Summary
CFTR F508del (c.1521_1523delCTT, p.Phe508delPhe) is the most common pathogenic allele underlying cystic fibrosis (CF), and its frequency varies in a geographic cline across Europe. We hypothesized that genetic variation associated with this cline is overrepresented in a large cohort (N > 5,000) of persons with CF who underwent whole-genome sequencing and that this pattern could result in spurious associations between variants correlated with both the F508del genotype and CF-related outcomes. Using principal-component (PC) analyses, we showed that variation in the CFTR region disproportionately contributes to a PC explaining a relatively high proportion of genetic variance. Variation near CFTR was correlated with population structure among persons with CF, and this correlation was driven by a subset of the sample inferred to have European ancestry. We performed genome-wide association studies comparing persons with CF with one versus two copies of the F508del allele; this allowed us to identify genetic variation associated with the F508del allele and to determine that standard PC-adjustment strategies eliminated the significant association signals. Our results suggest that PC adjustment can adequately prevent spurious associations between genetic variants and CF-related traits and are therefore effective tools to control for population structure even when population structure is confounded with disease severity and a common pathogenic variant.
Keywords: CFTR F508del, population structure, genome-wide association study
Introduction
Cystic fibrosis (CF; MIM: 219700) is the most common life-shortening recessive disorder in people of European descent, affecting >90,000 people worldwide. CF is monogenic, due to mutations in CF transmembrane regulator (CFTR; MIM: 602421).1,2 Both extensive allelic heterogeneity and genetic modifiers across the genome influence the phenotypic presentation of CF.3 While >2,000 pathogenic variants in CFTR have been reported and 360 verified as pathogenic by the Clinical and Functional Translation of CFTR project (CFTR2), approximately 82% of persons with CF carry ≥1 copy of the F508del allele (c.1521_1523delCTT, p.Phe508delPhe).2 The F508del variant results in little to no protein function due to impaired traffic of the protein product to the membrane and reduced stability within the membrane. F508del-allele frequency ranges from 1.4% among those with predominantly non-Finnish European ancestry to <0.4% among those with predominantly sub-Saharan African, Asian, or Native American ancestry.2,4 The F508del allele also follows a northwest-southeast cline in Europe, with a frequency of 87.2% among Danish people with CF but only 21.3% among Turkish people with CF.5 This suggests that CFTR F508del genotypes may be correlated with other genetic variants exhibiting a similar cline across Europe.
There is no consistent guidance on how genetic association studies of CF should account for CFTR genotype. While phenotypes such as pancreatic insufficiency are strongly predicted by CFTR genotype, others vary widely despite a shared CFTR genotype.6 F508del is the most common pathogenic variant and may cause severe disease, particularly in homozygotes and in the absence of modulator therapy. Studies of CF-related traits are sometimes restricted to F508del homozygotes to minimize phenotypic variation explained by CFTR genotype; however, such restrictions reduce the generalizability of the study to the larger CF community (H. Ling et al., 2020, N. Am. Cyst. Fibros. Conf., abstract).
Variants correlated with CFTR F508del because of shared ancestry may be spuriously associated with CF-related outcomes despite having no direct effect on the condition. While genome-wide association studies (GWASs) typically include principal components (PCs) as covariates to effectively control for population structure,7 subtle population structure, such as that within Britain, is not always adequately captured by PCs.8 Because CFTR F508del is both correlated with geography within Europe and with clinical outcomes for persons with CF, confidence that PC covariates eliminate this source of confounding in GWAS for CF modifiers is required.9
To better understand the relationship between CFTR F508del and variation across the genome among persons with CF, we examine population structure within the Cystic Fibrosis Genome Project (CFGP) using PC analysis (PCA) and association testing. We identify variants associated with F508del genotype among persons with CF and test whether including PCs as covariates can effectively eliminate these associations. We investigate whether the loci identified by this GWAS have previously been associated with population structure in Europe. Finally, we determine whether variants previously associated with CF-related phenotypes are also associated with F508del genotype, suggesting inadequate control for population structure linked to CFTR F508del in the original studies.
Subjects and methods
The CFGP data
The CFGP data consist of whole-genome sequence (WGS) data from persons with CF enrolled in cohort studies at three sites. Johns Hopkins University (JHU) contributed samples from the CF Twin-Sibling Study,10 which includes persons from 95% of known twins and sibling pairs affected by CF in the Cystic Fibrosis-Related Diabetes study.11 The University of North Carolina (UNC) contributed samples from the North American Cystic Fibrosis Genetic Modifier Study.3,12 The University of Washington/Seattle Children’s Hospital (UW) contributed samples from the Early Pseudomonas Infection Control Observational Study, representing patients who were both younger than 13 years between 2004 and 2006 and either never had a positive culture for Pseudomonas aeruginosa (Pa) or had been negative for Pa for at least 2 years (≥1 culture/year) prior to enrollment.13 All individuals in the CFGP were consented and enrolled in one or more of the individual studies, which were reviewed and received IRB approval at their respective institutions.
WGS on 5,199 samples was performed by the Broad Institute Sequencing Center using their PCR-free Illumina sequencing-by-synthesis protocol (supplemental methods). Briefly, cluster amplification was performed per manufacturer’s protocol by the Illumina cBot. Sequencing was performed on HiSeq X machines to produce 151 bp paired-end reads. These data were processed by a Picard data-processing pipeline to generate sample-level BAM14 and variant call format (VCF)15 files aligned to the GRCh38 reference genome.16 Of the original 5,199 samples, 5,134 passed all of the sequencing center’s quality control (QC) filters. A multi-sample VCF for these 5,134 samples was provided, generated using HaplotypeCaller in the Genome Analysis Toolkit (GATK; v.4.0.9.0)17, containing >120 M single-nucleotide variants (SNVs) and short insertion/deletions (indels).
Samples were excluded from analysis if they showed evidence for contamination (Freemix estimate ≥2%)18, high chimera rate (≥5%), low coverage (mean ≤29.5X, 20X coverage ≤85%, 10X coverage ≤95%), discordant pedigree versus empirical kinship estimates,19,20 sample duplication or identity errors, or low support for CF diagnosis. CFTR genotypes in the WGS data and the national Cystic Fibrosis Patient Registry data underwent expert review,21 identifying or verifying the pathogenic variants for all but 26 participants whose data were excluded from further analysis. A total of 4,966 samples (JHU: 1,643; UNC: 1,706; UW: 1,244) passed all sample-level QC filters. Of the 96 M SNVs and indels passing both the GATK Variant Quality Score Recalibration and GATK hard filters (QualByDepth [QD] >2; Quality [QUAL] >30; StrandOddsRatio [SOR] <3; FisherStrand [FS] <60; RMSMappingQuality [MQ] > 40; ReadPosRankSum > -8), we restricted analysis to the 5,490,867 (5,483,159 in analyses restricted to F508del carriers) autosomal biallelic SNVs with minor allele frequency (MAF) >5% and missingness rate <5%.
Relatedness and PCs
We estimated orthogonally partitioned genetic structure in our sample using the GENESIS22 package in R,23 generating PCs and a genetic relatedness matrix (GRM) using the 4,966 samples that passed QC and a subset of 177,718 variants after pruning the 5.5 M SNVs passing QC for linkage disequilibrium (LD; r2 < 0.1). We estimated empirical relatedness using KING-Robust20 to identify the “unrelated” set (kinship <2-9/2) for PC-AiR.9 We then adjusted the KING-Robust estimates for PCs to create a GRM using PC-Relate.19 With this approach, kinship coefficients are independent of the population-level structure captured by PCs, reducing the likelihood of over-correcting for relatedness or introducing spurious associations.19,22 We repeated the PC-AiR and -Relate processes a second time using the results from the first round to improve precision.22 The percent variance explained drops from the first to the second PC and so on, then plateaus; we define early PCs as those preceding and including the inflection point of a scree plot. For each early PC, we calculated the correlation between eigenvector values and each of the 5.5 M SNVs passing QC.
Testing for association with CFTR F508del genotype
To identify variants that may share similar population histories with the CFTR F508del allele, we performed a GWAS comparing persons with CF with one versus two copies of the F508del allele. This binary outcome was tested using the logistic mixed model with orthogonally partitioned structure (LMM-OPS) approach implemented in the GENESIS22 package in R.23 We excluded an additional 373 unique samples representing one identical twin per pair (n = 27) and those that did not carry a CFTR F508del allele (n = 349), leaving 4,593 samples for the GWAS. We performed single-variant association testing for each of the 5.5 M SNVs passing QC using two LMMs: a baseline model adjusted for site of recruitment as a fixed effect and the GRM as a random effect, and a PC-adjusted model that added the first four PCs as fixed effect covariates to the baseline model. Genome-wide significance was defined using the threshold p < 5 × 10-8; this value is slightly less stringent than the conservative Bonferroni-corrected threshold (p < 0.05/5.5 M = 9.1 × 10-9), which does not account for LD between variants.
Variants correlated with pathogenic or modifier variants due to shared population history may appear to be associated with the outcome if it also varies in frequency by ancestry,8 leading to an increase in test statistic values and a corresponding decrease in p values across the genome. We measure evidence of this shift using the genomic inflation factor (λ),7,24 defined as the squared median test statistic divided by 0.455. A λ value >1 indicates genomic inflation or an inflated ratio of observed-to-expected test statistics under the null chi-squared distribution.
Secondary-subset analyses
Because the frequency of the CFTR F508del allele varies across Europe and between populations, we analyzed subsets of the CFGP to determine whether the correlation between variation near CFTR and PCs is ancestry dependent. One subset (European) contained 4,567 individuals with >80% estimated European ancestry based on Somalier25 analysis, while the other (non-European) contained 270 individuals with <20% estimated European ancestry (supplemental methods). PCs and GRMs were calculated separately within each subset using the method described above. Within the European-ancestry subset, 5,456,627 SNVs passed the same QC, MAF, and missingness filters as described above, with 173,575 SNVs remaining after LD pruning for PCA and GRM estimations. Similarly, 6,441,286 SNVs passed QC, MAF, and missingness filters in the primarily non-European-ancestry subset, with 240,152 SNVs remaining after LD pruning.
We also performed an analysis of the CFGP Europeans restricted to a subset of the 5.4 M SNVs passing our QC that were also rare (MAF ≤5%) in a non-Finnish European reference panel (gnomAD v.3.1.1)4; only 37,943 SNVs remained. The CFGP Europeans share a similar ancestry composition with the non-Finnish Europeans, making this set over-represent SNVs with observed frequencies affected by ascertainment bias. New PCA and GRM analyses were performed using an LD-pruned set of 11,404 SNVs, the KING-Robust estimates from the original analysis of CFGP Europeans, and the PC-AiR/PC-Relate approach described above.
Trans-Omics for Precision Medicine (TOPMed) analysis
We performed a similar PCA in an independent dataset representing participants with predominantly European ancestry ascertained for different phenotypes as part of the TOPMed program. The purpose of this analysis was to differentiate population structure in the CFGP reflecting European ancestry in general versus that which is specifically correlated with CFTR variation.26 We selected samples representing participants who consented to general research and were reported as non-Hispanic White (n = 3,199) from five studies unrelated to CF: Cleveland Clinic Atrial Fibrillation Study (CCAF; n = 358); Mayo Clinic Venous Thromboembolism Study (Mayo VTE; n = 707); Defining the Time-Dependent Genetic and Transcriptomic Responses to Cardiac Injury among Patients with Arrhythmias (miRhythm; n = 67); Severe Asthma Research Program (SARP; n = 1017); and Vanderbilt Genetic Basis of Atrial Fibrillation (VU_AF, n = 1052). Samples were sequenced at one of three sequencing centers: the Broad Institute for Human Genetics (CCAF, miRhythm, and VU_AF), the Baylor College of Medicine Human Genome Sequencing Center (Mayo VTE), or the New York Genome Center (SARP). These data are available through the database of Genotypes and Phenotypes (dbGaP) under the following study accession numbers: CCAF (phs001189), Mayo VTE (phs001402), miRhythm (phs001434), SARP (phs001446), and VU_AF (phs001032).
WGS from TOPMed was performed at an average depth of 38× across using Illumina HiSeq X Ten technology. All sequencing centers used a common pipeline to align reads to human genome build GRCh38. Read alignment was harmonized, and variants were jointly called by the TOPMed Informatics Research Center. Variant QC was performed using a support vector machine classifier, with additional filtering for variants with excess heterozygosity and Mendelian discordance. Sample QC and identity resolution was performed by the TOPMed Data Coordinating Center using annotated and genetic sex concordance, concordance with previous genotyping, and expected and observed relatedness when possible. Details regarding the sequencing methods, variant calling, and QC can be found on the TOPMed website (https://topmed.nhlbi.nih.gov), in a common methods document in each study’s dbGaP directory, and in a published paper.26
We extracted the same 177,718 SNVs from the CFGP PCA in the TOPMed data then applied the same QC, MAF, and missingness filters; 166,628 SNVs remained. These 166,628 SNVs were used to estimate PCs and a GRM using PC-AiR and PC-Relate as described above. Finally, we estimated the correlation between the TOPMed PCs and each of the 6,221,456 autosomal biallelic SNVs passing the QC and 5% MAF and missingness filters described above.
Regions of interest
Certain regions of the human genome exhibit long-range LD,27,28 defined as the correlation between SNPs that extends across a physical distance that is atypical of most of the genome (e.g., across chromosomes). Long-range LD is explained by limited recombination, such as that observed at inversion polymorphisms, regions under selection, or within recently admixed populations. Variation within these regions can be associated with population structure or cause spurious associations in GWASs.7,28 Variation within four regions of the genome exhibiting long-range LD are strongly correlated with PCs representing participants with European ancestry,29 which we define using the bounds provided in the GWASTools R package:30 the 2q21.1-2q22.1 region containing LCT (MIM: 603202) associated with lactase persistence,31 the 6p22.3-6p21.2 region containing the major histocompatibility complex (MHC),32 and two inversion polymorphisms at 8p23 and 17q21.31.33 We investigated whether loci correlated with early PCs or associated with CFTR F508del genotype within the CFGP fall within these regions.
Prior genetic-modifier studies of CF-related traits may have been vulnerable to spurious associations due to genotype correlations with CFTR F508del. We identified 10 autosomal SNVs previously associated with CF-related traits by GWAS (p < 5 × 10-8)11,34, 35, 36 and determined whether they were also associated with F508del genotype in the CFGP due to confounding with F508del-associated population structure.
Results
Sample description
The CFGP data represent multiple studies that ascertained participants based on different clinical criteria and familial relationships, resulting in varied demographic characteristics and CFTR F508del genotype distributions across study sites (Table 1). The majority of participants (58%) were CFTR F508del homozygotes and 93% had at least one copy of F508del. Participants in the UNC cohort were more likely to have one copy of F508del (96%) or be F508del homozygotes (72%) compared with participants in the UW (92% carriers, 54% homozygotes) or JHU cohorts (91% carriers, 48% homozygotes). As the F508del carriers represent the majority of the sample, their descriptive statistics are nearly identical (Table S1). Although the F508del allele represents 75% of the pathogenic CFTR allele within the CFGP, dozens of other pathogenic variants were observed (Table S2).
Table 1.
Summary of the Cystic Fibrosis Genome Project participants
|
Institution |
John Hopkins University |
University of North Carolina |
University of Washington |
Total |
|---|---|---|---|---|
| Study(s) | Cystic fibrosis-related diabetes, twins and Sibs | Genetic-modifier study | Early pseudomonas infection control observational study | |
| Total | 1,809 | 1,783 | 1,347 | 4,939 |
| Birth year: mean (range) | 1991 (1943–2011) | 1982 (1946–2007) | 2000 (1992–2006) | 1900 (1943–2011) |
| Age diagnosed, years: mean (SD) | 2.4 (5.7) | 2.4 (4.5) | 0.9 (1.7) | 2.0 (4.5) |
| Genotype: N (%) | ||||
| CFTR F508del carriersa | 1,643 (90.8) | 1,706 (95.7) | 1,244 (92.4) | 4,593 (93.0) |
| CFTR F508del homozygotes | 875 (48.4) | 1,282 (71.9) | 722 (53.6) | 2,879 (58.3) |
| Male: N (%) | 946 (52.3) | 1,004 (56.3) | 673 (50.0) | 2,623 (53.1) |
| Empirical ancestry: N (%) | ||||
| African | 32 (1.8) | 25 (1.4) | 33 (2.4) | 90 (1.8) |
| Native American | 86 (4.8) | 46 (2.6) | 64 (4.8) | 196 (4.0) |
| East Asian | 4 (0.2) | 0 (0) | 1 (0.1) | 5 (0.1) |
| European | 1,681 (92.9) | 1,710 (95.9) | 1,247 (92.6) | 4,638 (93.9) |
| South Asian | 6 (0.3) | 2 (0.1) | 2 (0.1) | 10 (0.2) |
Values are provided for the 4,939 participants passing quality control and included in PCAs. Estimated ancestry defined as the ancestry group with the highest probability estimated by Somalier analysis. Details limited to carriers are presented in Table S1.
CFTR F508del homozygotes were included in the count of carriers.
Genetic structure within the CFGP
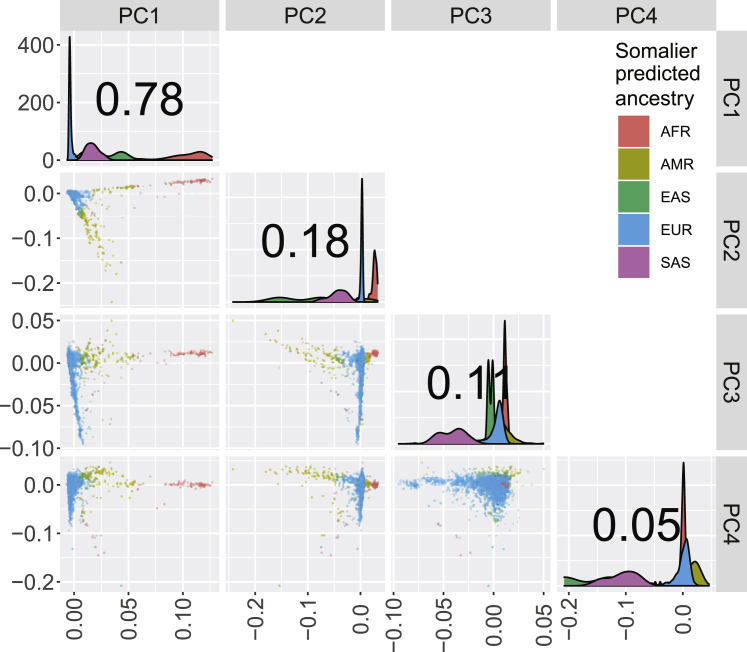
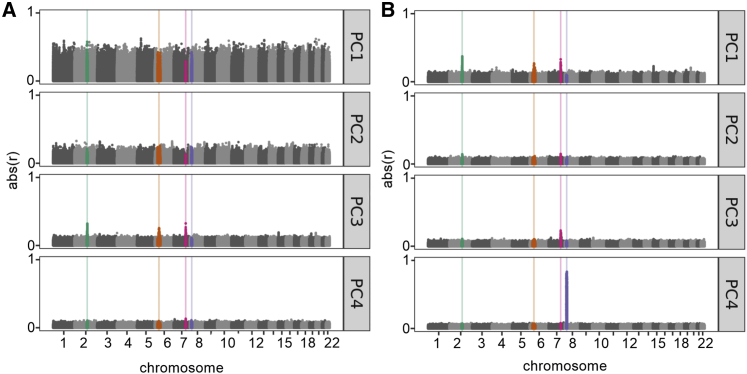
PCA identifies orthogonal axes of variation within multidimensional data, ranking them by the amount of variance explained. In human genetic data, these axes of variation often correspond with historical patterns of migration or population structure. Among the early PCs estimated in the CFGP data, PC3 captures variation between participants in the European subset (Figure 1). PC3 shows a higher Pearson correlation with variation at CFTR (rmax = 0.33) and with two of the four loci known to exhibit long-range LD in persons with European ancestry: the 2q21 region containing LCT (rmax = 0.32) and the 6p21.32 region containing the MHC locus (rmax = 0.26) (Figure 2A). Recalculating PCs while excluding variants within the four loci known to exhibit long-range LD did not substantially alter the correlations between variants at these loci and PC3. In an analysis of the subset of the CFGP with >80% European ancestry, variation at the CFTR, 2q21, and 6p21.32 loci is strongly correlated with PC1 (Figures 2B and S1), with the PC capturing the most genetic variation within this subset. Sensitivity analyses excluding variants with MAF >5% in non-Finnish Europeans echoed the signals at CFTR and the MHC but identified no additional influential loci (Figures S2 and S3). Similar analyses of the CFGP participants with <20% European ancestry do not show above-average correlation between these three loci and the early PCs, although the overall magnitude of correlation between variations across the genomes and early PCs is higher in this subset representing greater ancestral diversity (Figures S4 and S5). These subset analyses suggest that variation near CFTR, 2q21, and 6p21.32 have a greater than background influence on early PCs estimated in a sample of people with CF with European ancestry. Analysis of the TOPMed data suggests that PC-correlation peaks at 2q21 and possibly 6p21.32 and 8p23 are expected from a sample with primarily European ancestry, while the peak at CFTR is specific to datasets over-representing persons with CF (Figures S6 and S7).
Figure 1.
Population structure within the entire CFGP (n = 4,939)
Pairwise principal-component (PC) plots are shown for PCs 1–4 with frequency distributions and percentage of variance explained by each PC on the diagonal. Ancestry estimates indicate the ancestry with the highest estimated proportion using Somalier.25 Abbreviations: AFR, sub-Saharan African; AMR, Native American; EAS, East Asian; EUR, European; SAS, South Asian.
Figure 2.
Correlation between PCs and genomic position
(A and B) The correlation between PCs (Y axis) and genomic position (X axis) are shown for the (A) CFGP (n = 4,939) and (B) CFGP participants with estimated European ancestry >80% (n = 4,567). The number of PCs shown is the number used to calculate the genetic relatedness matrix and, for the total CFGP dataset, used in the PC-adjusted GWAS analysis. Color-coded regions include 7q21.31 (CFTR, pink) and three regions that have previously shown evidence of long-range LD: 2q21.1-2q22.1 (LCT, teal), 6p22.3-6p21.2 (the major histocompatibility complex, orange), and the 8p23 inversion polymorphism (purple).
Genome-wide association between SNVs and CFTR F508del genotype
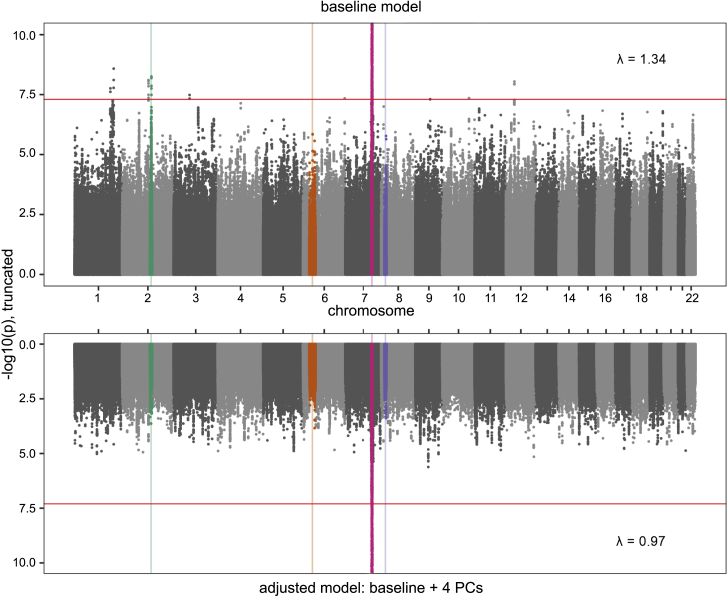
CFTR F508del genotype was significantly associated with 10 loci including CFTR (p < 1 × 10-300) under the baseline model (Figure 3; Table 2), which showed strong evidence for genomic inflation (λ = 1.34; Figure S8). After adding PCs 1–4 to the baseline model, the evidence for genomic inflation vanished (λ = 0.97) as did the significant evidence of association with F508del genotype outside the CFTR locus (Figure 3; Table S3). The evidence of association between F508del and the long-range LD regions in Europeans was not significant in the PC-adjusted model (p > 0.0001; Table S4). However, the top 10 peaks in the PC-adjusted model had similar evidence for association with F508del genotype under the baseline model, which suggests that subtle population structure linked to CFTR may not be captured by early PCs in studies of CF populations. None of the 10 autosomal SNVs previously associated with CF-related traits were nominally associated with F508del genotype in the PC-adjusted model, and only one reached nominal significance using the baseline model (rs546131 near APIP, p = 0.0426; Table S5), suggesting that the evidence of association between these 10 SNVs and CF-related phenotypes is not due to indirect association with CFTR F508del. Whereas variants across the genome are associated with F508del genotype under the baseline model, adjusting the model with early PC covariates adequately controls for the corresponding population structure.
Figure 3.
GWASs for CFTR F508del heterozygosity versus homozygosity
(Top) The baseline model adjusted for site and relatedness. (Bottom) The PC-adjusted model. Association signals are measured as -log10(p values). Plot is truncated at p = 1 × 10-10, as the peak at CFTR on chr7 reaches p < 1 × 10-300 under both models. The genome-wide significance level, p < 5 × 10-8, is indicated by the horizontal line.
Table 2.
Regions of the genome significantly associated with CFTR F508del heterozygosity versus homozygosity under the baseline model
| Region | Nearest gene | rsID | REF allele | ALT allele | AAFNFE | AAFCFGP | p |
|---|---|---|---|---|---|---|---|
| 1q31.3 | AL450352.1 | rs2813164 | A | G | 0.33 | 0.33 | 1.74 × 10-8 |
| 1q41 | PROX1-AS1 | rs853741 | G | A | 0.95 | 0.94 | 2.62 × 10-9 |
| 2q14.3 | AC062020 | rs1911632 | A | C | 0.17 | 0.20 | 7.84 × 10-9 |
| 2q22.1 | THSD7B | rs533344 | T | A | 0.72 | 0.70 | 5.58 × 10-9 |
| 3p14.1 | SUCLG2 | rs11127729 | T | C | 0.95 | 0.95 | 3.28 × 10-8 |
| 6q27 | AL611929.1 | rs9455973 | G | A | 0.08 | 0.10 | 4.56 × 10-8 |
| 7q31.2 | CFTR | rs7802924 | A | G | 0.09 | 0.85 | 1 × 10-300 |
| 9q21.32 | SLC28A3 | rs6559779 | A | G | 0.10 | 0.10 | 4.95 × 10-8 |
| 10q25.2 | AL136119.1 | rs1923653 | A | G | 0.94 | 0.92 | 4.47 × 10-8 |
| 12p11.1 | SYT10 | rs949473 | G | A | 0.14 | 0.16 | 8.99 × 10-9 |
Significant p value threshold: 5 × 10-8. The baseline association model is adjusted for site and a genetic relatedness matrix. Sequence positions of association peaks are provided on the GRCh38 map. Alternate allele frequencies (AAFs) are given for non-Finnish Europeans in gnomAD v.3.1.14 and within the CFGP.
Discussion
We have shown that PCA and GWAS can identify population structure driven by ascertainment for an allele strongly associated with disease status. Within the CFGP, PC3 is driven by three genomic loci harboring variants whose distribution is correlated with geography in Europe. GWAS reveals significant association between SNVs across the genome and CFTR F508del genotype, leading to substantial genomic inflation that can be eliminated by including PC covariates in the analysis. The loci driving PC3 were also associated with F508del genotype and include a region with prior evidence of long-range LD: rs533344 that falls within 750 KB of LCT (MIM: 603202) and MCM6 (MIM: 601806), a regulator of LCT). The frequency of the MCM6 alleles associated with greater lactase persistence is higher in populations with European ancestry and follows a similar distribution to CFTR F508del across Europe.2,31 The relatively strong contribution of CFTR variation to early PCs was only observed in CFGP analyses including participants with high European-ancestry probabilities; similar correlations between variations at the CFTR locus and early PCs were not observed in analyses of the CFGP non-Europeans or the TOPMed data.
GWAS comparing persons with CF with one versus two CFTR F508del alleles did not demonstrate genomic inflation or reveal significant association signals when early PCs were included as covariates. These results suggest that GWASs for genetic modifiers of CF can be reliably performed in datasets ascertained for CF in general or for CFTR F508del genotype specifically so long as the PCs included as covariates capture population structure within Europe. We suggest that investigators directly assess the association between PCs and European ancestry and/or CFTR genotype to verify that the PCs included as covariates in their study specifically capture this variation. This is consistent with the observation that prior GWASs of CF-related traits,11,34, 35, 36 which included PC covariates, did not suffer from genomic inflation or identify significant associations with variants correlated with CFTR variation in the CFGP.
Our study has several limitations. This study focused on the CFTR F508del allele as it is the most common pathogenic variant within the CFGP and does not investigate potential cross-chromosomal correlations with other relatively common (>1%) pathogenic variants in CFTR. For example, non-European populations are not well represented within the CFGP, preventing similar investigations of founder alleles including the S549R allele (c.1645A>C, p.Ser549Arg) common in the United Arab Emirates or the Y122X allele (c.366T>A, p.Tyr122X), which is more common on Réunion Island.37 Participants without a copy of the CFTR F508del allele were excluded from the GWAS presented. GWASs including these individuals could identify other loci correlated with CFTR genotype, but such analyses were not well powered in the CFGP given the small number of participants (n = 346) without a CFTR F508del allele. This analysis focuses exclusively on genetic stratification, and PCs may not be able to control for environmental stratification within the sample. Genuine genetic modifiers of CF may be correlated with population structure; GWASs including PCs capturing this structure will have reduced statistical power to detect such modifiers. Rare variants arose more recently and are limited in their geographic distribution relative to common variants, and their influence on association results may not be detected by PCA or genomic-inflation factors.38 Aggregate association testing of rare variants and CF-related outcomes may therefore yield spurious results uncontrolled by PC covariates.
Our approach for identifying loci associated with a strong genetic predictor of a trait could be extended to other traits where the frequency of the most common pathogenic allele varies by ancestry. Similar concerns may arise in genetic studies of traits meeting the following criteria: (1) there is a relatively common variant underlying a Mendelian condition; (2) multiple pathogenic alleles exist; (3) the severity or expression of the trait varies across these alleles; and (4) the frequency of the pathogenic allele varies by population or with geography. For example, sickle cell disease results from pathogenic variants in HBB, where homozygosity for the HbS allele (c.20A>T, p.Glu7Val) leads to sickle cell disease (MIM: 603903), while other HBB variants associated with specific geographic regions result in other hereditary anemias.39 Major-effect alleles for complex traits can also be correlated with population structure, including the APOE (MIM: 107741) ε4 allele (c.388T>C, p.Cys130Arg), which is common, varies in frequency across Europe, and is a major risk allele for late-onset Alzheimer’s disease (MIM: 104310).4,40 In these situations, we suggest that the relationship between pathogenic or major-effect alleles and population structure within Europe be evaluated to ensure that significant genotype-phenotype associations are not driven by an indirect relationship with the allele.
Acknowledgments
We thank the participants and their families for their contribution to this work. This work was supported by the Cystic Fibrosis Foundation (grant nos. BAMSHA18XX0, CUTTIN18XX1, and KNOWLE18XX0]. The authors wish to acknowledge the contributions of the consortium working on the development of the NHLBI BioData Catalyst ecosystem and the analysis pipeline support provided by National Institutes of Health (NIH) National Institute on Aging R01AG059737. We gratefully acknowledge the studies and participants who provided biological samples and data for the TOPMed program and provide detailed acknowledgments by study in the supplement.
Declaration of interests
M.B. is the editor-in-chief and J.X.C. (member of the Cystic Fibrosis Genome Project) is the deputy editor of HGG Advances. The authors declare no other competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2022.100117.
Web resources
Bioconductor: Open Source Software for Bioinformatics, https://bioconductor.org/.
The Broad Institute Whole Genome Sequencing service, http://genomics.broadinstitute.org/products/whole-genome-sequencing.
The Clinical and Functional Translation of CFTR (CFTR2), http://cftr2.org.
The Cystic Fibrosis Foundation Patient Registry, https://www.cff.org/Research/Researcher-Resources/Patient-Registry/.
GENetic Estimation and Inference in Structured samples (GENESIS), https://bioconductor.org/packages/release/bioc/html/GENESIS.html.
Genome Aggregation Database (gnomAD), https://gnomad.broadinstitute.org/.
The National Heart, Lung, and Blood Institute Trans-Omics for Precision Medicine (TOPMed), https://nhlbiwgs.org.
OMIM, http://www.omim.org/.
R Statistical Software, https://www.r-project.org/.
TOPMed analysis pipeline, https://github.com/UW-GAC/analysis_pipeline.
Supplemental information
Data and code availability
CFGP sequence data and phenotypic information have been placed in a restricted access database maintained by the Cystic Fibrosis Foundation and is available to approved researchers (see https://www.cff.org/researchers/whole-genome-sequencing-project-data-requests for details). TOPMed data are available to approved researchers through dbGaP under the following study accession numbers: CCAF (phs001189), Mayo VTE (phs001402), miRhythm (phs001434), SARP (phs001446), and VU_AF (phs001032). The analysis pipeline is derived from the TOPMed analysis pipeline, which is publicly available on GitHub (https://github.com/UW-GAC/analysis_pipeline). This pipeline utilizes tools within the GENESIS package, which is publicly available on Bioconductor (https://bioconductor.org/packages/release/bioc/html/GENESIS.html).
References
- 1.Bell S.C., Mall M.A., Gutierrez H., Macek M., Madge S., Davies J.C., Burgel P.R., Tullis E., Castanos C., Castellani C., et al. The future of cystic fibrosis care: a global perspective. Lancet Respir. Med. 2020;8:65–124. doi: 10.1016/S2213-2600(19)30337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopes-Pacheco M. CFTR modulators: the changing face of cystic fibrosis in the era of precision medicine. Front. Pharmacol. 2020;10:1662. doi: 10.3389/fphar.2019.01662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drumm M.L., Konstan M.W., Schluchter M.D., Handler A., Pace R., Zou F., Zariwala M., Fargo D., Xu A., Dunn J.M., et al. Genetic modifiers of lung disease in cystic fibrosis. N. Engl. J. Med. 2005;353:1443–1453. doi: 10.1056/NEJMoa051469. [DOI] [PubMed] [Google Scholar]
- 4.Karczewski K.J., Francioli L.C., MacArthur D.G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1530/ey.17.14.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mateu E., Calafell F., Ramos M.D., Casals T., Bertranpetit J. Can a place of origin of the main cystic fibrosis mutations be identified? Am. J. Hum. Genet. 2002;70:257–264. doi: 10.1086/338243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cutting G.R. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat. Rev. Genet. 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 8.Cook J.P., Mahajan A., Morris A.P. Fine-scale population structure in the UK Biobank: implications for genome-wide association studies. Hum. Mol. Genet. 2020;29:2803–2811. doi: 10.1093/hmg/ddaa157. [DOI] [PubMed] [Google Scholar]
- 9.Conomos M.P., Miller M.B., Thornton T.A. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol. 2015;39:276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collaco J.M., Blackman S.M., McGready J., Naughton K.M., Cutting G.R. Quantification of the relative contribution of environmental and genetic factors to variation in cystic fibrosis lung function. J. Pediatr. 2010;157:802–807.e3. doi: 10.1016/j.jpeds.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blackman S.M., Commander C.W., Watson C., Arcara K.M., Strug L.J., Stonebraker J.R., Wright F.A., Rommens J.M., Sun L., Pace R.G., et al. Genetic modifiers of cystic fibrosis-related diabetes. Diabetes. 2013;62:3627–3635. doi: 10.2337/db13-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartlett J.R., Friedman K.J., Ling S.C., Pace R.G., Bell S.C., Bourke B., Castaldo G., Castellani C., Cipolli M., Colombo C., et al. Genetic modifiers of liver disease in cystic fibrosis. JAMA. 2009;302:1076–1083. doi: 10.1001/jama.2009.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Treggiari M.M., Rosenfeld M., Mayer-Hamblett N., Retsch-Bogart G., Gibson R.L., Williams J., Emerson J., Kronmal R.A., Ramsey B.W., Group E.S. Early anti-pseudomonal acquisition in young patients with cystic fibrosis: rationale and design of the EPIC clinical trial and observational study. Contemp. Clin. Trials. 2009;30:256–268. doi: 10.1016/j.cct.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider V.A., Graves-Lindsay T., Howe K., Bouk N., Chen H.C., Kitts P.A., Murphy T.D., Pruitt K.D., Thibaud-Nissen F., Albracht D., et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017;27:849–864. doi: 10.1101/gr.213611.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Auwera G.A., O'Connor B.D. first edition. O'Reilly Media; 2020. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. [Google Scholar]
- 18.Jun G., Flickinger M., Hetrick K.N., Romm J.M., Doheny K.F., Abecasis G.R., Boehnke M., Kang H.M. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am. J. Hum. Genet. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conomos M.P., Reiner A.P., Weir B.S., Thornton T.A. Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet. 2016;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCague A.F., Raraigh K.S., Pellicore M.J., Davis-Marcisak E.F., Evans T.A., Han S.T., Lu Z., Joynt A.T., Sharma N., Castellani C., et al. Correlating cystic fibrosis transmembrane conductance regulator function with clinical features to inform precision treatment of cystic fibrosis. Am. J. Respir. Crit. Care Med. 2019;199:1116–1126. doi: 10.1164/rccm.201901-0145OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gogarten S.M., Sofer T., Chen H., Yu C., Brody J.A., Thornton T.A., Rice K.M., Conomos M.P. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics. 2019;35:5346–5348. doi: 10.1093/bioinformatics/btz567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R Core Team . R Foundation for Statistical Computating; 2017. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 24.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 25.Pedersen B.S., Bhetariya P.J., Brown J., Kravitz S.N., Marth G., Jensen R.L., Bronner M.P., Underhill H.R., Quinlan A.R. Somalier: rapid relatedness estimation for cancer and germline studies using efficient genome sketches. Genome Med. 2020;12:62. doi: 10.1186/s13073-020-00761-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price A.L., Weale M.E., Patterson N., Myers S.R., Need A.C., Shianna K.V., Ge D., Rotter J.I., Torres E., Taylor K.D., et al. Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet. 2008;83:132–135. doi: 10.1016/j.ajhg.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grinde K. University of Washington; 2019. Statistical Inference in Admixed Populations. PhD Thesis. [Google Scholar]
- 29.Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R., et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gogarten S.M., Bhangale T., Conomos M.P., Laurie C.A., McHugh C.P., Painter I., Zheng X., Crosslin D.R., Levine D., Lumley T., et al. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics. 2012;28:3329–3331. doi: 10.1093/bioinformatics/bts610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itan Y., Jones B.L., Ingram C.J., Swallow D.M., Thomas M.G. A worldwide correlation of lactase persistence phenotype and genotypes. BMC Evol. Biol. 2010;10:36. doi: 10.1186/1471-2148-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doytchinova I.A., Guan P., Flower D.R. Identifiying human MHC supertypes using bioinformatic methods. J. Immunol. 2004;172:4314–4323. doi: 10.4049/jimmunol.172.7.4314. [DOI] [PubMed] [Google Scholar]
- 33.Ma J., Amos C.I. Investigation of inversion polymorphisms in the human genome using principal components analysis. PLoS One. 2012;7:e40224. doi: 10.1371/journal.pone.0040224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright F.A., Strug L.J., Doshi V.K., Commander C.W., Blackman S.M., Sun L., Berthiaume Y., Cutler D., Cojocaru A., Collaco J.M., et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat. Genet. 2011;43:539–546. doi: 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corvol H., Blackman S.M., Boelle P.Y., Gallins P.J., Pace R.G., Stonebraker J.R., Accurso F.J., Clement A., Collaco J.M., Dang H., et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat. Comm. 2015;6:8382. doi: 10.1038/ncomms9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong J., Wang F., Xiao B., Panjwani N., Lin F., Keenan K., Avolio J., Esmaeili M., Zhang L., He G., et al. Genetic association and transcriptome integration identify contributing genes and tissues at cystic fibrosis modifier loci. PLoS Genet. 2019;15:e1008007. doi: 10.1371/journal.pgen.1008007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bobadilla J.L., Macek M., Jr., Fine J.P., Farrell P.M. Cystic fibrosis: a worldwide analysis of CFTR mutations--correlation with incidence data and application to screening. Hum. Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- 38.Zaidi A.A., Mathieson I. Demographic history mediates the effect of stratification on polygenic scores. Elife. 2020;9:e61548. doi: 10.7554/elife.61548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rees D.C., Williams T.N., Gladwin M.T. Sickle-cell disease. Lancet. 2010;376:2018–2031. doi: 10.1016/s0140-6736(10)61029-x. [DOI] [PubMed] [Google Scholar]
- 40.Corbo R.M., Scacchi R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE∗4 a 'thrifty' allele? Ann. Hum. Genet. 1999;63:301–310. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CFGP sequence data and phenotypic information have been placed in a restricted access database maintained by the Cystic Fibrosis Foundation and is available to approved researchers (see https://www.cff.org/researchers/whole-genome-sequencing-project-data-requests for details). TOPMed data are available to approved researchers through dbGaP under the following study accession numbers: CCAF (phs001189), Mayo VTE (phs001402), miRhythm (phs001434), SARP (phs001446), and VU_AF (phs001032). The analysis pipeline is derived from the TOPMed analysis pipeline, which is publicly available on GitHub (https://github.com/UW-GAC/analysis_pipeline). This pipeline utilizes tools within the GENESIS package, which is publicly available on Bioconductor (https://bioconductor.org/packages/release/bioc/html/GENESIS.html).