Abstract
The transferrin receptor 1 (TfR1) plays a key role in cellular iron uptake through its interaction with iron-bound Tf. TfR1 is often reported to be overexpressed in malignant cells, and this increase may be associated with poor prognosis in different types of cancer, which makes it an attractive target for antitumor therapy. The marine natural product Muriceidine A is a potent anticancer agent reported in our previous work. In this study, we designed and synthesized a series of Muriceidine A derivatives and described the systematic investigation into their cytotoxic activities against four tumor cells. Most of the derivatives showed stronger antitumor activity and we found that the introduction of electron-donating groups at position C-2 of unsaturated piperidine was beneficial to anticancer activity and unsaturated piperidine was responsible for the antiproliferative activity. Among these compounds, 12b (methyl at position C-2 of unsaturated piperidine) exhibited the strongest cytotoxicity against MDA-MB-231 cells. Further pharmacological research showed that 12b bound to Transferrin receptor 1 (TfR1) directly caused iron deprivation and ROS imbalance along with the degradations of several oncoproteins, especially FGFR1, through the proteasome pathway; thus, inducing cell cycle arrest and apoptosis in MDA-MB-231 breast cancer cells. Our findings indicate that 12b is a promising lead compound targeting TfR1 for triple negative breast cancer.
Keywords: Muriceidine A, structural optimization, transferrin receptor 1, iron deprivation, anticancer agent
1. Introduction
Iron is an important element that plays a crucial role in various physiological and pathological processes in all kinds of mammalian cells [1,2]. It also has the capacity to engage in redox cycling and free radical formation. The balance between intracellular iron acquisition, storage and usage should be tightly regulated, since disorders of iron metabolism can induce tumorigenesis [3,4]. Iron may accelerate tumor initiation by enhancing the formation of free radicals, as well as function as a nutrient that fosters tumor cell proliferation. Compared with non-malignant cells, the growth of cancer cells is more dependent on iron and more sensitive to iron deprivation, a phenomenon known as “iron addiction” [5]. Evidence shows iron deprivation can induce apoptosis and autophagy in a series of cell lines, especially in breast cancer and hepatocellular carcinoma cells [6,7], suggesting that disruption of iron homeostasis is a promising strategy for cancer therapy investigation [8,9].
The transferrin receptor 1 (TfR1), also known as cluster of differentiation 71 (CD71), is a type II transmembrane glycoprotein that binds transferrin (Tf) and performs a critical role in cellular iron uptake and cellular iron homeostasis through the interaction with iron-bound Tf [10,11]. In general, TfR1 is expressed at low levels in most normal cells [12]. Many types of cancer cells reprogramme iron metabolism in ways that result in net iron influx. They upregulate proteins that are involved in iron uptake, such as TfR1, and decrease the expression of iron efflux proteins. Increased expression is observed in malignant cells with a high rate of proliferation and high need for iron, including breast [13], colon [14], liver [15], and brain cancer cells. In particular, cancer cells resistant to conventional therapies might particularly over-express TfR1 [16], indicating a poor prognosis for patients [17]. Interestingly, TfR1 can support mitochondrial respiration and reactive oxygen species production, both of which are key players in tumor cell growth and survival [18]. Taken together, the high expression of TfR1 on malignant cells, and its key role in cancer cell iron metabolism make it an attractive target for cancer therapy.
As TfR1 is an appealing therapeutic target for a variety of cancers, inhibition of TfR1 function by existing anti-TfR1 antibodies can lead to iron deprivation, which in turn promotes cancer cell death. The murine anti-human TfR1 IgG antibody 7579 downregulates TfR1 surface levels on cancer cells and has shown inhibitory effects on proliferation as well as inducing apoptosis of human U87MG, U251, and A172 glioma cells in vitro [19]; JST-TFR09, an antibody to human TfR1, has a great affinity to TfR1 in adult T-cell leukemia/lymphoma cells. It could interfere with the binding between TfR1 and Tf and inhibited the iron intake of adult T-cell leukemia/lymphoma, which has been used in preclinical development [20]. In addition, miRNA drug targeting TfR1 has been reported to be a good drug in the clinical treatment of leukemia [21]. However, all these reports about targeting TfR1 mainly focus on biopharmaceutical development. Compared to biological drugs, small molecule drugs have many more advantages, such as good permeability, low immunogenicity, cheap, easy storage, and transportation. However, there is still no small molecule compound reported to target TfR1 [22]. This underlines the significance to search for new TfR1 chemotherapeutic compounds with anticancer effects.
Marine natural products (MNPs) are regarded as important sources for the discovery of lead compounds for anti-cancer drugs, due to their unique chemical structures and diverse biological activities [23]. Simplifying complex structures of natural products, while retaining the desired biological activity, is a valid and meaningful strategy in the drug development process [24,25]. This strategy has been successfully used in the lead optimization of MNPs and yielded a number of marketed drugs and drug candidates [26,27].
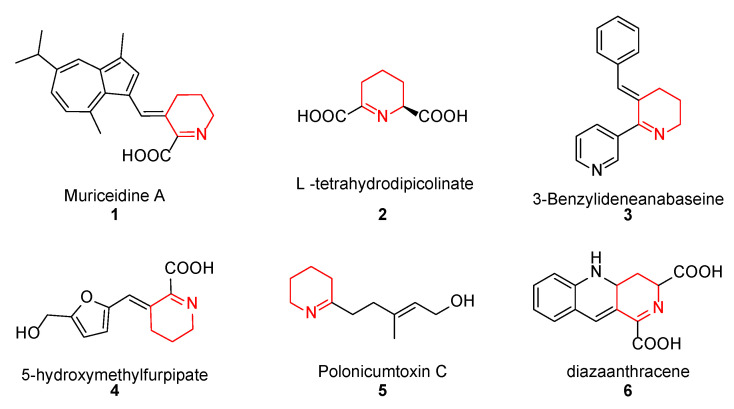
Muriceidine A (1, Figure 1), a new skeleton guaiazulene alkaloid originally isolated from the South China Sea gorgonian Muriceides collaris, exhibits potent cytotoxic activity against the tumor cell line K562 with an IC50 value of 8.37 μM [28]. Structurally, Muriceidine A is comprised of the conjunction of guaiazulene and unsaturated pipecolic acid (Δ1-pipecolic acid). Guaiazulene (GA) has unique physical and chemical properties due to its electronic distribution and appearance features [29]. GA has potential research value for its anti-inflammatory, anti-allergy, anti-cancer, and other activities in the field of medicinal chemistry [30]. Δ1-pipecolic acid and its substitution derivatives are considered privileged scaffolds in drug discovery and are widely introduced in many antitumor drugs (2–6, Figure 1) [31,32,33]. Therefore, the modification of antitumor pharmacy cores with unsaturated piperidine structures has attracted much attention among medicinal chemists.
Figure 1.
Chemical structure of some unsaturated piperidine derivatives.
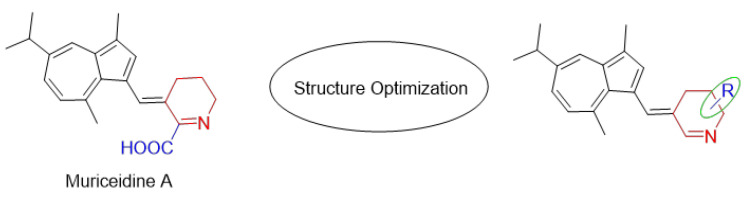
Encouraged by the antitumor activity of Muriceidine A and the virtues of unsaturated piperidine structures, it can be expected that the combination of unsaturated piperidine derivatives and guaiazulene may improve antitumor efficiency. Therefore, different substituent groups were introduced into the functional unsaturated piperidine moiety in the present work in the hope of obtaining agents with stronger antitumor activity (Figure 2). Herein, we describe the synthesis of five Muriceidine A derivatives (12a–12d, 16) and the systematic investigation into their cytotoxic activities against four tumor cells (MDA-MB-231, K562, Hela, and HCT-116). Moreover, the antitumor mechanisms of the most active derivative, 12b, were examined. It directly binds to TfR1, causing iron deprivation and ROS imbalance with degradations of several oncoproteins, especially FGFR1, through the proteasome pathway, thereby promoting cell apoptosis in MDA-MB-231 breast cancer cells.
Figure 2.
Chemical structure of Muriceidine A and designed compounds.
2. Materials and Methods
2.1. Chemistry
2.1.1. Materials and Methods
All reagents and solvents were commercially available. The structures of target compounds were confirmed by characterization with nuclear magnetic resonance (1H NMR and 13C NMR) and liquid chromatography-mass spectrometry (LC-MS). NMR spectra were recorded on the Bruker AVANCE instrument (300 MHz) at 25 °C using TMS as the internal standard in CDCl3. NMR spectra was shown in supplementary data (Figures S1–S11). Mass spectra were obtained using a Waters ZQ 2000 (Waters Instruments, Wilmslow, UK) electrospray ionization (ESI) single quadrupole mass spectrometer. Compounds were performed by flash column chromatography on silica gel (200–300 mesh) produced by Qingdao Marine Chemical Factory, Qingdao (China). Analytical thin-layer chromatography (TLC) was conducted on Fluka TLC plates (silica gel 60 F254, aluminum foil).
2.1.2. General Procedure A for the Synthesis of Intermediates 11a–e
T-Butyl hypochlorite (t-BuOCl): In a 25 mL round-bottom flask, t-BuOH (2 mL, 20.9 mmol), aqueous NaOCl solution (active chlorine ≥ 5%, 12 mL, 18.6 mmol), and acetic acid (2 mL, 34.3 mmol) were added in order and stirred at 0 °C for 5 min. The desired product was isolated from the lower aqueous phase as a yellow liquid. The organic fraction was directly used as a chlorine source in the next step without further purification.
Δ1-piperidine derivatives (11a–e) [34]: Compounds 9a–e (2.1 mmol) were dissolved in a 50 mL flask with 15 mL of anhydrous ether, respectively. The freshly prepared t-BuOCl was then added dropwise at 0 °C under an N2 atmosphere. The resulting mixture was stirred at 0 °C for 3 h to give 10a–e. TLC and ninhydrin colorimetry methods were used to track the reaction. When the reactants are completely converted, excessive solid KOH (500 mg, 8.9 mmol) was added in one batch, followed by 10 mL dry MeOH. The mixture was heated in an oil bath at 50 °C for 1h and then kept at room temperature overnight. The ether was removed by rotary evaporation at 20 °C which was further filtered to remove the solid and get the methanol solution of compounds 11a–e (Δ1-piperidine derivatives). TLC proved that compounds 11a–e existed as a near equimolar tautomeric mixture of enamine and imine. Due to the unstable nature of 11a–e, the obtained solution was used in the next step without further purification.
2.1.3. General Procedure B for the Synthesis of Final Products 12a–e
Since intermediate 8 can efficiently be synthesized from commercially available guaiazulene 7 by a Vilsmeier-Hacck reaction [35], the synthesis process of 8 will not be described in detail here. CH3ONa (93 mg, 0.29 mmol) was dissolved in methanol (5 mL) in a 100 mL flask. Then, the methanol solution of compound 11a–e (1 mmol) was added. The solution was stirred at room temperature for 5 min and then a 2 mL methanol solution of compound 8 (1 mmol) containing acetic acid (72 μL, 1.2 mmol) was added, and the mixture was added to the flask dropwise. Then the reaction was heated to 60 °C and stirred for 20 h. After cooling, the solution was evaporated to remove the methanol. The residue was dissolved in distilled water and extracted three times with EtOAc. The organic layers were combined and concentrated by evaporation to give the crude product which was further purified by column chromatography (eluent: CH2Cl2/CH3OH) to obtain the final products 12a–e.
3-((Guaiazulene-1-yl)ethylene) Δ1-piperidine (12a)
Following general procedure A and B, the crude residue obtained from piperidine was purified by flash-chromatography, using CH2Cl2/CH3OH 30:1 (v/v) as eluent, to furnish compound 12a as a red solid (yield 50%). 1H NMR (500 MHz, CDCl3): δ 8.39 (s, 1H), 8.33 (s, 1H), 8.23 (s, 1H), 7.87 (s, 1H), 7.63 (d, J = 11.0 Hz, 1H), 7.44 (d, J = 11.0 Hz, 1H), 3.70 (s, 2H), 3.46 (s, 1H), 3.19–3.11 (m, 4H), 2.94 (t, J = 5.8 Hz, 2H), 2.58 (s, 3H), 2.07 (d, J = 5.7 Hz, 2H), 1.38 (d, J = 6.8 Hz, 6H). 13C NMR (126 MHz, CDCl3): δ 165.88, 150.80, 149.10, 148.66, 144.28, 141.80, 141.70, 139.07, 137.75, 135.35, 134.68, 129.58, 123.29, 119.79, 77.33, 77.08, 76.82, 42.50, 38.13, 29.44, 24.36, 23.83, 20.00, 13.17. MS (ESI): [M + H]+ 392.44.
3-((Guaiazulene-1-yl)ethylene)-2-(methyl) Δ1-piperidine (12b)
Following general procedure A and B, the crude residue obtained from 2-methylpiperidine was purified by flash-chromatography, using CH2Cl2/CH3OH 30:1 (v/v) as eluent, to furnish compound 12b as a red solid (yield 35%). 1H NMR (500 MHz, CDCl3) δ: 8.53 (s, 1H), 8.42 (s, 3H), 8.21 (d, J = 1.7 Hz, 1H), 7.80 (s, 1H), 7.57 (dd, J = 10.9, 1.7 Hz, 1H), 7.34 (d, J = 11.0 Hz, 1H), 3.71 (t, J = 5.3 Hz, 2H), 3.13 (dt, J = 13.7, 6.8 Hz, 1H), 3.07 (s, 3H), 2.93 (t, J = 5.9 Hz, 2H), 2.63 (s, 3H), 2.59 (s, 3H), 2.01 (dt, J = 11.9, 6.0 Hz, 2H), 1.37 (d, J = 6.9 Hz, 6H). 13C NMR (126 MHz, CDCl3): δ 173.26, 166.19, 147.45, 147.26, 143.73, 142.90, 140.54, 138.96, 137.32, 135.20, 133.10, 128.54, 122.03, 120.01, 77.37, 77.12, 76.86, 43.41, 38.04, 29.21, 24.50, 24.39, 20.29, 19.77, 13.13. MS (ESI): [M + H]+ 306.35.
3-((Guaiazulene-1-yl)ethylene)-4-(hydroxymethyl) Δ1-piperidine (12c)
Following general procedure A and B, the crude residue obtained from 4-methanolpiperidine was purified by flash-chromatography, using CH2Cl2/CH3OH 20:1 (v/v) as eluent, to furnish compound 12c as a red solid (yield 25%). 1H NMR (500 MHz, CDCl3) δ: 8.22 (s, 1H), 8.20 (d, J = 1.6 Hz, 1H), 8.09 (s, 1H), 7.57 (d, J = 10.9 Hz, 1H), 7.35 (d, J = 10.9 Hz, 1H), 3.89 (dd, J = 11.0, 3.4 Hz, 1H), 3.85–3.69 (m, 4H), 3.64 (s, 1H), 3.13 (dt, J = 13.8, 6.8 Hz, 1H), 3.06 (s, 3H), 2.59 (s, 3H), 2.45 (d, J = 12.8 Hz, 1H), 1.86 (s, 1H), 1.38 (d, J = 6.9 Hz, 6H). 13C NMR (126 MHz, CDCl3): δ 166.09, 149.34, 148.43, 147.97, 143.96, 141.74, 138.48, 137.30, 135.10, 134.28, 130.06, 122.33, 120.74, 77.27, 77.02, 76.76, 61.01, 39.37, 38.04, 34.29, 29.57, 24.36, 21.15, 13.21. MS (ESI): [M + H]+ 321.66.
3-((Guaiazulene-1-yl)ethylene)-Δ1-pyrrolidine (12d)
Following general procedure A and B, the crude residue obtained from pyrrole was purified by flash-chromatography, using CH2Cl2/CH3OH 15:1 (v/v) as eluent, to furnish compound 12d as a red solid (yield 6%). 1H NMR (500 MHz, CDCl3): δ 8.61 (d, J = 12.0 Hz, 2H), 8.24 (s, 3H), 7.83 (s, 1H), 7.62 (d, J = 11.4 Hz, 1H), 7.41 (d, J = 10.9 Hz, 1H), 4.33–4.28 (m, 2H), 3.32–3.26 (m, 3H), 3.20–3.12 (m, 4H), 2.60 (s, 3H), 1.39 (d, J = 6.9 Hz, 6H). 13C NMR (126 MHz, CDCl3): δ 170.56, 140.18, 137.85, 137.49, 135.33, 134.06, 52.36, 38.09, 29.28, 27.70, 24.39, 13.23. MS (ESI): [M + H]+ 277.42.
Muriceidine A (12e)
Following general procedure A and B, the crude residue obtained from L-pipecolic acid was purified by flash-chromatography, using CH2Cl2/CH3OH 15:1 (v/v) as eluent, to furnish compound Muriceidine A as a red solid (yield 10%). 1H NMR (500 MHz, CDCl3) δ: 9.57 (s, 1H), 8.15 (s, 1H), 7.77 (s, 1H), 7.50 (d, J = 10.7 Hz, 1H), 7.32 (d, J = 10.7 Hz, 1H), 3.71 (s, 2H), 3.15 (s, 3H), 3.10 (m, 1H), 2.88 (s, 2H), 2.57 (s, 3H), 1.99 (s, 2H), 1.37 (d, J = 6.9 Hz, 6H). MS (ESI): [M + H]+ 336.21.
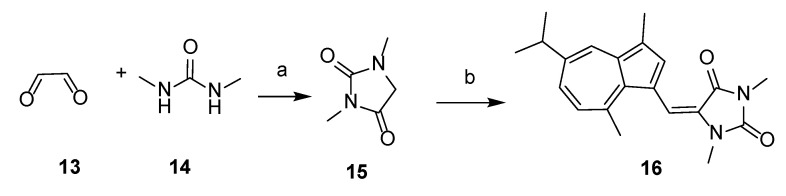
2.1.4. Synthesis of 5-((Guaiazulene-1-yl)ethylene)-1,3-dimethylhydantoin (16)
Intermediate 1,3-dimethylhydantoin (15) was obtained from glyoxal (13) and 1,3-dimethylurea (14) according to the method described in the literature [34]. Product 16 was then synthesized using the same method as procedure B, by reacting 15 with 3-formyl guaiazulene 8. The crude residue was purified by flash-chromatography, using CH2Cl2/CH3OH 15:1 (v/v) as eluent, to furnish compound 16 as a green solid (yield 24%). 1H NMR (600 MHz, CDCl3) δ 8.13 (d, J = 1.9 Hz, 1H), 7.60 (s, 1H), 7.42 (s, 1H), 7.39 (dd, J = 10.7, 1.8 Hz, 1H), 7.02 (d, J = 10.8 Hz, 1H), 3.16 (s, 3H), 3.05 (dt, J = 13.8, 6.9 Hz, 1H), 3.00 (s, 3H), 2.95 (s, 3H), 2.62 (s, 3H), 1.37 (d, J = 6.9 Hz, 7H). 13C NMR (151 MHz, CDCl3) δ 164.1, 156.4, 147.0, 142.1, 139.4, 139.4, 136.3, 136.0, 134.60, 128.4, 126.9, 125.1, 118.4, 113.3, 77.4, 77.2, 77.0, 38.0, 30.8, 27.7, 25.1, 24.6, 13.0. MS (ESI): [M + H]+ 337.52.
2.2. Biological Evaluation
2.2.1. Cell Culture and Reagents
All human cancer cell lines were provided by the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). The human triple negative breast cancer cell line MDA-MB-231 was cultured in L-15 complete medium (Gibco, Grand Island, NY, USA). SH-SY5Y, Siha, PC-3, HO-8910, MGC-803, NCI-H1975, and BEL-7402 cell lines were cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA). HeLa and MCF-7 cell lines were cultured in MEM medium. A549 cell line was cultured in F-12K medium (Gino, Hangzhou, China). The K562 cell line was cultured in IMDM medium (Gibco, Grand Island, NY, USA). The HCT116 cell line was cultured in high glucose DMEM medium (Gibco, Grand Island, NY, USA). Cell lines were supplemented with 10% FBS (Gibco, Grand Island, NY, USA), 2 mM L-glutamine, and 1% penicillin-streptomycin (Solarbio, Beijing, China) and propagated as monolayer cultures at 37 °C in a humidified 5% CO2 incubator.
2.2.2. Cell Viability Assay
The inhibition of cancer cell viability was assessed by the MTT assay. Cells were plated into a 96-well plate at a density of 5 × 103 cells per well. After attachment, cells were treated with 12b in different concentrations or 1% DMSO solution as the negative control group. After 72 h, 20 μL of MTT (Sigma-Aldrich, St. Louis, MO, USA) was added to each well. Four hours later, the formazan product was dissolved in DMSO and quantitated spectrophotometrically at a wavelength of 570 nm using a microplate reader (BioTek, Winooski, VT, USA). The IC50 value was defined as the concentration that inhibited cell viability by 50%.
2.2.3. Flow Cytometry Analysis of Cell Cycle
MDA-MB-231 cells were plated into 6-well plates at a density of 2.5 × 105 cells per well and were treated with 12b at indicated concentrations for 24 h. After that, the cells were collected and washed in PBS and fixed in ice-cold 70% (v/v) ethanol overnight. After washing twice with PBS, cells were stained with a solution containing 50 μg/mL PI (Sigma, St. Louis, MO, USA) and 10 μg/mL RNase A (Solarbio, Beijing, China) for 30 min in the dark at 4 °C. Cell cycle distribution analysis was performed using a MoFlo XDP flow cytometry system (Beckman Coulter, Boulevard Brea, CA, USA).
2.2.4. Extraction and Western Blotting
MDA-MB-231 cells were incubated with 12b at indicated concentrations, then washed with PBS and disrupted on ice for 40 min by a loading buffer, and boiled for 15 min.
The protein concentration of lysates was quantified by using a BCA Protein Assay Kit (Beyotime Biotechnology, Shanghai, China). Equal amounts of protein were separated using sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and then transferred to nitrocellulose membranes. The membranes were blotted with primary antibodies overnight at 4 °C followed by HRP-conjugated secondary antibodies for 1 h at room temperature. Proteins on the membranes were visualized with enhanced chemiluminescence by using chemiluminescence detection reagents.
Antibodies to detect Cleaved poly-(ADP-ribose) polymerase (C-PARP), Caspase 8, Cleaved Caspase 3 (C-Cas3), Survivin, γ-H2AX, Bcl-2, Mcl-1, P21, Akt, phosphor-Akt (Ser473), ERK1/2, phosphor-ERK1/2 (pT185/pY187), Stat3, phosphor-Stat3 (Ser727), PDGFR, FGFR1, VEGFR, EGFR, Src, and phosphor-Src (Tyr416) were purchased from Cell Signaling Technology (Boston, MA, USA). TfR1 antibody, the primary antibody β-Tubulin, and the secondary antibodies were purchased from Sangon Biotech (Shanghai, China).
2.2.5. Immunofluorescence
MDA-MB-231 cells were seeded in 384-well microplates with a glass bottom (Corning, Corning, NY, USA) at a density of 1 × 103 cells per well. After being treated with 12b at 4 μmol/L for 24 h, the cells were fixed with 4% paraformaldehyde (Sangon Biotech, Shanghai, China) in PBS for 30 min and permeabilized with 0.3% Triton X-100 (Sangon Biotech, Shanghai, China) for 15 min at room temperature. After blocking with 1% BSA in PBS, the cells were stained overnight with primary antibody FGFR1 in blocking buffer at 4 °C, then incubated with secondary FITC conjugated anti-rabbit IgG antibody (Solarbio, Beijing, China) for 1 h at room temperature. 4′,6-diamidino-2-phenylindole (DAPI, Solarbio Science, Beijing, China) was used for nuclear staining. The resulting images were captured using a laser scanning confocal microscope (Carl Zeiss, Jena, Germany).
2.2.6. Cellular Thermal Shift Assay (CETSA)
MDA-MB-231 cells were plated into 6-well plates at a density of 2.5 × 105 cells per well. After attaching overnight, cells were treated with 12b at 4 μmol/L for 2 h. Cells were harvested and resuspended in PBS and digested with 0.25% trypsin (Sigma-Aldrich, St. Louis, MO, USA). The cells were then resuspended with PBS and divided into several PCR tubes. Six aliquots were treated with DMSO and another six aliquots were treated with 12b. The tubes were heated at different temperatures by a PCR Thermal Cycler Dice (Analytikjena, Germany) for 3 min and then cooled at 25 °C for 3 min. The heated cells were treated with protease inhibitor PMSF (Solarbio, Beijing, China) and were freeze-thawed three times using liquid nitrogen to lyse cells. Cell lysates were centrifuged at 20,000× g at 4 °C for 20 min. The supernatants were harvested and loading buffer was added before boiling. Protein levels were assessed by Western blotting.
2.2.7. Ricinus Communis Agglutinin I (RCA I) Staining and Mass Spectrometric Analysis
Following the CETSA assay, equal amounts of protein were run on 8% SDS-PAGE gels, then transferred to nitrocellulose membranes. The membranes were incubated with the indicated RCA I antibody (Vector Laboratories, Inc., Burlingame, CA, USA) for 30 min at room temperature, followed by HRP-Streptavidin antibody (R&D Systems, Minneapolis, MN, USA) at room temperature for 1 h. After washing with TBST, signals were detected by chemiluminescence with the enhanced chemiluminescence (ECL) detection reagents.
The gel strips around 80 KD were cut and sent to OE Biotech (Shanghai, China). The 12b-binding proteins were detected by liquid chromatography-mass spectrometry/mass spectrometry. The 12b-interacting candidate proteins were identified and analyzed by using Mascot 2.3 software (Matrix Science, Chicago, IL, USA) and the UniProt-Homo database.
2.2.8. SiRNA and Transfections
MDA-MB-231 cells were plated into 6-well plates at a density of 2.5 × 105 cells per well and were transfected at 70–90% confluency with 100 nM Si-NC or Si-TfR1 (sense: 5′-GGCCAGCAAAGUUGAGAAATT-3′) using Lipofectamine 3000 Transfection Reagent (Thermofisher, Waltham, MA, USA) according to the manufacturer’s instructions. Small interfering RNAs (siRNAs) were purchased from Gene Pharma. Western blotting was used to detect the knockout efficiency of target genes 48 h later.
2.2.9. Detection of Cellular Fe2+ Ions Generation
To clarify Fe2+ ions generation via 12b treatment, CLSM measurement was performed. MDA-MB-231 cells were seeded in 24-well plates at the density of 5 × 104 per well, cultured for 24 h, and then 12b (2, 4 μM) was added into each well, respectively. Meanwhile, DMSO was prepared as a control group. After 24 h, the medium was removed and cells were washed with PBS three times. Then the intracellular Fe2+ ions probe FerroOrange was diluted to 1 μM in serum-free medium and was added to the cells. Cells were incubated at 37 °C for 30 min in the dark. Finally, the fluorescence images of cells were captured by using a confocal microscope (Ex: 543 nm, Em: 580 nm). FerroOrange was purchased from the Dojindo Molecular Technologies Company.
2.2.10. Reactive Oxygen Species (ROS) Level Detection
ROS levels were detected using 2,7-dichloro-dihydrofluorescein diacetate (H2DCFDA) (Biosharp Life Science, Hefei, China) and dihydroethidium (DHE) (Sigma-Aldrich). MDA-MB-231 cells were seeded in 24-well plates at the density of 5 × 104 per well, cultured for 24 h, and then treated with 12b at indicated concentrations. Resup (50 mg/mL) was used as a positive control group. After being cultured for 24 h, the cells were washed with PBS three times. Then, H2DCFDA as an intracellular ROS ion probe (10 μM) was added to the cells. The cells were stationary cultured at room temperature in the dark. Fluorescence images of cells were presented using confocal microscopy (Ex: 488 nm, Em: 530 nm). After 12b treatment for 24 h, cells were harvested and incubated with DHE (5 μM) for 30 min at 37 °C in the dark. DHE fluorescence (Ex: 535 nm, Em: 610 nm) was measured using a MoFlo XDP flow cytometry system (Beckman Coulter, Boulevard Brea, CA, USA).
2.2.11. Statistical Analysis
Statistical analyses were performed using the GraphPad Prism 5 software (GraphPad Inc, San Diego, CA, USA). Data were presented as mean values ± standard deviation. Statistical significance was analyzed by one-way ANOVA for at least three independent experiments. A p-value < 0.05 was considered statistically significant.
3. Results
3.1. Chemistry
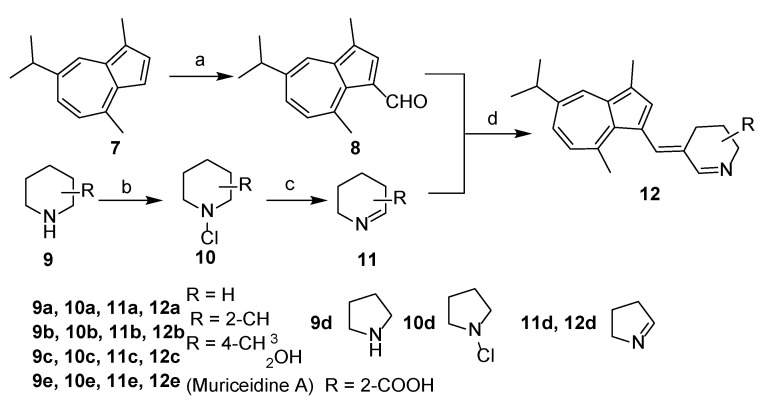
The overall synthetic routes of the title compounds are illustrated in Scheme 1 and Scheme 2. 3-formyl guaiazulene 8 was efficiently synthesized from commercially available guaiazulene 7 following a Vilsmeier-Hacck reaction [35] (Scheme 1). The Δ1-piperidine derivatives 11a–e were synthesized from piperidine derivatives 9a–e through chlorination and dehydrochlorination, following our previously reported methods (Scheme 1). Under the presence of sodium methoxide and acetic acid, 12a–e could be obtained by an aldol-condensation-like reaction between 8 and 11a–e in an 8–20% yield. (Scheme 1). Compound 16 was obtained by condensation of 3-formyl guaiazulene 8 and 1,3-dimethylhydantoin 15 in 24% yield (Scheme 2) [36].
Scheme 1.
Synthesis of 12a–e. Reagents and conditions: (a) POCl3, DMF, 0 °C to rt, 1 h, 95%; (b) t-BuOCl, Et2O, 0 °C, 3 h; (c) KOH, Et2O, MeOH, 50 °C, 1 h, rt, 10h; (d) AcOH, CH3ONa, MeOH, 60 °C, 20 h, 8–20%.
Scheme 2.
Synthesis of 16. Reagents and conditions: (a) H2SO4, 100 °C, 1 h, 80%; (b) AcOH, CH3ONa, MeOH, compound 8, 60 °C, 20 h, 24%.
3.2. Biological Activity
3.2.1. Antitumor Activity In Vitro
In this study, the MTT assay was used as a preliminary screen for the evaluation of in vitro cytotoxicity. The antitumor-viability effect of all target compounds (12a–12d, 16) was evaluated against four cancer cell lines: MDA-MB-231, K562, Hela, and HCT-116. Doxorubicin was used as a positive control. The results were expressed as IC50 values and summarized in Table 1.
Table 1.
Antitumor activity of target compounds 12a–d and 16 against four human cancer cell lines.
| Compounds | IC50 (μM) a | |||
|---|---|---|---|---|
| MDA-MB-231 b | K562 c | Hela d | HCT-116 e | |
| 12a | 13.93 ± 0.042 | 6.99 ± 0.035 | 8.73 ± 0.016 | >100 |
| 12b | 1.15 ± 0.010 | 2.41 ± 0.029 | 1.21 ± 0.010 | 4.63 ± 0.041 |
| 12c | 4.64 ± 0.017 | 1.91 ± 0.014 | 2.58 ± 0.020 | 3.22 ± 0.012 |
| 12d | 10.91 ± 0.021 | 58.41 ± 0.043 | >100 | >100 |
| 16 | >100 | >100 | >100 | >100 |
| Muriceidine A | 77.61 ± 0.063 | 8.37 ± 0.070 | 17.97 ± 0.024 | >100 |
| Doxorubicin f | 0.50 ± 0.040 | 0.39 ± 0.010 | 0.55 ± 0.027 | 0.61 ± 0.080 |
a Cell viability was determined by MTT assay (IC50 mean ± SD). b Human triple negative breast cancer cell line. c Human chronic myeloid leukemia cell line. d Human ovarian cancer cell line. e Human colon cancer cell line. f Doxorubicin was used as the positive control. The bold value shows the strongest antitumor activity of all these compounds in the four types of cells.
Generally, the antitumor activity of the tested compounds varied with different substitutions of Δ1-piperidine at different positions. Compounds (12b and 12c) showed more potent inhibitory activity against cancer cells with IC50 values < 10 μM than that of other compounds. It is notable that compound 12b was the most active derivative which showed potent activities against four cancer cell lines with IC50 values from 1.15 μM to 4.63 μM, and displayed the best activity against MDA-MB-231 cells with an IC50 value of 1.15 μM.
For the MDA-MB-231 cell line, compounds 12a (Δ1-piperidine), 12b (methyl at position C-2 of Δ1-piperidine), 12c (hydroxymethyl at position C-4 of Δ1-piperidine), and 12d (Δ1-pyrrolidine) showed higher inhibitory activities (IC50 = 13.93, 1.15, 4.64, and 10.91 μM, respectively) than Muriceidine A (IC50 = 77.61 μM). Among these compounds, compound 12b with a methyl at position C-2 of Δ1-piperidine (IC50 = 1.15 μM) was the most active compound and was 67.5-fold more potent than Muriceidine A. For the K562 cell line, compounds 12a, 12b, and 12c were the most active compounds with IC50 values of 6.99, 2.41, and 1.91 μM, respectively. Compound 12c was approximately 4.4-fold more potent than Muriceidine A (IC50 = 8.37 μM). For the Hela cell line, compounds 12a, 12b, and 12c exhibited significant cytotoxicity with IC50 values of 8.73, 1.21, and 2.58 μM, respectively. Notably, compound 12b had 15-fold higher cytotoxic activity than Muriceidine A (IC50 = 14.9 μM). For the HCT-116 cell line, only compounds 12b and 12c exhibited significant cytotoxicity with IC50 values of 4.63 and 3.22 μM, respectively.
Taken together, in comparing the IC50 values, we found that the Δ1-piperidine derivatives with electron-donating groups (H/Me) at C-2 had a profound effect on antiproliferative activity since the potency increased following the order Me > H > COOH, which indicated that the combination of Δ1-piperidine derivatives with electron-donating groups and guaiazulene was beneficial for cytotoxic activities. The replacement of methanol at C-4 in Δ1-piperidine can also significantly enhance the anti-proliferative activity. Interestingly, compared with compound 12b which possesses piperidine, compounds with pyrrolidine (12d) or 1,3-dimethylhydantoin (16) displayed decreased activity, suggesting that Δ1-piperidine was responsible for the antiproliferative activity.
3.2.2. Compound 12b Inhibits Various Cells’ Viability and Induces MDA-MB-231 Cells Apoptosis
As compound 12b displayed the best antiproliferative activity in the initial screening, the inhibitory activity of 12b on more types of human cancer cells was next to be examined. As shown in Figure 3A, 12b exhibited cytotoxic IC50 values in the range of 1.15–7 μM after 72 h treatment in various cancer cells with the lowest one towards MDA-MB-231 cells, and dose- and time-dependently suppressed the viabilities of these cells (Figure 3B). Thus, in the following studies, MDA-MB-231 cells were used to further investigate the anticancer effect and mechanism of 12b.
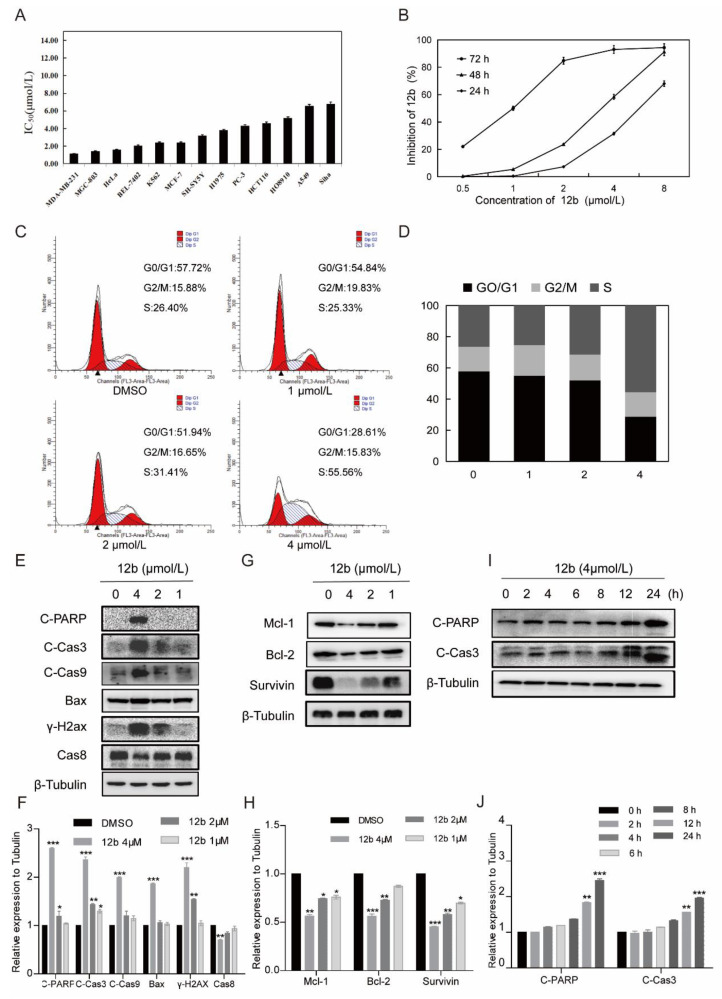
Figure 3.
12b inhibits various cells’ viability and induces MDA-MB-231 cells apoptosis. (A) IC50 values of 12b on various tumor cells. Cancer cells were treated with the indicated concentration of 12b for 72 h. Cell proliferation was determined by using an MTT assay. (B) Proliferation inhibition of 12b on MDA-MB-231 cells. Cells were treated with the indicated concentration of 12b for 24, 48, and 72 h. Cell proliferations were determined by MTT assay. (C) 12b affected cell cycle distribution. Cells were treated with 12b (1–4 μM) for 24 h, then were collected and stained by PI. The DNA content of cells was determined with the MoFlo XDP flow cytometry system. (D) Histograms showed the percentage of cells in G0/G1, G2/M, and S phase after treatment with 12b. (E–J) Effects of 12b on the expression of proteins related to apoptosis. MDA-MB-231 cells were treated with 12b (1, 2, 4 μM) for 24 h or treated with 12b (4 μM) for indicated times. Protein levels were analyzed by Western blotting. The relative band intensities of proteins to the β-Tubulin were analyzed by Image J software. Protein band densities were quantified by normalizing to the DMSO group. Data are presented as means ± SD. All experiments were performed in three replicates. * p < 0.05, ** p < 0.01, *** p < 0.001.
To investigate whether cell cycle arrest is associated with 12b-induced viability inhibition, we further analyzed the cell cycle distribution and found that MDA-MB-231 cells in S phase increased from 26.4% to 55.56% in the presence of 4 μM 12b (Figure 3C,D). These results suggest that compound 12b inhibits the viability of cancer cells by arresting the cell cycle in the S phase.
Next, we determined whether S phase arrest evoked by 12b resulted in cell apoptosis. Cysteine-aspartate-specific protease-9 (Caspase-9) and cysteine-aspartate-specific protease-3 (Caspase-3) are considered to be important components of the effector caspases. Moreover, cleavage and activation of caspase-9/caspase-3 are hallmarks of innate apoptosis [37]. In addition, poly (ADP-ribose) polymerase (PARP) is the substrate of Caspase-3, which has been reported as an early marker of apoptosis [38]. As shown in Figure 3E,I, the expression levels of the active cleaved fragments of Caspase-9, Caspase-3, and PARP were distinctly increased with 12b treatment. Moreover, the level of caspase-8, an important component of the death signal receptor pathway of apoptosis [39], decreased after exposure to 12b treatment, implying the activation of caspase-8. Bax, a member of the Bcl-2 family, can promote apoptosis mediated by mitochondria. We also observed obvious increases of Bax in the presence of 12b. In addition, DNA strand breaks have been demonstrated to play a crucial role in regulating the apoptotic pathway [40]. The levels of γ-H2AX, an essential marker of DNA double-strand breaks [41], were increased significantly in a dose-dependent manner. Cell apoptosis occurs frequently by overexpression of inhibitor of apoptosis (IAP) and Bcl-2 family antiapoptotic proteins. The IAP family such as survivin, and the Bcl-2 family including Bcl-2 and Mcl-1 proteins play important roles in suppressing apoptosis of cells mediated by mitochondria [42]. We then tested whether 12b affected the expression levels of these anti-apoptotic proteins. After treatment with 1 to 4 μM 12b for 24 h, the protein levels of survivin, Bcl-2, and Mcl-1 were obviously decreased (Figure 3G). Taken together, 12b induces cell apoptosis through the mitochondrial pathway combining with the death receptor pathway in MDA-MB-231 cells.
3.2.3. Compound 12b Induces Degradation of Receptor Protein in MDA-MB-231 Cells through the Proteasome Pathway
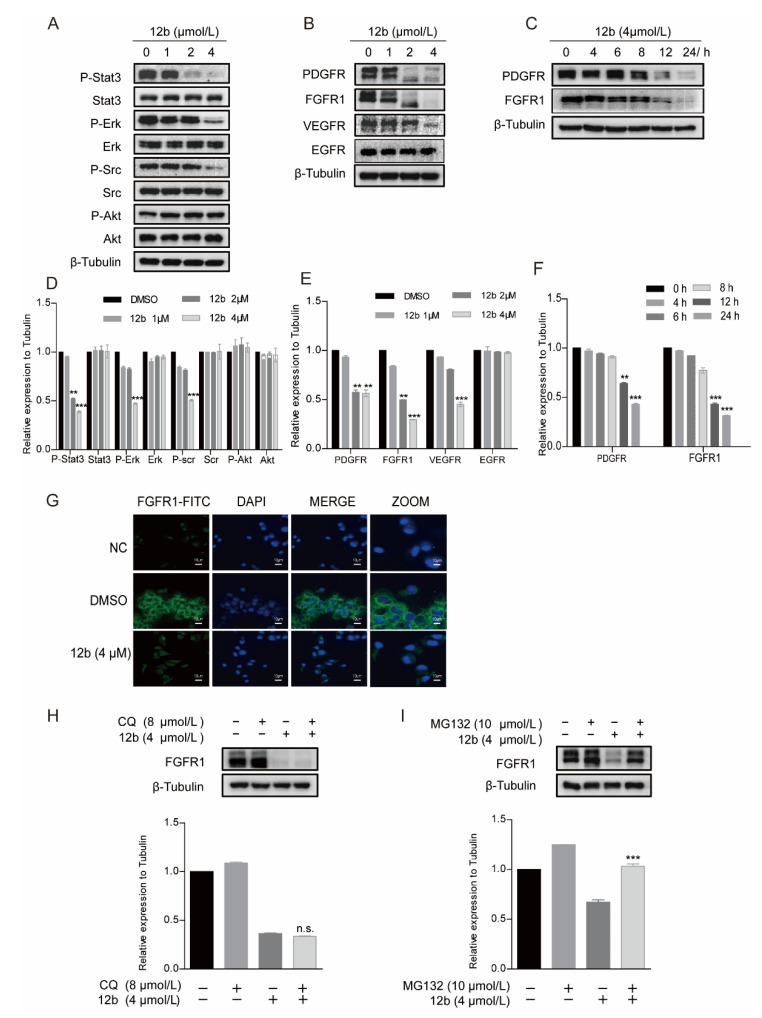
Cell apoptosis is regulated by cell signal transduction. To explore the mechanism underlying 12b-mediated cytotoxicity, we examined major signaling pathways including the expression and activation of AKT [43], ERK [44], and STAT3 in MDA-MB-231 cells. As shown in Figure 4A, after treatment with 1, 2, and 4 μM 12b for 24 h, the phosphorylation of STAT3, Src, and ERK decreased significantly. These results indicate that 12b-induced cell apoptosis is probably associated with the inactivation of cell proliferation upstream signaling. Thus, we further examined the kinase expression and activities upon 12b on some upstream tyrosine kinase receptors including PDGFR, FGFR1, VEGFR, and EGFR. The results show that the expression levels and activities of PDGFR, FGFR1, and VEGFR other than EGFR were reduced by 12b in a time and dose-dependent manner in MDA-MB-231 cells (Figure 4B,C). Increasing evidence supports that FGFR1, a typical kind of receptor tyrosine kinase (RTK), can play a more important part in TNBC progression as the expression of FGFR1 is an independent negative prognostic factor in it [45,46,47]. As FGFR1 expressed on the cell membrane is closely related to the malignant biological behavior of breast cancer, an immunofluorescence assay was used to further determine FGFR1 levels. Compared with the control group, 12b significantly reduced FGFR1 expression on the surface of cells (Figure 4G). These results suggest that 12b can selectively inhibit the expressions of some kinase receptor proteins including FGFR1 in MDA-MB-231 cells.
Figure 4.
12b induces degradation of receptor protein in MDA-MB-231 cells through the proteasome pathway. (A) MDA-MB-231 cells were treated with 12b (0–4 μM) for 24 h and the levels of proliferation-related proteins were detected by Western blotting. (B,C) MDA-MB-231 cells were treated with 12b (0–4 μM) for 24 h or 12b (4 μM) for the indicated times and the levels of tyrosine kinase receptors were detected by Western blotting. (D–F) The relative band intensities of proteins to the β-Tubulin were analyzed and densities were quantified by normalizing to the DMSO group. (G) MDA-MB-231 cells were treated with 12b (4 μM) for 24 h and were stained with primary antibody FGFR1, followed by FITC-antibody. DAPI was used for nuclear staining. The confocal microscope was used for immunofluorescence analysis. The scale bars are 10 μm. (H) MDA-MB-231 cells were treated with 12b (4 μM) or/and 8 μM Chloroquine for 12 h. (I) MDA-MB-231 cells were treated with 12b (4 μM) or/and 10 μM MG132 for 12 h. Protein levels were examined by Western blotting. Protein band densities were quantified by normalizing to β-Tubulin (below). Data are presented as means ± SD. All experiments were performed in three replicates. ** p < 0.01, *** p < 0.001.
We further explored the cause of the decrease in FGER1 protein as an example. As the reduction of protein level via degradation by lysosomal and proteasome is a common way, we first treated cells with protein lysosomal inhibitor Chloroquine before the addition of 12b and found that Chloroquine did not significantly reverse the protein levels of FGFR1 reduced by 12b (Figure 4H). Next, MG132 was used as a proteasome inhibitor to investigate the proteasome pathway. As shown in Figure 4I, we found significant suppressions of 12b-induced FGFR1 degradation by MG132 in MDA-MB-231 cells. These results indicate that the 12b-induced down-regulation of receptor proteins is related to proteasome degradation.
3.2.4. Compound 12b Can Bind to a Glycoprotein Containing α, β-D Galactose
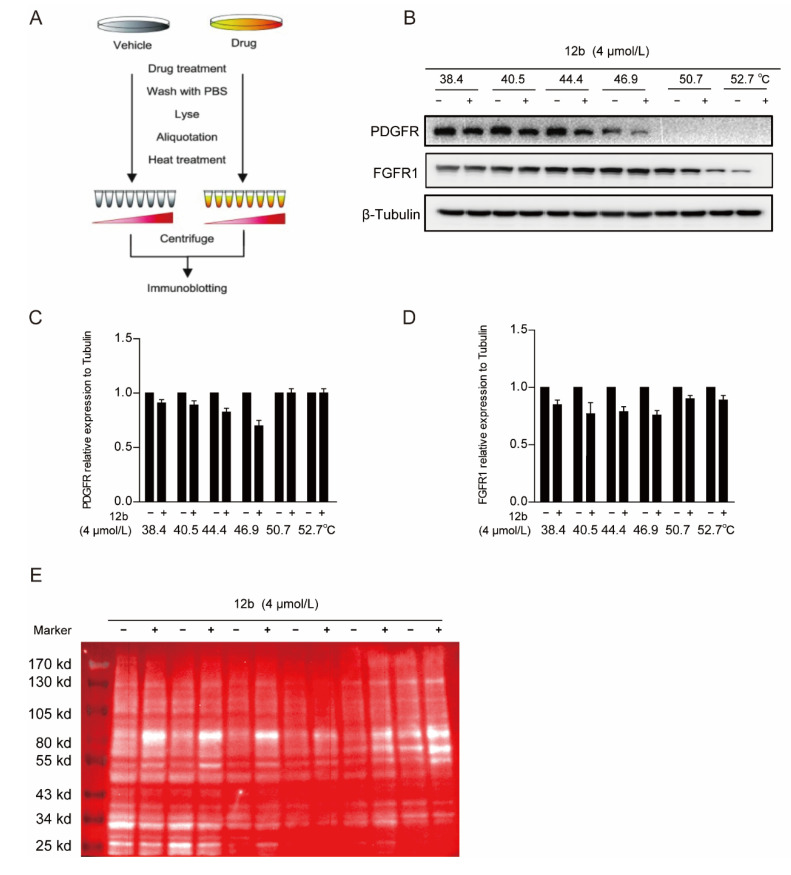
We next performed a cellular thermal shift assay (CETSA) to determine whether 12b reduced tyrosine kinase receptor expression of PDGFR and FGFR1 by binding to it directly in cells. CETSA is a method based on the ligand-induced thermal stability of protein to directly detect the interaction between a drug molecule and target protein (Figure 5A) [48]. We showed that FGFR1 and PDGFR were degraded with increased incubation temperature after 12b treatment, suggesting that 12b does not bind to the two tyrosine kinase receptors directly in MDA-MB-231 cells (Figure 5B–D).
Figure 5.
12b bind to a glycoprotein containing α, β-D galactose. (A,B) Effect of 12b on RTK proteins stabilization. MDA-MB-231 cells were treated with or without 12b (4 μM) for 2 h before being heated at different temperatures. Protein levels were analyzed by Western blotting. (C,D) The relative band intensity to β-Tubulin was analyzed by ImageJ software. Protein band densities were quantified by normalizing to DMSO. (E) The samples in (A) were taken for immunoblotting, membranes were incubated with the RCA I antibody, and HRP-Streptavidin antibody. Signals were detected by chemiluminescence.
Since 12b significantly inhibited multiple downstream signals in cells, we still suspected that it was related to cellular receptor proteins. Membrane receptor proteins tend to be highly glycosylated, which is one of the most common post-translational modifications. Ricinus Communis Agglutinin I (RCA I) as a kind of lectin can specifically bind to α, β-D galactose, which are widely found in most glycoprotein chains. Thus, we used RCA I to screen the binding of glycoproteins to 12b in MDA-MB-231 cells by a modified CESTA experiment and found that the glycoproteins around 80 KD were distinctly thermally stable after 12b treatment. These results indicate that 12b can bind to a glycoprotein containing α, β-D galactose around 80 KD (Figure 5E).
3.2.5. Compound 12b Disrupts the Iron Homeostasis and Cellular Oxidative Balance
To explore the 12b binding glycoprotein, the gel strips around 80 KD were cut and analyzed by liquid chromatography-mass spectrometry/mass spectrometry. As shown in Table 2, among these candidates, TfR1 was the only receptor glycoprotein in a high percentage sequence coverage.
Table 2.
Mass spectrometry analysis of proteins (scores > 80) interacting with 12b.
| Accession | Score * | Mass | Matches † | Sequences ‡ | emPAI § |
|---|---|---|---|---|---|
| sp|P38646|GRP75_HUMAN | 657 | 73,920 | 68 (35) | 40 (28) | 3.01 |
| sp|P23588|IF4B_HUMAN | 586 | 69,167 | 63 (33) | 30 (17) | 1.31 |
| sp|Q96G03|PGM2_HUMAN | 479 | 68,754 | 62 (28) | 39 (20) | 1.93 |
| sp|P18206|VINC_HUMAN | 386 | 124,292 | 39 (23) | 27 (19) | 0.68 |
| sp|Q14247|SRC8_HUMAN | 285 | 61,720 | 57 (23) | 31 (15) | 1.42 |
| sp|Q15291|RBBP5_HUMAN | 251 | 59,800 | 30 (10) | 20 (9) | 0.62 |
| sp|Q09666|AHNK_HUMAN | 218 | 629,213 | 44 (10) | 38 (10) | 0.05 |
| sp|P31040|SDHA_HUMAN | 186 | 73,672 | 36 (14) | 21 (13) | 0.84 |
| sp|Q86YZ3|HORN_HUMAN | 177 | 283,140 | 17 (9) | 15 (9) | 0.11 |
| sp|P02545|LMNA_HUMAN | 136 | 74,380 | 15 (9) | 11 (8) | 0.41 |
| tr|G3V0E5|TfR1_HUMAN | 116 | 76,259 | 18 (4) | 11 (3) | 0.13 |
| sp|P10515|ODP2_HUMAN | 101 | 69,466 | 20 (7) | 15 (7) | 0.38 |
| sp|P23229|ITA6_HUMAN | 96 | 127,724 | 9 (5) | 8 (5) | 0.13 |
| sp|O95810|CAVN2_HUMAN | 82 | 47,202 | 10 (6) | 10 (6) | 0.5 |
| sp|Q912a23|CR025_HUMAN | 80 | 43,597 | 8 (4) | 4 (3) | 0.24 |
| sp|Q96HN2|SAHH3_HUMAN | 80 | 67,705 | 11 (2) | 8 (2) | 0.1 |
* Score: A description of the correlation of this candidate protein. † Matches: Number of high confidence images of mass spectrometry (confidence interval > 95%). ‡ Sequences: Number of highly reliable peptides matched to the compound. § emPAI: Protein content. Bold values are parameters of TfR1 in mass spectrometry analysis.
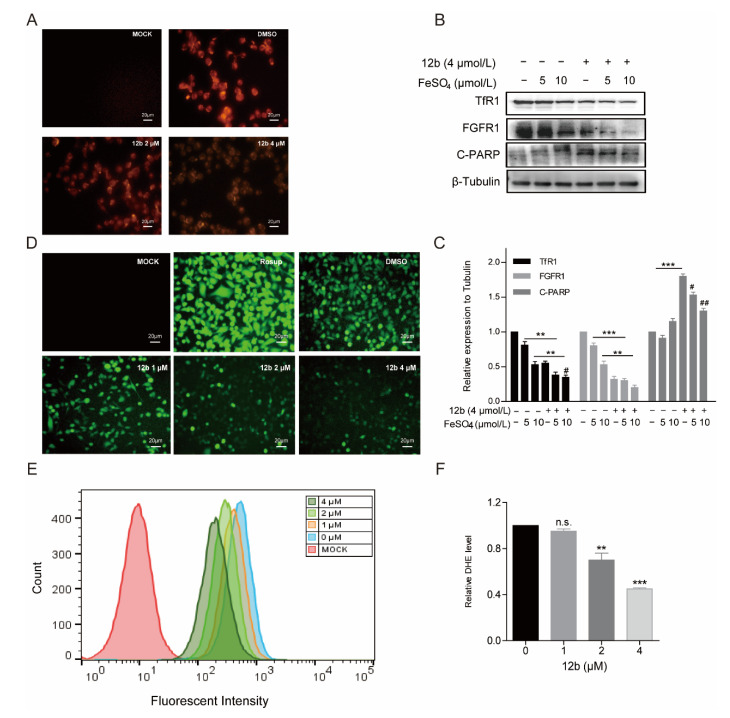
As TfR1 performs a key role in cellular iron uptake, we next wanted to corroborate if concentrations of iron in cells were decreased by 12b. The generation of Fe2+ ions was determined using a Fe2+ ions probe known as Ferrorange which reacts with Fe2+ ions to produce a bright fluorescent substance. Compared with the control groups, the orange fluorescence of cells treated with 12b was much weaker (Figure 6A), suggesting 12b attenuated the influx of Fe2+. The result suggests that TfR1 may be affected by 12b.
Figure 6.
12b disrupts the cellular oxidative balance. (A) CLSM images of MDA-MB-231 cells were collected to visualize the intracellular Fe2+ ions generation using the Fe2+ ions fluorescent probe Ferrorange. The scale bars are 20 μm. (B) MDA-MB-231 cells were pretreated with FeSO4 (0, 5, 10 μM) for 24 h and treated with or without 12b (4 μM). Protein levels were analyzed by Western blotting. (C) The protein band densities were quantified by normalizing to DMSO. (D) CLSM images of MDA-MB-231 cells treated under different conditions of 12b to evaluate ROS production based on DCF fluorescence intensity using the fluorescent probe H2DCFDA. Resup (50 mg/mL) was used as a positive control. The scale bars are 20 μm. (E,F) The intracellular ROS levels were measured through staining by flow cytometric assay using dihydroethidium (DHE). Data are presented as means ± SD. All experiments were performed in three replicates. ** p < 0.01, *** p < 0.001 versus DMSO; # p < 0.05, ## p < 0.01 versus the group of FeSO4 alone.
It has been reported that Transferrin receptors are down-regulated at high concentrations of iron [49]. Therefore, we pre-used FeSO4 to reduce the level of TfR1 and to investigate the effect of 12b on cell apoptosis. As expected, FeSO4 obviously decreased TfR1 expression. We found that 12b also reduced the expression of TfR1 accompanied by an increase in the apoptosis protein C-PARP, which was attenuated after 12b treatment combined with FeSO4 (Figure 6B,C).
Iron is involved in a number of cellular metabolic processes and the imbalance of iron homeostasis can directly or indirectly lead to the generation of ROS and induce cell damage or death [50]. To explore the effect of 12b on the intracellular oxidative environment, the ROS generation capacity was assessed. We compared the ROS yielding ability of different groups by means of CLSM and flow cytometry. A fluorescent probe H2DCFDA was chosen to test intracellular ROS generation by CLSM. After incubation with 12b, the fluorescence intensity of the cells became weaker in a dose-dependent manner (Figure 6D). Quantitative fluorescence analysis of DHE also demonstrated the result of attenuating intracellular ROS upon dosing with 12b (Figure 6E,F). These results demonstrate that 12b reduces the expression of TfR1, and subsequently impairs the influx of Fe2+ and with reduction of intracellular ROS generation, thus leading the cellular oxidative imbalance to cell apoptosis.
3.2.6. Compound 12b—Induced Apoptosis of MDA-MB-231 Cells Depends on TfR1
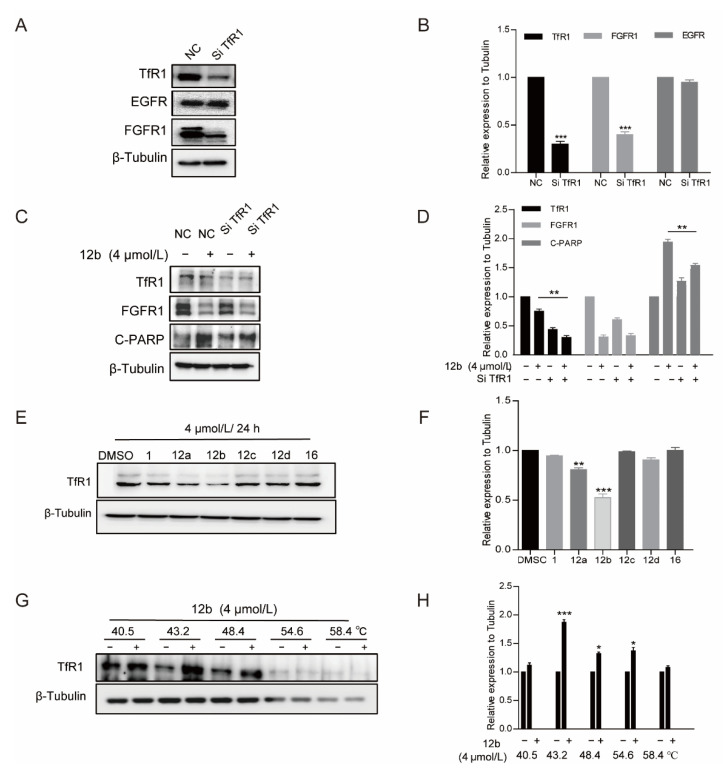
Further, we treated MDA-MB-231 cells with TfR1-targeting siRNA to deplete its expression to investigate the pro-apoptotic activity of 12b. It was observed that the expression of TfR1 was obviously decreased. Similarly, cells with TfR1 depletion produced less C-PARP than control cells upon 12b treatment for 24 h. Interestingly, we noted that FGFR1 was down-regulated while EGFR had no significant change in TfR1 knocked down cells, which is consistent with the reduction of FGFR1 but not EGFR caused by 12b in Figure 4B, supporting that TfR1 was affected by 12b. These results indicate that 12b required TfR1 to exert its activity (Figure 7A–D).
Figure 7.
12b-induced apoptosis of MDA-MB-231 cells depends on TfR1. (A,B) MDA-MB-231 cells were transfected with TfR1-targeting siRNA by using Lipo3000 transfection reagents. After 48 h, the transfection efficiency was examined by Western blotting. (C,D) Cells with/without TfR1-knockdown were treated with 12b (4 μM), protein levels were analyzed by Western blotting, and protein band densities were quantified by normalizing to β-Tubulin. (E,F) MDA-MB-231 cells were treated with 12a–12d and 16 (4 μM) for 24 h, the level of TfR1 was detected by Western blotting, The relative band intensities of proteins to the β-Tubulin were analyzed and densities were quantified by normalizing to DMSO group. (G,H) Effect of 12b on TfR1 stabilization. MDA-MB-231 cells were treated with or without 12b (4 μM) for 2 h before being heated at different temperatures. Protein levels were analyzed by Western blotting. The relative band intensity to β-Tubulin was analyzed. Protein band densities were quantified by normalizing to DMSO. Data are presented as means ± SD. All experiments were performed in three replicates. * p < 0.05, ** p < 0.01, *** p < 0.001 versus DMSO.
We then explored whether the difference in antiproliferative activity (Table 1) of compounds 12a–12d and 16 in MDA-MB-231 cells is related to TfR1, thus the cell viabilities and the expressions of TfR1 were evaluated by MTT assay and Western blot after treatment of a series of compounds. As shown in Figure 7E, the antitumor activities of these compounds were in line with the inhibition abilities on TfR1 expressions, and 12b reduced TfR1 expression most significantly accompanied by the most strong activity of proliferation inhibition. The data further indicate that 12b exerts antitumor effects through TfR1.
Finally, we used the CETSA method to verify the binding between TfR1 and 12b with the antibody of TfR1. As shown in Figure 7G, TfR1 was unstable with an increase in incubation temperature, which was reversed by 12b treatment, corroborating that 12b binds to TfR1 in MDA-MB-231 cells.
Together, these results demonstrate that TfR1 is a molecular target of 12b, mediating the deficiency of iron metabolism and degradation of FGFR1, thereby inducing cell toward apoptosis in MDA-MB-231 cells.
4. Discussion
Triple negative breast cancer (TNBC) exhibits an aggressive subtype and a poor prognosis because these cancers lack ER, PR, and Her2 receptors [51]. Therefore, TNBC does not respond to hormone therapies or Her2 targeted therapies [52]. Given the paucity of effective targeted treatments for this disease subtype, this remains an intense area of investigation. TfR1 is highly expressed in TNBCs, particularly under iron-deficient conditions. Overexpression of TfR1 is thought to meet the increased requirements of iron uptake as well as intracellular ROS levels which are necessary for cell growth [53]. Together with its extracellular accessibility, ability to internalize, and central role in cancer cell pathology, TfR1 inhibition represents a promising anticancer strategy to treat TNBCs.
Muriceidine A, a new compound first isolated by us from the South China Sea gorgonian Muriceides collaris was a potent anticancer agent toward K562 cells with stronger cytotoxicity than that in HL60, A549, and Hela cells at an IC50 value of 8.37 μM [28]. However, its activity was not very satisfactory and the mechanism was also unclear. In the present paper, we evaluated the cytotoxicities of several derivatives with different substituent groups introduced in the functional unsaturated piperidine moiety of Muriceidine A in various cancer cells. It was shown that 12a, 12b, and 12c showed stronger cytotoxicity against K562 cells, and the MDA-MB-231, Hela, and HCT-116 cell lines were sensitive to the three compounds. 12b (methyl at position C-2 of Δ1-piperidine) exhibited the strongest cytotoxicity and induced cell apoptosis in MDA-MB-231 cells and structure-activity relationship analysis revealed that Δ1-piperidine was essential for antiproliferative activity and electron-donating groups at position C-2 of Δ1-piperidine was beneficial to anticancer activity.
The mechanism of the antitumor activity of 12b was further examined. We found that 12b treatment of MDA-MB-231 cells significantly reduced the phosphorylation of signal transduction proteins such as stat3, Scr, and Erk, which suggested to us that receptor tyrosine kinases (RTKs) may play a crucial role in 12b. We found the levels of FGFR1, PDGFR, and VEGFR were down-regulated in a dose- and time-dependent manner except for EGFR. Furthermore, FGFR1 and PDGFR did not bind to 12b. Therefore, we turned to other methods to look for a target molecule. Using a modified CESTA experiment with RCA I as a binding tag analyzed by LC-MS/MS protein spectrum, an obvious receptor molecule TfR1 was fished. Increasing evidence supports FGFR1, a kind of typical RTK, of which expression is an independent negative prognostic factor in TNBC [45,46,47,54]. FGFR1 was selected as an example and we found that whether FeSO4 or siRNA inhibits the expression of TfR1, the levels of FGFR1 were reduced accompanied by decreases in Fe2+ concentration and cell apoptosis. It was noticed that EGFR was not changed after TfR1 was deleted, which is in line with the 12b treatment on MDA-MB-231 cells. Iron serves important functions for enzymes involved in DNA stability and cell cycle progression, ROS-generated enzymes in the electron transport chain, such as nicotinamide adenine dinucleotide phosphate hydride (NADPH) oxidase, P450 enzymes, lipoxygenases (LOX), and xanthine oxidase [55]. In line with these reports, 12b displayed an arrested cell cycle at the S phase and reduced the generation of intracellular ROS due to blocking the uptake of Fe2+. These results suggest that TfR1 might be a target of 12b. We further performed the CESTA experiment with the TfR1 antibody and it was shown that 12b could bind to TfR1. Furthermore, the anti-proliferation activities of Muriceidine A derivatives 12a–12d and 16 were proved consistent with the inhibition abilities on TfR1 expressions. Together, all these results corroborate that 12b exerts antitumor effects through targeting against TfR1.
The mechanism of TfR1 involved in tumorigenesis remains poorly characterized and TfR1 has only been reported to act as a mitochondrial regulator to activate the JNK signaling pathway of lung cancer cells [56]. In our study, the findings that depletion of FGFR1 in TfR1 siRNA-treated cells and 12b-induced down-regulation of FGFR1 are related to proteasome degradation suggest that TfR1 may be an upstream molecule of FGFR1 and promote the proteasome pathway degradation of FGFR1 after 12b treatment in MDA-MB-231 cells. However, the detailed molecular mechanism is not clear, which needs to be discovered in future research.
In all, 12b as a Muriceidine A derivative, was demonstrated to bind to TfR1 directly and cause iron deprivation as well as dysregulation of the intracellular oxidative environment, inducing degradation of FGFR1 oncoprotein through proteasome pathway and TfR1-related cell apoptosis in MDA-MB-231 breast cancer cells. These results support that 12b is a novel, promising, small molecule led compound targeting TfR1 against triple negative breast cancer cells worth further studying in the future, and the finding of the structure-activity relationship helps to design new more effective compounds against TfR1.
5. Conclusions
In this study, we have designed and synthesized a variety of new Muriceidine A derivatives and carried out systematic structure-activity relationship research. Most of the target derivatives showed stronger cytotoxicity than their parent compound, Muriceidine A, against the MDA-MB-231, K562, Hela, and HCT-116 cell lines. Particularly, we found that introduction of electron-donating groups at position C-2 of Δ1-piperidine was beneficial to anticancer activity. Additionally, the replacement of Δ1-piperidine with other unsaturated nitrogen heterocycle resulted in significantly decreased potency, suggesting that Δ1-piperidine was responsible for the antiproliferative activity. Among these compounds, 12b (methyl at position C-2 of Δ1-piperidine) exhibited the strongest cytotoxicity against MDA-MB-231 cells and was more potent than Muriceidine A with an IC50 value of 1.15 μM. 12b was demonstrated to bind to TfR1 directly and cause iron deprivation as well as dysregulation of the intracellular oxidative environment, in turn inducing degradation of FGFR1 oncoprotein through proteasome pathway and TfR1-related cell apoptosis in MDA-MB-231 breast cancer cells. These results support that 12b is a novel promising small molecule lead compound targeting TfR1 against triple negative breast cancer cells worth further studying in the future.
6. Patents
There is a patent resulting from the work reported in this manuscript. Public Announcement No.: CN110791543A.
Acknowledgments
The authors are grateful for financial support granted by the NSFC-Shandong Joint Fund (U1906212), the Shandong Provincial Natural Science Foundation (ZR2020KC002), the State Key Program of National Natural Science of China (82030074), Shandong Provincial Natural Science Foundation (major basic research projects, ZR2019ZD18, ZR2021ZD28), and the National Key Research and Development Program of China (2018YFC0310903).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antiox11050834/s1, Figure S1: 1H-NMR spectra of compound Muriceidine A; Figure S2: 1H-NMR spectra of compound 12a; Figure S3: 13C-NMR spectra of compound 12a; Figure S4: 1H-NMR spectra of compound 12b; Figure S5: 13C-NMR spectra of compound 12b; Figure S6: 1H -NMR spectra of compound 12c; Figure S7: 13C-NMR spectra of compound 12c; Figure S8: 1H-NMR spectra of compound 12d; Figure S9: 13C-NMR spectra of compound 12d; Figure S10: 1H-NMR spectra of compound 16; Figure S11: 13C-NMR spectra of compound 16.
Author Contributions
Conceptualization, J.L.; methodology, X.Q.; software, X.Q.; validation, Y.W., Z.M., X.M., X.L. and P.L.; formal analysis, Y.W. and Z.M.; investigation, Y.W., X.M., X.L. and J.L.; resources, J.L.; data curation, X.Q.; writing—original draft preparation, Y.W.; writing—review and editing, J.L. and G.L.; visualization, Y.W. and Z.M.; supervision, J.L.; project administration, G.L. and P.L.; funding acquisition, J.L. and G.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the NSFC-Shandong Joint Fund (U1906212), the Shandong Provincial Natural Science Foundation (ZR2020KC002), the State Key Program of National Natural Science of China (82030074), Shandong Provincial Natural Science Foundation (major basic research projects, ZR2019ZD18, ZR2021ZD28), and the National Key Research and Development Program of China (2018YFC0310903).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Torti S.V., Torti F.M. Iron and cancer: More ore to be mined. Nat. Rev. Cancer. 2013;13:342–355. doi: 10.1038/nrc3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xue X., Ramakrishnan S.K., Weisz K., Triner D., Xie L., Attili D., Pant A., Győrffy B., Zhan M., Carter-Su C., et al. Iron Uptake via DMT1 Integrates Cell Cycle with JAK-STAT3 Signaling to Promote Colorectal Tumorigenesis. Cell Metab. 2016;24:447–461. doi: 10.1016/j.cmet.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inoue S., Kawanishi S. Hydroxyl radical production and human DNA damage induced by ferric nitrilotriacetate and hydrogen peroxide. Cancer Res. 1987;47:6522–6527. [PubMed] [Google Scholar]
- 4.Dizdaroglu M., Rao G., Halliwell B., Gajewski E. Damage to the DNA bases in mammalian chromatin by hydrogen peroxide in the presence of ferric and cupric ions. Arch. Biochem. Biophys. 1991;285:317–324. doi: 10.1016/0003-9861(91)90366-Q. [DOI] [PubMed] [Google Scholar]
- 5.Torti S.V., Torti F.M. Iron: The cancer connection. Mol. Asp. Med. 2020;75:100860. doi: 10.1016/j.mam.2020.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoenfeld J.D., Sibenaller Z.A., Mapuskar K.A., Wagner B.A., Cramer-Morales K.L., Furqan M., Sandhu S., Carlisle T.L., Smith M., Abu Hejleh T., et al. O2− and H2O2-Mediated Disruption of Fe Metabolism Causes the Differential Susceptibility of NSCLC and GBM Cancer Cells to Pharmacological Ascorbate. Cancer Cell. 2017;31:487–500.e8. doi: 10.1016/j.ccell.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung M., Weigert A., Mertens C., Rehwald C., Brüne B. Iron Handling in Tumor-Associated Macrophages—Is There a New Role for Lipocalin-2? Front. Immunol. 2017;8:1171. doi: 10.3389/fimmu.2017.01171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ba Q., Hao M., Huang H., Hou J., Ge S., Zhang Z., Yin J., Chu R., Jiang H., Wang F., et al. Iron Deprivation Suppresses Hepatocellular Carcinoma Growth in Experimental Studies. Clin. Cancer Res. 2011;17:7625–7633. doi: 10.1158/1078-0432.CCR-10-3099. [DOI] [PubMed] [Google Scholar]
- 9.Pullarkat V., Meng Z., Donohue C., Yamamoto V.N., Tomassetti S., Bhatia R., Krishnan A., Forman S.J., Synold T. Iron chelators induce autophagic cell death in multiple myeloma cells. Leuk. Res. 2014;38:988–996. doi: 10.1016/j.leukres.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Daniels T.R., Bernabeu E., Rodríguez J.A., Patel S., Kozman M., Chiappetta D.A., Holler E., Ljubimova J.Y., Helguera G., Penichet M.L. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim. Biophys. Acta. 2012;1820:291–317. doi: 10.1016/j.bbagen.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniels T.R., Delgado T., Rodriguez J.A., Helguera G., Penichet M.L. The transferrin receptor part I: Biology and targeting with cytotoxic antibodies for the treatment of cancer. Clin. Immunol. 2006;121:144–158. doi: 10.1016/j.clim.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Kawabata H. Transferrin and transferrin receptors update. Free Radic. Biol. Med. 2019;133:46–54. doi: 10.1016/j.freeradbiomed.2018.06.037. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z., Wu H., Wei Z., Wang X., Shen P., Wang S., Wang A., Chen W., Lu Y. TRPM8: A potential target for cancer treatment. J. Cancer Res. Clin. Oncol. 2016;142:1871–1881. doi: 10.1007/s00432-015-2112-1. [DOI] [PubMed] [Google Scholar]
- 14.Carvalho C., Glynne-Jones R. Challenges behind proving efficacy of adjuvant chemotherapy after preoperative chemoradiation for rectal cancer. Lancet Oncol. 2017;18:e354–e363. doi: 10.1016/S1470-2045(17)30346-7. [DOI] [PubMed] [Google Scholar]
- 15.Crielaard B.J., Lammers T., Rivella S. Targeting iron metabolism in drug discovery and delivery. Nat. Rev. Drug Discov. 2017;16:400–423. doi: 10.1038/nrd.2016.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazan H.H., Urfali-Mamatoglu C., Gunduz U. Iron metabolism and drug resistance in cancer. BioMetals. 2017;30:629–641. doi: 10.1007/s10534-017-0037-7. [DOI] [PubMed] [Google Scholar]
- 17.Jian J., Yang Q., Huang X. Src regulates Tyr20 phosphorylation of transferrin receptor-1 and potentiates breast cancer cell survival. J. Biol. Chem. 2011;286:35708–35715. doi: 10.1074/jbc.M111.271585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong S.M., Hwang S., Seong R. Transferrin receptor regulates pancreatic cancer growth by modulating mitochondrial respiration and ROS generation. Biochem. Biophys. Res. Commun. 2016;471:373–379. doi: 10.1016/j.bbrc.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 19.Leoh L.S., Kim Y.K., Candelaria P.V., Martínez-Maza O., Daniels-Wells T.R., Penichet M.L. Efficacy and Mechanism of Antitumor Activity of an Antibody Targeting Transferrin Receptor 1 in Mouse Models of Human Multiple Myeloma. J. Immunol. 2018;200:3485–3494. doi: 10.4049/jimmunol.1700787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimosaki S., Nakahata S., Ichikawa T., Kitanaka A., Kameda T., Hidaka T., Kubuki Y., Kurosawa G., Zhang L., Sudo Y., et al. Corrigendum to “Development of a complete human IgG monoclonal antibody to transferrin receptor 1 targeted for adult T-cell leukemia/lymphoma” [Biochem. Biophys. Res. Commun. 485 (1) 2017 144–151] Biochem. Biophys. Res. Commun. 2020;530:486. doi: 10.1016/j.bbrc.2020.05.147. [DOI] [PubMed] [Google Scholar]
- 21.Schaar D.G., Medina D.J., Moore D.F., Strair R.K., Ting Y. miR-320 targets transferrin receptor 1 (CD71) and inhibits cell proliferation. Exp. Hematol. 2009;37:245–255. doi: 10.1016/j.exphem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Reichert J.M., Rosensweig C., Faden L.B., Dewitz M.C. Monoclonal antibody successes in the clinic. Nat. Biotechnol. 2005;23:1073–1078. doi: 10.1038/nbt0905-1073. [DOI] [PubMed] [Google Scholar]
- 23.Seo Y., Rho J.-R., Geum N., Yoon A.J.B., Shin J. Isolation of guaianoid pigments from the gorgonian Calicogorgia granulosa. J. Nat. Prod. 1996;59:985–986. doi: 10.1021/np960485y. [DOI] [PubMed] [Google Scholar]
- 24.Wang S., Dong G., Sheng C. Structural Simplification of Natural Products. Chem. Rev. 2019;119:4180–4220. doi: 10.1021/acs.chemrev.8b00504. [DOI] [PubMed] [Google Scholar]
- 25.Khan S., Al-Fadhli A.A., Tilvi S. Discovery of cytotoxic natural products from Red Sea sponges: Structure and synthesis. Eur. J. Med. Chem. 2021;220:113491. doi: 10.1016/j.ejmech.2021.113491. [DOI] [PubMed] [Google Scholar]
- 26.Liu J., Ma R., Bi F., Zhang F., Hu C., Venter H., Semple S., Ma S. Novel 5-methyl-2-phenylphenanthridium derivatives as FtsZ-targeting antibacterial agents from structural simplification of natural product sanguinarine. Bioorg. Med. Chem. Lett. 2018;28:1825–1831. doi: 10.1016/j.bmcl.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Steadman V.A., Pettit S.B., Poullennec K.G., Lazarides L., Keats A.J., Dean D.K., Stanway S.J., Austin C.A., Sanvoisin J.A., Watt G.M., et al. Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A. J. Med. Chem. 2017;60:1000–1017. doi: 10.1021/acs.jmedchem.6b01329. [DOI] [PubMed] [Google Scholar]
- 28.Li P., Liu X., Zhu H., Tang X., Shi X., Liu Y., Li G. Unusual Inner-Salt Guaiazulene Alkaloids and bis-Sesquiterpene from the South China Sea Gorgonian Muriceides collaris. Sci. Rep. 2017;7:7697. doi: 10.1038/s41598-017-08100-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becker D.A., Ley J.J., Echegoyen L., Alvarado R. Stilbazulenyl nitrone (STAZN): A nitronyl-substituted hydrocarbon with the potency of classical phenolic chain-breaking antioxidants. J. Am. Chem. Soc. 2002;124:4678–4684. doi: 10.1021/ja011507s. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L.-Y., Yang F., Shi W.-Q., Zhang P., Li Y., Yin S.-F. Synthesis and antigastric ulcer activity of novel 5-isoproyl-3,8-dimethylazulene derivatives. Bioorg. Med. Chem. Lett. 2011;21:5722–5725. doi: 10.1016/j.bmcl.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 31.Mitsakos V., Devenish S.R., O’Donnell P.A., Gerrard J.A., Hutton C.A. LC–MS and NMR characterization of the purple chromophore formed in the o-aminobenzaldehyde assay of dihydrodipicolinate synthase. Bioorg. Med. Chem. 2011;19:1535–1540. doi: 10.1016/j.bmc.2010.12.029. [DOI] [PubMed] [Google Scholar]
- 32.Murata M., Totsuka H., Ono H. Browning of furfural and amino acids, and a novel yellow compound, furpipate, formed from lysine and furfural. Biosci. Biotechnol. Biochem. 2007;71:1717–1723. doi: 10.1271/bbb.70129. [DOI] [PubMed] [Google Scholar]
- 33.Kem W.R., Mahnir V.M., Prokai L., Papke R.L., Cao X., LeFrancois S., Wildeboer K., Prokai-Tatrai K., Porter-Papke J., Soti F. Hydroxy metabolites of the Alzheimer’s drug candidate 3-[(2,4-dimethoxy)benzylidene]-anabaseine dihydrochloride (GTS-21): Their molecular properties, interactions with brain nicotinic receptors, and brain penetration. Mol. Pharmacol. 2004;65:56–67. doi: 10.1124/mol.65.1.56. [DOI] [PubMed] [Google Scholar]
- 34.Martínez-López D., Yu M.-L., García-Iriepa C., Campos P.J., Frutos L.M., Golen J.A., Rasapalli S., Sampedro D. Hydantoin-Based Molecular Photoswitches. J. Org. Chem. 2015;80:3929–3939. doi: 10.1021/acs.joc.5b00244. [DOI] [PubMed] [Google Scholar]
- 35.Al-Horani R.A., Desai U.R. Chemical Sulfation of Small Molecules—Advances and Challenges. Tetrahedron. 2010;66:2907–2918. doi: 10.1016/j.tet.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitagawa T., Okazaki T., Komatsu K., Takeuchi K. Solvolysis of [3-13C]-4-homoadamantyl tosylate. Limited degeneracy of 4-homoadamantyl cation via multiple Wagner-Meerwein rearrangement and vicinal hydride shifts under solvolytic conditions. J. Org. Chem. 1993;58:7891–7898. doi: 10.1021/jo00079a039. [DOI] [PubMed] [Google Scholar]
- 37.Bröker L.E., Kruyt F., Giaccone G. Cell death independent of caspases: A review. Clin. Cancer Res. 2005;11:3155–3162. doi: 10.1158/1078-0432.CCR-04-2223. [DOI] [PubMed] [Google Scholar]
- 38.Khan M., Zheng B., Yi F., Rasul A., Gu Z., Li T., Gao H., Qazi J.I., Yang H., Ma T. Pseudolaric Acid B induces caspase-dependent and caspase-independent apoptosis in U87 glioblastoma cells. Evid.-Based Complement. Altern. Med. 2012;2012:957568. doi: 10.1155/2012/957568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu J.W., Shi Y. FLIP and the death effector domain family. Oncogene. 2008;27:6216–6227. doi: 10.1038/onc.2008.299. [DOI] [PubMed] [Google Scholar]
- 40.Ranjha L., Howard S.M., Cejka P. Main steps in DNA double-strand break repair: An introduction to homologous recombination and related processes. Chromosoma. 2018;127:187–214. doi: 10.1007/s00412-017-0658-1. [DOI] [PubMed] [Google Scholar]
- 41.Ling X., Wu W., Fan C., Xu C., Liao J., Rich L.J., Huang R.-Y., Repasky E.A., Wang X., Li F. An ABCG2 non-substrate anticancer agent FL118 targets drug-resistant cancer stem-like cells and overcomes treatment resistance of human pancreatic cancer. J. Exp. Clin. Cancer Res. 2018;37:240. doi: 10.1186/s13046-018-0899-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castedo M., Perfettini J.-L., Roumier T., Andreau K., Medema R., Kroemer G. Cell death by mitotic catastrophe: A molecular definition. Oncogene. 2004;23:2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- 43.Guo Y.J., Pan W.W., Liu S.B., Shen Z.F., Xu Y., Hu L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020;19:1997–2007. doi: 10.3892/etm.2020.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Little A.S., Smith P.D., Cook S.J. Mechanisms of acquired resistance to ERK1/2 pathway inhibitors. Oncogene. 2013;32:1207–1215. doi: 10.1038/onc.2012.160. [DOI] [PubMed] [Google Scholar]
- 45.Chang J., Liu X., Wang S., Zhang Z., Wu Z., Zhang X., Li J. Prognostic value of FGFR gene amplification in patients with different types of cancer: A systematic review and meta-analysis. PLoS ONE. 2014;9:e105524. doi: 10.1371/journal.pone.0105524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng C.L., Thike A.A., Tan S.Y.J., Chua P.J., Bay B.H., Tan P.H. Expression of FGFR1 is an independent prognostic factor in triple-negative breast cancer. Breast Cancer Res. Treat. 2015;151:99–111. doi: 10.1007/s10549-015-3371-x. [DOI] [PubMed] [Google Scholar]
- 47.Turner N., Pearson A., Sharpe R., Lambros M., Geyer F., Lopez-Garcia M.A., Natrajan R., Marchio’ C., Iorns E., Mackay A., et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molina D.M., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C., Sreekumar L., Cao Y., Nordlund P. Monitoring Drug Target Engagement in Cells and Tissues Using the Cellular Thermal Shift Assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 49.Daniels-Wells T.R., Penichet M.L. Transferrin receptor 1: A target for antibody-mediated cancer therapy. Immunotherapy. 2016;8:991–994. doi: 10.2217/imt-2016-0050. [DOI] [PubMed] [Google Scholar]
- 50.Jomova K., Valko M. Advances in metal-induced oxidative stress and human disease. Toxicology. 2011;283:65–87. doi: 10.1016/j.tox.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Chacón R.D., Costanzo M.V. Triple-negative breast cancer. Breast Cancer Res. 2010;12((Suppl. S2)):S3. doi: 10.1186/bcr2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andreopoulou E., Schweber S.J., Sparano J.A., McDaid H.M. Therapies for triple negative breast cancer. Expert Opin. Pharmacother. 2015;16:983–998. doi: 10.1517/14656566.2015.1032246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zong L., Li J., Chen X., Chen K., Li W., Li X., Zhang L., Duan W., Lei J., Xu Q., et al. Lipoxin A4Attenuates Cell Invasion by Inhibiting ROS/ERK/MMP Pathway in Pancreatic Cancer. Oxidative Med. Cell. Longev. 2016;2016:6815727. doi: 10.1155/2016/6815727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elsheikh S.E., Green A.R., Lambros M.B.K., Turner N.C., Grainge M.J., Powe D., Ellis I.O., Reis-Filho J.S. FGFR1 amplification in breast carcinomas: A chromogenic in situhybridisation analysis. Breast Cancer Res. 2007;9:R23. doi: 10.1186/bcr1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crichton R. Cellular and Systemic Iron Homeostasis. John Wiley and Sons; Hoboken, NJ, USA: 2016. pp. 346–375. [DOI] [Google Scholar]
- 56.Malumbres M., Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article.