Abstract
Enterobacter cloacae A-11 is a transposon mutant of strain 501R3 that was deficient in cucumber spermosphere colonization and in the utilization of certain carbohydrates (D. P. Roberts, C. J. Sheets, and J. S. Hartung, Can. J. Microbiol. 38:1128–1134, 1992). In vitro growth of strain A-11 was reduced or deficient on most carbohydrates that supported growth of strain 501R3 but was unaffected on fructose, glycerol, and all amino acids and organic acids tested. Colonization by strain A-11 was significantly reduced (P ≤ 0.05) for cucumber and radish seeds compared to that of strain 501R3, but colonization of pea, soybean, sunflower, and sweet corn seeds was not reduced. Pea seeds released several orders of magnitude more total carbohydrates and amino acids than cucumber and radish seeds and approximately 4,000-fold more fructose. Fructose was the only carbohydrate detected in the seed exudates which supported wild-type levels of in vitro growth of strain A-11. Soybean, sunflower, and sweet corn seeds also released significantly greater amounts of fructose and total carbohydrates and amino acids than cucumber or radish seeds. The exogenous addition of fructose to cucumber and radish seeds at quantities similar to the total quantity of carbohydrates released from pea seeds over 96 h increased the populations of strain A-11 to levels comparable to those of strain 501R3 in sterile sand. Molecular characterization of strain A-11 indicated that the mini-Tn5 kanamycin transposon was inserted in a region of the genome with significant homology to pfkA, which encodes phosphofructo kinase. A comparison of strain A-11 with Escherichia coli DF456, a known pfkA mutant, indicated that the nutritional loss phenotypes were identical. Furthermore, the pfkA homolog cloned from E. cloacae 501R3 complemented the nutritional loss phenotypes of both E. coli DF456 and E. cloacae A-11 and restored colonization by strain A-11 to near wild-type levels. These genetic and biochemical restoration experiments provide strong evidence that the quantities of reduced carbon sources found in seed exudates and the ability of microbes to use these compounds play important roles in the colonization of the spermosphere.
Colonization of subterranean portions of plants can be an essential process for a number of beneficial microbial activities, including plant growth promotion, plant disease control, and bioremediation (2, 6). Much research has been conducted in attempts to understand the processes by which microbes colonize plants, and several bacterial traits have been correlated with the colonization of seeds and roots in specific systems (e.g., references 3, 4, 7–9, 12, 14, 18, 26, and 41). One trait, microbial growth, has been established as an essential process for colonization, and recent studies have been published concerning the nutritional requirements for microbial growth on subterranean portions of plants (24, 29–32, 35, 36). However, the contributing roles played by the catabolic pathways of beneficial microbes, and the nutrients supplied by the host plant, in growth and colonization are still unclear.
The plant-beneficial bacterium Enterobacter cloacae suppresses Pythium ultimum damping-off of cucumber and other crops Pythium ultimum by (22) and colonizes the spermospheres and rhizospheres of a number of plant species (13, 16, 17, 22, 28–31, 39). It is believed that seeds and roots support growth by bacteria such as E. cloacae through the release of complex mixtures of carbohydrates, amino acids, water-soluble and -insoluble organic acids, and other nutrients (5). The growth of strain A-11, a nutritional mutant of E. cloacae 501R3 (29), was reduced or deficient on almost all carbohydrates released by seeds and roots that supported the growth of strain 501R3 in vitro (29, 32). The colonization of seeds of a cucumber (but not of a pea) cultivar by strain A-11 was significantly reduced (P ≤ 0.05) relative to that of strain 501R3 in studies conducted in natural soil and in sterile sand (29, 32). We have characterized the mutation in strain A-11 at the molecular level in an attempt to understand the colonization behavior of this strain. We report here that glycolysis in E. cloacae A-11 is blocked by a mutation in pfkA and that this gene is most important for colonization of seeds that release limited quantities of reduced carbon sources. Portions of this work have been published previously (32).
MATERIALS AND METHODS
Bacterial strains and plasmids.
Descriptions of the bacterial strains and plasmids are in Table 1. Unless otherwise indicated, E. cloacae and Escherichia coli strains were grown to the stationary phase at 35°C and 250 rpm. The following media were used: Luria-Bertani (LB) broth or agar (19), M9 basal salts broth or agar (19), and M56 basal salts broth or agar (23) supplemented as previously described. The antibiotic levels used to maintain or select strains and plasmids were 100 μg/ml for rifampin (RIF) and streptomycin (STR), 50 μg/ml for kanamycin (KAN), and 12.5 μg/ml for tetracycline (TET).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| E. cloacae 501R3 | Spontaneous Rifr mutant of E. cloacae Ec CT501 | 29 |
| E. cloacae A-11 | Mini-Tn5 KAN mutant of 501R3; Rifr Kmr | 29 |
| E. coli DH5α | (φ80dlacZΔM15) Δ(lacZYA-argF) U169 glnV44 deoR gyrA96 recA1 relA91 endA1 thi-1 hsdR17 | 27 |
| E. coli DF456 | glnV44 rpsL104 pfkA300::Mu metB1 lacY Smr | 40 |
| Plasmids | ||
| pLAFR3 | Tcr, pLAFR1 derivative | 11 |
| pRK2013 | Kmr | 10 |
| pRK415 | Tcr | N. Keen |
| p82 | pfkA+ Tcr | This work |
| pSubB | pfkA+ Tcr | This work |
| pSubB-3 | Tcr | This work |
| pBB1415 | pfkA+ Tcr | This work |
| pBS1415 | pfkA+ Tcr | This work |
Kmr, Rifr, Smr, and Tcr designate resistance to kanamycin, rifampin, streptomycin, and tetracycline, respectively; pfkA+, phosphofructo kinase positive.
In vitro growth on reduced carbon and nitrogen sources.
Growth of bacterial strains on various reduced carbon compounds was compared spectrophotometrically in basal salts broth or on basal salts agar amended with 0.2% carbohydrate or with 0.5% l-amino acid or 0.5% organic acid as previously described (19, 29, 30). The carbohydrates used were arabinose, cellobiose, fructose, galactose, glucose, N-acetylglucosamine, lactose, maltose, mannitol, mannose, raffinose, rhamnose, ribose, salicin, stachyose, sucrose, trehalose, and xylose. The l-amino acids and organic acids tested were alanine, asparagine, proline, glutamate, glutamine, serine, pyruvate, and malate. The growth of E. cloacae 501R3 and A-11 on alanine, asparagine, cysteine, glutamate, glutamine, proline, and serine as sources of nitrogen were measured as previously described (30).
Seed colonization assays.
E. cloacae strains were grown, washed, resuspended, and applied to single cucumber (Cucumis sativum cv. Marketmore 76), radish (Raphinus sativus cv. Cherry Bomb), pea (Pisum sativum cv. Sugar snap), soybean (Glycine max cv. Chesapeake), sunflower (Helianthus giganteus), or sweet corn (Zea mays cv. Stowells Evergreen) seeds at approximately 104 CFU per seed as previously described (29). Seeds were buried in 4 g of a natural Galestown gravelly loamy sand soil (77.8% sand, 12.6% silt, 9.6% clay, and 0.6% organic matter) that had been previously equilibrated to −75 kPa or in 4 g of washed, sterile sand moistened with 4 ml of sterile distilled water (SDW) in 14-ml sterile snap cap tubes and incubated at 22°C. CFU were determined by spiral plating (Spiral Systems, Cincinnati, Ohio) the contents of the tubes onto LB agar containing 100 μg of cycloheximide/ml and the appropriate antibiotics for each bacterial strain.
Experiments were performed three times each with three replicates for each seed-strain combination at each of three sampling times (24, 45, and 96 h) for experiments comparing colonization of seeds by strains A-11 and 501R3. The mean log10 CFU per seed were determined and compared for strains 501R3 and A-11 at each time for each seed type by using Student’s t test (SAS Institute, Cary, N.C.). Data from all three experiments were combined prior to analysis. There was a significant experiment effect (P ≤ 0.05); however, there was no significant experiment×treatment effect for experiments conducted in natural soil and in sterile sand.
For biochemical restoration of seed colonization, E. cloacae strains were grown, washed, and resuspended in SDW or SDW containing 6% fructose. Suspensions (40 μl) were applied to individual cucumber or radish seeds in sterile sand, and populations were determined after 18 h as described above. The quantities of fructose added to the cucumber and radish seeds were similar to the quantity of carbohydrates exuded from pea seeds over the initial 96 h after the onset of imbibition. Experiments were performed twice with six replicates for each treatment. For genetic restoration of seed colonization, E. cloacae strains were added to cucumber seeds in sterile sand as described above and sampled at 18 h. Experiments were performed three times with three replicates for each treatment. Means were determined and compared by least significant differences (SAS) for both the biochemical and the genetic restoration experiments. Results from both sets of experiments were combined prior to analysis.
Analysis of aqueous seed extracts.
Seed extracts were made essentially as previously described (21, 29). Seeds (25 g) in a 250-ml Erlenmeyer flask were surface disinfested in 10% Clorox for 20 min followed by two 20-min rinses in SDW. Surface-disinfested seeds (2.5 g) were added to 10 ml of SDW and incubated at room temperature in the dark. At the sampling times (24, 45, and 96 h) the aqueous contents of the flasks were decanted and checked for microbial contamination by spotting 10-μl aliquots onto nutrient agar. SDW (10 ml) was subsequently added to the flasks, and the flasks were incubated as described above until the next sampling time. All noncontaminated samples from each sampling time for each seed type were pooled, frozen, and lyophilized to dryness.
Total carbohydrates in the samples were estimated by the anthrone assay (20) with glucose as the standard. Individual carbohydrates were identified and quantified as trifluoroacetyl derivatives by using gas chromatography in 24-, 45-, and 96-h samples as previously described (32, 38). Total amino acids in samples were estimated by the ninhydrin assay (37) with l-leucine as the standard.
Molecular techniques and bacterial matings.
DNA isolations, transformations, restriction digests, electrophoresis, ligations, and Southern blot hybridizations were performed as previously described (34). Complementation of strain A-11 was performed by mobilizing a total genomic DNA cosmid library of E. cloacae 501R3 in the cosmid vector pLAFR3 (constructed by J. Loper, Agricultural Research Service, Corvallis, Oreg.) into strain A-11 by triparental mating (10) and selecting directly for transconjugants which grew on M56 basal salts agar containing 0.2% N-acetylglucosamine, RIF, and TET. All other transconjugants were selected by growth on LB agar supplemented with RIF and TET, except matings into E. coli DF456, which were selected by growth on LB agar supplemented with STR and TET.
Nucleotide sequence was determined by the dideoxy chain termination method by fluorescence labelling with Amplitaq (Applied Biosystems Inc. [ABI]) run on an ABI model 373 automated sequencer. Overlapping sequences were generated by using a series of nested deletions of fragments subcloned into pGEM7Z(+) generated by digestion with ExoIII (15) or by the use of selected primers. Sequences were analyzed using the DNA analysis programs of DNAStar (Lasergene, Inc.). BLAST searches of databases (1) were conducted with translated proteins by using the Blastp program available on the National Center for Biotechnology Information web page (20a).
Nucleotide sequence accession number.
The nucleotide sequence and the sequences of the translated proteins have been deposited in GenBank under accession no. AF098509.
RESULTS AND DISCUSSION
Seed colonization.
E. cloacae A-11 increased slightly (17-fold) from 104 CFU per seed to approximately 105 CFU per seed over the initial 24 h after application to cucumber seeds in natural soil. Populations of strain A-11 remained at this level 45 and 96 h after application (Table 2). Populations of strain 501R3 increased continuously (200-fold) and more rapidly than those of strain A-11 over this 96-h period to approximately 106 CFU per seed. Populations of strain 501R3 were significantly greater (P ≤ 0.02) than those of strain A-11 at 96 h on cucumber seeds in natural soil. Similar results were obtained for experiments performed in radish spermospheres in natural soil. Populations of both strains increased over this 96-h period. However, populations of strain A-11 were significantly lower than those of strain 501R3 at 45 h (P ≤ 0.001) and at 96 h (P ≤ 0.01) after application. Similar results were also obtained when seed colonization studies were performed in sterile sand. Populations of strain A-11 grew more slowly and were significantly smaller (P ≤ 0.005) than those of strain 501R3 at 24 h in both cucumber and radish spermospheres (Table 2).
TABLE 2.
Growth of E. cloacae 501R3 and A-11 on various seeds in natural soil and in sterile sanda
| Seed | Time (h) | Growth in natural soil (log10 CFU/seed)
|
P | Growth in sterile sand (log10 CFU/seed)
|
P | ||
|---|---|---|---|---|---|---|---|
| 501R3 | A-11 | 501R3 | A-11 | ||||
| Cucumber | 24 | 5.17 | 5.22 | 0.92 | 7.21 | 4.49 | 0.0001* |
| 45 | 5.75 | 4.99 | 0.11 | 7.40 | 7.01 | 0.11 | |
| 96 | 6.35 | 5.25 | 0.02* | 7.73 | 7.90 | 0.46 | |
| Radish | 24 | 4.95 | 4.67 | 0.56 | 7.50 | 6.79 | 0.005* |
| 45 | 5.89 | 4.36 | 0.001* | 7.69 | 7.69 | 0.98 | |
| 96 | 6.54 | 5.36 | 0.01* | 8.17 | 7.94 | 0.35 | |
| Pea | 24 | 7.31 | 7.24 | 0.88 | 8.51 | 8.28 | 0.36 |
| 45 | 8.10 | 7.39 | 0.13 | 8.89 | 8.99 | 0.68 | |
| 96 | 8.22 | 7.35 | 0.07 | 9.30 | 9.08 | 0.37 | |
| Soybean | 24 | 7.01 | 6.89 | 0.59 | 8.11 | 8.17 | 0.81 |
| 45 | 7.52 | 7.13 | 0.09 | 8.38 | 8.40 | 0.92 | |
| 96 | 8.06 | 8.06 | 0.97 | 9.07 | 8.61 | 0.06 | |
| Sunflower | 24 | 6.68 | 6.16 | 0.27 | 8.32 | 8.37 | 0.83 |
| 45 | 6.03 | 5.80 | 0.63 | 8.60 | 8.62 | 0.92 | |
| 96 | 6.69 | 5.90 | 0.10 | 8.78 | 8.90 | 0.64 | |
| Sweet corn | 24 | 6.81 | 6.73 | 0.72 | 8.28 | 8.23 | 0.86 |
| 45 | 7.20 | 7.08 | 0.60 | 8.55 | 8.57 | 0.91 | |
| 96 | 7.46 | 7.29 | 0.54 | 9.39 | 8.95 | 0.08 | |
Similar populations (approximately 104 CFU per seed) of strains 501R3 and A-11 were applied to all seed cultivars. Populations of these strains were determined by spiral plating 24, 45, and 96 h after application. Asterisks indicate statistically significant (P ≤ 0.05) differences between the strains.
There was evidence of strain A-11 achieving slightly lower populations than strain 501R3 on pea, soybean, sunflower, and sweet corn seeds at 45 and 96 h after application. However, populations of strains A-11 and 501R3 increased significantly and were statistically similar (P ≤ 0.05) in pea, soybean, sunflower, and sweet corn spermospheres at 24, 45, and 96 h after application in both natural soil and sterile sand (Table 2). This suggests that compounds other than reduced carbon compounds are limiting to colonization in the spermospheres of these pea, soybean, sunflower, and sweet corn seed cultivars.
Analysis of seed exudate.
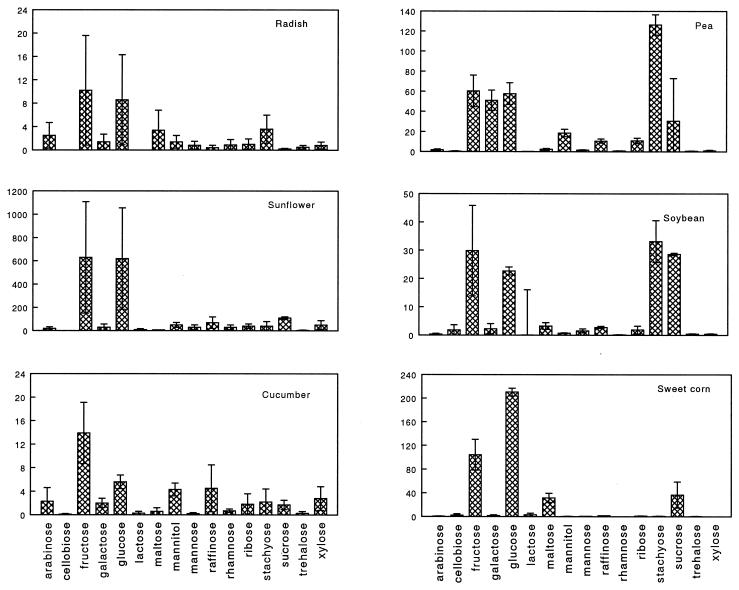
The cultivars of pea, soybean, and sweet corn seeds tested released approximately 1,000-fold more glucose equivalents, as determined by the anthrone assay, than radish or cucumber seeds, while sunflower seeds released approximately 20-fold more glucose equivalents (Table 3). This trend was confirmed by gas chromatography (Fig. 1). Fructose comprised a major portion of the carbohydrates released by each seed for all six seed cultivars over 96 h. However, there were approximately 2 to 3 orders of magnitude more fructose released by the cultivars of the sunflower, pea, soybean, and sweet corn seeds tested than by those of the cucumber and radish seeds. Other prevalent carbohydrates detected were glucose, galactose, sucrose, and stachyose. Significantly more total amino acids, as determined by the ninhydrin assay, were also released by pea, soybean, sunflower, and sweet corn seeds than by the cultivars of cucumber or radish seeds tested over this 96-h period (Table 3).
TABLE 3.
Analysis of aqueous seed extractsa
| Seed | Total carbohydrates (μg of glucose equivalents/seed) at:
|
Total amino acids (μg of leucine equivalents/seed) at:
|
||||
|---|---|---|---|---|---|---|
| 0–24 h | 24–45 h | 45–96 h | 0–24 h | 24–45 h | 45–96 h | |
| Cucumber | 1.0 ± 0.4 | 0.7 ± 0.1 | 1.6 ± 0.1 | UDb | 1.0 ± 1.0 | UD |
| Radish | 5.9 ± 0.8 | 1.8 ± 0.2 | 0.9 ± 0.1 | 2.4 ± 0.3 | 0.9 ± 0.1 | 0.9 ± 0.9 |
| Pea | 2,981.8 ± 184.0 | 409.2 ± 16.9 | 95.7 ± 6.2 | 336.8 ± 30.7 | 131.2 ± 0 | 83.1 ± 4.4 |
| Soybean | 987.7 ± 42.8 | 105.6 ± 7.5 | 54.5 ± 2.3 | 190.0 ± 44.3 | 12.9 ± 12.9 | 17.2 ± 17.2 |
| Sunflower | 52.1 ± 0.2 | 21.3 ± 1.5 | 43.8 ± 0.6 | 31.2 ± 31.2 | 9.8 ± 9.8 | 15.6 ± 15.6 |
| Sweet corn | 1,762.2 ± 193.3 | 1,664.7 ± 92.1 | 1,649.9 ± 86.8 | 92.9 ± 16.4 | 88.8 ± 20.5 | 120 ± 11.0 |
Extracts were collected at the indicated time intervals after the start of imbibition. Total carbohydrates were determined by the anthrone assay, and total amino acids were determined by the ninhydrin assay.
UD, undetectable.
FIG. 1.
Individual carbohydrates released by cucumber, radish, pea, soybean, sunflower, and sweet corn seeds. Quantities represented are summed from 0 to 24 h, 24 to 45, and 45 to 96 h samples. Error bars represent one standard deviation from the mean. Note that quantities are micrograms/seed for pea, soybean, and sweet corn seeds and nanograms/seed for cucumber, radish, and sunflower seeds.
Characterization of E. cloacae A-11.
Strain 501R3 grew on 13 of 16 carbohydrates detected by gas chromatography in the seed exudates (Fig. 1) when supplied as sole sources of reduced carbon. Strain 501R3 did not grow on d-lactose, l-rhamnose, or stachyose. In contrast, strain A-11 showed significant growth on only fructose. Growth on fructose by strain A-11 was similar to that by strain 501R3 (data not shown). Strains 501R3 and A-11 had similar growth on the l-amino acids alanine, asparagine, glutamine, glutamate, proline, and serine and on pyruvate and malate when supplied as sole sources of reduced carbon. Strains 501R3 and A-11 also had similar growth on glycerol supplied as a reduced carbon source and on the l-amino acids alanine, asparagine, cysteine, glutamate, glutamine, proline, and serine when supplied as sole sources of nitrogen (data not shown). This nutritional utilization profile is consistent with that of pfkA mutants of the closely related bacterium E. coli (33).
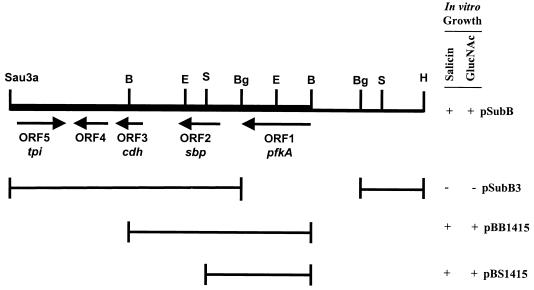
Cosmid p82, which was identified from a genomic library of strain 501R3, restored the ability of strain A-11 to grow on M56 basal salts agar amended with either 2% N-acetylglucosamine or 2% salicin. Plasmid pSubB, containing an 8.5-kb subcloned fragment from p82 (Fig. 2), restored the growth of strain A-11 on M56 minimal salts medium amended individually with salicin, N-acetylglucosamine and all 12 other carbohydrates tested. The 8.5-kb DNA fragment from pSubB was used as a probe in Southern hybridizations to genomic DNA from strains A-11 and 501R3 digested with EcoRI. Analysis of the blot (data not shown) indicated that transposon mini-Tn5 KAN was inserted within a 1.5-kb EcoRI fragment located within pSubB (Fig. 2). A 2.5-kb BamHI fragment which contained this EcoRI fragment was subsequently subcloned from pSubB in both orientations into pRK415, resulting in plasmids pBB1415a and pBB1415b. Both plasmids restored the growth of strain A-11 on salicin, N-acetylglucosamine, and all 12 other carbohydrates tested, indicating that this 2.5-kb BamHI fragment contained the gene of interest in its entirety. In contrast, pSubB-3, which consisted of a 2-kb BglII deletion of pSubB, did not restore the growth of strain A-11 on either salicin or N-acetylglucosamine (Fig. 2).
FIG. 2.
Physical maps of plasmid pSubB and pSubB-derived subclones. Abbreviations: B, BamHI; Bg, BglII; E, EcoRI; H, HindIII; S, SalI. Arrows indicate the direction of transcription. The thick line indicates the sequenced portion of pSubB. E. cloacae A-11 harboring the indicated plasmid was capable (+) or incapable (−) of in vitro growth on M56 minimal medium plus 0.2% salicin or N-acetylglucosamine.
Nucleotide sequence analysis of the 2.5-kb BamHI fragment from pBB1415 indicated the presence of two complete open reading frames (ORFs) and part of a third ORF, all with the same transcriptional orientation (Fig. 2). Each of these ORFs contained a predicted ATG start codon that was preceded within 6 to 10 bases by sequences resembling ribosome binding sites. The predicted start codon of ORF1 was located 212 bases from the BamHI site and encoded a predicted protein product of 320 amino acids. Two DNA inverted repeat sequences, representing potential transcriptional termination sites, were identified immediately downstream of the stop codon. The first repeat was located 12 bases from the stop codon and consisted of the 7-base inverted repeat sequence of GCCCGGT-N12-ACCGGGC. The second repeat was located 20 bases downstream from the first repeat and consisted of the sequence GCCGGGT-N12-ACCCGGC. Database searches revealed that the predicted protein for this ORF had an amino acid sequence identity of 88% to the pfkA gene product of E. coli (GenBank accession no. P06998). pfkA encodes phosphofructo kinase, a key enzyme in glycolysis catalyzing the conversion of fructose-6-phosphate to fructose-1,6-phosphate (33). To verify that the mutation in strain A-11 was located within pfkA, a 1.5-kb BamHI-SalI fragment containing the entire pfkA ORF and only the 5′ end of the sbp ORF was subcloned into pRK415. The resultant plasmid, pBS1415 (Fig. 2), restored the growth of strain A-11 on all 14 carbohydrates tested. In addition, pBS1415 was mobilized into E. coli DF456, a pfkA-deficient strain, restoring growth on N-acetylglucosamine.
The second ORF (ORF2) within the 2.5-kb BamHI fragment was positioned 1,384 bp from the BamHI site, beginning 202 bases from the termination codon of the pfkA homolog, and consisted of a predicted protein product of 329 amino acids. Database searches indicated 83% sequence identity to the sulfur binding protein of E. coli encoded by sbp (GenBank accession no. S40860). Analysis of the nucleotide sequence extending an additional 3.25 kb downstream from the 2.5-kb BamHI fragment indicated that pfkA and sbp are physically linked to a gene with 76% sequence identity to cdh of E. coli, which encodes CDP-diglyceride hydrolase (GenBank accession no. P06282) and an ORF (ORF4) of unknown function. Database searches for this ORF did not identify any matches of significance at the nucleotide or predicted amino acid sequence levels. A fifth ORF (ORF5) encodes a protein with 91% identity to triose phosphate isomerase of E. coli (GenBank accession no. P04790) (Fig. 2).
Genetic and biochemical restoration of seed colonization by strain A-11.
Populations of strain 501R3 increased from approximately 104 CFU per seed to greater than 107 CFU per seed in 18-h assays on surface-disinfested cucumber seeds in sterile sand (Table 4). Populations of strains A-11 and A-11(pRK415) showed no substantial increases after 18 h when approximately 104 CFU per seed were added to the cucumber seeds. Populations of strains A-11 and A-11(pRK415) were substantially smaller than those of strain 501R3 at this time. Plasmids pBB1415 and pBS1415 (Fig. 2) restored the ability of A-11 to grow on cucumber seeds to near the levels associated with the wild-type strain, strain 501R3 (Table 4).
TABLE 4.
Genetic restoration of colonization of cucumber spermosphere by E. cloacae A-11
| Straina | Mean colonization (log10 CFU/seed)b |
|---|---|
| 501R3 | 7.56 A |
| A-11(pBB1415) | 7.07 B |
| A-11(pBS1415) | 6.98 B |
| A-11 | 4.64 C |
| A-11(pRK415) | 2.75 D |
| LSDc | 0.29 |
Similar populations (approximately 104 CFU per seed) of all strains were applied to cucumber seeds in sterile sand.
Populations at 18 h after application of bacteria to the seed. Numbers followed by the same letters are not significantly different (P ≤ 0.05).
LSD, least significant difference (P ≤ 0.05).
Addition of fructose, a carbohydrate capable of supporting wild-type growth of strain A-11, to treatments containing strain A-11, also restored the ability of strain A-11 to grow on both cucumber and radish seeds to levels similar to those of strain 501R3 (Table 5). The quantity of fructose added to cucumber and radish seeds in these treatments was similar to the quantity of total carbohydrates exuded from pea seeds over the initial 96 h after the onset of imbibition.
TABLE 5.
Biochemical restoration of colonization of cucumber and radish spermospheres by E. cloacae A-11
| Strain and treatmenta | Mean colonization (log10 CFU/seed)b on:
|
|
|---|---|---|
| Cucumber seeds | Radish seeds | |
| 501R3 | 7.16 A | 7.10 A |
| A-11 + fructose | 7.45 A | 6.81 A |
| A-11 | 6.46 B | 6.37 B |
| LSDc | 0.33 | 0.31 |
Similar populations (approximately 104 CFU per seed) were added to cucumber and radish seeds in sterile sand for all treatments.
Populations at 18 h after application to cucumber seed. Numbers followed by the same letters in each column are not significantly different (P ≤ 0.05).
LSD, least significant difference (P ≤ 0.05).
Genetic and biochemical data presented here strongly suggest that the strain A-11 colonization phenotype is solely due to inactivation of pfkA rather than to the loss of other physically linked genes that appear to be involved in general carbohydrate catabolism (Fig. 2). A DNA fragment containing only pfkA restored the strain 501R3 phenotype to strain A-11 with regard to in vitro growth and seed colonization. Also, the presence of an inverted repeat indicative of a transcriptional termination signal downstream from pfkA suggests this gene is expressed independently of other genes. Finally, the exogenous addition of fructose in colonization experiments circumvented the impact of the mutation in pfkA on colonization by supplying strain A-11 with a reduced carbon source utilizable for growth. Fructose is not expected to biochemically complement any of the genes physically linked to pfkA. These observations link the loss of pfkA function, the loss of carbohydrate catabolic capabilities, and decreased colonization of seeds that release relatively small quantities of reduced carbon such as the cucumber and radish cultivars tested here.
Conclusion.
Nutrient-rich plant exudates are expected to support an abundance of microbial growth while nutrient-poor exudates are not. Since it has been established that growth is an essential component of microbial colonization processes (24, 29–31, 35), it can be assumed that the catabolic capabilities of a bacterium, with regard to specific reduced carbon compounds found in exudates, contribute directly to its ability to colonize a given host plant. Under nutrient-poor conditions a greater ability to catabolize compounds in exudates is expected to improve colonization. Although these assumptions are obvious and accepted by many, there is little evidence to support them. Our studies with E. cloacae 501R3 and the near-isogenic strain A-11 directly support these assumptions for microbial colonization of the spermosphere. Strain A-11 had decreased catabolic capabilities with regard to specific carbohydrates in seed exudates relative to those of strain 501R3. The colonization of cucumber and radish seeds by strain A-11 was also reduced relative to that by strain 501R3, but there were no differences between the two strains relative to the colonization of pea, soybean, sunflower, and sweet corn seeds. The exudates from these cultivars of pea, soybean, sunflower, and sweet corn seeds contained, on average, several orders of magnitude more carbohydrates and amino acids than the exudate from cucumber or radish seeds.
Work with additional carbohydrate utilization mutants of E. cloacae and other plant-beneficial bacteria needs to be performed to substantiate the findings presented here. In addition, mutants affected in the utilization of other nutrients such as nitrogen need to be analyzed. The acquisition of reduced carbon compounds and other nutrients is fundamentally important for growth and other desired microbial activities such as biocontrol in the spermosphere and rhizosphere (25). An understanding of the nutritional requirements of beneficial bacteria colonizing subterranean portions of plants and the impact of available nutrients on beneficial activities such as colonization and biocontrol is required if the behavior of beneficial bacteria in specific environments is to be accurately predicted.
ACKNOWLEDGMENTS
We acknowledge Kim Brandon, Ricky Brathwaite, and Sean Wu for excellent technical assistance.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang A, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson T A, Guthrie E A, Walton B T. Bioremediation in the rhizosphere. Environ Sci Technol. 1993;27:2630–2636. [Google Scholar]
- 3.Anderson A J, Habibzadegah-Tari P, Tepper C S. Molecular studies on the role of root surface agglutinin in adherence and colonization by Pseudomonas putida. Appl Environ Microbiol. 1988;54:375–380. doi: 10.1128/aem.54.2.375-380.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buell C R, Anderson A J. Genetic analysis of the aggA locus involved in agglutination and adherence of Pseudomonas putida, a beneficial fluorescent pseudomonad. Mol Plant-Microbe Interact. 1992;5:154–162. doi: 10.1094/mpmi-5-154. [DOI] [PubMed] [Google Scholar]
- 5.Curl E A, Truelove B. The rhizosphere. New York, N.Y: Springer-Verlag; 1986. [Google Scholar]
- 6.Davison J. Plant beneficial bacteria. Bio/Technology. 1988;6:282–286. [Google Scholar]
- 7.De Mot R, Proost P, Van Damme J, Vanderleyden J. Homology of the root adhesion of Pseudomonas fluorescens OE 28.3 with porin F of P. syringae. Mol Gen Genet. 1992;231:489–493. doi: 10.1007/BF00292721. [DOI] [PubMed] [Google Scholar]
- 8.de Weger L A, van der Vlugt C I M, Wijfges A H M, Bakker P A H M, Schippers B, Lugtenberg B. Flagella of a plant-growth-stimulating Pseudomonas fluorescens are required for colonization of potato roots. J Bacteriol. 1987;169:2769–2773. doi: 10.1128/jb.169.6.2769-2773.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Weger L A, Bakker P A H M, Schippers B, van Loosdrecht M C M, Lugtenberg B J J. Pseudomonas spp. with mutational changes in the O-antigenic side chain of their lipopolysaccharide and affected in their ability to colonize potato roots. In: Lugtenberg B J J, editor. NATO ASI Series. H36. Berlin, Germany: Springer-Verlag; 1989. pp. 197–202. [Google Scholar]
- 10.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 12.Gamliel A, Katan J. Chemotaxis of fluorescent pseudomonads towards seed exudates and germinating seeds in solarized soil. Phytopathology. 1992;82:328–332. [Google Scholar]
- 13.Hadar Y, Harman G E, Taylor A G, Horton J M. Effects of pregermination of pea and cucumber seeds and of seed treatment with Enterobacter cloacae on rots caused by Pythium spp. Phytopathology. 1983;73:1322–1325. [Google Scholar]
- 14.Heinrich D, Hess D. Chemotactic attraction of Azospirillum lipoferum by wheat roots and characterization of some attractants. Can J Microbiol. 1985;31:27–31. [Google Scholar]
- 15.Henikoff S. Unidirectional digestion with exonuclease III creates target breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 16.Kleeberger A, Castorph H, Klingmüller W. The rhizosphere microflora of wheat and barley with special reference to Gram-negative bacteria. Arch Microbiol. 1983;136:306–311. [Google Scholar]
- 17.Ladha J K, Barraquio W L, Watanabe I. Isolation and identification of nitrogen-fixing Enterobacter cloacae and Klebsiella planticola associated with rice plants. Can J Microbiol. 1983;29:1301–1308. doi: 10.1139/m83-141. [DOI] [PubMed] [Google Scholar]
- 18.Matthysse A G, McMahan S. Root colonization by Agrobacterium tumefaciens is reduced in cel, attB, attD, and attR mutants. Appl Environ Microbiol. 1998;64:2341–2345. doi: 10.1128/aem.64.7.2341-2345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 20.Morris D L. Quantitative determination of carbohydrates with Dreywood’s anthrone reagent. Science. 1948;107:254–255. doi: 10.1126/science.107.2775.254. [DOI] [PubMed] [Google Scholar]
- 20a.National Center for Biotechnology Information. 1 April 1999, revision date. National Center for Biotechnology Information. [Online.] http://www.ncbi.nlm.nih.gov/BLAST/. [15 October 1998, last date accessed.]
- 21.Nelson E B, Chao W L, Norton J M, Nash G T, Harman G E. Attachment of Enterobacter cloacae to hyphae of Pythium ultimum: possible role in the biological control of Pythium preemergence damping-off. Phytopathology. 1986;76:327–335. [Google Scholar]
- 22.Nelson E B. Biological control of Pythium seed rot and preemergence damping-off of cotton with Enterobacter cloacae and Erwinia herbicola applied as seed treatments. Plant Dis. 1988;72:140–142. [Google Scholar]
- 23.Nguyen N D, Gottgert M, Singh M, Klingmüller W. Nif− hybrids of Enterobacter cloacae: selection for nif− gene integration with nif− plasmids containing the Mu transposon. Mol Gen Genet. 1983;192:439–443. [Google Scholar]
- 24.O’Sullivan D J, O’Gara F. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev. 1992;56:662–676. doi: 10.1128/mr.56.4.662-676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulitz T C. Biochemical and ecological aspects of competition in biological control. In: Baker R, Dunn P E, editors. New directions in biological control: alternatives for suppressing agricultural pests and diseases. New York, N.Y: Alan R. Liss; 1990. pp. 713–724. [Google Scholar]
- 26.Polonenko D R, Dumbroff E B, Mayfield C I. Microbial responses to salt-induced osmotic stress. II. Population changes in the rhizoplane and rhizosphere. Plant Soil. 1981;63:415–426. [Google Scholar]
- 27.Provence D L, Curtiss R., III . Gene transfer in gram-negative bacteria. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 317–347. [Google Scholar]
- 28.Rennie J R, de Freitas J R, Ruschel A P, Vose P B. Isolation and identification of N2-fixing bacteria associated with sugar cane (Saccharum sp.) Can J Microbiol. 1982;28:462–467. [Google Scholar]
- 29.Roberts D P, Sheets C J, Hartung J S. Evidence for proliferation of Enterobacter cloacae on carbohydrates in cucumber and pea spermosphere. Can J Microbiol. 1992;38:1128–1134. [Google Scholar]
- 30.Roberts D P, Marty A M, Dery P D, Yucel I, Hartung J S. Amino acids as reduced carbon sources for Enterobacter cloacae during colonization of the spermospheres of crop plants. Soil Biol Biochem. 1996;28:1015–1020. [Google Scholar]
- 31.Roberts D P, Dery P D, Hartung J S. Peptide utilization and colonization of corn, radish and wheat spermospheres by Enterobacter cloacae. Soil Biol Biochem. 1996;28:1109–1111. [Google Scholar]
- 32.Roberts D P, Dery P D, Yucel I, Buyer J, Holtman M A, Kobayashi D Y. Genetic analysis of the seed colonization mutant Enterobacter cloacae A-11. In: Ogashi A, Kobayashi K, Homma Y, Kodama F, Kondo N, Akino S, editors. Plant growth-promoting rhizobacteria. Present status and future prospects. Proceedings of the Fourth International Workshop on Plant Growth-Promoting Rhizobacteria, Japan-OECD Joint Workshop, Sapporo, Japan, October 5–10, 1997. 1997. pp. 330–332. [Google Scholar]
- 33.Roehl R A, Vinopal R T. Lack of glucose phosphotransferase function in phosphofructokinase mutants of Escherichia coli. J Bacteriol. 1976;126:852–860. doi: 10.1128/jb.126.2.852-860.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Simons M, Van Der Bij A J, Brand I, de Weger L A, Wijffelman C A, Lugtenberg B J J. Gnotobiotic system for studying rhizosphere colonization by plant growth-promoting Pseudomonas bacteria. Mol Plant-Microbe Interact. 1996;9:600–607. doi: 10.1094/mpmi-9-0600. [DOI] [PubMed] [Google Scholar]
- 36.Simons M, Permentier H P, de Weger L A, Wijffelman C A, Lugtenberg B J J. Amino acid synthesis is necessary for tomato root colonization by Pseudomonas fluorescens strain WCS365. Mol Plant-Microbe Interact. 1997;10:102–106. [Google Scholar]
- 37.Spies J R. Colorimetric procedures for amino acids. Methods Enzymol. 1957;3:467–476. [Google Scholar]
- 38.Sullivan J E, Schewe L R. Preparation and gas chromatography of highly volatile trifluoroacetylated carbohydrates using N-methyl bis[trifluoroacetamide] J Chromatogr Sci. 1977;15:196–197. [Google Scholar]
- 39.Thomas-Bauzon D, Weinhard P, Villecourt P, Balandreau J. The spermosphere model. I. Its use in growing, counting, and isolating N2-fixing bacteria from the rhizosphere of rice. Can J Microbiol. 1982;28:922–928. [Google Scholar]
- 40.Thomson J, Gerstenberger P D, Goldberg D E, Gociar E, de Silva A O, Fraenkel D G. ColE1 hybrid plasmids for Escherichia coli genes of glycolysis and the hexose monophosphate shunt. J Bacteriol. 1979;137:502–506. doi: 10.1128/jb.137.1.502-506.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vesper S J. Production of pili (fimbriae) by Pseudomonas fluorescens and correlation with attachment to corn roots. Appl Environ Microbiol. 1987;53:1397–1403. doi: 10.1128/aem.53.7.1397-1405.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]