Abstract
A classically derived tryptophan-producing Corynebacterium glutamicum strain was recently significantly improved both by plasmid-mediated amplification of the genes for the rate-limiting enzymes in the terminal pathways and by construction of a plasmid stabilization system so that it produced more tryptophan. This engineered strain, KY9218 carrying pKW9901, produced 50 g of tryptophan per liter from sucrose after 80 h in fed-batch cultivation without antibiotic pressure. Analysis of carbon balances showed that at the late stage of the fermentation, tryptophan yield decreased with a concomitant increase in CO2 yield, suggesting a transition in the distribution of carbon flow from aromatic biosynthesis toward the tricarboxylic acid cycle via glycolysis. To circumvent this transition by increasing the supply of erythrose 4-phosphate, a direct precursor of aromatic biosynthesis, the transketolase gene of C. glutamicum was coamplified in the engineered strain by using low- and high-copy-number plasmids which were compatible with the resident plasmid pKW9901. The presence of the gene in low copy numbers contributed to improvement of tryptophan yield, especially at the late stage, and led to accumulation of more tryptophan (57 g/liter) than did its absence, while high-copy-number amplification of the gene resulted in a tryptophan production level even lower than that resulting from the absence of the gene due to reduced growth and sugar consumption. In order to assemble all the cloned genes onto a low-copy-number plasmid, the high-copy-number origin of pKW9901 was replaced with the low-copy-number one, generating low-copy-number plasmid pSW9911, and the transketolase gene was inserted to yield pIK9960. The pSW9911-carrying producer showed almost the same fermentation profiles as the pKW9901 carrier in fed-batch cultivation without antibiotic pressure. Under the same culture conditions, however, the pIK9960 carrier achieved a final tryptophan titer of 58 g/liter, which represented a 15% enhancement over the titers achieved by the pKW9901 and pSW9911 carriers.
l-Tryptophan, one of the limiting essential amino acids required in the diet of pigs and poultry, is the second least abundant of the common amino acids, generally constituting 1% or less of the average protein mass (18). Although l-tryptophan has much commercial potential as a supplement in animal feed, its application is hampered by high production costs; thus, a method for cost-effective production by fermentation is being sought.
We have been pursuing tryptophan production with Corynebacterium glutamicum, an amino acid-producing organism which is widely used for the industrial production of various amino acids (15). Recently, we attempted to metabolically engineer an existing tryptophan-producing mutant of C. glutamicum and reported the remarkable gains to be made in titer, yield (percent conversion from sugar), and productivity (product formation rate) (7, 11). The significant improvement was achieved both by a rational molecular approach to deregulating and/or overexpressing the terminal pathways leading to both tryptophan and serine, the other substrate of the final reaction in the tryptophan pathway, and by construction of a plasmid stabilization system based on the presence of the serine-biosynthetic gene on the plasmid and the gene’s absence from the chromosome. The stable recombinant strain, KY9218 carrying pKW9901, produced 50 g of tryptophan per liter after 80 h in fed-batch fermentor cultivation with antibiotic-free medium. To our knowledge, the titer exceeds any of those that have ever been reported for fermentative production of tryptophan from sugar by microorganisms. However, further improvement seems likely to be achieved if more carbon flux is redirected from central metabolism to the aromatic pathway.
The tryptophan-biosynthetic pathways in the engineered strain have already undergone extensive genetic improvements to efficiently channel carbon toward tryptophan production. Therefore, the principal factor limiting carbon flux toward tryptophan might be the potential of the strain to supply the direct precursors of aromatic biosynthesis, phosphoenolpyruvate (PEP) and erythrose 4-phosphate (E4P), into the aromatic pathway. Carbon flux distribution studies described herein imply that E4P is the first limiting metabolite for tryptophan biosynthesis in the engineered C. glutamicum. Thus, further yield improvement will most likely involve engineering the pentose phosphate pathway to increase the availability of E4P.
Studies with glucose 6-phosphate dehydrogenase- or transketolase-defective C. glutamicum mutants suggest that E4P can be formed from sugar by two routes, the oxidative pentose phosphate pathway and the nonoxidative pentose phosphate pathway (4, 8), although little information has been available about the contribution of either pathway to E4P synthesis in amino acid-producing Corynebacterium. However, supplying carbon for E4P synthesis through the nonoxidative pathway may be more advantageous than supplying it via the oxidative pathway because the latter pathway inevitably involves the release of 1 mol of CO2, accompanied by oxidation of 1 mol of hexose. Therefore, we undertook a strategy to increase the potential of the producer to supply E4P through the nonoxidative pentose phosphate pathway.
Transketolase is a key enzyme of the nonoxidative pentose phosphate pathway. Together with aldolase, transketolase creates a reversible link between glycolysis and the pentose phosphate pathway, thereby enabling the cells to shuttle ribose 5-phosphate and glycolytic intermediates between the two pathways. In a previous article, we reported that a single transketolase was responsible for aromatic biosynthesis in C. glutamicum (8). Furthermore, we cloned the transketolase gene from the organism and showed that the overexpressed transketolase could function in directing carbon toward E4P formation in low producers of the aromatic amino acids (9, 12). From a practical point of view, much work remains to translate these results into the highly engineered hyperproducer described above; however, such work should have a great impact in advancing the field of biotechnology, since many research reports and reviews concerning metabolic engineering methods have been published without demonstrating the practical usefulness of such methods. In this study, we have attempted transketolase modification to further improve the tryptophan-producing recombinant C. glutamicum strain with the highest titer so far reported, with our research being guided by analysis of carbon balances.
MATERIALS AND METHODS
Bacterial strains and plasmids.
C. glutamicum KY9218 (7) is a 3-phosphoglycerate dehydrogenase (PGD)-deficient serine auxotroph of KY10894, which is a tryptophan-producing mutant derived through multiple rounds of mutagenesis from a phenylalanine and tyrosine double-auxotrophic strain, KY9456.
Plasmid pKW9901 (7) contains the desensitized 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (DS) gene of the phenylalanine producer C. glutamicum KY10694, the PGD gene of the wild-type strain C. glutamicum ATCC 31833, and the tryptophan-biosynthetic gene cluster in the C. glutamicum multicopy vector pCG116, which carries the streptomycin and spectinomycin resistance genes as selectable markers. The last gene cluster was cloned from the tryptophan producer C. glutamicum KY10894 and has undergone mutational alterations of its encoded anthranilate synthase (ANS) and anthranilate phosphoribosyltransferase, rendering them insensitive to tryptophan inhibition (11). Plasmids pLTK65 and pCTK60 (9) contain the transketolase gene of the wild-type strain C. glutamicum ATCC 31833 in vectors pCLEK4 and pCSEK20 (6), respectively. The copy numbers of pCLEK4 and pCSEK20 are 3 to 5 and 7 to 10, respectively, in C. glutamicum cells, and both have the kanamycin resistance gene from the Escherichia coli vector pGA22 (1). Low-copy-number vector pCS43 (13) is a derivative of C. glutamicum endogenous plasmid pCG4 (14), which carries the streptomycin and spectinomycin resistance genes.
Media.
Complete medium BY, minimal medium MM, and enriched minimal medium MMYE were those previously described (5). Solid plates were made by the addition of 1.6% (wt/vol) Bacto-Agar (Difco). RCGA medium (14) was used for regeneration of C. glutamicum protoplasts. When required, supplements or antibiotics were added as described previously (5). TS1 and TP2 media (5) were used for second-seed culture and production, respectively, in jar fermentations.
Cultivations for tryptophan production in 2-liter jar fermentors.
A 2.4-ml sample of the first-seed culture grown for 24 h at 30°C on BYG medium (containing 1.0% glucose in medium BY) was inoculated into 120 ml of TS1 medium in a 1-liter flask. After 24 h of cultivation at 30°C on a rotary shaker, the second-seed broth was transferred into a 2-liter jar fermentor containing 550 ml of TP2 medium. After the sugar initially added was consumed, a solution containing 60% sucrose and 540 mg of tyrosine per liter was continuously fed until the total amount of sugar in the medium reached 25%. The culture was agitated at 800 rpm and aerated at 1 liter/min at 30°C, and pH was maintained at 6.1 with NH4OH. Cultivations of all recombinant strains except the pLTK65 and pCTK60 carriers were performed in the absence of antibiotics. The recombinant strains carrying pLTK65 and that carrying pCTK60 were cultivated under the presence of kanamycin (100 μg/ml).
Recombinant DNA techniques.
Plasmid DNA was isolated by the alkaline lysis method (17) and, if necessary, purified by CsCl-ethidium bromide equilibrium density gradient centrifugation (14). DNA digestion and ligation were carried out by standard procedures (17). Transformation was done according to the protoplast method (14).
Enzyme assays.
Crude cell extracts were prepared by sonic disruption of cells grown in MMYE medium supplemented with phenylalanine and tyrosine (100 μg/ml each) as described previously (5). Protein quantity was determined by the method of Bradford (2). Transketolase activities in crude cell extracts were measured as described previously (8). Other enzyme activities were measured by the method of Srinivasan and Sprinson (19) for DS and by the method of Sugimoto and Shiio (20) for ANS. All assays were carried out at 30°C.
Analysis.
Cell growth, sugar concentration, and tryptophan titer were determined as described previously (11). Cell dry weight was determined by centrifuging 10-ml samples, washing the cell pellet twice with water, and drying it for 24 h at 100°C. The carbon content of biomass was calculated based on the measured elemental composition of C. glutamicum: C, 46.6%; H, 6.46%; O, 31.0%; N, 11.8%; and ash, 3.02% (22). On-line analysis of the CO2 evolution rate was carried out with an exhaust oxygen carbon dioxide meter (model EX-1562; Able Co., Ltd.).
RESULTS
Carbon flux distribution during tryptophan fermentation.
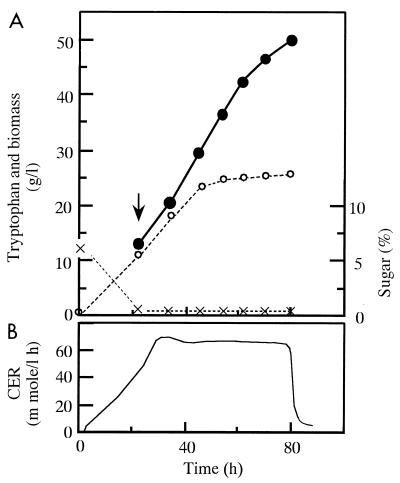
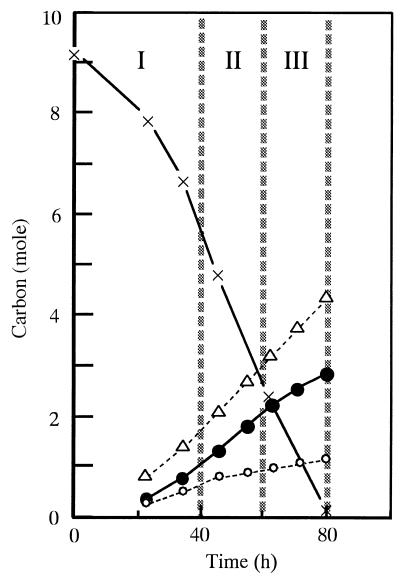
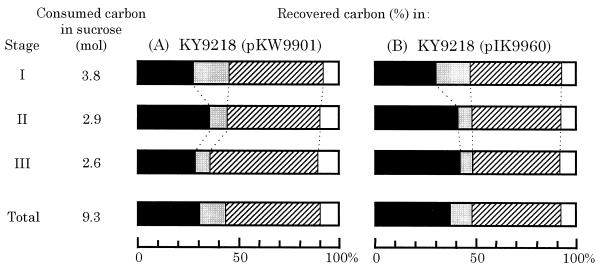
C. glutamicum KY9218 carrying pKW9901 has the ability to produce 50 g of tryptophan per liter after 80 h in fed-batch fermentor cultivation with sucrose medium to which sugar was added at an initial concentration of 6%, with a subsequent increase to 25% (Fig. 1). In this fermentation, the carbon originating from sucrose was mostly diverted to biomass, CO2, and tryptophan. In order to determine the carbon flux distribution in more detail, the fermentation was analyzed by examination of carbon balances. Figure 2 illustrates the profiles of both consumption of the total carbon supplied in sucrose and recovery of carbon in tryptophan, biomass, and CO2 throughout the course of the fermentation. It was calculated that of the total supplied carbon (9.3 mol), 31 and 12% were used for synthesis of tryptophan and biomass, respectively, and as much as 48% was converted to CO2 (Fig. 3A). After summation, the carbon balance added up to 90%. The remaining 10% represented the outflow of carbon to a wide variety of by-products such as other amino acids, keto acids, and organic acids. The major by-products were amino acids such as proline, valine, and alanine, which made up about 6% of the total supplied carbon. The fermentation was divided into three separate stages as portrayed in Fig. 2 (stages: I, 0 to 40 h; II, 40 to 60 h; and III, 60 to 80 h) and analyzed for carbon balances in each stage (Fig. 3A). The average yield of tryptophan (percent conversion of carbon) in stage I was relatively low (28%), probably because the carbon in the sugar that had been consumed was preferentially used to build up biomass in the early stage, when the required amino acids, phenylalanine and tyrosine, were available. In stage II, when growth reached a plateau, the average yield of tryptophan increased to 35%. During this stage and the following stage (stage III), the limitation of phenylalanine and tyrosine prevented overgrowth of cells and most of the carbon in the consumed sucrose was directed to tryptophan and CO2. However, it is noteworthy that the average yield of tryptophan in stage III decreased to 29%, accompanied by increased CO2 yield, which eventually led to a decrease in the overall tryptophan yield from sugar. This slow transition of carbon flux distribution from tryptophan biosynthesis to CO2 evolution in stage III seemed to show the increased carbon flux into the tricarboxylic acid cycle through glycolysis, reflecting a lower proportion of carbon flowing through the pentose phosphate pathway.
FIG. 1.
Tryptophan fermentation by strain KY9218 carrying pKW9901 in fed-batch jar-fermentor cultivation. (A) Profiles of tryptophan (●), biomass (○), and sugar (×). The arrow indicates the point at which feeding with a 60% sucrose solution began. (B) Carbon dioxide evolution rate (CER) during the culture. All data represent mean values from three independent cultures.
FIG. 2.
Consumption of total supplied carbon in sucrose (×) and recovery of carbon in tryptophan (●), biomass (○), and CO2 (▵) during the course of the tryptophan fermentation shown in Fig. 1. The roman numerals represent three stages of tryptophan production (I, 0 to 40 h, II, 40 to 60 h, and III, 60 to 80 h).
FIG. 3.
Carbon balances during the three stages of the tryptophan fermentation by KY9218 carrying pKW9901 (A) and KY9218 carrying pIK9960 (B). Stages correspond to those in Fig. 2. Recovered carbon in tryptophan (solid areas), biomass (shaded areas), CO2 (hatched areas), and by-products (open areas) indicates the relative carbon balances for each stage, expressed as conversion of carbon from sucrose to each metabolite (moles percent).
Effect of introduction of transketolase plasmids on tryptophan production.
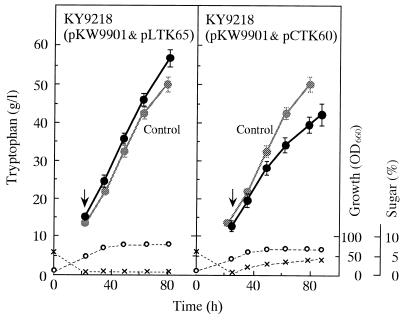
To increase the potential of KY9218 carrying pKW9901 to supply E4P through the nonoxidative pentose phosphate pathway, the recombinant producer was transformed with two kinds of plasmids that contain the transketolase gene of C. glutamicum. One is the low-copy-number plasmid pLTK65 and the other is the high-copy-number plasmid pCTK60, both of which can coexist with the resident plasmid pKW9901 in host cells. The presence of pLTK65 and pCTK60 in strain KY9218 carrying pKW9901 elevated the level of transketolase activities about three- and sevenfold, respectively. Parent strain KY9218 carrying pKW9901 and its transketolase plasmid carriers were tested for tryptophan production in jar fermentors (Fig. 4). The pLTK65 carrier displayed a 14% yield increase relative to its parent and produced 57 g of tryptophan per liter, provided that the cultivation was carried out in the presence of kanamycin to maintain pLTK65 in cells. Three independent cultures showed that the effect was reproducible although not always at such a high level. In contrast, the pCTK60 carrier produced lower levels of tryptophan than even the parent. In this case, growth and sugar consumption were significantly reduced. Since the vector itself did not have any detrimental action on the cells (data not shown), the growth impairment suggested that high expression of the transketolase gene affected the physiology of the cells of the tryptophan producer employed.
FIG. 4.
Tryptophan fermentation by strain KY9218 carrying pLTK65 or pCTK60 together with pKW9901 in fed-batch jar-fermentor cultivation. Symbols: ●, tryptophan; ○, biomass; ×, sugar. For comparison, the profiles of tryptophan production by strain KY9218 carrying pKW9901 are shown as controls. Arrows indicate the points at which feeding with a 60% sucrose solution began. Data represent mean values from three independent cultures. The standard deviations from the means are indicated as error bars only for tryptophan titers. The absence of error bars indicates that the error was smaller than the symbol size. OD660, optical density at 660 nm.
Construction of pSW9911 and pIK9960.
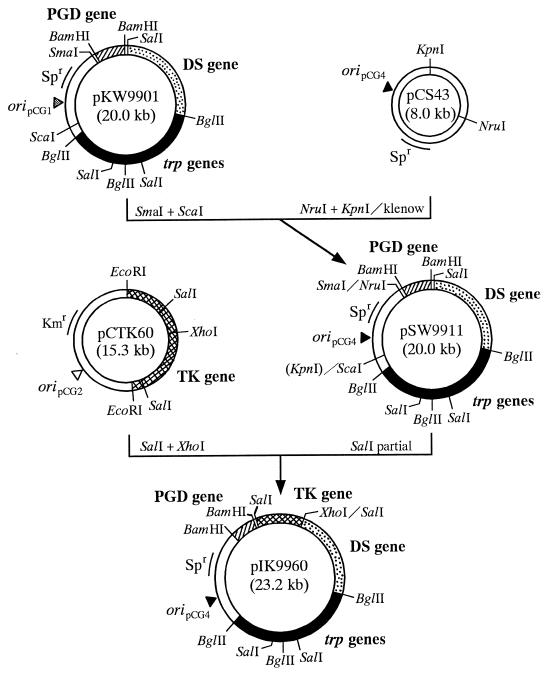
To ensure plasmid stability in the absence of antibiotic pressure, the transketolase gene on pCTK60 and all the cloned genes on pKW9901 were assembled into one vector as depicted in Fig. 5. To circumvent the detrimental action of high expression of transketolase on cells, low-copy-number vector pCS43 was used. First, the pKW9901-derived 14.3-kb SmaI-ScaI fragment containing the DS gene, the tryptophan-biosynthetic gene cluster, and the PGD gene was ligated with pCS43 to generate low-copy-number plasmid pSW9911. Next, the pCTK60-derived 3.2-kb SalI-XhoI fragment containing the intact transketolase gene was ligated with pSW9911 to generate pIK9960.
FIG. 5.
Construction of low-copy-number plasmid pIK9960 containing the transketolase gene as well as the DS gene, the PGD gene, and the tryptophan-biosynthetic gene cluster. Symbols: stippled bars, C. glutamicum KY10694 chromosomal DNA fragment containing the DS gene; solid bars, C. glutamicum KY10894 chromosomal DNA fragment containing the tryptophan-biosynthetic gene cluster (trp genes); hatched bars, C. glutamicum ATCC 31833 chromosomal DNA fragment containing the PGD gene; cross-hatched bar, C. glutamicum ATCC 31833 chromosomal DNA fragment containing the transketolase (TK) gene; open bars, vector pCG116 (pCG1 origin), pCSEK20 (pCG2 origin), or pCS43 (pCG4 origin). Spr, spectinomycin resistance; Kmr, kanamycin resistance; Ori, origin.
Tryptophan production by strain KY9218 carrying pSW9911 or pIK9960.
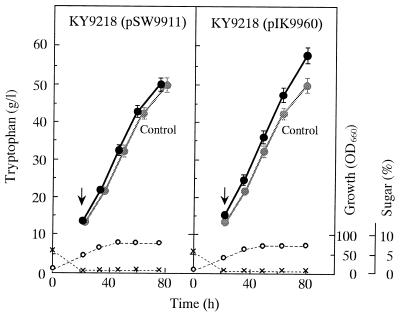
The newly constructed low-copy-number plasmids, pSW9911 and pIK9960, were introduced into the PGD-deficient serine-requiring host KY9218, and their effects on both enzyme activities and tryptophan production were examined. While the original high-copy-number plasmid pKW9901 conferred on the host around 10-fold increases in DS and ANS activities, pSW9911 gave the same host 3- to 4-fold increases in the two enzyme activities (data not shown). In the case of the pIK9960 carrier, the level of transketolase was also elevated about 3-fold relative to the host. These three recombinant strains were then tested for tryptophan production in fed-batch fermentor cultivation without the addition of antibiotics (Fig. 6). The pSW9911 carrier showed almost the same fermentation profiles as the pKW9901 carrier and accumulated 50 g of tryptophan per liter after 76 h cultivation, indicating that low gene dosage (three- to fourfold) was sufficient for removal of the bottlenecks in the overall terminal pathways leading to not only tryptophan but also serine. Under the same culture conditions, the pIK9960 carrier produced 58 g of tryptophan per liter, a yield increase of 15% relative to the pKW9901 or pSW9911 carrier. The increase was highly reproducible and statistically significant. Throughout cultivation, each plasmid was stably maintained in the host. Analysis of carbon balances showed that tryptophan yield in stage III was significantly improved in the pIK9960-carrying new strain (Fig. 3B).
FIG. 6.
Tryptophan fermentation by strain KY9218 carrying pSW9911 or pIK9960 in fed-batch jar-fermentor cultivation. Symbols: ●, tryptophan; ○, biomass; ×, sugar. For comparison, the profiles of tryptophan production by strain KY9218 carrying pKW9901 are shown as controls. Arrows indicate the points at which feeding with a 60% sucrose solution began. Data represent mean values from three independent cultures. The standard deviations from the means are indicated as error bars only for tryptophan titers. The absence of error bars indicates that the error was smaller than the symbol size. OD660, optical density at 660 nm.
DISCUSSION
We have shown here that a moderate increase in transketolase activity resulted in further improvement of tryptophan production in the recombinant hyperproducing C. glutamicum strain. This enhanced tryptophan production occurred because increased activity of transketolase directed more carbon to E4P formation through the nonoxidative pentose phosphate pathway and contributed to increased availability of E4P. In comparing the fermentation profile (Fig. 6) and the carbon balances (Fig. 3) of the newly engineered strain with those of the parental recombinant strain, it should be noted that the greatest effect of amplified transketolase on tryptophan production is at the late stage of fermentation. This kinetic feature of tryptophan production supports our initial conjecture that in the parent, tryptophan production at the late stage was limited by the availability of E4P. Presumably, metabolic flux through the pentose phosphate pathway would be reduced at the late stage of fermentation, when cell growth is decelerating, leading to a decreased supply of E4P. In contrast, increased flux through glycolysis would cause augmentation of carbon flux into the tricarboxylic acid cycle, resulting in a relative increase in CO2 evolution. These hypotheses are in agreement with our observation that the decreased tryptophan yield (35% in stage II versus 29% in stage III) was accompanied by increased CO2 yield (46% in stage II versus 52% in stage III) at the late stage of fermentation in the parent. This probable alteration of flux distribution between glycolysis and the pentose phosphate pathway seems to be related to the modulation of carbon flux through central metabolism as suggested by Vallino and Stephanopoulos (21), who predicted lower flux through the pentose phosphate pathway during the decelerating growth phase than during the exponential growth phase in C. glutamicum.
The presence of the high-copy-number plasmid pCTK60 in strain KY9218 carrying pKW9901 caused decreased tryptophan production compared to strain KY9218 carrying pKW9901 alone, accompanied by reduced growth and sugar consumption. Considering that phosphorylated derivatives of sugars are thought to be inhibitory to bacterial growth (10), it seems likely that overexpression of transketolase activity redirected a higher proportion of glycolytic intermediates into the pentose phosphate pathway and resulted in accumulation of a pentose phosphate(s) intracellularly to a detrimental level. To explore this possibility, we attempted qualitative analysis of pentose phosphates, but high-level accumulation was detected neither intracellularly nor extracellularly (data not shown). Nevertheless, this result does not necessarily indicate that growth inhibition is unrelated to formation of pentose phosphates because it is possible that small increases have large effects.
Recently, several different approaches to optimizing flux distribution within central metabolism to achieve maximum product yield have been proposed for E. coli and in some cases experimentally investigated. The approaches include network analysis of carbon flux and energy levels, but the representative example was shown by Patnaik and Liao (16), who achieved 3-deoxy-d-arabinoheptulosonic acid 7-phosphate production with near theoretical yield by increasing the supply of the aromatic precursors, E4P and PEP, via simultaneous overexpression of transketolase and PEP synthase as well as DS in E. coli. Although the work has illustrated the strategies useful in increasing carbon flow to aromatics, the practical applicability of such strategies to industrial fermentation strains has not yet been evaluated because in those experiments, nongrowth conditions (high-cell-density resuspension cultures) were used to circumvent the problem of growth impairment caused by specific genetic modifications. Considering that fermentation processes are associated with cell growth, genetic modifications leading to growth impairment are practically undesirable. In this sense, not only a positive effect on metabolic flow but also its physiological consequences should be the important consideration in the creation of industrial fermentation strains. In the present study, the problem of growth impairment caused by high-copy amplification of the transketolase gene could be overcome by lower gene dosage, and further yield improvement was ultimately achieved without affecting the growth kinetics. This is one of few examples of successful metabolic engineering with practical significance and thus should provide valuable insight into the construction of industrially useful production strains.
In conclusion, our present study as well as recent work with E. coli (3, 16) shows that transketolase is the pivotal enzyme in determining the capacity of a host to supply E4P in both E. coli and C. glutamicum, thus suggesting the general applicability of the approach to increasing E4P availability in microorganisms with similar central pathways.
REFERENCES
- 1.An G, Friesen J D. Plasmid vehicles for direct cloning of Escherichia coli promoters. J Bacteriol. 1979;140:400–407. doi: 10.1128/jb.140.2.400-407.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Flores N, Xiao J, Berry A, Bolivar F, Valle F. Pathway engineering for the production of aromatic compounds in Escherichia coli. Nat Biotechnol. 1996;14:620–623. doi: 10.1038/nbt0596-620. [DOI] [PubMed] [Google Scholar]
- 4.Ihnen E D, Demain A L. Glucose-6-phosphate dehydrogenase and its deficiency in mutants of Corynebacterium glutamicum. J Bacteriol. 1969;98:1151–1158. doi: 10.1128/jb.98.3.1151-1158.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikeda M, Katsumata R. Metabolic engineering to produce tyrosine or phenylalanine in a tryptophan-producing Corynebacterium glutamicum strain. Appl Environ Microbiol. 1992;58:781–785. doi: 10.1128/aem.58.3.781-785.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikeda M, Katsumata R. A novel system with positive selection for the chromosomal integration of replicative plasmid DNA in Corynebacterium glutamicum. Microbiology. 1998;144:1863–1868. doi: 10.1099/00221287-144-7-1863. [DOI] [PubMed] [Google Scholar]
- 7.Ikeda M, Nakanishi K, Kino K, Katsumata R. Fermentative production of tryptophan by a stable recombinant strain of Corynebacterium glutamicum with a modified serine-biosynthetic pathway. Biosci Biotechnol Biochem. 1994;58:674–678. doi: 10.1271/bbb.58.674. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda M, Okamoto K, Katsumata R. A transketolase mutant of Corynebacterium glutamicum. Appl Microbiol Biotechnol. 1998;50:375–378. doi: 10.1007/s002530051382. [DOI] [PubMed] [Google Scholar]
- 9.Ikeda M, Okamoto K, Katsumata R. Cloning of the transketolase gene and the effect of its dosage on aromatic amino acid production in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 1999;51:201–206. doi: 10.1007/s002530051382. [DOI] [PubMed] [Google Scholar]
- 10.Josephson B L, Fraenkel D G. Sugar metabolism in transketolase mutants of Escherichia coli. J Bacteriol. 1974;118:1082–1089. doi: 10.1128/jb.118.3.1082-1089.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katsumata R, Ikeda M. Hyperproduction of tryptophan in Corynebacterium glutamicum by pathway engineering. Bio/Technology. 1993;11:921–925. [Google Scholar]
- 12.Katsumata, R., and M. Ikeda. February 1997. Process for producing l-tryptophan, l-tyrosine or l-phenylalanine. U.S. patent 5,605,818.
- 13.Katsumata R, Mizukami T, Ozaki A, Kikuchi Y, Kino K, Oka T, Furuya A. Gene cloning in glutamic acid bacteria: the system and its applications. In: Neijssel O M, et al., editors. Proceedings 4th European Congress on Biotechnology. Vol. 4. Amsterdam, The Netherlands: Elsevier; 1987. pp. 767–776. [Google Scholar]
- 14.Katsumata R, Ozaki A, Oka T, Furuya A. Protoplast transformation of glutamate-producing bacteria with plasmid DNA. J Bacteriol. 1984;159:306–311. doi: 10.1128/jb.159.1.306-311.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinoshita S, Nakayama K. Amino acids. In: Rose A H, editor. Primary products of metabolism. London, England: Academic Press; 1987. pp. 209–261. [Google Scholar]
- 16.Patnaik R, Liao J C. Engineering of Escherichia coli central metabolism for aromatic metabolite production with near theoretical yield. Appl Environ Microbiol. 1994;60:3903–3908. doi: 10.1128/aem.60.11.3903-3908.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 18.Somerville R L. Tryptophan: biosynthesis, regulation, and large-scale production. In: Herrmann K M, Somerville R L, editors. Amino acids: biosynthesis and genetic regulation. Reading, Mass: Addison-Wesley; 1983. pp. 351–378. [Google Scholar]
- 19.Srinivasan P R, Sprinson D B. 2-Keto-3-deoxy-d-arabo-heptonic acid 7-phosphate synthetase. J Biol Chem. 1959;234:716–722. [PubMed] [Google Scholar]
- 20.Sugimoto S, Shiio I. Enzymes of the tryptophan synthetic pathway in Brevibacterium flavum. J Biochem. 1977;81:823–833. doi: 10.1093/oxfordjournals.jbchem.a131546. [DOI] [PubMed] [Google Scholar]
- 21.Vallino J J, Stephanopoulos G. Flux determination in cellular bioreaction network: application to lysine fermentations. In: Sikdar S K, Bier M, Todd P, editors. Frontiers in bioprocessing. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 205–219. [Google Scholar]
- 22.Vallino J J, Stephanopoulos G. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction. Biotechnol Bioeng. 1993;41:633–646. doi: 10.1002/bit.260410606. [DOI] [PubMed] [Google Scholar]