Abstract
This article provides a comprehensive review of biosensing with DNAzymes, providing an overview of different sensing applications while highlighting major progress and seminal contributions to the field of portable biosensor devices and point-of-care diagnostics. Specifically, the field of functional nucleic acids is introduced, with a specific focus on DNAzymes. The incorporation of DNAzymes into bioassays is then described, followed by a detailed overview of recent advances in the development of in vivo sensing platforms and portable sensors incorporating DNAzymes for molecular recognition. Finally, a critical perspective on the field, and a summary of where DNAzyme-based devices may make the biggest impact are provided.
1. Introduction
The story of catalytic nucleic acids began in the late 1980s with the seminal work of Thomas Cech and Sidney Altman, who were awarded the Nobel Prize in Chemistry for their parallel discoveries of the catalytic properties of ribonucleic acids (RNA).1,2 Prior to these discoveries, conventional belief confined the roles of deoxyribonucleic acids (DNA) and RNA in biological systems strictly to genetic information storage and transfer.3 The idea that RNA could play a role in biocatalysis was revolutionary, as the role of catalysis in biological systems had been understood to be performed solely by protein enzymes since the crystallization of urease in 1926.4
The creation of a DNA-based enzyme by Gerald Joyce and Ronald Breaker in 1994 was a major breakthrough for two main reasons: first, no DNA-based enzymes had ever been discovered in natural systems; and, second, DNA has fewer chemical functionalities in comparison to proteins and RNA, both of which have been selected by Mother Nature to build powerful enzymes to support the activities of life.5 Fast forwarding to 2021, we have yet to find a naturally occurring DNA enzyme; however, a large number of artificial DNA sequences with catalytic activities have been reported over the past 27 years.6 These man-made enzymes are now known as deoxyribozymes, or simply DNAzymes, as this latter term is more easily understood and accepted by people outside the nucleic acid field. The term of “DNA enzymes” was also used in the past to describe DNAzymes, however, the same term has already been used to indicate protein enzymes that act on DNA substrates, such as restriction enzymes. Therefore, we recommend not to use it to describe DNAzymes.
The discovery of DNAzymes has been made possible thanks to the invention of the “in vitro selection” or systematic evolution of ligands by exponential enrichment (SELEX) technique.7–9 This is a relatively simple but very powerful technique because, when designed and implemented properly, it allows a researcher to find rare sequences of nucleic acids with a pre-programmed function from just a few drops of a reaction solution, which nevertheless contain as many as quadrillions of sequence variants. This technique has been used to isolate numerous aptamers (ligand-binding nucleic acids), ribozymes (RNA molecules with catalytic activities) and DNAzymes. Aptamers, ribozymes and DNAzymes are collectively called functional nucleic acids (FNAs) to differentiate them from the classic functions of DNA and RNA as genetic information storage and transfer molecules, respectively. This review exclusively deals with DNAzymes. Aptamers and ribozymes have been the subject of many excellent review articles and will not be discussed herein.10–17
The initial DNAzyme described by Breaker and Joyce was an RNA-cleaving DNAzyme (RCD), catalyzing the transesterification reaction of the phosphodiester bond of RNA.5 Since its selection, many DNAzymes have been demonstrated to catalyze other chemical reactions, such as DNA cleavage,18 RNA ligation,19 DNA phosphorylation,20 DNA capping,21 DNA ligation,22,23 porphyrin metalation,24 thymine dimer repair,25 nucleopeptide formation,26 tyrosine and serine dephosphorylation,27 tyrosine phosphorylation,28 ester and amide bond hydrolysis,29,30 and very recently alkyne–azide ‘click’ cycloaddition.31 The diverse activities of DNAzymes discovered over the past 27 years have been the subject of several excellent review articles,32–35 which readers are encouraged to explore.
A particularly active area of research on DNAzymes is the exploration of DNAzymes as important components of biosensors, broadly defined here as any analytical device that uses a biological component to allow detection of a specific analyte of interest. Biosensors consist of two key components: a molecular recognition element (MRE), and a signal reporting element (SRE), along with sample input and readout components. Conventional biosensors have mostly employed protein-based recognition elements (including protein enzymes and antibodies) as MREs. Most protein enzymes have been evolved to work with specific substrates (ligands) and are difficult to adapt or modify for different ligands. Antibodies can be generated for most molecular targets through injecting the target into an animal to initiate an immune response. However, producing antibodies for low molecular weight targets (i.e., haptens), and generating monoclonal variants with higher selectivity take considerable time and suffer from high cost, and difficulties with scale-up and batch-to-batch consistency. With increasing demand for biosensor technology, functional nucleic acids, particularly DNA aptamers and DNAzymes, have become desirable alternatives as they can often overcome the disadvantages with traditional protein-based MREs.
DNAzymes possess several physical and chemical properties that make them attractive as key components of biosensors. Target-responsive DNAzymes can serve as excellent MREs because such DNAzymes can be obtained through in vitro selection in a test tube to deliver high affinity for a target of interest, high selectivity against interfering targets, and rapid target binding. The same selection can be repeated to obtain DNAzymes that are selective for a broad range of other targets. DNAzymes can be rationally designed to allow for the exploitation of the complementarity of DNA for controlled capture and release, as well as for incorporation of isothermal nucleic acid-based signal amplification strategies, such as rolling circle amplification and loop-mediated isothermal amplification.36,37 DNA is a highly stable polymer that is easy to synthesize and offers a long shelf-life and low production cost. An obvious challenge for DNAzymes is their instabilities in the presence of nucleases – this issue can be alleviated by conducting DNAzyme selection directly in biological samples that contain nucleases to obtain DNAzymes that are resistant to nuclease digestion, as well as by adding protection tags at both ends of the DNAzymes to prevent exonuclease digestion.38,39 As described below, DNAzyme selection can be conducted in a sample matrix containing not only the analyte but also potential interferants to minimize their negative impact on the performance of the DNAzyme.
Fig. 1 depicts fourteen seminal reports that have advanced the progressive development of biosensors made of DNAzymes. The earliest biosensing work involving a DNAzyme was described in 2000 when the Lu group developed a simple fluorescence assay for the detection of Pb2+ ion using the 17E DNAzyme, an RNA-cleaving DNAzyme with a robust catalytic activity in the presence of Pb2+.40 Since then, a variety of biosensing strategies or devices have been described, such as the first colorimetric assay for Pb2+ using gold nanoparticles in 2003,41 the first electrochemical biosensor for Pb2+ in 2007,42 the first DNAzyme-based lateral flow device in 2010,43 the first personal glucose meter-based sensor for Pb2+ in 2011,44 the first amplified DNAzyme assay in 2013,45 a bacterial litmus test for E. coli in 2014,46 the first paper-based device for E. coli detection in 2017,47 a colorimetric paper device for H. pylori in 2019,48 integration of DNAzymes with CRISPR technology49 to achieve point-of-care diagnostics (POCD), and diagnosis of urinary tract infections (UTI) using a DNAzyme programmed electrochemical sensor,50 among many other examples.51–54
Fig. 1.
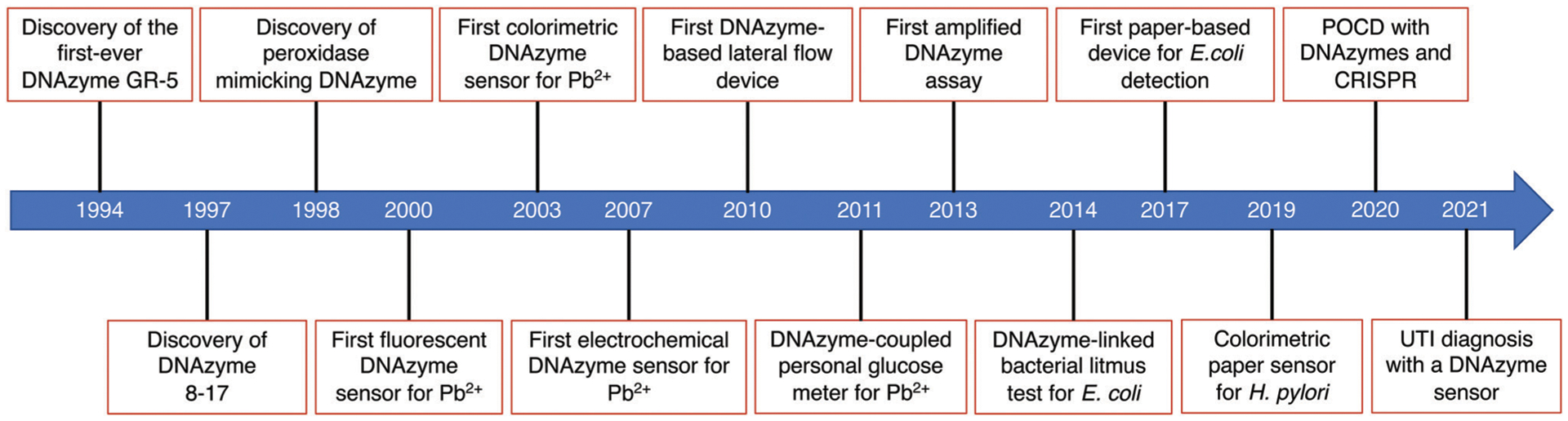
Timeline summary of important events in the development of DNAzyme based biosensors.
This article will be exclusively dedicated to reviewing the design and applications of DNAzyme-based biosensors. We will begin this review by providing some background information on DNAzymes in Section 2, including well-studied examples of DNAzymes for biosensing applications and how these DNAzymes are generated. We will then discuss strategies to expand the range of targets amenable to DNAzymes through the rational design of ligand responsive DNAzymes (termed aptazymes) by coupling existing aptamers and DNAzymes. The design of DNAzymes that act as reporter molecules for biosensors will also be discussed in this section. In Section 3, we will discuss the development of DNAzymes for sensing applications, with a particular focus on various DNAzyme-utilizing strategies designed for the detection of metal ions, small molecules, proteins, nucleic acids and bacteria. Section 4 will describe the integration of DNAzymes into portable biosensor devices, with emphasis on sensors that can be used at the point of care, including optical sensors, sensors based on personal glucose meters and lateral flow devices, and paper-based sensors. In Section 5, we will offer our views on the challenges and future directions of DNAzyme biosensors.
2. DNAzyme basics
2.1. RNA-cleaving DNAzymes as the pre-eminent DNAzyme system for biosensing applications
In theory any DNAzyme system can be exploited for biosensing applications, and as discussed above, there are many such systems to choose from. However, DNAzyme biosensors demonstrated to date have largely focussed on the RCD system, for two key reasons. First, it is relatively easy to isolate diverse, target-responsive, catalytically efficient RCDs from random-sequence pools,33,55,56 where the resulting RCDs can act as MREs for diverse targets and provide a rapid response time.57,58 Second, the outcome of RNA cleavage is the production of two shorter nucleic acid segments (Fig. 2A), offering a convenient way to design a signal output module, as described in Section 3. There has been tremendous progress made with exploiting RCDs for biosensing, as highlighted by the many biosensors reported for detection of metal ions, small molecules, proteins and more recently, bacterial and mammalian targets, many of which will be discussed in this review. In the next section, we will highlight a few well-known and well-characterized RCDs.
Fig. 2.
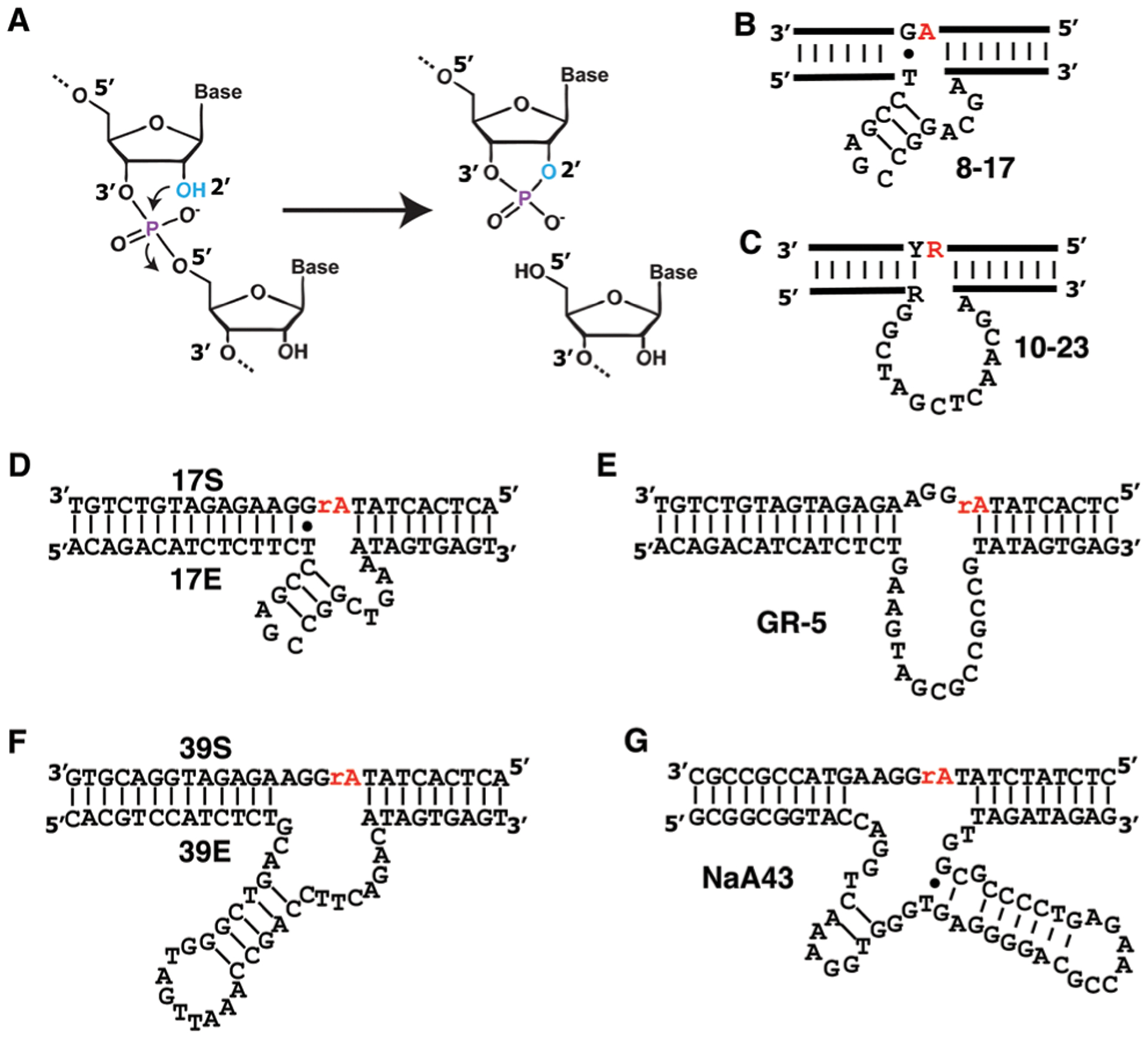
The RNA-cleaving DNAzyme (RCD) system. (A) Chemical transformation of RNA cleavage. Nearly all RCDs whose cleavage mechanism has been elucidated use the 2′-hydroxyl group (blue OH) next to the scissile phosphodiester bond to attack the phosphodiester bond, producing a 5′-cleavage fragment with 2′,3′-cyclic phosphate and 3′ fragment with 5′-OH. RNA-cleaving DNAzymes known as 8–17 (B), 10–23 (C), 17E/17S (D), GR-5 (E), 39E/39S (F) and NaA43 (G). Y and R in panel C: pyrimidine and purine, respectively. Both the 10–23 and 8–17 DNAzymes were originally isolated to cleave an all-RNA substrate; these two DNAzymes cleave the phosphodiester bond following the red ribonucleotide. 17E, GR-5, 39E and NaA43 were isolated to cleave an adenine ribonucleotide (rA) embedded in an otherwise all-DNA sequence.
2.2. Well-known examples of RNA-cleaving DNAzymes
2.2.1. The 8–17 DNAzyme.
The 8–17 DNAzyme (Fig. 2B) was first reported in 1997 by the Joyce group, along with another well-known RCD named the 10–23 DNAzyme (Fig. 2C).59 The 8–17 DNAzyme was later found in several other in vitro selection studies.60–64 The classic 8–17 DNAzyme (Fig. 2B) has a catalytic core of 14 nucleotides, 9 in the hairpin element and 5 in the single-stranded element.59 The DNAzyme interacts with its substrate using two Watson–Crick base-pairing binding arms. The cleavage site was originally identified as A–G59 and later expanded to N–G (N = A, U, C or G).65 Subsequent studies have further revealed that: (1) the 8–17 DNAzyme is capable of cleaving 14 of the 16 possible dinucleotide junctions; (2) only four residues in the catalytic core – A and G in the hairpin, C and G in the single-stranded element – are absolutely conserved and essential for catalysis; and (3) nucleotide substitution, addition, and deletion are well tolerated at other locations.62 Many 8–17 DNAzyme variants exhibit outstanding activities for purine–purine junctions (kobs of >1 min−1), excellent activities for pyrimidine–purine (kobs between 0.1–1 min−1) and purine–pyrimidine (kobs between 0.001–0.1 min−1) junctions, but poor activities for pyrimidine–pyrimidine junctions (kobs between 0.00001–0.001 min−1).66 Recently, a crystal structure of the 8–17 DNAzyme has been reported,67 following the report of the crystal structure of the RNA-ligating 9DB1 DNAzyme.68 These structural studies are important as they provide high-resolution details of DNAzyme structures to aid in the understanding of catalytic mechanisms, roles of metal-ion cofactors, and essential nucleotides within the sequences of DNAzymes. Detailed structural information might also help improve the rational design of biosensors and optimization of reaction conditions.
A noteworthy member of the 8–17 DNAzyme family is the 17E DNAzyme whose sequence and secondary structure is provided in Fig. 2D. 17E was initially selected in the presence of Zn2+ and was found to have a very strong activity in the presence of 100 nM Zn2+, high activity with 100 nM Mn2+ and Co2+, and moderate activities with 100 nM Cd2+, Ni2+, Mg2+, Ca2+, and Sr2+.61 A later study revealed that 17E has the highest activity in the presence of Pb2+: the Kd values of 17E for Pb2+ and Zn2+ (the next best metal ion) were 13.5 and 970 μM, respectively.40 This surprising discovery opened up the flood-gates for using the 17E DNAzyme as a model DNAzyme for specific metal-ion sensing by the Lu group and other researchers. More details on 17E as a lead or zinc sensor are provided below.
2.2.2. The 10–23 DNAzyme.
The 10–23 DNAzyme, discovered along with the 8–17 DNAzyme, has the ability to cleave all-RNA substrates with high efficiency, and has the advantage that it can be designed to target virtually any RNA substrate at a purine–pyrimidine junction, via the design of two binding arms of 6–10 nucleotides that hybridize to the RNA on each side of the cleavage site.59 The 10–23 DNAzyme is the most efficient DNAzyme known to date as it exhibits a kcat of ~10 min−1 in the presence of high Mg2+ concentrations (10 mM or above). Even under simulated physiological conditions (Mg2+ concentration below 5 mM), this DNAzyme still has a kcat of ~0.1 min−1.69 Because of these traits, the 10–23 DNAzyme is regarded as an effective, sequence-specific RNase and has been widely examined as a molecular tool to control levels of various cellular RNA molecules in biological systems. Interested readers may refer to the many excellent reviews on this topic.70–72
2.2.3. The GR-5 DNAzyme.
Another excellent candidate RCD for Pb2+ sensing is GR-5 (Fig. 2E), the first DNAzyme ever discovered.5 Compared to 17E, GR-5 offers a much higher selectivity and an improved detection limit.73 Therefore, for Pb2+ sensing, GR-5 is a better MRE. This was attributed to the fact that the DNAzyme was selected in the presence of Pb2+,5 while 17E was selected in the presence of Zn2+.61 Interestingly, the Liu group was able to demonstrate that GR-5 and 17E are in fact very closely linked, by showing that GR5 can be mutated to obtain the 17E DNAzyme. Several nucleotides were found to play key roles in the mutation process from GR5 to 17E, consequently accounting for the change in metal specificity.74
2.2.4. The 39E DNAzyme.
39E is another notable RCD because it is capable of cleaving the substrate 39S (Fig. 2F) and shows high sensitivity and specificity for UO22+, an important environmental contaminant that can negatively impact human health.75,76 The Lu group selected 39E in 2007 and the DNAzyme exhibits a catalytic rate of ~1 min−1.75 In the same study, they also designed a highly sensitive and selective fluorescent UO2 2+ sensor based on this DNAzyme and showed that the sensor was capable of achieving a detection limit of 45 pM with 1-million-fold selectivity over 19 other metal ions.75 The same DNAzyme has now been studied for sensing applications using other signal transduction mechanisms. 39E will be featured as the key component of many biosensors to be discussed in this review.
2.2.5. The NaA43 DNAzyme.
One of the latest RCDs is NaA43, a Na+-dependent DNAzyme, which is the first nucleic acid enzyme that is specific for a monovalent metal ion (Fig. 2G).77 NaA43 exhibits a catalytic rate constant (kobs) of 0.11 min−1 at 20 °C in the presence of 400 mM Na+, and a selectivity of greater than 10 000-fold over the next best ion when it was tested against 22 different monovalent and divalent metal ions.77 With excellent catalytic activity and superb selectivity, NaA43 can be used as the recognition element to design sensors for Na+ sensing.77 The discovery of NaA43 is very interesting, perhaps even surprising, as it was thought that DNA, or nucleic acids in general, may not have sufficiently diverse functionalities to create a tight binding site for a simple monovalent metal ion.
2.3. In vitro selection of RNA-cleaving DNAzymes
2.3.1. General strategies for selecting RNA-cleaving DNAzymes.
RCDs are typically isolated from a random-sequence DNA library using a common bead-based selection strategy (Fig. 3A). For example, GR-5 was selected using this method.5 For this selection strategy, a DNA pool is typically appended with a biotinylated chimeric DNA/RNA substrate sequence containing a single ribonucleotide (R) as the cleavage site so that the library can be immobilized onto streptavidin-coated beads via the high-affinity biotin–streptavidin interaction. Upon incubation with a target of interest, catalytically active sequences cleave the attached substrate, releasing themselves from the beads. The reaction solution is then separated from the beads, and the DNA in the solution is amplified by PCR. The biotinylated forward primer is usually designed to contain the ribonucleotide so that the double-stranded DNA amplicons from the PCR can be immobilized on the streptavidin-coated beads again. Washing the beads with an alkaline solution helps remove the complementary strand and regenerate the candidate DNAzymes in the single-stranded form for the next round of selection.
Fig. 3.
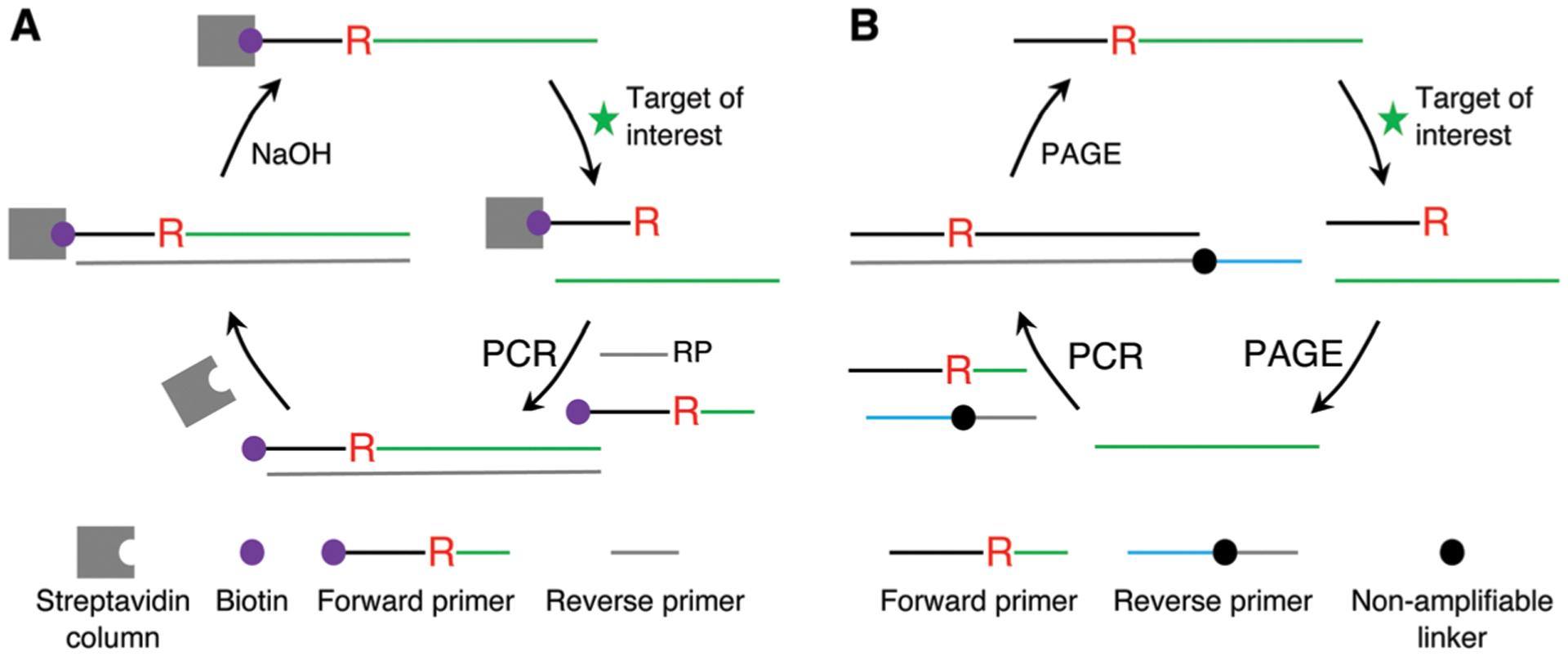
Two common strategies for selecting RNA-cleaving DNAzymes. (A) Column or bead-based selection strategy. (B) Gel based strategy.
An alternative method for the selection of RCDs involves the use of denaturing polyacrylamide gel electrophoresis (PAGE) to isolate the cleavage product (Fig. 3B). For example, the 39E DNAzyme was identified by this method.75 The method is based on the principle that the two cleavage products produced by the cleavage event have different sizes and thus different gel mobility. Therefore, the potential DNAzymes can be separated as the cleavage product, cut out from the gel, eluted and amplified by PCR. The PCR can be conducted with a set of forward and reverse primers to achieve two important outcomes: the forward primer has the needed RNA unit, whereas the reverse primer contains a non-amplifiable linker in the middle of the sequence so that the DNA fragment placed ahead of it cannot be copied during PCR. As a result, the amplicons from the PCR step contain two strands of unequal length, which permits isolation of the DNAzyme-containing strand by PAGE. The purified DNA construct is then used for the next round of selection.
2.3.2. In vitro selection of metal-ion dependent RNA-cleaving DNAzymes.
DNAzymes are particularly well suited for metal-ion binding because metal ions are important for the creation of stable structures of DNA, a negatively charged polymer, and they are known to function as catalytic cofactors for nucleic acid enzymes.78 In fact, nearly all DNAzymes known to date can be classified as metalloenzymes because their catalysis requires the assistance of metal ions. The RCDs featured in Section 2.2 above are all metalloenzymes.
Many of the RCDs reported to date are not only metalloenzymes but are also specific or highly selective for a given metal ion, making them highly attractive as MREs for biosensing applications. As a result, metal ion sensing has become a key focus of DNAzyme-based sensing and Section 3.1.1 to follow will be entirely dedicated to metal ion sensing using RCDs.
Most metal-specific RCDs have been derived from random pools simply by incorporating the metal ion of interest into the selection buffer. One good example is the first DNAzyme study where GR-5 was discovered. In this case, each selection round only had a positive selection step with Pb2+ but did not have a counter selection step for selecting against other metal ions.5 Subsequent analysis of GR-5’s metal ion specificity revealed that this DNAzyme was highly specific for Pb2+.73 The same approach has been applied successfully for the isolation of many other metal-ion-specific RCDs. These include the 39E/39S DNAzyme that is extremely selective for UO2 2+,75 and one of the latest DNAzymes NaA43 that is highly specific for Na+.77
It is also possible to incorporate a counter selection step using a mixture of unintended or potentially interfering metal ions, along with a positive selection step with the intended metal ion.79–81 For example, the Liu group published a study where counter selection with Pb2+, Zn2+ and Cu2+, and positive selection with Ni2+ overcame the issue of having initially failed to select Ni2+-specific RCDs.81 The group has also applied a similar approach to derive a Cu2+-specific DNAzyme following the observation that the initially enriched DNA pool established with only positive selection with Cu2+ also exhibited strong cleavage activity with a metal-ion mixture of Cd2+, Zn2+, and Pb2+.80 The incorporation of a counter selection step with these 3 metal ions led to the isolation of a number of DNAzymes with significantly improved selectivity for Cu2+. These studies suggest that counter-selection with other metal ions can be used as a productive strategy for deriving DNAzymes with high metal-ion specificity.
DNAzyme selections may also produce DNAzymes with unexpected but still highly desirable metal-ion specificities. An excellent example is 17E, which was initially selected from a DNA pool in the presence of Zn2+, but was later discovered to have the highest activity in the presence of Pb2+.61
Since DNA molecules have limited functional groups capable of binding metal ions strongly and selectively, particularly when compared to proteins or organic chelants used for sensors, it is important to introduce modified nucleotides into DNAzymes either during or after in vitro selection. An example of this method is the incorporation of modified nucleotides into the initial library by template-directed extension, which has allowed DNAzymes to be selected for Zn2+ and Hg2+ ions.82,83 A similar strategy has been used to select DNAzymes that can operate independent of divalent metal ions and DNAzymes with improving resistance to nuclease degradation. The limitations of using modified nucleotides are the requirement for expensive engineered polymerases to incorporate the modified nucleotides and difficulty in obtaining PCR amplification of some modified nucleotides.38 To avoid these issues, chemically modified DNA substrates are often used without the need to use engineered polymerases or PCR in the selection. The modification is fixed at a specific region of the library. Thus, normal PCR can be performed to amplify the library during selection. For thiophilic metal ions including Cd2+ and Cu2+, a single phosphorothioate modification at the cleavage junction in the library was successfully applied to perform new selections.79,80 Using this method, the Cu10 and Cd16 DNAzymes were obtained with high activity. For transition metal ions such as Zn2+ and Ni2+, an imidazole group near the cleavage junction was inserted and a Zn2+-specific DNAzyme and a Ni2+-specific DNAzyme were obtained with high selectivity.81,84 Selections have also been performed with fluorophore/quencher labels next to the cleavage junction for signal generation.85,86
2.3.3. In vitro selection of RNA-cleaving DNAzymes that are dependent on small-molecule or protein targets.
Despite the great success in the selection of RCDs that respond to metal ions as analytes, only one in vitro selection experiment has been described for isolating an RCD that was activated by a defined, non-metal, small-molecule target. A group of RCDs, specifically HD1, HD2 and HD3 (Fig. 4), that are dependent on the amino acid histidine was described in a study published by Roth and Breaker in 1998.87 In order to select histidine-dependent rather than metal-dependent RCDs, the RNA cleavage step was conducted in a reaction mixture containing the desired target, l-histidine, as well as EDTA, which was included to chelate any contaminating metal ions. After 11 rounds of selection, one of the isolated DNA molecules exhibited an RNA-cleaving activity only in the presence of histidine, with a kobs of 1.5 × 10−3 min−1 in 50 mM l-histidine. The authors then built a mutagenized pool based on this sequence, subjected this pool to two parallel reselections in a reaction mixture containing either 50 mM histidine or 5 mM histidine/50 mM HEPES. After five rounds of reselection, HD1 and HD2 were examined as the representative mutants, which were more active than the original DNAzyme. HD1 exhibited saturation kinetics with a kobs of 4.7 × 10−3 min−1 and Kd of ≈25 mM. HD2 showed a better catalytic activity (kobs of 0.2 min−1 at 100 mM histidine) than HD1 but the binding site was not saturated even at 100 mM histidine (the maximum soluble concentration). Further selection using a mutagenized pool based on HD2 using 0.1 mM histidine led to the discovery of the DNAzyme HD3. HD3 exhibited a saturation kinetic profile, with a kobs of 0.2 min−1 and Kd of ≈25 mM.
Fig. 4.
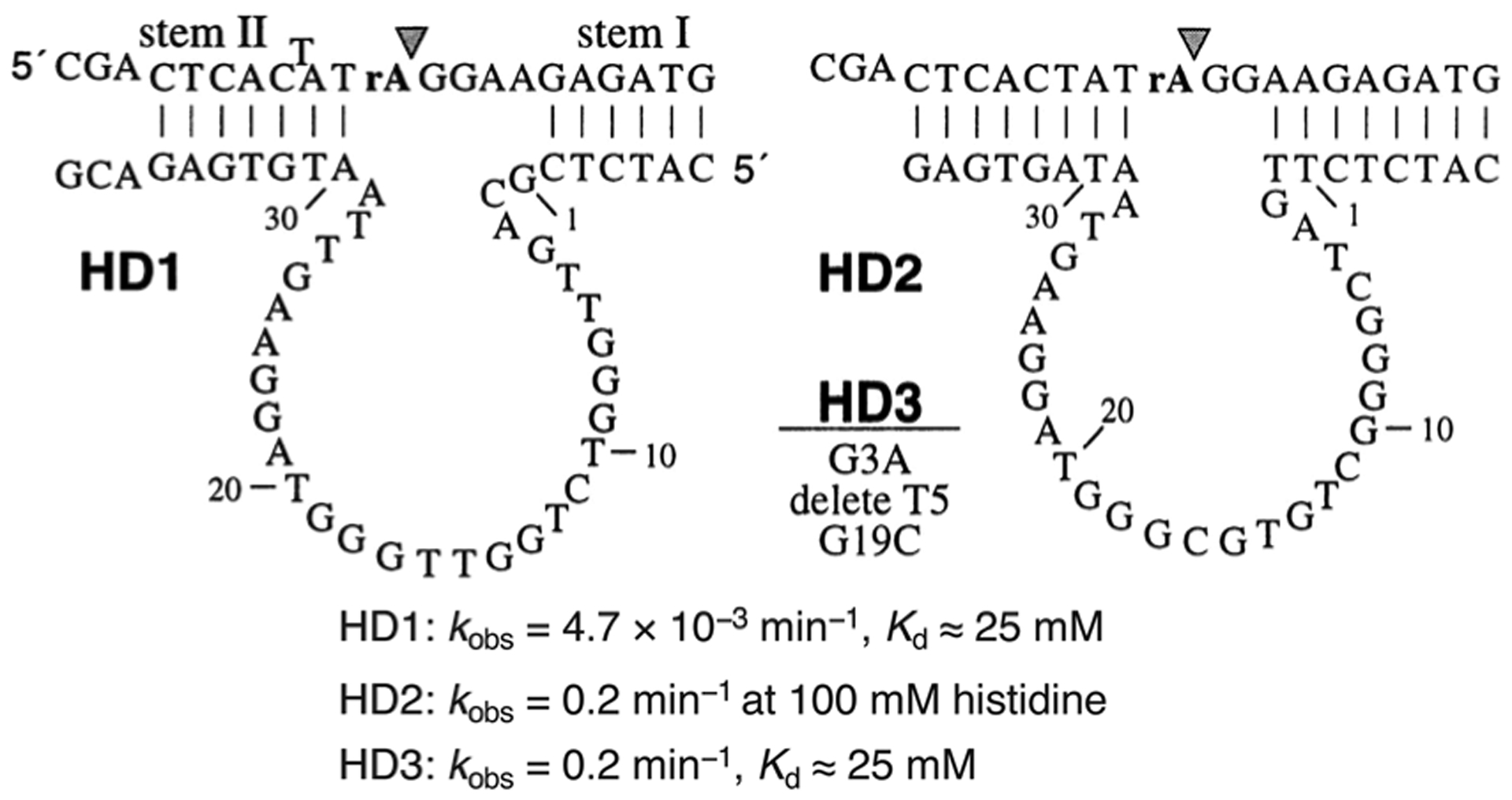
Histidine-dependent DNAzymes HD1, HD2 and HD3. Reprinted (adapted) with permission from A. Roth and R. R. Breaker, An amino acid as a cofactor for a catalytic polynucleotide, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6027–6031. Copyright (2007) National Academy of Science.87
This study is important for future studies aimed at generating RCDs that bind small molecules. First, this study shows small-molecule binding DNAzymes do exist in random-sequence DNA pools. Second, thoughtful selection strategies, such as the inclusion of metal-chelating agents in the selection buffer (to disfavor the selection of metal-ion-dependent RCDs) and utilizing multiple cycles of reselection (to derive DNAzymes with better catalytic rates and higher affinity for the target of interest) may have to be implemented in order to obtain high-performing RCDs for sensing non-metal small-molecule targets.
More small-molecule binding DNAzymes are expected to exist in random DNA libraries based on three additional lines of evidence. First, efficient, metabolite-dependent RNA-cleaving ribozymes exist, as exemplified by the discovery of a natural ribozyme responding to a specific metabolite, glucosamine-6-phosphate.88 Second, allosteric RCDs have been successfully engineered from aptamers and RNA-cleaving DNAzymes (more on this in Section 2.4). Third, many so-called “kinase DNAzymes” – DNAzymes that phosphorylate themselves at their 5′-end in the presence of specific nucleoside 5′-triphosphates (NTPs or dNTPs), such as GTP – have been reported.20 These kinase DNAzymes must be able to bind an NTP (or dNTP) in order to get selected. Since metal ions may have to be included in the selection buffer in addition to the specific analyte of interest (to assist the folding of potential RCDs), the success of an RCD selection in the presence of an analyte of interest as well as metal ions does not absolutely depend on the recognition of the DNAzyme for the analyte, because simple metallo-DNAzymes without the assistance of the analyte can produce the cleavage fragment to be selected. This was observed multiple times in several unpublished studies conducted in our laboratories. It certainly represents a worthy future effort for DNAzyme engineers to devise more efficient methods for the selection of RNA-cleaving DNAzymes that can only be activated by a defined non-metal target, and such methods will significantly expand the use of RCDs for sensing a much broader range of analytes.
To date, there is not a single published study where a DNAzyme has been selected to recognize a defined protein target, although there were unsuccessful, and hence unpublished efforts towards selecting protein-dependent RNA-cleaving DNAzymes in our labs. These failures may once again have been linked to the requirement of including metal ions in the selection buffer, giving metal-promoted RCDs a selective advantage that resulted in these being identified in the selection. Interestingly, proteins have been identified as the targets of RCDs that are selective for cells such as E. coli and C. difficile (see next section), and hence it should be possible to identify protein-selective RCDs using well-designed selection strategies.
2.3.4. In vitro selection of RNA-cleaving DNAzymes that are activated by undefined cellular targets.
An alternative strategy for selection of DNAzymes is based on taking advantage of small differences in the molecular composition of complex mixtures used for positive and counter selection steps, which has had considerable success in selecting target-responsive RCDs.48,85,86,89–91 Instead of using a defined target for the DNAzyme selection, a crude mixture of targets derived from a specific type of bacterial or human cell was used as the target to induce the cleavage of candidate RCDs. The idea behind this approach is to select RCDs that respond specifically to an initially unknown target, which is nevertheless unique to the cell of interest, for example, a specific bacterial pathogen. The key to the success of this method is to apply stringent positive and counter selection steps to achieve high recognition specificity. For example, to derive RCDs that are specific to bacterium A (intended bacterium) but non-responsive to bacterium B (unintended bacterium), the DNA library will be first incubated with the cellular mixture from bacterium B and any cleaved molecules at this step will be discarded. The uncleaved fraction of the library will then be incubated with the cellular mixture from bacterium A and the cleavage product at this step will be isolated, amplified and used for the next cycle of selection, with each round having both positive and counter selection steps (Fig. 5). Upon successful selection of the DNAzymes with high recognition specificity, efforts are then made to identify the activating target molecule. To date, several successful studies have been conducted to derive RCDs for bacterial48,85,86,89,90 and human cancer cells,92–94 as described in more detail below.
Fig. 5.
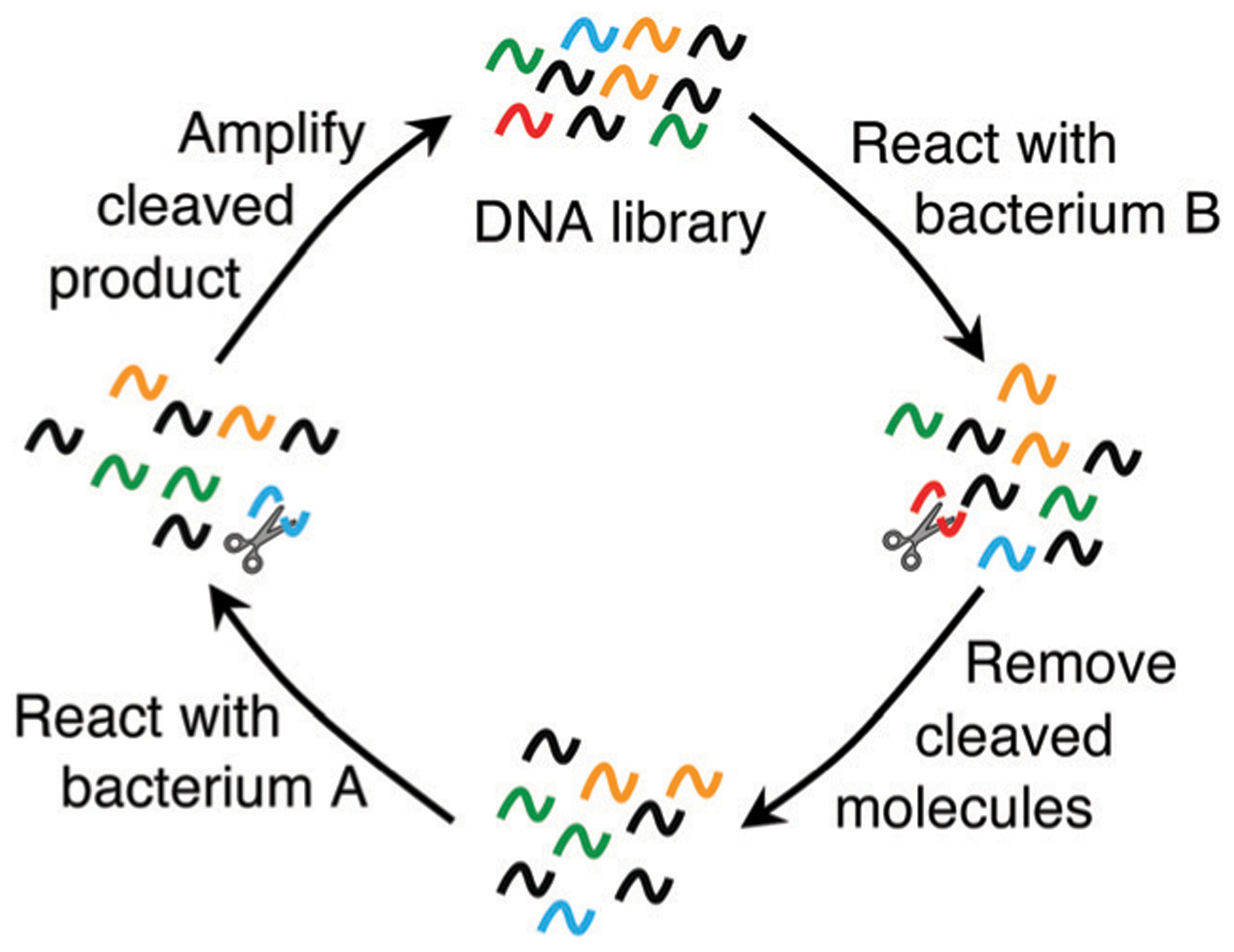
In vitro selection of RCDs for recognizing an unknown bacterial target using double selection strategy: counter selection (or negative selection) with the cellular mixture from a control bacterium (bacterium B) and positive selection with the cellular mixture from the intended bacterium (bacterium A). Reprinted (adapted) from M. Liu, D. Chang, and Y. Li, Discovery and Biosensing Applications of Diverse RNA-Cleaving DNAzymes, Acc. Chem. Res., 2017, 50, 2273–2283.95 Copyright 2017 American Chemical Society.
In the first study of this type, Ali et al. reported the selection of an RCD, called RFD-EC1, that was catalytically active in the presence of E. coli.85 The reason that this DNAzyme was named “RFD” is because the DNAzyme cleaves a single ribonucleotide at a cleavage site that is flanked by two nucleotides modified with a fluorophore (fluorescein-dT) and a quencher (dabcyl-dT), making it an RNA-cleaving Fluorogenic DNAzyme. RFD-EC1 was selected from a random pool with counter-selection and positive-selection steps that used the crude extracellular mixture (CEM) of Bacillus subtilis (CEM-BS) and E. coli (CEM-EC), respectively. The counter-selection step involved incubation of the DNA pool with CEM-BS and isolation of the uncleaved sequences by PAGE. These sequences were then incubated with CEM-EC in the positive selection step; the cleaved sequences were then isolated using PAGE and amplified to produce the next pool to seed the next cycle of selection. RFD-EC1 was discovered after 20 rounds of selection. This DNAzyme was found to be activated by a protein target whose identity is unknown at this moment. Interestingly, even though RFD-EC1 was derived using the CEM from only one type of bacterium (B. subtilis) as a counter-selection target, further tests indicate that it did not exhibit cross reactivities with many other Gram-positive and Gram-negative bacteria, an excellent trait for biosensing applications. Because RFD-EC1 was encoded with fluorescence-signaling properties, it could be used to develop simple ‘mix-and-read’ assays for bacterial detection. For example, an optimized assay with RFD-EC1 was shown to achieve a detection limit of 1000 colony-forming units (CFU) without a culturing step and 1 CFU following as short as 4 hours of bacterial culturing in a growth medium.96
RFD-EC1 was not selected to recognize a specific strain of E. coli and therefore, it can be activated by many different strains of this bacterial species.97 However, the same method can be applied to select for strain-specific DNAzymes as well, as demonstrated by the discovery of RFD-CD1 that is highly specific for a BI/027 strain of Clostridium difficile.86 RFD-CD1 was derived using the CEM prepared from the BI/027 strain of C. difficile as the positive selection target, and the combined CEM from CD630 (a non-BI/027 strain of C. difficile), E. coli and B. subtilis as the counter-selection target mixture. RFD-CD1 was discovered after 25 vigorous cycles of counter-selection and positive-selection and is only active with the BI/027 strain of C. difficile. The RFD-CD1 is also an exceptional example in that the target, a truncated version of TcdC (a transcription factor) that is unique to the BI/027 strain of C. difficile, has been identified.86
Subsequent studies using similar selection methods have led to the discovery of several other RFDs for other bacterial targets. The list includes DHp3T4 for Helicobacter pylori (HP), which shows selectivity against 20 other Gram-positive and Gram-negative bacteria,48 RFD-KP6 for Klebsiella pneumoniae (KP),89 and VAE-2 for Vibrio anguillarum (VA).90 VAE-2 differs from the other four DNAzymes by the lack of the fluorophore and quencher labels flanking the cleavage site on the substrate. Instead, selection was performed with no labels present so as to avoid the possibility of generating an aptazyme that required these labels for activity.
In addition to bacterial targets, there are two examples which have employed a similar selection method to generate aptazymes for cancer cells. In the first example, SELEX was used to generate RFDs using the cell lysate of MDA-MB-231 breast cancer cells for positive selection, and a mixture of lysates from both healthy and alternate cancer cell lines for counter selection.93 This resulted in a highly selective aptazyme for detection of breast cancer lines, denoted as AA12–5, with a detection limit of 5000 cells per mL and the ability to detect over 90% of malignant breast tumors. In a second study,92 an RFD was selected against K562 cancer cell lysate to produce a diagnostic test for acute myeloid leukemia, again using a mixture of lysates from healthy and alternate cancer cell lines for counter selection. The resulting RFD-aptazyme, denoted as A1–3, was able to detect K562 cell lysates spiked in human serum. These two studies demonstrate that the selection of aptazymes using cell lysates can be applied to mammalian cells and can produce aptazymes that are able to distinguish between healthy and diseased cell phenotypes, opening the door to the development of aptazymes for a range of human diseases.
2.4. Rational design of target responsive DNAzymes
2.4.1. Allosteric DNAzymes containing communication modules.
The scope of DNAzyme biosensors can be significantly expanded through the development of allosteric DNAzymes. Many protein enzymes are allosterically controlled; the catalytic activity of these enzymes is significantly regulated by the binding of an effector (or allosteric modulator). Control by allostery was also successfully demonstrated for nucleic acid enzymes, first for ribozymes98–100 and then for DNAzymes.101 By the classic definition, an allosteric enzyme has an effector binding site different from the enzyme’s active site.
The use of a small-molecule binding aptamer to allosterically control the activity of a nucleic acid enzyme via a “communication module” was pioneered by Ronald Breaker, whose group has rationally designed many allosteric ribozymes that are responsive to biological cofactors or metabolites.98–100 Several allosteric DNAzymes that respond to small molecules via a communication module have also been reported, which typically use a short duplex element to link an aptamer to a DNAzyme. The duplex element, which plays a structural role for both the aptamer and the DNAzyme, is deliberately weakened so that the catalytic activity of the DNAzyme is significantly reduced. The binding of the analyte to the aptamer strengthens the duplex, leading to the recovery of DNAzyme catalytic activity. All allosteric DNAzymes designed this way are regulated by the same ATP binding DNA aptamer (which also binds AMP or adenosine). The list includes a ligase DNAzyme (Fig. 6A),101 an RNA-branching DNAzyme,102 and two different RNA-cleaving DNAzymes,103,104 one of which can function in 50% ethanol (Fig. 6B).104 The latter work demonstrates that allosteric DNAzymes, and DNAzymes in general, can maintain remarkable structural stability and perform recognition functions in high-content organic solutions. This is a research area that warrants further attention as it may be possible to develop DNAzyme biosensors that recognize targets that are only soluble in organic solvents.
Fig. 6.
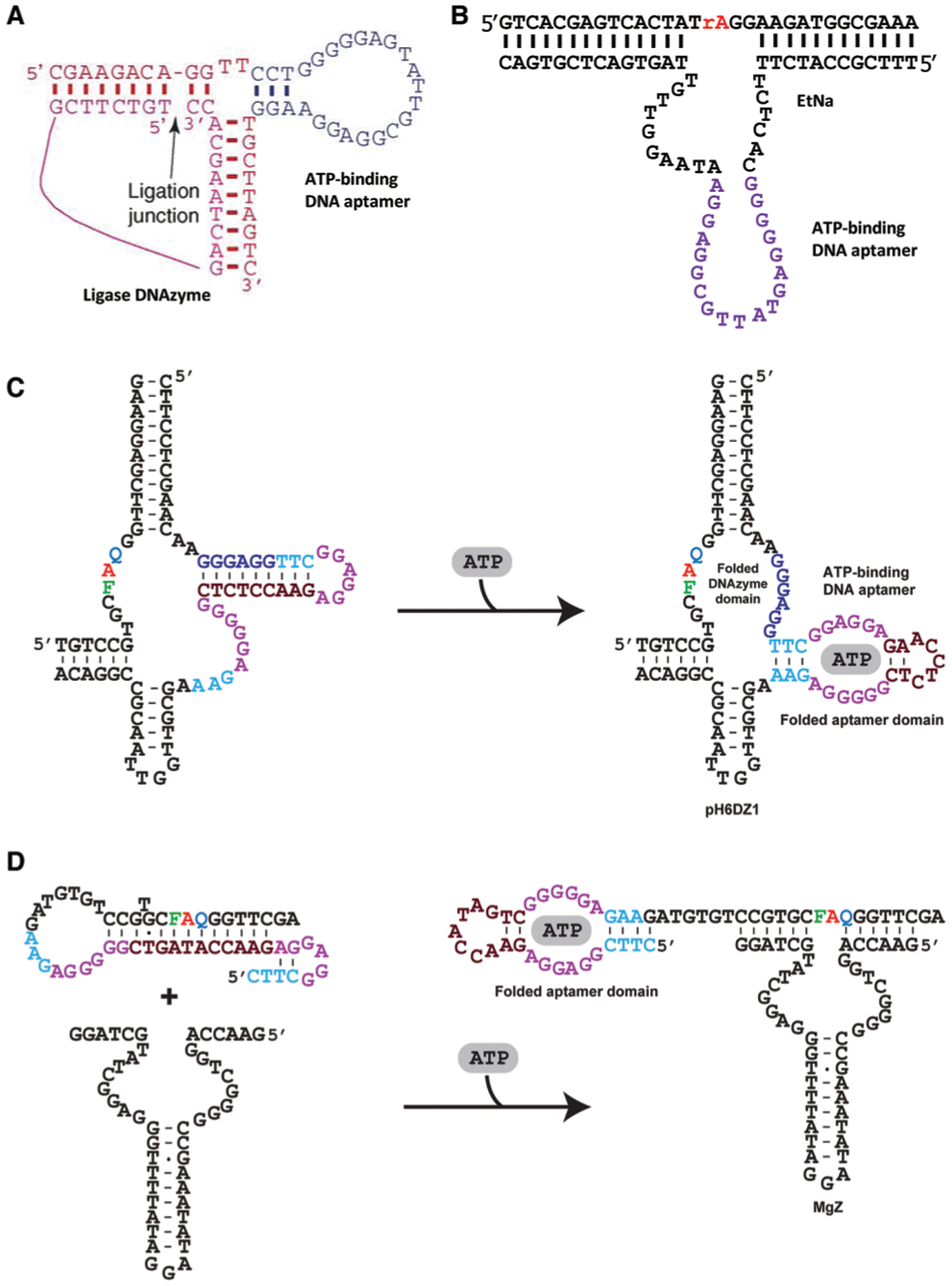
Representative allosteric DNAzymes and aptazymes using an ATP binding DNA aptamer. (A) An allosteric ligase DNAzyme. Reprinted (adapted) from M. Levy and A. D. Ellington, ATP-Dependent Allosteric DNA Enzymes, Chem. Biol., 2002, 9, 417–426, with permission from Elsevier.101 (B) Allosteric EtNa (an RCD) activated by AMP in 50% ethanol. rA: adenine ribonucleotide. Reprinted (adapted) with permission from T. Yu, W. Zhou and J. Liu, Ultrasensitive DNAzyme-Based Ca2+ Detection Boosted by Ethanol and a Solvent-Compatible Scaffold for Aptazyme Design, ChemBioChem, 2018, 19, 31–36. Copyright 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.104 (C) An aptazyme built with pH6DZ1 (an RCD). (D) An aptazyme built with MgZ (an RCD). For panels (C and D): Red A is adenine ribonucleotide; F: fluorescein-dT; Q: DABCYL-dT. Panels (C and D) are reprinted (adapted) from M. Liu, D. Chang, and Y. Li, Discovery and Biosensing Applications of Diverse RNA-Cleaving DNAzymes, Acc. Chem. Res., 2017, 50, 2273–2283.95 Copyright 2017 American Chemical Society.
2.4.2. DNA aptazymes.
The creativity of functional nucleic acid researchers has significantly expanded the concept of allosteric DNAzymes to include “DNA aptazymes”, which covers any functional DNA system where the catalytic activity of a DNAzyme is regulated by the binding of the analyte to an aptamer. There are many published strategies for the design of DNA aptazymes; some are very simple and others involve elaborate designs, particularly those that incorporate signal amplification. An example of a simple system is an aptazyme designed with the 10–23 DNAzyme and the adenosine-binding DNA aptamer105 in which the binding of adenosine to the aptamer works to stabilize the DNAzyme-substrate complex. In the second example, a DNAzyme known as pH6DZ1 is linked to the ATP-binding DNA aptamer in a way that allows part of the aptamer to form a pairing element with a few catalytically important nucleotides of the RCD. Upon ATP binding, these nucleotides become unpaired, restoring the cleavage activity of the DNAzyme (Fig. 6C).106 A third example is a structure-switching based aptazyme made of a duplex formed between a DNA sequence containing the 8–17 DNAzyme, the ATP binding DNA aptamer and a complementary DNA sequence that binds simultaneously to the DNAzyme and the aptamer domain.107 The binding of ATP leads to the dissociation of the complementary DNA strand first from the aptamer and then the DNAzyme, freeing up the DNAzyme for substrate binding and cleavage.
Two simple aptazymes have also been described based on a fluorogenic DNAzyme known as MgZ for the detection of ATP and ADP, respectively, via the use of the anti-ATP DNA aptamer (Fig. 6D) and an anti-ADP RNA aptamer;108 these two aptazymes were designed to use the target-aptamer binding to regulate the accessibility of the substrate by the DNAzyme. The same design was also used to create another aptazyme from the anti-ATP DNA aptamer and an l-RNA-cleaving DNAzyme known as LRT-BD1.109
More complex aptazyme systems have also been designed to detect protein targets. In one study, the Pb2+-dependent RNA-cleaving DNAzyme and prostate specific antigen (PSA)-binding DNA aptamer were combined into one sequence to create a hairpin structure that inhibits the substrate binding and catalytic activity. The binding of PSA to the aptamer domain frees up the DNAzyme to carry out catalysis.110 In another study, an aptazyme was designed to detect human thrombin111 that involves the use of the 8–17 DNAzyme and two different thrombin binding DNA aptamers. Aptamer 1 is combined with a cis-acting version of 8–17 DNAzyme that is configured into a hairpin to lock the catalytic activity of the DNAzyme. The aptamer 2 carries a sequence extension that is required to create a stable 8–17 DNAzyme/substrate complex. In the presence of thrombin, both aptamers bind the protein; the binding of aptamer 1 leads to unlocking of the hairpin, and the binding of aptamer 2 places its sequence extension in close proximity. These events work to create a stable DNAzyme-substrate complex for catalytic activity.
DNAzymes have also been utilized to create aptazyme-like systems whose activity is regulated by a nucleic acid target, which are typically designed to take advantage of Watson–Crick interactions for the formation of competitive duplex structures. A good example is the design of a three-way 10–23 DNAzyme system in which one of the two substrate binding arms of the 10–23 DNAzyme is modified so that it is only able to form a very weak binding interaction with the RNA substrate unless a nucleic acid sequence targeted for detection is present; this target is designed to bind both the substrate and the DNAzyme, creating a stable substrate/DNAzyme complex to allow robust RNA cleavage.112 This design has been used to develop nucleic acid regulated 8–17 DNAzyme and bipartite RNA-cleaving DNAzymes.113 Another example is the “catalytic molecular beacon”.114 A molecular beacon is a hairpin structure made of an unpaired loop sequence closed with a stem (a short Watson–Crick duplex). The catalytic molecular beacon places a substrate binding sequence of an RNA-cleaving DNAzyme as part of the stem so that the DNAzyme cannot bind the substrate. The DNA target to be detected is designed to be complementary to the loop of the molecular beacon. In the presence of the DNA target, the formation of a strong duplex between the target and the loop opens the stem, allowing for the binding of the DNAzyme and its substrate and enabling the cleavage by the DNAzyme.
An interesting aspect of catalytic molecular beacons, as well as other DNAzymes, has been their use in the field of DNA computing.115,116 In these cases, stem-loop modules were combined with fluorogenic substrates with different recognition arms such that an input DNA sequence could alter the configuration of the catalytic beacon or substrate, as described above, to activate catalytic activity (YES gate, a turn-on sensor for the input sequence) or inhibit the activity of the catalytic beacon (NOT gate – a form of turn-off sensor). Multiple catalytic beacons could also be combined to have dual inputs, allowing AND and XOR gates.115 Over the past 7 years there have been many advances in DNAzyme-based logic gates, computing and robotics, and interested readers are encouraged to read several excellent reviews on this topic.117–119
2.5. DNAzymes as reporter molecules
DNAzymes have also been utilized as reporter elements in a wide variety of assays. The most widely used reporter element is the peroxidase mimicking DNAzyme (PMD), although RCDs such as 8–17, 10–23, and Mg2+-dependent DNAzymes have also been utilized, particularly in the form of catalytic beacons, and multicomponent nucleic acid enzymes (MNAs).120 There have been multiple reviews covering basic aspects of PMDs,121 as well as their use for various spectroscopic and electrochemical assays;122–125 and the use of both PMDs and RCDs for amplified detection of nucleic acids and proteins.120 Given the number of recent reviews on these reporting systems,120–125 this topic will not be covered here, except for those cases where a DNAzyme acts as a MRE to produce a signaling output containing a PMD or RCD.
3. DNAzymes for sensing applications
3.1. Direct Sensing of Analytes using DNAzymes
3.1.1. Detection of metal ions.
Metal ions play essential roles in many complex biological systems from the cellular to the ecosystem level, and therefore sensitive detection of specific metal ions is of interest for a variety of applications. The detection of metal ions in environmental samples, biological matrices, and living systems has been the focus of many research groups. Typically, metal-ion detection is achieved by laboratory-based methods that utilize complicated instruments such as inductively coupled plasma mass spectrometry, atomic absorption spectroscopy, or various electrochemical methods.126,127 The major issue with many of these assays is that they are too complicated for untrained users and too costly for use in resource-limited regions. Other aspects of assays for such samples, including sample collection, transfer to the lab, complex sample preparation steps and analysis by trained personnel make such approaches highly labor intensive and time consuming. Hence, there is a significant need for easy-to-use and inexpensive assays for metal-ion detection.
3.1.1.1. Detection of metal ions in environmental, food and clinical samples.
The presence of metal ions in living systems is essential for proper functioning and homeostasis, but when the concentrations of essential or trace metal ions go unchecked they can wreak havoc at the molecular level with devastating consequences to the organism.128 One source where humans and other animals are exposed to metal ions is through exposure to the environment, particularly fresh water sources. In fact, the contamination of environmental water sources by metals is so problematic that governing agencies such as the World Health Organization (WHO: international), the United States Environmental Protection Agency (EPA: US), the European Drinking Water Directive (EDWD: European Union), and Health Canada (HC: Canada) have set strict allowable limits to maintain safe potability.129 Similar regulating bodies have set guidelines for safe levels of metal ions in food and beverage sources, where bioaccumulation of metal ions can be particularly problematic.
To monitor metal ions to ensure they are below regulatory limits, DNAzyme-based sensors have been reported for the detection of many metal ions in complex matrices, such as Pb2+, UO2 2+, Hg2+, Tl3+, Cd2+, Cr3+, Cr6+, Ag+, Cu2+, Ca2+, Mg2+, and Na+.53,130 As discussed previously, the activities of DNAzymes generally require metal ions as cofactors, a property which has been exploited in diverse strategies for the detection of metal and non-metal analytes. The detection of metal-ion cofactors via their ability to control DNAzyme catalysis has proven to be especially widespread and has been demonstrated in multiple complex matrices.
By far the most commonly reported DNAzyme-based metal-ion sensors have been those developed for the detection of lead ions. The first-ever DNAzyme, GR-5, was selected with the use of Pb2+ as the metal-ion cofactor.5 However, the first major advancement in DNAzyme-based metal sensing was the development of a fluorescent sensor for Pb2+ detection using the 17S/17E system, a landmark study published by the Lu group in 2000.40 The sensor was designed such that Pb2+-dependent cleavage of the embedded RNA site in the substrate by the DNAzyme led to the generation of a fluorescent signal owing to decreased proximity of a fluorophore and a quencher covalently placed on the DNAzyme and substrate pair, as illustrated in Fig. 7A.
Fig. 7.
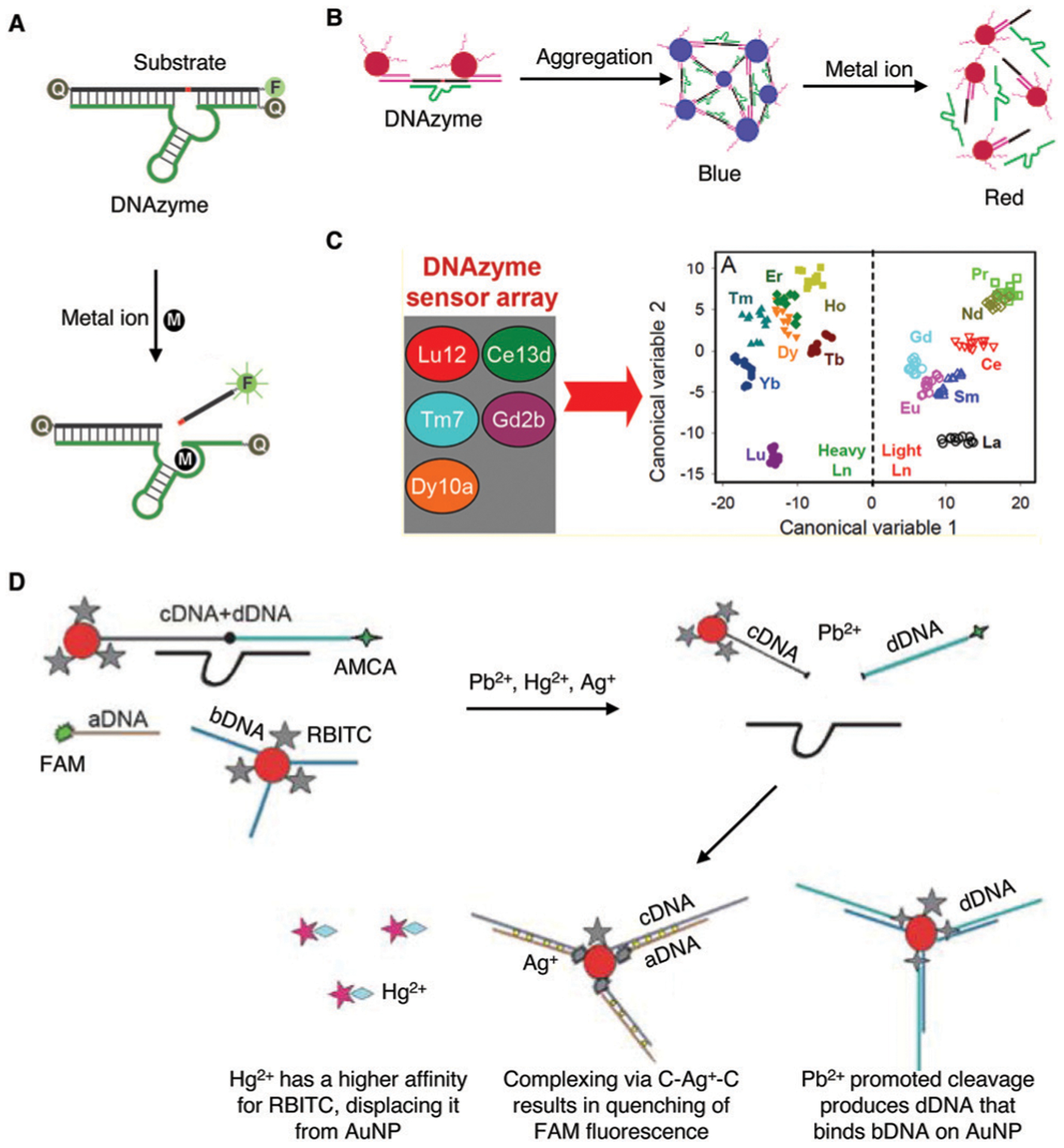
Representative DNAzyme sensors for metal-ion detection. (A) Florescence-based sensors for metal ions using a pair of fluorophore and quencher. Reprinted (adapted) with permission from J. Liu, A. K. Brown, X. Meng, D. M. Cropek, J. D. Istok, D. B. Watson and Y. Lu, A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2056–2061.75 (B) Colorimetric sensor based on gold nanoparticle aggregation and cleavage promoted disassembly. Reprinted (adapted) with permission from J. Liu and Y. Lu, A Colorimetric Lead Biosensor Using DNAzyme-Directed Assembly of Gold Nanoparticles, J. Am. Chem. Soc., 2003, 125, 6642–6643. Copyright 2003 American Chemical Society.41 (C) A multiplex DNAzyme sensor assay involving five DNAzymes. Reprinted (adapted) with permission from P. J. J. Huang, M. Vazin, J. J. Lin, R. Pautler and J. Liu, Distinction of Individual Lanthanide Ions with a DNAzyme Beacon Array, ACS Sens., 2016, 1, 732–738. Copyright 2016 American Chemical Society.208 (D) Use of a DNAzyme, fluorescent dyes and gold nanoparticles for the detection of Pb(II), Hg(II) and Ag(I) in a one-pot reaction. Reprinted (adapted) with permission from Y. Deng, Y. Chen and X. Zhou, Simultaneous sensitive detection of lead(II), mercury(II) and silver ions using a new nucleic acid-based fluorescence sensor, Acta Chim. Slov., 2018, 65, 271–277. Copyright 2018 Yuan Deng, Yinran Chen, Xiaodong Zhou.207
Another common metal contaminant in environmental water sources, which can negatively impact human health, is uranium in the form of the uranyl ion (UO22+).76 In 2007, the Lu group reported the selection of 39E and used this DNAzyme for the design of a fluorescent sensor using the same strategy as illustrated in Fig. 7A. This sensor exhibited impressive parts-per-trillion sensitivity (45 pM) and one million-fold selectivity over 19 other metal ions.75 The detection sensitivity was several orders of magnitude lower than the 130 nM maximum concentration limit set by the US EPA for uranyl in drinking water.
DNAzymes have also been used to detect other metal ions, including calcium,131 magnesium,132 nickel,81,133 sodium,77,134 and zinc.135 While the fluorescent sensor based on the 39E DNAzyme has very high sensitivity, sensors based on other DNAzymes may not have enough sensitivity to meet the needs for practical applications. To overcome this limitation, the Lu and Xiong groups took advantage of the CRISPR-Cas12a system, which has been shown to allow signal amplification through indiscriminate cleavage of any nontarget ssDNA with a fluorophore and quencher at the two ends near the target DNA, using the DNAzyme to regulate the CRISPR-Cas12a process.49 In the absence of a metal ion, the DNA activator is embedded into the binding arms of the DNAzyme and is not able to activate the CRISPR-Cas12a process. On the other hand, the presence of a metal ion will promote DNAzyme-based cleavage, releasing the DNA activator to activate CRISPR-Cas12a, which provides signal amplification. Using this method, they have demonstrated a decrease in the LOD of Na+ by an Na+-specific NaA43 DNAzyme to 0.10 mM, sensitive enough for POCD of Na+ in plasma to help evaluate the pathophysiological conditions hyponatremia and hypernatremia. On the other hand, the detection of copper is a special case: while nearly all of the DNAzymes used for metal-ion detection are RCDs, the detection system for copper often uses a Cu2+-dependent DNA-cleaving DNAzyme (CuDD) selected by Breaker and coworkers in 1996.136
Other optical and non-optical reporting methods have also been demonstrated for DNAzyme-based metal-ion detection. For example, back in 2003, the Lu group described the first colorimetric assay for Pb2+ detection using gold nanoparticle (AuNP) assembly and disassembly.41 The DNAzyme and the AuNP-tagged substrate are first assembled into blue colored aggregates; in the presence of lead ion, the DNAzyme cleaves its substrate, causing the AuNP aggregate to disassemble, due to a difference in melting temperature of the DNA enzyme/substrate strand and DNAzyme-cleaved products, resulting in a change of color from blue to red (Fig. 7B). This work has been the inspiration for many subsequent studies, as demonstrated by the accumulation of over 1300 citations.41 Since its report, multiple groups have utilized variations of DNAzyme–AuNP interactions for their sensor designs, including recent examples for the detection of lead,137–141 mercury,142 and uranyl ions143 in environmental water sources. The same group also described incorporation of DNAzymes into a colorimetric dipstick assay in 201043 and into an assay using a personal glucose meter in 2011,44 both of which can provide an easy-to-use, easy-to-interpret method for the detection of metal ions and other targets. Detailed descriptions of the use of these devices to detect metal ions and other analytes are provided in Section 4.5.
In the two decades following the selection of the first-ever DNAzyme GR-5, many optical,137,138,140,144–153 electrochemical,139,141,154–173 and other174–185 types of sensors that utilized the GR-5 or 8–17 (specifically 17S/17E) DNAzymes, or other DNAzymes for the detection of lead and other metal ions have been described and reviewed.53,56,58,130,186–190 Specifically, McGhee et al.130 and Zhou et al.186 provided comprehensive summaries of DNAzyme-based biosensors for diverse metal ions. Herein, Table 1 lists various methods, published since 2017, that utilize DNAzymes for the detection of metal ions in complex matrices, such as lake water, soil, and milk.
Table 1.
Recent advances in DNAzyme-based biosensors for metal-ion detection in complex matrices
| Analyte | DNAzyme | Sensor class | Limit of detection (LOD) | Sample type |
|---|---|---|---|---|
| Pb2+ | GR-5,138,140,146 17E152 | CR | 32,138 50,146 and 59.39 pM,140 100 nM152 | Water,138,140,152 soil146 |
| Pb2+ | GR-5 | OF | 15 fM148 | Water |
| Pb2+ | 8–17,144 GR-5145,153 | FR | 50,144 160,153 and 500 pM145 | Water |
| Pb2+ | GR-5 | OM | 2.12178 and 10 nM177 | Water |
| Pb2+ | GR-5,155,171,173 17E,160,169 8–17139,155,157,159,160,167,168,170 | EC | 4.8,155 17.4,170 and 290 fM,157 2,160 8,159 l5,169 20,173 95,139 and 330 pM,171 1.7141 and 10 nM,167 38 fg mL−1168 |
Water |
| Pb2+ | 17E,172,179 8–17154,183 | ECL | 200 fM,154 3,179 4.73,183 140 pM172 | Water,172,179 soil154 |
| Pb2+ | 17E | EB | 830 pM180 | Water |
| Pb2+ | GR-5 | LAPS | 0.01 ppb185 | Water |
| Pb2+ | GR-5 | QCM | 300 pM182 | Water |
| Pb2+ | 8–17 | RRS | 500 pM184 | Water |
| Pb2+ | 8–17 | SERS | 70 fM175 | Water |
| Hg2+ | E6191–193 MZ,194 | CR,142,192 FR,195 EC193,194 | 4.2,193 and 23 fM,194 5,142 30,195 and 33 pM,192 | Water,142 herbs192,195 |
| UO22+ | 39E | CR,143,196,197 FR,143,198 SERS199 | 72,199 and 100 fM (FR),143 20 (CR),143 190,198 and 330 pM,197 0.08 μg L−1 196 | Water |
| Ca2+ | EtNa | FR | 11,200 and 17 μM131 | Milk,200 water131 |
| Mg2+ | E6 | FR | 300 pM132 | Water |
| Ni2+ | Ni03l,81 NC133 | FR,81 ECL133 | 12.9 μM,81 3.1 nM133 | Water |
| Na+ | Ce13d | FR | 0.4 mM134 | Water |
| Zn2+ | 17E | CR | 3.5 nM135 | Water |
| Cu2+ | CuDD,201–204 10–23,205 CCB206 | CR,201,202 FR,203 EC204,205 | 50 fmol,203 330 fM,205 457 pM,206 1.31,201 5,204 8 nM202 | Water |
| Pb2+, Ag+, Hg2+ | 8–17 | FR | Pb2+: 480 pM, Ag+: 230 pM, Hg2+: 170 pM207 | Water |
Abbreviations: CCB: Cu2+-dependent DNAzyme, CR: colorimetric, CuDD: Cu2+-dependent DNA-cleaving DNAzyme, EB: electronic balance, EC: electrochemical, ECL: electrochemiluminescence, FR: fluorometric, LAPS: light addressable potentiometric sensor, NC: DNAzyme sequence source not cited but included, OF: optofluidic, OM: optical measurement, PEC: photoelectrochemical, RRS resonance Rayleigh scattering, SERS: surface enhanced Raman spectroscopy, QCM: quartz crystal microbalance.
Another recent innovation came in the design of an l-DNAzyme for the detection of Pb2+ in environmental samples.209 DNA naturally exists in the d-form, therefore l-form DNAzymes afford enhanced stability in complex sample matrices, because they are more resistant to degradation by natural nucleases present in many biological samples. Hence, this work demonstrated a viable strategy for developing DNAzyme-based biosensors and devices with exceptionally high biostability.
The high specificity of metal-ion-responsive DNAzymes makes it possible to detect several metal ions at once, or with the same assay. One such recent example demonstrated the multiplex detection of lanthanides (Ln3+) using five different lanthanide-dependent DNAzymes.208 The five DNAzymes used were Lu12, Ce13d, Tm7, Gd2b, and Dy10a, which shared common substrate and substrate binding arms.208,210–213 By analyzing the DNAzyme reactivity patterns when subjected to different metal ions, lanthanides could be detected and distinguished (Fig. 7C). This work demonstrated the feasibility of using multiple DNAzymes with varying reactivity to produce pattern-based recognition of multiple analytes.
Another example for multiplex metal-ion detection has been reported that is capable of detecting Pb2+, Hg2+ and Ag+ ions in a one-pot reaction.207 It employed the 8–17 DNAzyme immobilized on AuNPs, three fluorescent dyes: aminomethylcoumarin acetic acid (AMCA), 5-carboxyfluorescein (FAM), and rhodamine B isothiocyanate (RBITC), and quenching or recovery of the fluorescence of these dyes to detect these 3 metal ions, as illustrated in Fig. 7D. The Pb2+-promoted cleavage leads to the release of two DNA fragments: one of which (dDNA, carrying AMCA) hybridized with bDNA placed on AuNPs, resulting in fluorescence quenching (for Pb2+ detection), and the other (linked to AuNPs) hybridizes with FAM-labeled aDNA via a C–Ag+–C complex (for Ag+ detection). Hg2+ is detected based on its preferential interaction with RBITC, which is originally associated with the DNA–AuNP complex but is displaced by Hg2+.
3.1.1.2. Detection of metal ions in cells.
Because of the high sensitivity and selectivity of metal-ion-dependent DNAzymes, they have been widely applied as sensors for environmental monitoring and medical applications, as described above. In addition, DNAzymes exhibit high biocompatibility and the ability to detect biologically relevant targets. Thus, DNAzyme sensors have also been developed to detect those targets inside biological systems. Over the past ~10 years, as shown in Fig. 8, a major focus of the field has been to design DNAzyme sensors for imaging in living cells, by developing novel delivery methods, and engineering novel DNAzymes with higher biostability and ability for quantification. Built upon this success, the field has shifted from cellular imaging to in vivo imaging in animals.
Fig. 8.
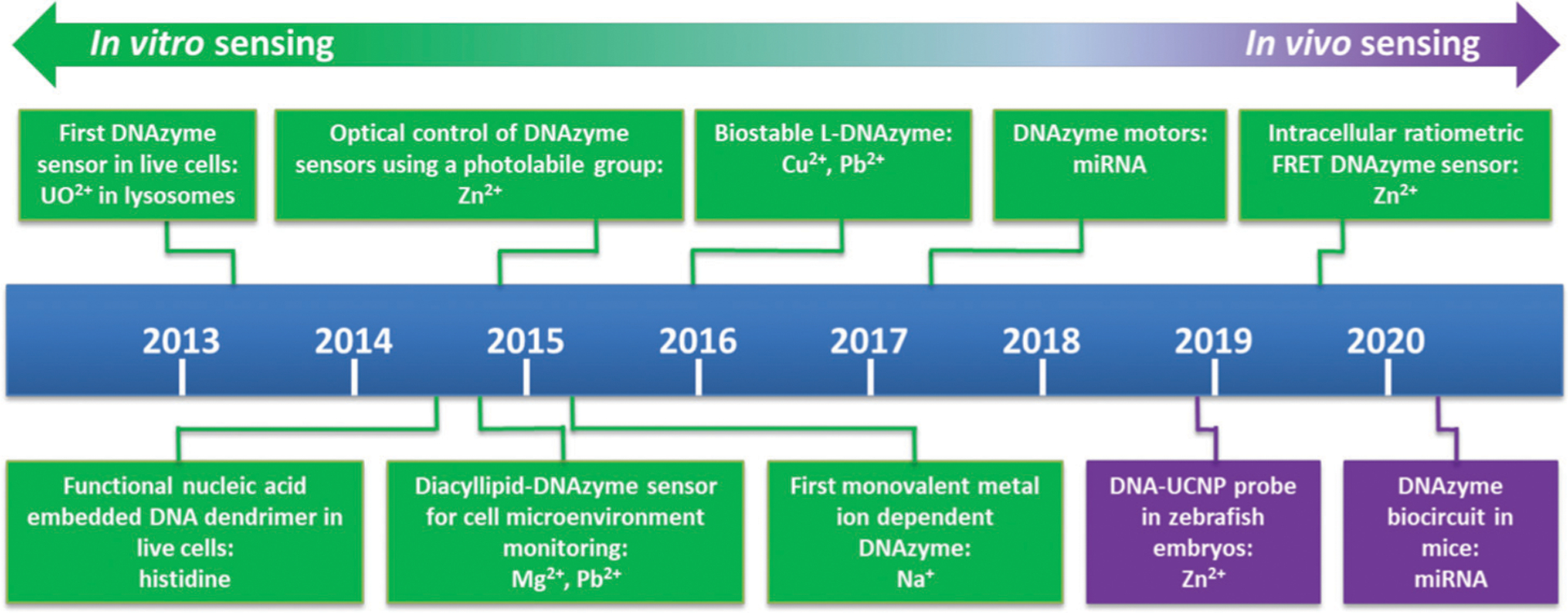
Timeline of the major milestones of DNAzyme sensors for cellular and in vivo sensing. Major sensor types and sensing target(s) are listed.53,77,214–222
In 2013, the Lu group reported the first intracellular DNAzyme sensor for imaging uranyl ion in living cells by selecting and then conjugating the uranyl-specific 39E DNAzyme to AuNPs.214 As shown in Fig. 9A, the substrate strand, modified with Cy3 at the 3′ end and Black Hole Quencher-2 (BHQ-2) at the 5′ end, is hybridized to 39E conjugated to AuNPs, which serves both as a cellular delivery agent and an effective quencher of the fluorophore. In the presence of uranyl, the substrate strand is cleaved, resulting in a short Cy3-containing DNA strand with a melting temperature (21 °C), which is significantly lower than the original substrate (60 °C). This DNA strand separates from the AuNP and the other cleavage product carrying BHQ-2, leading to enhanced fluorescence. The sensor has been used to image uranyl in HeLa cells based on both fluorescence imaging and flow cytometry.
Fig. 9.
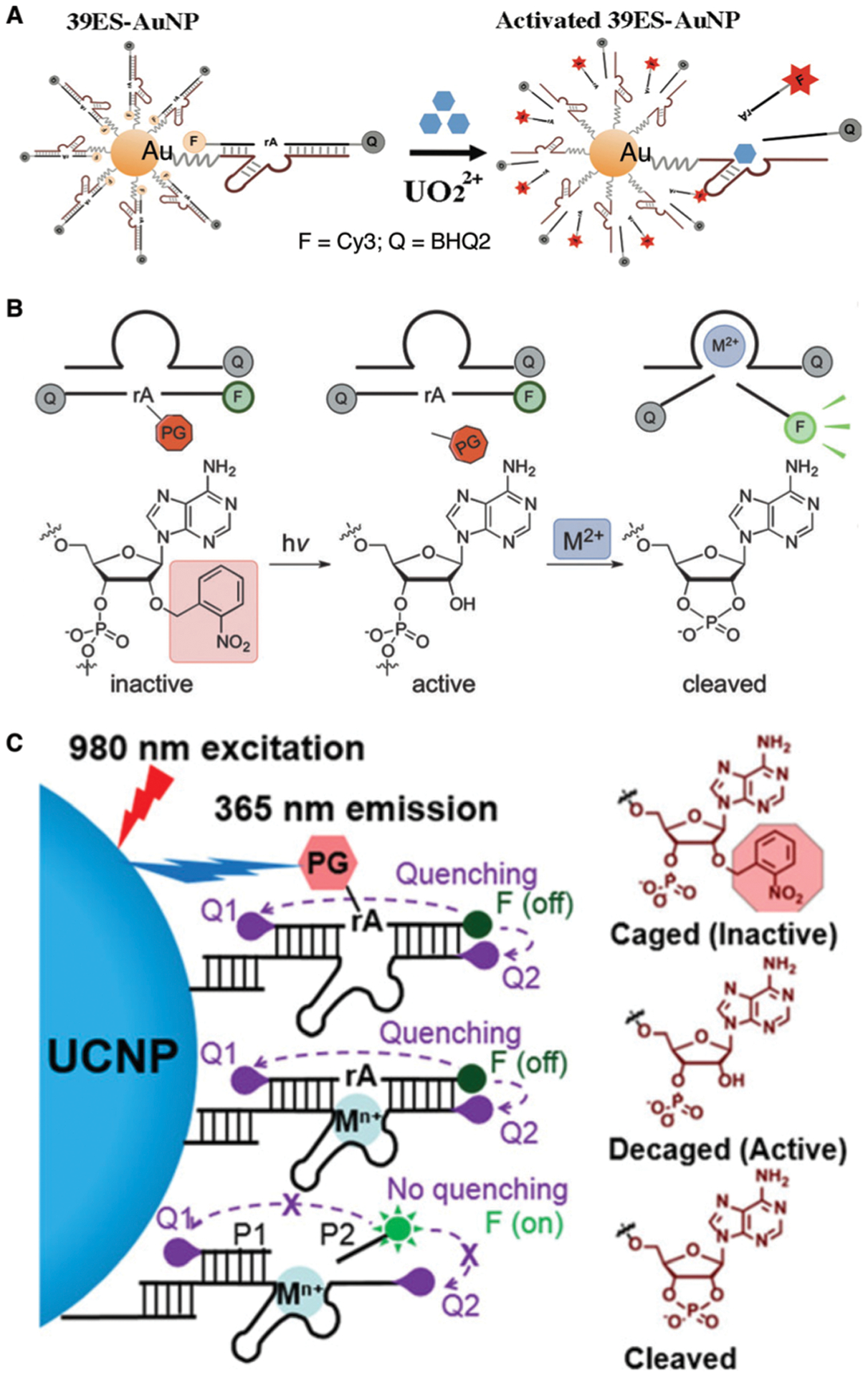
DNAzyme sensors for metal-ion imaging in living cells. (A) The first intracellular DNAzyme based sensor for uranyl ion detection in living cells using 39E immobilized onto gold nanoparticles. Reprinted (adapted) with permission from P. Wu, K. Hwang, T. Lan and Y. Lu, A DNAzyme-Gold Nanoparticle Probe for Uranyl Ion in Living Cells, J. Am. Chem. Soc., 2013, 135, 5254–5257. Copyright 2013 American Chemical Society.214 (B) A DNAzyme sensor for metal-ion imaging in living cells with blockage of the 2′-OH of the scissile ribonucleotide with a light-sensitive nitrobenzyl group. Reprinted (adapted) with permission from K. Hwang, P. Wu, T. Kim, L. Lei, S. Tian, Y. Wang and Y. Lu, Photocaged DNAzymes as a General Method for Sensing Metal Ions in Living Cells, Angew. Chem., Int. Ed., 2014, 53, 13798–13802. Copyright 2014 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.217 (C) Zn2+ sensing in vivo using photocaged Zn2+-selective DNAzyme conjugated on lanthanide-doped upconversion nanoparticles (UCNPs). Reprinted (adapted) with permission from Z. Yang, K. Y. Loh, Y.-T. Chu, R. Feng, N. S. R. Satyavolu, M. Xiong, S. M. Nakamata Huynh, K. Hwang, L. Li, H. Xing, X. Zhang, Y. R. Chemla, M. Gruebele and Y. Lu, Optical Control of Metal Ion Probes in Cells and Zebrafish Using Highly Selective DNAzymes Conjugated to Upconversion Nanoparticles, J. Am. Chem. Soc., 2018, 140, 17656–17665. Copyright 2018 American Chemical Society.219
In comparison with sensors or imaging agents which are based on binding only, such as aptamers, target responsive DNAzymes rely on both binding and catalytic activity, which provides enhanced sensitivity and selectivity. However, DNAzymes can become active during cellular delivery and uptake, which can negatively affect sensor performance (due to high signal-to-background ratios). To overcome this limitation, the Lu group introduced a photocaged nitrobenzyl group to the 2′-OH of the scissile ribonucleotide in the substrate strand (Fig. 9B).217 In this way, the DNAzyme remains inactive during cellular delivery and uptake, and more importantly, this approach allows for temporal and spatial control of DNAzyme activity via controlling the timing and location of the applied light.
While the nitrobenzyl is a common and effective photo-caging/decaging group, it requires the use of high energy UV (~365 nm) irradiation that not only can damage the cells but also has limited cell penetration depth. To overcome this issue, the Lu group designed a DNAzyme sensor that can be photo-thermally activated by near-IR radiation, which can increase the local temperature by application of 808 nm light.223 In this case, 8–17 DNAzyme was coupled to gold nanoshells and a short DNA sequence was used to sequester the catalytic activity of the DNAzyme via hybridization with the catalytic core. By applying light at a specific location of the cell, the local temperature increased, resulting in the dehybridization of the blocking DNA to activate the DNAzyme sensor. Similarly, the Kuang group has recently constructed a chiral satellite assembly from three different DNAzymes and spiny platinum modified with gold nanorods and upconversion nanoparticles, which can be activated by handedness-dependent circular polarized light. Their system used Zn2+-, Mg2+-, and Cu2+-dependent DNAzymes, and was capable of performing simultaneous quantitative analysis of these three metal ions in living cells.224
While near-IR radiation can overcome the limitations of the UV light for DNAzyme activation in vivo, light activation is generally still invasive to the cellular processes. To activate DNAzymes in a less invasive manner, a magnetic field-activated binary DNAzyme was reported to take advantage of nano-magnetic actuation, which enabled sensing of a specific mRNA analyte via application of a magnetic field remotely.225 In addition to spatial and temporal control using external stimuli, endogenous and orthogonal control using a homing restriction enzyme I-SceI that is expressed inside cells allowed the DNAzyme sensor to adopt its active confirmation by cleaving a double-stranded DNA segment that blocks formation of the active configuration in the absence of I-SceI.226
One significant challenge in applying DNAzyme sensors in living cells is to deliver them into the cells, as the negatively charged DNA molecules are not normally taken up by cells, thus requiring various carriers to deliver DNAzymes to cells. These include gold nanoparticles,214 gold nanoshells,223 cationic liposomes,217 MnO2 nanosheets,227 DNA nanostructures,228,229 and cell penetrating peptides,230 as reviewed elsewhere.130,220 Another challenge is biostability of the DNAzyme sensors. To increase the stability, non-natural l-DNA has been used, which exhibits similar reactivity to the d-DNA enantiomer but cannot be degraded by native nucleases.218 Furthermore, framework nucleic acids have been used to encapsulate DNAzyme sensors, which can not only protect sensors from nuclease degradation and nonspecific protein binding, but can also improve delivery of the DNAzymes into cells without the need of other carriers.229 To demonstrate the utility of these DNAzyme sensors to detect metal ions in living cells, most studies involved the addition of high concentrations of metal ions in the reaction mixture. To detect metal ions at physiological concentrations, the Lu and Jiang groups used catalytic hairpin assembly (CHA) to amplify the cleavage product of the DNAzyme inside HeLa cells to amplify the fluorescent signal, resulting in higher sensitivity and thus detection of endogenous metal ions.231
DNAzyme sensors can not only detect various targets in cells, but also monitor the cell microenvironment. The Tan group developed cell membrane-anchored DNAzyme sensors, consisting of diacyllipid–DNAzyme conjugates that allow real-time monitoring of both exogenous and cell-extruded metal-ion targets. Diacyllipid–DNAzyme conjugates can efficiently self-assemble onto the cell membrane based on the hydrophobic interaction between the lipophilic tail and the cellular phospholipid layer. In addition, the diacyllipid–DNA conjugate-based cell-membrane modification strategy can be extended to engineer different DNA sensors on the cell surface for real-time analysis of various targets, such as metal ions, metabolites, proteins and extracellular vesicles, in the cellular microenvironment, providing potentially powerful tools for biological and biomedical research.216
Thanks to the above progress towards increasing cellular delivery efficiency, biostability and external and internal controls, successful strategies have been described for detection of several metal ions, such as UO22+,214,228 Zn2+,217,220,223,224,232–234 Pb2+,228,232 Na+,77,231 Mg2+,224,226,233,235 and Cu2+,224,235 as well as other important non-metal targets, a subject that will be discussed in Section 3.2.
3.1.1.3. Detection of metal ions in vivo.
Efforts have also been made towards the development of DNAzyme sensors for imaging of metal ions in living animals. The Lu group reported such an example using a photocaged Zn2+-selective DNAzyme conjugated to lanthanide-doped upconversion nanoparticles (UCNPs).219 As shown in Fig. 9C, the UCNP is capable of upconverting 980 nm excitation light into 365 nm emission. Then, the UV photon efficiently photodecaged the substrate strand containing a nitrobenzyl group at the 2′-OH of the adenosine ribonucleotide, allowing enzymatic cleavage by the Zn2+-selective DNAzyme. Because the released strand contained a FAM fluorophore, which was initially quenched by BHQ1 and Dabcyl quenchers, the cleavage event resulted in an increase in fluorescence emission. The DNAzyme–UCNP probe enabled Zn2+ sensing by exciting in the near IR (NIR) biological imaging window in both living cells and zebrafish embryos and detecting in the visible region.219 Although DNAzyme–UCNP probes have been applied in zebrafish embryos and larvae, the signal detection still relies on the transparency of early fish larvae and is limited when applied to other non-transparent animal models. This limitation could be overcome by either using other fluorophores with NIR emission or other imaging techniques, such as photoacoustic imaging236 or MRI,237 which have deeper tissue penetration.
3.1.2. Detection of small molecules and proteins using allosteric DNAzymes or DNA aptazymes.
As discussed in Section 2.4, various allosteric DNAzymes and DNA aptazymes have been designed for the detection of small molecules, most of which have integrated the well-known ATP-binding DNA aptamer (which recognizes ATP, ADP, AMP or adenosine) as the model aptamer to demonstrate various sensor design concepts. Early studies did not focus on expanding sensors to other targets or on the analytical performance of the sensors. However, more attention has been paid to these two aspects in recent years. For example, some recent studies have employed the histidine-dependent RCD discussed in Section 2.3.387 for the detection of l-histidine.238,239
More recently, Yan et al. have reported an aptazyme that places the Pb2+-dependent RCD and the PSA-binding DNA aptamer into a hairpin structure so that it can be used for PSA detection.111 The sensor takes advantage of size-dependent DNA adsorption by graphene oxide (GO): GO has stronger affinity for the longer, uncleaved substrate than for the shorter cleavage fragments. In the absence of PSA, the fluorophore-tagged substrate is absorbed by GO and efficiently quenched, but in the presence of PSA, the substrate is cleaved into shorter cleavage products that do not absorb efficiently to GO, resulting in a loss of quenching and a higher level of fluorescence (Fig. 10A). The method was shown to have an LOD 0.76 pg mL−1 and a linear dynamic range of 1–100 pg mL−1 in human serum.
Fig. 10.
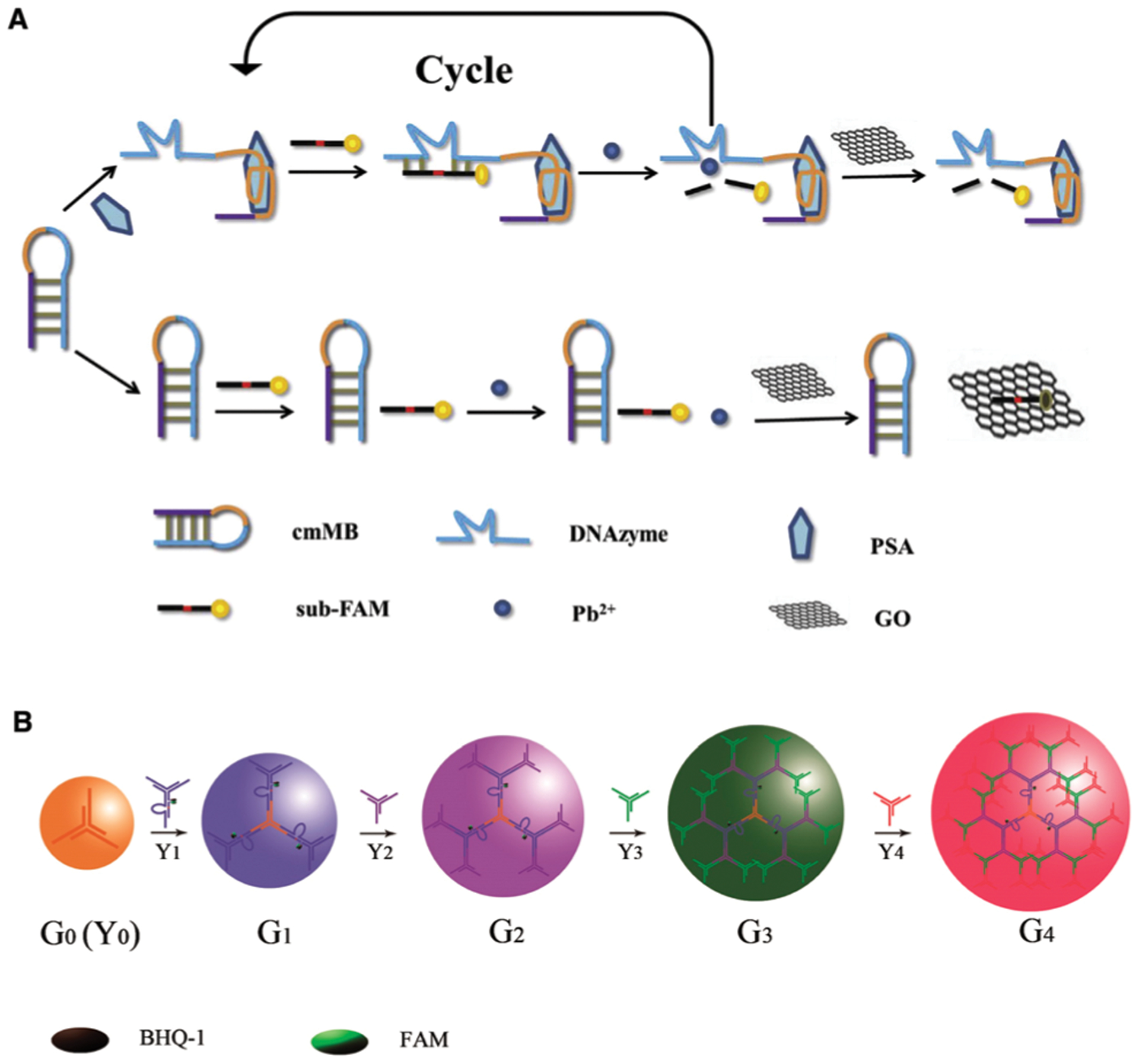
DNAzyme sensors for protein and small-molecule detection. (A) A DNAzyme sensor for PSA detection which incorporates an aptamer, an RCD and GO. Reprinted (adapted) from Y. Yan, C. Ma, Z. Tang, M. Chen and H. Zhao, A novel fluorescent assay based on DNAzyme-assisted detection of prostate specific antigen for signal amplification, Anal. Chim. Acta, 2020, 1104, 172–179, with permission from Elsevier.111 (B) A DNAzyme sensor for in situ monitoring of histidine. Reprinted (adapted) with permission from H.-M. Meng, X. Zhang, Y. Lv, Z. Zhao, N.-N. Wang, T. Fu, H. Fan, H. Liang, L. Qiu, G. Zhu and W. Tan, DNA Dendrimer: An Efficient Nanocarrier of Functional Nucleic Acids for Intracellular Molecular Sensing, ACS Nano, 2014, 8, 6171–6181.215 Further permissions related to the material excerpted should be directed to the American Chemical Society.
Allosteric DNAzymes and DNA aptazymes have also been examined for small-molecule detection in living cells. For example, the Tan and Zhang groups reported a DNA dendrimer scaffold as an efficient nanocarrier to deliver DNAzymes and to conduct in situ monitoring of histidine in living cells. The DNA dendrimers are assembled step by step using a series of Y-shaped DNA structures, as shown in Fig. 10B. Upon full assembly, the DNA dendrimers, with the DNAzyme unit tagged with a fluorophore-quencher pair, maintained the catalytic activity toward histidine in the cellular environment. Most notably, the dendrimers exhibit excellent biocompatibility, cell membrane permeability, and significantly enhanced intracellular stability over the free DNAzyme.215 The Wang group took advantage of spherical nucleic acid structures and developed an aptazyme sensor for amplified ATP probing in living cells.240 In this sensor, the authors modified AuNPs with substrate strands hybridized to ATP-recognizing aptazyme strands. ATP binding results in the cleavage of the substrate and the release of the fluorophore-labeled substrate strands from the AuNPs, resulting in fluorescence enhancement. Furthermore, the process is repeated so that each copy of the target can cleave multiple substrate strands, thus achieving a detection sensitivity that is 2 or 3 orders of magnitude higher than that of aptamer-only ATP sensors in living cells.
3.1.3. Detection of bacteria using aptazymes
3.1.3.1. Aptazyme-based bacterial detection without amplification.
Several assays have utilized the inherent fluorogenic nature of RFD-EC1 to produce solution-based tests based on cleavage-induced fluorescence dequenching. An early example involved the E. coli induced cleavage of RFD-EC1 for monitoring bacterial inhibition by antibiotics and for studying bacterial competition as a result of cohabitation.241 The assay could distinguish membrane-targeting pore-forming antibiotics from other types of antibiotics owing to the DNAzyme activation by a cytoplasmic protein target.
The Hong and Lu groups adapted the RFD-EC1 DNAzyme using the cleavage-based dequenching method to rapidly identify discrete E. coli colonies in water samples and differentiate them against non-E. coli colonies reported to cause false positives in other tests using simultaneous enzyme-based agar media.242 The presence of E. coli could be detected over 90 non-E. coli strains which had been isolated from waste water sourced from multiple locations. This RFD system provided an inherent advantage over traditional verification methods, as it was not inhibited by interferents in the media and wastewater samples.
The fluorescence dequenching method has also been used in conjunction with the VA DNAzyme, which was configured as a catalytic beacon carrying terminal F and Q moieties. This method could detect 4000 CFU mL−1 of V. anguillarum in fish tissue and feeding water samples when using carefully optimized buffer conditions.90 As noted above, the RFD fluorescence dequenching method has also been applied to detection of both breast cancer cells, with a detection limit of 5000 cells per mL, and K562 cancer cell to produce a diagnostic test for acute myeloid leukemia.
An interesting example using the dequenching method was reported by Zhao and co-workers, who presented a new droplet microfluidic platform termed ‘Integrated Comprehensive Droplet Digital Detection’ (IC 3D) that was able to selectively detect bacteria directly from milliliters of diluted blood at single-cell sensitivity in one step, which could have significant benefits for rapid detection of sepsis within 4 h.97 Here, the EC1 DNAzyme was encapsulated within aqueous droplets formed in a stream of organic solvent (Fig. 11A), and a high-throughput 3D particle counter system was used to detect EC1 fluorescence from each droplet. This method provided absolute quantification of both stock and clinical isolates of E. coli in spiked blood over a range covering 1 to 104 bacteria per mL.
Fig. 11.
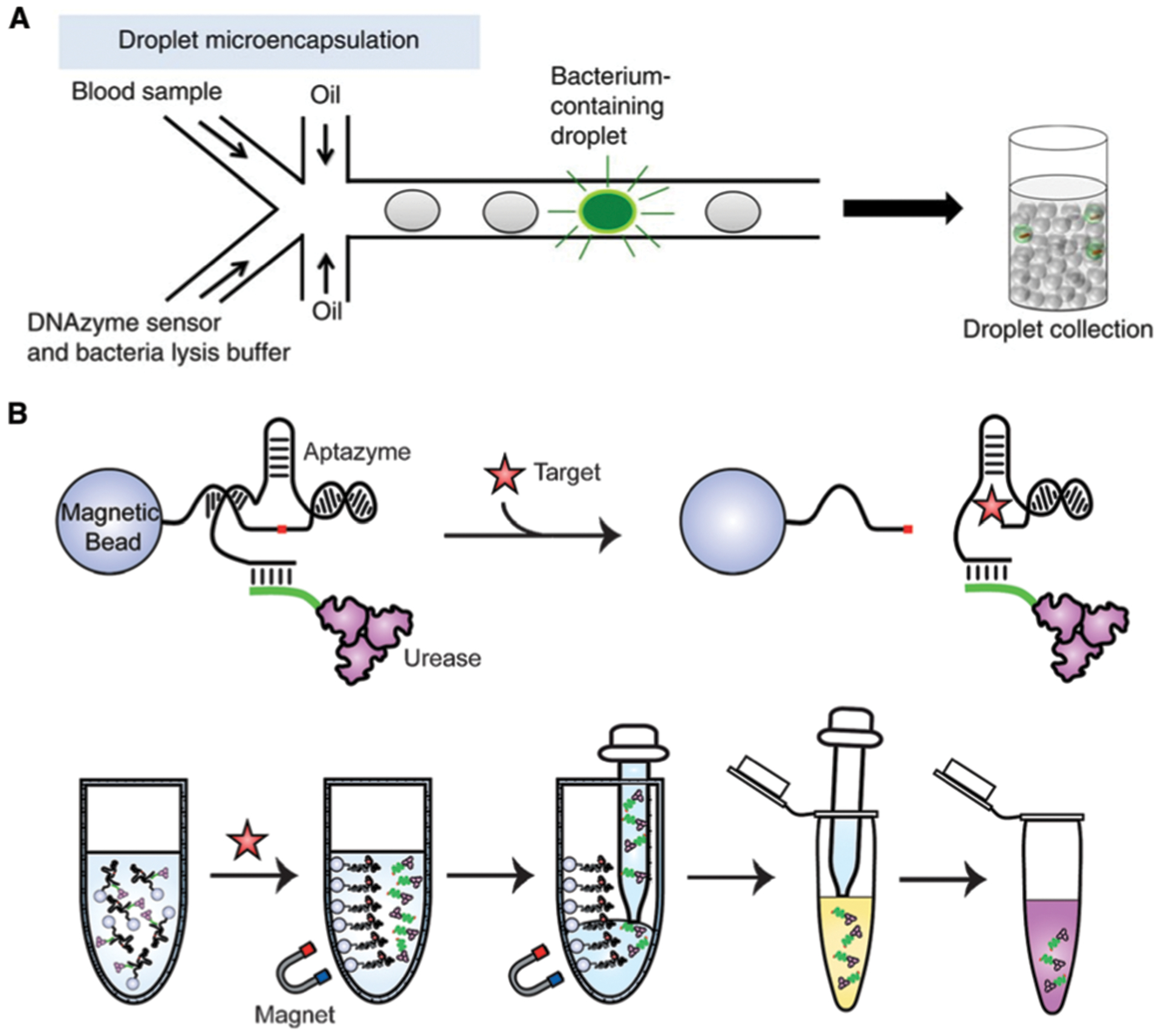
Aptazyme-based E. coli detection without amplification. (A) Integrated Comprehensive Droplet Digital Detection (IC 3D). Reprinted (adapted) with permission from D. K. Kang, M. M. Ali, K. Zhang, S. S. Huang, E. Peterson, M. A. Digman, E. Gratton and W. Zhao, Rapid detection of single bacteria in unprocessed blood using Integrated Comprehensive Droplet Digital Detection, Nat. Commun., 2014, 5, 1–10. Copyright 2014 Kang et al.97 (B) Bacterial litmus test. Reprinted (adapted) with permission from K. Tram, P. Kanda, B. J. Salena, S. Huan and Y. Li, Translating Bacterial Detection by DNAzymes into a Litmus Test, Angew. Chem., Int. Ed., 2014, 53, 12799–12802. Copyright 2014 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.46
An alternate method to generate a fluorescence signal from EC1 was based on the adsorption of the DNAzyme onto a graphene surface to strongly quench the emission output.243 Exposure of the adsorbed EC1 to E. coli-containing samples resulted in the release of the DNAzyme, followed by the cleavage-mediated production of a higher fluorescent signal. This method was able to detect 105 CFU mL−1 E. coli cells in 30 min and was capable of single cell detection when combined with a 10 h culture step.
Another key innovation involving EC1 was the development of a simple colorimetric assay for E. coli using a modified litmus assay.46 In this work, the EC1 DNAzyme was immobilized onto magnetic beads and the sequence was extended to allow for the hybridization of a complementary DNA probe that was conjugated to urease, which can efficiently catalyze the hydrolysis of urea into carbon dioxide and ammonia, thereby increasing solution pH. In the presence of the target, cleavage of the substrate led to the specific release of the EC1–urease complex from the magnetic beads, after which the cleaved EC1–urease complex was transferred to a separate reaction tube which contained urea and a pH indicator dye (Fig. 11B). With increased urease concentration, representative of increased bacteria in the original reaction, a concentration-dependent color change was observed over 2 h, with a detection limit of 500 CFU mL−1 of E. coli. A similar assay format was developed using acetylcholinesterase (AChE) in place of urease, where the AChE was released from magnetic beads upon EC1 cleavage by E. coli, followed by AChE-catalyzed hydrolysis of acetylthiocholine and subsequent enhancement of the emission intensity of DNA-bound silver nanoclusters by the produced thiocholine.244 This method could detect 60 CFU mL−1 of E. coli in 75 min.
A very recent example utilizing EC1 involved the development of a two-channel electrochemical chip that uses an electroactive version of EC1 that releases a DNA barcode from a “release” electrode via E. coli-induced RNA cleavage, which is subsequently recaptured on a nearby “capture” electrode to generate a ratiometric amperometric signal that can identify the presence of the bacterial target.50 By taking advantage of the high surface area from the nanostructured electrodes it was possible to achieve the analytical sensitivity and specificity needed for clinical analysis. This e-RCD assay was capable of detecting 10 CFU (equivalent to 1000 CFU mL−1) of E. coli and demonstrated clinical sensitivity of 100% and specificity of 78% for detection of bacterial infections from 41 patient urine samples, with an assay time of less than one hour.
3.1.3.2. Aptazyme-based bacterial detection incorporating amplification.
Given that many of the current aptazymes target bacteria or cancer cells, the ability to achieve ultra-low detection limits is particularly important. For example, in the case of drinking water, many regulatory bodies require assays be able to detect a single culturable organism in a volume of 100 mL, with limits for recreational water at between 126 (USA EPA’s Ambient Water Quality Criteria for Bacteria) and 500 (European Directive 2006/7/EC) CFU per 100 mL.245 To achieve such detection limits, several groups have integrated an aptazyme as the MRE with isothermal amplification (ITA) strategies, where activation of the aptazyme by the target causes the aptazyme to initiate the ITA process, which then produces a colorimetric or fluorometric output. While there are a multitude of ITA methods that have been used for amplification of nucleic acid targets,246–251 there are currently only three ITA methods that have been utilized to amplify aptazyme-target interactions; hybridization chain reaction (HCR), rolling circle amplification (RCA) and catalytic hairpin amplification (CHA). Examples of each amplification strategy are given below.
The Brennan and Li groups developed an RCA strategy for E. coli detection using RFD-EC1 and a DNA[2] catenane (D2C; made of two interlocked single-stranded DNA rings as shown in Fig. 12A).252 The component rings in the D2C are designed to form a strong linking duplex between them so that neither can act as the circular template for RCA because of the topological constraint. One of the rings is also designed to be the substrate for RFD-EC1 so the DNAzyme can cleave this ring to release the topological constraint and allow the other ring to act as a template for RCA. It was shown that this sensor was able to achieve a detection limit of 10 CFU mL−1. Furthermore, the use of circular DNA makes the system highly resistant to nucleases, making it functional in blood samples.
Fig. 12.
Aptazyme-based E. coli detection incorporating RCA. (A) Enabling RCA via releasing topological constraint of DNA[2] catenane (D2C) by EC1. Reprinted (adapted) with permission from M. Liu, Q. Zhang, Z. Li, J. Gu, J. D. Brennan and Y. Li, Programming a topologically constrained DNA nanostructure into a sensor, Nat. Commun., 2016, 7, 12074. Copyright 2016 Liu et al.252 (B) Cross-amplification by RCA and substrate cleavage by RCD produced by RCA. Reprinted (adapted) with permission from M. Liu, Q. Zhang, D. Chang, J. Gu, J. D. Brennan and Y. Li, A DNAzyme Feedback Amplification Strategy for Biosensing, Angew. Chem., Int. Ed., 2017, 56, 6142–6146. Copyright 2017 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.253
The same groups also developed an approach to create a cross-feedback mechanism between RCA and an RCD (Fig. 12B).253 It begins with the creation of an amplifiable primer/circular DNA complex (complex I) by the combined action of the EC1 DNAzyme and phi29 DNA polymerase. RCA is initiated with complex I, which in turn generates copies of MgZ (an RCD) that can cleave Complex II to produce more complex I. Therefore, the production of MgZ from the initial RCA feeds back into the RCA reaction to generate additional DNA assemblies for additional RCA, producing an exponential amplification cascade. The reaction products could be detected either through binding of an intercalating fluorescent dye, or via generation of PW17 (a PMD) to achieve colorimetric detection. The sensor was capable of detecting 10 CFU mL−1 in under 30 min and single cell detection in 2 h, an improvement in LOD of 3 orders of magnitude over conventional non-amplified methods.
Another study integrated RFD-EC1 with HCR to achieve signal amplification (Fig. 13A).254 EC1 was immobilized to magnetic beads (MBs), and upon target activation and cleavage, a residual DNA strand remained on the beads, which was used to initiate HCR to grow a DNA assembly on the MBs containing multiple biotin moieties that then captured avidin-labelled glucose oxidase (GOx) as a reporter element. GOx catalyzed production of peroxide led to etching of plasmonic silver nanoparticles, and a corresponding color change. This method afforded a LOD of 50 CFU mL−1 of E. coli in 20 min.
Fig. 13.
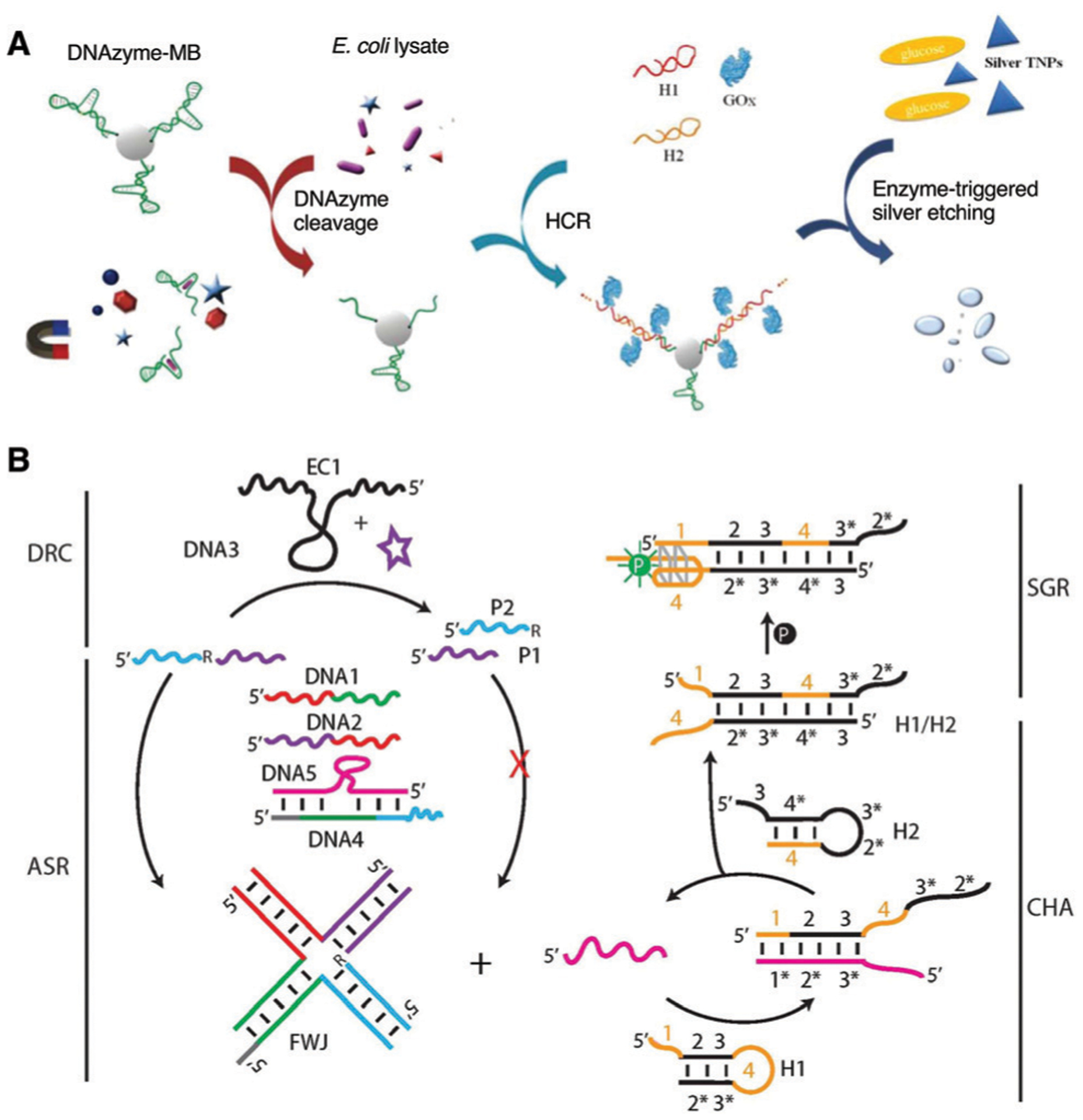
Detection of E. coli using RFD-EC1 as part of (A) hairpin chain reaction (HCR) and (B) catalytic hairpin assembly (CHA). Panel A is reprinted (adapted) from F. Yu, Y. Li, M. Li, L. Tang and J. J. He, DNAzyme-integrated plasmonic nanosensor for bacterial sample-to-answer detection, Biosens. Bioelectron., 2017, 89, 880–885, with permission from Elsevier.254 Panel B is Reprinted (adapted) with permission from Z. Zhou, J. D. Brennan and Y. Li, A Multi-component All-DNA Biosensing System Controlled by a DNAzyme, Angew. Chem., Int. Ed., 2020, 59, 10401–10405. Copyright 2020 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.255
Most recently, a multi-component all-DNA biosensing system has been developed that incorporates RFD-EC1 and CHA to achieve signal amplification.255 As shown in Fig. 13B, the presence of E. coli will activate the DNAzyme-mediated RNA cleavage (DRC), and this reaction will impact the assembly-mediated strand release reaction (ASR) designed to create a DNA four-way junction (4WJ): the cleavage inhibits a downstream CHA reaction that results in a split G-quadruplex reassembly (SGR), which binds protoporphyrin IX to provide a fluorescence readout. Using this method, a limit of detection of 50 CFU mL−1 was observed with a reaction time of 90 min. While slower than the HCR method described above, this method does not require any separation steps and the use of unstable protein enzymes, and in theory can be easily adapted to other DNAzymes.
3.2. Indirect sensing using DNAzymes
Although many DNAzymes have been reported, the targets that are directly recognized by these DNAzymes are still relatively limited. As such, there has been considerable interest in devising strategies to allow the use of the existing, well-characterized DNAzymes for sensing analytes they do not directly bind. Various approaches have been demonstrated for different types of targets, including metal ions, toxic gases, small molecules, nucleic acids, and proteins. These include sequestering the metal-ion cofactor of a DNAzyme, targeting the precursor of the target that the DNAzyme binds, and the use of a DNAzyme as reporter molecule for the binding interaction between another MRE and its target. We will discuss selected examples of these strategies in this section, which is once again organized by target type.
3.2.1. Detection of toxic gases.
The CuDD DNAzyme, selected by the Breaker Lab,256 has been used to detect hydrogen sulfide (H2S). Hydrogen sulfide is a toxic gas with a distinctive rotten egg smell, that is produced as a byproduct of industries such as papermaking and petroleum refining.257 The DNAzyme sensor is based on a loss of DNAzyme activity owing to sequestering of Cu2+ by H2S via the formation of CuS.257 The DNAzyme was labeled with a fluorophore and quencher pair so that it could function as a fluorescent sensor. Without H2S, Cu2+ is available to the DNAzyme, which cleaves its substrate, generating a strong fluorescence signal due to the release of the quencher. In the presence of H2S, Cu(II) is converted to CuS, which prevents the cleavage, resulting in low levels of fluorescence. This method was able to achieve an LOD of 0.2 μM.
Two DNAzyme biosensors have been reported for the detection of radon (222Rn), a radioactive, odorless and colorless gas classified as a first-class environmental carcinogen. Radon exists naturally in environmental sources and is produced as a combustion product of coal and natural gas.258 No DNAzyme has been reported to recognize 222Rn, but radon is known to decay with a half-life of 3.8 days into its stable daughter product, 210Pb, which, upon conversion to Pb2+ via adsorption into acetic acid, can be detected with the Pb2+-dependent RCD GR-5.259 In the first sensor, GR-5 was designed to cleave a hairpin structure (MB) containing the cleavage site in the loop region.259 In the presence of Pb2+, MB was cleaved by the DNAzyme, and the cleavage fragments then adsorbed to the surface of AuNPs, protecting the nanoparticles from salt induced agglomeration. In the absence of Pb2+, the MB remained intact, and salt-induced agglomeration of the AuNPs led to a distinct color change. By this method, radon was detected in the linear dynamic range of 6.22 × 102–1.02 × 105 Bq h m−3 with an LOD of 186.48 Bq h m−3. The second sensor, described by the same group, again used GR-5 for Pb2+ recognition but used a PMD for colorimetric signal generation.260 In the presence of Pb2+, a fragment produced by GR-5 facilitated the opening of hairpin 1 (H1). H1 was also partially complementary to hairpin 2 (H2). Successive opening of H1 and H2 led to the assembly of a double-stranded DNA product that was branched with repeating units of the PMD to facilitate colorimetric detection. The linear dynamic range of this sensor was reported as 5.14 × 103–1.65 × 105 Bq h m−3, with a limit of detection of 1.62 × 103 Bq h m−3. Both sensors were compared to the RAD7 radon detector, and the results were strongly correlated.
The three examples discussed in this section provide good evidence that DNAzymes can be utilized for monitoring air quality. The detection of toxic gases is essential for environmental monitoring and human health as exposure to these gases can affect infrastructure, wildlife, and human life.261 Other naturally occurring toxic gases which may be of interest for further development include nitrogen dioxide and sulphur dioxide.261
3.2.2. Indirect detection of metal ions.
As discussed above, many excellent metallo-DNAzymes have been derived by in vitro selection and used as MREs for metal-ion detection. However, highly functional DNAzymes have yet to be created for some metal ions. One example is the mercury ion (Hg2+). While a Hg2+-selective DNAzyme, denoted as 10–13, containing 2 modified nucleobases, has been reported, the concentration response to Hg2+ shows an unusual bell-shape, suggesting a competition between activation and inhibition by Hg2+.83 Fortunately, Hg2+ can be specifically recognized via T–Hg2+–T formation. Several DNAzyme-based assays and portable biosensors have taken advantage of this and employed an RCD or PMD for signal transduction and/or signal amplification. These sensors can be classified as colorimetric,142,192,195,262–266 photoelectrochemical,267 or electrochemical,193,194 (see Section 4 for more details on portable sensor devices) all of which demonstrated limits of detection that were well below the regulatory limit of 10 nM set by the EPA.186 For biosensors that employed RCDs in their sensing strategy, the DNAzyme was used to control or propagate some form of cyclic nucleic acid-based isothermal amplification. The exception was the sensor described by Yun et al., which featured a DNAzyme motor on AuNPs.195 The enzyme strand is designed such that it contains a flexible arm and a catalytic domain that is only active upon the formation of T–Hg2+–T. The activated DNAzyme then binds and cleaves each of the many fluorescently labeled substrate strands immobilized also on AuNPs. Due to the short length of the substrate strand, the signal of the fluorophore is quenched when it is in close proximity to the AuNP. Using this method, a limit of detection of 30 pM was demonstrated, and the detection of mercury in herbs was demonstrated.
3.2.3. Detection of small molecules.
DNAzymes have also been used to design sensing systems for small molecules that are not directly recognized by the DNAzymes. For example, Gu et al. described a turn-off DNAzyme biosensor for detection of l-histidine, in which the absence of l-histidine induced a colorimetric signal (Fig. 14).268 The sensing molecule consists of the CuDD DNAzyme and a PMD within a single DNA sequence. The core principle of this design is the sequestration of Cu2+ by l-histidine. In the absence of l-histidine, Cu2+ is free to form a complex with the DNAzyme, inducing self-cleavage, releasing the PMD for signal transduction (Fig. 14). In the presence of l-histidine, Cu2+ is sequestered, which prevents the DNA cleavage reaction that is needed to activate the PMD. This method allowed for the detection of l-histidine at concentrations as low as 50 nM, as well as for its recovery at micromolar levels in spiked human urine samples.
Fig. 14.
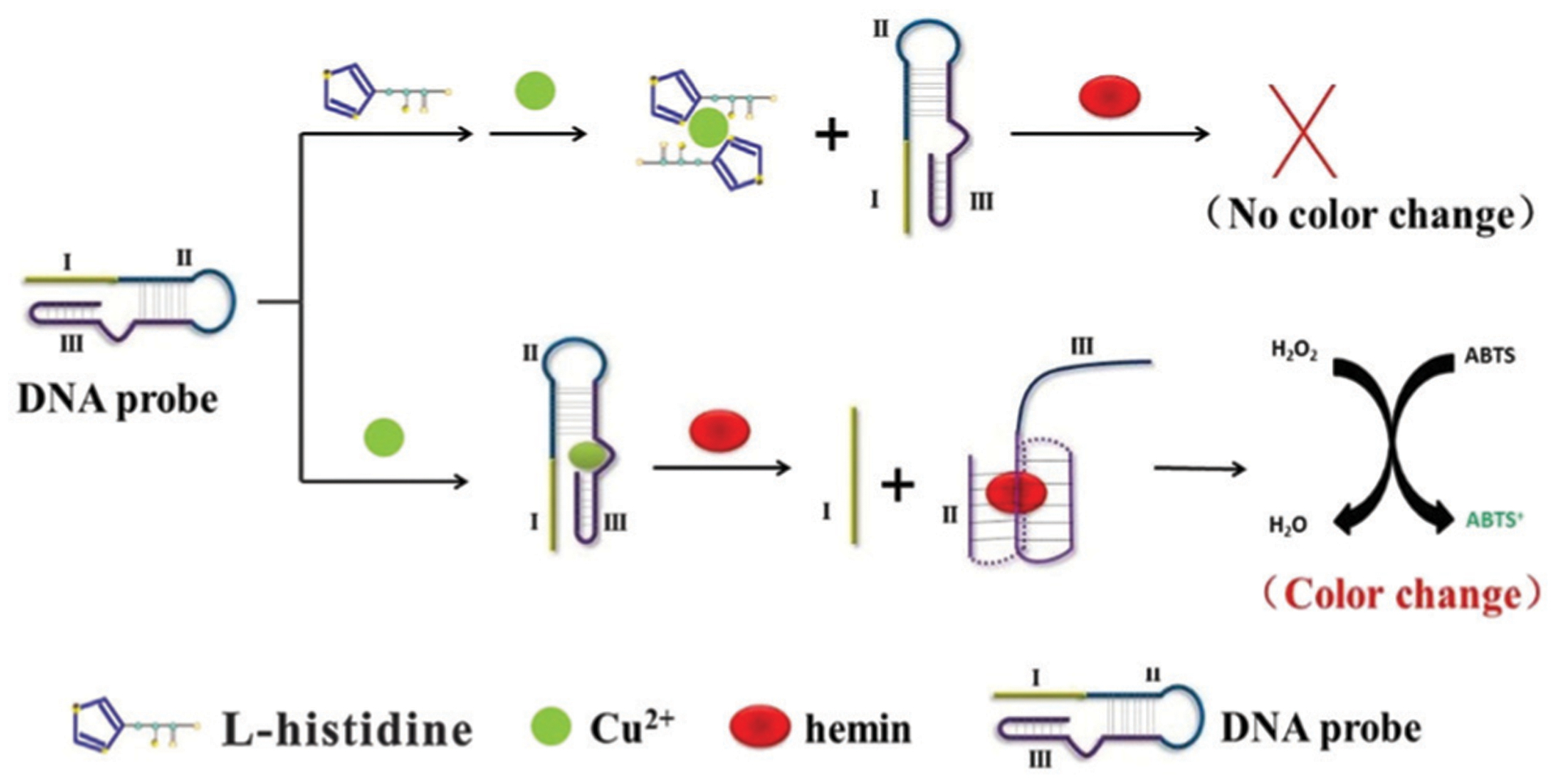
Detection of l-histidine using DNAzymes that do not bind l-histidine directly. Reprinted (adapted) from P. Gu, G. Zhang, Z. Deng, Z. Tang, H. Zhang, F. Y. Khusbu, K. Wu, M. Chen and C. Ma, A novel label-free colorimetric detection of l-histidine using Cu2+-modulated G-quadruplex-based DNAzymes, Spectrochim. Acta, Part A, 2018, 203, 195–200, with permission from Elsevier.268
Similarly, through the use of the CuDD DNAzyme, Shen et al. constructed a fluorometric biosensor for the detection of histidine, but this method also incorporated nucleic acid-based signal amplification.269 When present, histidine bound to and sequestered the available copper ions. As a result, the DNAzyme was unable to cleave its substrate strand. The intact substrate strand then went on to induce HCR. Subsequently, fluorescence was emitted via berberine, in response to the recognition of HCR products by a triplex-forming oligonucleotide. This fluorescent strategy led to a detection limit of 2.0 nM and demonstrated the detection of nanomolar concentrations of histidine in diluted human urine samples.
3.2.4. Protein sensing.
The direct detection of protein targets by DNAzymes that bind the protein has yet to be reported. However, several studies have shown that proteins can be detected with high selectivity using DNAzymes in combination with other MREs. The DNAzymes used for these purposes include RCDs, CuDD, and PMDs. Below we will highlight some recent examples.
The RCD 8–17 DNAzyme has been used to detect antibodies, which can be used as biomarkers to monitor immune response and indicate infection.270 Specifically, Li et al. described a biosensor for antibody detection using the anti-digoxigenin (anti-DIG) antibody as a model antibody and the 8–17 DNAzyme as a reporter (Fig. 15A).271 The two ends of the enzyme strand of 8–17 DNAzyme are modified with digoxigenin (DIG) and the substrate strand contains a fluorophore (FAM) and quencher (BHQ). In the absence of the antibody, the DNAzyme proceeds to cleave its substrate, producing a detectable fluorescent signal. In the presence of the antibody, the target binds the two ends of the DNAzyme, forcing it into a stretched conformation that is unable to bind and cleave the substrate.
Fig. 15.
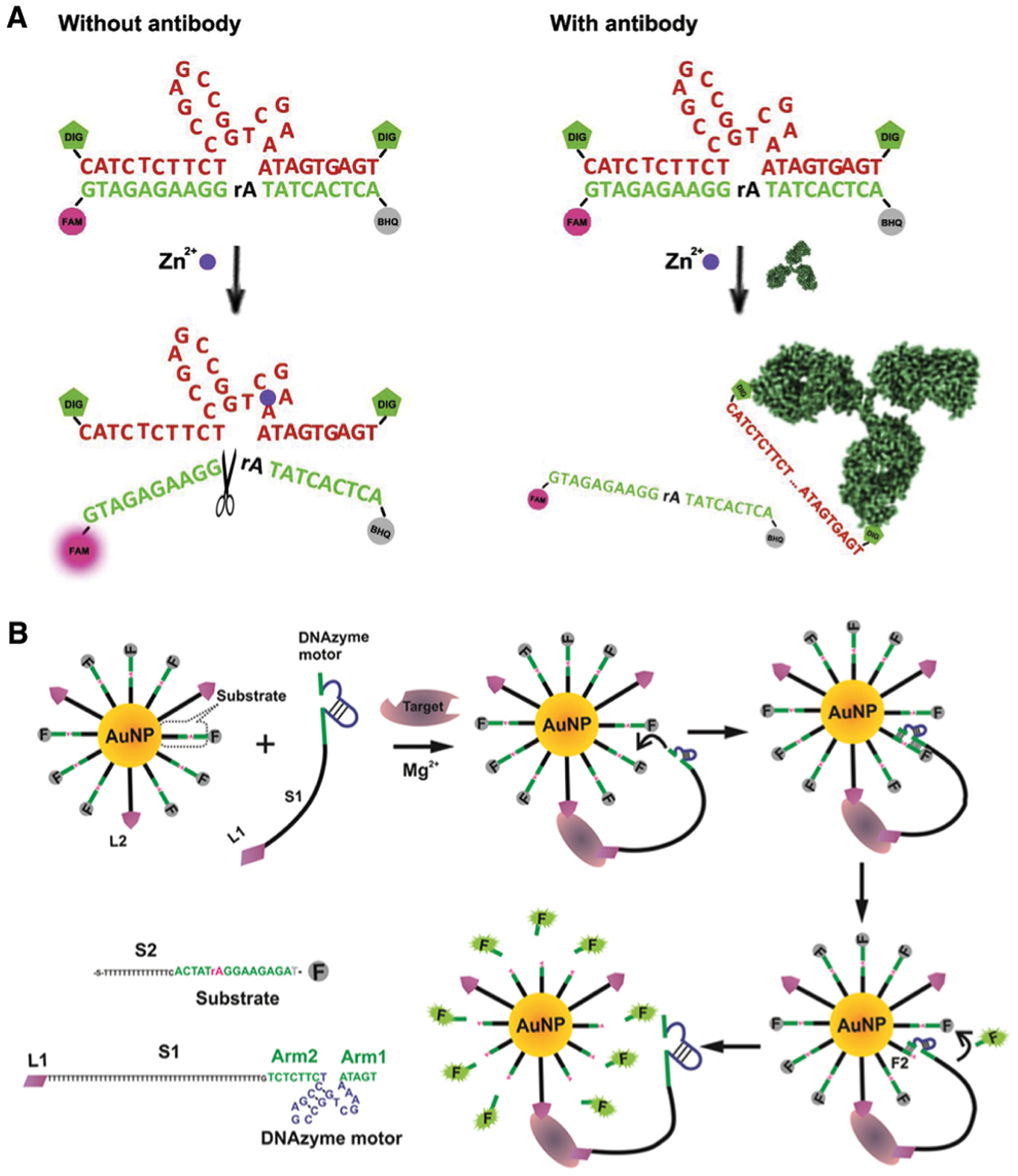
Protein detection using an RCD modified with an affinity probe. (A) Antibody detection. Reprinted (adapted) from C. Li, J. Ma, H. Shi, X. Hu, Y. Xiang, Y. Li and G. Li, Design of a stretchable DNAzyme for sensitive and multiplexed detection of antibodies, Anal. Chim. Acta, 2018, 1041, 102–107, with permission from Elsevier.271 (B) Protein detection using an RCD walker, AuNP and a pair of DNA aptamers that bind the same target. Reprinted (adapted) with permission from J. Chen, A. Zuehlke, B. Deng, H. Peng, X. Hou and H. Zhang, A Target-Triggered DNAzyme Motor Enabling Homogeneous, Amplified Detection of Proteins, Anal. Chem., 2017, 89, 12888–12895.272 Further permissions related to the material excerpted should be directed to the American Chemical Society.
The 8–17 DNAzyme has also been used to construct a self-powered, aptamer-containing, target-triggered DNAzyme motor for protein detection. The system contains two components: (1) the 8–17 DNAzyme that is combined with a protein-binding DNA aptamer and (2) AuNPs decorated with a second aptamer for the same protein target as well as fluorescent substrate strands of the DNAzyme. Binding of the target to the two aptamers induces hybridization between the DNAzyme and its substrate on the AuNP, and this event triggers the repetitive cleavage of the substrate strands on the AuNP, enabling amplified detection of the target (Fig. 15B). Two anti-thrombin DNA aptamers, TBA15 and TBA29, were used to demonstrate the concept.272 However, the same strategy should work with any protein target with a pair of aptamers that bind two different epitopes of the target.
RCDs have also been used to detect the activities of enzymes. For example, the 8–17 DNAzyme has been used to set up a DNAzyme motor to detect the RNase H activity of HIV.273 In this case, both the 8–17 DNAzyme and its fluorescent substrate were placed on a AuNP, but the enzyme strand was pre-locked with the formation of a DNA/RNA duplex to inhibit its binding to the substrate. In the presence of RNase H, the RNA in the locking duplex was hydrolyzed, freeing the DNAzyme for substrate cleavage and signal generation.
The 8–17 DNAzyme has also been used to detect two other proteins as the target: nuclear factor kappa-B and folate receptor.274,275 In both cases the designed DNA probe contains the 8–17 DNAzyme and an element that can be degraded by a nuclease in the absence of the target. When the target is available, it binds the element, which blocks the cleavage of the 8–17 DNAzyme sequence by the exonuclease and allows it to cleave a fluorescent substrate for signal generation.
Another RCD, the MgZ DNAzyme, has been employed to detect human 8-oxoguanine DNA glycosylase (hOGG1),276 an important enzyme that is crucial for DNA repair and prevention of mutations linked to human cancers. The sensor used a DNA molecule that contained 8-oxoguanine, which can be cleaved by hOGG1, producing 5′ phosphorylated DNA that can be circularized by a DNA ligase. The circular DNA is then subjected to RCA, which generates RCA products containing repetitive units of the MgZ DNAzyme for signal generation.
CuDD has also been used to detect enzymes. In one study, the detection of AChE was achieved colorimetrically via the use of CuDD, HCR and AuNP.277 The design is based on the sequestration of Cu2+ ion by thiocholine, the product generated by AChE catalyzed hydrolysis of acetylthiocholine. Therefore, in the presence of AChE, Cu2+ is not available to the DNAzyme, which is unable to cleave its substrate. The intact substrate can then act as an initiator to trigger HCR, producing the DNA product that induces the assembly of AuNPs, accompanied by a sharp color-change from red to blue.
In another study, the CuDD was used to create a DNAzyme spider to monitor telomerase activity in cancer cells.278 Three precursor oligonucleotides were combined with dNTPs and telomerase extracted from HeLa cells. One of the precursor oligonucleotides, D-TSP, contained a primer site, which was used for extension of the sequence by telomerase. The extended D-TSP was then able to combine with the other two oligonucleotides, Y1 and Y2, to form the DNAzyme spider containing CuDD. The spider was able to hybridize to substrate embedded molecular beacons on the surface of a gold electrode. An electrochemical signal was generated when the cleaved fragment that was 5′-labeled with the redox indicator ferrocene accumulated on the surface of the gold electrode. Using this method, telomerase from as few as 10 HeLa cells was detected. Additionally, telomerase activity was detected in two other cell lines (A549 and MCF-7).
PMDs have also been used to detect proteins or enzyme activities. Xu et al. developed an interesting biosensing system termed “DNAzyme-catalyzed tyramide depositing reaction (DCTDR)” for in situ amplified imaging of membrane protein status.279 A PMD was split into two sequences and each was linked to a HER2-binding DNA aptamer. The binding of HER2 with the two DNA probes induced the formation of the whole PMD on the cell surface to catalyze the covalent deposition of fluorescent tyramide on the membrane proteins to facilitate their imaging.279 In another study, a PMD was used to set up an electrochemiluminescence (ECL) assay for the detection of the activity of adenine methyltransferase (Dam MTase). A double-stranded DNA molecule was designed to contain a restriction site for a specific endonuclease and its cleavage released a single-stranded DNA molecule to trigger HCR, leading to the formation of a large DNA assembly containing multiple PMD units, which suppressed the ECL signal. However, the methylation of the restriction site by Dam MTase prevented DNA cleavage and subsequent HCR, leading to the recovery of ECL.
3.2.5. Nucleic acid sensing.
DNAzymes can also be incorporated into detection systems to detect nucleic acid molecules, including specific DNA sequences and microRNAs (miRNAs). For example, Willner’s group have described several assays that utilize amplified cascade systems triggered by the target DNA, in which DNA target triggers isothermal amplification to produce a fluorescence or colorimetric output with detection limits as low as 1 aM of target DNA.280–285 In a related approach the 8–17 DNAzyme has been used to design an amplified biosensing platform termed the “hybridization-triggered DNAzyme cascade (HTDC)” assay for DNA detection.286 The presence of the target DNA induces the 8–17 DNAzyme to hybridize with its fluorogenic substrate strand. Following hybridization, the DNAzyme cleaves its substrate, producing a fluorescence enhancement. The DNAzyme then enters another cycle of the hybridization triggered DNAzyme cascade for signal amplification (Fig. 16A).
Fig. 16.
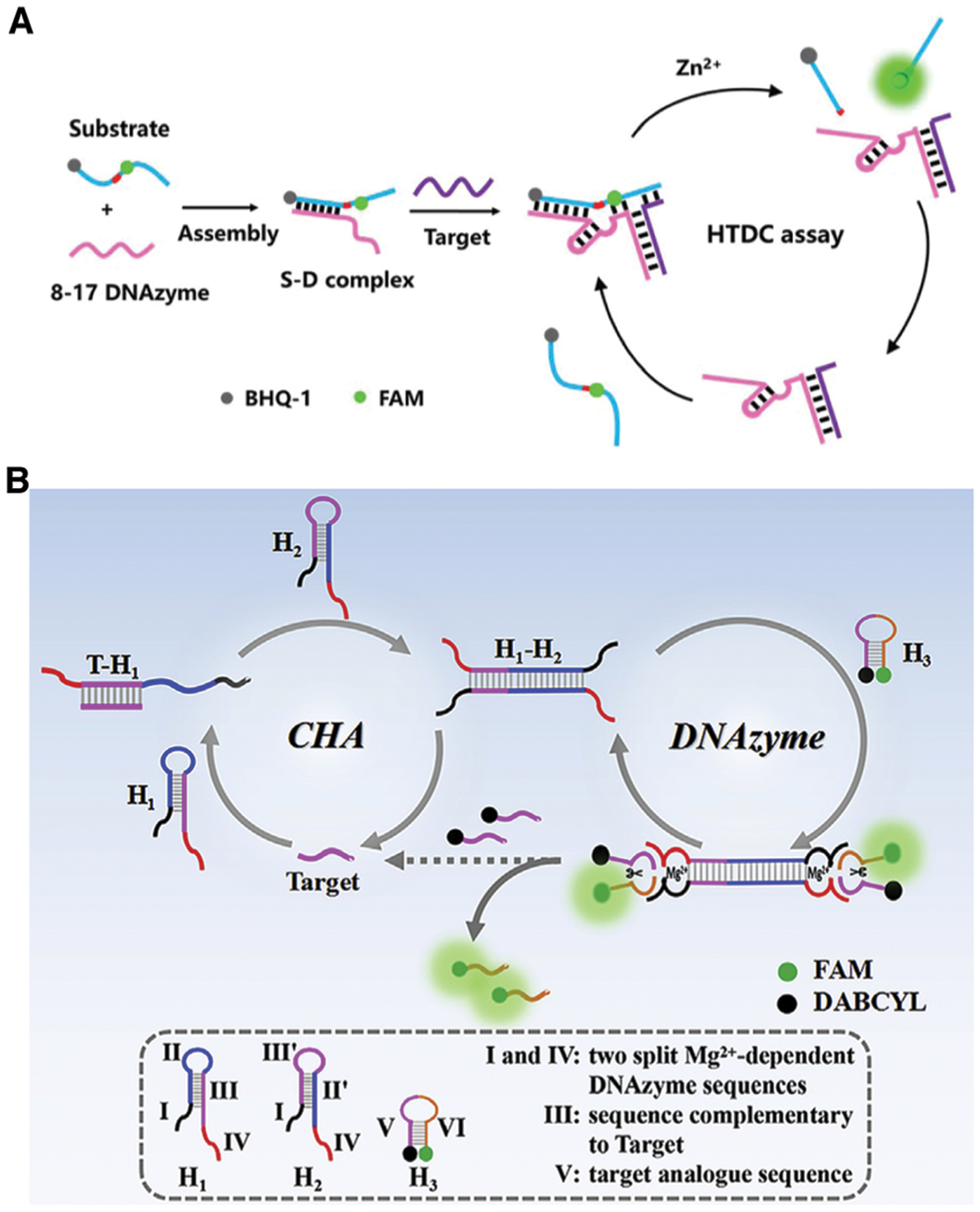
DNA detection using RCDs. (A) Hybridization-triggered DNAzyme cascade (HTDC) assay. Reprinted (adapted) from H. Wang, D. He, R. Wu, H. Cheng, W. Ma, J. Huang, H. Bu, X. He and K. Wang, A hybridization-triggered DNAzyme cascade assay for enzyme-free amplified fluorescence detection of nucleic acids, Analyst, 2019, 144, 143–147, with permission from The Royal Society of Chemistry.286 (B) Catalytic hairpin assembly-mediated double-end DNAzyme feedback amplification. Reprinted (adapted) from X. Liu, X. Zhou, X. Xia and H. Xiang, Catalytic hairpin assembly-based double-end DNAzyme cascade-feedback amplification for sensitive fluorescence detection of HIV-1 DNA, Anal. Chim. Acta, 2020, 1096, 159–165, with permission from Elsevier.287
An RCD has also been incorporated into an interesting CHA reaction for the detection of HIV-1 DNA.287 The system used 3 distinct DNA hairpins (H1, H2, and H3). By virtue of being structurally analogous, H1 and H2 were able to hybridize in the presence of the target analyte, and induced CHA (Fig. 16B). CHA led to the formation of a duplex DNA containing overhanging elements at each end that can create an active RCD, which is capable of cleaving fluorogenic substrate H3. One of the cleavage products can feedback to the CHA reaction loop as the target.
Several examples have been reported that use DNA or RNA-directed assembly of PMDs to produce colorimetric outputs as a method for detection of genetic material, typically producing detection limits in the nanomolar range.288–291 Nucleic acid directed assembly of split RCDs has also been reported for detection of both miRNA, bacterial and viral DNA.292,293 In these assays, a split Mg(ii)-dependent DNAzyme is assembled by the two halves binding to the gene target, causing activation of DNAzyme activity. This results in a cleavage reaction to release a caged PMD, producing a colorimetric output.
A recent example has utilized a combination of isothermal amplification via recombinase–polymerase amplification (RPA) followed by target mediated assembly of an MNA to detect antimicrobial resistance genes via cleavage mediated assembly of DNA-modified gold nanoparticles.294 Detection of the mecA gene in uncultured nasal, groin, axilla, and wound swabs from patients provided 90% clinical sensitivity and 95% clinical specificity and represents one of the first examples of DNAzyme-based assays being validated using patient samples.
A final example involves the visual detection of SARS-CoV-2 RNA using RT-PCR-induced generation of a PMD.295 In this example, a pilot scale validation was performed using patient nasopharyngeal swab samples and produced clinical sensitivity and specificity of 100% over 34 samples. Again, this example shows that DNAzyme-based assays are beginning to move toward clinical testing, which is a key step toward commercialization of such assays for patient testing.
A DNAzyme-based motor constructed on a 20 nm AuNP was reported for miRNA detection in individual cancer cells (Fig. 17A). The AuNPs were decorated with hundreds of substrate strands serving as DNA tracks and dozens of DNAzyme molecules silenced by locking strands. Intracellular binding of a target miRNA initiated the autonomous walking of the motor on the AuNP, enabling amplified detection of the miRNA.222 Recently, Chen et al. reported a Na+-specific DNAzyme-based DNAzyme motor differentiating cell subtypes of non-small cell lung cancer by simultaneously sensing intracellular miRNA-21 and miRNA-205, avoiding the need to add exogenous cofactors.296
Fig. 17.
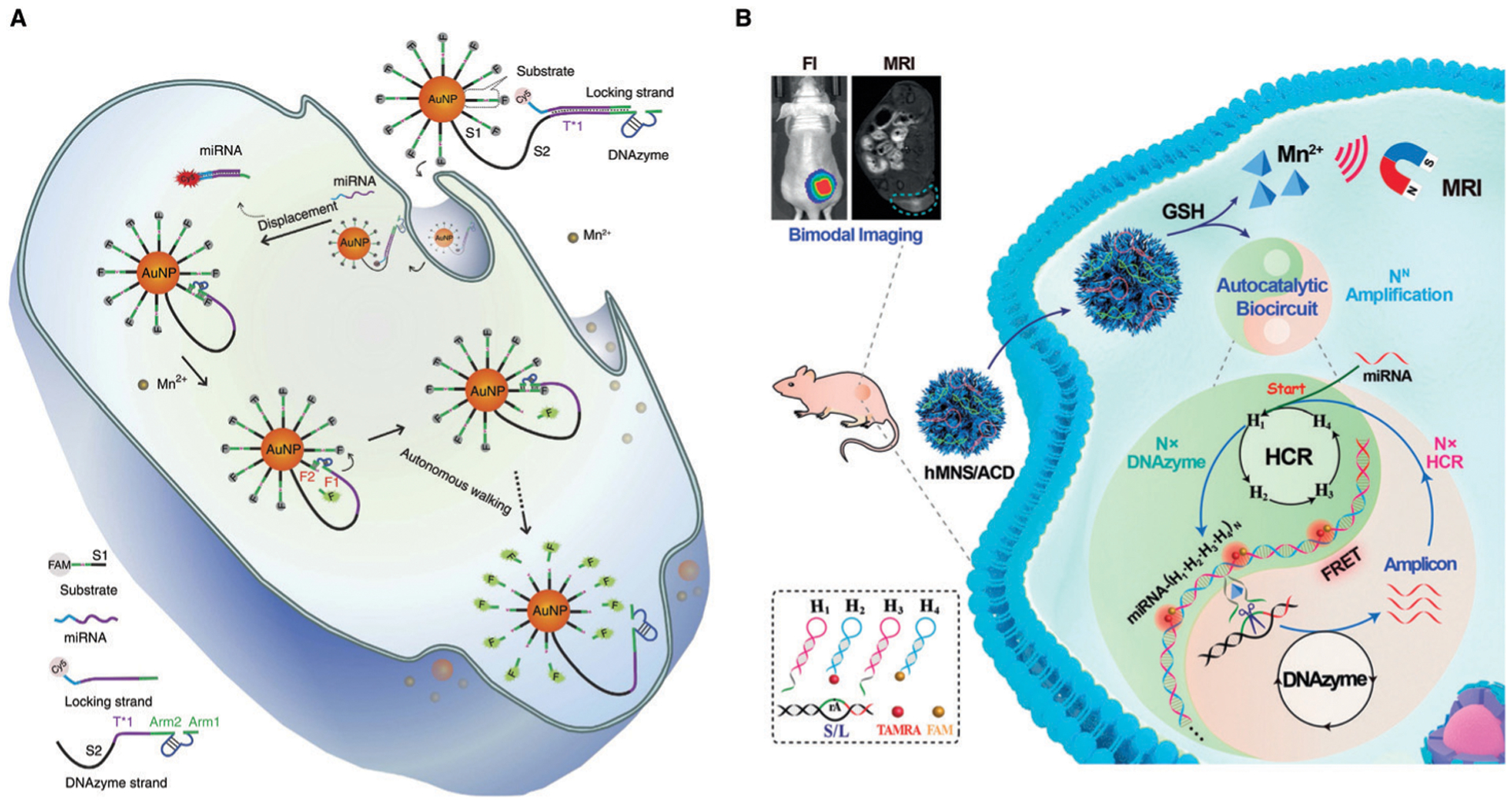
Intracellular microRNA detection involving RCDs. (A) An RCD walker on AuNPs. Reprinted (adapted) with permission from H. Peng, X. F. Li, H. Zhang and X. C. Le, A microRNA-initiated DNAzyme motor operating in living cells, Nat. Commun., 2017, 8, 1–13. Copyright 2017, Peng et al.222 (B) A DNAzyme-containing biocircuit constructed with a honeycomb MnO2 nanosponge (hMNS). Reprinted (adapted) with permission J. Wei, H. Wang, Q. Wu, X. Gong, K. Ma, X. Liu and F. Wang, A Smart, Autocatalytic, DNAzyme Biocircuit for in vivo, Amplified, MicroRNA Imaging, Angew. Chem., Int. Ed., 2020, 59, 5965–5971. Copyright 2020 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.221
A DNAzyme-containing biocircuit has been described for miRNA imaging in mice.221 As shown in Fig. 17B, the system consists of a honeycomb MnO2 nanosponge (hMNS) decorated with 4 hairpin probes and a substrate for an RCD. The hMNS element, responsive to glutathione (GSH), not only delivers DNA probes, but also supplies Mn2+ (the cofactor for the DNAzyme) and MRI agent. Upon entering cells, hMNS is disassembled by endogenous GSH, producing Mn2+ to assist DNAzyme catalysis and release the DNA molecules. Intracellular miRNA-21 triggers HCR, producing DNA nanowires containing tandem DNAzyme units. These DNAzymes then cleave the substrate to generate new triggers to feed back into the HCR cycle. Through cross-feedback of HCR and DNAzyme cleavage, the biocircuit amplifies the limited signal resulting from the initial miRNA recognition, providing sensitive localization of miRNA in vivo.
4. Portable devices incorporating DNAzymes
4.1. Overview of DNAzyme-based sensor devices
The examples provided in the previous sections primarily focused on solution-based assays, and many utilized complicated multi-step procedures and sophisticated instrumentation. While they are quite useful for laboratory investigations of the roles that metal ions, proteins and other targets play in biological systems, they are neither user-friendly nor portable, limiting their utility for on-site and real-time applications. In this section, we focus on the integration of DNAzyme-based assays with highly sensitive and specific portable devices that satisfy the World Health Organization ASSURED criteria, which calls for devices which are affordable, sensitive, selective, user-friendly, rapid and robust, equipment-free and deliverable to end users.297,298
We begin by describing a selection of devices based on colorimetric and SERS detection platforms that use portable readers, which have shown good promise for portable detection using various molecular recognition strategies.129 We then describe recent progress on integrating DNAzyme MREs with commercially available portable devices, including the ANDalyze portable fluorescence reader, the personal glucose meter (PGM) and the lateral flow device. We conclude the section with a description of DNAzyme-sensors based on microscale paper analytical devices (μPADs) that utilize colorimetric, fluorimetric, electrochemical or electrochemiluminescence (ECL) outputs in conjunction with portable readers. Examples utilizing both unamplified and amplified detection schemes are presented, along with μPADs that include integrated sample preparation and/or clean-up components to make such devices more user-friendly.
4.2. Portable devices based on visual reading of capillary flow
Several groups have recently developed equipment-free “distance-based” assays for the detection of metal ions based on visually interpretable movement of a substance through a capillary casing. The schematic representation of one of these strategies is shown in Fig. 18A. Jiang et al. reported a portable visual capillary sensor which allowed for quantitative lead detection based on the activity of a DNA crosslinked hydrogel.177 As illustrated in Fig. 18A, branched polymer chains (polyacrylamide–nucleic acids) were assembled into a hydrogel network in the presence of the GR-5 DNAzyme substrate and enzyme strands. In this way, the DNAzyme controlled the physical transition of the substance from solution to hydrogel and back, regulating form in response to lead. In the absence of Pb2+, the hydrogel remained intact, whereas in the presence of Pb2+ cleavage of the substrate strand led to the disassembly of the hydrogel network, and a physical transition to the solution state. In this device (Fig. 18A, right), the distance the solution traveled through the capillary tube was directly proportional to Pb2+ concentration. In the absence of Pb2+, the gel could not migrate through the capillary tube. This method allowed for the detection of lead in under an hour at levels above 10 nM.
Fig. 18.
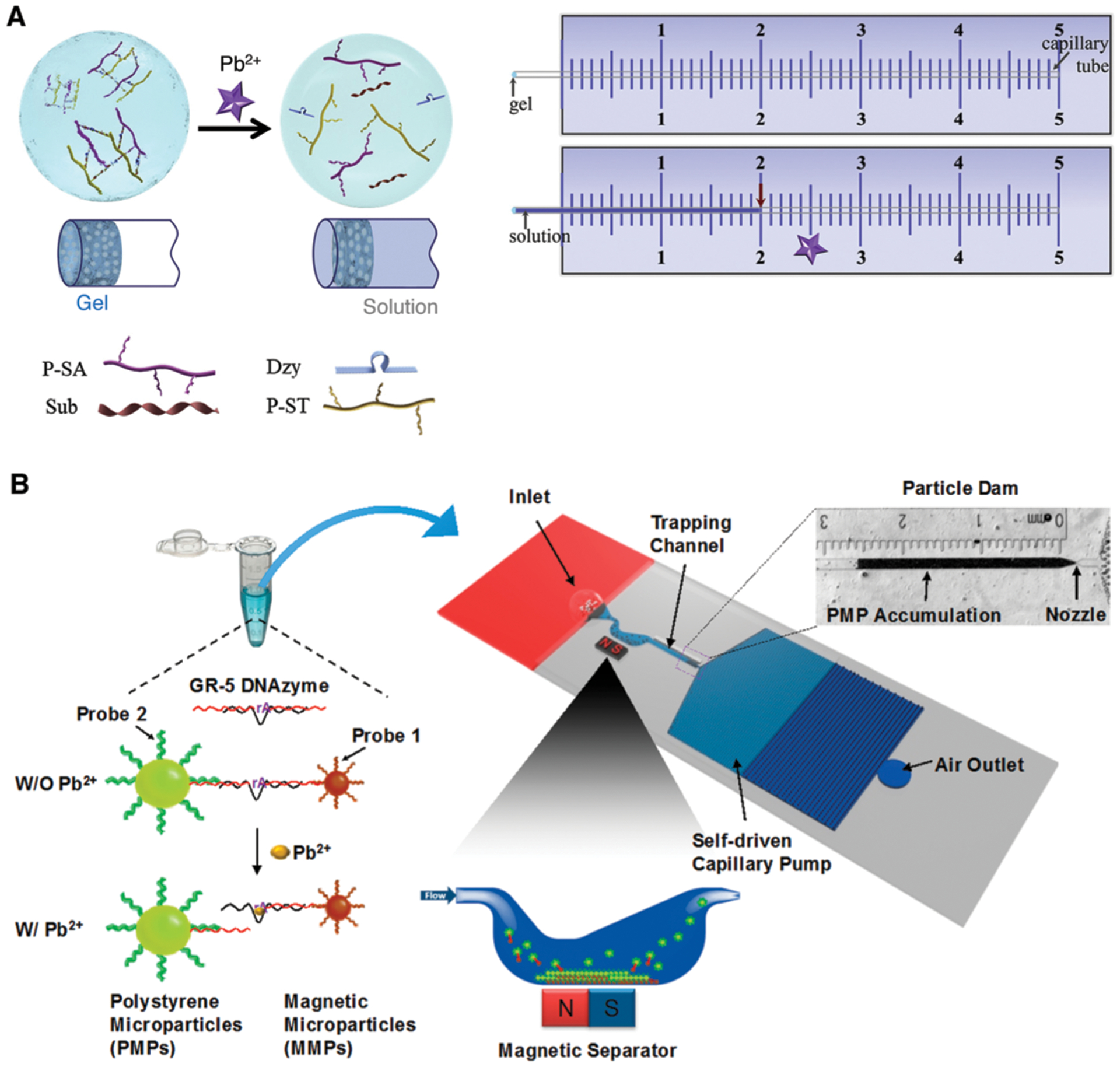
Portable DNAzyme devices for the detection of Pb2+ based on visual reading of capillary flow. (A) A portable hydrogel capillary sensor. Reprinted (adapted) from C. Jiang, Y. Li, H. Wang, D. Chen and Y. Wen, A portable visual capillary sensor based on functional DNA crosslinked hydrogel for point-of-care detection of lead ion, Sens. Actuators, B, 2020, 307, 127625, with permission from Elsevier.177 (B) A portable sensor with a microfluidic particle dam. Reprinted (adapted) with permission from G. Wang, L. T. Chu, H. Hartanto, W. B. Utomo, R. A. Pravasta and T.-H. Chen, Microfluidic Particle Dam for Visual and Quantitative Detection of Lead Ions, ACS Sens., 2020, 5, 19–23. Copyright 2020, American Chemical Society.178
A second distance-based capillary device for the detection of Pb2+ was described by Wang et al. and is shown schematically in Fig. 18B.178 In this design, the GR-5 DNAzyme was utilized to hybridize DNA-modified polystyrene microparticles (PMPs) and magnetic microparticles (MMPs) together. In the absence of Pb2+, the MMPs–GR-5–PMPs complex was isolated via magnetic separation. In the presence of Pb2+, the substrate strand of the GR-5 DNAzyme was cleaved, releasing the PMPs from MMPs–GR-5–PMPs complex. The PMPs were then free to migrate to the particle dam region via capillary flow, where they accumulated within the microchannel at a distance that was proportional to Pb2+ concentration. This device was able to detect as low as 2.12 nM Pb2+, and demonstrated recoveries ranging from 68.1–77.8% of spiked Pb2+ in tap water.
4.3. Portable colorimetric devices
Another common method for DNAzyme-based sensing is to develop simple devices that utilize a portable spectrometer to detect color changes. The examples below show two different approaches to generating a color change, one based on a PMD and the other on aggregation of gold nanoparticles.
The first example demonstrates a RCA-mediated sensing strategy for the detection of Hg2+ using a 3D-printed RCA chip for on-site detection with a hand-held spectrophotometer.265 Briefly, the device was composed of an RCA reaction chamber, a flow trap, and a colorimetric detection chamber. These features were connected to an external Peltier temperature module and an automated reagent supply system (Fig. 19A). When no Hg2+ was present, the polyT primer was able to bind to the circular template, initiating RCA (top right). RCA generated repeating units of a PMD, which in the presence of hemin and H2O2 catalyzed the conversion of ABTS (clear) to ABTS+ (dark green). When Hg2+ was present, the polyT primer preferentially interacted with free Hg2+ to form T–Hg2+–T stabilized duplex DNA, which prevented RCA and the subsequent generation of a colorimetric signal. The device was able to detect as little as 3.6 μg L−1 in spiked tap water.
Fig. 19.
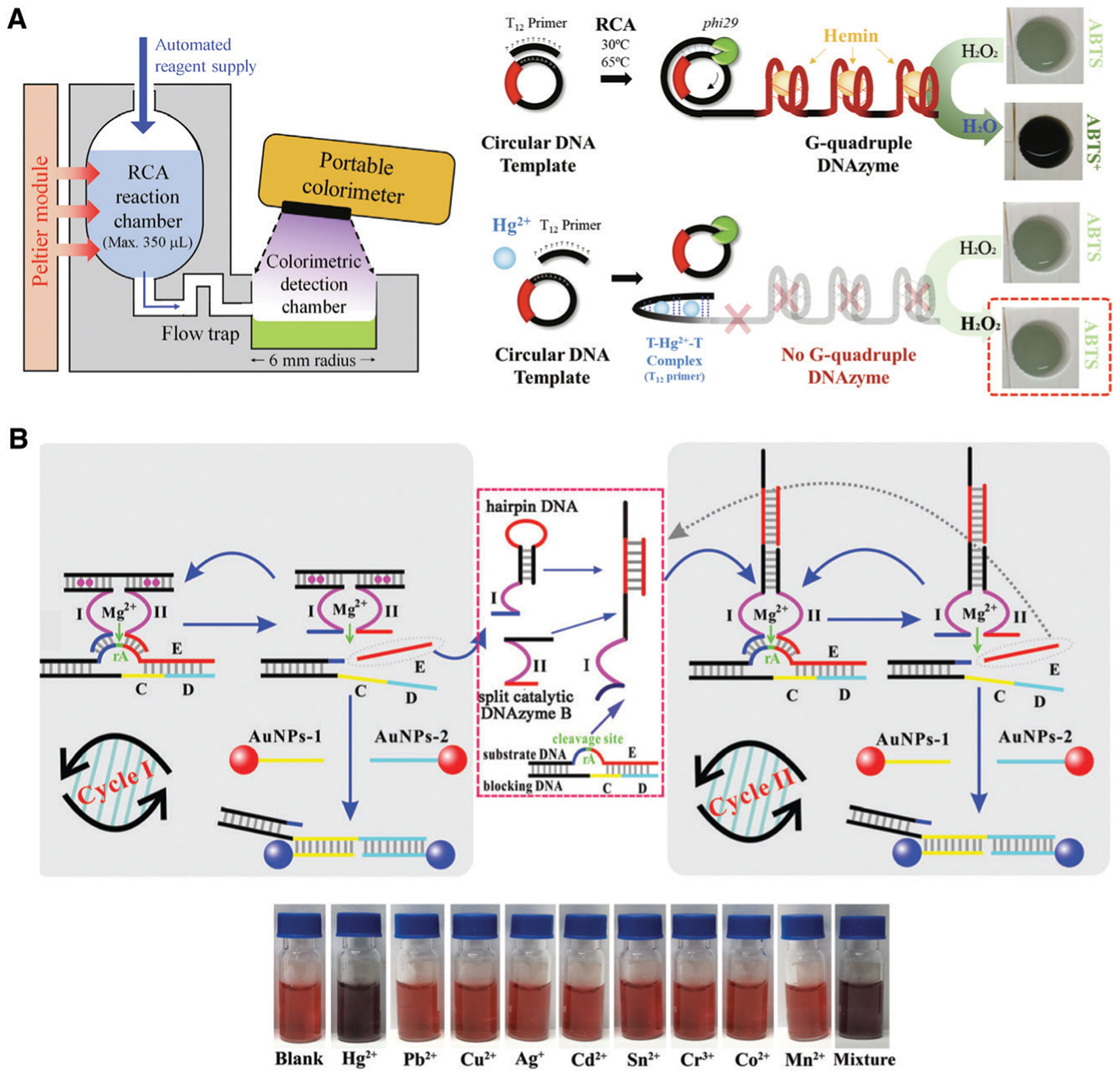
Portable colorimetric devices for the detection of mercury. (A) Schematic representation of the sensing strategy for a 3D-printed rolling circle amplification chip for on-site colorimetric detection of mercury. Reprinted (adapted) from J. W. Lim, T.-Y. Kim, S.-W. Choi and M.-A. Woo, 3D-printed rolling circle amplification chip for on-site colorimetric detection of inorganic mercury in drinking water, Food Chem., 2019, 300, 125177, with permission from Elsevier.265 (B) Naked-eye colorimetric detection of Hg2+ using a AuNP assay. Reprinted (adapted) from J. Chen, J. Pan and S. Chen, A naked-eye colorimetric sensor for Hg2+ monitoring with cascade signal amplification based on target-induced conjunction of split DNAzyme fragments, Chem. Commun., 2017, 53, 10224–10227, with permission from The Royal Society of Chemistry.142
The second example was chosen to represent a body of research that can be broadly applied for portable detection with a hand-held UV-Visible spectrophotometer, which utilizes AuNPs for colorimetric signal generation for the detection of metal ions. As noted above, the Liu group have taken advantage of T–Hg2+–T interaction to convert a uranyl-specific 39E DNAzyme into a highly sensitive colorimetric sensor for Hg2+, with a LOD of 2.4 nM.299 To improve the sensitivity even further, Chen et al. incorporated a split Mg2+-dependent DNAzyme into a catalytic hairpin assembly to generate AuNP agglomeration (Fig. 19B).142 This system consisted of two cycles. During cycle I, and in the presence of Hg2+, the split Mg2+ DNAzyme was assembled as a five-oligonucleotide construct. Upon substrate cleavage, one half of the substrate acted as the catalyst to initiate cycle II, the split enzyme construct was regenerated, and following the addition of two AuNP labeled oligonucleotides, the formation of a double-stranded construct brought the AuNP–DNA probes into close enough proximity so as to change their surface plasmon resonance. Cycle II began when the catalyst strand from cycle I bound to a hairpin DNA, opening it up and exposing the sequence from one half of the split DNAzyme. In a similar way to the first cycle, Pb2+-dependent cleavage led to the release of the catalyst strand, and the formation of a double-stranded construct from two AuNP–DNA probes. The sensing system allowed for the detection of as little as 10 pM of Hg2+, with high specificity over other common metal ions and demonstrated recoveries of spiked Hg2+ in river water that ranged from 88–106%.
4.4. Portable devices using surface enhanced Raman spectroscopy
Surface-enhanced Raman spectroscopy (SERS) has demonstrated potential for the development of portable devices as it can be easily miniaturized, is fast and very sensitive. Wang and coworkers recently reported two portable microfluidic SERS-based biosensors of the detection of uranyl.199,300 Sensing of uranyl using the first portable SERS-based sensor utilized the uranyl DNAzyme labelled with the SERS reporter molecule 5′-rhodamine B (RhB).199 Following DNAzyme cleavage, the 5′-RhB labeled fragment traveled to three separate SERS bio-chips (3D-ZnO–Ag mesoporous nanosheet arrays) which had been modified with complementary capture DNA. A strong signal was generated by the increased proximity of the RhB reporter molecule to the substrate surface of the SERS biochip. This device produced an impressive detection limit of 3.17 fM and was used to detect spiked uranyl in river and tap water with recoveries that ranged from 95.2 to 106.3%.
In 2020 the same group reported a similar biosensing system, however the microfluidic device was changed as shown in Fig. 20.300 The biggest difference between the 2019 and 2020 devices was that for the newer device, reaction and detection occurred in the same zone. Though both devices were recyclable, the newer device could be regenerated using only PBS buffer containing supplemental oligonucleotides, as compared to the older device which required washing with dilute nitric acid, and regeneration of the entire SERS substrate. The newer device showed a detection limit of 0.72 pM UO22+, and demonstrated recoveries of 97.6–108.2% of spiked uranyl from tap and river water.
Fig. 20.
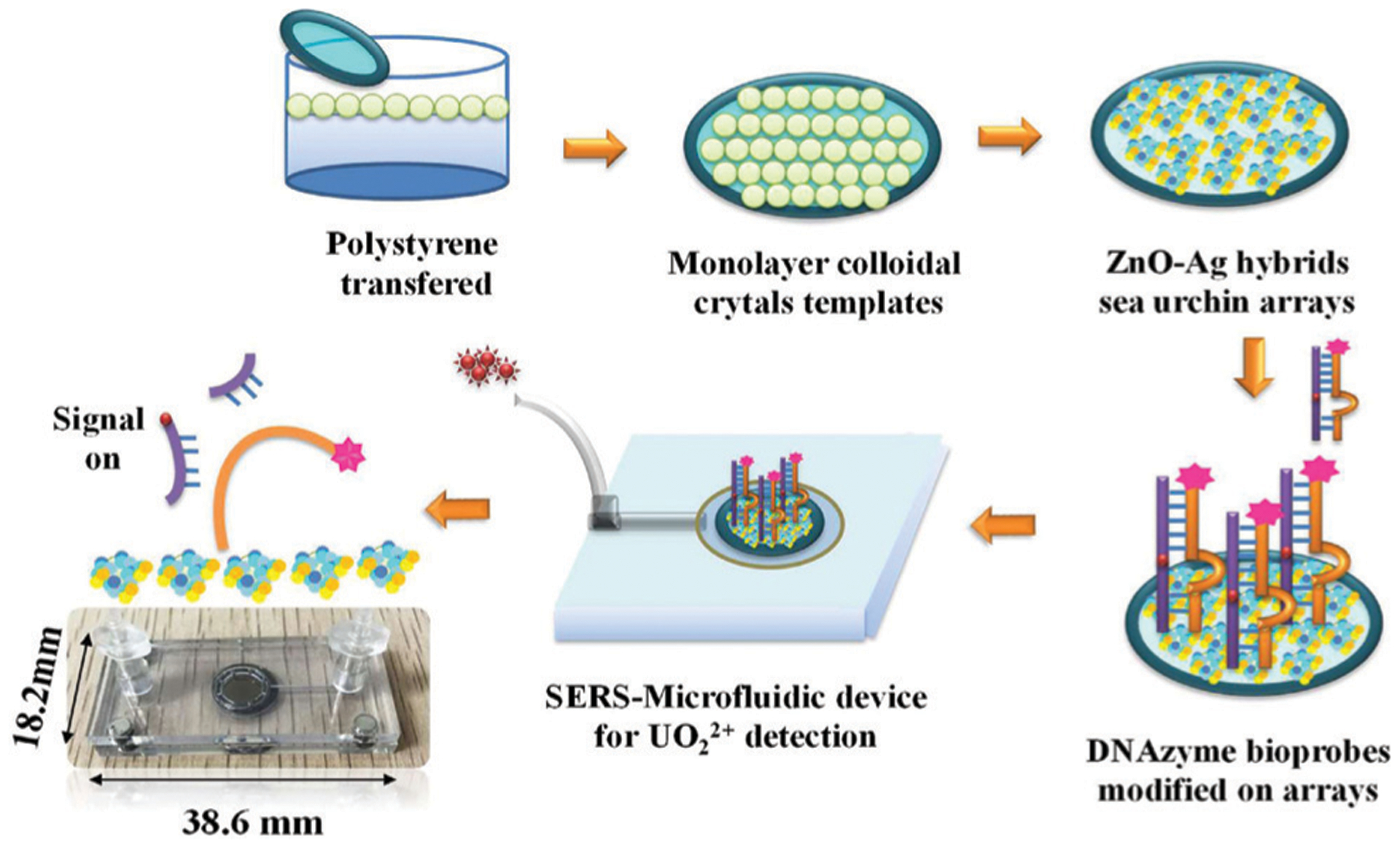
Schematic representation of the detection of uranyl ion using a microfluidic SERS device. Reprinted (adapted) from X. He, X. Zhou, Y. Liu and X. Wang, Ultrasensitive, recyclable and portable microfluidic surface-enhanced Raman scattering (SERS) biosensor for uranyl ions detection, Sens. Actuators, B, 2020, 311, 127676, with permission from Elsevier.300
4.5. Point-of-need devices based on commercial biosensor platforms
The examples outlined above represent prototypes of portable devices that have mainly been demonstrated in a laboratory setting and have not yet been produced in mass quantities using scalable manufacturing processes. As such, these devices are not yet routinely available for consumer use and will likely require additional development to allow mass manufacturing. To accelerate the translation of DNAzyme-based devices to the market, one approach that has been examined is the adaptation of widely available sensor platforms, mainly based on commercially available readers, such as the personal glucose meter (PGM), or simple devices similar to the home pregnancy test (lateral flow device), which are already manufactured at large scale and could be easily adapted for different analytes without the need to redevelop manufacturing processes. Four examples of such device platforms are provided below.
4.5.1. Smart thermometer.
The Lu group designed a uranyl-specific sensor based on the 39E DNAzyme as a MRE and a commercially available target-responsive smart thermometer as a detection platform (Fig. 21A).301 The sensing principle is shown schematically in panel a. In this design, oligonucleotide conjugated magnetic beads and phospholipase A2 (PLA2) form a construct based on the assembly of the uranyl DNAzyme. In the presence of the target, the PLA2–DNA conjugate was released from the enzyme/functional DNA immobilized magnetic beads. Following removal of the magnetic beads, the remaining PLA2–DNA conjugate catalyzed the release of a NIR dye from liposomes. Detection by the smart thermometer was dependent on the excitation of the NIR dye, and the subsequent increase in solution temperature due to released heat from the dye. Panels b–d show the device setup, the smart thermometer, and the sensor region of the smart thermometer probe tip, respectively. The limit of detection of this device was reported as 25.7 nM.
Fig. 21.
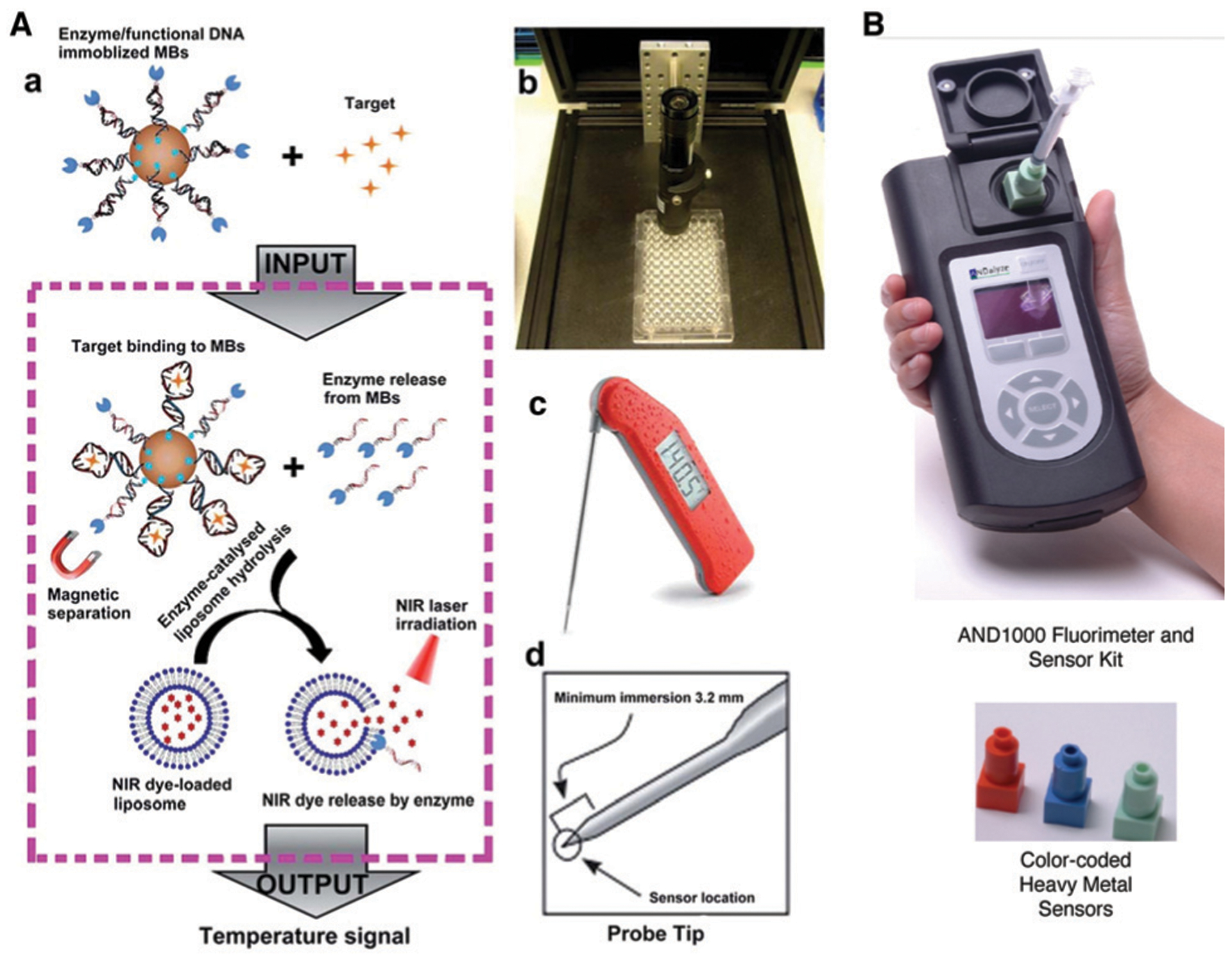
(A) Detection of target analyte using a smart thermometer. Reprinted (adapted) with permission from J. Zhang, H. Xing and Y. Lu, Translating molecular detections into a simple temperature test using a target-responsive smart thermometer, Chem. Sci., 2018, 9, 3906–3910. Copyright 2018, The Royal Society of Chemistry.301 (B) ANDalyze Portable Fluorescence Reader and disposable sensors. Copyright permission granted by ANDalyze.
4.5.2. ANDalyze portable fluorescence reader.
As noted in Section 3, many DNAzyme sensors use fluorescence outputs, specifically catalytic beacons and fluorogenic RNA-cleaving DNAzymes. Work in the early 2000s showed that simple fluorescence detection strategies could be combined with portable hand-held devices.40,75,141 This detection strategy has been commercialized by ANDalyze Inc. via the incorporation of various fluorogenic DNAzymes, particularly those for metal ions, into consumable cartridges (Fig. 21B). With the use of a simple hand-held fluorometer, this technology has been demonstrated for specific detection of lead, uranium, copper, mercury, zinc, and cadmium. This test can provide quantitative results in under a minute using a milliliter of water and in most cases can detect the metal ions at or below the maximum contamination levels in drinking water, defined by the US EPA.
4.5.3. Personal glucose meter for metal ion sensing.
The personal glucose meter is among the most widely used POC device, and possesses advantages including low cost, portability, ease-of-use, and rapid results.302 However, despite its success and widespread use, the PGM can only detect glucose and commercially available electrodes are not available to detect other analytes. Instead of developing a new portable device or associated electrodes with different selectivity, which would be costly to develop and unfamiliar to users, the Lu group proposed to adapt the standard PGM system and expand it to detect many non-glucose targets. In this way, the many years of research and development and scaled-up manufacturing methods could be easily adapted to help accelerate lab-based assays to commercial point-of-need (PON) devices. To expand the applications of PGMs to non-glucose targets, the key requirement is to develop a general way to correlate the presence and concentrations of non-glucose targets with a detectable glucose signal, or to develop a new PGM-based signaling strategy by using other enzymatic reactions that can be detected with standard PGMs. With the development of this field in recent years, many targets including metal ions, small organic molecules, nucleic acids, proteins, and cancer cells have been detected by commercially available PGMs.44,302,303 Here, we focus on PGMs using DNAzymes as MREs.
The first report that utilized a DNAzyme as a MRE and a PGM for detection was reported by the Lu group in 2011.44 In this work, the UO22+-dependent DNAzyme (39E) and the substrate (39S) were used. As shown in Fig. 22A, the biotin–DNA was immobilized on magnetic beads, and DNA–invertase conjugates were connected by 39S via 12-base-pair hybridization. 39S was further hybridized with 39E (modified with invertase at the 5′ end). In the absence of UO22+, 39S could not be cleaved by 39E, the DNA–invertase conjugate would not be released from magnetic beads and no glucose could be detected by the PGM. In the presence of UO22+, the cleavage of 39S allowed release of the DNA–invertase conjugate, which converted sucrose into glucose that was detectable using a PGM. A detection limit of 9.1 nM and a detection range of 0–200 nM UO22+ was achieved. This method is based on the target-induced release of invertase from a functional–DNA–invertase conjugate. Aptamers have also been used to quantify other non-glucose targets, including cocaine, adenosine, and interferon-gamma of tuberculosis. Since then, this concept has been further extended for the detection of other metal ions, including Pb2+,304–306 UO22+,307 Hg2+,308 Cu2+,309 Cd2+,310 and Na+.311
Fig. 22.
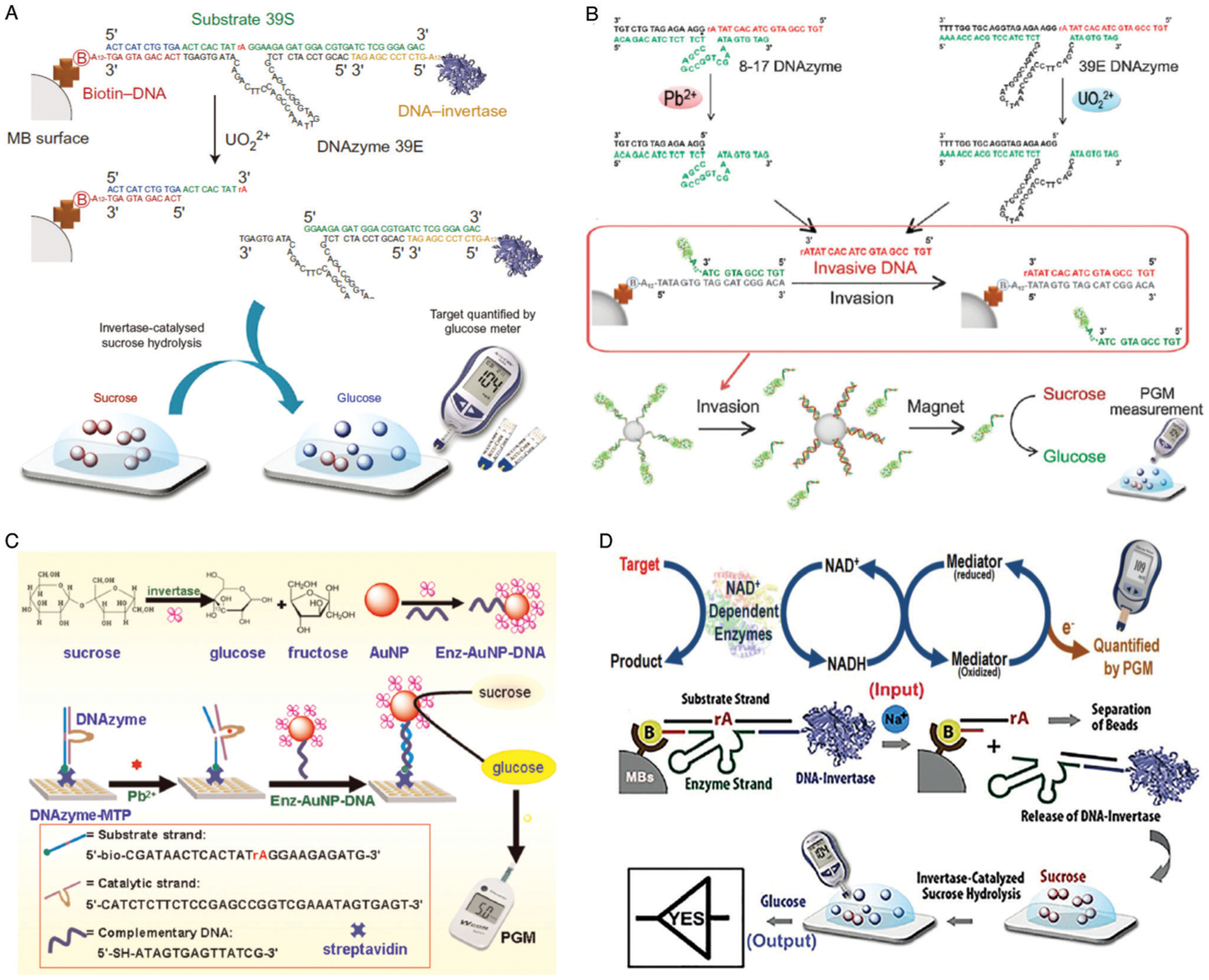
DNAzyme based devices using a PGM for the detection of metal ions. (A) Schematic of method using a PGM to detect UO22+. Figure reproduced with permission from the corresponding author of Xiang, Y. & Lu, Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 3, 697–703 (2011), as per the Nature policy for self-archiving and licence to publish.44 (B) Schematic representation of the detection of Pb2+ and UO22+ using a PGM. Reprinted (adapted) with permission from Y. Xiang, and Y. Lu, An invasive DNA approach toward a general method for portable quantification of metal ions using a personal glucose meter, Chem. Commun. 2013, 49, 585–587. Copyright 2013, The Royal Society of Chemistry.307 (C) Schematic illustration of low-cost and highly efficient DNA biosensor for Pb2+ detection using 8–17 DNAzyme-modified microplate and PGM. Reprinted (adapted) from J. Zhang, Y. Tang, L. M. Teng, M. H. Lu, and D. P. Tang. Low-cost and highly efficient DNA biosensor for heavy metal ion using specific DNAzyme-modified microplate and portable glucometer-based detection mode. Biosens. Bioelectron. 2015, 68, 232–238, with permission from Elsevier.304 (D) Nicotinamide adenine dinucleotide (NADH)/PGM system for target detection using NADH-dependent enzymes and a “YES” logic gate for Na+ detection based on the Na+–DNAzyme-invertase conjugate on magnetic beads through biotinylated using glucose as the signal output. Figures reprinted (adapted) with permission from J. Zhang, Y. Xiang, M. Wang, A. Basu, and Y. Lu, Dose-Dependent Response of Personal Glucose Meters to Nicotinamide Coenzymes: Applications to Point-of-Care Diagnostics of Many Non-Glucose Targets in a Single Step, Angew. Chem., Int. Ed. 2016, 55, 732–736. Copyright 2016 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.312 and from J. J. Zhang, and Y. Lu, Biocomputing for Portable, Resettable, and Quantitative Point-of-Care Diagnostics: Making the Glucose Meter a Logic-Gate Responsive Device for Measuring Many Clinically Relevant Targets, Angew. Chem., Int. Ed. 2018, 57, 9702–9706. Copyright 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.311
Lu’s group found that the performance of the method reported in 2011 decreased when it was applied to detect other heavy metal ions (i.e., Pb2+), probably because enzymes used in the sensing mixture could compete with DNAzymes in binding metal ions through accessible amino acid residues of those proteins. To make the methodology more generally applicable to different metal ions, a new approach was reported to separate the metal-ion-induced cleavage of DNAzymes’ substrates from the target-induced release of invertase-DNA conjugates (Fig. 22B).307 To detect Pb2+ and UO22+, the 8–17 DNAzyme and UO22+-dependent DNAzyme were used, respectively. Upon addition of the respective metal ions, the DNAzymes catalyzed the cleavage of the DNA substrates at the 3′ phosphoester bond of the ribonucleotide. The cleaved substrates were released and the released oligonucleotides in turn served as an invasive DNA to compete with the DNA–invertase conjugates in hybridizing with the biotinylated DNA on the magnetic beads, inducing the release of the DNA–invertase conjugates. The released DNA–invertase conjugates were collected by magnetic separation and used to transform sucrose into glucose, which was measured by a PGM to determine the Pb2+ and UO22+ concentrations in the samples. Using this method, the detection limit of 16 nM and 5.0 nM for Pb2+ and UO22+ were achieved, respectively.
To develop a simple and low-cost DNA sensing platform for Pb2+ detection, as shown in Fig. 22C, Tang’s group immobilized the 8–17 DNAzyme onto a streptavidin-functionalized micro-titer plate based on avidin–biotin chemistry.304 Invertase and single-stranded DNA-labeled AuNPs were utilized as the signal-production tags. Upon Pb2+ introduction into the microplate, the substrate strands were cleaved, resulting in the detachment of the catalytic strand from the microplate. The newly generated single-stranded DNA on the microplate hybridizes with the oligonucleotides on the gold nanoparticles. The AuNP-bound invertase hydrolyzes sucrose to glucose, which can be quantitatively monitored using a PGM. Based on this method, the LOD of Pb2+ was as low as 1.0 pM.
One potential issue of using PGM for detection is the presence of high concentrations of endogenous glucose in biological samples, such as blood. To address this issue, the Lu group developed a method to remove the endogenous glucose using hexokinase before the target-induced conversion of sucrose into glucose.312 More importantly, they took advantage of the fact that some PGMs have dose-dependent responses to nicotinamide co-enzymes (Fig. 22D), which can be readily expanded for the detection of a broad range of targets using various NADH-dependent enzymes.312 As a result, a PGM-based biocomputing platform was further developed based on a combination of glucose and NADH-based signal transduction strategies.311 As shown in Fig. 22D, a YES gate was developed for sodium ions using a Na+-specific DNAzyme (43E) and its substrate (43S). A DNA sandwich structure was assembled on magnetic beads by connecting the DNA–invertase conjugate to a biotinylated capture DNA (biotin–DNA) through simultaneous hybridization with 43S. Upon addition of Na+, the DNAzyme could catalyze the cleavage of 43S and subsequently release the DNA–invertase conjugates. The released DNA–invertase conjugates then catalyzed the hydrolysis of sucrose to glucose, producing a detectable PGM signal. Using this YES gate, the target Na+ could be quantified in the range from 8.0 mM to 120 mM in human serum, with a detection limit of 1.9 mM.
4.5.4. PGMs for indirect sensing of non-metal analytes.
The examples outlined above utilized metal-dependent DNAzymes for direct detection of metals. However, these DNAzymes can also be utilized as part of PGM sensor systems for indirect detection of non-metal analytes. For example, assays utilizing PGMs have been developed for the detection of multiple analyte classes based on modulation of DNAzyme function by aptamers for small molecule detection,44 antibodies for small molecule, protein and bacteria detection,313,314 or DNA hybridization for cDNA detection.53,302,315 The Lu group’s demonstration that many PGMs can give a dose-dependent responses to nicotinamide coenzymes, such as NADH (see above) allowed the group to develop one-step homogeneous assays of many non-glucose targets that are difficult to recognize by DNAzymes, aptamers, or antibodies, and without the need for conjugation and multiple steps of sample dilution, separation, or fluid manipulation.312 The methods are based on the target-induced consumption or production of NADH through cascade enzymatic reactions. Using these methods, they were able to monitor glucose and l-lactate levels in human plasma. Since a large number of commercially available enzymatic assay kits utilize NADH in their reactions, this platform allows the transformation of almost all of these clinical lab tests into POC tests that use a PGM.
The Lu group continued to innovate with the report of a multiparameter sensor which integrated logic gates and multiple DNAzyme-conjugated protein enzyme complexes for the detection of analytes in complex biological samples.311 For the specific application of food safety, Yang et al. recently reported the use of a PGM-sensor for the detection of aflatoxin B1 in bread samples (Fig. 23A).316 Briefly, an aptamer for AFB1 was immobilized on the surface of a gold electrode via gold–thiol chemistry. The substrate strand of the 8–17 DNAzyme, which was terminally modified with invertase, was hybridized to the aptamer. In the presence of AFB1 the aptamer preferentially bound to the target, allowing the walker DNA (the enzyme strand of the 8–17 DNAzyme) to hybridize and cleave the substrate strand, ultimately releasing the invertase labeled cleavage fragment. Finally, invertase catalyzed the conversion of sucrose to glucose for detection by the PGM. The device demonstrated a limit of detection of 10 pM and excellent specificity over other agricultural toxins. Furthermore, the device was used to detect AFB1 in extract from moldy bread at the nanomolar concentration level.
Fig. 23.
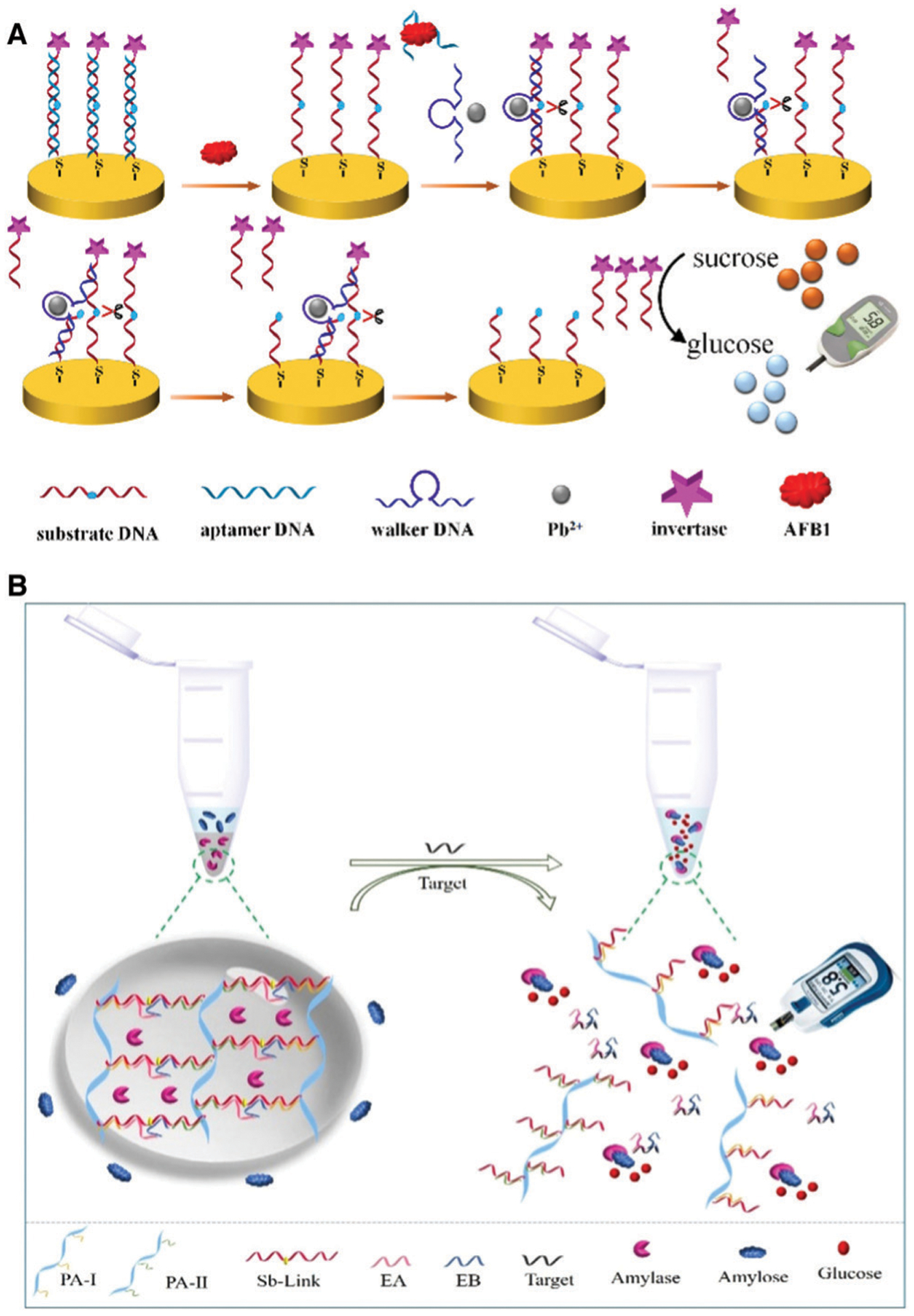
DNAzyme based devices for the detection of non-metal targets. (A) A PGM-sensor for the detection of aflatoxin B1. Reprinted (adapted) from X. Yang, D. Shi, S. Zhu, B. Wang, X. Zhang and G. Wang, Portable Aptasensor of Aflatoxin B1 in Bread Based on a Personal Glucose Meter and DNA Walking Machine, ACS Sens., 2018, 3, 1368–1375. Copyright 2018, American Chemical Society.316 (B) Detection of microRNAs by PGM and oligonucleotide cross-linked hydrogel. Reprinted (adapted) from Y. Si, L. Li, N. Wang, J. Zheng, R. Yang and J. Li, Oligonucleotide Cross-Linked Hydrogel for Recognition and Quantitation of MicroRNAs Based on a Portable Glucometer Readout, ACS Appl. Mater. Interfaces, 2019, 11, 7792–7799. Copyright 2019, American Chemical Society.317
Another example was reported by Si et al., who described the development of an oligonucleotide cross-linked hydrogel for the detection of multiple microRNAs by a PGM (Fig. 23B).317 Platforms which detect small RNAs are especially desirable given their diverse role in human health and disease. In this device, the hydrogel was first prepared by encapsulating amylose in a network of DNA-branched polyacrylamides linked by DNAzyme fragments. In the presence of the target, the hydrogel disassembled by the progressive cleavage of the substrate linkage sequences in the presence of Pb2+ by the target RNA-stabilized DNAzyme. Subsequently, amylase was released into the solution where it was converted by amylose to glucose. Indirect quantitative detection of several miRNAs was achieved by the measurement of glucose by the PGM. For example, the device could detect as low as 0.325 fmol of miR-21. As a demonstration of the practical application of this device, the detection of miR-21, miR-122, and miR-155 from HeLa, HepG2, MCF-7, and LO2 cell extracts was shown.
4.5.5. Lateral flow devices.
Lateral flow assays have been widely used for rapid and inexpensive detection of a wide range of analytes. Originally these assays were based on antibody-based binding assays, with devices for the detection of human chorionic gonadotropin (HCG, the pregnancy hormone) becoming a major commercial success.318 In more recent years, LFD devices have been reported based on nucleic acids as recognition elements, including devices based on hybridization of cDNA with or without amplification steps,319 devices using DNA aptamers as MREs,15,320 and as described below, devices using DNAzymes as MREs. In all cases, the LFD is comprised of four main components: (1) a sample pad for introduction of an analyte solution, which can also aid in separation to remove interferants and adjust buffer conditions; (2) a conjugate pad which contains the MRE labelled with a suitable detection moiety (gold or latex particle or fluorescent label); (3) a nitrocellulose membrane on which test and control lines are printed to capture the analyte-MRE conjugate and a control DNA sequence, respectively; and (4) an absorption pad that provides a sink for liquid flow to allow unidirectional flow of liquids without backflow. Such devices provide an intense control line to validate proper function of the LFD, and a test line that will vary in intensity with analyte concentration, providing a semi-quantitative output.
The first report that utilized a DNAzyme as a MRE for a lateral flow assay was reported in 2010 by the Lu group.43 In this work, the 17E DNAzyme and 17S substrate were used for detection of Pb2+. As shown in Fig. 24A, the 17S was modified to carry a biotin at one end and a thiol at the other, which allowed binding to a gold nanoparticle. In the absence of Pb2+, the 17S was captured by a streptavidin control line and was unable to flow to the upstream test line. In the presence of Pb2+, the 17S cleaved to remove the biotinylated region, and thus flowed past the streptavidin region to a test line containing cDNA to capture the 17S–AuNP conjugate. When the cleavage reaction was done directly on the LFD a detection limit of 5 μM Pb2+ was achieved in 10 min. Running the cleavage reaction in solution for 15 min prior to introduction to the LFD reduced the LOD to 0.5 μM.
Fig. 24.
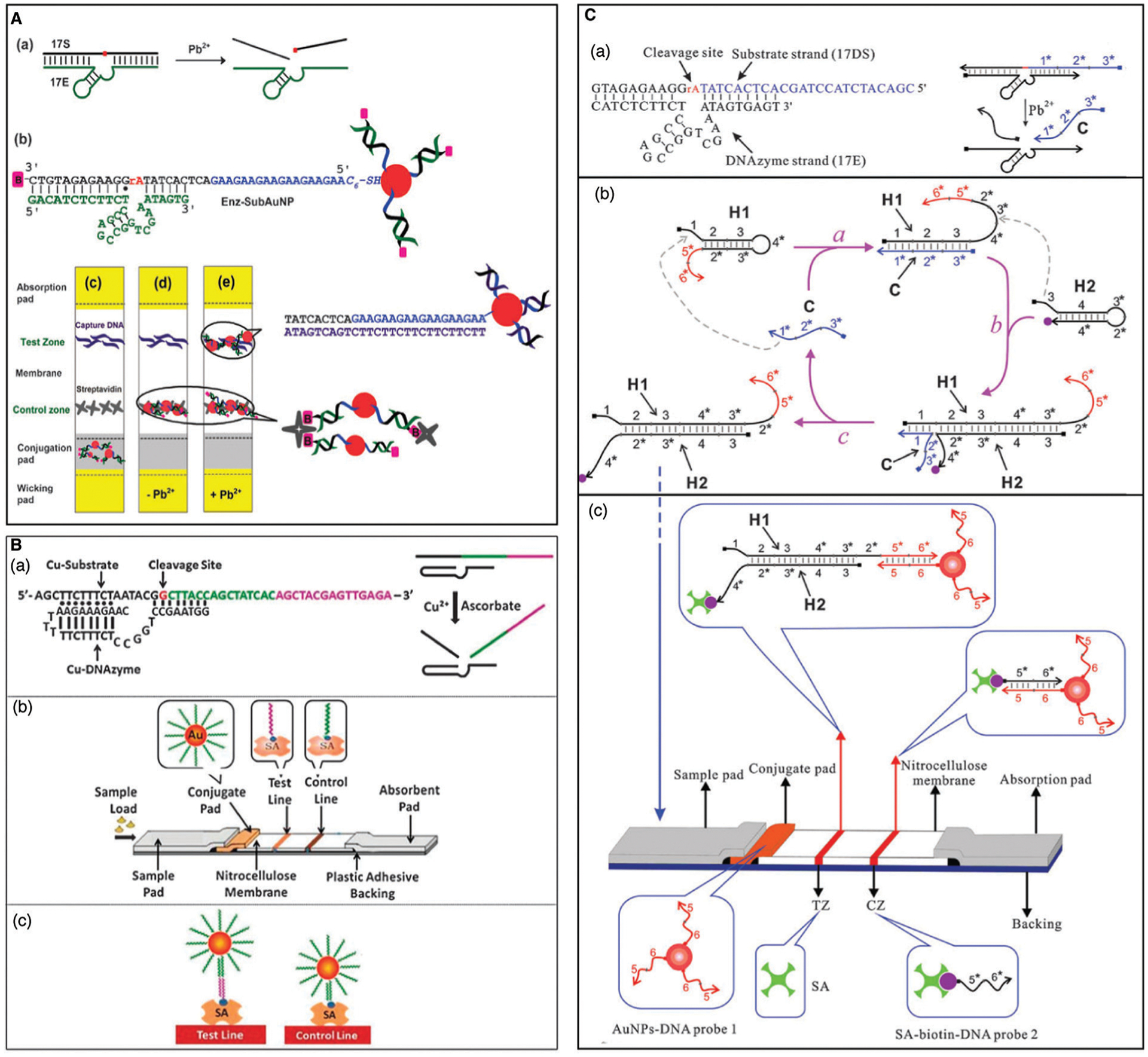
DNAzyme based lateral flow devices (LFDs). (A) An LFD for the detection of Pb2+ using 8–17 DNAzyme. Reprinted (adapted) from D. Mazumdar, J. Liu, G. Lu, J. Zhou, Y. Lu, Easy-to-use dipstick tests for detection of lead in paints using non-cross-linked gold nanoparticle–DNAzyme conjugates, Chem. Commun., 2010, 46, 1416–1418, with permission from The Royal Society of Chemistry.43 (B) LFD-based Detection of Cu2+ using Cu–DNAzyme. Reprinted (adapted) from Z. Fang, J. Huang, P. Lie, Z. Xiao, C. Ouyang, Q. Wu, G. Liu, L. Zeng, Lateral flow nucleic acid biosensor for Cu2+ detection in aqueous solution with high sensitivity and selectivity. Chem. Commun., 2010, 46, 9043–9045, with permission from The Royal Society of Chemistry.321 (C) A biosensor for Pb2+ detection using 8–17 DNAzyme and catalytic hairpin assembly. Reprinted (adapted) from J. Chen, X. Zhou, L. Zeng, Enzyme-free strip biosensor for amplified detection of Pb2+ based on a catalytic DNA circuit, Chem. Commun., 2013, 49, 984–986, with permission from The Royal Society of Chemistry.322
That same year, Fang and co-workers reported an alternative method to generate a LFD using the Cu2+-dependent DNAzyme.321 As shown in Fig. 24B, a reaction was first carried out in solution to allow Cu2+-dependent cleavage of a modified version of a substrate to liberate a DNA strand with a 3′ sequence complementary to cDNA bound to a AuNP and a 5′ sequence complementary to cDNA on a test line, such that the cleaved DNA strand acted as a bridging sequence to bind AuNP to the test line. Unbound AuNP was able to bind to cDNA on a control line for test validation. Using this approach, the LFD could produce a detection limit of 10 nM of Cu2+ with a reaction time of 30 min.
More recently, an isothermal amplification step has been implemented to improve the LOD of DNAzyme-based lateral flow devices.322 This example used the 17E DNAzyme for Pb2+ mediated cleavage of a modified 17S substrate. The cleaved fragment acted as a catalyst strand to initiate catalytic hairpin assembly (Fig. 24C), causing input hairpins H1 and H2 to assemble into a H1–H2 complex that could act as a bridging DNA to link AuNP to a test line. The LFD could detect 10 pM of Pb2+ using a CHA reaction time of 90 min.
While the LFD platform is simple and inexpensive, it is clear that there is much room for improvement. As noted in Section 3, there are multiple methods that can be used for amplifying DNAzyme reactions, which could be implemented with LFDs. In addition, at this time LFDs have only been used in conjunction with metal-dependent DNAzymes. Examples incorporating rationally designed aptazymes or bacteria-selective aptazymes remain to be reported.
4.6. Paper based devices
A relatively recent technology that has been proposed to allow integration of multiple assay steps, including sample preparation, separation, amplification and detection, is paper-based diagnostic devices, also commonly referred to as microscale paper-based analytical devices (μPADs), paper microfluidics or bioactive paper. Paper-based diagnostic devices provide a platform for portable, low-cost, low-volume, disposable, and simple sensors,323–327 which meet the World Health Organization ASSURED criteria.298 Paper-based diagnostic devices have been extensively reviewed, and interested readers are referred to recent reviews covering the fabrication of such devices,328 common detection methods,329 applications in environmental analysis330,331 and clinical diagnostics,332–334 commercialization of these devices335 as well as specialized reviews on the development of paper based diagnostic devices for nucleic acid detection,336 and paper sensors incorporating functional nucleic acids as MREs.51
4.6.1. Paper based sensors using metal-dependent DNAzymes.
Paper-based devices have been utilized for both visual and electrochemiluminescence based detection of metal ions. In this section, we highlight three recent studies using μPADS for detection of metal ions, and one using a metal-dependent DNAzyme for miRNA detection. The first study reported on the detection of lead ions using a dual-mode lab-on-paper device.179 This device incorporated both semiquantitative and quantitative methods for the detection of Pb2+ within the range of 0.1–2000 nM and 0.01–2000 nM respectively. The complex paper device was fabricated using multiple paper pads printed with microfluidic wells and channels to enable dual-mode visual and ECL detection. For ECL detection, one of the pads was screen printed with a carbon working electrode, and another was screen printed with an Ag/AgCl reference electrode and carbon counter electrode. The chemistry of the device is represented schematically in Fig. 25A. Semiquantitative indirect colorimetric detection of Pb2+ was achieved through the formation of a polyDAB precipitate following the reaction of 3,3′-diaminobenzidine (DAB) and hydrogen peroxide with horseradish peroxidase functionalized Au nanocubes (Au/HRP) immobilized on the surface of HKUST-1/rGO/Au particles (where HKUST-1 is a metal organic framework) via the DNAzyme. In the presence of Pb2+ ions, the Au/HRP was able to permeate through the device into the ECL regions, where the Au/HRP was immobilized on the surface of CdS quantum dots, leading to an enhanced ECL signal in the presence of luminol and hydrogen peroxide. Using this method, the detection of spiked Pb2+ in mineral and lake water was demonstrated with recoveries that ranged between 96.20 and 104.12%.
Fig. 25.
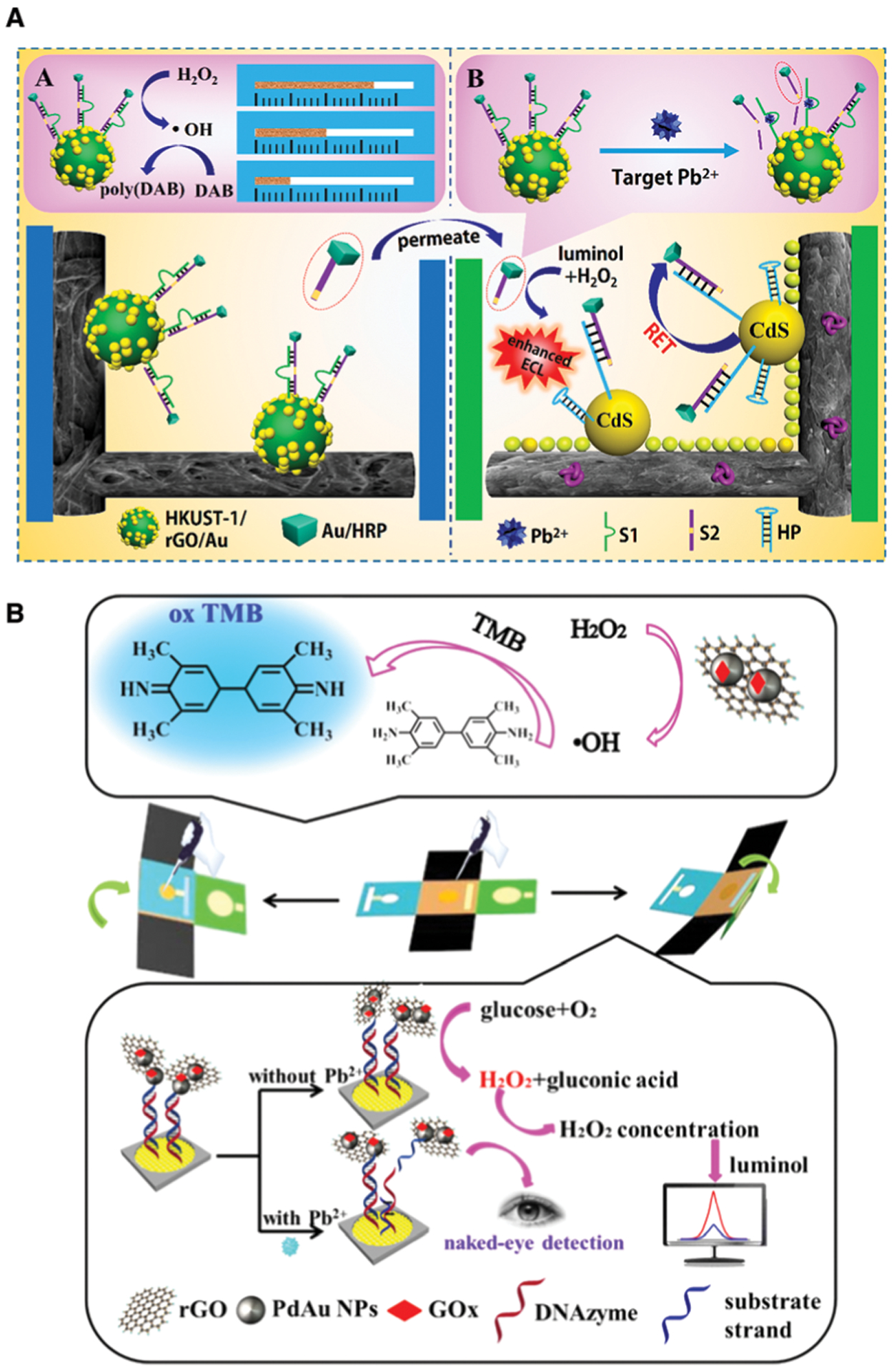
DNAzyme based devices for the detection of Pb2+. (A) Distance-based visualized analysis (panel A) and ratiometric electrochemiluminescence assay (panel B) with a dual-mode lab-on-paper device. Reprinted (adapted) with permission from Y. Zhang, J. Xu, S. Zhou, L. Zhu, X. Lv, J. Zhang, L. Zhang, P. Zhu and J. Yu, DNAzyme-Triggered Visual and Ratiometric Electrochemiluminescence Dual-Readout Assay for Pb(ii) Based on an Assembled Paper Device, Anal. Chem., 2020, 92, 3874–3881. Copyright 2020 American Chemical Society.179 (B) Dual-mode colorimetric and electrochemiluminescence analysis using an integrated lab-on-paper device. Reprinted (adapted) with permission J. Xu, Y. Zhang, L. Li, Q. Kong, L. Zhang, S. Ge and J. Yu, Colorimetric and Electrochemiluminescence Dual-Mode Sensing of Lead Ion Based on Integrated Lab-on-Paper Device, ACS Appl. Mater. Interfaces, 2018, 10, 3431–3440. Copyright 2018, American Chemical Society.172
A second μPAD for detection of Pb2+ is illustrated in Fig. 25B. In this case, an integrated lab-on-paper device, assembled using strategic paper folding, was designed for the dual-mode colorimetric and electrochemiluminescence detection of Pb2+ in tap and river water.172 The device, which consisted of a central square surrounded by a single square on each side, contained detection, reference, and channel tabs, as well as working, counter, and reference electrodes. To assemble the device, it was simply folded so that the channel tab overlapped the detection tab. Importantly, the DNAzyme was assembled on the surface of the device via the interaction of AuNP–DNAzyme and rGO–PdAu–GOx-substrate constructs. In the presence of Pb2+, the rGO–PdAu–GOx construct was released via cleavage of the substrate strand. Visual detection occurred when the rGO–PdAu–GOx construct was able to catalyze the reaction of hydrogen peroxide with TMB, leading to the generation of a blue coloured product. ECL detection was achieved in the absence of Pb2+, when the free rGO–PdAu–GOx construct catalyzed the conversion of glucose and O2 to gluconic acid and H2O2, which reacts with luminol to generate a signal. Therefore, with this device the visual signal increased and the ECL signal decreased with increasing Pb2+ concentrations. Concentrations as low as 0.14 nM of Pb2+ were detected, and recoveries of spiked Pb2+ in tap and river water ranged from 96.13 to 99.52%.
A third example, shown in Fig. 26, further demonstrates the versatility of foldable paper devices. Using a similar strategy to the sensors described for Pb2+, a foldable paper device for the detection of Hg2+ and Ni2+ was described by Huang et al.133 In this case, the paper was designed like a T (Fig. 26A), but still contained detection zones, and a paper working electrode (PWE), reference electrodes and a counter electrode. Strategic folding of the device is shown in Fig. 26B. The sensing strategy for ECL detection is shown in Fig. 26C. Briefly, the nucleic acid recognition sequences (polyT for Hg2+ or Ni2+–DNAzyme) and ECL reporters were immobilized to the surface of the paper device through complementary oligonucleotide binding. In the absence of the targets, the ECL signal was strongly generated. In the presence of the target metal analytes, the hybridization reaction was disrupted, initiating a cascade effect which led to a weaker ECL signal. The device was able to detect Hg2+ and Ni2+ with limits of detection of 3.1 nM and 3.8 pM, respectively, and demonstrated efficient recovery of spiked metal ion in lake water.
Fig. 26.
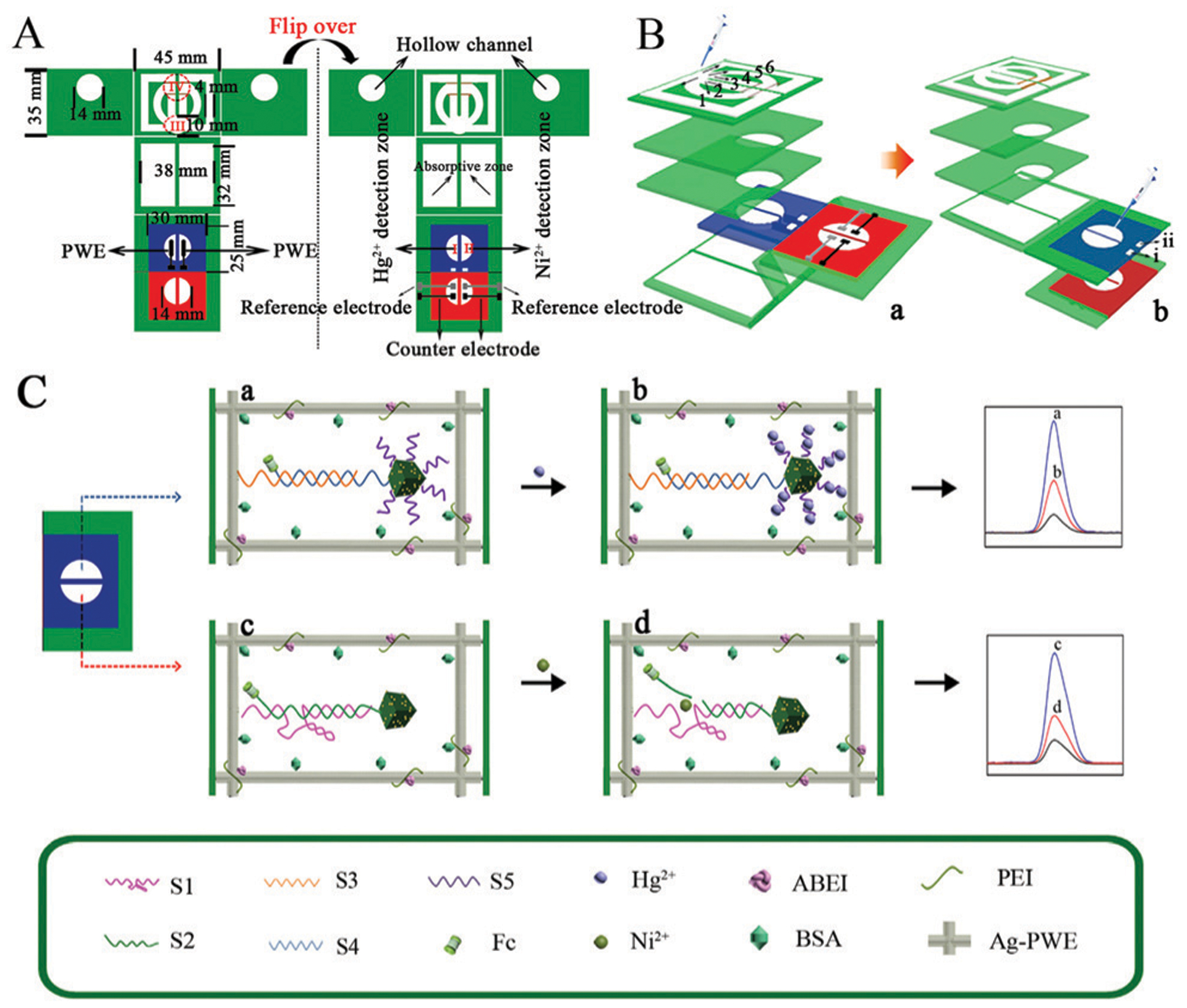
An electrochemiluminescence paper device for the detection of Hg2+ and Ni2+. Reprinted (adapted) with permission from Y. Huang, L. Li, Y. Zhang, L. Zhang, S. Ge and J. Yu, Auto-cleaning paper-based electrochemiluminescence biosensor coupled with binary catalysis of cubic Cu2 O–Au and polyethyleneimine for quantification of Ni2+ and Hg2+, Biosens. Bioelectron., 2019, 126, 339–345. Copyright 2018 Elsevier B.V.133
A final example, reported by the Liu group, used a paper device to detect a nucleic acid target, but in this case described the electrochemical paper-based detection of microRNA using a Mg2+-dependent DNAzyme (Fig. 27).337 Briefly, sensing was initiated when the target miRNA (miR-21) was incubated with a DNA probe (P1), KF polymerase, and nicking endonuclease (Nt.BbvCI). In this cyclic process, the miR-21 hybridized to the P1, and initiated the extension of the sequence by KF polymerase, then the nicking endonuclease cut the double-stranded product releasing a Mg2+-dependent DNAzyme strand. When this solution was added to the paper, the DNAzyme strand hybridized to the immobilized substrate strands, and cleavage was facilitated. The electrochemical signal was generated by the adsorption of the ferrocene labeled cleavage fragment onto a carbon nanotube modified working electrode (CNTs-WE). To show the potential of the device, a limit of detection of 1 fM was described, and the detection of miR-21 in spiked human serum was demonstrated with recoveries ranging from 93–102%.
Fig. 27.
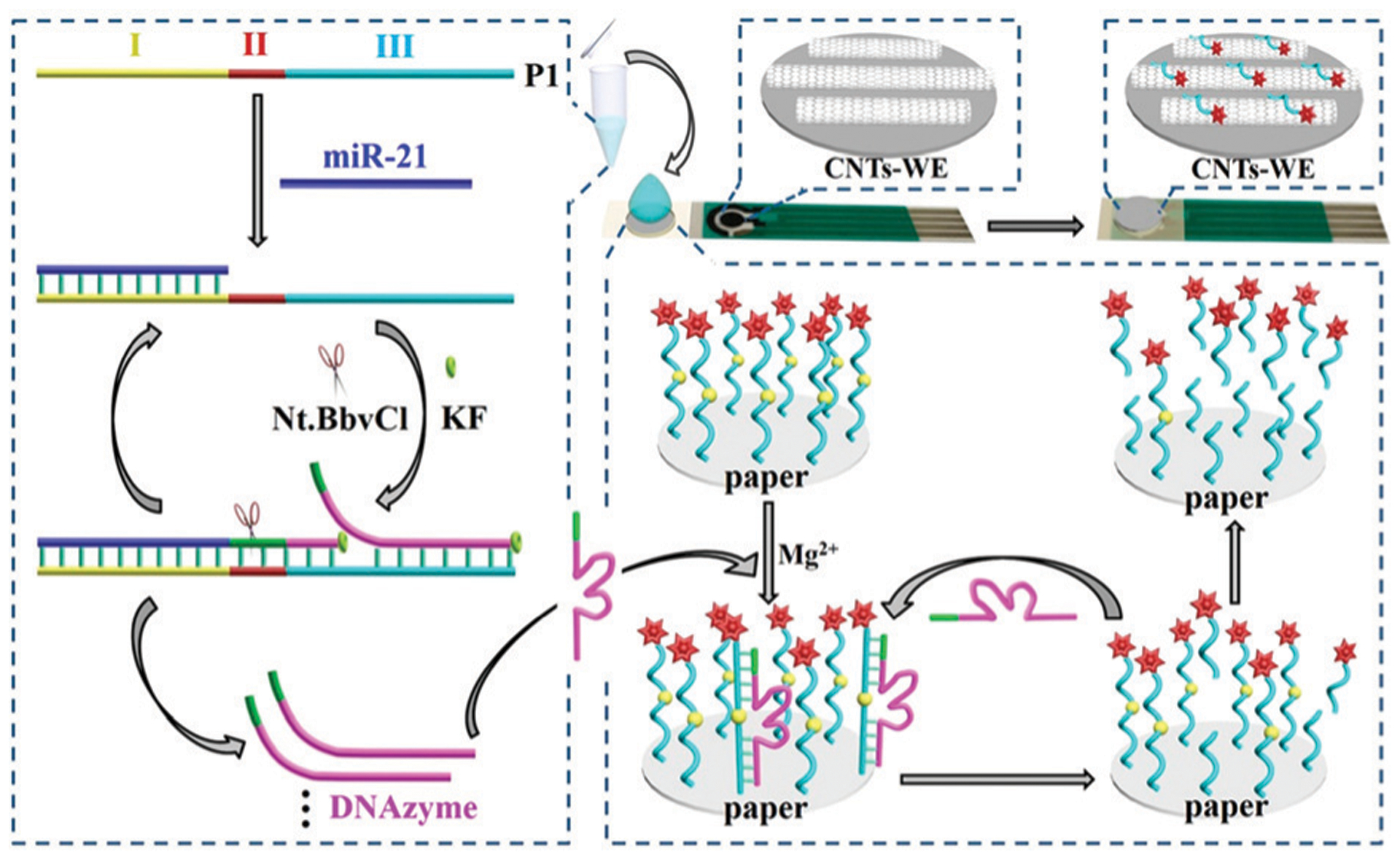
A paper device for microRNA detection. The recognition of miR-21 is shown on the left and electrochemical response to the released DNAzymes is shown on the right, with CNTs-WE before and after adsorption of Fc-SDNA (upper right). Reprinted (adapted) with permission from X. Liu, X. Li, X. Gao, L. Ge, X. Sun and F. Li, A Universal Paper-Based Electrochemical Sensor for Zero-Background Assay of Diverse Biomarkers, ACS Appl. Mater. Interfaces, 2019, 11, 15381–15388. Copyright 2019 American Chemical Society.337
4.6.2. Paper-based sensors using bacteria-selective aptazymes.
At this time, there are relatively few examples of paper-based sensors that utilize aptazymes as MREs. The first report, in 2017, utilized the RFD-EC1 aptazyme that was printed into a paper-based 96 well paper plate formed by printing of hydrophobic wax barriers to generate the 96 well outline (Fig. 28A).47 The aptazyme was printed into the wells using an ink containing pullulan, a polysaccharide that forms an oxygen impermeable film that has been reported to stabilize biological moieties for periods of up to 2 years.338–340 The film also contained lysozyme, which lysed the E. coli cells to increase the concentration of the target protein, thereby improving detection limits by an order of magnitude. The paper sensor produced a fluorescence signal in 5 minutes with a limit of detection of 100 cells per mL, could be used in a variety of sample matrixes (i.e., milk, apple juice), without sample enrichment, and remained stable for at least 6 months when stored at ambient temperature.
Fig. 28.
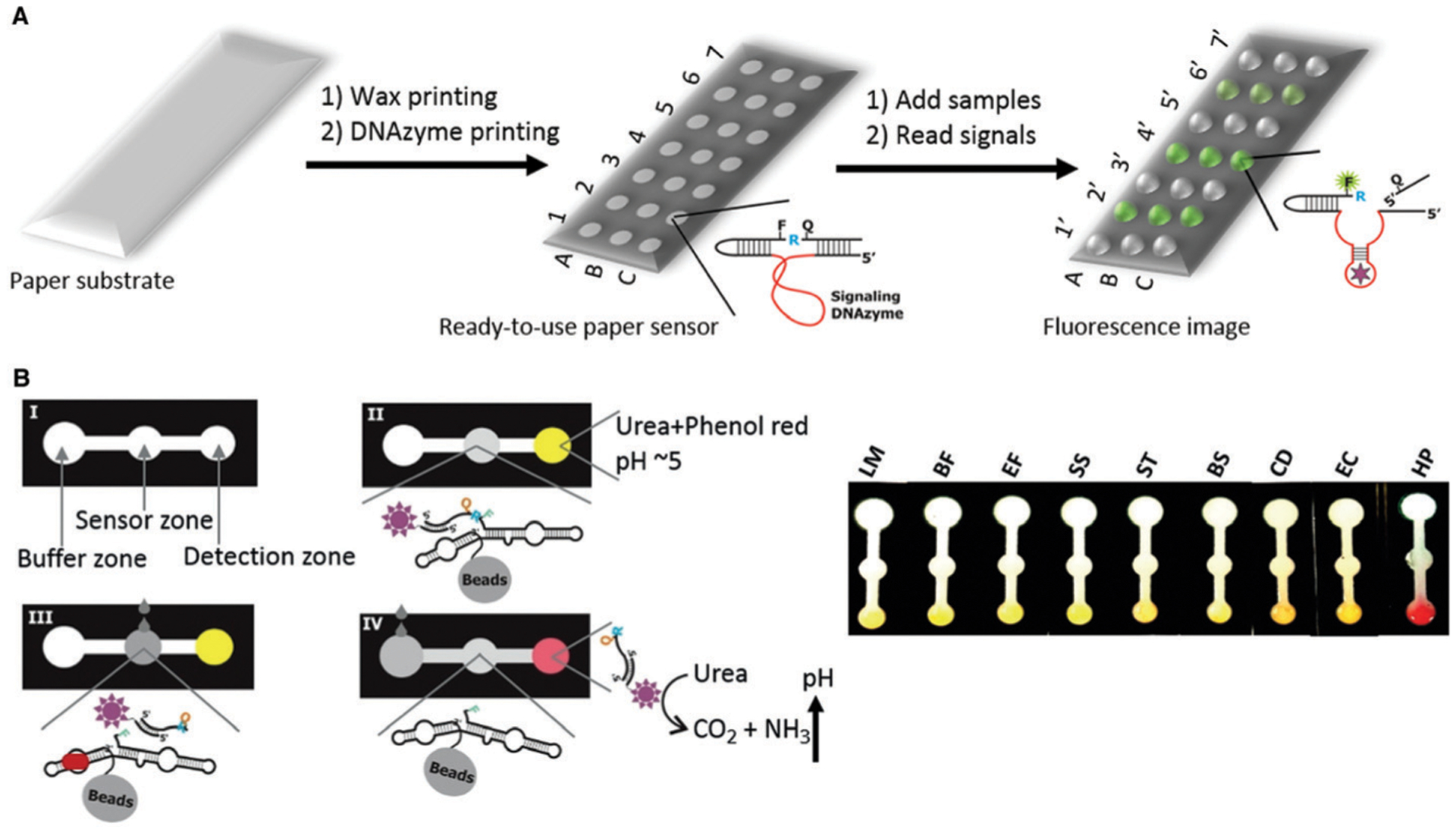
DNAzyme based paper sensors for bacterial detection. (A) Paper plate sensor for bacterial detection. Reprinted (adapted) with permission from M. M. Ali, C. L. Brown, S. Jahanshahi-Anbuhi, B. Kannan, Y. Li, C. D. M. Filipe and J. D. Brennan, A Printed Multicomponent Paper Sensor for Bacterial Detection, Sci. Rep., 2017, 7, 12335. Copyright 2017 Ali et al.47 (B) A colorimetric paper-based sensor using the HP DNAzyme and a modified version of the urease-based litmus test for the detection of H. pylori. HP: H. pylori; others are control bacteria. Reprinted (adapted) with permission from M. M. Ali, M. Wolfe, K. Tram, J. Gu, C. D. M. M. Filipe, Y. Li and J. D. Brennan, A DNAzyme-Based Colorimetric Paper Sensor for Helicobacter pylori, Angew. Chem., Int. Ed., 2019, 1, 9907–9911. Copyright 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.48
A similar format was used to detect K. pneumoniae using the RFD-KP6 aptazyme.89 In this case, the paper-based microwell plates were used for mix-and-read fluorescence assays to evaluate 20 strains of clinical bacterial isolates, and produced a signal for samples containing K. pneumoniae, regardless of their source or drug resistance, with a limit of detection of 105 CFU mL−1. The relatively poor LOD is likely based on a combination of poorer performance for the RFD-KP6 aptazyme (relative to RFD-EC1) and the use of intact rather than lysed cells.
An issue with the assays described above is the use of a fluorescence readout, which requires a reader and thus does not meet the equipment-free ASSURED criteria. To address this issue, a colorimetric paper-based sensor was developed using the HP DNAzyme and a modified version of the urease-based litmus test for the detection of H. pylori in human stool samples (Fig. 28B).48 The simple paper device was wax printed to form three zones, for buffer, sensing and detection. The RFD was modified with urease and immobilized on agarose beads before being deposited in the sensor zone via encapsulation in a pullulan film. In the presence of the bacteria, cleavage of the RFD allowed the urease modified oligonucleotide fragment to migrate to the detection zone, in which the reagents for the modified litmus assay had been deposited. The presence of the target bacteria led to a color change that provided a LOD of 104 CFU mL−1 when assessed with spiked stool samples and remained stable for at least 4 months when stored at room temperature. An advantage of the paper-based litmus test was the elimination of the magnetic separation step used in solution assays, although the use of a pH change for signal development required careful control of both initial pH and buffer capacity to ensure good sensitivity. Use of alternative enzymes such as AChE, as described in Section 3.1.3.2, may be able to overcome this limitation.
4.6.3. Paper sensors utilizing isothermal amplification for detection of bacteria.
An emerging area of paper-based diagnostics is the integration of signal amplification methods as part of the sensor platform. Several reviews have now appeared describing progress in this area341,342 although the vast majority of reports describe detection of nucleic acids rather than DNAzyme or aptazyme-initiated ITA on paper-based devices. At this time, there are only a few such reports using aptazymes, both of which are based on the use of room-temperature rolling circle amplification, which was demonstrated on paper devices in 2016 (Fig. 29A).343 The basic approach involves printing a capture DNA sequence that is able to capture a specific nucleic acid sequence (miRNA, cDNA, or DNA released by a DNAzyme or aptamer) in a manner that leaves an overhang. This overhang can then bind to a circular template (also printed in an RCA reaction zone, along with other reagents such as phi29 and dNTPs required for RCA) and act as a primer for RCA. The concatenated DNA output can be detected by binding of fluorophore or AuNP labelled DNA, or by producing a PMD that allows a peroxidase-based colorimetric output (Fig. 29B).
Fig. 29.
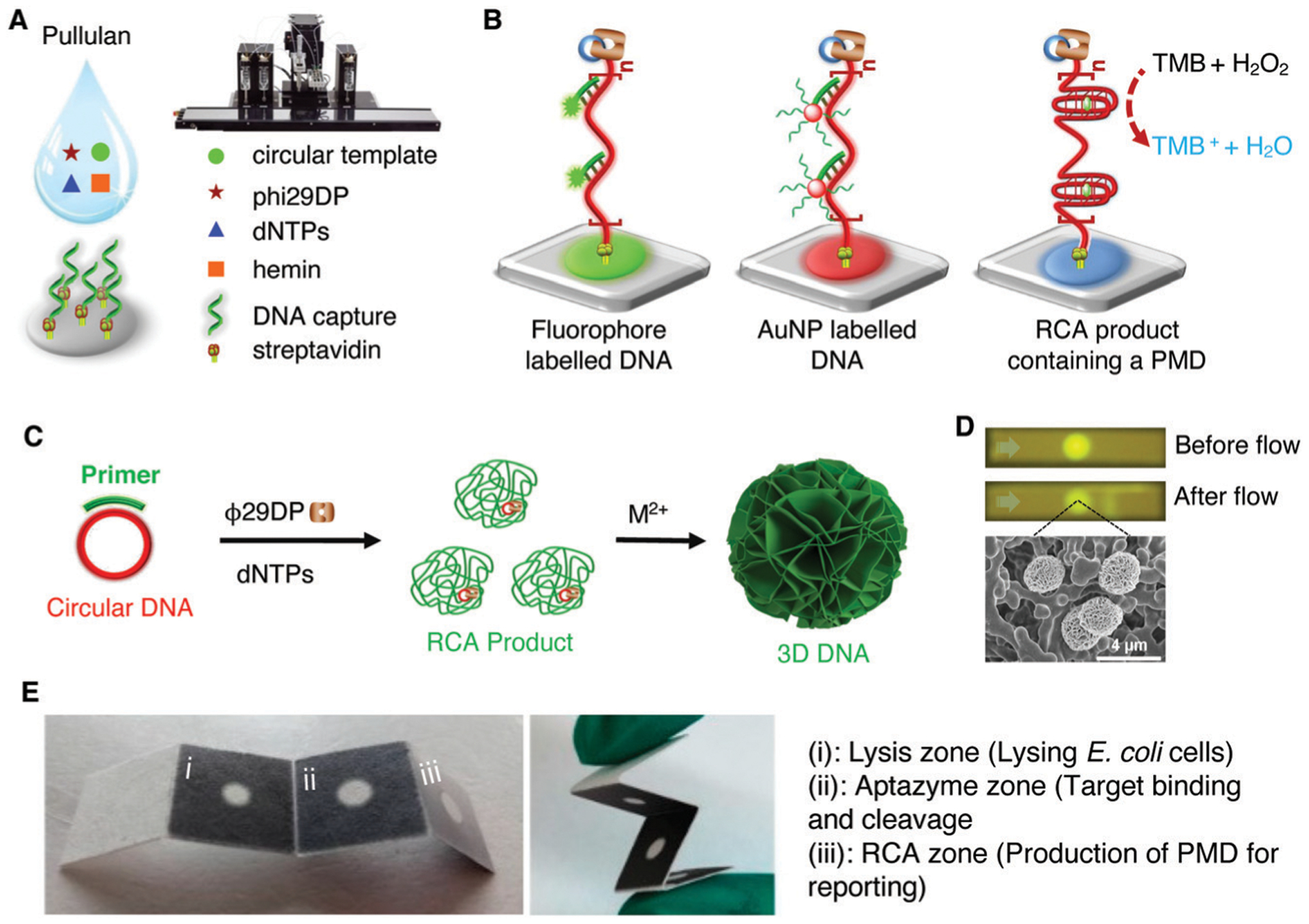
Paper sensors incorporating DNAzymes and RCA. (A) Printed paper device capable of performing RCA on paper. (B) Detection of RCA products by binding of fluorophore or AuNP labelled DNA, or by producing a PMD in the RCA product. A and B reprinted (adapted) with permission from M. Liu, C. Y. Hui, Q. Zhang, J. Gu, B. Kannan, S. Jahanshahi-Anbuhi, C. D. M. Filipe, J. D. Brennan and Y. Li, Target-Induced and Equipment-Free DNA Amplification with a Simple Paper Device, Angew. Chem., Int. Ed., 2016, 55, 2709–2713. 2016 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.343 (C) Printed paper sensor using DNAzyme containing 3D DNA created from RCA. (D) 3D DNA-coated nitrocellulose paper strips before and after liquid flow. Reprinted (adapted) with permission from M. Liu, Q. Zhang, B. Kannan, G. A. Botton, J. Yang, L. Soleymani, J. D. Brennan and Y. Li, Self-Assembled Functional DNA Superstructures as High-Density and Versatile Recognition Elements for Printed Paper Sensors, Angew. Chem., Int. Ed., 2018, 57, 12440–12443. Copyright 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.344 (E) Foldable 3D DNA paper sensor for E. coli detection. Reprinted (adapted) with permission from Y. Sun, Y. Chang, Q. Zhang and M. Liu, An Origami Paper-Based Device Printed with DNAzyme-Containing DNA Superstructures for Escherichia coli Detection, Micromachines, 2019, 10, 531. Copyright 2019 Sun et al.345
Following the initial report of RCA on paper, the Brennan and Li groups investigated the potential of using the large RCA reaction products as a means of size-based immobilization of DNA-based MREs onto paper devices. In the first report, micrometer-sized functional nucleic acid (FNA) superstructures (denoted as 3D DNA) were produced by RCA and examined as a unique class of biorecognition elements to produce highly functional bioactive paper surfaces.344 3D DNA containing repeating sequences of either a DNA aptamer or DNAzyme (MgZ or EC1) was created from long-chain products of rolling circle amplification followed by salt aging (Fig. 29C) and was used as a bioink for creating paper sensors via inkjet printing. The printed 3D DNA superstructures were found to remain immobilized at their initial positions after flow (Fig. 29D), indicating that 3D DNA can be physically adsorbed on paper surfaces. 3D DNA paper sensors showed resistance to degradation by nucleases, suppressed nonspecific protein adsorption, and provided a much higher surface density of functional DNA relative to monomeric FNAs, making such species ideally suited for development of paper-based biosensors.
In a follow up report, 3D DNA was further used to develop a four-panel device, which was created such that it could be assembled flat and then folded for detection (Fig. 29E).345 Upon cleavage of the EC1 DNAzyme units within the superstructure, the cleavage fragment migrated to the RCA zone thereby initiating the generation of PW17, producing a color change in the presence of TMB, hemin, and H2O2. The device could detect E. coli with a limit of 103 CFU mL−1 in 35 minutes. Additionally, the recovery of spiked E. coli K12 in orange juice and milk was demonstrated.
4.6.4. Sample preconcentration and sensing on paper.
By taking advantage of the complementarity of DNA, the Li and Filipe groups developed a highly sensitive RNA-cleaving DNAzyme sensor which took advantage of a strategy called surface-to-surface product enrichment for signal generation (Fig. 30A).346 In this method, the intact RCD RFD-EC1 was immobilized on agarose beads, and a reporter moiety was conjugated to the cleavage fragment. Target detection was achieved by the addition of a small circular paper disk, which had been coated with a capture probe that was complementary to the cleavage fragment, into the reaction tube. The cleaved fragment was captured by the paper whereas intact DNAzyme remained on the agarose bead that settled to the bottom of the tube. Complementary hybridization of the released strand carrying a urease enzyme or fluorophore to the detection zone allowed semi-quantitative detection of E. coli with detection limits as low as 100 CFU mL−1 and 10 CFU mL−1, respectively.
Fig. 30.
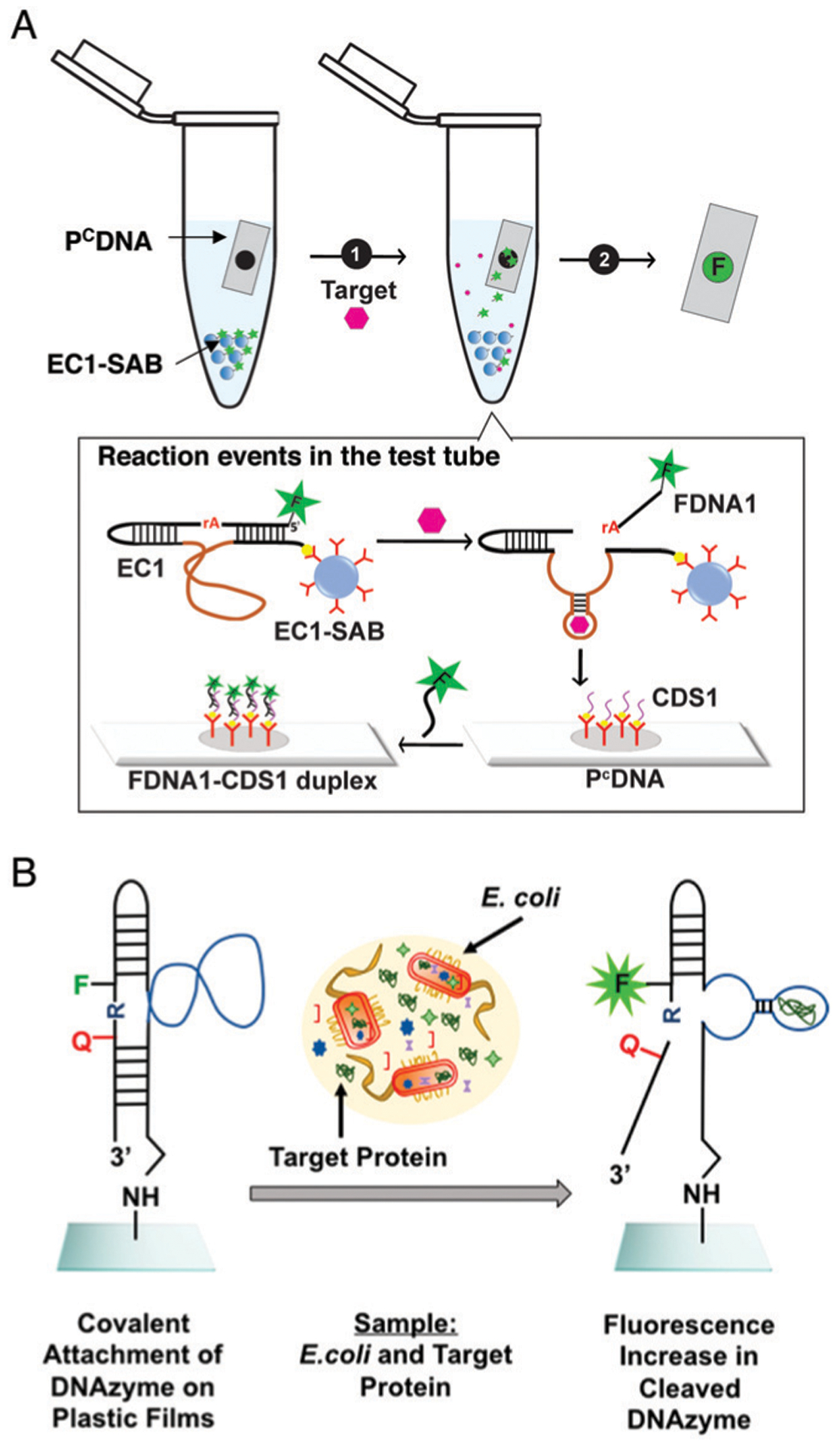
(A) Surface-to-surface product enrichment assay. EC1-SAB: DNAzyme EC1 immobilized on streptavidin-containing agarose beads; PCDNA: paper strip with a capture DNA sequence (named CDS1); FDNA: the fluorescent cleavage fragment of EC1. Reprinted (adapted) with permission from S. E. Samani, D. Chang, E. M. McConnell, M. Rothen-broker, C. D. M. Filipe and Y. Li, Highly Sensitive RNA-Cleaving DNAzyme Sensors from Surface-to-Surface Product Enrichment, ChemBioChem, 2020, 21, 632–637. Copyright 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.346 (B) A foodwrap sensor DNAzyme sensor capable of E. coli. Amine-terminated RFD-EC1 is covalently attached to thin, flexible, and transparent epoxy films. The cleavage of the fluorogenic substrate by the DNAzyme in the presence of the target produced by live E. coli cells produces a detectable signal. Reprinted (adapted) with permission from H. Yousefi, M. M. Ali, H. M. Su, C. D. M. Filipe and T. F. Didar, Sentinel Wraps: Real-Time Monitoring of Food Contamination by Printing DNAzyme Probes on Food Packaging, ACS Nano, 2018, 12, 3287–3294. Copyright 2018 American Chemical Society.348
4.6.5. Food wrap sensors.
A final example of DNA aptazyme-based bacterial detection is based on printing of DNAzymes onto plastic surfaces rather than paper to allow for fluorimetric sensing of bacteria directly on the surface of meat. Real-time detection of food-borne pathogens affects food safety at all points in the food supply chain.347 In order to address the challenge of monitoring food safety for consumers, Yousefi et al. reported the incorporation of RFD-EC1 into food packaging348 via immobilization of the DNAzyme on a cyclo-olefin polymer (COP) film via an amine linkage (Fig. 30B).349 When meat spiked with E. coli was wrapped in the DNAzyme-modified film, a fragment carrying a quencher was released and a fluorescence signal was generated in cases where 103 CFU mL−1 or higher of bacteria was present, while the immobilized DNAzyme remained stable for up to 14 days when no E. coli was present. While an interesting first step, the current detection limits and strain selectivity are insufficient. For example, current US guidelines call for detection limits of 1 CFU of E. coli H7:O157 per 325 g of boneless or ground beef,350 and RFD-EC1 is unable to identify the key pathogenic (H7:O157) strain of E. coli, which is critical as this is the strain that is responsible for the majority of infections from ground beef.
5. Summary and perspectives
In contrast to naturally occurring ribozymes, DNAzymes were first identified by Breaker and Joyce in 1994 through in vitro selection from a random DNA library consisting of approximately 1014 DNA sequences. Since then, a variety of DNAzymes have been identified. The functionalities of DNAzymes depend on both the sequences of the DNAzymes and additional cofactors such as metal ions or proteins. Many chemical reactions including RNA cleavage, oxidative or hydrolytic DNA cleavage, DNA or RNA ligation, DNA phosphorylation, and peroxidase reactions based on G-quadruplexes, have now been carried out by DNAzymes. Most of the DNAzymes can catalyze their respective reactions with multiple turnovers, which allows for sensitive detection when used as sensors. Moreover, DNAzymes possess several other properties, including good chemical stability, easy modification with signaling molecules, and synthetic accessibility, as well as ease of integration into colorimetric, fluorimetric and electrochemical assays, with or without DNA amplification steps, making them well suited as sensors in simple and portable devices, such as hand-held readers, lateral-flow devices, paper-based devices, personal glucose meters, and even a thermometer. As a result, a wide variety of DNAzymes have been successfully applied for environmental monitoring, food safety and medical diagnostics, as well as for biomedical imaging of metal ions, bacteria and other targets, as summarized in this review.
Despite the enormous progress in this field, few of the reported biosensors have reached the level of commercialization enjoyed by other types of sensors, such as enzyme and antibody-based sensors. To advance the field further and to reach the full potential of the DNAzymes, we need to meet several challenges.
First, while DNAzyme sensors have been obtained for a variety of metal ions, there remain a substantial number of metal ions for which DNAzymes have yet to be obtained. For example, while there are numerous DNAzyme sensors for divalent metal ions, far fewer DNAzymes have been reported for monovalent metal ions, especially for applications in cells and other biological systems, owing to the challenges associated with selective detection of monovalent ions in the presence of high concentrations of Na+ or K+. In addition, while DNAzyme sensors for diamagnetic metal ions (those that contain no unpaired electrons, such as Pb2+ and Zn2+) have been reported, DNAzyme sensors for paramagnetic metal ions (those that contain an unpaired electron, such as Fe2+) need further development. Finally, since most metal ions have multiple oxidation states, DNAzyme sensors capable of differentiating between different oxidation states of the same metal ion (e.g., Fe2+ vs. Fe3+) would be very useful.
Second, there is a need to develop better DNAzymes that can detect a wider range of targets. To achieve this goal, we need to engineer DNAzymes with higher activity and specificity toward their targets in order to meet the demands of high sensitivity, selectivity and fast testing speed for real samples. While it is possible to create artificial nucleic acid enzymes that exhibit catalytic rate enhancements that match those of their protein counterparts,351 most DNAzymes reported to date are orders of magnitude slower than protein enzymes. Improved selection methods using lower target concentrations and shorter reaction times along with more stringent counter-selection steps may help DNAzymes match the performance of protein enzymes. Selections incorporating modified nucleotides may also help, as this may provide access to additional functional groups that are normally found in the 20 natural amino acids used to build protein enzymes, thereby expanding the reactivity of DNAzymes. This may require the engineering of new polymerases that can accommodate the modified nucleotides during PCR so that highly active and selective DNAzymes can be enriched. Such efforts, combined with additional fundamental studies of DNAzyme structure and activity, including determination of pre- and post-catalytic 3D structures via crystallography,67,68 may help expand the range of DNAzyme reactions to include such classes as redox reactions, and the range of targets to include anions, organic metabolites and specific proteins, which have not yet been successful targets for DNAzyme selection. Natural riboswitches already exist for an anion (fluoride), however the fluoride ion is encapsulated by Mg2+ and phosphates, which is an atypical situation.352 However, this does suggest that it may be possible to select DNAzymes that bind to non-conventional targets such as anions through adaptive strategies.
Third, a number of studies have expanded the targets of RNA-cleaving DNAzymes beyond metal ions through coupling the metal-ion-dependent DNAzyme activity with other molecules. For example, several studies have taken advantage of sequestration of Cu2+ by other molecules such as histidine268,269 and thiocholine277 as an indirect method to use Cu2+-dependent DNAzyme activity for detection. An issue for this method is that many molecules can sequester metal ions like Cu2+ and when they are all present in the same sample, it would be difficult to detect these molecules selectively. To address this issue, in vitro selection of aptazymes with rigorous counter selection against competing targets is required.
Fourth, in the case of bacterial selective aptazymes, there remain several challenges to be overcome to advance this field. There are currently only seven examples of directly selected aptazymes, and only one example of an aptazyme that can selectively detect a multidrug resistant version of a bacterium, and no examples where an aptazyme can discriminate between a pathogenic and non-pathogenic strains of the same bacterium. Additional work is needed to better design selection protocols to address this issue. In addition, in almost all cases, the actual target (putatively a protein) is unknown, and as such it is difficult to predict whether a given DNAzyme will demonstrate sufficient selectivity against other bacteria. Furthermore, the selection method does not provide a high degree of control over DNAzyme performance (kobs, KM, background cleavage rate), and hence improved selection strategies will be needed to obtain faster and more selective aptazymes.
Fifth, it is important to identify DNAzymes that can work well in real sample matrices. Most reported DNAzymes only work in relatively pure solutions, under well controlled conditions of pH, ionic strength, buffering agent and temperature. As a result, a small change in reaction conditions can result in large changes in activity. In addition, many sample matrixes, particularly clinical samples, may contain a range of nucleases that can rapidly degrade DNAzymes, or nucleic acid-binding proteins that can interfere with DNAzyme function. Several methods can be employed to help move toward use of DNAzymes in complex samples. For example, screening of assay buffers can help optimize DNAzyme activity, as demonstrated by the Liu group, who showed that a colorimetric gold nanoparticle-based sensor for the detection of DNA had 15.7-fold better sensitivity when performed in a MES buffer compared to a HEPES buffer.353 An alternative strategy to address interferences in real samples is to select DNAzymes directly in environmental or biological samples, which could remove DNAzymes that do not work well in real samples. This is analogous to selection of bacteria-selective DNAzymes using cell lysates, which have been shown to produce aptazymes that can perform in complex matrices such as urine or stool samples.48,50 In addition, rational design of DNAzymes whose activities can tolerate fluctuations of conditions, such as temperatures,354 should be explored. It is important to note that while numerous studies have evaluated DNAzyme performance using biological samples spiked with the target of interest, at this time there is only one study that has reported a clinical trial utilizing patient samples.
Sixth, related to sampling in real matrices is the ability to use DNAzymes for in vivo imaging applications, where initial studies in cells have not yet translated to studies of animals or humans. To overcome this limitation, more methodologies need to be developed in order to improve delivery and targeting efficiency, and biostability. In addition, novel signalling, such as the photoacoustic method that has been demonstrated for aptamer-based imaging236 and MRI that has been demonstrated in in vitro studies, could be applied.237 Finally, isothermal signal amplification methods that do not require externally added enzymes, such as catalytic hairpin assembly that has been demonstrated in living cells,231 need to be developed.
Seventh, there are only a few examples at this time that demonstrate the use of DNAzyme-based assays or devices for testing of patient samples.294,295 As noted above, most studies have evaluated the performance of DNAzymes in biological samples that have been spiked with the target of interest and have demonstrated that DNAzymes can operate in such matrixes with minimal interference. However, the few studies with real patient samples (including urine, nasal swabs and scab exudate) have shown that it is also possible to detect target analytes even in these complex samples, with the ability to achieve clinical sensitivity and specificity ranging from 80 to 100%. While such clinical studies are still relatively rare, they are a critical step in translation of DNAzyme-based assays toward commercial products, and it is expected that many more such studies will appear in the coming years.
Finally, while there are many examples of devices that integrate DNAzymes, commercialization of DNAzymes and DNAzyme devices remains a challenge. At this time, DNAzyme-based technologies and sensors are being commercialized by Innovogene Biosciences Inc. (www.innovogene.com; ANDzymes and Urasensor tests), ANDalyze (http://andalyze.com; DNAzyme-based metal ion sensors) and GlucoSentient Inc. (http://glucosentient.com; DNAzyme-based PGM devices). Further commercialization of DNAzyme-based sensors will require both the validation of these sensors, particularly those intended for clinical use, as well as reducing the complexity of some of the currently reported devices, which can be challenging to manufacture in a scalable manner. In addition, the cost of some portable devices, including complicated microchips and microfluidic systems, also makes it difficult for such devices to be produced inexpensively so that they can meet the ASSURED criteria. Further development of sensors based on established commercial platforms, such as LFDs and PGMs, may help to alleviate these issues and result in a pathway to the development of easily manufactured and commercially viable point-of-need sensors.
As described herein, designing portable and miniaturized devices using DNAzymes as MREs is a growing area, which undoubtedly will benefit biosensing given the ability to utilize multiple assay formats, with or without signal amplification, and integrate these assays into a range of different devices. By integrating additional features, such as sample collection and pre-treatment, amplification, and multiplexed readouts it may be possible to produce commercially viable sample-to-readout devices suitable for use in resource-limited regions. With increased research on DNAzyme-based devices we expect that commercially viable PON sensors should be reaching the marketplace in the near future.
Acknowledgements
Funding for DNAzyme and biosensor research in JDB and YL’s labs is provided by Natural Sciences and Engineering Research Council of Canada (NSERC), the Canadian Institutes of Health Research (CIHR), Ontario Ministry of Research and Innovation and Canada Foundation for Innovation (CFI), while the research in Yi Lu’s lab is funded by the US National Institute of Health (GM124316, MH110975 and GM141931). JDB holds the Canada Research Chair in Point-of-Care Diagnostics.
Biographies

Erin M. McConnell
Erin M. McConnell is a postdoctoral fellow whose research interests lie at the intersection of identifying functional nucleic acids, and building DNA-based tools to understand and monitor human health. Specifically, she is interested in how DNAzymes can be used to make highly sensitive and selective biosensors. Erin received her PhD in chemistry under the supervision of Prof. Maria C. DeRosa at Carleton University, where she studied applications for aptamers within the central nervous system, and just recently completed a postdoctoral fellowship under the supervision of Prof. Yingfu Li, at McMaster University, building DNAzyme-based platforms for detecting pathogenic bacteria in water.

Ioana Cozma
Ioana Cozma was born in Romania and immigrated to Canada in 2003. She obtained her Bachelor’s degree in 2013 from Queen’s University and her MD in 2016 from McMaster University. She is currently completing her residency training in the field of Anesthesiology at McMaster University. In 2018, she concurrently entered the Clinician Investigator Program to launch her research career under the supervision of Dr Yingfu Li. Cozma’s research interests lie at the intersection of functional nucleic acids and clinical applications, specifically focusing on using DNAzymes to identify new biomarkers of disease and developing novel diagnostic and therapeutic techniques.

Quanbing Mou
Quanbing Mou completed his BS degree in Polymer Materials and Engineering from Sichuan University in 2012. Then he got his PhD degree under the supervision of Dr Deyue Yan, Dr Xinyuan Zhu, and Dr Chuan Zhang in School of Chemistry and Chemical Engineering at Shanghai Jiao Tong University in 2018. Currently, he is a postdoctoral researcher in Dr Yi Lu’s group at University of Illinois at Urbana-Champaign working on modified oligonucleotides for bioapplications.

John D. Brennan
John Brennan holds the Canada Research Chair in Point-of-Care Diagnostics and is Director of the Biointerfaces Institute at McMaster University. He received his BSc, MSc and PhD degrees from the University of Toronto under Dr Ulrich J. Krull. After 2 years of postdoctoral research in Dr Arthur Szabo’s group at the National Research Council of Canada he joined McMaster University, where he is now a Professor. His group interests are in bioanalytical chemistry and functional nucleic acids, with a main focus on biosensors.

Yi Lu
Yi Lu is a Jay and Ann Schenck Professor in the Department of Chemistry at the University of Illinois at Urbana-Champaign. He received his BS degree from Peking University in 1986 and his PhD degree from UCLA in 1992 under Dr Joan S. Valentine. After 2 years of postdoctoral research in Dr Harry B. Gray’s group at California Institute of Technology, he started his independent career at University of Illinois at Urbana-Champaign in 1994. His group interests are in bioinorganic, biomaterial, and bioanalytical chemistry.

Yingfu Li
Yingfu Li received his BSc in chemistry from Anhui University in 1983, his MSc in applied chemistry from China Agriculture University in 1989, and his PhD in biochemistry and chemistry from Simon Fraser University in 1997 under the guidance of Prof. Dipankar Sen. He spent the next two years at Yale University as a postdoctoral fellow with Prof. Ronald Breaker. In 1999, he joined the Department of Biochemistry and Biomedical Sciences and the Department of Chemistry and Chemical Biology at McMaster University where he is now a Professor. His group studies DNAzymes, aptamers, biosensors, nanotechnology, and noncoding RNA.
Footnotes
Conflicts of interest
Yi Lu is a co-founder of ANDalyze Inc. and GlucoSentient Inc.; Yingfu Li is a co-founder of Innovogene Biosciences Inc.
References
- 1.Altman S, Nat. Struct. Biol, 2000, 7, 827–828. [DOI] [PubMed] [Google Scholar]
- 2.Shampo MA, Kyle RA and Steensma DP, Mayo Clin. Proc, 2012, 87, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crick F, Nature, 1970, 227, 561–563. [DOI] [PubMed] [Google Scholar]
- 4.Kappaun K, Piovesan AR, Carlini CR and Ligabue-Braun R, J. Adv. Res, 2018, 13, 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breaker RR and Joyce GF, Chem. Biol, 1994, 1, 223–229. [DOI] [PubMed] [Google Scholar]
- 6.Ponce-Salvatierra A, Boccaletto P and Bujnicki JM, Nucleic Acids Res, 2021, 49, D76–D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellington AD and Szostak JW, Nature, 1990, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 8.Tuerk C and Gold L, Science, 1990, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 9.Robertson DL and Joyce GF, Nature, 1990, 344, 467–468. [DOI] [PubMed] [Google Scholar]
- 10.Breaker RR, Chem. Rev, 1997, 97, 371–390. [DOI] [PubMed] [Google Scholar]
- 11.Wilson DS and Szostak JW, Annu. Rev. Biochem, 1999, 51, 435. [DOI] [PubMed] [Google Scholar]
- 12.Brody EN and Gold L, Rev. Mol. Biotechnol, 2000, 74, 5–13. [DOI] [PubMed] [Google Scholar]
- 13.Cech TR, Biochem. Soc. Trans, 2002, 30, 1162–1166. [DOI] [PubMed] [Google Scholar]
- 14.Micura R and Höbartner C, Chem. Soc. Rev, 2020, 49, 7331–7353. [DOI] [PubMed] [Google Scholar]
- 15.Reid R, Chatterjee B, Das SJ, Ghosh S and Sharma TK, Anal. Biochem, 2020, 593, 113574. [DOI] [PubMed] [Google Scholar]
- 16.Yu H, Alkhamis O, Canoura J, Liu Y and Xiao Y, Angew. Chem., Int. Ed, 2021, 2–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaban SM and Kim DH, Sensors, 2021, 21, 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandra M, Sachdeva A and Silverman SK, Nat. Chem. Biol, 2009, 5, 718–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flynn-Charlebois A, Wang Y, Prior TK, Rashid I, Hoadley KA, Coppins RL, Wolf AC and Silverman SK, J. Am. Chem. Soc, 2003, 125, 2444–2454. [DOI] [PubMed] [Google Scholar]
- 20.Li Y and Breaker RR, Proc. Natl. Acad. Sci. U. S. A, 1999, 96, 2746–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Liu Y and Breaker RR, Biochemistry, 2000, 39, 3106–3114. [DOI] [PubMed] [Google Scholar]
- 22.Cuenoud B and Szostak JW, Nature, 1995, 375, 611–614. [DOI] [PubMed] [Google Scholar]
- 23.Sreedhara A, Li Y and Breaker RR, J. Am. Chem. Soc, 2004, 126, 3454–3460. [DOI] [PubMed] [Google Scholar]
- 24.Li Y and Sen D, Nat. Struct. Mol. Biol, 1996, 3, 743–747. [DOI] [PubMed] [Google Scholar]
- 25.Chinnapen DJ-F and Sen D, Proc. Natl. Acad. Sci. U. S. A, 2004, 101, 65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pradeepkumar PI, Höbartner C, Baum DA and Silverman SK, Angew. Chem., Int. Ed, 2008, 47, 1753–1757. [DOI] [PubMed] [Google Scholar]
- 27.Chandrasekar J and Silverman SK, Proc. Natl. Acad. Sci. U. S. A, 2013, 110, 5315–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh SM, Sachdeva A and Silverman SK, J. Am. Chem. Soc, 2013, 135, 14928–14931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandsen BM, Hesser AR, Castner MA, Chandra M and Silverman SK, J. Am. Chem. Soc, 2013, 135, 16014–16017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou C, Avins JL, Klauser PC, Brandsen BM, Lee Y and Silverman SK, J. Am. Chem. Soc, 2016, 138, 2106–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu K, Lat PK, Yu H-Z and Sen D, Nucleic Acids Res, 2020, 48, 7356–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Achenbach J, Chiuman W, Cruz R and Li Y, Curr. Pharm. Biotechnol, 2004, 5, 321–336. [DOI] [PubMed] [Google Scholar]
- 33.Schlosser K and Li Y, Chem. Biol, 2009, 16, 311–322. [DOI] [PubMed] [Google Scholar]
- 34.Silverman SK, Trends Biochem. Sci, 2016, 41, 595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison D, Rothenbroker M and Li Y, Small Methods, 2018, 1700319, 1700319. [Google Scholar]
- 36.Liu M, Yin Q, McConnell EM, Chang Y, Brennan JD and Li Y, Chem. – Eur. J, 2018, 24, 4473–4479. [DOI] [PubMed] [Google Scholar]
- 37.Zhu L, Xu Y, Cheng N, Xie P, Shao X, Huang K, Luo Y and Xu W, Sens. Actuators, B, 2017, 242, 880–888. [Google Scholar]
- 38.Huang PJJ and Liu J, ChemistryOpen, 2020, 9, 1046–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hollenstein M, Molecules, 2015, 20, 20777–20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J and Lu Y, J. Am. Chem. Soc, 2000, 122, 10466–10467. [Google Scholar]
- 41.Liu J and Lu Y, J. Am. Chem. Soc, 2003, 125, 6642–6643. [DOI] [PubMed] [Google Scholar]
- 42.Xiao Y, Rowe AA and Plaxco KW, J. Am. Chem. Soc, 2007, 129, 262–263. [DOI] [PubMed] [Google Scholar]
- 43.Mazumdar D, Liu J, Lu G, Zhou J and Lu Y, Chem. Commun, 2010, 46, 1416–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang Y and Lu Y, Nat. Chem, 2011, 3, 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y, Zhou L and Tang Z, Nat. Commun, 2013, 4, 1–8. [DOI] [PubMed] [Google Scholar]
- 46.Tram K, Kanda P, Salena BJ, Huan S and Li Y, Angew. Chem., Int. Ed, 2014, 53, 12799–12802. [DOI] [PubMed] [Google Scholar]
- 47.Ali MM, Brown CL, Jahanshahi-Anbuhi S, Kannan B, Li Y, Filipe CDM and Brennan JD, Sci. Rep, 2017, 7, 12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ali MM, Wolfe M, Tram K, Gu J, Filipe CDMM, Li Y and Brennan JD, Angew. Chem., Int. Ed, 2019, 1, 9907–9911. [DOI] [PubMed] [Google Scholar]
- 49.Xiong Y, Zhang J, Yang Z, Mou Q, Ma Y, Xiong Y and Lu Y, J. Am. Chem. Soc, 2020, 142, 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pandey R, Chang D, Smieja M, Hoare T, Li Y and Soleymani L, Nat. Chem, DOI: 10.1038/s41557-021-00718-x. [DOI] [PubMed] [Google Scholar]
- 51.Liu R, McConnell EM, Li J and Li Y, J. Mater. Chem. B, 2020, 8, 3213–3230. [DOI] [PubMed] [Google Scholar]
- 52.Zhou W, Ding J and Liu J, Theranostics, 2017, 7, 1010–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lake RJ, Yang Z, Zhang JJ and Lu Y, Acc. Chem. Res, 2019, 52, 3275–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McConnell EM, Cozma I, Morrison D and Li Y, Anal. Chem, 2020, 92, 327–344. [DOI] [PubMed] [Google Scholar]
- 55.Silverman SK, Nucleic Acids Res, 2005, 33, 6151–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlosser K and Li Y, ChemBioChem, 2010, 11, 866–879. [DOI] [PubMed] [Google Scholar]
- 57.Navani NK and Li Y, Curr. Opin. Chem. Biol, 2006, 10, 272–281. [DOI] [PubMed] [Google Scholar]
- 58.Liu J, Cao Z and Lu Y, Chem. Rev, 2009, 109, 1948–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santoro SW and Joyce GF, Proc. Natl. Acad. Sci. U. S. A, 1997, 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Faulhammer D and Famulok M, Angew. Chem., Int. Ed. Engl, 1996, 35, 2837–2841. [Google Scholar]
- 61.Li Y, Zheng J, Kwon W and Lu AH, Nucleic Acids Res, 2000, 28, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cruz RP, Withers JB and Li Y, Chem. Biol, 2004, 11, 57–67. [DOI] [PubMed] [Google Scholar]
- 63.Schlosser K and Li Y, Biochemistry, 2004, 43, 9695–9707. [DOI] [PubMed] [Google Scholar]
- 64.Schlosser K and Li Y, J. Mol. Evol, 2005, 61, 192–206. [DOI] [PubMed] [Google Scholar]
- 65.Brown AK, Li J, Pavot CM-B and Lu Y, Biochemistry, 2003, 42, 7152–7161. [DOI] [PubMed] [Google Scholar]
- 66.Schlosser K, Gu J, Sule L and Li Y, Nucleic Acids Res, 2008, 36, 1472–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu H, Yu X, Chen Y, Zhang J, Wu B, Zheng L, Haruehanroengra P, Wang R, Li S and Lin J, Nat. Commun, 2017, 8, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ponce-Salvatierra A, Wawrzyniak-Turek K, Steuerwald U, Höbartner C and Pena V, Nature, 2016, 529, 231–234. [DOI] [PubMed] [Google Scholar]
- 69.Santoro SW and Joyce GF, Biochemistry, 1998, 37, 13330–13342. [DOI] [PubMed] [Google Scholar]
- 70.Dass CR, Choong PFM and Khachigian LM, Mol. Cancer Ther, 2008, 7, 243–251. [DOI] [PubMed] [Google Scholar]
- 71.Burgess DJ, Nat. Rev. Cancer, 2012, 12, 509. [DOI] [PubMed] [Google Scholar]
- 72.Khachigian LM, Cancer Res, 2019, 79, 879 LP–888. [DOI] [PubMed] [Google Scholar]
- 73.Lan T, Furuya K and Lu Y, Chem. Commun, 2010, 46, 3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ren W, Huang PJ, He M, Lyu M, Wang S, Wang C and Liu J, ChemBioChem, 2020, 21, 1293–1297. [DOI] [PubMed] [Google Scholar]
- 75.Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB and Lu Y, Proc. Natl. Acad. Sci. U. S. A, 2007, 104, 2056–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin Y-W, Biomolecules, 2020, 10, 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Torabi S-F, Wu P, McGhee CE, Chen L, Hwang K, Zheng N, Cheng J and Lu Y, Proc. Natl. Acad. Sci. U. S. A, 2015, 112, 5903–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ward WL, Plakos K and DeRose VJ, Chem. Rev, 2014, 114, 4318–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang P-JJ and Liu J, Nucleic Acids Res, 2015, 43, 6125–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang PJJ and Liu J, Anal. Chem, 2016, 88, 3341–3347. [DOI] [PubMed] [Google Scholar]
- 81.Ren W, Jimmy Huang PJ, de Rochambeau D, Moon WJ, Zhang J, Lyu M, Wang S, Sleiman H and Liu J, Biosens. Bioelectron, 2020, 165, 112285. [DOI] [PubMed] [Google Scholar]
- 82.Santoro SW, Joyce GF, Sakthivel K, Gramatikova S and Barbas CF, J. Am. Chem. Soc, 2000, 122, 2433–2439. [DOI] [PubMed] [Google Scholar]
- 83.Hollenstein M, Hipolito C, Lam C, Dietrich D and Perrin DM, Angew. Chem., Int. Ed, 2008, 47, 4346–4350. [DOI] [PubMed] [Google Scholar]
- 84.Huang PJ, Rochambeau D, Sleiman HF and Liu J, Angew. Chem., Int. Ed, 2020, 59, 3573–3577. [DOI] [PubMed] [Google Scholar]
- 85.Ali MM, Aguirre SD, Lazim H and Li Y, Angew. Chem., Int. Ed, 2011, 50, 3751–3754. [DOI] [PubMed] [Google Scholar]
- 86.Shen Z, Wu Z, Chang D, Zhang W, Tram K, Lee C, Kim P, Salena BJ and Li Y, Angew. Chem., Int. Ed, 2016, 55, 2431–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roth A and Breaker RR, Proc. Natl. Acad. Sci. U. S. A, 1998, 95, 6027–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Winkler WC, Nahvi A, Roth A, Collins JA and Breaker RR, Nature, 2004, 428, 281–286. [DOI] [PubMed] [Google Scholar]
- 89.Ali MM, Slepenkin A, Peterson E and Zhao W, ChemBioChem, 2019, 20, 906–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gu L, Yan W, Wu H, Fan S, Ren W, Wang S, Lyu M and Liu J, Anal. Chem, 2019, 91, 7887–7893. [DOI] [PubMed] [Google Scholar]
- 91.McConnell EM, Morrison D, Rey Rincon MA, Salena BJ and Li Y, TrAC, Trends Anal. Chem, 2020, 124, 115785. [Google Scholar]
- 92.Geng X, Zhang M, Wang X, Sun J, Zhao X, Zhang L, Wang X and Shen Z, Anal. Chim. Acta, 2020, 1123, 28–35. [DOI] [PubMed] [Google Scholar]
- 93.He S, Qu L, Shen Z, Tan Y, Zeng M, Liu F, Jiang Y and Li Y, Anal. Chem, 2015, 87, 569–577. [DOI] [PubMed] [Google Scholar]
- 94.Gao F, Liu F, Zheng J, Zeng M and Jiang Y, Anal. Sci, 2015, 31, 815–822. [DOI] [PubMed] [Google Scholar]
- 95.Liu M, Chang D and Li Y, Acc. Chem. Res, 2017, 50, 2273–2283. [DOI] [PubMed] [Google Scholar]
- 96.Aguirre SD, Ali MM, Salena BJ and Li Y, Biomolecules, 2013, 3, 563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kang DK, Ali MM, Zhang K, Huang SS, Peterson E, Digman MA, Gratton E and Zhao W, Nat. Commun, 2014, 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tang J and Breaker RR, Chem. Biol, 1997, 4, 453–459. [DOI] [PubMed] [Google Scholar]
- 99.Breaker RR, Nature, 2004, 432, 838–845. [DOI] [PubMed] [Google Scholar]
- 100.Breaker RR, Curr. Opin. Biotechnol, 2002, 13, 31–39. [DOI] [PubMed] [Google Scholar]
- 101.Levy M and Ellington AD, Chem. Biol, 2002, 9, 417–426. [DOI] [PubMed] [Google Scholar]
- 102.Alila KO and Baum DA, Chem. Commun, 2011, 47, 3227. [DOI] [PubMed] [Google Scholar]
- 103.Mei SHJ, Liu Z, Brennan JD and Li Y, J. Am. Chem. Soc, 2003, 125, 412–420. [DOI] [PubMed] [Google Scholar]
- 104.Yu T, Zhou W and Liu J, ChemBioChem, 2018, 19, 31–36. [DOI] [PubMed] [Google Scholar]
- 105.Wang DY, Lai BHY and Sen D, J. Mol. Biol, 2002, 318, 33–43. [DOI] [PubMed] [Google Scholar]
- 106.Shen Y, Chiuman W, Brennan JD and Li Y, ChemBioChem, 2006, 7, 1343–1348. [DOI] [PubMed] [Google Scholar]
- 107.Achenbach JC, Nutiu R and Li Y, Anal. Chim. Acta, 2005, 534, 41–51. [Google Scholar]
- 108.Chiuman W and Li Y, PLoS One, 2007, 2, e1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tram K, Xia J, Gysbers R and Li Y, PLoS One, 2015, 10, e0126402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yan Y, Ma C, Tang Z, Chen M and Zhao H, Anal. Chim. Acta, 2020, 1104, 172–179. [DOI] [PubMed] [Google Scholar]
- 111.Yun W, Li N, Wang R, Yang L, Chen L and Tang Y, Anal. Chim. Acta, 2019, 1064, 104–111. [DOI] [PubMed] [Google Scholar]
- 112.Wang DY and Sen D, J. Mol. Biol, 2001, 310, 723–734. [DOI] [PubMed] [Google Scholar]
- 113.Wang DY, Nucleic Acids Res, 2002, 30, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stojanovic MN, de Prada P and Landry DW, ChemBioChem, 2001, 2, 411–415. [DOI] [PubMed] [Google Scholar]
- 115.Stojanovic MN, Mitchell TE and Stefanovic D, J. Am. Chem. Soc, 2002, 124, 3555–3561. [DOI] [PubMed] [Google Scholar]
- 116.Pei R, Macdonald J and Stojanovic MN, Methods Mol. Biol, 2012, 848, 419–437. [DOI] [PubMed] [Google Scholar]
- 117.Stojanovic MN, Stefanovic D and Rudchenko S, Acc. Chem. Res, 2014, 47, 1845–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Orbach R, Willner B and Willner I, Chem. Commun, 2015, 51, 4144–4160. [DOI] [PubMed] [Google Scholar]
- 119.Arredondo D, Lakin MR, Stefanovic D and Stojanovic MN, DNA- and RNA-Based Computing Systems, Wiley, 2021, pp. 293–306. [Google Scholar]
- 120.Peng H, Newbigging AM, Wang Z, Tao J, Deng W, Le XC and Zhang H, Anal. Chem, 2018, 90, 190–207. [DOI] [PubMed] [Google Scholar]
- 121.Ida J, Chan S, Glökler J, Lim Y, Choong Y and Lim T, Molecules, 2019, 24, 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lin X, Sun X, Luo S, Liu B and Yang C, TrAC, Trends Anal. Chem, 2016, 80, 132–148. [Google Scholar]
- 123.Chiorcea-Paquim A-M and Oliveira-Brett A, Chemosensors, 2016, 4, 13. [Google Scholar]
- 124.Funabashi H, Electrochemistry, 2016, 84, 290–295. [Google Scholar]
- 125.Liu Z-L, Tao C-A and Wang J-F, Chin. J. Anal. Chem, 2020, 48, 153–163. [Google Scholar]
- 126.Mishra GK, Sharma V and Mishra RK, Biosensors, 2018, 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Malik LA, Bashir A, Qureashi A and Pandith AH, Environ. Chem. Lett, 2019, 17, 1495–1521. [Google Scholar]
- 128.Lu Y, Angew. Chem., Int. Ed, 2006, 45, 5588–5601. [DOI] [PubMed] [Google Scholar]
- 129.Dalmieda J and Kruse P, Sensors, 2019, 19, 5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McGhee CE, Loh KY and Lu Y, Curr. Opin. Biotechnol, 2017, 45, 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhou W, Saran R, Huang P-JJ, Ding J and Liu J, ChemBioChem, 2017, 18, 518–522. [DOI] [PubMed] [Google Scholar]
- 132.Wang J, Wang X, Tang H, He S, Gao Z, Niu R, Zheng Y and Han S, Sens. Actuators, B, 2018, 272, 146–150. [Google Scholar]
- 133.Huang Y, Li L, Zhang Y, Zhang L, Ge S and Yu J, Biosens. Bioelectron, 2019, 126, 339–345. [DOI] [PubMed] [Google Scholar]
- 134.Zhou W, Ding J and Liu J, Nucleic Acids Res, 2016, 44, 10377–10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhu L, Li G, Shao X, Huang K, Luo Y and Xu W, Microchim. Acta, 2020, 187, 26. [DOI] [PubMed] [Google Scholar]
- 136.Carmi N, Shultz LA and Breaker RR, Chem. Biol, 1996, 3, 1039–1046. [DOI] [PubMed] [Google Scholar]
- 137.Yun W, Cai D, Jiang J, Zhao P, Huang Y and Sang G, Biosens. Bioelectron, 2016, 80, 187–193. [DOI] [PubMed] [Google Scholar]
- 138.Rong M, Li J, Hu J, Chen A, Wu W and Lyu J, J. Chem. Technol. Biotechnol, 2018, 93, 3254–3263. [Google Scholar]
- 139.Song X, Wang Y, Liu S, Zhang X, Wang J, Wang H, Zhang F, Yu J and Huang J, Microchim. Acta, 2019, 186, 559. [DOI] [PubMed] [Google Scholar]
- 140.Huang Z, Chen J, Luo Z, Wang X and Duan Y, Anal. Chem, 2019, 91, 4806–4813. [DOI] [PubMed] [Google Scholar]
- 141.Wang XY, Niu CG, Guo LJ, Hu LY, Wu SQ, Zeng GM and Li F, J. Fluoresc, 2017, 27, 643–649. [DOI] [PubMed] [Google Scholar]
- 142.Chen J, Pan J and Chen S, Chem. Commun, 2017, 53, 10224–10227. [DOI] [PubMed] [Google Scholar]
- 143.Yun W, Wu H, Liu X, Zhong H, Fu M, Yang L and Huang Y, Sens. Actuators, B, 2018, 255, 1920–1926. [Google Scholar]
- 144.Pan J, Li Q, Zhou D and Chen J, New J. Chem, 2019, 43, 5857–5862. [Google Scholar]
- 145.Zhang T, Liu C, Zhou W, Jiang K, Yin C, Liu C, Zhang Z and Li H, J. Anal. Methods Chem, 2019, 2019, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang H-B, Ma L-H, Fang B-Y, Zhao Y-D and Hu X-B, Colloids Surf., B, 2018, 169, 305–312. [DOI] [PubMed] [Google Scholar]
- 147.Fu T, Ren S, Gong L, Meng H, Cui L, Kong R-M, Zhang X-B and Tan W, Talanta, 2016, 147, 302–306. [DOI] [PubMed] [Google Scholar]
- 148.Fu L, Lu Q, Liu X, Chen X, Wu X and Xie S, Talanta, 2020, 213, 120815. [DOI] [PubMed] [Google Scholar]
- 149.Li D, Yuan X, Li C, Luo Y and Jiang Z, J. Lumin, 2020, 221, 117056. [Google Scholar]
- 150.Chen J, Zhang Y, Cheng M, Mergny J-L, Lin Q, Zhou J and Ju H, Microchim. Acta, 2019, 186, 786. [DOI] [PubMed] [Google Scholar]
- 151.Peng D, Li Y, Huang Z, Liang RP, Qiu JD and Liu J, Anal. Chem, 2019, 91, 11403–11408. [DOI] [PubMed] [Google Scholar]
- 152.Li C, Zhao Z, Liu Y, Lv L, Qi B, Lin H, He L and Sun S, Surf. Rev. Lett, 2018, 25, 1–5. [Google Scholar]
- 153.Wu J, Lu Y, Ren N, Jia M, Wang R and Zhang J, Sensors, 2019, 19, 2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ni J, Zhang H, Chen Y, Luo F, Wang J, Guo L, Qiu B and Lin Z, Sens. Actuators, B, 2019, 289, 78–84. [Google Scholar]
- 155.Cai W, Xie S, Zhang J, Tang D and Tang Y, Biosens. Bioelectron, 2018, 117, 312–318. [DOI] [PubMed] [Google Scholar]
- 156.Huang X, Li J, Zhang Q, Chen S, Xu W, Wu J, Niu W, Xue J and Li C, J. Electroanal. Chem, 2018, 816, 75–82. [Google Scholar]
- 157.Liao X, Luo J, Wu J, Fan T, Yao Y, Gao F and Qian Y, J. Electroanal. Chem, 2018, 829, 129–137. [Google Scholar]
- 158.Ma R-N, Wang L-L, Zhang M, Jia L-P, Zhang W, Shang L, Jia W-L and Wang H-S, Sens. Actuators, B, 2018, 257, 678–684. [Google Scholar]
- 159.Wang L, Wen Y, Li L, Yang X, Jia N, Li W, Meng J, Duan M, Sun X and Liu G, Biosens. Bioelectron, 2018, 115, 91–96. [DOI] [PubMed] [Google Scholar]
- 160.Yu Y, Yu C, Niu Y, Chen J, Zhao Y, Zhang Y, Gao R and He J, Biosens. Bioelectron, 2018, 101, 297–303. [DOI] [PubMed] [Google Scholar]
- 161.Tan Y, Qiu J, Cui M, Wei X, Zhao M, Qiu B and Chen G, Analyst, 2016, 141, 1121–1126. [DOI] [PubMed] [Google Scholar]
- 162.Zhang C, Lai C, Zeng G, Huang D, Tang L, Yang C, Zhou Y, Qin L and Cheng M, Biosens. Bioelectron, 2016, 81, 61–67. [DOI] [PubMed] [Google Scholar]
- 163.Zhou Y, Tang L, Zeng G, Zhang C, Xie X, Liu Y, Wang J, Tang J, Zhang Y and Deng Y, Talanta, 2016, 146, 641–647. [DOI] [PubMed] [Google Scholar]
- 164.Xue S, Jing P and Xu W, Biosens. Bioelectron, 2016, 86, 958–965. [DOI] [PubMed] [Google Scholar]
- 165.Liu S, Wei W, Sun X and Wang L, Biosens. Bioelectron, 2016, 83, 33–38. [DOI] [PubMed] [Google Scholar]
- 166.Zeng G, Zhu Y, Zhang Y, Zhang C, Tang L, Guo P, Zhang L, Yuan Y, Cheng M and Yang C, Environ. Sci.: Nano, 2016, 3, 1504–1509. [Google Scholar]
- 167.Guo X, Li M, Zhao R, Yang Y, Wang R, Wu F, Jia L, Zhang Y, Wang L, Qu Z, Wang F, Zhu Y, Hao R, Zhang X and Song H, Nanomedicine, 2019, 21, 102035. [DOI] [PubMed] [Google Scholar]
- 168.Ji R, Niu W, Chen S, Xu W, Ji X, Yuan L, Zhao H, Geng M, Qiu J and Li C, Biosens. Bioelectron, 2019, 144, 111560. [DOI] [PubMed] [Google Scholar]
- 169.Lai C, Zhang Y, Liu X, Liu S, Li B, Zhang M, Qin L, Yi H, Li M, Li L, Fu Y, He J and Chen L, Anal. Bioanal. Chem, 2019, 411, 7499–7509. [DOI] [PubMed] [Google Scholar]
- 170.Yu Z, Li N, Hu X, Dong Y, Lin Y, Cai H, Xie Z, Qu D and Li X, Synth. Met, 2019, 254, 164–171. [Google Scholar]
- 171.Xiao S, Chen L, Xiong X, Zhang Q, Feng J, Deng S and Zhou L, J. Electroanal. Chem, 2018, 827, 175–180. [Google Scholar]
- 172.Xu J, Zhang Y, Li L, Kong Q, Zhang L, Ge S and Yu J, ACS Appl. Mater. Interfaces, 2018, 10, 3431–3440. [DOI] [PubMed] [Google Scholar]
- 173.Zhou Y, Zhang J, Tang L, Peng B, Zeng G, Luo L, Gao J, Pang Y, Deng Y and Zhang F, Talanta, 2017, 165, 274–281. [DOI] [PubMed] [Google Scholar]
- 174.Skotadis E, Tsekenis G, Chatzipetrou M, Patsiouras L, Madianos L, Bousoulas P, Zergioti I and Tsoukalas D, Sens. Actuators, B, 2017, 239, 962–969. [Google Scholar]
- 175.Tian A, Liu Y and Gao J, Talanta, 2017, 171, 185–189. [DOI] [PubMed] [Google Scholar]
- 176.Zhang Y, Zhang L, Kong Q, Ge S, Yan M and Yu J, Anal. Bioanal. Chem, 2016, 408, 7181–7191. [DOI] [PubMed] [Google Scholar]
- 177.Jiang C, Li Y, Wang H, Chen D and Wen Y, Sens. Actuators, B, 2020, 307, 127625. [Google Scholar]
- 178.Wang G, Chu LT, Hartanto H, Utomo WB, Pravasta RA and Chen T-H, ACS Sens, 2020, 5, 19–23. [DOI] [PubMed] [Google Scholar]
- 179.Zhang Y, Xu J, Zhou S, Zhu L, Lv X, Zhang J, Zhang L, Zhu P and Yu J, Anal. Chem, 2020, 92, 3874–3881. [DOI] [PubMed] [Google Scholar]
- 180.Huang Y, Lin C, Luo F, Qiu B, Guo L, Lin Z and Chen G, ACS Sens, 2019, 4, 2465–2470. [DOI] [PubMed] [Google Scholar]
- 181.Du XL, Kang TF, Lu LP and Cheng SY, Anal. Methods, 2018, 10, 51–58. [Google Scholar]
- 182.Zhang Q, Cui H, Xiong X, Chen J, Wang Y, Shen J, Luo Y and Chen L, Analyst, 2018, 143, 549–554. [DOI] [PubMed] [Google Scholar]
- 183.Bin Liang W, Zhuo Y, Zheng YN, Xiong CY, Chai YQ and Yuan R, ACS Appl. Mater. Interfaces, 2017, 9, 39812–39820. [DOI] [PubMed] [Google Scholar]
- 184.Ren W, Zhang Y, Fan YZ, Dong JX, Li NB and Luo HQ, J. Hazard. Mater, 2017, 336, 195–201. [DOI] [PubMed] [Google Scholar]
- 185.Jia Y and Li F, Anal. Chem, 2018, 90, 5153–5161. [DOI] [PubMed] [Google Scholar]
- 186.Zhou W, Saran R and Liu J, Chem. Rev, 2017, 117, 8272–8325. [DOI] [PubMed] [Google Scholar]
- 187.Liang G, Man Y, Li A, Jin X, Liu X and Pan L, Micro-chem. J, 2017, 131, 145–153. [Google Scholar]
- 188.Jiao L, Zhong N, Zhao X, Ma S, Fu X and Dong D, TrAC, Trends Anal. Chem, 2020, 127, 115892. [Google Scholar]
- 189.Wang H, Wang H, Willner I and Wang F, Top. Curr. Chem, 2020, 378, 20. [DOI] [PubMed] [Google Scholar]
- 190.Willner I, Shlyahovsky B, Zayats M and Willner B, Chem. Soc. Rev, 2008, 37, 1153. [DOI] [PubMed] [Google Scholar]
- 191.Breaker RR and Joyce GF, Chem. Biol, 1995, 2, 655–660. [DOI] [PubMed] [Google Scholar]
- 192.Deng P, Zheng S, Yun W, Zhang W and Yang L, Spectrochim. Acta, Part A, 2019, 210, 335–340. [DOI] [PubMed] [Google Scholar]
- 193.Cai W, Xie S, Zhang J, Tang D and Tang Y, Biosens. Bioelectron, 2017, 98, 466–472. [DOI] [PubMed] [Google Scholar]
- 194.Li Y, Chang Y, Ma J, Wu Z, Yuan R and Chai Y, Anal. Chem, 2019, 91, 6127–6133. [DOI] [PubMed] [Google Scholar]
- 195.Yun W, Zhong H, Zheng S, Wang R and Yang L, Sens. Actuators, B, 2018, 277, 456–461. [Google Scholar]
- 196.Huang C, Fan X, Yuan Q, Zhang X, Hou X and Wu P, Talanta, 2018, 185, 258–263. [DOI] [PubMed] [Google Scholar]
- 197.Zhang H, Cheng X, Chen L, Mo F, Xu L and Fu F, Anal. Chim. Acta, 2017, 956, 63–69. [DOI] [PubMed] [Google Scholar]
- 198.Feng M, Gu C, Sun Y, Zhang S, Tong A and Xiang Y, Anal. Chem, 2019, 91, 6608–6615. [DOI] [PubMed] [Google Scholar]
- 199.He X, Wang S, Liu Y and Wang X, Sci. China: Chem, 2019, 62, 1064–1071. [Google Scholar]
- 200.Yu T, Zhou W and Liu J, Anal. Methods, 2018, 10, 1740–1746. [Google Scholar]
- 201.Park Y, Lee CY, Park KS and Park HG, Chem. – Eur. J, 2017, 23, 17379–17383. [DOI] [PubMed] [Google Scholar]
- 202.Xu W, Tian J, Luo Y, Zhu L and Huang K, Sci. Rep, 2017, 7, 43362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhu P, Shang Y, Tian W, Huang K, Luo Y and Xu W, Food Chem, 2017, 221, 1770–1777. [DOI] [PubMed] [Google Scholar]
- 204.Wu J, Yu Y, Wei S, Xue B and Zhang J, Int. J. Electrochem. Sci, 2017, 12, 11666–11676. [Google Scholar]
- 205.Qing M, Xie S, Cai W, Tang D, Tang Y, Zhang J and Yuan R, Anal. Chem, 2018, 90, 11439–11445. [DOI] [PubMed] [Google Scholar]
- 206.Xu S, Dai B, Xu J, Jiang L and Huang H, Electroanalysis, 2019, 31, 2330–2338. [Google Scholar]
- 207.Deng Y, Chen Y and Zhou X, Acta Chim. Slov, 2018, 65, 271–277. [DOI] [PubMed] [Google Scholar]
- 208.Huang PJJ, Vazin M, Lin JJ, Pautler R and Liu J, ACS Sens, 2016, 1, 732–738. [Google Scholar]
- 209.Liang H, Xie S, Cui L, Wu C and Zhang X, Anal. Methods, 2016, 8, 7260–7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Huang P-JJ, Lin J, Cao J, Vazin M and Liu J, Anal. Chem, 2014, 86, 1816–1821. [DOI] [PubMed] [Google Scholar]
- 211.Huang P-JJ, Vazin M, Matuszek Z and Liu J, Nucleic Acids Res, 2015, 43, 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Huang P-JJ, Vazin M and Liu J, Anal. Chem, 2014, 86, 9993–9999. [DOI] [PubMed] [Google Scholar]
- 213.Huang P-JJ, Vazin M and Liu J, Biochemistry, 2016, 55, 2518–2525. [DOI] [PubMed] [Google Scholar]
- 214.Wu P, Hwang K, Lan T and Lu Y, J. Am. Chem. Soc, 2013, 135, 5254–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Meng H-M, Zhang X, Lv Y, Zhao Z, Wang N-N, Fu T, Fan H, Liang H, Qiu L, Zhu G and Tan W, ACS Nano, 2014, 8, 6171–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Qiu L, Zhang T, Jiang J, Wu C, Zhu G, You M, Chen X, Zhang L, Cui C, Yu R and Tan W, J. Am. Chem. Soc, 2014, 136, 13090–13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hwang K, Wu P, Kim T, Lei L, Tian S, Wang Y and Lu Y, Angew. Chem., Int. Ed, 2014, 53, 13798–13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Cui L, Peng R, Fu T, Zhang X, Wu C, Chen H, Liang H, Yang CJ and Tan W, Anal. Chem, 2016, 88, 1850–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yang Z, Loh KY, Chu Y-T, Feng R, Satyavolu NSR, Xiong M, Nakamata Huynh SM, Hwang K, Li L, Xing H, Zhang X, Chemla YR, Gruebele M and Lu Y, J. Am. Chem. Soc, 2018, 140, 17656–17665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hwang K, Mou Q, Lake RJ, Xiong M, Holland B and Lu Y, Inorg. Chem, 2019, 58, 13696–13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wei J, Wang H, Wu Q, Gong X, Ma K, Liu X and Wang F, Angew. Chem., Int. Ed, 2020, 59, 5965–5971. [DOI] [PubMed] [Google Scholar]
- 222.Peng H, Li XF, Zhang H and Le XC, Nat. Commun, 2017, 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang W, Satyavolu NSR, Wu Z, Zhang J-R, Zhu J-J and Lu Y, Angew. Chem., Int. Ed, 2017, 56, 6798–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Gao R, Xu L, Hao C, Xu C and Kuang H, Angew. Chem., Int. Ed, 2019, 58, 3913–3917. [DOI] [PubMed] [Google Scholar]
- 225.Bakshi SF, Guz N, Zakharchenko A, Deng H, Tumanov AV, Woodworth CD, Minko S, Kolpashchikov DM and Katz E, J. Am. Chem. Soc, 2017, 139, 12117–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Lin Y, Yang Z, Lake RJ, Zheng C and Lu Y, Angew. Chem., Int. Ed, 2019, 58, 17061–17067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Fan H, Zhao Z, Yan G, Zhang X, Yang C, Meng H, Chen Z, Liu H and Tan W, Angew. Chem., Int. Ed, 2015, 54, 4801–4805. [DOI] [PubMed] [Google Scholar]
- 228.Zhou W, Liang W, Li D, Yuan R and Xiang Y, Biosens. Bioelectron, 2016, 85, 573–579. [DOI] [PubMed] [Google Scholar]
- 229.Fu X, Ke G, Peng F, Hu X, Li J, Shi Y, Kong G, Zhang X-B and Tan W, Nat. Commun, 2020, 11, 1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Xing Z, Gao S, Han H, Zhang J, Chen X, Yang Y and Li Q, J. Controlled Release, 2015, 213, e146–e147. [DOI] [PubMed] [Google Scholar]
- 231.Wu Z, Fan H, Satyavolu NSR, Wang W, Lake R, Jiang J-H and Lu Y, Angew. Chem., Int. Ed, 2017, 56, 8721–8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Cui M-R, Li X-L, Xu J-J and Chen H-Y, ACS Appl. Mater. Interfaces, 2020, 12, 13005–13012. [DOI] [PubMed] [Google Scholar]
- 233.Xiong M, Yang Z, Lake RJ, Li J, Hong S, Fan H, Zhang X-B and Lu Y, Angew. Chem., Int. Ed, 2020, 59, 1891–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yang C, Yin X, Huan S-Y, Chen L, Hu X-X, Xiong M-Y, Chen K and Zhang X-B, Anal. Chem, 2018, 90, 3118–3123. [DOI] [PubMed] [Google Scholar]
- 235.Hao C, Xu L, Sun M, Zhang H, Kuang H and Xu C, Chem. – Eur. J, 2019, 25, 12235–12240. [DOI] [PubMed] [Google Scholar]
- 236.Zhang J, Smaga LP, Satyavolu NSR, Chan J and Lu Y, J. Am. Chem. Soc, 2017, 139, 17225–17228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Xu W, Xing H and Lu Y, Analyst, 2013, 138, 6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Han Q, Mo F, Wu J, Wang C, Chen M and Fu Y, Sens. Actuators, B, 2020, 302, 127191. [Google Scholar]
- 239.He J-L, Zhang Y, Yang C, Huang S-Y, Wu L, Mei T-T, Wang J and Cao Z, Anal. Methods, 2019, 11, 2204–2210. [Google Scholar]
- 240.Yang Y, Huang J, Yang X, Quan K, Wang H, Ying L, Xie N, Ou M and Wang K, Anal. Chem, 2016, 88, 5981–5987. [DOI] [PubMed] [Google Scholar]
- 241.Qu L, Ali MM, Aguirre SD, Liu H, Jiang Y and Li Y, PLoS One, 2014, 9, e115640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zaouri N, Cui Z, Peinetti AS, Lu Y and Hong P-Y, Environ. Sci.: Water Res. Technol, 2019, 5, 2260–2268. [Google Scholar]
- 243.Liu M, Zhang Q, Brennan JD and Li Y, MRS Commun, 2018, 8, 687–694. [Google Scholar]
- 244.Zheng L, Qi P and Zhang D, Sens. Actuators, B, 2018, 276, 42–47. [Google Scholar]
- 245.Edberg SC, Rice EW, Karlin RJ and Allen MJ, J. Appl. Microbiol, 2000, 88, 106S–116S. [DOI] [PubMed] [Google Scholar]
- 246.Yan L, Zhou J, Zheng Y, Gamson AS, Roembke BT, Nakayama S and Sintim HO, Mol. BioSyst, 2014, 10, 970. [DOI] [PubMed] [Google Scholar]
- 247.Zhao Y, Chen F, Li Q, Wang L and Fan C, Chem. Rev, 2015, 115, 12491–12545. [DOI] [PubMed] [Google Scholar]
- 248.Obande GA and Banga Singh KK, Infect. Drug Resist, 2020, 13, 455–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Mayboroda O, Katakis I and O’Sullivan CK, Anal. Biochem, 2018, 545, 20–30. [DOI] [PubMed] [Google Scholar]
- 250.Bodulev OL and Sakharov IY, Biochem, 2020, 85, 147–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Craw P and Balachandran W, Lab Chip, 2012, 12, 2469. [DOI] [PubMed] [Google Scholar]
- 252.Liu M, Zhang Q, Li Z, Gu J, Brennan JD and Li Y, Nat. Commun, 2016, 7, 12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Liu M, Zhang Q, Chang D, Gu J, Brennan JD and Li Y, Angew. Chem., Int. Ed, 2017, 56, 6142–6146. [DOI] [PubMed] [Google Scholar]
- 254.Yu F, Li Y, Li M, Tang L and He JJ, Biosens. Bioelectron, 2017, 89, 880–885. [DOI] [PubMed] [Google Scholar]
- 255.Zhou Z, Brennan JD and Li Y, Angew. Chem., Int. Ed, 2020, 59, 10401–10405. [DOI] [PubMed] [Google Scholar]
- 256.Carmi N, Balkhi SR and Breaker RR, Proc. Natl. Acad. Sci. U. S. A, 1998, 95, 2233–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Yue G, Huang D, Luo F, Guo L, Qiu B, Lin Z and Chen G, J. Lumin, 2019, 207, 369–373. [Google Scholar]
- 258.Li S, Liu H, Yang G, Liu S, Liu R and Lv C, J. Environ. Radioact, 2018, 195, 60–66. [DOI] [PubMed] [Google Scholar]
- 259.Liu H, Chen Y, Song C, Tian G, Li S, Yang G and Lv C, Anal. Bioanal. Chem, 2018, 410, 4227–4234. [DOI] [PubMed] [Google Scholar]
- 260.Yang G, Song C, Shi Q, Liu H, Li S, Liu R, Liu S and Lv C, J. Pharm. Biomed. Anal, 2018, 159, 459–465. [DOI] [PubMed] [Google Scholar]
- 261.Khan M, Rao M and Li Q, Sensors, 2019, 19, 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Gribas AV and Sakharov IY, Anal. Lett, 2018, 51, 1280–1290. [Google Scholar]
- 263.Li C, Dai P, Rao X, Shao L, Cheng G, He P and Fang Y, Talanta, 2015, 132, 463–468. [DOI] [PubMed] [Google Scholar]
- 264.Ren W, Zhang Y, Huang WT, Li NB and Luo HQ, Biosens. Bioelectron, 2015, 68, 266–271. [DOI] [PubMed] [Google Scholar]
- 265.Lim JW, Kim T-Y, Choi S-W and Woo M-A, Food Chem, 2019, 300, 125177. [DOI] [PubMed] [Google Scholar]
- 266.Xie M, Zhang K, Zhu F, Wu H and Zou P, RSC Adv, 2017, 7, 50420–50424. [Google Scholar]
- 267.Zhang B, Meng H, Wang X, Li J, Chang H and Wei W, Sens. Actuators, B, 2018, 255, 2531–2537. [Google Scholar]
- 268.Gu P, Zhang G, Deng Z, Tang Z, Zhang H, Khusbu FY, Wu K, Chen M and Ma C, Spectrochim. Acta, Part A, 2018, 203, 195–200. [DOI] [PubMed] [Google Scholar]
- 269.Shen R, Zou L, Wu S, Li T, Wang J, Liu J and Ling L, Spectrochim. Acta, Part A, 2019, 213, 42–47. [DOI] [PubMed] [Google Scholar]
- 270.Xu X, Ng SM, Hassouna E, Warrington A, Oh S-H and Rodriguez M, Future Neurol, 2015, 10, 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Li C, Ma J, Shi H, Hu X, Xiang Y, Li Y and Li G, Anal. Chim. Acta, 2018, 1041, 102–107. [DOI] [PubMed] [Google Scholar]
- 272.Chen J, Zuehlke A, Deng B, Peng H, Hou X and Zhang H, Anal. Chem, 2017, 89, 12888–12895. [DOI] [PubMed] [Google Scholar]
- 273.Wang W, Shu M, Nie A and Han H, Sens. Actuators, B, 2020, 304, 127380. [Google Scholar]
- 274.Ming J, Jiang TF, Wang YH and Lv ZH, Sens. Actuators, B, 2016, 228, 605–611. [Google Scholar]
- 275.Ming J, Jiang TF, Wang YH and Lv ZH, Sens. Actuators, B, 2017, 252, 450–457. [Google Scholar]
- 276.Kong XJ, Wu S, Cen Y, Yu RQ and Chu X, Biosens. Bioelectron, 2016, 79, 679–684. [DOI] [PubMed] [Google Scholar]
- 277.Zou L, Li X, Li T and Ling L, Sens. Actuators, B, 2018, 267, 272–278. [Google Scholar]
- 278.He JL, Zhang Y, Mei TT, Tang L, Huang SY and Cao Z, Biosens. Bioelectron, 2019, 144, 111692. [DOI] [PubMed] [Google Scholar]
- 279.Xu L, Liu S, Yang T, Shen Y, Zhang Y, Huang L, Zhang L, Ding S, Song F and Cheng W, Theranostics, 2019, 9, 1993–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wang F, Freage L, Orbach R and Willner I, Anal. Chem, 2013, 85, 8196–8203. [DOI] [PubMed] [Google Scholar]
- 281.Wang F, Elbaz J, Teller C and Willner I, Angew. Chem., Int. Ed, 2011, 50, 295–299. [DOI] [PubMed] [Google Scholar]
- 282.Wang F, Elbaz J, Orbach R, Magen N and Willner I, J. Am. Chem. Soc, 2011, 133, 17149–17151. [DOI] [PubMed] [Google Scholar]
- 283.Lu C-H, Wang F and Willner I, J. Am. Chem. Soc, 2012, 134, 10651–10658. [DOI] [PubMed] [Google Scholar]
- 284.Elbaz J, Moshe M, Shlyahovsky B and Willner I, Chem. – Eur. J, 2009, 15, 3411–3418. [DOI] [PubMed] [Google Scholar]
- 285.Freage L, Wang F, Orbach R and Willner I, Anal. Chem, 2014, 86, 11326–11333. [DOI] [PubMed] [Google Scholar]
- 286.Wang H, He D, Wu R, Cheng H, Ma W, Huang J, Bu H, He X and Wang K, Analyst, 2019, 144, 143–147. [DOI] [PubMed] [Google Scholar]
- 287.Liu X, Zhou X, Xia X and Xiang H, Anal. Chim. Acta, 2020, 1096, 159–165. [DOI] [PubMed] [Google Scholar]
- 288.Darius AKL, Ling NJ and Mahesh U, Mol. BioSyst, 2010, 6, 792. [DOI] [PubMed] [Google Scholar]
- 289.Deng M, Zhang D, Zhou Y and Zhou X, J. Am. Chem. Soc, 2008, 130, 13095–13102. [DOI] [PubMed] [Google Scholar]
- 290.Jiang X, Zhang H, Wu J, Yang X, Shao J, Lu Y, Qiu B, Lin Z and Chen G, Talanta, 2014, 128, 445–449. [DOI] [PubMed] [Google Scholar]
- 291.Shahbazi N, Hosseinkhani S, Khajeh K and Ranjbar B, Biopolymers, 2017, 107, 1–9. [DOI] [PubMed] [Google Scholar]
- 292.Xiang L, Zhang F, Chen C and Cai C, Microchim. Acta, 2019, 186, 511. [DOI] [PubMed] [Google Scholar]
- 293.Dhar BC, Reed AJ, Mitra S, Rodriguez Sanchez P, Nedorezova DD, Connelly RP, Rohde KH and Gerasimova YV, Biosens. Bioelectron, 2020, 165, 112385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Abdou Mohamed MA, Kozlowski HN, Kim J, Zagorovsky K, Kantor M, Feld JJ, Mubareka S, Mazzulli T and Chan WCW, ACS Nano, 2021, 15, 9379–9390. [DOI] [PubMed] [Google Scholar]
- 295.Anantharaj A, Das SJ, Sharanabasava P, Lodha R, Kabra SK, Sharma TK and Medigeshi GR, Front. Mol. Biosci, 2020, 7, 586254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Chen K, Huang Q, Fu T, Ke G, Zhao Z, Zhang X and Tan W, Anal. Chem, 2020, 92, 7404–7408. [DOI] [PubMed] [Google Scholar]
- 297.Mahato K, Srivastava A and Chandra P, Biosens. Bioelectron, 2017, 96, 246–259. [DOI] [PubMed] [Google Scholar]
- 298.Kosack CS, Page A-LL and Klatser PR, Bull. W. H. O, 2017, 95, 639–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Liu J and Lu Y, Angew. Chem., Int. Ed, 2007, 46, 7587–7590. [DOI] [PubMed] [Google Scholar]
- 300.He X, Zhou X, Liu Y and Wang X, Sens. Actuators, B, 2020, 311, 127676. [Google Scholar]
- 301.Zhang J, Xing H and Lu Y, Chem. Sci, 2018, 9, 3906–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Lisi F, Peterson JR and Gooding JJ, Biosens. Bioelectron, 2020, 148, 111835. [DOI] [PubMed] [Google Scholar]
- 303.Zhang J, Lan T and Lu Y, TrAC, Trends Anal. Chem, 2020, 124, 115782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Zhang J, Tang Y, Teng L, Lu M and Tang D, Biosens. Bioelectron, 2015, 68, 232–238. [DOI] [PubMed] [Google Scholar]
- 305.Liao J-Y and Li H, Chem. Lett, 2014, 43, 1599–1600. [Google Scholar]
- 306.Fu L, Zhuang J, Lai W, Que X, Lu M and Tang D, J. Mater. Chem. B, 2013, 1, 6123. [DOI] [PubMed] [Google Scholar]
- 307.Xiang Y and Lu Y, Chem. Commun, 2013, 49, 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Huang S, Wang W, Cheng F, Yao H and Zhu J-J, Sens. Actuators, B, 2017, 242, 347–354. [Google Scholar]
- 309.Ming J, Fan W, Jiang T-F, Wang Y-H and Lv Z-H, Sens. Actuators, B, 2017, 240, 1091–1098. [Google Scholar]
- 310.Zeng L, Gong J, Rong P, Liu C and Chen J, Talanta, 2019, 198, 412–416. [DOI] [PubMed] [Google Scholar]
- 311.Zhang J and Lu Y, Angew. Chem., Int. Ed, 2018, 57, 9702–9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Zhang J, Xiang Y, Wang M, Basu A and Lu Y, Angew. Chem., Int. Ed, 2016, 55, 732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Xiang Y and Lu Y, Anal. Chem, 2012, 84, 4174–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Huang H, Zhao G and Dou W, Biosens. Bioelectron, 2018, 107, 266–271. [DOI] [PubMed] [Google Scholar]
- 315.Xiang Y and Lu Y, Anal. Chem, 2012, 84, 1975–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Yang X, Shi D, Zhu S, Wang B, Zhang X and Wang G, ACS Sens, 2018, 3, 1368–1375. [DOI] [PubMed] [Google Scholar]
- 317.Si Y, Li L, Wang N, Zheng J, Yang R and Li J, ACS Appl. Mater. Interfaces, 2019, 11, 7792–7799. [DOI] [PubMed] [Google Scholar]
- 318.Mak WC, Beni V and Turner APF, TrAC, Trends Anal. Chem, 2016, 79, 297–305. [Google Scholar]
- 319.Liu Y, Zhan L, Qin Z, Sackrison J and Bischof JC, ACS Nano, 2021, 15, 3593–3611. [DOI] [PubMed] [Google Scholar]
- 320.Chen A and Yang S, Biosens. Bioelectron, 2015, 71, 230–242. [DOI] [PubMed] [Google Scholar]
- 321.Fang Z, Huang J, Lie P, Xiao Z, Ouyang C, Wu Q, Wu Y, Liu G and Zeng L, Chem. Commun, 2010, 46, 9043–9045. [DOI] [PubMed] [Google Scholar]
- 322.Chen J, Zhou X and Zeng L, Chem. Commun, 2013, 49, 984–986. [DOI] [PubMed] [Google Scholar]
- 323.Martinez AW, Phillips ST, Whitesides GM and Carrilho E, Anal. Chem, 2010, 82, 3–10. [DOI] [PubMed] [Google Scholar]
- 324.Li X, Ballerini DR and Shen W, Biomicrofluidics, 2012, 6, 011301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Yetisen AK, Akram MS and Lowe CR, Lab Chip, 2013, 13, 2210. [DOI] [PubMed] [Google Scholar]
- 326.Parolo C and Merkoçi A, Chem. Soc. Rev, 2013, 42, 450–457. [DOI] [PubMed] [Google Scholar]
- 327.Liana DD, Raguse B, Gooding JJ and Chow E, Sensors, 2012, 12, 11505–11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Xia Y, Si J and Li Z, Biosens. Bioelectron, 2016, 77, 774–789. [DOI] [PubMed] [Google Scholar]
- 329.Fu L-M and Wang Y-N, TrAC, Trends Anal. Chem, 2018, 107, 196–211. [Google Scholar]
- 330.Meredith NA, Quinn C, Cate DM, Reilly TH, Volckens J and Henry CS, Analyst, 2016, 141, 1874–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Kung C-T, Hou C-Y, Wang Y-N and Fu L-M, Sens. Actuators, B, 2019, 301, 126855. [Google Scholar]
- 332.Sher M, Zhuang R, Demirci U and Asghar W, Expert Rev. Mol. Diagn, 2017, 17, 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Hu J, Wang S, Wang L, Li F, Pingguan-Murphy B, Lu TJ and Xu F, Biosens. Bioelectron, 2014, 54, 585–597. [DOI] [PubMed] [Google Scholar]
- 334.Yamada K, Shibata H, Suzuki K and Citterio D, Lab Chip, 2017, 17, 1206–1249. [DOI] [PubMed] [Google Scholar]
- 335.Akyazi T, Basabe-Desmonts L and Benito-Lopez F, Anal. Chim. Acta, 2018, 1001, 1–17. [DOI] [PubMed] [Google Scholar]
- 336.Dong T, Wang GA and Li F, Anal. Bioanal. Chem, 2019, 411, 4401–4414. [DOI] [PubMed] [Google Scholar]
- 337.Liu X, Li X, Gao X, Ge L, Sun X and Li F, ACS Appl. Mater. Interfaces, 2019, 11, 15381–15388. [DOI] [PubMed] [Google Scholar]
- 338.Jahanshahi-Anbuhi S, Pennings K, Leung V, Liu M, Carrasquilla C, Kannan B, Li Y, Pelton R, Brennan JD and Filipe CDM, Angew. Chem., Int. Ed, 2014, 53, 6155–6158. [DOI] [PubMed] [Google Scholar]
- 339.Kannan B, Jahanshahi-Anbuhi S, Pelton RH, Li Y, Filipe CDM and Brennan JD, Anal. Chem, 2015, 87, 9288–9293. [DOI] [PubMed] [Google Scholar]
- 340.Jahanshahi-Anbuhi S, Kannan B, Leung V, Pennings K, Liu M, Carrasquilla C, White D, Li Y, Pelton RH, Brennan JD and Filipe CDM, Chem. Sci, 2016, 7, 2342–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Liu L, Yang D and Liu G, Biosens. Bioelectron, 2019, 136, 60–75. [DOI] [PubMed] [Google Scholar]
- 342.Mukama O, Nie C, de J, Habimana D, Meng X, Ting Y, Songwe F, Al Farga A, Mugisha S, Rwibasira P, Zhang Y and Zeng L, Anal. Biochem, 2020, 600, 113762. [DOI] [PubMed] [Google Scholar]
- 343.Liu M, Hui CY, Zhang Q, Gu J, Kannan B, Jahanshahi-Anbuhi S, Filipe CDM, Brennan JD and Li Y, Angew. Chem., Int. Ed, 2016, 55, 2709–2713. [DOI] [PubMed] [Google Scholar]
- 344.Liu M, Zhang Q, Kannan B, Botton GA, Yang J, Soleymani L, Brennan JD and Li Y, Angew. Chem., Int. Ed, 2018, 57, 12440–12443. [DOI] [PubMed] [Google Scholar]
- 345.Sun Y, Chang Y, Zhang Q and Liu M, Micromachines, 2019, 10, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Samani SE, Chang D, McConnell EM, Rothenbroker M, Filipe CDM and Li Y, ChemBioChem, 2020, 21, 632–637. [DOI] [PubMed] [Google Scholar]
- 347.Griesche C and Baeumner AJ, TrAC, Trends Anal. Chem, 2020, 128, 115906. [Google Scholar]
- 348.Yousefi H, Ali MM, Su HM, Filipe CDM and Didar TF, ACS Nano, 2018, 12, 3287–3294. [DOI] [PubMed] [Google Scholar]
- 349.Yousefi H, Su HM, Ali M, Filipe CDM and Didar TF, Adv. Mater. Interfaces, 2018, 5, 1–8. [Google Scholar]
- 350.USDA, Microbiological Testing of AMS Purchased Meat, Poultry and Egg Commodities, https://www.ams.usda.gov/resources/microbiological-testing, accessed 29 July 2020.
- 351.Emilsson GM, Nakamura S, Roth A and Breaker RR, RNA, 2003, 9, 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Ren A, Rajashankar KR and Patel DJ, Nature, 2012, 486, 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Jimmy Huang P-J, Yang J, Chong K, Ma Q, Li M, Zhang F, Moon WJ, Zhang G and Liu J, Chem. Sci, 2020, 11, 6795–6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Nagraj N, Liu J, Sterling S, Wu J and Lu Y, Chem. Commun, 2009, 4103. [DOI] [PMC free article] [PubMed] [Google Scholar]


