Abstract
The use of juvenile Artemia as feed in aquaculture and in the pet shop industry has been getting more attention during the last decade. In this study, the use of selected bacterial strains to improve the nutritional value of dry food for Artemia juveniles and to obtain control of the associated microbial community was examined. Nine bacterial strains were selected based on their positive effects on survival and/or growth of Artemia juveniles under monoxenic culture conditions, while other strains caused no significant effect, significantly lower rates of survival and/or growth, or even total mortality of the Artemia. The nine selected strains were used to preemptively colonize the culture water of Artemia juveniles. Xenic culture of Artemia under suboptimal conditions yielded better survival and/or growth rates when they were grown in the preemptively colonized culture medium than when grown in autoclaved seawater. The preemptive colonization of the culture water had a drastic influence on the microbial communities that developed in the culture water or that were associated with the Artemia, as determined with Biolog GN community-level physiological profiles. Chemotaxonomical characterization based on fatty acid methyl ester analysis of bacterial isolates recovered from the culture tanks was performed, and a comparison with the initially introduced strains was made. Finally, several modes of action for the beneficial effect of the bacterial strains are proposed.
Because of convenience in production and their suitable biochemical composition, the brine shrimp Artemia is the most frequently used live food in the larviculture of economically important crustaceans and fishes. Among Artemia at different life stages that are appropriate for use in aquaculture, the use of juvenile and adult Artemia has been getting more attention over the last decade (3).
It has been demonstrated that bacteria have a beneficial effect in the culture of the obligate suspension feeder Artemia, as the addition of bacterial strains to axenic cultures of Artemia fed other foods revealed that some bacterial strains may improve the survival and growth rates of Artemia (2). Similarly, the culture of Artemia under nonsterile conditions usually results in higher biomass production (BP) than that under axenic conditions, showing that the nutritional value of the food partly depends on the spontaneous colonization of the food particles by harmless bacteria (2, 4). Colonization by bacteria may even be essential when using inexpensive agricultural waste products like rice bran to support Artemia culture (2, 4). However, this may as well depend on the quality of the provided food, since other authors were able to culture Artemia axenically on autoclaved rice bran (5).
Several attempts to culture Artemia on a diet consisting solely of bacteria failed (2, 19). However, Gorospe and Nakamura (5) found a Pseudomonas sp. that was able to delay the death of the Artemia when no other food was given, and they assumed that it was used as a food. Rico-Mora and Voltolina (18) came to the same conclusions regarding the use of several strains isolated from a diatom culture. Yasuda and Taga (23) found an Acinetobacter strain which was by itself able to support the mass culture of Artemia. A Flexibacter strain, lnp3, provided as the only food source supported survival and growth (88% survival rate and 5-mm body length, respectively) of nauplii to preadults in 8 days, although seven times more bacterial biomass than algal biomass was required to yield similar growth (9).
Bacteria are reported to contribute to the nutritional value of foods by being a major source of protein and amino acids (6, 20). The results of Intriago and Jones (9) suggest that the bacteria also assisted in the digestion of the unicellular algae, although convincing evidence was not provided.
Agricultural byproducts, such as rice bran, corn bran, soybean pellets, lactoserum, etc., are used as cheap food sources for the intensive culture of Artemia up to the adult stage as a cost-effective alternative to algae (3). Under these intensive culture conditions, opportunistic bacteria develop, and unfavorable colonization of the culture medium and the Artemia may occur (22). As no further microbial control is usually performed, this may lead to a low production of Artemia biomass or result in the transfer of pathogenic bacteria via the Artemia to the predator (4, 13). In this perspective, Yasuda and Taga (23) anticipated that bacteria would be found to be useful not only as food for Artemia but also as biological controllers of fish disease and activators of the rate of nutrient regeneration.
The present study examines the application of bacterial strains in the culture of Artemia juveniles with a twofold goal, the improvement of the nutritional value of food for Artemia, leading to a higher biomass production of the culture and the control of deleterious bacteria associated with Artemia through preemptive colonization of the culture medium. In the first stage of this study, bacterial isolates were selected based on their positive effect on the Artemia culture under monoxenic conditions. In the second stage, xenic cultures were performed in media preemptively colonized by the selected bacterial strains, and their effect on the zootechnical performance and the microbial community was assessed.
MATERIALS AND METHODS
Bacterial strains.
Eighteen arbitrarily chosen bacterial strains were examined under monoxenic culture conditions. All the strains originated from previous well-performing Artemia cultures, except Pseudomonas fluorescens LMG1244 (ATCC 17571; isolated from polluted seawater in Denmark), Vibrio alginolyticus LMG4409T (ATCC 17749; isolated from spoiled horse mackerel), Vibrio proteolyticus Q113 (isolated from a well-performing culture of sea bass in Spain), and V. proteolyticus CW8T2 (isolated from the artificial feed used in a sea bass hatchery in Spain).
Monoxenic culture of Artemia juveniles. (i) Axenic hatching.
As axenic Artemia nauplii were required for the experiments, high-quality cysts (EG grade; INVE Aquaculture Inc., Baasrode, Belgium) were disinfected according to a modification of the procedure of Provasoli and Shiraishi (16). About 0.5 g of cysts was suspended in 30 ml of autoclaved seawater and shaken for approximately 2 min. Floating cysts were removed from the surface and discarded. Subsequently, the seawater was removed and replaced by 30 ml of an aqueous solution of merthiolate (1 g/liter), and the tube was shaken for approximately 10 min. The liquid was replaced by merthiolate and the shaking was done two more times. The merthiolate was then discarded, and the cysts were rinsed five times with 30 ml of autoclaved artificial seawater containing 33 g of Instant Ocean synthetic sea salt (Aquarium Systems Inc., Sarrebourg, France)/liter. An aliquot of the disinfected cysts was subsequently transferred to test tubes containing 2 ml of marine broth 2216 (Difco Laboratories, Detroit, Mich.) and hatched for 24 h. If the disinfection procedure was not efficient or if any bacterial contamination had occurred, it manifested itself by an increased turbidity of the marine broth compared to that in uninoculated test tubes. In such cases, the hatched Artemia nauplii were not used further in the experiments.
(ii) Growth conditions.
The hatched nauplii were diluted in autoclaved seawater, and 20 nauplii were transferred to sterile 50-ml Falcon tubes (Becton Dickinson Labware, Lincoln Park, N.J.) containing 30 ml of autoclaved seawater. The Artemia were fed daily with 4.9 mg (days 0 and 1) or 5.6 mg (days 2 to 6) of gamma-irradiated food (10 kGy) suspended in autoclaved deionized water. This food was regularly checked for its sterility by plating 100 μl of the food suspension on marine agar plates.
(iii) Inoculation of the bacterial strains.
Pure cultures of the bacterial strains were grown overnight in marine broth at 28°C, transferred to centrifugation tubes, and centrifuged at approximately 8,500 × g for 10 min. The supernatant was discarded and the pellet was resuspended in autoclaved nine-salt solution (NSS) (14). The bacterial density was determined spectrophotometrically at 550 nm, assuming that an optical density of 1.000 corresponded to 1.2 × 109 cells/ml according to the McFarland standard (BioMérieux, Marcy l’Etoile, France). Immediately after the transfer of the nauplii to the tubes, the bacterial suspension was added at a density of approximately 5 × 106 cells/ml. Several test runs were performed, each with four replicates per treatment.
Uninoculated tubes remaining axenic throughout the growth period acted as a control. At regular intervals, sterility controls of the axenic control treatment were performed by plating 100 μl of the undiluted culture medium on marine agar.
(iv) Evaluation of the addition of the individual strains.
After 3 or 6 days, depending on the experiment, the surviving Artemia in the Falcon tubes were counted, and their body lengths were determined as described by Verschuere et al. (22). The individual dry weights (IDW [in micrograms]) were calculated from the body length (in millimeters) according to the method of Abreu-Grobois et al. (1). The BP in the Falcon tube was calculated based on the number of surviving Artemia (ranging from 0 to 20) and their IDW as follows:
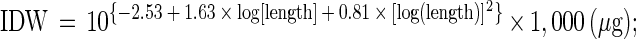 |
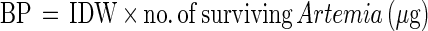 |
The survival rates, the body lengths, and the BP of the Artemia were compared statistically to the corresponding control treatment with the t test (if the experiment was performed only once) or with analysis of variance (general factorial procedure) in which both the treatment and the experiments were considered fixed factors, taking into consideration the interaction between both factors whenever it occurred. The latter statistical comparison was performed with SPSS for Windows release 7.5.2 (SPSS, Inc.).
The xenic culture of Artemia in PCCM.
To examine the effect of the bacterial colonization of the culture medium on the Artemia culture performance, seawater was preemptively colonized by the nine bacterial strains selected in the previous experiments. This type of culture water is hereafter referred to as preemptively colonized culture medium (PCCM). Alternatively, culture water was preemptively colonized by opportunistic bacteria through a prolonged recirculation over a biofilter exposed to the ambient air. The seawater was allowed to be colonized spontaneously by bacteria incidentally present in the environment and able to proliferate under the prevailing conditions.
(i) Preparation of PCCM.
Autoclaved natural seawater was inoculated with all nine strains that were selected based on the results of the monoxenic cultures (Table 1). They were individually grown in marine broth, centrifuged, resuspended, and quantified as described above. Each strain was added under sterile conditions at a density of approximately 106 cells/ml together with 0.1 g of the gamma-irradiated food/liter. This culture medium was incubated for 2 or 3 days at 28°C, and the selected bacterial strains were allowed to adapt to the conditions occurring in the Artemia culture. Before use, the food particles still in suspension were allowed to settle, and only the supernatant was used for the culture of the Artemia.
TABLE 1.
Effects of the addition of the individual bacterial strains on survival, body length, and biomass production of Artemia juveniles in monoxenic culture
| Strain | No. of experiments | Effect of strain on Artemiab:
|
||
|---|---|---|---|---|
| Survival | Body length | Biomass | ||
| LVS1 | 3 | 0 | +* | 0 |
| LVS2 | 3 | 0 | +** | +** |
| LVS3 | 5 | +* | +** | +** |
| LVS4 | 4 | 0 | +** | +** |
| LVS5 | 4 | 0 | +** | +** |
| LVS6 | 4 | 0 | +** | +** |
| LVS7 | 4 | 0 | +** | +** |
| LVS8 | 5 | +** | +** | +** |
| LVS9 | 6 | +* | +* | +** |
| Kwestam3A | 1 | 0 | −** | 0 |
| KA | 1 | 0 | −** | 0 |
| T20kleinB | 1 | 0 | −** | 0 |
| V. proteolyticus Q113 | 1 | −* | +** | 0 |
| P. fluorescens LMG1244 | 1 | −* | −** | −** |
| V. alginolyticus LMG4409T | 1 | 0 | −** | −** |
| V. proteolyticus CW8T2a | 9 | −** | −** | −** |
| Art8stam4Aa | 1 | −** | −** | −** |
| Art8stam1Ba | 1 | −** | −** | −** |
| Raw seawatera | 2 | −** | −** | −** |
Total mortality resulted.
+, positive effect; 0, no effect; −, negative effect. Statistical significance: *, significant positive or negative effect at a 5% level; **, significant positive or negative effect at a 1% level.
(ii) Preemptive colonization through a biofilter.
Another aliquot of microfiltered (0.22-μm pore size) natural seawater was preemptively colonized through prolonged recirculation over a biofilter exposed to the ambient air. Seawater (approximately 13 liters) was recirculated for 3 weeks over 1.5 liters of aerated activated carbon at a flow rate of 9 liters/h. The activated carbon was not sterilized initially. The temperature was kept at 28°C, and 0.1 g of the gamma-irradiated food/liter was added daily to the seawater. Remaining food particles were also allowed to settle before use.
Autoclaved natural seawater was used as a control culture medium for the Artemia, allowing opportunistic bacteria to develop during the culture of the brine shrimp.
(iii) Hatching and growth of Artemia.
An aliquot of cysts was disinfected for approximately 30 min with NaOCl according to the method of Van Stappen (21) and thoroughly rinsed with autoclaved NSS. The cysts were then divided in three equal parts and hatched in the different culture media for 24 h at 28°C according to the method of Merchie (12). After hatching, the Artemia nauplii were harvested and distributed in conic culture tanks containing 800 ml of the corresponding culture medium and kept at 29 to 30°C in a temperature-controlled water bath. The Artemia were transferred to this stagnant culture system at densities ranging from 10 to 20 nauplii/ml, which is considered suboptimal under the given circumstances with regard to growth and survival (3). The culture lasted 6 days, and the Artemia were fed daily with the gamma-irradiated food at a rate of 0.1 g/liter.
Twelve hours after the transfer of the nauplii, the initial animal density was determined. This time span of 12 h was necessary to allow any remaining cysts to hatch. At 36, 84, and 132 h after the transfer of the nauplii, survival rates and body lengths were determined. The survival rate in the xenic culture experiments was expressed as the percentage of surviving Artemia divided by the initial density. The body lengths were measured as described above. Three identical test runs were performed.
Microbial community analysis with Biolog.
To compare microbial communities in a fast and sensitive way without identification of the individual strains or species, the Biolog system was used as an explorative technique to examine the microbiota of the culture waters and those associated with the Artemia. The community-level physiological profiles were obtained with Biolog GN microtiter plates (Biolog Inc., Hayward, Calif.).
Samples of both the culture waters and the Artemia were taken 12 and 84 h after the transfer of the freshly hatched Artemia nauplii to the conic culture tanks. Before the inoculation of the Biolog microtiter plates, the inoculum densities of the different samples of culture water or associated bacteria were equalized. The bacterial inoculum density of the culture waters was quantified with ATP measurements, and the waters were subsequently diluted to an ATP content of 5 pg/ml as described by Verschuere et al. (22). For the analysis of the microbial communities associated with the Artemia, 10 ml of the culture water with the Artemia was filtered on an autoclaved filter (150-μm mesh size). The Artemia collected on the filter were rinsed twice with 10 ml of autoclaved NSS, resuspended in 20 ml of sterile NSS, and put in a stomacher blender (400SN; Seward Medical, London, United Kingdom) for 5 min to dislodge surface and intestinal bacteria. These samples were then diluted in sterile NSS to a density corresponding to 1 Artemia/ml based on the corresponding survival data.
The diluted samples were inoculated in Biolog GN plates and in six wells of a Biolog MT plate. The latter was done to overcome the problem of the low reproducibility of the control well, as identified by Kersters et al. (10). The Biolog plates were then incubated at 28°C, and the optical density at 590 nm (OD590) in each well was read with a biokinetics reader (EL312e) and the KinetiCalc enzyme immunoassay application software release 2.03 (Bio-Tek Instruments Inc., Winooski, Vt.) after 24, 30, 36, and 48 h of incubation. The results for the culture waters after 24 h and those for the Artemia after 48 h of incubation are given.
Before further data processing was done, the OD590 of each well was compensated for the average values of the corresponding control wells in the MT plate, yielding the net OD590 for each carbon source. Differences among the Biolog fingerprints were then assessed by using principal component analysis (PCA) executed with two components and a varimax rotation with Kaizer normalization and performed with SPSS for Windows release 7.5.2 (10, 22).
The similarity of community-level physiological profiles among the different replicates of a single treatment and between the different treatments was quantified with the Pearson correlation coefficient (17, 22) as follows:
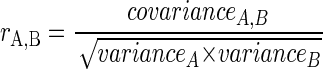 |
(iv) Plate counts.
The microbiota of the culture water and the blended Artemia were sampled 12 and 84 h after the transfer of the Artemia nauplii and were plated on marine agar to quantify the total amount of culturable marine heterotrophic bacteria. To do so, 100 μl of an appropriate 10-fold dilution was spread on the agar plates, incubated at 28°C, and counted after 5 days of incubation.
(v) FAME analysis of recovered bacterial isolates and the originally introduced strains.
The microbiota associated with Artemia cultured in PCCM was further examined. A total of 76 bacterial isolates associated with the Artemia grown in PCCM were recovered on marine agar. The recovered isolates originated from the three experiments and from each of the replicates. After two subsequent purification steps, the isolates were inoculated on marine agar plates by using the quadrant streak method. Following a 24-h incubation at 28°C, cells were harvested for fatty acid methyl ester (FAME) extraction. The FAME extracts were analyzed, and the chromatographical profiles of the isolates were clustered with the nine initially introduced strains by using the Microbial Identification System (Microbial ID, Newark, Del.) according to the method described by Osterhout et al. (15).
RESULTS
Monoxenic culture of Artemia juveniles.
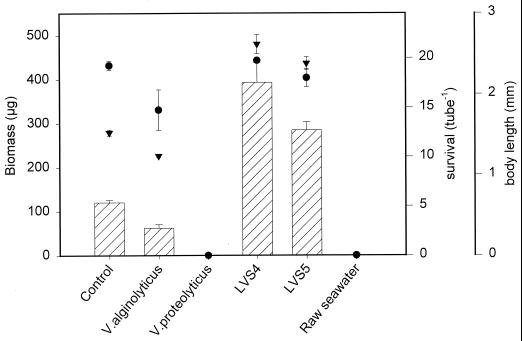
In Fig. 1, survival, body length, and biomass production results are shown for the Artemia cultures treated with no bacteria (control); with the bacterial strains LVS4, LVS5, V. alginolyticus LMG4409T, and V. proteolyticus CW8T2; and with raw seawater (not autoclaved). There was no significant effect of the treatments on the survival of Artemia, except for V. proteolyticus CW8T2 and the raw seawater, for which total mortality occurred. The higher biomass production observed with LVS4 and LVS5 could be attributed to a better growth of the Artemia. V. alginolyticus LMG4409T affected the growth of the Artemia, causing a significantly lower biomass production than the control. Similar experiments were done with all the strains.
FIG. 1.
Survival rate (●), body length (▾), and biomass production (bars) of the brine shrimp Artemia cultured under monoxenic conditions (V. alginolyticus LMG4409T, V. proteolyticus CW8T2, LVS4, LVS5, and raw seawater). The results shown are from one test run with four replicates per treatment. Error bars indicate the standard errors.
Table 1 presents an overview of the effects of the 18 tested bacterial strains and the raw seawater on survival, body length, and biomass production of the Artemia with an overall evaluation. The effect of a strain was considered significant when the P value was less than 0.05. Different categories of strains could be distinguished: nine strains showed a positive effect on the Artemia, either on the survival rate or on body length. All those strains gave rise to improved growth, while only three strains (LVS3, LVS8, and LVS9) caused better survival rates. These nine strains were retained for further experiments. Four strains (Kwestam3A, KA, T20kleinB, and V. proteolyticus Q113) caused a significantly lower survival or growth rate, although the biomass production was not significantly affected. Two strains (P. fluorescens LMG1244 and V. alginolyticus LMG4409T) gave rise to a significantly lower rate of growth and/or survival, with a significantly lower biomass production as a consequence. Finally, three strains (V. proteolyticus CW8T2, Art8stam4A, and Art8stam1B) and the raw seawater—the latter allowing opportunistic bacteria to develop—caused total mortality of the Artemia. It should be noted, however, that on several occasions, a significant interaction between the experiment and the treatment occurred, showing that the effect of the strain was not always of the same magnitude, although overall, a significantly positive or negative effect of the treatment could be found. This could be at least partially explained by the different harvesting times, as explained above.
The xenic culture of Artemia in PCCM.
Subsequently, three identical test runs were performed. In Table 2, the initial Artemia densities and the zootechnical results of the cultures are given for the three test runs. At 36 h after transfer of the nauplii, a positive effect of the PCCM on the survival and/or the growth rate of the Artemia was observed in all test runs. A similar observation was made after 84 h. After 132 h, the only culture tanks that still contained living Artemia were those preemptively colonized with the selected bacterial strains.
TABLE 2.
Ranges of the initial densities, survival rates, and body lengths of Artemia over the course of three experiments
| Treatment typea | Result obtained after the indicated period of timeb
|
||||||
|---|---|---|---|---|---|---|---|
| 12 h
|
36 h
|
84 h
|
132 h
|
||||
| Density (10 ml−1) | Survival (%) | Body length (mm) | Survival (%) | Body length (mm) | Survival (%) | Body length (mm) | |
| First test run | |||||||
| Control | 105–165 | 45 ± 27 | 0.94 ± 0.15 | 2.2 ± 2.6 | – | 0 | – |
| Biofilter | 91–138 | 23 ± 23 | 1.020 ± 0.047 | 2.5 ± 5.1 | – | 0 | – |
| PCCM | 144–183 | 90 ± 23* | 1.200 ± 0.058* | 73 ± 10** | 1.73 ± 0.31** | 57 ± 18** | 2.60 ± 0.83** |
| Second test run | |||||||
| Control | 147–168 | 88.2 ± 8.6 | 1.030 ± 0.027 | 15.7 ± 4.8 | 1.230 ± 0.093 | 0 | – |
| Biofilter | 138–179 | 36 ± 21** | 1.113 ± 0.056* | 0 | – | 0 | – |
| PCCM | 129–172 | 80 ± 24 | 1.170 ± 0.047** | 38 ± 32 | 1.630 ± 0.066** | 11.1 ± 7.5* | 2.43 ± 0.33** |
| Third test run | |||||||
| Control | 197–220 | 70 ± 15 | 0.958 ± 0.032 | 38 ± 17 | 1.42 ± 0.11 | 0 | – |
| Biofilter | 161–203 | 46 ± 28 | 1.015 ± 0.052 | 2.1 ± 3.3 | 1.21 ± 0.15 | 0 | – |
| PCCM | 181–195 | 91 ± 29* | 1.040 ± 0.012** | 70 ± 22* | 1.408 ± 0.077 | 13.8 ± 7.3** | 1.81 ± 0.14** |
Control, treatment in which Artemia were cultured in autoclaved culture water; biofilter, treatment in which Artemia were cultured in culture water extensively recirculated over a biofilter; PCCM, treatment in which Artemia were cultured in preemptively colonized culture medium.
*, significantly different from the control at a 5% level; **, significantly different from the control at a 1% level; –, not determined due to almost total mortality.
The plate counts of the culture waters revealed significant differences (P < 0.05) after 12 h between the control (7.50 ± 0.21, 7.72 ± 0.41, and 7.153 ± 0.090 log CFU/ml for test runs 1, 2, and 3, respectively) and the other treatments, i.e., the PCCM (8.00 ± 0.31, 8.06 ± 0.17, and 7.963 ± 0.085 log CFU/ml for runs 1, 2, and 3) and the biofilter-treated culture water (7.88 ± 0.15, 8.10 ± 0.24, and 7.62 ± 0.14 log CFU/ml for runs 1, 2, and 3). The average bacterial density in the culture waters after 84 h amounted to 8.34 log CFU/ml, but no significant differences among the different treatments could be observed.
After 12 h, no significant differences among the plate counts done for the Artemia were observed. On average, the bacterial colonization of the Artemia amounted to 5.52 log CFU/Artemia. However, after 84 h, the surviving Artemia of the control treatment were significantly more colonized (6.83 ± 0.11 and 5.74 ± 0.22 log CFU/ml for experiments 2 and 3, respectively) than the Artemia grown in PCCM (5.52 ± 0.19 and 5.20 ± 0.29 log CFU/ml).
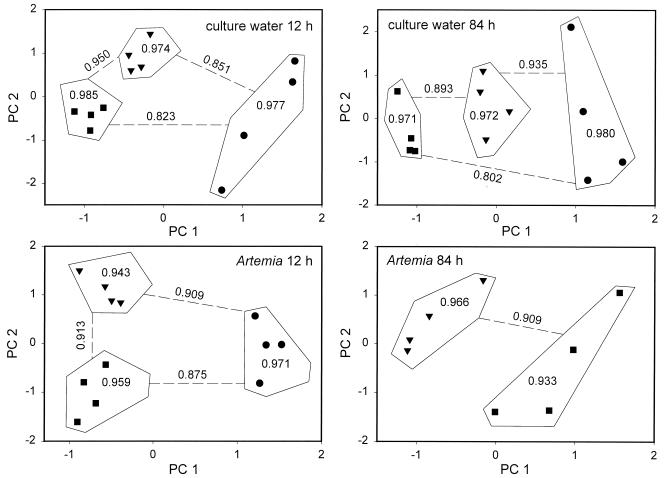
In Fig. 2, the comparison of the Biolog profiles with PCA is shown for the culture waters and the microbial communities associated with the Artemia for the third experiment. A clear difference among the results for the different treatments (control, biofilter, and PCCM) could be observed, for the Artemia as well as for the culture waters and for both sampling times (12 and 84 h). This shows that manipulation of the microbial communities of the Artemia culture water and those associated with the Artemia is possible through preemptive colonization of the culture water. A similar separation according to the treatments was found in the two other experiments (data not shown).
FIG. 2.
Results of PCA on the Biolog profiles of the Artemia culture waters and the microbial communities associated with the Artemia for the third experiment, 12 and 84 h respectively, after the transfer of the nauplii to the culture tanks. ▾, control; ■, PCCM; ●, biofilter-treated culture water. The Pearson correlation coefficients among the replicates of the treatment (within the outlining) and between the treatments (along the dashed lines) are given.
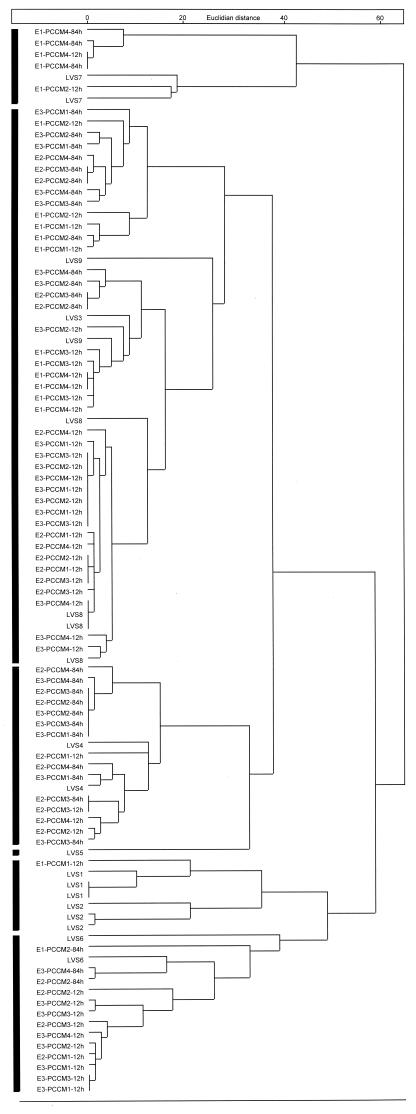
A chemotaxonomical comparison between all bacterial isolates recovered from the Artemia cultured in PCCM and the initially introduced nine strains based on FAME analysis was made. The results of the cluster analysis are shown in a dendrogram (Fig. 3). Several clusters can be distinguished (from top to bottom), as shown by the black bars. (i) A first cluster includes LVS7 and five recovered isolates. LVS7 has a very characteristic colony type that was not observed in any of the recovered isolates, suggesting that it is very improbable that the five recovered isolates were similar to LVS7. (ii) The second big cluster includes 42 isolates and LVS3, -8, and -9. In previous experiments, these three strains were shown to be chemotaxonomically quite closely related to each other, which may explain their appearance in the same cluster. (iii) A third cluster contains LVS4 and 15 related isolates. Furthermore, several recovered isolates had an orange colony type identical to that of LVS4. (iv) No isolates related to LVS5 were recovered from the Artemia. (v) A fifth cluster contains LVS1, LVS2, and one recovered isolate. (vi) The last cluster includes LVS6 and 13 recovered isolates. It was also visually observed that many recovered isolates showed the same characteristic yellow colony type as LVS6.
FIG. 3.
FAME analysis of recovered bacterial isolates of Artemia grown in PCCM and of the originally introduced strains (LVS1 to LVS9). The code indicates the origin of the isolate: the number following E identifies the experiment (1 to 3), and the number following PCCM identifies the conic culture tank (numbered 1 to 4) and is followed by the incubation time at which the strain was isolated (12 or 84 h).
Considering the rather low reproducibility of the FAME analyses and the high chemotaxonomical similarity of some introduced strains, it is impossible to demonstrate unequivocally which of the initially introduced strains could dominate the microbiota associated with Artemia cultured in PCCM. However, some preliminary conclusions can be drawn. It seems clear that LVS1, -2, -5, and -7 are not able to colonize the Artemia dominantly, as only one recovered isolate showed a high similarity to those strains. At least some of the isolates may be similar to LVS3, -4, -6, -8, and -9. The large number of recovered isolates closely related to LVS8 is remarkable. Although some conclusions could be made, more definitive information is needed before concluding which of the introduced strains can successfully colonize the Artemia.
DISCUSSION
Nine bacterial strains were selected based on their contribution to the growth and/or the survival of Artemia juveniles in monoxenic cultures (Table 1). As described in the introduction, it has been substantiated in the literature that bacteria can provide a nutritional contribution to the food for such cultures. This study shows that selected bacterial strains can increase the zootechnical performances of xenic Artemia cultures (Table 2). The preemptive colonization of the culture water either by PCCM or by the biofilter treatment had a drastic influence on the microbial communities that developed in the culture water or were associated with the Artemia (Fig. 2). Although not definitive, the FAME characterization of the bacterial isolates recovered from the Artemia grown in PCCM indicated that strains chemotaxonomically related to five of the nine introduced strains were able to colonize dominantly the body of the Artemia and to minimize the development of other (opportunistic) bacteria (Fig. 3). Further confirmation of the clonal strain identity could be obtained by using high-resolution DNA fingerprinting techniques.
For Artemia culturists, attention so far has been focused only on the contribution of bacteria to the nutritional quality of the food, without further consideration of their possible role as biological control agents of the microbial environment. In the present study, it is shown that the preemptive colonization of the culture medium led not only to an improvement of the nutritional value of the food for the Artemia but also to a manipulation of the ambient and associated microbiota, resulting in higher survival and/or growth rates of the animals.
Generally, the observed survival rates in the xenic cultures of the Artemia were low, even when they were cultured in PCCM (Table 2). This can be explained by the high initial Artemia densities (10,000 to 20,000/liter), possibly causing a water quality deterioration, leading to lower growth and survival rates than under optimal conditions (3). Furthermore, in our experience, the feeding rate was also very high, possibly causing overfeeding and accentuating water quality deterioration. It was expected that the effect of PCCM would be more pronounced when suboptimal culture conditions were applied.
Several modes of action may be responsible for the positive effects of the selected strains. Bacteria may serve as a direct source of nutrients for shrimp and may also contribute to the digestion of the provided food (7). Bacteria may be a major source of protein and amino acids for Artemia (6). Uchida et al. (20) reported that the surface attachment of bacteria to Ulva fronds resulted in the formation of protein-rich detrital particles and increased its nutritional quality for Artemia. In the mass culture of the rotifer Brachionus plicatilis, vitamin B12-producing bacteria are present and provide a complementary source of vitamin B12 that is a limiting nutrient for the culture on Baker’s yeast (24). Lysed bacteria may deliver enzymes that remain active in the gut (i.e., acquired bacterial enzymes), and this may provide the host with additional digestive abilities (11). Similarly, extracellular enzymes may be produced by the bacteria, helping in the breakdown of refractory compounds of the food (8). It can also be hypothesized that dissolved nutrients normally unavailable to the Artemia are converted to bacterial biomass with an appropriate particle size and consequently become available as a food source to the suspension feeder (nutrient recycling). One of these nutritional modes of actions is most probably the origin of the observed effects in the monoxenic cultures (Fig. 1 and Table 1).
In the xenic cultures, however, other modes of action cannot be excluded. The added selected bacteria may remove toxic metabolic substances that could adversely affect the growth and survival of the Artemia, especially under the suboptimal conditions of the xenic experiments (stagnant culture). Bacteria that are well adapted to the conditions prevailing in the intensive Artemia culture may also be able to prevent the proliferation of opportunistic pathogens. The fact that most of the isolates showed a high chemotaxonomical similarity to four of the initially introduced strains (Fig. 3) indicates that those strains could proliferate in the gut or on the surface of the Artemia. Through competition for available resources (nutrients, space, adhesion sites, etc.) or through antagonism (production of toxic substances), the selected bacterial strains allowed to colonize the culture water preemptively may prevent potentially deleterious strains from developing or surviving in the culture system. Further research is necessary to elucidate the exact mode of action of the observed beneficial effects and to understand the possibilities and the limitations of microbial control in aquaculture.
ACKNOWLEDGMENTS
We thank L. Verdonck for providing bacterial strains, W. Mondelaers (Department of Subatomic and Radiation Physics, University of Ghent) for the gamma irradiation of the food, and J. Swings.
This research was partly supported by project 3G006396 of the Nationaal Fonds voor Wetenschappelijk Onderzoek (NFWO).
REFERENCES
- 1.Abreu-Grobois F A, Briseño-Dueñas R, Herrera M A, Malagon M L. A model for growth of Artemia franciscana based on food ratio-dependent gross growth efficiency. Hydrobiologia. 1991;212:27–37. [Google Scholar]
- 2.D’Agostino A. The vital requirements of physiology and nutrition. In: Persoone G, Sorgeloos P, Roels O, Jasper E, editors. The brine shrimp Artemia: ecology, culture, use in aquaculture. Vol. 2. Wetteren, Belgium: Universa Press; 1980. pp. 55–82. [Google Scholar]
- 3.Dhont J, Lavens P. Tank production and use of ongrown Artemia. In: Lavens P, Sorgeloos P, editors. Manual of the production and use of live food for aquaculture. Food and Agriculture Organization Fisheries Technical Paper No. 361. Rome, Italy: Food and Agriculture Organization; 1996. pp. 164–195. [Google Scholar]
- 4.Douillet P. Effect of bacteria on the nutrition of the brine shrimp Artemia fed on dried diets. In: Sorgeloos P, Bengston D A, Decleir W, Jaspers E, editors. Artemia: research and its applications. Ecology, culturing and use in aquaculture. Vol. 3. Wetteren, Belgium: Universa Press; 1987. pp. 295–308. [Google Scholar]
- 5.Gorospe J, Nakamura K. Associated bacterial microflora in Artemia-rice bran culture. Isr J Aquacult Bamidgeh. 1996;48:99–107. [Google Scholar]
- 6.Gorospe J, Nakamura K, Abe M, Higashi S. Nutritional contribution of Pseudomonas sp. in Artemia culture. Fish Sci. 1996;62:914–918. [Google Scholar]
- 7.Harris J M. The presence, nature, and role of gut microflora in aquatic invertebrates: a synthesis. Microb Ecol. 1993;25:195–231. doi: 10.1007/BF00171889. [DOI] [PubMed] [Google Scholar]
- 8.Hood M A, Meyers S P, Colmer A R. Bacteria of the digestive tract of the white shrimp Penaeus setiferus. Bacteriol Proc. 1971;71:48–53. [Google Scholar]
- 9.Intriago P, Jones D A. Bacteria as food for Artemia. Aquaculture. 1993;113:115–127. [Google Scholar]
- 10.Kersters I, Van Vooren L, Verschuere L, Vauterin L, Wouters A, Mergaert J, Swings J, Verstraete W. Utility of the Biolog system for the characterization of heterotrophic microbial communities. Syst Appl Microbiol. 1997;20:439–447. [Google Scholar]
- 11.Martin M M. The role of ingested enzymes in the digestive processes of insects. In: Anderson J M, Raynon A D M, Walton D W A, editors. Invertebrate-microbial interactions. Cambridge, United Kingdom: Cambridge University Press; 1984. pp. 155–177. [Google Scholar]
- 12.Merchie G. Use of nauplii and metanauplii. In: Lavens P, Sorgeloos P, editors. Manual on the production and use of live food for aquaculture. Food and Agriculture Organization Fisheries Technical Paper No. 361. Rome, Italy: Food and Agriculture Organization; 1996. pp. 137–163. [Google Scholar]
- 13.Muroga K, Higashi M, Keitoku H. The isolation of intestinal microflora of farmed red seabream (Pagrus major) and black seabream (Acanthopagrus schlegeli) at larval and juvenile stages. Aquaculture. 1987;65:79–88. [Google Scholar]
- 14.Olsson J C, Westerdahl A, Conway P L, Kjelleberg S. Intestinal colonization potential of turbot (Scophthalmus maximus)- and dab (Limanda limanda)-associated bacteria with inhibitory effects against Vibrio anguillarum. Appl Environ Microbiol. 1992;58:551–556. doi: 10.1128/aem.58.2.551-556.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osterhout G J, Shull V H, Dick J D. Identification of clinical isolates of gram-negative nonfermentative bacteria by an automated cellular fatty acid identification system. Appl Environ Microbiol. 1991;29:1822–1830. doi: 10.1128/jcm.29.9.1822-1830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Provasoli L, Shiraishi K. Axenic cultivation of the brine shrimp Artemia. Biol Bull. 1959;117:347–355. [Google Scholar]
- 17.Randerson P F. Ordination. In: Fry J C, editor. Biological data analysis: a practical approach. New York, N.Y: Oxford University Press; 1993. pp. 173–217. [Google Scholar]
- 18.Rico-Mora R, Voltolina D. Effects of bacterial isolates from Skeletonema ostatum cultures on the survival of Artemia franciscana nauplii. J Invertebr Pathol. 1995;66:203–204. [Google Scholar]
- 19.Seki H. Studies in microbial participation to food cycle in the sea. I. Participation in the microcosm at static condition. J Oceanogr Soc Jpn. 1964;20:20–32. [Google Scholar]
- 20.Uchida M, Nakata K, Maeda M. Conversion of Ulva fronds to a hatchery diet of Artemia nauplii utilizing the degrading and attaching abilities of Pseudoalteromonas espejiana. J Appl Phycol. 1997;9:541–549. [Google Scholar]
- 21.Van Stappen G. Use of cysts. In: Lavens P, Sorgeloos P, editors. Manual on the production and use of live food for aquaculture. Food and Agriculture Organization Fisheries Technical Paper No. 361. Rome, Italy: Food and Agriculture Organization; 1996. pp. 107–136. [Google Scholar]
- 22.Verschuere L, Dhont J, Sorgeloos P, Verstraete W. Monitoring Biolog patterns and r/K-strategists in the intensive culture of Artemia juveniles. J Appl Microbiol. 1997;83:603–612. [Google Scholar]
- 23.Yasuda K, Taga N. A mass-culture method for Artemia salina using bacteria as food. Mer. 1980;18:55–62. [Google Scholar]
- 24.Yu J P, Hino A, Hirano R, Hirayama K. Vitamin B12-producing bacteria as a nutritive complement for a culture of the rotifer Brachionus plicatilis. Nippon Suisan Gakkaishi. 1988;54:1873–1880. [Google Scholar]