used for the initial placement of 1000 returned conformations, and the top 100 conformations ranked by the London dG scoring function were further refined through energy minimization, followed by rescoring using Generalized Born/Volume Integral (GB/VI) solvation energy. The induced-fit protocol was applied for energy minimization: the ligand was fully flexible during the conformation sampling process, and the side chains of the receptor were also allowed to move. Finally, 20 top-ranked poses were retained, and the most representative pose was retrieved by visual inspection for further analysis (See detailed methods in Supporting Information).
2.18. Cellular thermal shift assays (CETSA)
3T3-L1-derived adipocytes were collected and freeze–thawed three times using liquid nitrogen. The lysates were divided into three aliquots, with one aliquot being incubated with 20 μmol/L EQST, the second with 20 μmol/L BVT.2733, and the third with DMSO as a negative control. After 1 h at room temperature, the samples were heated individually at different temperatures (40–72 °C) for 3 min, followed by cooling for 3 min on ice before centrifuging at 11,000 × g for 10 min at 4 °C (Laborzentrifugen, Sigma, Osterode am Harz, Germany). The supernatant was then analyzed by Western blotting.
2.19. Statistical analyses
Statistical analyses were performed with GraphPad Prism (version 8.4.3). Independent Student's t-tests were used to compare the means of numerical variables. Data are presented as the mean ± standard error of mean (SEM). Statistical significance was defined as P values < 0.05. Experiments involving measurements of obesity indexes were performed in a blinded fashion with randomly grouped individuals. Imaging and histology were also performed and analyzed in a blinded fashion prior, to the decoding of the sample identities.
3. Results
3.1. EQST inhibited terminal differentiation of preadipocytes
A screen of marine natural product compounds showed that equisetin (EQST, Fig. 1A) exhibited apparently strong lipid-lowering activity in 3T3-L1 preadipocyte cells. We first evaluated the cytotoxicity of EQST, CCK-8 assay result demonstrated that EQST did not show cytotoxicity to 3T3-L1 preadipocyte cells (Supporting Information Fig. S1). To investigate the lipid-lowering effects of EQST, we continuously exposed differentiating 3T3-L1 preadipocytes to different doses of EQST, in experiments which used a reported inhibitor of adipogenesis36 as a positive control (cordycepin, CPN). Quantification at the end point of the 3T3-L1 preadipocyte differentiation procedure showed that both total lipid content and TG content were significantly decreased in the cells given EQST (1 and 10 μmol/L) compared to untreated model cells (Fig. 1B–D). Moreover, we found that EQST dose-dependently suppressed both the mRNA and protein abundances of known pan-adipocyte markers including PPARγ and FABP437 (Fig. 1E and G). Genes that function in lipogenesis (Srebp1, Acc, Fas)38 and in lipid droplet formation (Fsp27 and Plin5)39,40 were also down-regulated in response to EQST (Fig. 1F).
In addition, we found that EQST dramatically increased the expression of UCP1 in 3T3-L1-derived mature adipocytes (Fig. 1E and G). UCP1 is a mitochondrial protein that functions in thermogenesis, specifically by uncoupling oxidative phosphorylation to enable non-shivering thermogenesis in brown and beige adipocytes41, 42, 43. These observations together indicate that EQST can somehow suppress the expression of key adipogenic factors while also promoting the expression of a gene for increased thermogenic energy expenditure.
3.2. EQST inhibited HFD-induced obesity and enhanced energy expenditure in HFD-induced obese mice
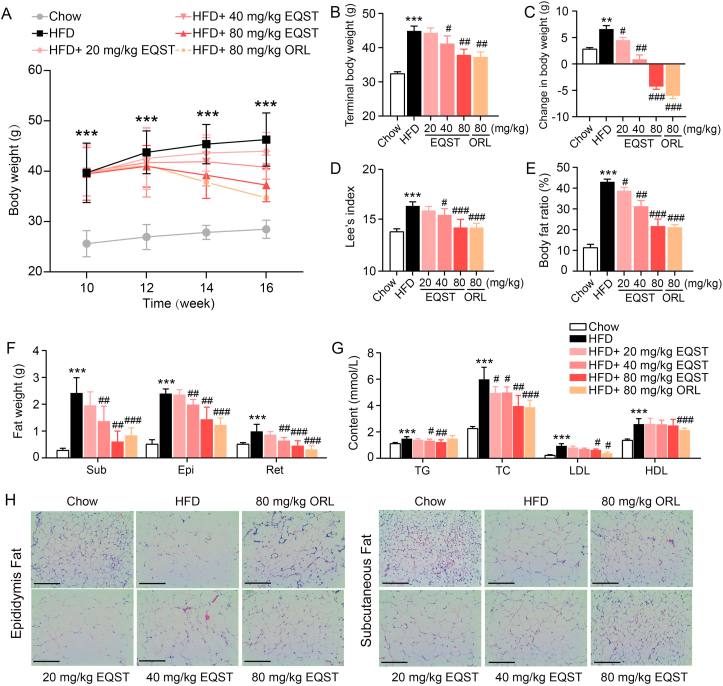
We next investigated the potential in vivo anti-obesity effect(s) of EQST. C57BL/6J mice were given a high-fat diet (HFD) for 10 weeks to establish a classic diet-induced obese (DIO) model, after which mice were given EQST daily (i.g.) for 6 weeks (n = 8). Orlistat, an approved drug for long-term treatment of obesity44, was used as a positive control. At Week 16, the EQST-treated mice all weighed significantly less than the vehicle control group, and our testing of three EQST concentrations (20, 40, and 80 mg/kg) showed that this effect on weight gain was in dose-dependent manner (Fig. 2A–D). It should be noted that EQST treatment did not affect food intake (Supporting Information Fig. S2), heart rate, or blood pressure (Supporting Information Table S2), which led us to speculate that EQST may act directly on adipose tissue. Pursuing this, experiments using Sprague–Dawley (SD) rats to determine the tissue distribution of EQST showed that a 10 mg/kg dose of EQST given to rats by oral administration for 7 consecutive days led to EQST accumulation of 0.38 ± 0.13 μg/g in adipose tissue, a significantly higher level than detected in lung (0.26 ± 0.08 μg/g) or skin (0.24 ± 0.05 μg/g) (Supporting Information Table S3). The detected EQST in adipose tissue supports the plausibility that the observed anti-adipogenesis effects could result from the EQST's lipid-lowering effects in adipocytes.
Figure 2.
EQST administration inhibited HFD-induced obesity. C57BL/6J mice under HFD feeding were treated with different doses of EQST, Orlistat or vehicle (n = 8). (A) Change curves of body weight. (B) Terminal body weight. (C) Total weight change. (D) Terminal Lee's index. (E) Terminal body fat percentage. (F) Wet weights of subcutaneous, epididymal, and retroperitoneal adipose tissue. (G) Serum levels of triglyceride (TG), total cholesterol (TC), low-density lipoprotein (LDL) and high-density lipoprotein (HDL). (H) Representative H&E staining of dissected epididymal adipose tissue (left) and subcutaneous adipose tissue (right) (scar bar = 100 μm). Data are presented as mean ± SD, n = 8. ∗∗P < 0.01, ∗∗∗P < 0.001 compared to Chow group. #P < 0.05, ##P < 0.01, ###P < 0.001 compared to HFD group. P value was assessed by two-tailed Student's t-test. ORL: Orlistat group.
We also found that the EQST-treated mice had significantly reduced body fat compared to the vehicle control DIO animals (Fig. 2E), and a closer examination revealed significant reductions in the weight of subcutaneous, epididymal, and retroperitoneal white adipose tissue (sWAT, eWAT, and rWAT; Fig. 2F). Potentially helping to explain their retarded body weight gain, the EQST-treated mice showed significant reduced content for blood lipids including TG, TC, and LDL (Fig. 2G). H&E staining showed that EQST treatment significantly reduced the sizes of individual adipocytes within both eWAT and sWAT in DIO (Fig. 2H). Finally, EQST reduced the expression levels of PPARγ and FABP4 in subcutaneous fat (Supporting Information Fig. S3C) and visceral fat (Supporting Information Fig. S4) extracted from the DIO mice. These data from HFD-induced obese mice demonstrate that EQST acts to retard body weight gain and show that EQST affects adipose tissues by suppressing lipid accumulation.
We also monitored oxygen uptake (VO2) kinetics for the DIO mice and found that mice treated with EQST had significantly higher whole-body energy expenditure (Fig. 3A and B, and Fig. S3A) than vehicle control mice. Moreover, mice in EQST group had a lower respiratory quotient (RQ, VCO2/VO2) than did control mice, suggesting that EQST treatment may induce a fuel switch toward fatty acid oxidation (Fig. S3B). Immunoblotting result showed that EQ treatment significantly increased the expression of UCP1 in sWAT of DIO mice (Fig. S3C). We also extracted proteins from sWAT of the DIO mice and performed multiple reaction monitoring (MRM), and MRM-based quantitation showed that EQST treatment significantly increased the sWAT accumulation of proteins known to function in fatty acid oxidation, lipid metabolism (e.g., CPT2, ETFDH, HADH, HADHA, HADHB45), the electron transport chain (e.g., UQCRC1, SDHA46), and the TCA cycle (e.g., SDHA, ACO247) (Fig. 3C). These in vivo results suggest that EQST can both inhibit adipogenesis and induce elevated energy expenditure, apparently by ameliorating obesity-linked metabolic impairment.
Figure 3.
EQST administration promoted energy expenditure of HFD-induced obese mice. (A) Whole-body energy expenditure (VO2, mL/kg/h) of HFD-induced obese mice. Control, n = 8; EQST, n = 6. P value by two-way repeated measures ANOVA. (B) EE (energy expenditure) of mice in (A). (C) Analysis for various lipolysis proteins by MRM from subcutaneous adipose in Vehicle- or EQST-administrated mice (n = 3). Data are presented as mean ± SD. ∗∗P < 0.01, ∗∗∗P < 0.005 vs. Model. P value was assessed by two-tailed Student's t-test.
3.3. EQST targets 11β-HSD1
We next attempted to identify potential targets of EQST to help explore the mechanism(s) underlying EQST's anti-obesity effects. Briefly (Fig. 4A), we used a reverse docking method to examine EQST's apparent capacity to interact with proteins having solved structures in the PDB database: 40 candidate target proteins had binding propensity (PLB) scores higher than 9 (docking score less than −9 kcal/mol). The GeneCards database (https://www.genecards.org/) lists a total of 466 putative obesity-related proteins, and a Venn analysis indicated that 8 of these overlapped with our reverse docking candidates, among which 11β-HSD1 had the highest GeneCards relevance score for the obesity phenotype (Supporting Information Table S4).
Figure 4.
EQST targets 11β-HSD1 and inhibits 11β-HSD1 enzyme activity. (A) Retrieval process of potential targets of EQST. (B) 11β-HSD1 enzyme activity in EQST treated 3T3-L1-derived mature adipocyte, n = 3. (C) EQST inhibits exogenous 11β-HSD1 enzyme activity in HepG2 cells, n = 3. (D, E) Lipid content post 3T3-L1 differentiation with or without 40 nmol/L 11-dehydrocortisone treatment, n = 3; (D) Quantitative analysis of dissolved oil red O, n = 3; (E) Triglyceride (TG) content, n = 3. Cortisol assay was performed following 24 h treatment of EQST at variable doses. The activity of 11β-HSD1 in non-treated adipocytes was taken as 100%, n = 3. (F) Representative oil red O-stained images (scar bar = 100 μm, n = 3). (G) Quantitative analysis of dissolved oil red O. (H) TG content post differentiation of 3T3-L1 transfected with scramble shRNA (shNC) or with shRNA against 11β-HSD1 (shHSD1), n = 3. (I) qPCR analysis of crucial factors (Pparg, Fabp4 and Ucp1) of post differentiation of 3T3-L1 transfected with scramble shRNA (shNC) or with shRNA against 11β-HSD1 (shHSD1), normalized by Gapdh, n = 3. (J) qPCR analysis of lipogenesis genes (Acc, Fas) and lipid droplet formation genes (Fsp27, Plin5) of post differentiation of 3T3-L1 transfected with scramble shRNA (shNC) or with shRNA against 11β-HSD1 (shHSD1), normalized by Gapdh, n = 3. All data are presented as mean ± SD of at least three independent experiments; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.005 by two-tailed Student's t-test. C: control rgoup; EQ: EQST group.
The full name of 11β-HSD1 is 11β-hydroxysteroid dehydrogenase I; this enzyme functions in lipid metabolism and adipogenesis, specifically (in humans) by mediating the conversion of the inactive corticosteroid cortisone into its active form (cortisol). Note that inhibition of 11β-HSD1 has been reported as beneficial for treating obesity48. We assayed potential inhibitory impacts of EQST on 11β-HSD1 in experiments using 3T3-L1-derived mature adipocytes, EQST did not affect 11β-HSD1 protein abundance (Supporting Information Fig. S5), and we found that EQST inhibited the enzyme activity of 11β-HSD1 at a dose of 1 and 10 μmol/L (Fig. 4B). Human-liver-derived HepG2 cells express little or no 11β-HSD149, so we constructed a human cell model based on exogenous expression of 11β-HSD1 in HepG2 cells. Treatment of 11β-HSD1 expressing (or empty vector) HepG2 cells with EQST (10 μmol/L) showed that EQST does inhibit 11β-HSD1 enzyme activity in human cells (Fig. 4C).
In mice, 11β-HSD1 transforms the inactive substrate 11-dehydrocortisone (11-DHC) into the active product corticosterone (CORT)48. Excessive 11-DHC levels promote the expression of 11β-HSD1 in vitro50, while excessive CORT inhibits the expression of 11β-HSD1 in vitro51; these regulatory outcomes are based on substrates or products binding at the 11β-HSD1 active site52. We exposed 3T3-L1 preadipocytes that had been cultured with or without 40 nmol/L 11-DHC to EQST and examined total lipid and TG content: the presence of an inhibitory 11-DHC concentration blocked any EQST-induced alteration of lipid content and of TG content (Fig. 4D and E). We then exposed 3T3-L1 preadipocytes that had been cultured with or without 40 nmol/L CORT to EQST and examined both total lipid and TG levels. Our findings that the presence of an inhibitory CORT concentration blocked any EQST-induced alteration of lipid or TG content supports direct involvement of 11β-HSD1 in EQST's anti-obesity effects (Supporting Information Fig. S6A and S6B); that is EQST's capacity to inhibit 11β-HSD1's transformation of 11-DHC to CORT can explain the reduction of lipid accumulation we observed earlier in 3T3-L1 preadipocytes. We also investigated the 11β-HSD1 inhibition effects of EQST in human primary adipocytes, in experiments using the previously reported human 11β-HSD1 inhibitor53 as a positive control (PF-915275). Human primary adipocytes were exposed to 10 μmol/L EQST or 10 μmol/L PF-915275 for 24 h. Compared to untreated model cells, the total lipid content was significantly decreased in the cells given EQST (10 μmol/L) and PF-915275 (10 μmol/L) (Supporting Information Fig. S7).
To offer additional direct evidence for the involvement of 11β-HSD1, we constructed an shRNA-mediated 11β-HSD1-knockdown 3T3-L1 cell line (Supporting Information Fig. S8A and S8B). Compared with non-target control cells, there is a significant reduction in lipid accumulation and the expression of genes function in adipogenesis in 11β-HSD1 knockdown cells (Fig. 4F, G, Fig. S8C and S8D). And we found that EQST exerted no inhibitory effect on lipid accumulation during adipogenesis in 11β-HSD1-knockdown 3T3-L1 cells (Fig. 4F–H, Supporting Information Fig. S7). Further, the 11β-HSD1-knockdown 3T3-L1 cells exposed to EQST actually had higher expression levels of adipogenesis-related genes than the vehicle control cells [e.g., Pparg, Fabp4, Acc, Fas, Fsp27, Plin5 (Fig. 4I and J)]. Finally, it was notable that EQST did not induce UCP1 expression in the 11β-HSD1-knockdown 3T3-L1 cells (Fig. 4I), suggesting that 11β-HSD1 somehow regulates energy expenditure. Collectively, these results demonstrate the direct involvement of 11β-HSD1 in the EQST-induced adipogenesis inhibitory effects.
3.4. 11β-HSD1 is required for EQST's in vivo anti-obesity effects
Building from findings with cell cultures, we also generated homozygous 11β-HSD1 gene knockout C57BL/6J mice. Compared with the wild-type mice, knockout of 11β-HSD1 decreased the body weight of mice under 4 weeks HFD-feeding, whereas EQST's anti-obesity effects disappeared in 11β-HSD1 knockout mice (Supporting Information Fig. S9). To further explore the relationship between EQST and 11β-HSD1, we performed a rescue experiment. For 4 weeks, we fed 11β-HSD1 knockout mice a HFD supplemented with excess corticosterone (100 μg/mL) and prophylactically co-administered with EQST (80 mg/kg) or positive control (orlistat, 80 mg/kg) to establish a 11β-HSD1-bypassed DIO model (n = 6).
With these 11β-HSD1 knockout mice, there was no difference in body weight between the vehicle and EQST-treated groups, although there was an obvious anti-obesity effect from the Orlistat treatment (Fig. 5A–C). We also found that the weights of different fat deposits in the 11β-HSD1 knockout mice (Fig. 5D and E) and the serum levels of biochemical indicators (TG, TC, LDL, HDL, and FFA; Fig. 5F) were effectively improved by orlistat treatment; no such effects resulted from EQST treatment of these animals. These results suggest that EQST cannot inhibit the obesity induced by excessive exogenous glucocorticoids in 11β-HSD1 knockout mice, which confirmed that 11β-HSD1 is required for the EQST's anti-obesity effects (Figure 4, Figure 5).
Figure 5.
11β-HSD1 is required for of EQST's in vivo anti-obesity effects. 11β-HSD1-knockout mice were allowed free access to HFD and drinking water supplemented with corticosterone (100 μg/mL, n = 6). (A) Terminal body weight. (B) Total weight change. (C) Terminal Lee's index. (D) Terminal body fat percentage. (E) Wet weight of subcutaneous, epididymal, and retroperitoneal adipose tissue. (F)Serum levels of triglyceride (TG), total cholesterol (TC), low density lipoprotein (LDL), high density lipoprotein (HDL). All data are presented as mean ± SD of at least three independent experiments; ns, P > 0.05; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.005 vs. the Chow group; #P < 0.05, ##P < 0.01, ###P < 0.005 vs. the HFD group. P value was assessed by two-tailed Student's t-test. EQ: EQST group; ORL: Orlistat group.
3.5. EQST exerts stronger anti-obesity efficacy than the known 11β-HSD1 inhibitor BVT.2733
Molecular docking of EQST with the 11β-HSD1 protein (Fig. 6A and Supporting Information Figs. S10–S13) and of the known 11β-HSD1 inhibitor BVT.273354 with its target revealed notable distinctions with potential implications for binding affinity and potency. BVT.2733 is a murine selective 11β-HSD1 inhibitor, which is effective in reducing body weight, reducing adipocyte size and stimulating energy expenditure of DIO animal model55. EQST is predicted to interact with Asp259, Ser170, and Tyr183 (Fig. 6A, Supporting Information Figs. S10 and S12), while BVT.2733 is predicted to undergo interactions with Ala172, Ser170, and Tyr177 (Fig. 6A, Supporting Information Figs. S11 and S13). These apparently distinct binding modes led us to conduct cellular assays to explore pharmacodynamic differences in their inhibition of 11β-HSD1. To avoid the influence of dynamic changes on 11β-HSD1 during adipocyte differentiation56,57, we used 3T3-L1-derived mature adipocytes, which were treated with 10 μmol/L EQST or 10 μmol/L BVT.2733. Both EQST and BVT.2733 were effective in inhibiting 11β-HSD1 activity, although 10 μmol/L EQST inhibited 11β-HSD1 activity more potently than BVT.2733 (Fig. 6B). 11β-Hydroxysteroid dehydrogenase type 2 (11β-HSD2) is another 11β-HSD1 isozyme, which functions to transform active GCs into inactive GCs26,48. High selective inhibition over 11β-HSD1 is required for the development of 11β-HSD1 inhibitors since that non-selective 11β-HSD2 inhibition may increase active GCs content and lead to long-term side effects58, 59, 60. To assess the selectivity of EQST for 11β-HSD1, we continuously exposed differentiating 3T3-L1 preadipocytes to 10 μmol/L EQST or 10 μmol/L BVT.2733, after which 3T3-L1-derived mature adipocyte were given 160 nmol/L CORT for 24 h and assayed for 11-DHC content. No significant 11β-HSD2 inhibition was detected upon 10 μmol/L EQST treatment, whereas 10 μmol/L BVT.2733 significantly inhibited the 11β-HSD2 enzyme activity (Fig. 6C). Upon extracting proteins from EQST- and BVT.2733-treated 3T3-L1-derived mature adipocytes and performing cellular thermal shift assays (CETSA), we found that the EQST-11β-HSD1 complex was more thermostable than the BVT.2733-11β-HSD1 complex, helping to explain EQST's higher 11β-HSD1 inhibition potency (Fig. 6D).
Figure 6.
EQST exerts stronger anti-obesity effect than the known 11β-HSD1 inhibitor BVT.2733. (A) Binding mode between EQST (left) and BVT.2733 (right) with 11β-HSD1. EQST is predicted to interact with Asp259, Ser170, and Tyr183; BVT.2733 is predicted to undergo interactions with Ala172, Ser170, and Tyr177. (B) 11β-HSD1 enzyme activity in EQST or BVT.2733 treated 3T3-L1-derived mature adipocyte, n = 3. (C) 11β-HSD2 enzyme activity in EQST or BVT.2733 treated 3T3-L1-derived mature adipocyte, n = 3. (D) CETSA analysis of the binding strength of EQST and BVT.2733 to 11β-HSD1 in mature adipocytes, n = 3. (E, F) Lipid lowering effect in EQST or BVT.2733 treated 3T3-L1 preadipocyte. (E) Representative oil red O-stained images (scar bar = 100 μm, n = 3). (F) Quantitative analysis of dissolved oil red O, n = 3. (G) Triglycerides (TG) content, n = 3. (H–J) Anti-obesity effect of EQST or BVT.2733 in vivo. HFD-induced obese mice were treated with vehicle, 40 mg/kg EQST or 40 mg/kg BVT.2733, respectively (n = 6). (H) Change curves of body weight. (I) Total weight change. (J) Terminal body fat percentage. All data are presented as mean ± SD; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.005 vs. the Model group; ###P < 0.005 vs. the Model group. ns, not significant. P value was assessed by two-tailed Student's t-test. EQ: EQST group; BVT: BVT.2733 group.
We then treated differentiating 3T3-L1 preadipocytes with EQST or BVT.2733 and found that EQST was more effective in reducing lipid content than BVT.2733 at the same dose of 10 μmol/L (Fig. 6E–G). Further, we constructed a prophylactical drug delivery model to compare their anti-obesity in vivo efficacy. We fed C57BL/6J mice a HFD and co-administered 40 mg/kg EQST or 40 mg/kg BVT.2733 for 4 weeks. The body weights and body fat percentages of the EQST-treated animals were significantly lower than in the vehicle-treated DIO mice (Fig. 6H–J and Supporting Information Fig. S14A). EQST also outperformed BVT.2733 regarding serum lipid profiles (Fig. S14B). These data indicate that EQST exerts more potent antiob-esity effects than the known anti-obesity agent and 11β-HSD1 inhibitor BVT.2733.
4. Discussion
Natural products represent a rich source of diverse molecules for drug development. EQST is a meroterpenoid which was first isolated from a marine sponge-derived fungus for its antibacterial activity29. EQST can also be used as an HIV-1 integrase inhibitor61,62. Here, we demonstrate that the marine natural product EQST is an 11β-HSD1 inhibitor that confers potent anti-obesity activity by acting on adipose tissue.
eWAT and sWAT are the main adipose tissues which undergo adipocyte hypertrophy in the early stages of HFD exposure63; over-loaded lipids distributed in the blood can also lead to ectopic fat deposition1, which leads to metabolic syndrome in obese individuals64. Reducing body weight and improving fat distribution are regarded as highly relevant aspects for developing anti-obesity drugs65. In our study, EQST administration effectively inhibited adipogenesis in vitro and suppressed adipocyte size of eWAT and sWAT in mice, indicating that EQST reduced fat weight by alleviating adipose tissue expansion through suppressing adipocyte hypertrophy. Moreover, EQST accumulated in adipose tissue and did not affect food intake, blood pressure or heart rate, indicating that EQST exerts its anti-obesity effects on adipose tissue directly, rather than acting on the CNS or on the cardiovascular system.
Elevated TCA cycle activity has been linked to increased expression of UCP1 and to the reduction of adipocyte size in brown adipose tissue (BAT), eWAT, and sWAT; this can help promote WAT browning and prevent lipid deposition to the liver and kidneys66. Knockout or downregulation of genes function in lipid droplets formation also help increase UCP1 expression and induces the appearance of brown fat-like phenotype, which promote WAT browning67,68. EQST significantly inhibited lipid droplets formation genes (Fsp27 and Plin5) expression in vitro, EQST treatment also significantly increased UCP1 expression and induced the expression of markers associated with fat burning and elevated TCA cycle activity in sWAT of HFD-induced DIO mice, indicating that EQST promotes energy expenditure in sWAT via promoting WAT browning. Increased expression of PPARγ is known to promote the polarization of adipose tissue macrophages (ATM)69, and our observation of EQST-induced PPARγ repression hints that EQST may exert some anti-inflammatory effects on adipose tissues. More studies will be needed to assess potential impacts of EQST on regulating adipose tissue macrophages. There is a close relationship between UCP1, BAT, and WAT browning70, and it should be informative to conduct additional experiments to explore EQST's impacts for enhancing BAT function and/or promoting WAT browning. For example, primary adipose tissue could be isolated and treated with EQST, followed by analysis seeking phenotypes related to beige adipocytes, beige-progenitor cells70, and possibly even mitochondrial dynamics71.
11β-HSD1 plays a role in metabolic disease by producing locally excessive active glucocorticoids (GCs) in adipose tissue. The metabolism of GCs in obese individuals is unbalanced, resulting from the local upregulation of the expression and activity of 11β-HSD1 in WAT of obese individuals14,72, 73, 74, 75, 11β-HSD1 is also upregulated during the differentiation process of 3T3-L1 preadipocytes56,57. This upregulation results in elevated tissue GCs levels, and excess GCs in adipose tissue have been shown to cause adiposity and insulin resistance76 specifically by inducing PPARγ-dependent adipogenesis77. Given that the effects of GCs in tissue are known to depend on the enzymatic activity of 11β-HSD148,76, the inhibition of 11β-HSD1 has been considered a potential anti-obesity therapeutic strategy78, 79, 80, 81, 82, 83, 84. We found that EQST could inhibit the 11β-HSD1's promotion of lipogenesis induced by excess substrate 11-DHC. And EQST-induced down-regulation of genes integral to adipogenesis and lipid biosynthesis all disappeared in the 11β-HSD1-knockdown cells we examined. Knockout of 11β-HSD1 has been proved to be beneficial to metabolic diseases78, 79, 80,85, whereas enhanced metabolic adaptability of EQST observed in wild-type mice disappeared in 11β-HSD1-KO obese mice supplemented with excessive GC, confirming that 11β-HSD1 is required for the anti-obesity effect of EQST. GCs and 11β-HSD1 also regulate brown adipocyte function. GCs transcriptionally upregulating miR-19b, suppress the expression of beta-1-adrenergic receptors (ADRB1)19, which mediates adrenergic stimulation of UCP186. GCs also transcriptionally upregulate miR-27b and suppress the expression of zinc finger transcriptional co-regulator PR domain-containing protein 16 (PRDM16)18, which plays role in the beige transdifferentiation from white adipocyte and the brown preadipocyte differentiation70,87. Moreover, in hypertrophic adipocytes, increased 11β-HSD1 expression contributes to mitochondrial dysfunction with reduced fatty acid oxidation (FAO) and aerobic respiration, thus inhibiting energy expenditure13. Reducing GCs content by inhibiting the expression of 11β-HSD1 has been proved to promote browning by upregulating UCP1 and other BAT-specific proteins88,89. The observed abolition of EQST-enhanced UCP1 expression after 11β-HSD1-knockdown suggested that EQST upregulates UCP1 in an 11β-HSD1-dependent manner, yet more studies will be required to determine whether EQST and 11β-HSD1 influence WAT browning and if so to characterize their underlying mechanisms.
We also found that EQST displayed a higher-affinity 11β-HSD1 inhibition and better anti-obesity efficacy than the known 11β-HSD1 inhibitor BVT.2733, which has been reported to improve metabolism disorders54,88,90, 91, 92, 93 and to target adipose tissue54,91, but was not appropriate for further development because of the low 11β-HSD1 inhibition potent in human26. Structurally, EQST was predicted to form hydrogen bond interactions with 11β-HSD1's Ser170 and Tyr183 residues; both Ser170 and Tyr183 are known to be key active site residues for cortisone binding with 11β-HSD152 and the formation of H-bond interactions with Ser170 and Tyr183 has been previously proposed as a possible approach for enhancing the activity of 11β-HSD1 inhibitors94. BVT.2733 was predicted to form hydrogen bond interactions with 11β-HSD1 residues Ser170 and Tyr177; Tyr177 has been proposed to play a role in 11β-HSD1's substrate and inhibitor binding but unlikely a hydrogen bond donor95, this may give reason to the BVT.2733's weaker combination with 11β-HSD1 and weaker 11β-HSD1 inhibitory effect even if BVT.2733 was inserted deeper in the binding pocket of 11β-HSD1 than EQST (Supporting Information Fig. S8). 11β-HSD2, another isozyme of 11β-HSD, functions in the inactivation of active GCs48. Non-selective 11β-HSD2 inhibition may increase active GCs content and lead to long-term side effects because of the reduction of GCs inactivation in tissues59,60. Therefore, high selective inhibition over 11β-HSD1 is required for the development of 11β-HSD1 inhibitors58,59. The observed unchanged 11β-HSD2 activity after EQST treatment in vitro suggested that EQST displayed a better-selectivity 11β-HSD1 inhibition. These results may help to explain EQST's anti-obesity effects.
EQST exhibits favorable medicinal potential since that EQST achieve antibacterial effect by inhibiting de novo synthesis of fatty acids96 and inhibiting quorum sensing30, and external application of EQST can effectively promote wound healing by inhibiting bacteria31. Our results also demonstrate that EQST confers potent anti-obesity activity. EQST can also be used to synthesize cancer migration inhibitors fusarin A by chemical synthesis97. Thus, the development of EQST may provide more clues for drug development. With the establishment of chemical synthesis of EQST98 and the discovery of key biosynthetic enzymes of EQST99, more resources for EQST-related research would be provided, which will gradually promote the development of EQST and more marine natural products.
5. Conclusions
Our results establish (i) that EQST inhibited HFD-induced obesity in mice by acting on adipose tissue; (ii) that EQST targets and blocks the enzymatic activity of 11β-HSD1; and (iii) that EQST outperformed the 11β-HSD1 inhibitor and anti-obesity agent BVT.2733. Consequently, EQST is a favorable potential candidate for the development of anti-obesity treatments. Our research may provide more information for the development of novel 11β-HSD1 inhibitors.
Acknowledgments
This study was supported by the following grants: CXYJ-2021-04 from the Research Foundation of Capital Institute of Pediatrics (China); 81573436 from National Natural Science Foundation of China; 2018ZX09711-001-001-016 from the Found of the National New Drug Innovation Major Project of China.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.01.006.
Contributor Information
Wenhan Lin, Email: whlin@bjmu.edu.cn.
Peng Guo, Email: guopeng_chcip@163.com.
Author contributions
Peng Guo, Wenhan Lin, Jing Yuan designed the project. Zhenlu Xu, Dongyun Liu, Guihong Qi performed most of the experiments and analyzed data. Xue Ren analyzed in vitro data. Zhenlu Xu, Dongyun Liu, Guihong Qi, Yue Zhou performed in vivo experiments. Haibo Liu performed target fishing and molecular docking. Peng Guo, Zhenlu Xu and Dongyun Liu wrote the manuscript. Dong Liu provided compound of EQST. Kui Zhu, Zhongmei Zou, Chongming Wu contributed to the manuscript editing. All authors contributed and reviewed the results and approved the final version of the manuscript.
Conflicts of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kusminski C.M., Bickel P.E., Scherer P.E. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat Rev Drug Discov. 2016;15:639–660. doi: 10.1038/nrd.2016.75. [DOI] [PubMed] [Google Scholar]
- 2.Cowherd R., Lyle R., McGehee R. Molecular regulation of adipocyte differentiation. Semin Cell Dev Biol. 1999;10:3–10. doi: 10.1006/scdb.1998.0276. [DOI] [PubMed] [Google Scholar]
- 3.Jakab J., Miskic B., Miksic S., Juranic B., Cosic V., Schwarz D., et al. Adipogenesis as a potential anti-obesity target: a review of pharmacological treatment and natural products. Diabetes Metab Syndr Obes. 2021;14:67–83. doi: 10.2147/DMSO.S281186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu T., Gou L., Yan S., Huang T. Inhibition of acetyl-CoA carboxylase by PP-7a exerts beneficial effects on metabolic dysregulation in a mouse model of diet-induced obesity. Exp Ther Med. 2020;20:521–529. doi: 10.3892/etm.2020.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thyagarajan B., Foster M.T. Beiging of white adipose tissue as a therapeutic strategy for weight loss in humans. Horm Mol Biol Clin Invest. 2017;31:20170016. doi: 10.1515/hmbci-2017-0016. [DOI] [PubMed] [Google Scholar]
- 6.Rui L. Brown and beige adipose tissues in health and disease. Compr Physiol. 2017;7:1281–1306. doi: 10.1002/cphy.c170001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma P., He P., Xu C.Y., Hou B.Y., Qiang G.F., Du G.H. Recent developments in natural products for white adipose tissue browning. Chin J Nat Med. 2020;18:803–817. doi: 10.1016/S1875-5364(20)60021-8. [DOI] [PubMed] [Google Scholar]
- 8.Kuryłowicz A., Puzianowska-Kuźnicka M. Induction of adipose tissue browning as a strategy to combat obesity. Int J Mol Sci. 2020;21:6241. doi: 10.3390/ijms21176241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joharapurkar A., Dhanesha N., Shah G., Kharul R., Jain M. 11β-Hydroxysteroid dehydrogenase type 1: potential therapeutic target for metabolic syndrome. Pharmacol Rep. 2012;64:1055–1065. doi: 10.1016/s1734-1140(12)70903-9. [DOI] [PubMed] [Google Scholar]
- 10.Blaschke M., Koepp R., Streit F., Beismann J., Manthey G., Seitz M.T., et al. The rise in expression and activity of 11β-HSD1 in human mesenchymal progenitor cells induces adipogenesis through increased local cortisol synthesis. J Steroid Biochem Mol Biol. 2021;210:105850. doi: 10.1016/j.jsbmb.2021.105850. [DOI] [PubMed] [Google Scholar]
- 11.Li J., Zhang N., Huang X., Xu J., Fernandes J.C., Dai K., et al. Dexamethasone shifts bone marrow stromal cells from osteoblasts to adipocytes by C/EBPα promoter methylation. Cell Death Dis. 2013;4:e832. doi: 10.1038/cddis.2013.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G., Hernandez-Ono A., Crooke R.M., Graham M.J., Ginsberg H.N. Antisense reduction of 11β-hydroxysteroid dehydrogenase type 1 enhances energy expenditure and insulin sensitivity independent of food intake in C57BL/6J mice on a Western-type diet. Metabolism. 2012;61:823–835. doi: 10.1016/j.metabol.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koh E.H., Kim A.R., Kim H., Kim J.H., Park H.S., Ko M.S., et al. 11beta-HSD1 reduces metabolic efficacy and adiponectin synthesis in hypertrophic adipocytes. J Endocrinol. 2015;225:147–158. doi: 10.1530/JOE-15-0117. [DOI] [PubMed] [Google Scholar]
- 14.Bujalska I.J., Kumar S., Stewart P.M. Does central obesity reflect “Cushing’s disease of the omentum” ?. Lancet. 1997;349:1210–1213. doi: 10.1016/S0140-6736(96)11222-8. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y., Park F., Pietrusz J.L., Jia G., Singh R.J., Netzel B.C., et al. Suppression of 11β-hydroxysteroid dehydrogenase type 1 with RNA interference substantially attenuates 3T3-L1 adipogenesis. Physiol Genom. 2008;32:343–351. doi: 10.1152/physiolgenomics.00067.2007. [DOI] [PubMed] [Google Scholar]
- 16.Bujalska I.J., Gathercole L.L., Tomlinson J.W., Darimont C., Ermolieff J., Fanjul A.N., et al. A novel selective 11β-hydroxysteroid dehydrogenase type 1 inhibitor prevents human adipogenesis. J Endocrinol. 2008;197:297–307. doi: 10.1677/JOE-08-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bujalska I.J., Durrani O.M., Abbott J., Onyimba C.U., Khosla P., Moosavi A.H., et al. Characterisation of 11β-hydroxysteroid dehydrogenase 1 in human orbital adipose tissue: a comparison with subcutaneous and omental fat. J Endocrinol. 2007;192:279–288. doi: 10.1677/JOE-06-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong X., Yu J., Bi J., Qi H., Di W., Wu L., et al. Glucocorticoids transcriptionally regulate miR-27b expression promoting body fat accumulation via suppressing the browning of white adipose tissue. Diabetes. 2015;64:393–404. doi: 10.2337/db14-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv Y.F., Yu J., Sheng Y.L., Huang M., Kong X.C., Di W.J., et al. Glucocorticoids suppress the browning of adipose tissue via miR-19b in male mice. Endocrinology. 2018;159:310–322. doi: 10.1210/en.2017-00566. [DOI] [PubMed] [Google Scholar]
- 20.Park S.B., Park J.S., Jung W.H., Park A., Jo S.R., Kim H.Y., et al. Identification of a novel 11β-HSD1 inhibitor from a high-throughput screen of natural product extracts. Pharmacol Res. 2015;102:245–253. doi: 10.1016/j.phrs.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 21.Zhong C.X., Wang S.Z., Dang L., Xie D.L., Liu J., Ren F.Z., et al. Progress in 11β-HSD1 inhibitors for the treatment of metabolic diseases: a comprehensive guide to their chemical structure diversity in drug development. Eur J Med Chem. 2020;191:112134. doi: 10.1016/j.ejmech.2020.112134. [DOI] [PubMed] [Google Scholar]
- 22.Classen-Houben D., Schuster D., Da Cunha T., Odermatt A., Wolber G., Jordis U., et al. Selective inhibition of 11β-hydroxysteroid dehydrogenase 1 by 18α-glycyrrhetinic acid but not 18β-glycyrrhetinic acid. J Steroid Biochem. 2009;113:248–252. doi: 10.1016/j.jsbmb.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Livingstone D., Walker B. Is 11β-hydroxysteroid dehydrogenase type 1 a therapeutic target? Effects of carbenoxolone in lean and obese Zucker rats. J Pharmacol Exp Therapeut. 2003;305:167–172. doi: 10.1124/jpet.102.044842. [DOI] [PubMed] [Google Scholar]
- 24.Feng Y., Huang S., Dou W., Zhang S., Chen J., Shen Y., et al. Emodin, a natural product, selectively inhibits 11β-hydroxysteroid dehydrogenase type 1 and ameliorates metabolic disorder in diet-induced obese mice. Br J Pharmacol. 2010;161:113–126. doi: 10.1111/j.1476-5381.2010.00826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong X., Fu J., Yin X., Cao S., Li X., Lin L., et al. Emodin: a review of its pharmacology, toxicity and pharmacokinetics. Phytother Res. 2016;30:1207–1218. doi: 10.1002/ptr.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barf T., Vallgårda J., Emond R., Häggström C., Kurz G., Nygren A., et al. Arylsulfonamidothiazoles as a new class of potential antidiabetic drugs. Discovery of potent and selective inhibitors of the 11β-hydroxysteroid dehydrogenase type 1. J Med Chem. 2002;45:3813–3815. doi: 10.1021/jm025530f. [DOI] [PubMed] [Google Scholar]
- 27.Scott J., Bowker S., Deschoolmeester J., Gerhardt S., Hargreaves D., Kilgour E., et al. Discovery of a potent, selective, and orally bioavailable acidic 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) inhibitor: discovery of 2-[(3S)-1-[5-(cyclohexylcarbamoyl)-6-propylsulfanylpyridin-2-yl]-3-piperidyl]acetic acid (AZD4017) J Med Chem. 2012;55:5951–5964. doi: 10.1021/jm300592r. [DOI] [PubMed] [Google Scholar]
- 28.Gregory S., Hill D., Grey B., Ketelbey W., Miller T., Muniz-Terrera G., et al. 11β-Hydroxysteroid dehydrogenase type 1 inhibitor use in human disease-a systematic review and narrative synthesis. Metabolism. 2020;108:154246. doi: 10.1016/j.metabol.2020.154246. [DOI] [PubMed] [Google Scholar]
- 29.Vesonder R.F., Tjarks L.W., Rohwedder W.K., Burmeister H.R., Laugal J.A. Equisetin, an antibiotic from Fusarium equiseti NRRL 5537, identified as a derivative of N-methyl-2,4-pyrollidone. J Antibiot (Tokyo) 1979;32:759–761. doi: 10.7164/antibiotics.32.759. [DOI] [PubMed] [Google Scholar]
- 30.Zhang M., Wang M., Zhu X., Yu W., Gong Q. Equisetin as potential quorum sensing inhibitor of Pseudomonas aeruginosa. Biotechnol Lett. 2018;40:865–870. doi: 10.1007/s10529-018-2527-2. [DOI] [PubMed] [Google Scholar]
- 31.Chen S., Liu D., Zhang Q., Guo P., Ding S., Shen J., et al. A marine antibiotic kills multidrug-resistant bacteria without detectable high-level resistance. ACS Infect Dis. 2021;7:884–893. doi: 10.1021/acsinfecdis.0c00913. [DOI] [PubMed] [Google Scholar]
- 32.Burmeister H.R., Bennett G.A., Vesonder R.F., Hesseltine C.W. Antibiotic produced by Fusarium equiseti NRRL 5537. Antimicrob Agents Chemother. 1974;5:634–639. doi: 10.1128/aac.5.6.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart P., Krozowski Z. 11β-Hydroxysteroid dehydrogenase. Vitam Horm. 1999;57:249–324. [PubMed] [Google Scholar]
- 34.Bujalska I., Kumar S., Hewison M., Stewart P. Differentiation of adipose stromal cells: the roles of glucocorticoids and 11β-hydroxysteroid dehydrogenase. Endocrinology. 1999;140:3188–3196. doi: 10.1210/endo.140.7.6868. [DOI] [PubMed] [Google Scholar]
- 35.Trott O., Olson A. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi G., Zhou Y., Zhang X., Yu J., Li X., Cao X., et al. Cordycepin promotes browning of white adipose tissue through an AMP-activated protein kinase (AMPK)-dependent pathway. Acta Pharm Sin B. 2019;9:135–143. doi: 10.1016/j.apsb.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mota de Sá P., Richard A., Hang H., Stephens J. Transcriptional regulation of adipogenesis. Compr Physiol. 2017;7:635–674. doi: 10.1002/cphy.c160022. [DOI] [PubMed] [Google Scholar]
- 38.Shimano H., Sato R. SREBP-regulated lipid metabolism: convergent physiology-divergent pathophysiology. Nat Rev Endocrinol. 2017;13:710–730. doi: 10.1038/nrendo.2017.91. [DOI] [PubMed] [Google Scholar]
- 39.Walther T., Chung J., Farese R. Lipid droplet biogenesis. Annu Rev Cell Biol. 2017;33:491–510. doi: 10.1146/annurev-cellbio-100616-060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olzmann J., Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol. 2019;20:137–155. doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin C.S., Klingenberg M. Isolation of the uncoupling protein from brown adipose tissue mitochondria. FEBS Lett. 1980;113:299–303. doi: 10.1016/0014-5793(80)80613-2. [DOI] [PubMed] [Google Scholar]
- 42.Kozak L.P., Britton J.H., Kozak U.C., Wells J.M. The mitochondrial uncoupling protein gene. Correlation of exon structure to transmembrane domains. J Biol Chem. 1988;263:12274–12277. [PubMed] [Google Scholar]
- 43.Song N., Chang S., Li D., Villanueva C., Park K. Induction of thermogenic adipocytes: molecular targets and thermogenic small molecules. Exp Mol Med. 2017;49:e353. doi: 10.1038/emm.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narayanaswami V., Dwoskin L. Obesity: current and potential pharmacotherapeutics and targets. Pharmacol Ther. 2017;170:116–147. doi: 10.1016/j.pharmthera.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houten S., Violante S., Ventura F., Wanders R. The biochemistry and physiology of mitochondrial fatty acid β-oxidation and its genetic disorders. Annu Rev Physiol. 2016;78:23–44. doi: 10.1146/annurev-physiol-021115-105045. [DOI] [PubMed] [Google Scholar]
- 46.Cogliati S., Lorenzi I., Rigoni G., Caicci F., Soriano M. Regulation of mitochondrial electron transport chain assembly. J Mol Biol. 2018;430:4849–4873. doi: 10.1016/j.jmb.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 47.Tang M., Liu B., Wang S., Xu Y., Han P., Li P., et al. The role of mitochondrial aconitate (ACO2) in human sperm motility. Syst Biol Reprod Med. 2014;60:251–256. doi: 10.3109/19396368.2014.915360. [DOI] [PubMed] [Google Scholar]
- 48.Chapman K., Holmes M., Seckl J. 11β-Hydroxysteroid dehydrogenases: intracellular gate-keepers of tissue glucocorticoid action. Physiol Rev. 2013;93:1139–1206. doi: 10.1152/physrev.00020.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Staab C., Stegk J., Haenisch S., Neiss E., Köbsch K., Ebert B., et al. Analysis of alternative promoter usage in expression of HSD11B1 including the development of a transcript-specific quantitative real-time PCR method. Chem Biol Interact. 2011;191:104–112. doi: 10.1016/j.cbi.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 50.Wu L., Qi H., Zhong Y., Lv S., Yu J., Liu J., et al. 11β-Hydroxysteroid dehydrogenase type 1 selective inhibitor BVT.2733 protects osteoblasts against endogenous glucocorticoid induced dysfunction. Eedocr J. 2013;60:1047–1058. doi: 10.1507/endocrj.ej12-0376. [DOI] [PubMed] [Google Scholar]
- 51.Wei B., Zhu Z., Xiang M., Song L., Guo W., Lin H., et al. Corticosterone suppresses IL-1β-induced mPGE2 expression through regulation of the 11β-HSD1 bioactivity of synovial fibroblasts in vitro. Exp Ther Med. 2017;13:2161–2168. doi: 10.3892/etm.2017.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hosfield D.J., Wu Y., Skene R.J., Hilgers M., Jennings A., Snell G.P., et al. Conformational flexibility in crystal structures of human 11β-hydroxysteroid dehydrogenase type I provide insights into glucocorticoid interconversion and enzyme regulation. J Biol Chem. 2005;280:4639–4648. doi: 10.1074/jbc.M411104200. [DOI] [PubMed] [Google Scholar]
- 53.Siu M., Johnson T.O., Wang Y., Nair S.K., Taylor W.D., Cripps S.J., et al. N-(Pyridin-2-yl) arylsulfonamide inhibitors of 11β-hydroxysteroid dehydrogenase type 1: discovery of PF-915275. Bioorg Med Chem Lett. 2009;19:3493–3497. doi: 10.1016/j.bmcl.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 54.Wang L., Liu J., Zhang A., Cheng P., Zhang X., Lv S., et al. BVT.2733, a selective 11β-hydroxysteroid dehydrogenase type 1 inhibitor, attenuates obesity and inflammation in diet-induced obese mice. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anagnostis P., Katsiki N., Adamidou F., Athyros V., Karagiannis A., Kita M., et al. 11β-Hydroxysteroid dehydrogenase type 1 inhibitors: novel agents for the treatment of metabolic syndrome and obesity-related disorders?. Metab Clin Exp. 2013;62:21–33. doi: 10.1016/j.metabol.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Jessen B.A., Stevens G.J. Expression profiling during adipocyte differentiation of 3T3-L1 fibroblasts. Gene. 2002;299:95–100. doi: 10.1016/s0378-1119(02)01017-x. [DOI] [PubMed] [Google Scholar]
- 57.Napolitano A., Voice M.W., Edwards C.R., Seckl J.R., Chapman K.E. 11β-Hydroxysteroid dehydrogenase 1 in adipocytes: expression is differentiation-dependent and hormonally regulated. J Steroid Biochem Mol Biol. 1998;64:251–260. doi: 10.1016/s0960-0760(97)00200-8. [DOI] [PubMed] [Google Scholar]
- 58.Chuanxin Z., Shengzheng W., Lei D., Duoli X., Jin L., Fuzeng R., et al. Progress in 11β-HSD1 inhibitors for the treatment of metabolic diseases: a comprehensive guide to their chemical structure diversity in drug development. Eur J Med Chem. 2020;191:112134. doi: 10.1016/j.ejmech.2020.112134. [DOI] [PubMed] [Google Scholar]
- 59.Andrews R., Rooyackers O., Walker B. Effects of the 11β-hydroxysteroid dehydrogenase inhibitor carbenoxolone on insulin sensitivity in men with type 2 diabetes. J Clin Endocrinol Metab. 2003;88:285–291. doi: 10.1210/jc.2002-021194. [DOI] [PubMed] [Google Scholar]
- 60.Kotelevtsev Y., Brown R., Fleming S., Kenyon C., Edwards C., Seckl J., et al. Hypertension in mice lacking 11β-hydroxysteroid dehydrogenase type 2. J Clin Invest. 1999;103:683–689. doi: 10.1172/JCI4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hazuda D., Blau C., Felock P., Hastings J., Pramanik B., Wolfe A., et al. Isolation and characterization of novel human immunodeficiency virus integrase inhibitors from fungal metabolites. Antivir Chem Chemother. 1999;10:63–70. doi: 10.1177/095632029901000202. [DOI] [PubMed] [Google Scholar]
- 62.De Clercq E. Current lead natural products for the chemotherapy of human immunodeficiency virus (HIV) infection. Med Res Rev. 2000;20:323–349. doi: 10.1002/1098-1128(200009)20:5<323::aid-med1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 63.Wang Q., Tao C., Gupta R., Scherer P. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med. 2013;19:1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun K., Kusminski C.M., Scherer P.E. Adipose tissue remodeling and obesity. J Clin Invest. 2011;121:2094–2101. doi: 10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L., Waltenberger B., Pferschy-Wenzig E., Blunder M., Liu X., Malainer C., et al. Natural product agonists of peroxisome proliferator-activated receptor gamma (PPARγ): a review. Biochem Pharmacol. 2014;92:73–89. doi: 10.1016/j.bcp.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mills E., Pierce K., Jedrychowski M., Garrity R., Winther S., Vidoni S., et al. Accumulation of succinate controls activation of adipose tissue thermogenesis. Nature. 2018;560:102–106. doi: 10.1038/s41586-018-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toh S., Gong J., Du G., Li J., Yang S., Ye J., et al. Up-regulation of mitochondrial activity and acquirement of brown adipose tissue-like property in the white adipose tissue of fsp27 deficient mice. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen W., Chang B., Wu X., Li L., Sleeman M., Chan L. Inactivation of Plin4 downregulates Plin5 and reduces cardiac lipid accumulation in mice. Am J Physiol Endocrinol Metab. 2013;304:E770–E779. doi: 10.1152/ajpendo.00523.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li C., Ying W., Huang Z., Brehm T., Morin A., Vella A., et al. IRF6 regulates alternative activation by suppressing PPARγ in male murine macrophages. Endocrinology. 2017;158:2837–2847. doi: 10.1210/en.2017-00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu J., Boström P., Sparks L., Ye L., Choi J., Giang A., et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou Z., Torres M., Sha H., Halbrook C., Van den Bergh F., Reinert R., et al. Endoplasmic reticulum-associated degradation regulates mitochondrial dynamics in brown adipocytes. Science. 2020;368:54–60. doi: 10.1126/science.aay2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rask E., Olsson T., Soderberg S., Andrew R., Livingstone D.E., Johnson O., et al. Tissue-specific dysregulation of cortisol metabolism in human obesity. J Clin Endocrinol Metab. 2001;86:1418–1421. doi: 10.1210/jcem.86.3.7453. [DOI] [PubMed] [Google Scholar]
- 73.Rask E., Walker B.R., Soderberg S., Livingstone D.E., Eliasson M., Johnson O., et al. Tissue-specific changes in peripheral cortisol metabolism in obese women: increased adipose 11β-hydroxysteroid dehydrogenase type 1 activity. J Clin Endocrinol Metab. 2002;87:3330–3336. doi: 10.1210/jcem.87.7.8661. [DOI] [PubMed] [Google Scholar]
- 74.Paulmyer-Lacroix O., Boullu S., Oliver C., Alessi M.C., Grino M. Expression of the mRNA coding for 11β-hydroxysteroid dehydrogenase type 1 in adipose tissue from obese patients: an in situ hybridization study. J Clin Endocrinol Metab. 2002;87:2701–2705. doi: 10.1210/jcem.87.6.8614. [DOI] [PubMed] [Google Scholar]
- 75.Desbriere R., Vuaroqueaux V., Achard V., Boullu-Ciocca S., Labuhn M., Dutour A., et al. 11β-Hydroxysteroid dehydrogenase type 1 mRNA is increased in both visceral and subcutaneous adipose tissue of obese patients. Obesity (Silver Spring) 2006;14:794–798. doi: 10.1038/oby.2006.92. [DOI] [PubMed] [Google Scholar]
- 76.Akalestou E., Genser L., Rutter G. Glucocorticoid metabolism in obesity and following weight loss. Front Endocrinol. 2020;11:59. doi: 10.3389/fendo.2020.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han L., Wang B., Wang R., Gong S., Chen G., Xu W. The shift in the balance between osteoblastogenesis and adipogenesis of mesenchymal stem cells mediated by glucocorticoid receptor. Stem Cell Res Ther. 2019;10:377. doi: 10.1186/s13287-019-1498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seckl J., Walker B. 11β-Hydroxysteroid dehydrogenase type 1 as a modulator of glucocorticoid action: from metabolism to memory. Trends Endocrinol Metabol. 2004;15:418–424. doi: 10.1016/j.tem.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 79.Morton N., Holmes M., Fiévet C., Staels B., Tailleux A., Mullins J., et al. Improved lipid and lipoprotein profile, hepatic insulin sensitivity, and glucose tolerance in 11β-hydroxysteroid dehydrogenase type 1 null mice. J Biol Chem. 2001;276:41293–41300. doi: 10.1074/jbc.M103676200. [DOI] [PubMed] [Google Scholar]
- 80.Morton N., Paterson J., Masuzaki H., Holmes M., Staels B., Fievet C., et al. Novel adipose tissue-mediated resistance to diet-induced visceral obesity in 11β-hydroxysteroid dehydrogenase type 1-deficient mice. Diabetes. 2004;53:931–938. doi: 10.2337/diabetes.53.4.931. [DOI] [PubMed] [Google Scholar]
- 81.Zhang R., Kan J., Lu S., Tong P., Yang J., Xi L., et al. S100A16-induced adipogenesis is associated with up-regulation of 11 β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) Biosci Rep. 2019;39 doi: 10.1042/BSR20182042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu Y., Sun Y., Zhu T., Xie Y., Yu J., Sun W., et al. 11β-Hydroxysteroid dehydrogenase type 1 promotes differentiation of 3T3-L1 preadipocyte. Acta Pharmacol Sin. 2007;28:1198–1204. doi: 10.1111/j.1745-7254.2007.00602.x. [DOI] [PubMed] [Google Scholar]
- 83.Park J., Bae S., Choi S., Son Y., Park S., Rhee S., et al. A novel 11β-HSD1 inhibitor improves diabesity and osteoblast differentiation. J Mol Endocrinol. 2014;52:191–202. doi: 10.1530/JME-13-0177. [DOI] [PubMed] [Google Scholar]
- 84.Morton N.M. Obesity and corticosteroids: 11β-hydroxysteroid type 1 as a cause and therapeutic target in metabolic disease. Mol Cell Endocrinol. 2010;316:154–164. doi: 10.1016/j.mce.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 85.Kotelevtsev Y., Holmes M., Burchell A., Houston P., Schmoll D., Jamieson P., et al. 11β-Hydroxysteroid dehydrogenase type 1 knockout mice show attenuated glucocorticoid-inducible responses and resist hyperglycemia on obesity or stress. Proc Natl Acad Sci U S A. 1997;94:14924–14929. doi: 10.1073/pnas.94.26.14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Riis-Vestergaard M.J., Richelsen B., Bruun J.M., Li W., Hansen J.B., Pedersen S.B. Beta-1 and not beta-3 adrenergic receptors may be the primary regulator of human brown adipocyte metabolism. J Clin Endocrinol Metab. 2020;105:e994–1005. doi: 10.1210/clinem/dgz298. [DOI] [PubMed] [Google Scholar]
- 87.Cristancho A., Lazar M. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu J., Kong X., Wang L., Qi H., Di W., Zhang X., et al. Essential roles of 11β-HSD1 in regulating brown adipocyte function. J Mol Endocrinol. 2013;50:103–113. doi: 10.1530/JME-12-0099. [DOI] [PubMed] [Google Scholar]
- 89.Soumano K., Desbiens S., Rabelo R., Bakopanos E., Camirand A., Silva J.E. Glucocorticoids inhibit the transcriptional response of the uncoupling protein-1 gene to adrenergic stimulation in a brown adipose cell line. Mol Cell Endocrinol. 2000;165:7–15. doi: 10.1016/s0303-7207(00)00276-8. [DOI] [PubMed] [Google Scholar]
- 90.Xie Y., Zhu T., Zhong Y., Liu J., Yu J., Zha J.M., et al. Mechanism of BVT.2733 and pioglitazone in the improvement of insulin resistance. Chin J Intern Med. 2008;47:938–941. [PubMed] [Google Scholar]
- 91.Liu J., Wang L., Zhang A., Di W., Zhang X., Wu L., et al. Adipose tissue-targeted 11β-hydroxysteroid dehydrogenase type 1 inhibitor protects against diet-induced obesity. Endocr J. 2011;58:199–209. doi: 10.1507/endocrj.k10e-318. [DOI] [PubMed] [Google Scholar]
- 92.Shao S., Zhang X., Zhang M. Inhibition of 11β-hydroxysteroid dehydrogenase type 1 ameliorates obesity-related insulin resistance. Biochem Biophys Res Commun. 2016;478:474–480. doi: 10.1016/j.bbrc.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 93.Huang M., Liu J., Sheng Y., Lv Y., Yu J., Qi H., et al. 11β-Hydroxysteroid dehydrogenase type 1 inhibitor attenuates high-fat diet induced cardiomyopathy. J Mol Cell Cardiol. 2018;125:106–116. doi: 10.1016/j.yjmcc.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 94.Qian H., Chen J., Pan Y., Chen J. Molecular modeling studies of 11β-hydroxysteroid dehydrogenase type 1 inhibitors through receptor-based 3D-QSAR and molecular dynamics simulations. Molecules. 2016;21:1222. doi: 10.3390/molecules21091222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim K., Wang Z., Busby J., Tsuruda T., Chen M., Hale C., et al. The role of tyrosine 177 in human 11β-hydroxysteroid dehydrogenase type 1 in substrate and inhibitor binding: an unlikely hydrogen bond donor for the substrate. Biochim Biophys Acta. 2006;1764:824–830. doi: 10.1016/j.bbapap.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 96.Larson E., Lim A., Pond C., Craft M., Čavužić M., Waldrop G., et al. Pyrrolocin C and equisetin inhibit bacterial acetyl-CoA carboxylase. PLoS One. 2020;15 doi: 10.1371/journal.pone.0233485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu J., Caro-Diaz E., Lacoske M., Hung C., Jamora C., Theodorakis E. Fusarisetin A: scalable total synthesis and related studies. Chem Sci. 2012;3:3378–3386. doi: 10.1039/C2SC21308G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Burke L., Dixon D., Ley S., Rodríguez F. Total synthesis of the fusarium toxin equisetin. Org Biomol Chem. 2005;3:274–280. doi: 10.1039/b411350k. [DOI] [PubMed] [Google Scholar]
- 99.Sims J.W., Fillmore J.P., Warner D.D., Schmidt E.W. Equisetin biosynthesis in Fusarium heterosporum. Chem Commun (Camb) 2005;(2):186–188. doi: 10.1039/b413523g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.