Abstract
We induced mutants of Gibberella fujikuroi deficient in gibberellin (GA) biosynthesis by transformation-mediated mutagenesis with the vector pAN7-1. We recovered 24 GA-defective mutants in one of nine transformation experiments performed without the addition of a restriction enzyme. Each mutant had a similar Southern blot pattern, suggesting the integration of the vector into the same site. The addition of a restriction enzyme by restriction enzyme-mediated integration (REMI) significantly increased the transformation rate and the rate of single-copy integration events. Of 1,600 REMI transformants, two produced no GAs. Both mutants had multiple copies of the vector pAN7-1 and one had a Southern blot pattern similar to those of the 24 conventionally transformed GA-deficient mutants. Biochemical analysis of the two REMI mutants confirmed that they cannot produce ent-kaurene, the first specific intermediate of the GA pathway. Feeding the radioactively labelled precursors ent-kaurene and GA12-aldehyde followed by high-performance liquid chromatography and gas chromatography-mass spectrometry analysis showed that neither of these intermediates was converted to GAs in the mutants. Southern blot analysis and pulsed-field gel electrophoresis of the transformants using the bifunctional ent-copalyl diphosphate/ent-kaurene synthase gene (cps/ks) and the flanking regions as probes revealed a large deletion in the GA-deficient REMI transformants and in the GA-deficient transformants obtained by conventional insertional transformation. We conclude that transformation procedures with and without the addition of restriction enzymes can lead to insertion-mediated mutations and to deletions and chromosome translocations.
The rice pathogenic fungus Gibberella fujikuroi (mating population C) is a well-known producer of gibberellins (GAs). GAs are responsible for the growth aberrations observed in rice plants infected with G. fujikuroi (bakanae disease). Although GA formation has also been observed in Sphaceloma manihoticola (30), Neurospora crassa (20), Phaeosphaeria sp. (19), and some other plant pathogens (38), G. fujikuroi is unique because of the enormous quantities of GAs that it can secrete.
Chemically, GA metabolism in G. fujikuroi is relatively well understood (9, 12, 23), but molecular genetic analysis of this pathway has only recently begun (16, 25, 39–41, 43). We have isolated and characterized the genes coding for enzymes that are involved in the initial steps of GA biosynthesis: HMG-CoA reductase (43), FPP synthase (16), and GGDP synthase (25).
Several molecular approaches have been used to identify specific genes in the GA pathway in G. fujikuroi. A PCR approach using oligonucleotide primers, based on conserved amino acid sequences encoded by corresponding plant genes, led to the cloning of a gene from G. fujikuroi coding for the bifunctional copalyl diphosphate/ent-kaurene synthase (CPS/KS) (40). A second approach is to assume transcriptional regulation of genes involved in the GA pathway and then to screen for differential mRNA expression. Differential cDNA screening led to the cloning of a pathway-specific cytochrome P450 monooxygenase gene and the flanking genes, which have all been shown to be involved in GA biosynthesis in G. fujikuroi (39). The disadvantage of this approach is that it also yields housekeeping genes that are induced under GA production conditions, e.g., nitrogen starvation. Furthermore, some GA biosynthesis genes might not be differentially expressed and cannot be cloned by this technique. Therefore, we developed an insertional mutagenesis strategy to identify GA-deficient mutants. Insertional mutagenesis via integrative transformation has been successfully used to tag genes in N. crassa (18), Aspergillus nidulans (37), Colletotrichum lindemuthianum (8), Coprinus cinereus (14), and Magnaporthe grisea (36).
In many cases, insertional mutagenesis has been extended to include restriction enzyme-mediated integration (REMI). Initially, the method was developed for Saccharomyces cerevisiae (33), but it has also been used successfully for other fungi, including Dictyostelium sp. (21), Ustilago maydis (3), Cochliobolus heterostrophus (22), M. grisea (34, 36), A. nidulans (32), and Penicillium paxilli (17).
Our objectives in this study were (i) the recovery of GA-deficient mutants by insertional mutagenesis, (ii) the isolation of the “tagged” genes, and (iii) the identification of the biochemical pathway lesions in the mutants. We obtained pathway-specific mutants by transformation, but all of the transformants obtained with or without restriction enzymes have major deletions in their genomes.
MATERIALS AND METHODS
Strains and plasmids.
The transformation experiments were performed with the G. fujikuroi wild-type strain IMI 58289 (Imperial Mycological Institute, Egham, United Kingdom). Plasmid pAN7-1 (28), which carries the hygromycin B resistance gene, was used to transform G. fujikuroi.
Media and culture conditions.
For DNA isolation, the fungal strains were grown in 100 ml of CM liquid medium optimized for Fusarium spp. (27) for 3 days at 28°C on a rotary shaker at 200 rpm. For a standard GA assay of REMI transformants, strains were grown in test tubes containing 5 ml of 10% ICI medium (11).
DNA isolation.
Genomic DNA was isolated according to the method of Cenis (5). The mycelium was harvested by filtration through sterile filter paper, washed with sterile distilled water, frozen in liquid nitrogen, and lyophilized for 24 h. The lyophilized mycelium was ground to a fine powder. Plasmid DNA was extracted by using Jetprep columns (Genomed, Bad Oeynhausen, Germany) following the manufacturer’s protocol.
Southern blot analysis.
The restricted genomic DNA was transferred to Hybond N+ filters (Amersham, Braunschweig, Germany). The 32P-labelled probes were prepared by the random oligomer-primer method (31). Filters were hybridized under high-stringency conditions at 65°C in 5× Denhardt’s solution containing 5% dextran sulfate (31). Filters were washed in a mixture of 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 0.1% SDS, and 1× SSPE at hybridization temperature.
DNA transformation.
Transformation of G. fujikuroi was conducted with circular or linearized plasmid pAN7-1 (28) with or without addition of restriction endonucleases. Protoplasts were obtained as described previously (41). To mixtures containing 50 μl of protoplasts at a concentration of 1 × 108 per ml in STC (1.3 M sorbitol, 10 mM Tris-HCl [pH 7.5], 10 mM CaCl2), 10 μg of plasmid DNA and 50 μl of polyethylene glycol (PEG) (25% PEG 6000 in STC) were added. For REMI transformation, 10 μg of plasmid DNA was incubated with a restriction endonuclease (HindIII or XbaI) in 50 μl for 2 h and then mixed with 50 μl of 2× STC. The restriction mixture (100 μl) and 50 μl of PEG 6000 were added to 50 μl of protoplasts. The transformation mixture was incubated on ice for 25 min, and then an additional 2 ml of PEG 6000 was added. The mixture was mixed and kept at room temperature for 10 min before 4 ml of STC buffer was added. The transformation mixture was added to 100 ml of liquified regeneration medium (0.05% yeast extract [Difco, Detroit, Mich.], 0.7 M sucrose, 2% agar] containing 125 μg of hygromycin B (Calbiochem, Bad Soden, Germany)/ml. Individual transformants appeared after 3 to 4 days at 28°C. They were transferred to CM agar supplemented with 125 μg of hygromycin B/ml. For further purification, single microconidium colonies were isolated and tested again for hygromycin resistance.
Molecular analysis of the transformants.
The number of copies of integrated pAN7-1 in G. fujikuroi transformants was determined by Southern blot hybridization. Genomic DNA from the transformants and the wild-type IMI 58289 were digested with HindIII and EcoRI, which cut once and twice, respectively, in pAN7-1. Hybridization signals of HindIII-digested DNA were analyzed to determine the copy number of integrated vector.
GA assays. (i) TLC.
For standard analysis of GA formation, transformant and wild-type strains were incubated in 5 ml of 10% ICI medium for 7 days on a rotary shaker (200 rpm) at 28°C. After separation of the mycelium, 10 μl of the culture fluid was analyzed by thin-layer chromatography (TLC) (using chloroform:ethyl acetate:acetic acid [90:60:5]). For the detection of GAs, plates were air dried, exposed over HCl for 30 min, and heated at 120°C for 10 min. GAs were visualized with UV light (365 nm).
(ii) GC-MS conditions.
Samples for gas chromatography-mass spectrometry (GC-MS) analysis were converted to methyl esters by dissolution in methanol (200 μl), the addition of excess ethereal diazomethane, and then evaporation to dryness under N2. Samples were converted to trimethylsilyl (TMSi) ethers by heating with N-trimethyl-N-trimethylsilyltrifluoracetamide at 90°C for 30 min. The derivatized samples were analyzed by GC-MS by using a VG7070 device (V.G. Analytical, Wythenshawe, Manchester, United Kingdom), as described previously (15).
(iii) Provision of labelled substrates.
ent-[1,7,12,18-14C4]kaurene and [1,7,12,18-14C4]GA12-aldehyde were prepared from R-[2-14C]mevalonic acid as described by Graebe et al. (13). [17-3H,13C]GA4 was a gift from B. O. Phinney (University of California, Los Angeles).
(iv) GC-MS analysis of cultures.
Cultures of G. fujikuroi were grown for 10 days in 300-ml shake flasks containing 100 ml of 10% ICI medium (11) at 25°C and 180 rpm. Mycelium and culture medium were separated by filtration. The mycelium was extracted with methanol and analyzed by GC-MS as described above. The culture medium was acidified to pH 2.5 with 1 N HCl and partitioned against ethyl acetate (3× equal volume). The ethyl acetate phase was taken to dryness in vacuo and analyzed by GC-MS. For quantitative determination of ent-kaurene in the mycelium or of the GA3 in the medium, ent-[14C4]kaurene or [17-2H2]GA3 (a gift of L. N. Mander, Australian National University, Canberra, Australia), respectively, was added as an internal standard prior to extraction. Samples were analyzed by GC-MS with selected ion monitoring by using a Hewlett-Packard 5970 gas chromatograph coupled to a mass selective detector. In the case of ent-kaurene, ions in the molecular ion cluster were monitored to determine specific radioactivity and the ent-kaurene content was calculated from isotope dilution (4). For quantification of GA3, the molecular ions for 17-2H2-labelled and unlabelled GA3, at an m/z of 506 and 504, respectively, were monitored, and the GA3 content was determined from the peak area ratios by reference to a calibration curve.
(v) Culture feeding experiments.
Cultures were grown in 40% ICI medium (ICI medium with 40% of the nitrogen source) (15) at 25°C in shake flasks at 180 rpm. After 2 days, the mycelium was transferred to 0% ICI medium (no nitrogen). The labelled substrates were added, and the incubation was continued for 2 days. After filtration, the acidified culture medium was partitioned against ethyl acetate, and the dried extract was taken up in water and adjusted to pH 7 to 8 with KOH. Samples were purified on a QAE Sephadex A25 column and a C18 cartridge (Waters, Taunton, Mass.) as described previously (6) and then analyzed by high-performance liquid chromatography (HPLC) with on-line radiomonitoring, using equipment and general conditions described elsewhere (24). We confirmed metabolite identity by analyzing fractions containing radioactivity by GC-MS after derivatization. Nonpolar metabolites were extracted from the mycelium with methanol and analyzed by TLC or HPLC without purification. In some cases, the GA biosynthesis inhibitors, AMO-1618 (200 μM) (2) and/or paclobutrazol (100 μM) (29), were added to cultures of strain IMI 58289 to reduce formation of nonlabelled products that would interfere in the GC-MS analysis.
PFGE.
Protoplasts at a concentration of 2 × 108 per ml were embedded in 1.2% agarose solution (InCert Biozym, Oldendorf, Germany) containing 1.2 M sorbitol and 50 mM EDTA (pH 8.0) to get 0.6% agarose plugs (45). For lysing the cells, the plugs were incubated with a mixture of 0.5 M EDTA, 0.1 M Tris-HCl (pH 8.0), 1% sarcosyl, and 2 mg of proteinase K (Sigma, Deisenhofen, Germany)/ml for 48 h at 50°C and 100 rpm, changing the buffer after 24 h. The plugs were washed three times for 5 min and three times for 1 h with a mixture of 0.5 M EDTA and 0.1 Tris-HCl (pH 8.0) at 4°C and 100 rpm. They were stored in 50 mM EDTA (pH 8.0) at 4°C. Pulsed-field gel electrophoresis (PFGE) was performed with a CHEF-DR III device (Bio-Rad, Munich, Germany). Gels were run with an agarose (FASTLANE, FMC; Biozym, Oldenburg, Germany) concentration of 0.8% in 1× TAE buffer (31) at 14°C. Each block ran for 24 h at 2 V/cm, and the switch times and the angles were 1,200 s and 96° (block 1), 1,500 s and 100° (block 2), 1,800 s and 106° (block 3), and 2,100 s and 110° (block 4). For Southern analysis the gels were incubated with 0.25 M HCl for 30 min and then blotted with LKB 2016 Vacu-Gene (Pharmacia, Freiburg, Germany) for 3 h onto Hybond N+ filters (Amersham) by using 0.4 M NaOH as a transfer buffer.
RESULTS
Transformation frequency.
Protoplasts of the wild-type G. fujikuroi IMI 58289 were transformed conventionally by circular or linearized plasmid DNA of the vector pAN7-1 and by a modified REMI procedure in which 20, 50, and 100 U of one of the restriction enzymes HindIII and XbaI, which linearize the vector, were added to each transformation mixture (Table 1). Using circular plasmid DNA without restriction enzyme, the transformation rate was very low. Addition of XbaI and HindIII to the circular plasmid, and especially the addition of HindIII at 50 U, significantly increased transformation efficiency. Reproducibly high numbers of transformants were also obtained with HindIII-linearized pAN7-1 (20 U), with or without heat-inactivation of the restriction enzyme. Larger amounts of HindIII reduced the transformation frequency. XbaI had no significant effect on transformation frequency using linearized plasmid.
TABLE 1.
Transformation rates with different plasmid treatments in transformation of G. fujikuroi IMI 58289
| Enzyme | Quantitya (U) | Circular DNA (T/μg)b | Linear DNA (T/μg) |
|---|---|---|---|
| None | 0.1 | ||
| HindIII | 20c | 4.1 | |
| 20 | 2.1d | 3.6 | |
| 50 | 6.2d | 0.9 | |
| 100 | NDe | 0.3d | |
| XbaI | 20c | 0.1 | |
| 20 | ND | 0.3 | |
| 50 | 1.3 | 0.3 | |
| 100 | 0.1d | 0.3d |
The indicated quantity of enzyme was mixed with the DNA and then used directly (circular DNA) or after a 2-h incubation at 37°C (linear DNA) for transformation.
Average number of transformants per microgram of DNA.
The enzyme was heat inactivated for 30 min at 80°C.
These are data from a single experiment.
ND, not done.
Integration events.
The number of copies of integrated pAN7-1 was determined by Southern blot hybridization. A REMI event was defined as an integration of the linearized plasmid into identical restriction sites in the genome (1). In two of the 46 REMI transformants, the vector integrated at the HindIII site in the genome and could be recovered after HindIII restriction (data not shown). In all other cases, either one or both HindIII sites were lost after the integration event or the integration occurred at sites other than the restriction sites in the genomic DNA. Of the 46 analyzed transformants obtained by REMI, 38 carried one copy of the integrated plasmid or tandem insertion events in one integration site. With circular and linearized plasmids without addition of restriction enzymes, we obtained 24 and 29 single-copy transformants out of 50 analyzed, respectively. Therefore, the addition of restriction enzyme apparently resulted in an increase in single-copy integration events.
Screening for gib− mutants.
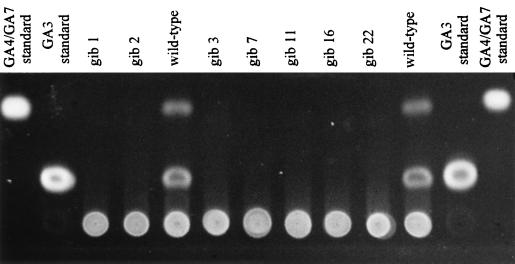
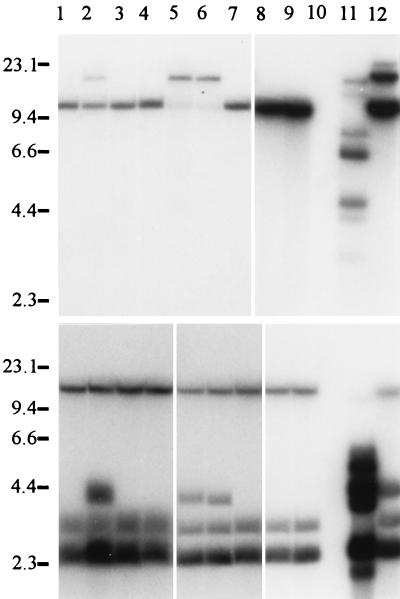
We screened approximately 1,600 transformants obtained by REMI and 371 transformants obtained by transformation with linearized or circular vector DNA without the addition of restriction enzyme for their ability to produce GAs. Among the REMI transformants, two strains (gib1 and gib2) no longer produced GAs. Both mutants were obtained by using HindIII as the restriction enzyme. None of the transformants we obtained by conventional transformation in eight independent experiments had a gib− phenotype. However, in one transformation experiment using the linearized vector pAN7-1, 24 transformants lacking GAs were obtained from different transformation mixtures after screening by TLC (Fig. 1). All of the gib− transformants had normal growth and sporulation characteristics, and no obvious morphological differences were observed. The REMI mutants carry at least two (gib1) and six (gib2) copies of the transformation vector pAN7-1 (Fig. 2). The gib1 mutant lost at least one HindIII site, since the intact plasmid could not be recovered after HindIII restriction. Mutant gib2 has a strongly hybridizing 6.8-kb HindIII fragment (vector size) that could result from a tandem integration of the vector in one location or several independent REMIs (1). Fifteen of the 24 GA-deficient mutants obtained following transformation without a restriction enzyme had single-copy insertions, and the remaining nine had two copies of the vector. In Southern blots of HindIII and EcoRI digests, the 24 GA-defective mutants and the REMI mutant gib1 have nearly identical patterns. In Fig. 2, the hybridization patterns for 11 of the 24 conventional and the two REMI transformants are shown.
FIG. 1.
Detection of GAs in culture filtrates of the wild type and of several mutant strains. Shown is a TLC plate after exposure to HCl and heat treatment observed under UV light. Volumes of 10 μl of culture filtrate of the wild type and of different gib− mutant strains were loaded in each lane as indicated.
FIG. 2.
Integration pattern of the transformation vector obtained by Southern blot analysis. Genomic DNAs of the wild type and the gib− mutants were digested with HindIII (upper part of the figure; one restriction site in pAN7-1) and EcoRI (lower part of the figure; two restriction sites in pAN7-1). The blot was probed with linearized pAN7-1. Lanes: 1, gib11; 2, gib10; 3, gib9; 4, gib8; 5, gib7; 6, gib6; 7, gib5; 8, gib4; 9, gib3; 10, wild type; 11, gib2; 12, gib. The heavy band at 6.8 kb in lane 11 in the upper part marks the vector size. The heavy band at 2.5 kb in all lanes of the lower part of the figure corresponds to an internal EcoRI fragment of the vector. Sizes are indicated on the left in kilobases.
Biochemical analysis of the GA-deficient REMI mutants.
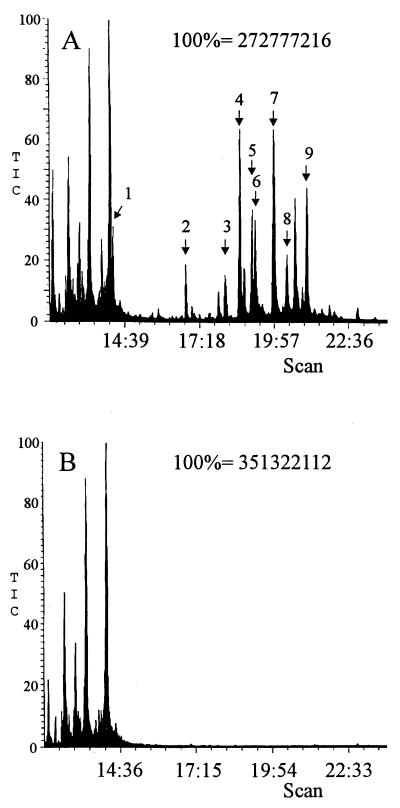
Cultures of gib1, gib2, and IMI 58289 were analyzed by GC-MS. The total ion chromatogram of the culture media of IMI 58289 showed at least 11 compounds, which were absent in the corresponding chromatograms of the REMI mutants (Fig. 3). Most of these compounds were identified as products of the GA biosynthesis pathway by comparison of their mass spectra with those of published spectra (10). Under conditions in which GA3 accumulated to 120 mg per liter of culture medium from the recipient strain, <10−4 mg of GA3 per liter was present in the media from the two mutant strains. Furthermore, 45 mg of ent-kaurene per kilogram of mycelium (dry weight) was measured in mycelium of IMI 58289, but less than 1 μg/kg occurred in the mycelium of the gib− mutants. These results suggest that the first step in the GA pathway, the conversion of GGDP to ent-kaurene (Fig. 4), was affected in both mutants.
FIG. 3.
Total ion chromatograms from GC-MS of extracts from the culture media from the wild type (A) and REMI mutant gib2 (B). Arrows in panel A indicate retention times of intermediates in the GA pathway identified in the culture medium of the wild type by GC-MS analysis as follows: 1, ent-kaurene; 2, GA9; 3, GA25 + GA24; 4, GA14; 5, GA4; 6, GA7; 7, GA13 + GA36; 8, iso-GA3 + GA16; 9, GA3.
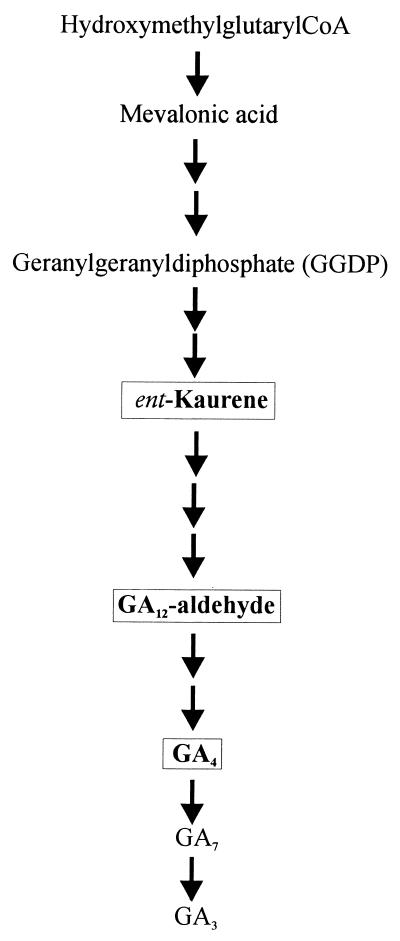
FIG. 4.
Simplified scheme of the GA-biosynthetic pathway. Fed intermediates are boxed.
Feeding experiments.
The radioactively labelled intermediates of the GA pathway, ent-[14C4]kaurene, [14C4]GA12-aldehyde, and [17-3H2,13C]GA4, were incubated with the mutant and wild-type cultures to determine if later steps of the pathway were also affected (Fig. 4). To reduce the high level of nonlabelled ent-kaurene in the mycelium of the wild-type strain that would interfere with the metabolism of the labelled substrate, we added AMO-1618, an inhibitor of ent-kaurene formation from GGDP in plants and fungi (2), prior to the addition of the labelled ent-[14C4]kaurene. With this treatment, the metabolism of the labelled substrate by strain IMI 58289 could easily be detected by HPLC. In cultures of both mutants incubated with ent-[14C4]kaurene, only the substrate was recovered in the mycelium, with no indication of metabolism, even when the cultures were treated with AMO-1618.
After feeding of [14C4]GA12-aldehyde to cultures of the mutants and IMI 58289, the wild-type strain synthesized labelled GA13, GA4, GA7, and GA3. In neither mutant could conversion of [14C4]GA12-aldehyde to the metabolites obtained with the recipient strain be detected.
In mutant cultures treated with [17-3H,13C]GA4, a late intermediate in the pathway to GA3 (Fig. 4), a small peak with a retention time corresponding to that of GA3 was present, but the amount of this compound was too small for identification by GC-MS. In the wild-type strain, the expected metabolism of [17-3H,13C]GA4 was obtained.
The results of these feeding experiments show that ent-kaurene formation and all of the downstream steps in the GA pathway are either blocked or greatly reduced in activity in both mutants.
Southern blot analysis of the GA-defective mutants.
Since neither of the GA-deficient REMI transformants produces ent-kaurene, the first intermediate of the GA-biosynthetic pathway, we initially thought that the vector had integrated into the cps/ks locus coding for the bifunctional CPS/KS. We probed a Southern blot of the mutants with the cps/ks gene (40), but none of the 26 gib− mutants obtained by REMI or conventional mutagenesis showed any hybridization signal (data not shown).
Further analysis using the flanking DNA fragments on the left (a 13-kb SalI fragment and the following 9.4-kb SalI fragment) and the right (a 6-kb SalI fragment and an 8-kb EcoRI fragment) sides of the cps/ks locus (39) as probes revealed the deletion to be at least 36 kb both in the REMI-derived GA-deficient transformants and in the GA mutants obtained by conventional transformation.
PFGE.
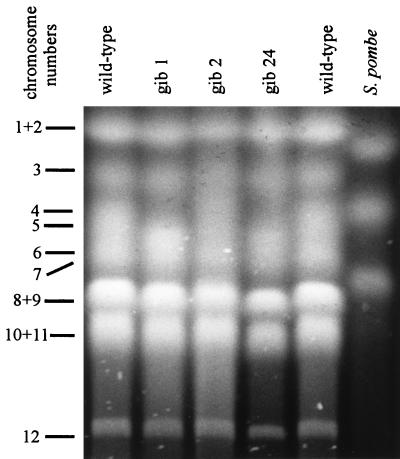
We performed PFGE with protoplasts from the two REMI strains, gib1 and gib2, and with gib24, which was generated by conventional transformation. The large deletion detected by Southern blot analysis is also visible at the chromosomal level (Fig. 5). In two of the three mutant strains tested, gib1 and gib24, chromosome 4 (according to the numbering in reference 44) is missing, whereas the chromosome 5 band seems to be intensified. In gib2, the situation is different. The native chromosome 4 also is missing, but instead a larger chromosome can be detected below chromosome 3, presumably due to a chromosome translocation event (Fig. 5).
FIG. 5.
PFGE of DNA from the wild type and the mutant strains gib1, gib2, and gib24. Numbering of the chromosomes follows that in reference 44. Chromosomes of Schizosaccharomyces pombe were used as size markers (5.7, 4.7, and 3.5 Mb).
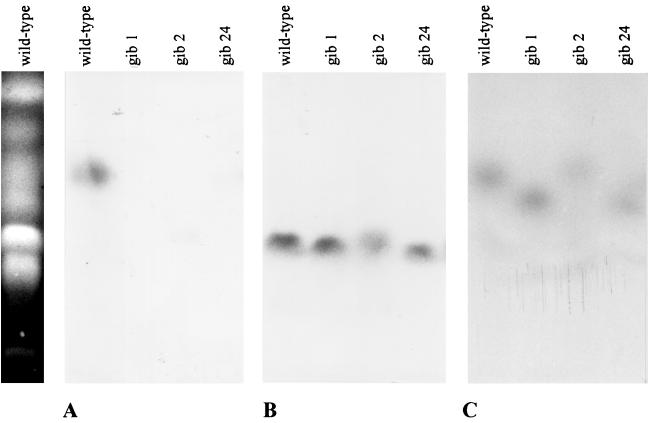
The gel was blotted and hybridized with different probes (Fig. 6). With the cps/ks gene as probe, we found signals only in the lanes with the wild type on chromosome 4. As controls, we hybridized the same filter with the ggs1 (25) and the niaD (41) genes of G. fujikuroi. The niaD gene is located on the double band of chromosomes 8 and 9, giving hybridizing signals of the same size for the lanes with the wild type and the gib− mutants (Fig. 6). The ggs1 gene is located on chromosome 4, which carries the large deletion in the GA-deficient mutants. In contrast to the cps/ks gene and the flanking regions, the ggs1 gene was not lost by the deletion. In gib1 and gib24, the hybridization signal was found on a smaller chromosome of approximately the same size as chromosome 5 which is not present in the wild-type strain. In gib2, the probe hybridized to a larger chromosome apparently obtained by a chromosome translocation event (Fig. 6C). The difference between the wild-type chromosome 4 and the altered chromosome 4 of gib1 and gib24 is about 0.3 to 0.4 Mb.
FIG. 6.
Southern blot analysis of the chromosomes of the wild type and the mutants gib1, gib2, and gib24, using the cps/ks (A), niaD (B), and ggs1 (C) genes as probes.
DISCUSSION
In other fungal systems, random insertional mutagenesis has been used to obtain mutants following the insertion of a plasmid into the genome. We evaluated the feasibility of insertional mutagenesis in G. fujikuroi by transformation with and without the addition of restriction enzymes for isolating mutants that are deficient in one of the presumed 9 to 13 genes required for the conversion of GGDP to GA3 (23).
We also tried to isolate gib− mutants by UV and chemical mutagenesis and found only two mutants in a screen of approximately 12,000 survivors (data not shown). Conventional mutagenesis often causes point mutations that yield leaky mutants, which produce small amounts of the target secondary metabolite (26). Insertional mutagenesis also enables the cloning of the mutagenized gene directly using the integrated plasmid as a tag (3, 8, 22).
The presence of target sites for integration, rather than the availability of foreign DNA, is the rate-limiting factor for plasmid integration (17). Modification of the conventional transformation procedure by the addition of restriction enzymes to the transformation mixtures (REMI) increases both the transformation frequency and the number of single-copy integration events in many fungi (1, 3, 17, 22, 34) and can potentially randomize integration sites (36).
We have shown that both the linearization of plasmid pAN7-1 and the presence of HindIII during transformation significantly increased the transformation rate (Table 1) and the percentage of single-copy integrations in G. fujikuroi. In only two of the analyzed transformants were both recognition sites for the restriction enzyme retained. In the other transformants, one or both restriction sites were lost during the integrations, presumably by degradation of overhanging ends prior to religation. In comparison, for P. paxilli (17) and U. maydis (3), about 50% of the analyzed REMI transformants regenerated the recognition sites. The reason for the low rate of intact restriction sites in G. fujikuroi is not clear.
Among 1,600 REMI transformants analyzed, two GA-deficient mutants (gib1 and gib2) were identified by TLC. Analysis of intermediates by GC-MS and feeding experiments showed that both gib1 and gib2 are blocked at the first step unique to the GA pathway, the biosynthesis of ent-kaurene. Southern blot hybridization using the cps/ks gene as probe revealed that the gib− phenotype is not due to an integration event but to a deletion. Interestingly, 24 gib− mutants obtained by transformation with linearized plasmid also had large deletions (at least 36 kb) at the cps/ks locus.
The results of the PFGE experiments are consistent with the Southern blot analysis showing that the loss of ability to produce GAs in all of the transformants is due to a large deletion in chromosome 4. The cps/ks gene probe hybridized only to chromosome 4 from the wild-type strain. The failure of the mutants to convert ent-kaurene or GA12-aldehyde indicates that several biochemical activities are missing in the mutants and is consistent with the hypothesis that all of the GA pathway genes are clustered on chromosome 4. The fact that even GA4, one of the last intermediates of GA biosynthesis, is not metabolized to the same extent as in the wild-type strain demonstrates that apparently all genes of the pathway are located in the deleted region.
It is surprising that the 24 GA-deficient mutants obtained by conventional transformation and the REMI transformant gib1 had nearly identical hybridization patterns. We suggest that plasmid integration into a recombination hot spot in or near the GA gene cluster on chromosome 4 results in chromosome rearrangements, both deletions and translocations. At a minimum, plasmid insertion appears to result in the loss of 300 to 400 kb of DNA, while in gib2, the chromosomal anomaly is even more complex and involves both a deletion and a significant rearrangement (Fig. 6). Thus, the gib− phenotype results from a deletion in chromosome 4 and not the disruption of individual genes.
Nonrandom integration into a recombination hot spot has also been reported in A. nidulans after conventional transformation (7) and in M. grisea after a REMI approach (36), where two insertions in two mutants out of 5,538 occurred in the same gene coding for an acyltransferase (36). In C. heterostrophus, transformation with and without restriction enzymes led to a 100-kb deletion (42) and a translocation (45) at the TOX1 locus, respectively (45). Large deletions were also obtained in transformation-mediated mutants of the paxilline pathway (46) and in REMI-mediated pathogenicity mutants of M. grisea (36).
In conclusion, we think that transformation, with or without addition of restriction enzymes, can lead not only to mutations based on plasmid insertion, but also to deletions and other chromosome rearrangements. The recovery of nontagged mutants with large deletions in the region of the GA biosynthesis genes was unexpected but provides additional evidence that genes coding for enzymes in the GA pathway are clustered near the cps/ks gene (43). Among these genes are several cytochrome P-450–monooxygenase genes and a second GA-specific geranylgeranyl diphosphate synthase (ggs2) gene. Other genes in this region are still not identified. We plan to use these deletion mutants to identify missing genes in the cluster by complementation with cosmids and differential cDNA screening comparing the expression patterns between the wild type and the deletion mutants.
ACKNOWLEDGMENTS
We thank B. O. Phinney for the gift of [3H,13C]GA4 and E. Cerda-Olmedo (University of Seville, Spain) for providing the wild-type strain IMI 58289.
The work was supported by the DFG (Br1245 1-3). IACR receives grant-aided support from the Biotechnology and Biological Science Research Council of the United Kingdom.
REFERENCES
- 1.Akamatsu H, Itoh Y, Kodama M, Otani H, Kohmoto K. AAL-toxin-deficient mutants of Alternaria alternata tomato pathotype by restriction enzyme-mediated integration. Phytopathology. 1997;87:967–972. doi: 10.1094/PHYTO.1997.87.9.967. [DOI] [PubMed] [Google Scholar]
- 2.Barnes M F, Light E N, Lang A. The action of plant growth retardants on terpenoid biosynthesis: inhibition of gibberellic acid production in Fusarium moniliforme by CCC and AMO-1618; action of these retardants on sterol biosynthesis. Planta (Berlin) 1969;88:172–182. doi: 10.1007/BF01391123. [DOI] [PubMed] [Google Scholar]
- 3.Bölker M, Böhnert H U, Braun K H, Görl J, Kahmann R. Tagging pathogenicity genes in Ustilago maydis by restriction enzyme-mediated integration (REMI) Mol Gen Genet. 1995;248:547–552. doi: 10.1007/BF02423450. [DOI] [PubMed] [Google Scholar]
- 4.Bowen D H, MacMillan J, Graebe J E. Determination of specific radioactivity of [14C]-compounds by mass spectrometry. Phytochemistry. 1972;11:2253–2257. [Google Scholar]
- 5.Cenis J L. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1993;20:2380. doi: 10.1093/nar/20.9.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croker S J, Hedden P, Lenton J R, Stoddart J L. Comparison of gibberellins in normal and slender barley seedlings. Plant Physiol. 1990;94:194–200. doi: 10.1104/pp.94.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diallinas G, Scazzocchio C. A gene coding for the uric acid-xanthine permease of Aspergillus nidulans: inactivational cloning, characterization, and sequence of a cis-acting mutation. Genetics. 1989;122:341–350. doi: 10.1093/genetics/122.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dufresne M, Bailey J A, Dron M, Langin T. ck1, A serine/threonine protein kinase-encoding gene, is involved in pathogenicity of Colletotrichum lindemutianum on common bean. Mol Plant-Microbe Interact. 1998;11:99–108. doi: 10.1094/MPMI.1998.11.2.99. [DOI] [PubMed] [Google Scholar]
- 9.Frankenberger W T, Arshad M, editors. Phytohormones in soil. New York, N.Y: Marcel Dekker, Inc.; 1995. [Google Scholar]
- 10.Gaskin P, MacMillan J. GC-MS of gibberellins and related compounds: methodology and a library of reference spectra. Bristol, United Kingdom: Cantock’s Press; 1992. [Google Scholar]
- 11.Geissman T A, Verbiscar A J, Phinney B O, Cragg G. Studies on the biosynthesis of gibberellins from (−)-kaurenoic acid in cultures of Gibberella fujikuroi. Phytochemistry. 1966;5:933–947. [Google Scholar]
- 12.Graebe J E. Gibberellin biosynthesis and control. Annu Rev Plant Physiol. 1987;38:419–465. [Google Scholar]
- 13.Graebe J E, Hedden P, Gaskin P, MacMillan J. Biosynthesis of gibberellins A12, A15, A24, A36, and A37 in a cell-free system from Cucurbita maxima. Phytochemistry. 1974;13:1433–1440. [Google Scholar]
- 14.Granado J D, Kertesz-Chaloupkova K, Aebi M, Kues U. Restriction enzyme-mediated DNA integration in Coprinus cinereus. Mol Gen Genet. 1997;256:28–36. doi: 10.1007/s004380050542. [DOI] [PubMed] [Google Scholar]
- 15.Hedden P, Hoad G V, Gaskin P, Lewis M J, Green J R, Fuber M, Mander L N. Kaurenoids and gibberellins, including the newly characterized gibberellin A88, in developing apple seeds. Phytochemistry. 1993;32:231–237. [Google Scholar]
- 16.Homann V, Mende K, Arntz C, Ilardi V, Macino G, Morelli G, Böse G, Tudzynski B. The isoprenoid pathway: cloning and characterization of fungal FPPS genes. Curr Genet. 1996;30:232–239. doi: 10.1007/s002940050126. [DOI] [PubMed] [Google Scholar]
- 17.Itoh Y, Scott B. Effect of de-phosphorylation of linearized pAN7-1 and of addition of restriction enzyme on plasmid integration in Penicillium paxilli. Curr Genet. 1997;32:147–151. doi: 10.1007/s002940050259. [DOI] [PubMed] [Google Scholar]
- 18.Kang S, Metzenberg R L. Insertional mutagenesis in Neurospora crassa: cloning and molecular analysis of the preg+ gene controlling the activity of the transcriptional activator NUC-1. Genetics. 1993;133:193–202. doi: 10.1093/genetics/133.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawaide H, Sassa T. Accumulation of gibberellin A1 and the metabolism of gibberellin A9 to gibberellin A1 in a Phaeosphaeria sp. L 487 culture. Biosci Biotechnol Biochem. 1993;57:1403–1405. [Google Scholar]
- 20.Kawanabe Y, Yamane H, Murayama T. Identification of gibberellin A3 in mycelia of Neurospora crassa. Agric Biol Chem. 1983;47:1693–1694. [Google Scholar]
- 21.Kuspa A, Loomis W F. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci USA. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu S, Lyngholm L, Yang G, Bronson C, Yoder O. Tagged mutations at the Tox1 locus of Cochliobolus heterostrophus by restriction enzyme-mediated integration. Proc Natl Acad Sci USA. 1994;91:12649–12653. doi: 10.1073/pnas.91.26.12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacMillan J. Biosynthesis of the gibberellin plant hormones. Nat Prod Rep. 1997;14:221–243. [Google Scholar]
- 24.MacMillan J, Ward D A, Phillips A L, Sánchez-Beltrán M J, Gaskin P, Lange T, Hedden P. Gibberellin biosynthesis from gibberellin A(12)-aldehyde in endosperm and embryos of Marah macrocarpus. Plant Physiol. 1997;113:1369–1377. doi: 10.1104/pp.113.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mende K, Homann V, Tudzynski B. The geranylgeranyl diphosphate synthase gene of Gibberella fujikuroi: isolation and expression. Mol Gen Genet. 1997;255:96–105. doi: 10.1007/s004380050477. [DOI] [PubMed] [Google Scholar]
- 26.Nelson M A, Morelli G, Carattoli A, Romano N, Macino G. Molecular cloning of a Neurospora crassa carotenoid biosynthetic gene (albino-3) regulated by blue light and the products of white collar genes. Mol Cell Biol. 1989;9:1271–1276. doi: 10.1128/mcb.9.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pontecorvo G V, Poper J A, Hemmonns L M, MacDonald K D, Buften A W. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 28.Punt P J, Oliver R P, Dingemanse M A, Pouwels P H, van den Hondel C A M J J. Transformation of Aspergillus nidulans based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56:117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- 29.Rademacher W. Inhibition of gibberellin production by plant growth retardants in the fungi Gibberella fujikuroi and Sphaceloma manihoticola. Plant Physiol. 1992;100:625–629. doi: 10.1104/pp.100.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rademacher W, Graebe J E. Gibberellin A4 produced by Sphaceloma manihoticola, the cause of the superelongation disease of cassava (Manihot esculenta) Biochem Biophys Res Commun. 1979;91:35–40. doi: 10.1016/0006-291x(79)90579-5. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Sanchez O, Navarro R E, Aguirre J. Increased transformation frequency and tagging of developmental genes in Aspergillus nidulans by restriction enzyme-mediated integration (REMI) Mol Gen Genet. 1998;258:89–94. doi: 10.1007/s004380050710. [DOI] [PubMed] [Google Scholar]
- 33.Schiestl R H, Petes T D. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi Z, Christian D, Leung H. Enhanced transformation in Magnaporthe grisea by restriction enzyme mediated integration of plasmid DNA. Phytopathology. 1995;85:329–333. [Google Scholar]
- 35.Spector C, Phinney B O. Gibberellin biosynthesis genetic studies in Gibberella fujikuroi. Physiol Plant. 1968;21:127–136. [Google Scholar]
- 36.Sweigard J A, Carroll A M, Farrall L, Chumley F G, Valent B. Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol Plant-Microbe Interact. 1998;11:404–412. doi: 10.1094/MPMI.1998.11.5.404. [DOI] [PubMed] [Google Scholar]
- 37.Tilburn J, Roussel F, Scazzocchio C. Insertional inactivation and cloning of the wa gene of Aspergillus nidulans. Genetics. 1990;126:81–90. doi: 10.1093/genetics/126.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tudzynski B. Fungal phytohormones in pathogenic and mutualistic associations. In: Esser K, Lemke P A, editors. The mycota. Berlin, Germany: Springer-Verlag; 1997. pp. 167–184. [Google Scholar]
- 39.Tudzynski B, Hölter K. The gibberellin biosynthetic pathway in Gibberella fujikuroi: evidence for a gene cluster. Fungal Genet Biol. 1998;25:157–170. doi: 10.1006/fgbi.1998.1095. [DOI] [PubMed] [Google Scholar]
- 40.Tudzynski B, Kawaide H, Kamiya Y. The gibberellin biosynthesis in Gibberella fujikuroi: cloning and characterization of the copalyl diphosphate synthase gene. Curr Genet. 1998;34:234–240. doi: 10.1007/s002940050392. [DOI] [PubMed] [Google Scholar]
- 41.Tudzynski B, Mende K, Weltring K M, Kinghorn J R, Unkles S E. The Gibberella fujikuroi niaD gene encoding nitrate reductase: isolation, sequence, homologous transformation and electrophoretic karyotype location. Microbiology. 1996;142:533–539. doi: 10.1099/13500872-142-3-533. [DOI] [PubMed] [Google Scholar]
- 42.Turgeon B G, Kodama M, Yang G, Rose M S, Su S W, Yoder O C. Function and chromosomal location of the Cochliobolus heterostrophus TOX1 locus. Can J Bot. 1995;73:1071–1076. [Google Scholar]
- 43.Woitek S, Unkles S E, Kinghorn J R, Tudzynski B. 3-Hydroxy-3-methylglutaryl-CoA reductase of Gibberella fujikuroi: isolation and characterization. Curr Genet. 1997;31:38–47. doi: 10.1007/s002940050174. [DOI] [PubMed] [Google Scholar]
- 44.Xu J-R, Yan K, Dickman M B, Leslie J F. Electrophoretic karyotypes distinguish the biological species of Gibberella fujikuroi (Fusarium section Liseola) Mol Plant-Microbe Interact. 1995;8:74–84. [Google Scholar]
- 45.Yang G. The molecular genetics of T-toxin biosynthesis by Cochliobolus heterostrophus. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1995. [Google Scholar]
- 46.Young C, Itoh Y, Johnson R, Garthwaite I, Miles C O, Munday-Finch S C, Scott B. Paxilline-negative mutants of Penicillium paxilli generated by heterologous and homologous plasmid integration. Curr Genet. 1998;33:368–377. doi: 10.1007/s002940050349. [DOI] [PubMed] [Google Scholar]