Abstract
The 4-methyl steranes serve as molecular fossils and are used for studying both eukaryotic evolution and geological history. The occurrence of 4α-methyl steranes in sediments has long been considered evidence of products of partial demethylation mediated by sterol methyl oxidases (SMOs), while 4β-methyl steranes are attributed entirely to diagenetic generation from 4α-methyl steroids since possible biological sources of their precursor 4β-methyl sterols are unknown. Here, we report a previously unknown C4-methyl sterol biosynthetic pathway involving a sterol methyltransferase rather than the SMOs. We show that both C4α- and C4β-methyl sterols are end products of the sterol biosynthetic pathway in an endosymbiont of reef corals, Breviolum minutum, while this mechanism exists not only in dinoflagellates but also in eukaryotes from alveolates, haptophytes, and aschelminthes. Our discovery provides a previously untapped route for the generation of C4-methyl steranes and overturns the paradigm that all 4β-methyl steranes are diagenetically generated from the 4α isomers. This may facilitate the interpretation of molecular fossils and understanding of the evolution of eukaryotic life in general.
Introduction
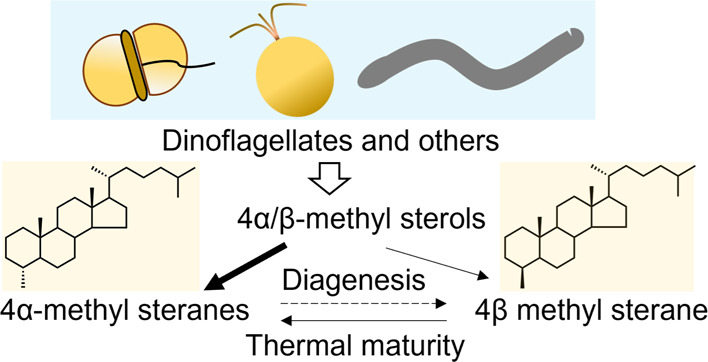
Sterols are essential eukaryotic lipids but are absent from most bacteria.1 Steranes retain the hydrocarbon skeletons of sterols and are stable in sedimentary rocks for long periods of time.2 Thus, they are well preserved as molecular fossils in ancient sediments and petroleum and are used for studying both eukaryotic evolution and geological history. 4-Methyl steranes occur widely in sediments3 and are important because their 4-methyl structures can be linked to specific biological inputs, thus enabling a more detailed interpretation of geological records.4 The 4-methylated steranes with an alpha configuration (i.e., with an equatorial methyl group) are assumed to be formed diagenetically from 4α-methyl sterols produced by ancient organisms, such as dinoflagellates.5 As key intermediates in sterol biosynthesis,6 4α-methyl sterols are derived from lanosterol or cycloartenol via sequential removal of the methyl groups at the C14 and C4β positions catalyzed by sterol methyl oxidases (SMOs)6 (Figure S1). In the case of lanosterol as the protosterol, 14-alpha-demethylase (EC 1.14.14.154) acts initially on lanosterol followed by removal of the 4β-methyl by an SMO1 (EC 1.14.18.10); the resulting 4α-methyl sterols may be converted into end products by the reaction catalyzed by an SMO2 (EC 1.14.18.11).7 Thus, 4α-methyl sterols may accumulate in natural systems3 through partial demethylation by blockage of SMO2 activity (Figure S1). However, this route has not yet been experimentally validated. Some bacteria are also known to produce 4α-methyl sterols exclusively via a sterol C4 demethylation mechanism that is distinct from that of eukaryotes.8 In contrast, the origin of 4β-sterenes is still under debate given the possibility that the organisms responsible for the biosynthesis of the parent sterols of diagenetically formed 4β-sterenes during ancient times no longer exist or have remained hitherto undetected.9 The conversion of stanols to sterenes in sediments via steroid ketones has long been recognized; thus, it is widely accepted that 4β-methyl steranes found in sediments are derived from 4α-methyl steroids during early diagenesis.10,11
Sterol surveys have indicated that 4α,23,24-trimethyl sterols are specific to dinoflagellates, which are regarded as major sources of 4α-methyl steranes in sediments.12,13 Dinoflagellates have left a rich sedimentary record in the form of fossil cysts that closely follows the record of dinosteranes in sediments as old as the early Cambrian (∼520 Ma).14 The symbiosis between dinoflagellate algae of the Symbiodiniaceae family and coral hosts is also very ancient and can be traced to a period from the Middle Ordovician to the late Permian (450–251 Ma).15 The dinoflagellate Breviolum minutum was first isolated from the Caribbean coral Montastraea faveolata, one of three modern species of the widely known generalist Montastraea annularis,16 which originated during the late Miocene (6.5–5.6 Ma).17 Modern species of M. annularis may date back to 2.9–3.5 Ma before the Plio-Pleistocene extinction event, in which approximately 80% of Caribbean reef coral species disappeared.18 These species ecologically dominate many modern reefs in the Caribbean region.19 Thus, they offer relatively continuous records through the Quaternary into the late Neogene and reliable biomarkers for evolutionary studies.
A clear understanding of the methylation mechanisms underlying the complex stereochemical consortia of A-ring methylated sterols in Symbiodiniaceae and the possible inputs of 4α/4β-methyl steranes from coral-relating algae is paramount, given that it would provide an experimentally validated biosynthetic pathway for the precursors of 4α-methyl steranes and prove the biological sources of 4β-methyl steranes. Moreover, these widespread reef-building corals occur at spatial and temporal scales that may influence the 4α/4β-methyl sterane ratio, thereby influencing the interpretation of geological history and the discovery of crude oil and gas.
A sterol A-ring methylase-1 (STRM-1)20 has been shown to catalyze methylation of the sterol nucleus at the C4 position in the nematode Caenorhabditis elegans,(21) which could not connect with fossils. As nematodes are sterol auxotrophic, they probably obtained the gene encoding STRM-1 through horizontal gene transfer, although its origin remains obscure. The results presented here demonstrate that coral dinoflagellates are biological sources for both stereoisomers in sediments, proceeding by a methylation pathway (catalyzed by an enzyme encoded by the STRM-1 ortholog), which is entirely distinct from the partial demethylation pathway mediated by SMOs. Moreover, the phylogeny of STRMs and molecular clock estimates revealed an early origin of BmSTRM-type enzymes (94.5 Ma) in widespread alveolates (including dinoflagellates) and haptophytes (comprising a major proportion of the globally distributed phytoplankton community and exerting large-scale impacts on ocean biogeochemistry22). These findings question the generality of the catalytic mechanism and ubiquitous biogenesis of 4β-methyl sterols and refute the current paradigm that all 4β-methyl steranes are diagenetically generated from the 4α isomers.
Results
In Vivo Substrate Feeding Reveals a Previously Unknown Mechanism of C4-Methyl Sterol Biosynthesis in the Dinoflagellate
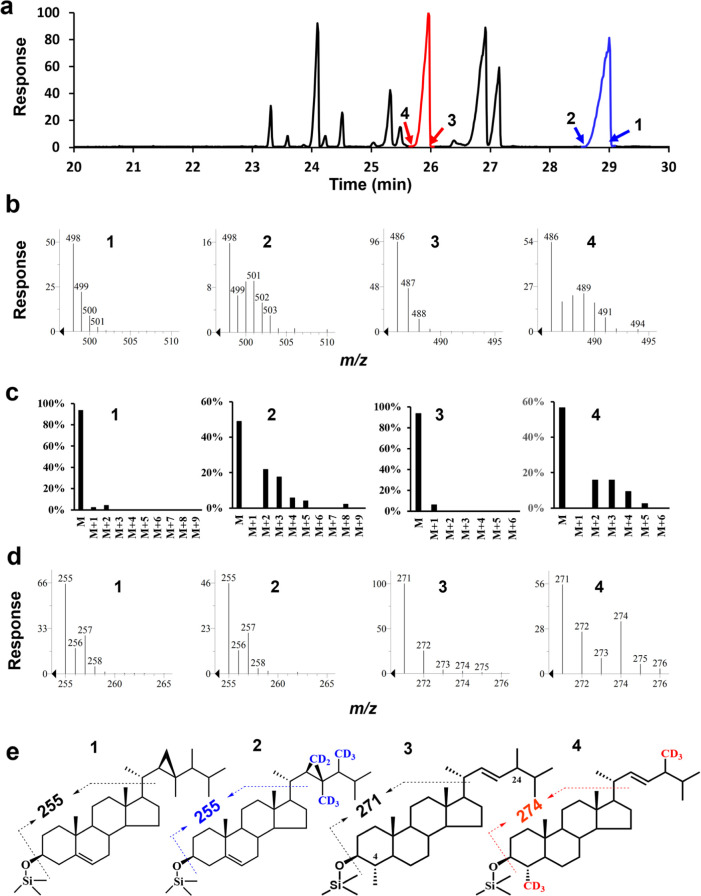
A mixture of both 4-methyl sterols and 4-desmethyl sterols was recovered from cultured Breviolum minimum cells fed with [2H3-methyl]-methionine. All deuterium atoms in the methionine would be incorporated into AdoMet and, thereby, lead to labeling of sterols by AdoMet-dependent methyltransferase-mediated methylation. As deuterium-labeled organic molecules elute slightly ahead of their unlabeled counterparts,23 the mass spectra of labeled sterols can be obtained from the leading edges of the sterol elution peaks (Figure 1a-1,a-3) and unlabeled sterols from the trailing edges (Figure 1a-2,a-4). Sterols were well separated in the chromatograms and include gorgosterol, a 4-desmethyl sterol eluting from 28.602 min (scan 1759) to 29.046 min (scan 1799), and 4α,24-dimethyl-5α-cholest-22E-en-3β-ol, a 4-methyl sterol eluting from 25.717 min (scan 1499) to 25.994 min (scan 1524) (Figure 1a). Mass spectra of these compounds are shown in Figure S2. Scan 1795 (29.001 min) provided spectra of unlabeled gorgosterol with a molecular mass of 498 Da (Figure 1a-1,b-1) while scan 1768 (28.702 min) provided labeled gorgosterol, with the molecular mass extending beyond 498 Da and peaking at 501 Da (Figure 1a-2,b-2). Isotopic distribution analysis (Figure 1c-1,c-2) indicated that these sterols have identical nuclei (Figure 1d-1,d-2), with the ion at 255 Da in common (Figure 1e-1,e-2). Eight deuterium atoms were incorporated into fully labeled gorgosterol, yielding a molecular mass peak of M + 8 (i.e., 506 Da; Figure 1e-2), suggesting three methylation events, all on the side chain (Figure 1e-2), and that gorgosterol was derived from lanosterol.
Figure 1.
Sterol profiles of B. minutum incubated with [2H3-methyl]-methionine. (a) GC trace of sterol trimethylsilyl ethers. The red peak is 4,24-dimethyl-5α-cholest-22-en-3β-ol, while the blue peak is gorgosterol. The arrows indicate mass spectra of the nondeuterated (1,3) and deuterated (2,4) sterols. (b) Partial mass spectra of substances at the leading and trailing edges of the peaks, indicating differences in their degrees of deuterium incorporation of unlabeled gorgosterol (1), labeled gorgosterol (2), unlabeled 4,24-dimethyl-5α-cholest-22E-en-3β-ol (3), and labeled 4,24-dimethyl-5α-cholest-22E-en-3β-ol (4). (c) Isotopic pattern deconvolution of the mass spectra of unlabeled gorgosterol (1), labeled gorgosterol (2), unlabeled 4α,24-dimethyl-5α-cholest-22E-en-3β-ol (3), and labeled 4α,24-dimethyl-5α-cholest-22E-en-3β-ol (4). (d) Partial mass spectra of the nuclei of unlabeled gorgosterol (1), labeled gorgosterol (2), unlabeled 4α,24-dimethylcholest-22E-en-3β-ol (3), and labeled 4α,24-dimethylcholest-22E-en-3β-ol (4). (e) Structures of unlabeled gorgosterol (1), labeled gorgosterol (2), unlabeled 4,24-dimethyl-5α-cholest-22E-en-3β-ol (3), and labeled 4α,24-dimethylcholest-22E-en-3β-ol (4).
In contrast, the molecular mass of unlabeled 4α,24-dimethyl-5α-cholest-22E-en-3β-ol (scan 1522, 26.0 min; Figure 1a-3,b-3) was 3 Da lighter (486 vs 489 Da) than the labeled form (scan 1503, 25.761 min; Figure 1a-4,b-4). The labeled sterol (Figure 1c-4) included an additional five deuterium atoms relative to its non-labeled counterpart (Figure 1c-3), suggesting that two methyl groups were added to the sterol by two methylation reactions. In addition, the nuclei of non-labeled sterol (m/z 271; Figure 1d-3,e-3) were three Da lighter than their labeled counterpart (m/z 274; Figure 1d-4,e-4), suggesting a methylation event on the nucleus (Figure 1e-4). Thus, the second methylation occurs on the side chain, and AdoMet is the methyl donor.
Therefore, in this dinoflagellate, the 4-methyl sterols are end products of a biosynthetic pathway involving a previously unknown sterol methyl transferase, which can be labeled with deuterium from [2H3-methyl] AdoMet rather than intermediates generated via inactivation of one of the SMOs (which could not be labeled with deuterium) (Figure S1).
In Vitro Enzymatic Assays Show that BmSTRM Catalyzes the Biosynthesis of Both C4α- and C4β-Methyl Sterols
To elucidate the molecular details of the biosynthesis of 4-methyl sterols, we sought orthologs of the C. elegans STRM-1 (CeSTRM) gene20 in the B. minutum genome. A gene, symbB.v1.2.040208.t1 (designated BmSTRM), encodes a protein with high similarity to CeSTRM, with a conserved AdoMet-binding domain and other highly conserved regions in well-studied C24 sterol methyltransferases. Expression of codon-optimized BmSTRM in Escherichia coli yielded a 48 kDa protein (Figure 2a). However, we could not detect reproducible activity with the microsomes of the bacteria expressing BmSTRM. Many rounds of optimization revealed that the in vitro activity of BmSTRM is very unstable and that even a single cycle of freezing and thawing would lead to absolute inactivation. Like CeSTRM,20 BmSTRM requires AdoMet as a cofactor and has a substrate preference for A-ring-saturated 3-ketosteroids, such as cholestanone (1), cholest-5-en-3-one (2), and 5α-cholest-7-en-3-one (3) (see Figure S3 for structures). In contrast, cholest-4-en-3-one (4), cholest-1-en-3-one (5), cholesterol (6), cholest-4-en-3β-ol (7), and 5α-cholestanol (8) (see Figure S3 for structures) are not substrates of this enzyme.
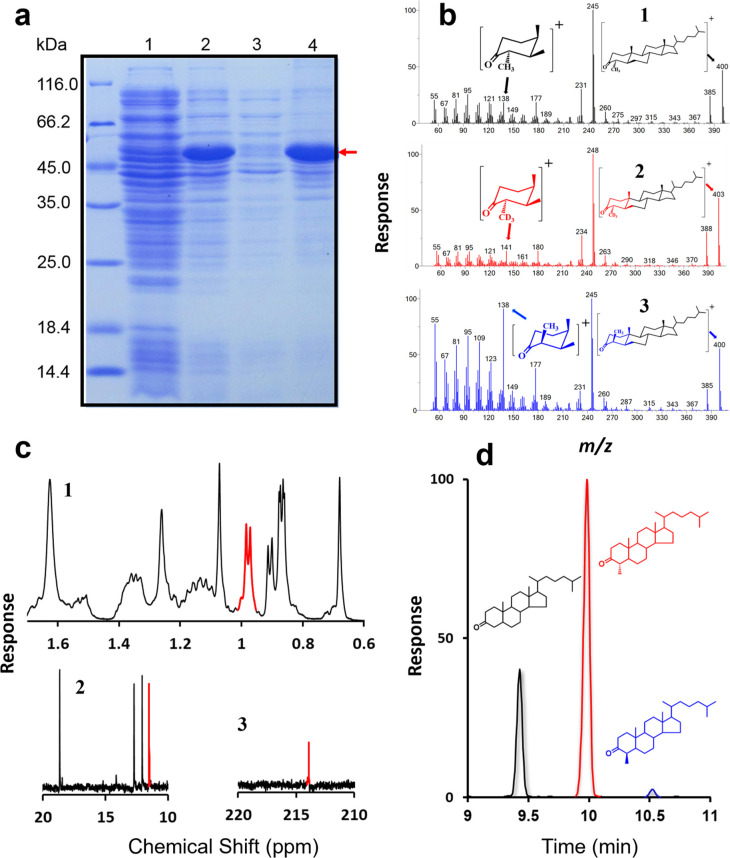
Figure 2.
In vitro assays of BmSTRM enzymatic activity. (a) Visualization, by gel imaging, of BmSTRM protein expressed in E. coli and separated in a precast 12% Bis-Tris gel. Lane 1, control with no induction by IPTG; Lane 2, proteins of broken cells induced by IPTG; Lanes 3–4, soluble (3) and insoluble fractions (4) of proteins of BmSTRM-expressing E. coli. (b) Comparison of mass spectra of 4α-methylcholestanone products obtained from reactions with BmSTRM, cholestanone, and AdoMet (1) or [2H3-methyl] AdoMet (2). Comparison of the mass spectra of 4α- and 4β-methylcholestanone (3). (c) PNMR spectroscopy and 13C NMR analysis of purified BmSTRM products. (1) PNMR spectroscopic analysis of the 4α-methyl group (with a doublet signal centered at 0.98 ppm), (2) 13C NMR analysis of the diagnostic signals of the 4α-methyl group at 11.5 ppm, and (3) 13C NMR analysis of the attachment point of the keto group at 214 ppm. (d) GC trace of products of the in vitro BmSTRM assay with a reaction mixture of cholestanone and 750 μM AdoMet.
In vitro enzymatic assays showed that cholestan-3-one (5 in Figure S1) was converted to a product with a molecular weight of 400 Da and with a fragmentation pattern highly similar to the reference pattern of 4-methylcholestan-3-one but not to other nucleus-methylated sterols (Figure 2b-1, see 6–8 in Figure S2 for mass spectra). When AdoMet was replaced by [2H3-methyl] AdoMet, the end product was 3 Da heavier (Figure 2b-1), confirming that AdoMet was a methyl donor for the BmSTRM-catalyzed reaction. A comparison of mass spectra showed that replacement of AdoMet with [2H3-methyl] AdoMet increased the product’s molecular weight by 3 Da from 138 Da (Figure 2b-1; note the left black arrow) to 141 Da (Figure 2b-2; note the left red arrow). Analysis of the fragmentation pattern indicated that this ion arose from disassociation of the A-ring containing the newly introduced 4-methyl group from AdoMet. Therefore, BmSTRM can catalyze methylation at the C4 position using AdoMet as a cofactor, supporting the results of the isotopic feeding study.
BmSTRM tautomerization activities were examined through a series of reactions with cholestan-3-one (see Figure S2 for its full mass spectrum) as the substrate with phosphate buffer containing 20% deuterated water (D2O) (Figure S4). The 3-keto steroid cholestan-3-one is subject to keto–enol tautomerism, as previously documented,24 but to a limited degree in control conditions (enzyme-free) either with (yielding 1.3% of the M + 1 isotopolog) or without addition of AdoMet (yielding 1.5% of the M + 1 isotopolog) (Figure S4a). Addition of BmSTRM increased deuterium incorporation, raising the proportion of the M + 1 isotopolog in the products to 5.0% and 19.3% (equivalent to the percentage of D2O in the buffer system) in the absence or presence of AdoMet, respectively (Figures S4a and 4b). Thus, the enzyme enhanced keto–enol tautomerization with or without the methyl donor AdoMet, while AdoMet significantly enhanced deuterium incorporation into the product. Moreover, only the M + 1 isotopolog was predominantly enriched (Figure S4a), indicating that only one deuterium atom is incorporated and that BmSTRM methylates the substrate at only one position (Figure S4c).
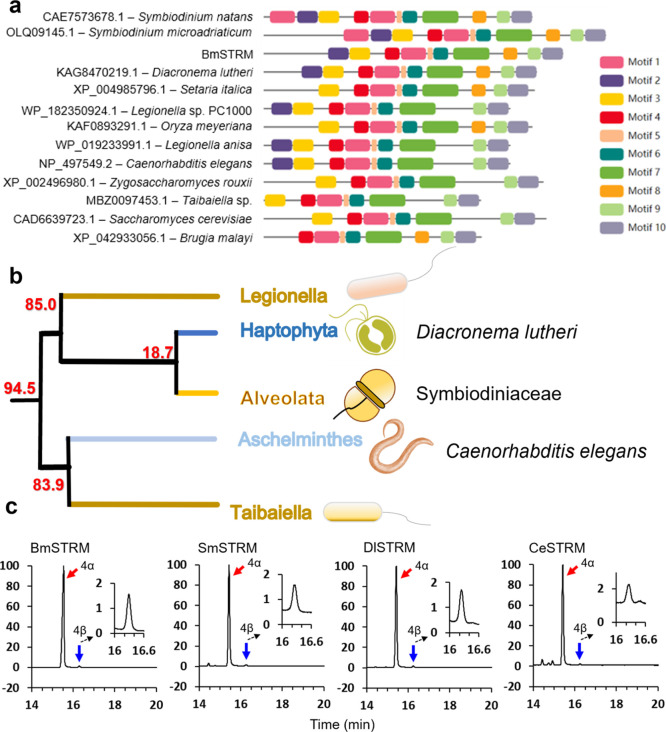
Figure 4.
Phylogenetic analysis and enzymatic activity of STRMs from S. microadriaticum, D. lutheri, and C. elegans. (a) Illustration of conserved motifs among the representative species. Different colors of the boxes refer to different motifs from 1 to 10. (b) Origin and diversification of the STRM gene family. Average divergence time are indicated for nodes of interest (million years, Ma). See Figure S6 for details. (c) Comparison of the enzymatic activity of BmSTRM and the STRMs of S. microadriaticum (SmSTRM), D. lutheri (DlSTRM), and C. elegans (CeSTRM). Chromatograms of products generated by BmSTRM (1), SmSTRM (2), DlSTRM (3), and CeSTRM (4) in a full-scan mode, with arrows indicating peaks of 4α-methylcholestanol (lophenol; the red arrows) and 4β-methylcholestanol (the blue arrows). Note: the dotted lines indicate enlarged chromatograms of the 4β isomer. See Figure S7 for the mass spectra of 4α- and 4β-methylcholestanol produced by SmSTRM, DlSTRM, and CeSTRM.
BmSTRM’s postulated activity was further validated by an in vitro assay in which the substrate amount of cholestan-3-one increased (from 20 to 100 μM). The reaction mixture was separated by thin-layer chromatography (TLC), which resulted in two major bands: one with a similar Rf to that of cholestan-3-one (ca. 0.5) and another with a lower Rf (ca. 0.3). As the 4-methyl sterols migrated more slowly than the 4-desmethyl counterparts,20 the slower band was hypothesized to represent a 4-methylated product of cholestan-3-one. The slower band was purified by high-performance liquid chromatography (HPLC), which yielded a single peak at approximately 23 min. Proton nuclear magnetic resonance (PNMR) spectroscopic analysis revealed the presence of a 4α-methyl group (with a doublet signal centered at 0.98 ppm and a coupling constant of 6.5 Hz; Figure 2c-1 and Table S1). This was corroborated by 13C NMR, which yielded diagnostic signals of the 4α-methyl group at 11.5 ppm (Figure 2c-2 and Table S2) and C3, the attachment point of the keto group, at 214 ppm (Figure 2c-3). Together with the keto–enol tautomerism results, BmSTRM-induced enolization is specific to the C3(4) double bond and results in methylation of the substrate at the C4 position (Figure S4c). We propose that BmSTRM accelerates keto–enol tautomerization (steps 1 and 2 in Figure S4c) of the sterol substrate, leading to the incorporation of deuterium (steps 3 and 4 in Figure S4c) and catalyzing the subsequent electrophilic addition of a methyl group from AdoMet to the C3(4) double bond of the enol tautomer (steps 5 and 6 in Figure S4c).
Interestingly, the gas chromatography–mass spectrometry (GC–MS) analysis of the HPLC-purified sterols with longer GC elution times revealed a minor compound that eluted later and had both the same molecular mass and a very similar mass spectrum to the major product (Figure S5). The minor isomer accounted for approximately 3% of the total products (highlighted in blue in Figure 2d) in in vitro assays using 750 μM of AdoMet and cholestan-3-one as the substrate. An isotopic assay with [2H3-methyl] AdoMet confirmed that the minor product was labeled in the same manner as the major product (full mass spectrum 9 in Figure S2), indicating that both were products of the BmSTRM-catalyzed reaction and not artifacts. A 138-Da fragment was more abundant in the mass spectrum of the minor product (Figure 2b-3; note the left blue arrow) compared to the spectrum of the 4α-methyl product (Figure 2b-1; note the left black arrow). This corroborates its identification as a 4β-methyl sterol, as the high intensity of a fragment with this m/z value is diagnostic of 4β-methyl sterols. This is because a 4β-methyl substituent at the axial position of an A-ring fragment (Figure 2b-3) is more stable than an equatorial 4α-methyl (Figure 2b-1). Although we failed to separate the two isomers by optimizing the HPLC conditions, these chromatographic and spectroscopic results support the identification of the major and minor components as 4α-methylcholestan-3-one and 4β-methylcholestan-3-one, respectively. Therefore, the activity of BmSTRM is highly unstable in vitro and produces both isomers. It is thus distinct from the previously proposed function of CeSTRM (which only generates the 4α isomers20).
Dinoflagellate B. minutum Produces Both C4α- and C4β-Methyl Sterols
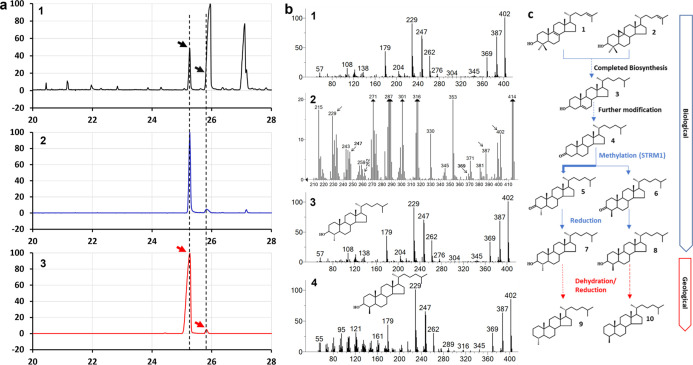
To confirm the production of 4β-methyl sterols in vivo, the sterols isolated from a larger scale culture of B. minutum were separated by HPLC. Fractions (1-min) of the eluents were collected, dried, and subjected to GC–MS analysis. In fraction 41, two sterols with a molecular weight of 402 Da (as expected for both 4α- and 4β-methyl cholestanol) were identified (Figure 3a-1). A peak at a retention time of 25.27 min was well separated and yielded a mass spectrum identical to that of 4α-methylcholestanol (Figure 3b-1). In addition, a minor component eluting at 25.84 min was detected, with similar key fragments and distribution patterns to those of 4α-methylcholestanol, a molecular mass ion of 402 Da, and the same major fragments (387, 369, 262, 247, and 229 Da; Figure 3b-2). However, we cannot unambiguously identify this sterol as the 4β-methyl sterol because interference from other sterols prevented acquisition of a clean mass spectrum. To gain more insights into the identity of this compound, external references for comparison of its retention time were prepared. The 4-methylated products of BmSTRM-catalyzed reactions in the in vitro feeding assay (using cholestan-3-one as substrate) were reduced to their corresponding alcohols to produce 4α- (Figure 3b-3) and 4β-methylcholestanol (Figure 3b-4), respectively. Fraction 41 includes two substances (Figure 3a-2) with retention times identical to the reference standards (Figure 3a-3). Therefore, both 4-methylated isomers are present in the dinoflagellate. This represents a novel biochemical pathway for sterol biosynthesis, and a putative 4-methyl sterane formation pathway is constructed (Figure 3c).
Figure 3.
GC–MS chromatographic and spectroscopic analysis of 4-methycholestanol obtained from B. minutum. (a) Chromatograms of fraction 41 obtained from HPLC separation of sterols (see Results for details) of B. minutum in a full-scan mode, with arrows indicating peaks of 4α-methylcholestanol (lophanol) and 4β-methylcholestanol (1); fraction 41 in SIM mode using m/z 402 (2); and generated reference standards (4-methylated products of the BmSTRM-catalyzed reaction in the in vitro feeding assay) in the SIM mode using m/z 402 (3). Note: the dotted lines indicate identical retention times in chromatograms. (b) Mass spectra of the following substances in fraction 41: 4α-methylcholestanol (1); the fraction’s component with a GC–MS retention time of 25.27 min and diagnostic fragments of 4β-methylcholestanol indicated by arrows (2); and authentic 4α-methylcholestanol and 4β-methylcholestanol derived from C4-methylated products of the BmSTRM-catalyzed reaction in the in vitro feeding assay with cholestanone as substrate (3 and 4, respectively). (c) Deduced route of 4-methylsterane generation. Proportions of the 4-methylated isomers indicate that sterols in fossil records in immature sediments have biogenic sources and conserved configurations rather than originating from geological processes. (1) lanosterol; (2) cycloartenol; (3) cholesterol; (4) cholestanone; (5) 4α-methylcholestanone; (6) 4β-methylcholestanone; (7) 4α-methylcholestanol; (8) 4β-methylcholestanol; (9) 4α-methylsterane; and (10) 4β-methylsterane.
BmSTRM-type Methylase is Distributed in Alveolates and Haptophytes
To investigate the prevalence of STRM-mediated biosynthesis of both C4α- and C4β-methyl sterols, the methyltransferase domain was used to search STRM gene sequences across the entire eukaryote (fungi, algae, protists, embryophytes, and animals) and bacterial databases. In total, more than 1000 orthologs were obtained (Figure S6), while 10 conserved motifs were identified among these candidates (Figure 4a). The phylogeny of STRMs revealed a high level of sequence conservation among alveolates (including Symbiodiniaceae) and haptophytes (comprising a major proportion of the phytoplankton community, i.e., globally distributed and exerting large-scale impacts on ocean biogeochemistry22) (Figure 4b). We thus randomly selected and characterized the STRMs from species of alveolates (i.e., Symbiodinium microadriaticum, SmSTRM) and haptophytes (i.e., Diacronema lutheri, formerly known as Pavlova lutheri, Chrysophyta; DlSTRM) and revisited the catalytic activity of CeSTRM.
As expected, both SmSTRM and DlSTRM catalyze methylation at the C4 position, generating both the C4α- and C4β-methyl sterols (Figures 4c and S7). Surprisingly, in contrast to the general notion that CeSTRM produces only C4α-methyl sterols,20 we found that CeSTRM can also catalyze the biosynthesis of both isomers (Figures 4c and S7). While BmSTRM-type methylase may be species-specific, it is clearly not specific to B. minutum alone but shared by representatives of a number of important plankton taxa (e.g., alveolates and haptophytes). The molecular clock estimates the appearance of BmSTRM-type enzymes at 94.5 Ma after the oldest dinoflagellate fossils (∼520 Ma)14 and before the oldest record of B. minutum (6.5–5.6 Ma),16 suggesting an early origin and widespread catalytic mechanisms (Figures 4b and S6).
Discussion
A small proportion of the vast number of microalgae that retain significant abundances of 4-methyl sterols (e.g., dinoflagellates and a few diatoms and haptophytes) have been subjected to sterol profiling.25 All 4-methyl sterols have been identified as 4α isomers,25,26 but the presence of 4β-methyl steranes in sediments has long been more enigmatic because their precursors, the 4β-methyl sterols, have rarely been detected in any organism. The sole report is the discovery of a previously unknown 4β-methyl sterol in marigold flowers more than 50 years ago,27 which should be treated very cautiously due to flaws in the chromatographic and spectroscopic techniques applied at the time.27 Moreover, no enzyme has been previously shown to catalyze the production of 4β-methyl sterols, potentially due to some degree of plasticity of the enzyme. The biosynthetic pathway for 4-methyl sterols has not been identified in any organisms that could be connected with fossils.
In contrast, in laboratory-controlled conditions mimicking natural geological processes, 4α-methylcholest-4-ene (derived from 4α-methylcholestanol; Structure 4 in Figure S1), has been converted to both 4α- (Structure 5 in Figure S1) and 4β-methylcholestane (Structure 6 in Figure S1) by reduction of the C4 double bond.28 The conversion of stanols to sterenes in sediments via steroid ketones has long been recognized; thus, it is widely accepted that 4β-methyl steranes found in sediments are derived from 4α-methyl steroids during early diagenesis. Thermal breakdown occurs with increasing temperature and pressure during deeper burial of sediments where the abundance of less stable compounds (e.g., 4β-methyl steranes) is converted into their stable isomerization products (4α-methyl steranes).10 The 4α/4β-methyl sterane ratio can thus serve as an indicator of thermal maturity, which indicates the extent of the conversion of sedimentary organic material into gas, petroleum, and other products. Therefore, the 4α/4β-methyl sterane ratio has been used to interpret geological history10,11 and help distinguish actual oil and gas source rocks from merely potential source rocks.29
B. minutum STRM catalyzes the methylation of sterols at the C4 position, yielding both 4α- and 4β-methyl sterols rather than only 4α-methyl sterols. Sterol profiling further supports the occurrence of both C4-methylated isomers in the dinoflagellate. Characterization of the STRMs from the randomly selected species of alveolates and haptophytes revealed a potential prevalence of BmSTRM-type methylase in these biogeochemically important protists. Surprisingly, we found that, in contrast with the previously documented feature of CeSTRM,20 the stereochemical mechanism of CeSTRM is the same as BmSTRM, mediating the biosynthesis of both C4α- and C4β-methyl sterols and suggesting an origin of such catalytic mechanisms in the common ancestor of alveolates, haptophytes, and nematodes. Otherwise, convergent evolution likely occurred and generated an enzyme with the same function. The former hypothesis is more likely when considering the high amino acid sequence similarity among the STRMs of these organisms.
The discovery and characterization of BmSTRM imply that 4β-methyl steranes in sediments could have biological sources. This finding calls for an amendment to the current paradigm of the genesis of 4-methyl steranes (Figure S8). Given the ubiquitous occurrence of alveolates and haptophytes on spatiotemporal scales,16,17 the hypothesis that all 4β-methyl steranes are diagenetically derived from 4α-methyl steroids10 is open to question. Although the abundance of 4β-methyl sterols found here is low, we cannot deny the possibility that organisms with a large proportion of 4β-methyl sterols remain undiscovered. Alternatively, such organisms may have existed in ancient times, but the biosynthesis capacity of 4β-methyl sterols may have diminished or been lost due to the marginal significance of these sterols in the life process.9 As STRM orthologs are widespread in eukaryotes, the discovery of BmSTRM and the protocol for the activity assay provided in this study (i.e., BmSTRM activity is highly unstable in vitro) suggest the need to revisit currently accepted notions regarding the function of STRMs in steroid biosynthesis. This may have implications for the interpretation of molecular fossils and understanding of the thermal evolution of source rocks, and thereby the search for commercial crude oil and gas. However, to what extent the biogenesis of 4β-methyl sterols contributes to source rocks remains an open question and requires careful analyses of the minor sterols of microalgae and further genomic investigations of sterol biosynthetic pathways.
Materials and Methods
Strains and Growth Conditions
B. minutum strain NIES-3808 was obtained from the National Institute for Environmental Studies (Japan). It was routinely cultured in 250 mL conical flasks with 100 mL of the L1 medium (pH 8.2)30 containing ampicillin (100 mg·L–1), kanamycin (50 mg·L–1), and streptomycin (50 mg·L–1). The alga was cultured at 25 °C and a constant 50 μmol·photons·m–2·s–1 light intensity.31
Chemical Application
Cultures were started with an initial density of 2 × 105 cells·mL–1 in the L1 medium and harvested at a density of ca. 1 × 106 cells·mL–1, then washed with sterile seawater, and inoculated into a fresh medium in the presence or absence of l-methionine-(methyl-D3) (0.2 g·L–1; Sigma-Aldrich) at 25 °C. Cells with a final biomass of ca. 5 g were collected by centrifugation (7000g for 10 min), washed, and resuspended in 10 mM Tris–HCl, pH 7.3. Sterols were extracted and measured as previously reported.32
Chemicals and Reagents
Cholest-4-en-3-one, cholest-5-en-3-one, cholesterol, cholestan-3-one, and the solvents and reagents required for TLC, HPLC, and GC–MS analyses were purchased from Sigma-Aldrich (Shanghai, China). Cholest-4-en-3-ol and cholest-7-en-3-one were purchased from Steraloids (Newport, RIUS; https://www.steraloids.com/contact-us), and a methyltransferase colorimetric assay kit was purchased from NeoBioscience Technology (Shenzhen, China).
Design and Optimization of Plasmids for Expression
BmSTRM was designed and expressed in E. coli following previously reported protocols with minor modifications.20,33 Briefly, BmSTRM’s topological structure was predicted by multiple algorithms (DAS, TMpred, and TMHMM). The hydrophobic transmembrane regions at the N-terminal (M1-L70) were deleted, and the remaining region (truncated BmSTRM, tBmSTRM) was synthesized and codon-optimized based on the codon frequency in E. coli. The synthetic tBmSTRM gene was cloned into the pCzn1, pGEX-4T-1, pET-22b, and pET-32a expression vectors, allowing the production of tBmSTRM fused to a His-tag, tBmSTRM fused to a GST-tag, tBmSTRM with an N-terminal pelB signal sequence for potential periplasmic localization (plus optional C-terminal His-tag sequence), and recombinant tBmSTRM with a thioredoxin tag. Approximately 109 cells expressing each of these constructs were collected by centrifugation, and soluble and insoluble proteins were extracted for SDS–PAGE analysis. Among all the expression vectors, pGEX-4T-1 was finally selected for subsequent experiments due to its high expression efficiency for soluble enzymes.
Preparation of Bacterial Material for Recombinant Enzyme and Non-radiolabeled Sterol Production Assays
STRM variants were produced using the E. coli Arctic Express system harboring corresponding expression vectors. Cells were grown at 37 °C in the LB medium to an OD600 of 1.0 and cooled to 4 °C. Then, expression of the recombinant protein was induced by overnight incubation after adding IPTG to 100 μM. For cells from 1 L culture pellets, cells were collected by centrifugation and suspended in 25 mL of protein solubilization buffer (Bio-Rad, 1632145). The cells were lysed by sonication and centrifuged at 10,000g for 15 min to remove debris, and then the supernatant’s STRM activity was assayed immediately.
Enzyme Assays
In vitro enzymatic assays were performed using the methyltransferase colorimetric assay kit, following the manufacturer’s instructions. Briefly, sterol substrates were dissolved in dimethyl sulfoxide (DMSO) and then added (individually, in triplicate) with AdoMet to the reaction mixture supplied with the kit to final concentrations of 50 μM and 150 μM, respectively. After incubation at 35 °C for 2 h, methyltransferase activities were determined by measuring the increase in absorption at 515 nm. AdoHcy (S-adenosyl-l-homocysteine) and DMSO were used as positive and negative controls, respectively. The enzyme activities with different substrates were normalized to that of cholestan-3-one.
For the detailed characterization of STRM’s catalytic activity, cholestan-3-one was mixed, individually or simultaneously, with AdoMet and STRM in a phosphate buffer prepared with 20% D2O. Protein preparation (490 μL) was added and vortexed for at least 20 s to dissolve the substrate. After 2 h of incubation, the sterols were extracted using n-hexane and dried. Sterols were analyzed by GC–MS using carbon tetrachloride as a diluent to prevent possible hydrogen exchange in the GC injection port. To obtain abundant enzymatic products for chromatographic and spectroscopic analyses, an optimized method was developed. Briefly, induced bacterial cells were lysed, and the resulting lysate was used directly in the assays. Each sterol substrate was dispersed by adding Tween-20 to 100 μM in the reaction mixture. The concentration of AdoMet (p-toluenesulfonate salt) was set at a high level (750 μM) to increase the amounts of end products. The sterol substrates and products in the assay mixture were extracted after overnight incubation (at 35 °C) with n-hexane following saponification with 10% KOH/methanol. The resulting extract was dried and used for further processing. For bulk incubation, a 4 L culture of E. coli harboring the STRM expression vector was grown, induced, and collected. The cells were lysed and centrifuged at 10,000 × g to remove cell debris. The supernatant was mixed with 100 μM of the sterol substrate and 750 μM of AdoMet. After overnight incubation, the mixture was saponified, and sterols were recovered through liquid–liquid extraction using n-hexane.
Sterol Separation by TLC
Extracted sterols were dissolved in a small amount of chloroform, and the sterol solution was applied to glass TLC plates coated with silica gel (Sigma-Aldrich, MI). After dehydration, the plates were developed with a mixture of toluene and diethyl ether in an 85:15 ratio (v/v). The fully developed plates were left in a fume hood for ca. 2 h until all solvents had evaporated, sprayed with dye prepared by dissolving 5 mg of primuline (Sigma-Aldrich, USA) in 100 mL of acetone/water (80/20, v/v), and then dehydrated at 100 °C for 10 min in an oven. Finally, the entire plate was sprayed, and fluorescent bands of sterols on the TLC plates were visualized using a UV transilluminator.
HPLC for Sterol Separation
Total sterols generated in the reactions were separated on a silica gel TLC plate. Each band was scraped from the plates and extracted four times with acetone. The acetone extracts were pooled and dried. A Shimadzu LC20 system equipped with a UV diode array detector (set at 210 nm) and a reverse-phase column (Agilent Zorbax SB-C18) was used for HPLC with methanol/water in a 95:5 ratio as the mobile phase.
Sterol Extraction and Identification by GC–MS Analysis
Sterol extraction and GC–MS measurements were conducted following earlier reports.32,34 Collected data were analyzed with Agilent GC–MS D Productivity ChemStation and AMDIS (Automated Mass Spectral Deconvolution and Identification System) software. The sterol spectra were compared with entries in the commercial NIST/EPA/NIH mass spectral library (NIST 08) for identification. The isotopic patterns were deconvoluted with Excel spreadsheets to calculate the positions and extents of the sterols’ isotope labeling.35 Keto sterols generated in the reactions to produce substances for NMR analysis were reduced by overnight incubation with sodium borohydride in isopropanol solution, as previously reported.36
Phylogenic Tree Reconstruction
The methyltransferase domain (PFAM13649) was used to search STRM candidates across all eukaryotes and bacteria by HMMER search.37 Multiple sequence alignment and conserved motif finding were performed using MUSCLE38 and Gblock,39 respectively. A phylogenetic tree was built using RAxML (1000 iterations with the maximum likelihood method).40 The R8S script was employed to estimate temporal divergence based on the molecular evolution rate and stable fossil nodes.41 A strict clock model was used to avoid horizontal gene transfer and other events that affected the divergence times. Multiple time constraints (fossil records for no less than three species within the same genus) were incorporated to evaluate our results using fossil cross-validation. The fossil records used in this study are relevant ones that were previously applied to estimate the divergence times of eukaryotes,42−46 including the divergence times for (1) Triticum and Oryza (from 42.0 to 52.0 Ma),47 (2) Triticum and Brachypodium (from 27.0 to 38.0 Ma),48 (3) Zingiber and Ensete (from 50.6 to 87.0 Ma),49 (4) Emiliania and Chrysochromulina (from 188.2 to 417.0 Ma),46 and (5) Ostreococcus and Setaria (from 970.0 to 1244.0 Ma).50 The fossil records are available at Timetree.51
Acknowledgments
This work was supported in part by grants from the National Key R&D Program of China (2021YFA0909600), the Intergovernmental Project of National Key R&D Program of China (2021YFE0110100), the National Natural Science Foundation of China (32060061), the Key R&D Program of Hainan Province (ZDYF2022XDNY140), and the Project of the State Key Laboratory of Marine Resource Utilization in South China Sea (MRUKF2021003).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c01401.
Proposed route of sterane generation; full mass spectra of the sterols discussed in this study; relative activities of BmSTRM with indicated sterol substrates; keto–enol tautomerization of BmSTRM; GC–MS analysis of 4-methyl steroid ketones used for NMR analysis; phylogenetic analysis and molecular clock estimate of sterol methyltransferases; mass spectra of 4α- and 4β-methylcholestanol produced by SmSTRM and CeSTRM; proposed route for the generation of C4-methyl steranes in this study; PNMR analysis of the products of the in vitro BmSTRM enzymatic assay; and 13C NMR analysis of the products of the in vitro BmSTRM enzymatic assay (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Pollier J.; et al. A widespread alternative squalene epoxidase participates in eukaryote steroid biosynthesis. Nat. Microbiol. 2018, 4, 226. 10.1038/s41564-018-0305-5. [DOI] [PubMed] [Google Scholar]
- Kodner R. B.; Summons R. E.; Pearson A.; King N.; Knoll A. H. Sterols in a unicellular relative of the metazoans. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 9897–9902. 10.1073/pnas.0803975105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goad L. J. The biosynthesis of plant sterols. Lipids Lipid Polym. Higher Plants, [Pap. Symp.] 1977, 146–168. 10.1007/978-3-642-66632-2_8. [DOI] [Google Scholar]
- Knapp F. F.; Trowbridge S. T.; Schroepfer G. J. Concerning the role of 4.beta.-methyl sterols in cholesterol biosynthesis. J. Am. Chem. Soc. 1975, 97, 3522–3524. 10.1021/ja00845a044. [DOI] [PubMed] [Google Scholar]
- Robinson N.; Eglinton G.; Brassell S. C.; Cranwell P. A. Dinoflagellate origin for sedimentary 4α-methylsteroids and 5α(H)-stanols. Nature 1984, 308, 439–442. 10.1038/308439a0. [DOI] [Google Scholar]
- Darnet S.; Schaller H. Metabolism and biological activities of 4-methyl-sterols. Molecules 2019, 24, 451. 10.3390/molecules24030451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summons R. E.; Bradley A. S.; Jahnke L. L.; Waldbauer J. R. Steroids, triterpenoids and molecular oxygen. Philos. Trans. R. Soc., B 2006, 361, 951–968. 10.1098/rstb.2006.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. K.; et al. C-4 sterol demethylation enzymes distinguish bacterial and eukaryotic sterol synthesis. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 5884–5889. 10.1073/pnas.1802930115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein I.; Albrecht P. The occurence of nuclear methylated steranes in a shale. J. Chem. Soc., Chem. Commun. 1975, 957–958. 10.1039/C39750000957. [DOI] [Google Scholar]
- Wolff G. A.; Lamb N. A.; Maxwell J. R. The origin and fate of 4-methyl steroids-II. Dehydration of stanols and occurrence of c30 4-methyl steranes. Org. Geochem. 1986, 10, 965–974. 10.1016/s0146-6380(86)80034-1. [DOI] [Google Scholar]
- Wolff G. A.; Lamb N. A.; Maxwell J. R. The origin and fate of 4-methyl steroid hydrocarbons. I. Diagenesis of 4-methyl sterenes. Geochim. Cosmochim. Acta 1986, 50, 335–342. 10.1016/0016-7037(86)90187-0. [DOI] [Google Scholar]
- Goodwin N. S.; Mann A. L.; Patience R. L. Structure and significance of C30 4-methyl steranes in lacustrine shales and oils. Org. Geochem. 1988, 12, 495–506. 10.1016/0146-6380(88)90159-3. [DOI] [Google Scholar]
- Summons R. E.; Volkman J. K.; Boreham C. J. Dinosterane and other steroidal hydrocarbons of dinoflagellate origin in sediments and petroleum. Geochim. Cosmochim. Acta 1987, 51, 3075–3082. 10.1016/0016-7037(87)90381-4. [DOI] [Google Scholar]
- Moldowan J. M.; Talyzina N. M. Biogeochemical evidence for dinoflagellate ancestors in the Early Cambrian. Science 1998, 281, 1168–1170. 10.1126/science.281.5380.1168. [DOI] [PubMed] [Google Scholar]
- Stanley G. D.; van de Schootbrugge B. The evolution of the coral-algal symbiosis. Ecol. Stud. 2009, 7–19. 10.1007/978-3-540-69775-6_2. [DOI] [Google Scholar]
- Knowlton N.; Weil E.; Weigt L. A.; Guzmán H. M. Sibling Species in Montastraea annularis , coral bleaching, and the coral climate record. Science 1992, 255, 330. 10.1126/science.255.5042.330. [DOI] [PubMed] [Google Scholar]
- Budd A. F. Tracing the long-term evolution of a species complex: Examples from the Montastraea “annularis” complex. Palaeoworld 2010, 19, 348–356. 10.1016/j.palwor.2010.09.001. [DOI] [Google Scholar]
- Budd A. F.; Klaus J. S. The origin and early evolution of the Montastraea ″annularis″ species complex (Anthozoa: Scleractinia). J. Paleontol. 2001, 75, 527–545. 10.1017/s0022336000039640. [DOI] [Google Scholar]
- Budd A. F. Neogene paleontology in the northern Dominican Republic: 11. The family Faviidae (Anthozoa: Scleractinia) ; part 1. The genera Montastraea. J. Gen. Microbiol. 2009, 19, 40–54. [Google Scholar]
- Zhou W.; et al. A nematode sterol C4α-methyltransferase catalyzes a new methylation reaction responsible for sterol diversity. J. Lipid Res. 2020, 61, 192–204. 10.1194/jlr.ra119000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannich J. T.; et al. Methylation of the sterol nucleus by STRM-1 regulates dauer larva formation in Caenorhabditis elegans. Dev. Cell 2009, 16, 833–843. 10.1016/j.devcel.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Tsuji Y.; Yoshida M.. Advances in Botanical Research; Hirakawa Y., Ed.; Academic Press, 2017; Vol. 84; pp 219–261. [Google Scholar]
- Caban M.; Stepnowski P. The application of isotopically labeled analogues for the determination of small organic compounds by GC/MS with selected ion monitoring. Anal. Methods 2020, 12, 3854–3864. 10.1039/D0AY00723D. [DOI] [PubMed] [Google Scholar]
- Le P. H.; Nes W. D. Sterols: Tritium-labeling and selective oxidations. Chem. Phys. Lipids 1986, 40, 57–69. 10.1016/0009-3084(86)90062-9. [DOI] [Google Scholar]
- Volkman J. Sterols in microorganisms. Appl. Microbiol. Biotechnol. 2003, 60, 495–506. 10.1007/s00253-002-1172-8. [DOI] [PubMed] [Google Scholar]
- Volkman J. K.; Barrett S. M.; Dunstan G. A.; Jeffrey S. W. Geochemical significance of the occurrence of dinosterol and other 4-methyl sterols in a marine diatom. Org. Geochem. 1993, 20, 7–15. 10.1016/0146-6380(93)90076-n. [DOI] [Google Scholar]
- Pyrek J. S. A new 4β-methyl-sterol from marigold flowers. Chem. Comm. 1969, 107–108. 10.1039/C29690000107. [DOI] [Google Scholar]
- Gagosian R. B.; Smith S. O.; Lee C.; Farrington J. W.; Frew N. M. Steroid transformations in recent marine sediments. Phys. Chem. Earth 1980, 12, 407–419. 10.1016/0079-1946(79)90122-8. [DOI] [Google Scholar]
- Curiale J. A.; Larter S. R.; Sweeney R. E.; Bromley B. W.. Thermal History of Sedimentary Basins; Naeser N. D., McCulloh T. H., Eds.; Springer: New York, 1989; pp 53–72. [Google Scholar]
- Guillard R. R. L.Culture of Marine Invertebrate Animals Smith W. L., Chanley M. H., Eds.; Springer: US, 1975; pp 29–60. [Google Scholar]
- Jiang J.; Lu Y. Metabolite profiling of Breviolum minutum in response to acidification. Aquat. Toxicol. 2019, 213, 105215. 10.1016/j.aquatox.2019.05.017. [DOI] [PubMed] [Google Scholar]
- Lu Y.; et al. Clade-specific sterol metabolites in dinoflagellate endosymbionts are associated with coral bleaching in response to environmental cues. mSystems 2020, 5, e00765 10.1128/msystems.00765-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.; et al. Role of an ancient light-harvesting protein of PSI in light absorption and photoprotection. Nat. Commun. 2021, 12, 679. 10.1038/s41467-021-20967-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.; et al. Regulation of the cholesterol biosynthetic pathway and its integration with fatty acid biosynthesis in the oleaginous microalga Nannochloropsis oceanica. Biotechnol. Biofuels 2014, 7, 81. 10.1186/1754-6834-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine J. B.; Pithawalla Y. B.; Naworal J. D.; Thomas C. E. Carbohydrate pyrolysis mechanisms from isotopic labeling. J. Anal. Appl. Pyrolysis 2007, 80, 297–311. 10.1016/j.jaap.2007.03.007. [DOI] [Google Scholar]
- Nes W. R.; Joseph J. M.; Landrey J. R.; Behzadan S.; Conner R. L. Steric effects at C-20 and C-24 on the metabolism of sterols by Tetrahymena pyriformis. J. Lipid Res. 1981, 22, 770–777. 10.1016/s0022-2275(20)37348-x. [DOI] [PubMed] [Google Scholar]
- Eddy S. R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Höhler D.; Pfeiffer W.; Ioannidis V.; Stockinger H.; Stamatakis A. RAxML Grove: An empirical phylogenetic tree Database. Bioinformatics 2021, 38, 1741. 10.1093/bioinformatics/btab863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson M. J. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 2003, 19, 301–302. 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Arakaki M.; et al. Contemporaneous and recent radiations of the world’s major succulent plant lineages. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 8379–8384. 10.1073/pnas.1100628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiz-Palacios O.; Schneider H.; Heinrichs J.; Savolainen V. Diversification of land plants: insights from a family-level phylogenetic analysis. BMC Evol. Biol. 2011, 11, 341. 10.1186/1471-2148-11-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G.; et al. From museums to genomics: old herbarium specimens shed light on a C3 to C4 transition. J. Exp. Bot. 2014, 65, 6711–6721. 10.1093/jxb/eru395. [DOI] [PubMed] [Google Scholar]
- Berney C.; Pawlowski J. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc. R. Soc. B 2006, 273, 1867–1872. 10.1098/rspb.2006.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey L. W.; Lahr D. J. G.; Knoll A. H.; Katz L. A. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 13624–13629. 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Z.; et al. Multi-locus plastid phylogenetic biogeography supports the Asian hypothesis of the temperate woody bamboos (Poaceae: Bambusoideae). Mol. Phylogenet. Evol. 2016, 96, 118–129. 10.1016/j.ympev.2015.11.025. [DOI] [PubMed] [Google Scholar]
- Foster C. S. P.; et al. Evaluating the impact of genomic data and priors on Bayesian estimates of the angiosperm evolutionary timescale. Syst. Biol. 2017, 66, 338–351. 10.1093/sysbio/syw086. [DOI] [PubMed] [Google Scholar]
- Zeng L.; et al. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat. Commun. 2014, 5, 4956. 10.1038/ncomms5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munakata H.; Nakada T.; Nakahigashi K.; Nozaki H.; Tomita M. Phylogenetic position and molecular chronology of a colonial green flagellate, Stephanosphaera pluvialis (Volvocales, Chlorophyceae), among unicellular algae. J. Eukaryotic Microbiol. 2016, 63, 340–348. 10.1111/jeu.12283. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Stecher G.; Suleski M.; Hedges S. B. TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017, 34, 1812–1819. 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.