Abstract
The transport of cellobiose in mixed ruminal bacteria harvested from a holstein cow fed an Italian ryegrass hay was determined in the presence of nojirimycin-1-sulfate, which almost inhibited cellobiase activity. The kinetic parameters of cellobiose uptake were 14 μM for the Km and 10 nmol/min/mg of protein for the Vmax. Extracellular and cell-associated cellobiases were detected in the rumen, with both showing higher Vmax values and lower affinities than those determined for cellobiose transport. The proportion of cellobiose that was directly transported before it was extracellularly degraded into glucose increased as the cellobiose concentration decreased, reaching more than 20% at the actually observed levels of cellobiose in the rumen, which were less than 0.02 mM. The inhibitor experiment showed that cellobiose was incorporated into the cells mainly by the phosphoenolpyruvate phosphotransferase system and partially by an ATP-dependent and proton-motive-force-independent active transport system. This finding was also supported by determinations of phosphoenolpyruvate phosphotransferase-dependent NADH oxidation with cellobiose and the effects of artificial potentials on cellobiose transport. Cellobiose uptake was sensitive to a decrease in pH (especially below 6.0), and it was weakly but significantly inhibited in the presence of glucose.
Ruminants can utilize structural carbohydrates in plant tissues, including cellulose and hemicellulose, through microbial activities in the rumen, although these components cannot be degraded by mammalian digestive enzymes. Cellobiose, one of the major products of cellulose degradation (38), can be metabolized by many cellulolytic and noncellulolytic species of ruminal bacteria (14, 32, 44). Transport of a nutritional substrate, the first energy-requiring process in a substrate-limiting environment, can often be rate limiting in the nutrient metabolisms of bacteria (7, 26). In order to understand the final steps in fiber utilization, it is necessary to investigate the dominant cellobiose transport system in the rumen as a whole.
There are many reports on the carbohydrate transport systems of each species of ruminal bacterium (24), but only a few species (i.e., Fibrobacter succinogenes [22], Ruminococcus flavefaciens [12], and Streptococcus bovis [25]) have been studied with regard to cellobiose transport. These studies have shown that the phosphoenolpyruvate-dependent phosphotransferase system (PEP-PTS) and active transport systems can be used for the cellular uptake of cellobiose. In addition to the problem of the scarcity of studies, there is the difficulty of accurately estimating the dominant transport systems in the rumen from the results of transport studies of each species, because it has been a long time since the first isolation of many of the ruminal type strains (4, 14) and also because it is difficult to determine the actual population size of a specific strain in the rumen.
It is not clear that cellobiose indeed can be transported into the ruminal microbial cells in its intact form, since β-glucosidase activities have been found in some ruminal bacteria (5, 35, 52) as well as in the fluids of actual and artificial rumens (8, 27, 51). Our study determined the relative contribution of cellobiose transport in the apparent utilization of cellobiose and characterized the major cellobiose transport systems in the rumen as a whole by using mixed ruminal bacteria taken from a cow fed a forage diet.
MATERIALS AND METHODS
Animal and diet.
A ruminally fistulated, nonlactating holstein cow (body weight, 470 kg) was fed 6.0 kg of first-cut, prebloom Italian ryegrass hay every day at 9:00 a.m., which was equivalent to the energy requirement for maintenance (28). The chemical composition of the hay (on a dry-matter basis), analyzed by the proximate method (10) and detergent analysis (48, 49), was as follows: neutral detergent fiber, 56.2%; acid detergent fiber, 31.1%; crude protein, 15.4%; ether extract, 3.5%; and crude ash, 10.5%. Water was freely given.
Preparation of mixed ruminal bacteria.
Liquid and solid portions of the ruminal content, taken by a suction pump and by hand grasp, respectively, were taken 2 h after feeding the cow through a fistula. Equal amounts (wt/wt) of these portions were mixed, ground with a homogenizer (model MN-2; Nihon Seiki Co., Tokyo, Japan) at 250 W for 1 min, and squeezed through four-layered gauze. The squeezed fluid was left undisturbed for 30 min at 38°C to separate the feed particles (53). The fluid obtained from a middle portion of the undisturbed sample was slowly centrifuged (at 750 × g for 10 min at 10°C) to remove protozoa (32) and then centrifuged again (at 10,000 × g for 15 min at 10°C) to harvest the mixed ruminal bacteria, in which no protozoa were microscopically detected. Anaerobic conditions were maintained through the whole procedure by using an N2 gas stream.
Measurements of cellobiose and glucose transports.
The mixed ruminal bacteria were washed twice and resuspended in NKMP buffer (15). The transport was initiated by the addition of cellobiose or glucose containing [3H]cellobiose (329 MBq/μmol) or d-[U-14C]glucose (11.2 MBq/μmol), respectively, to the cell suspension. The reaction was terminated by dispersing the suspension (100 μl) into ice-cold NKMP buffer (2.0 ml). After the cell suspension was sent through a membrane filter (0.45-μm pore size) and washed with 2.0 ml of ice-cold NKMP buffer, the radioactivity of the cells was measured by using a liquid scintillation counter (Tri-Carb 166TR; Packard Instrument Co., Meriden, Conn.). Transport rates were calculated from the difference between the levels of uptake at 38 and 0°C at 60 s, which nearly matched the result obtained with a regression coefficient from the values at 10, 30, and 60 s at 38°C. In the inhibitor experiments, the cells were incubated with 3,5-di-tert-butyl-4-hydroxybenzilidene-malononitrile (SF6847), triphenylmethyl phosphonium (TPMP) bromide, iodoacetate, chlorhexidine, and/or harmaline for 10 min prior to the addition of cellobiose (0.01 mM). Some inhibitors were dissolved in ethanol (up to 4% in final concentration), which exhibited no significant effect on cellobiose uptake.
Determinations of PEP-PTS and cellobiase activities.
The activity of the PEP-PTS in the presence of 0.01 mM cellobiose was assayed spectrophotometrically (15) and contrasted with the activity in the reaction mixture without cellobiose. The cell-associated cellobiase activity was measured by examining the generation of glucose from cellobiose in NKMP buffer at 38°C by the washed cells for 1 min. Both chlorhexidine (200 μm) and iodoacetate (2 mM) were added to the buffer 10 min prior to the addition of cellobiose in order to inhibit the uptake of cellobiose (as shown in Results) and glucose (15) into the cells. The extracellular cellobiase in the rumen was estimated by comparing the activities of a centrifuged ruminal fluid (at 10,000 × g for 15 min) to those of the cell fraction. Glucose in NKMP buffer with the washed cells was measured spectrophotometrically by an enzymatic method using hexokinase and glucose-6-phosphate dehydrogenase (18), but glucose and other sugars in the ruminal fluid were analyzed with a high-performance liquid chromatograph (HPLC) equipped with a pulsed electrochemical detector and a pellicular anion-exchange column as described previously (15), because some colored elements in the ruminal fluid could have disturbed the spectrophotometric assays.
Formation of artificial membrane potentials and determination of PMF.
The artificial potentials were generated as described previously (15). An artificial proton gradient (change in pH [ΔpH]) was created by diluting the acetate-loaded cells 50-fold in a potassium buffer. An artificial electrical potential (Δψ) was generated by loading the valinomycin-treated cells with potassium and diluting them 50-fold in a bis-Tris buffer. A chemical sodium gradient (ΔpNa) was created by diluting the potassium-loaded cells 50-fold in a sodium-potassium buffer. The proton motive force (PMF) was also determined as described previously (15). After the cells were incubated with 3H2O, [carboxyl-14C]inulin, [7-14C]benzoic acid, or [phenyl-3H]tetraphenyl phosphonium ([3H]TPP+) bromide, the radioactivities of the cells and extracellular water, being separated from each other by centrifugation through a silicon oil (a mixture of KF-961 and CH510; Shin-Etsu Chemical Co. and Toray Dow Corning, respectively, both in Tokyo, Japan), were counted by liquid scintillation. The intracellular volume (2.7 μl/mg of protein) was estimated by measuring the difference between levels of 3H2O and [carboxyl-14C]inulin. The ΔpH and Δψ were calculated by determining the levels of uptake of [7-14C]benzoic acid and [3H]TPP+ by using the Henderson-Hasselbalch and the Nernst equations, respectively. Nonspecific binding of [3H]TPP+ was estimated from examining the cells treated with 0.1% toluene.
Other analyses.
The protein content in the mixed ruminal bacteria was measured by the method of Lowry et al. (21); it was 166 μg/ml of the suspension when the solution’s optical density at 600 nm was 1.0. Intracellular sodium was measured as described previously (15) with an atomic absorption spectrophotometer (model Z-8000; Hitachi Ltd., Tokyo, Japan) with a correction for extracellular contamination. Intracellular ATP was measured with a luminometer (model 1250; LKB-Pharmacia, Turku, Finland) with a luciferin-luciferase mix (Sigma Chemical Co., St. Louis, Mo.) after extraction with 14% ice-cold perchloric acid (46). The difference between two averages was tested by the Student t test (43). Every measurement was repeated at least three times.
Materials.
Radiolabelled cellobiose and glucose were prepared by Amersham International, Little Chalfont, Buckinghamshire, United Kingdom. Cellobiose was randomly tritiated by a catalytic-exchange procedure with tritium gas and purified by HPLC to 99.8%. All other radiolabelled chemicals were supplied by DuPont Co., Wilmington, Del. d-Gluconic-δ-lactone and TPMP bromide were obtained from Nakarai Tesque, Kyoto, Japan. Nojirimycin(-1-sulfonic salt) and SF6847 were obtained from Seikagaku Corp., Tokyo, Japan, and Wako Pure Chemicals, Osaka, Japan, respectively. Nojirimycin was stored at −20°C and dissolved just before use. Chlorhexidine (diacetate salt) and harmaline were obtained from Sigma Chemical Co. All other chemicals were of the highest grade available from various commercial sources.
RESULTS
Monosaccharides were detected in the rumen in concentrations up to 0.49, 0.02, 0.02, and 0.12 mM for glucose, galactose, arabinose, and xylose, respectively, but cellobiose could not be detected at any time under these feeding conditions. The lower limits obtained by HPLC analysis of sugars in the ruminal fluid were 0.01 mM for the monosaccharides and 0.02 mM for cellobiose because of a high background noise. The diurnal fluctuation of the ruminal pH ranged between 6.3 and 7.2 (data not shown).
Values for kinetic parameters of the cell-associated cellobiase of the ruminal bacteria were 0.25 mM for the Km and 26 nmol/min/mg of protein for the Vmax. The extracellular cellobiase activity in the ruminal fluid was estimated to have a Km of 1.0 mM and a Vmax of 23 nmol/min/mg of protein in the corresponding bacterial cells. Several chemicals known to be β-glucosidase inhibitors (9, 30, 33) were examined for their effects on cell-associated cellobiase activity in the presence of 0.1 mM cellobiose. The cellobiase activity was almost completely inhibited (>95%) by 10 and 100 μM nojirimycin, and a similar degree of inhibition was also shown at a lower concentration of cellobiose (0.02 mM) with 10 μM nojirimycin. Gluconic-δ-lactone (10 and 100 μM), on the other hand, showed only a partial inhibition (40 to 60%), and iodoacetoamide (10 and 100 μM) did not significantly suppress (<10%) the cellobiase activities of the ruminal bacteria (data not shown).
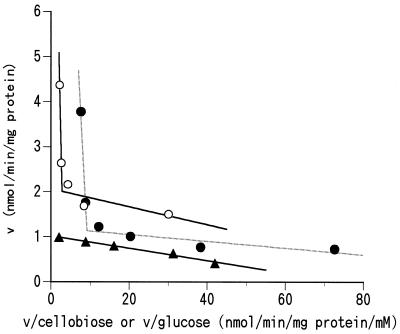
Figure 1 shows the Eadie-Hofstee plots of levels of uptake of cellobiose with and without 20 μM nojirimycin and of glucose into the cells at various substrate concentrations. The levels of uptake of cellobiose without nojirimycin and of glucose showed biphasic kinetics indicative of high-affinity–low-velocity and low-affinity–high-velocity systems. The high-affinity system for cellobiose and glucose uptake showed 7.4 and 20 μM for the Km and 1.2 and 2.1 nmol/min/mg of protein for the Vmax, respectively. The kinetic constants of the low-affinity system were difficult to correctly determine because of isotope dilution, but they likely were 1 to 3 mM (for cellobiose) or 3 to 5 mM (for glucose) for the Km and more than 10 nmol/min/mg of protein for the Vmax for both sugars. The uptake of cellobiose in the presence of nojirimycin, on the other hand, showed only one system, having a Km of 14 μM and a Vmax of 1.0 nmol/min/mg of protein.
FIG. 1.
Eadie-Hofstee plots of cellobiose uptake with (▴) and without (●) nojirimycin-1-sulfonate (20 μM) and of glucose uptake (○) into mixed ruminal bacteria.
The effects of inhibitors on cellobiose uptake with 20 μM nojirimycin are shown in Table 1. The table also shows the bioenergetic properties of the cells in the presence of the inhibitors. A proton-conducting uncoupler, SF6847 (13), and harmaline, which is known to be an inhibitor of sodium-dependent transport systems (6, 42), did not affect cellobiose uptake. Harmaline did not change any of the bioenergetic properties, while SF6847 decreased the ΔpH by about 80% compared with that of the control. A lipophilic ion, TPMP+, which is known to dissipate the membrane potential (37), showed no effect on cellobiose uptake at 1 mM, but it significantly decreased the uptake at 10 mM. The Δψ decreased significantly with both 1 and 10 mM TPMP bromide, by 23 and 55%, respectively, but only with 10 mM TPMP bromide was there a significant decrease in the intracellular ATP (by 42%) compared with the level in the control. Iodoacetate, an inhibitor of glyceraldehyde-3-phosphate dehydrogenase (17), showed a significant inhibition of cellobiose uptake when 2 mM was added, and it also showed a slight but significant inhibition when 500 μM was added. Generation of intracellular ATP was inhibited by 68 and 28% when 2 mM and 500 μM iodoacetate were added, respectively. Iodoacetate also significantly decreased the ΔpH at both concentrations. Chlorhexidine, an inhibitor of the PEP-PTS (23), showed a more than 80% inhibitory effect on cellobiose uptake, and it almost completely inhibited uptake when it was added with 2 mM iodoacetate.
TABLE 1.
Effects of various inhibitors on cellobiose transport and bioenergetic properties of the cellsa
| Inhibitor | Concn | Inhibition of cellobiose transport (%) | Value for bioenergetic property
|
||
|---|---|---|---|---|---|
| −Z ΔpH (mV) | Δψ (mV) | Intracellular ATP (mM) | |||
| No inhibitor | 0 | 17 | 102 | 0.78 | |
| SF6847 | 10 μM | 3 | 3** | 105 | 0.75 |
| TPMP bromide | 1 mM | 8 | 14 | 78** | 0.78 |
| 10 mM | 25** | 17 | 45** | 0.45** | |
| Iodoacetate | 500 μM | 11* | 7** | 96 | 0.56** |
| 2 mM | 43** | 5** | 110 | 0.25** | |
| Chlorhexidine | 200 μM | 83** | 19 | 98 | 0.76 |
| Harmaline | 4 mM | 3 | 16 | 102 | 0.76 |
| Chlorhexidine + iodoacetate | 200 μM | 97** | ND | ND | ND |
| 2 mM | |||||
In the presence of 0.01 mM cellobiose and 20 μM nojirimycin-1-sulfonate. * and ** indicate significant inhibition (P < 0.05 and 0.01, respectively). ND, not determined.
The effects of various artificial potentials on cellobiose uptake in the presence of 20 μM nojirimycin were investigated. However, none of the artificially generated Z ΔpH (45 mV), Δψ (93 mV), or Z ΔpNa (61 mV) showed significant promotion of cellobiose uptake compared with that of the control, which had no artificial potentials (data not shown).
The PEP-dependent oxidation of NADH, an index of the PEP-PTS, with 20 μM nojirimycin occurred at the rate of 0.35 nmol/min/mg of protein, while no oxidation was shown in the presence of 200 μM chlorhexidine (data not shown).
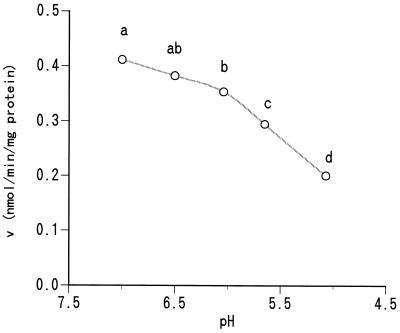
The effects of medium pH on cellobiose uptake are shown in Fig. 2. The uptake rate decreased as the pH declined, especially below 6.0, and the rate at pH 5.0 was approximately half the level at pH 7.0.
FIG. 2.
Effect of pH on cellobiose transport by mixed ruminal bacteria. Cells were incubated in the presence of cellobiose (0.01 mM) and nojirimycin-1-sulfate (20 μM) in buffers containing 25 mM Na2HPO4 and 50 mM MES (2-[N-morpholino]ethanosulfonic acid), which were adjusted at various pH levels. Circles with different letters indicate significant differences (P < 0.05).
The effects of addition (0.04 mM) of various sugars (i.e., cellobiose, glucose, mannose, galactose, fructose, xylose, arabinose, sucrose, and maltose) on cellobiose uptake (0.01 mM) were investigated. Cellobiose itself inhibited uptake of 3H-labeled cellobiose to a degree (56%) similar to the theoretical value (63%), which was calculated from the kinetic parameters of cellobiose uptake. Other than cellobiose, glucose alone among the nine investigated sugars showed a weak but significant inhibitory effect (13%) on cellobiose uptake (data not shown).
DISCUSSION
Cellobiase activities were detected both extracellularly and in a cell-associated form in this study. Williams et al. (51) also found that β-glucosidase activities were present in both the liquid and microbial fractions in the rumen when 5.8 mM cellobiose was used as a substrate, and both activities were in the range from 10 to 70 nmol/min/mg of protein, with some postprandial variations. In this study, the Vmax values were also similar between the extracellular and cell-associated cellobiases, showing approximately 25 nmol/min/mg of protein. When there is a low concentration of cellobiose, however, extracellular cellobiase activity is lower because it has a lower affinity than that of cell-associated cellobiase, comprising less than one-fifth of the total cellobiase activity in the rumen when the cellobiose concentration is below 0.02 mM. Cellobiose concentration in the rumen actually remained very low in this study (less than 0.02 mM) irrespective of postprandial time, which was possibly due to a higher degradation rate than the production rate of cellobiose suggested by Kitts and Underkofler (16).
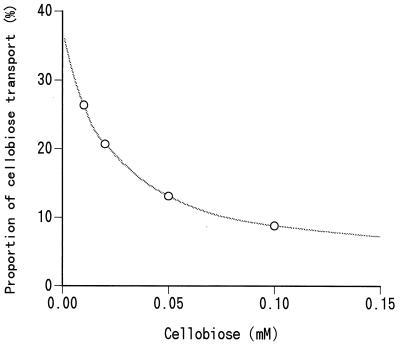
The apparent uptake of cellobiose by the mixed ruminal bacteria that occurred in the absence of any β-glucosidase inhibitors seemed to happen in a large part via the glucose transporter after extracellular hydrolysis by the cellobiases in the rumen. Glucose transport consisted of two different systems having the same kinetic parameters as those shown in a previous study of the mixed ruminal bacteria from a cow fed a forage concentrate diet (15). The high-affinity system of glucose transport in the previous study consisted mainly of the glucose PEP-PTS and partially of the sodium symport system, while the low-affinity system appeared to be a facilitated diffusion. Cellobiose uptake without nojirimycin in this study also showed biphasic kinetics (in hexose equivalents) with constants similar to those of glucose transport. Cellobiose uptake in the presence of nojirimycin, on the other hand, is considered to occur mostly via a real cellobiose transporter of the cells, because the cellobiase activity could be almost completely inhibited by nojirimycin. Since the cellobiose transporter has a much higher affinity for cellobiose than both the extracellular and cell-associated cellobiases, although it has a lower Vmax value, the contribution of the real cellobiose transporter in the apparent use of cellobiose becomes greater as the cellobiose concentration decreases even in the absence of any β-glucosidase inhibitors. The proportion of cellobiose that could be directly transported before being degraded extracellularly into glucose is shown in Fig. 3, the data in which were calculated from the kinetic parameters of cellobiose transport with nojirimycin and those of the cellobiases. When the cellobiose concentration in the rumen is less than 0.02 mM, more than 20% of the cellobiose could be incorporated into the cells in its intact form by the real cellobiose transporter.
FIG. 3.
Proportion of cellobiose that is incorporated into ruminal bacteria before it is degraded extracellularly into glucose, calculated from the kinetic parameters of the cellobiose transport system and cellobiases.
In the inhibitor experiment, chlorhexidine showed the strongest inhibitory effect on cellobiose transport, which suggests that the PEP-PTS is the most dominant system for cellobiose transport in the rumen. Although chlorhexidine possibly affects some properties other than the PEP-PTS (1, 25), no effect on the bioenergetic properties (i.e., intracellular ATP and PMF) was detected for the mixed ruminal bacteria in this study. Moreover, the PEP-dependent oxidation of NADH, which showed a rate similar to that of cellobiose transport (0.35 versus 0.41 nmol/min/mg of protein) and was mostly inhibited by chlorhexidine, also supports the inclusion of the PEP-PTS in cellobiose transport. Other than chlorhexidine, iodoacetate and a higher concentration of TPMP bromide (10 mM) significantly inhibited cellobiose transport. Since intracellular ATP generation was the common property inhibited by these inhibitors, we believe that an ATP-dependent active transport system functions as a cellobiose transport system to some degree. On the other hand, the PMF seems to be concerned little with cellobiose transport, because transport was not affected by the addition of SF6847 and TPMP bromide (1 mM), in the presence of which significant reductions in the ΔpH and Δψ, respectively, occurred without there being a decline in ATP production. Furthermore, the absence of PMF involvement in cellobiose transport by the mixed ruminal bacteria is supported by the fact that neither artificially generated ΔpH nor Δψ promoted cellobiose transport. The sodium symport system is not thought to be concerned with cellobiose transport either, because cellobiose uptake was not affected by harmaline and also not promoted by an artificial ΔpNa at all.
The cellobiose PEP-PTS has been reported in several genetic studies of nonruminal species such as Escherichia coli (34), Bacillus stearothermophilus (19), and Erwinia spp. (3, 11). An ATP-dependent active transport system of cellobiose has also been observed in some clostridia (29, 31, 45). For ruminal species, however, the PEP-PTS has been shown only in S. bovis (25), and no ATP-dependent transport system which was also independent of PMF has been found so far. On the other hand, inhibitor experiments with R. flavefaciens and F. succinogenes showed that these species had some active transport in which both PMF and ATP were involved (12, 22). The presence of a sodium symport system with cellobiose was also suggested for F. succinogenes (22). Because more than 25 species can ferment cellobiose in the rumen (14, 32, 44), other ruminal bacteria not yet investigated may use the PEP-PTS and ATP-dependent active transport system for uptake of cellobiose. Similarly, PMF-dependent active transport and sodium symport systems would not have much significance in cellobiose utilization among many cellobiose-utilizing species in the whole rumen.
Cellobiose transport is regarded as energetically more efficient than glucose transport. When we consider the PEP-PTS, 1 mol of PEP and 1 mol of ATP are needed to produce 2 mol of glucose phosphate from 1 mol of cellobiose when cellobiose is transported by the cellobiose PEP-PTS, while 2 mol of PEP is required when cellobiose is extracellularly hydrolyzed and transported by the glucose PEP-PTS. When PEP and ATP are compared, the change in Gibbs free energy of PEP to pyruvate is higher than that of ATP to ADP (−62 versus −35 kJ/mol at pH 7.0), and only 2 mol of PEP is generated from 1 mol of hexose via the Embden-Meyerhof-Parnas pathway. On the other hand, if we assume that cellobiose and glucose are transported by active transport systems, intracellular phosphorylation by cellobiose phosphorylase, which has been observed in several ruminal bacteria (2, 47, 50), may conserve ATP compared with the ATP conserved after phosphorylation of two glucose molecules by glucokinase (41). A higher growth efficiency of cellobiose-grown cells than that of glucose-grown cells was actually observed for Ruminococcus albus (47), which might be attributed to a lower energy requirement of the substrate transporter. Such an efficient transport of cellobiose would be more beneficial when the cellobiose concentration is low, when the competition for this substrate might be intense. This conclusion is consistent with there being a higher proportion of true cellobiose transport in the apparent utilization of cellobiose when there is a low level of cellobiose, as shown in this study. When a substrate is abundant, however, the decisive quality of its transport system may shift from efficiency to velocity. Since glucose transport has a high-velocity system in the rumen, presumably a facilitated diffusion (15), transport of glucose after the extracellular degradation of cellobiose may be advantageous when the cellobiose concentration is high.
In our study, cellobiose transport activity decreased as the medium pH declined. A decline in ruminal pH usually occurs when an animal is fed a diet rich in soluble carbohydrates and starch (14), when the ruminal microbes possibly access a sufficient amount of soluble sugars other than cellobiose. Although some cellulolytic species show a preference for cellobiose over other sugars (12, 47), these bacteria could hardly survive a low ruminal pH (40). Cellobiose transport in the rumen, therefore, seems to have relatively little significance when the pH is low, although it is not certain that the proportion of true cellobiose transport indeed decreases at a low ruminal pH, because β-glucosidase activity in the rumen also decreases at a pH below 5.5 (8).
Cellobiose uptake was inhibited by glucose in this study. The inhibitory effect of glucose on cellobiose transport was also shown for R. flavefaciens (12). Since the dominant transport system for both glucose and cellobiose in the mixed ruminal bacteria was the PEP-PTS, the inhibition might be due to competition for the same component of the system (presumably enzyme II) (36). No sugar other than glucose, however, inhibited cellobiose uptake in this study, although sucrose and maltose inhibit cellobiose utilization by catabolite regulatory mechanisms in several noncellulolytic ruminal bacteria (39). To understand the reason for this discrepancy, further studies of the other species capable of fermenting cellobiose are required. In addition, since transport and utilization of cellodextrins (i.e., cellotriose, cellotetraose, and cellopentaose) were observed in R. albus (20) and Clostridium thermocellum (45), further investigation is also needed to clarify the presence and extent of cellodextrin transport in the rumen.
REFERENCES
- 1.Attia-Ismail S A, Martin S A, Callaway T R. Effects of chlorhexidine diacetate on ruminal microorganisms. Curr Microbiol. 1998;36:348–352. doi: 10.1007/s002849900321. [DOI] [PubMed] [Google Scholar]
- 2.Ayers W A. Phosphorylation of cellobiose and glucose by Ruminococcus flavefaciens. J Bacteriol. 1958;76:515–517. doi: 10.1128/jb.76.5.515-517.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barras F, Chambost J P, Chippaux M. Cellobiose metabolism in Erwinia: genetic study. Mol Gen Genet. 1984;197:486–490. doi: 10.1007/BF00329947. [DOI] [PubMed] [Google Scholar]
- 4.Bryant M P. Bacterial species of the rumen. Bacteriol Rev. 1959;23:125–153. doi: 10.1128/br.23.3.125-153.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchanan C J, Mitchell W J. Two β-glucosidase activities in Fibrobacter succinogenes S85. J Appl Bacteriol. 1992;73:243–250. doi: 10.1111/j.1365-2672.1992.tb02984.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen C-C, Tsuchiya T, Yamane Y, Wood J M, Wilson T H. Na+ (Li+)-proline cotransport in Escherichia coli. J Membr Biol. 1985;84:157–164. doi: 10.1007/BF01872213. [DOI] [PubMed] [Google Scholar]
- 7.Chen G, Russell J B, Sniffen C J. A procedure for measuring peptides in rumen fluid and evidence that peptide uptake can be a rate-limiting step in ruminal protein degradation. J Dairy Sci. 1987;70:1211–1219. doi: 10.3168/jds.S0022-0302(87)80133-9. [DOI] [PubMed] [Google Scholar]
- 8.Conchie J. β-Glucosidase from rumen liquor. Preparation, assay and kinetics of action. Biochem J. 1954;58:552–560. doi: 10.1042/bj0580552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conchie J, Levvy G A. Inhibition of glycosidases by aldonolactones of corresponding configuration. Biochem J. 1957;65:389–395. doi: 10.1042/bj0650389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunniff P, editor. Official methods of analysis of AOAC International. 16th ed. Gaithersburg, Md: AOAC International; 1996. [Google Scholar]
- 11.Hassouni M E, Henrissat B, Chippaux M, Barras F. Nucleotide sequences of the arb genes, which control β-glucoside utilization in Erwinia chrysanthemi: comparison with the Escherichia coli bgl operon and evidence for a new β-glucohydrolase family including enzymes from eubacteria, archeabacteria, and humans. J Bacteriol. 1992;174:765–777. doi: 10.1128/jb.174.3.765-777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helaszek C T, White B A. Cellobiose uptake and mechanism by Ruminococcus flavefaciens. Appl Environ Microbiol. 1991;57:64–68. doi: 10.1128/aem.57.1.64-68.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heytler P G. Uncouplers of oxidative phosphorylation. Methods Enzymol. 1979;55:462–472. doi: 10.1016/0076-6879(79)55060-5. [DOI] [PubMed] [Google Scholar]
- 14.Hungate R E. The rumen and its microbes. New York, N.Y: Academic Press; 1966. [Google Scholar]
- 15.Kajikawa H, Amari M, Masaki S. Glucose transport by mixed ruminal bacteria from a cow. Appl Environ Microbiol. 1997;63:1847–1851. doi: 10.1128/aem.63.5.1847-1851.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitts W D, Underkofler L A. Hydrolytic products of cellulose and the cellulolytic enzymes. J Agric Food Chem. 1954;2:639–645. [Google Scholar]
- 17.Krebs E G. Glyceraldehyde-3-phosphate dehydrogenase from yeast. Methods Enzymol. 1955;1:408–414. doi: 10.1016/s0076-6879(82)89059-9. [DOI] [PubMed] [Google Scholar]
- 18.Kunst A, Draeger B, Ziegenhorn J. UV-methods with hexokinase and glucose-6-phosphate dehydrogenase. In: Bergmeyer H U, editor. Methods of enzymatic analysis. 3rd ed. VI. Weinheim, Germany: Verlag Chemie GmbH; 1984. pp. 163–172. [Google Scholar]
- 19.Lai X, Ingram L O. Cloning and sequencing of a cellobiose phosphotransferase system operon from Bacillus stearothermophilus XL-65-6 and functional expression in Escherichia coli. J Bacteriol. 1993;175:6441–6450. doi: 10.1128/jb.175.20.6441-6450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lou J, Dawson K A, Strobel H J. Cellobiose and cellodextrin metabolism by the ruminal bacterium Ruminococcus albus. Curr Microbiol. 1997;35:221–227. doi: 10.1007/s002849900242. [DOI] [PubMed] [Google Scholar]
- 21.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 22.Maas L K, Glass T L. Cellobiose uptake by the cellulolytic ruminal anaerobe Fibrobacter (Bacteroides) succinogenes. Can J Microbiol. 1991;37:141–147. doi: 10.1139/m91-021. [DOI] [PubMed] [Google Scholar]
- 23.Marsh P D, Keevil C W, McDermid A S, Williamson M I, Ellwood D C. Inhibition by the antimicrobial agent chlorhexidine of acid production and sugar transport in oral streptococcal bacteria. Arch Oral Biol. 1983;28:233–240. doi: 10.1016/0003-9969(83)90152-8. [DOI] [PubMed] [Google Scholar]
- 24.Martin S A. Nutrient transport by ruminal bacteria: a review. J Dairy Sci. 1994;72:3019–3031. doi: 10.2527/1994.72113019x. [DOI] [PubMed] [Google Scholar]
- 25.Martin S A, Russell J B. Transport and phosphorylation of disaccharides by the ruminal bacterium Streptococcus bovis. Appl Environ Microbiol. 1987;53:2388–2393. doi: 10.1128/aem.53.10.2388-2393.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matin A, Veldkamp H. Physiological basis of the selective advantage of a Spirillum sp. in a carbon-limited environment. J Gen Microbiol. 1978;105:187–197. doi: 10.1099/00221287-105-2-187. [DOI] [PubMed] [Google Scholar]
- 27.Morrison I A, Mousdale S A. Effect of the level of feeding on the digestion of hay and carbohydrase activities in an artificial rumen. Enzyme Microb Technol. 1992;14:284–288. [Google Scholar]
- 28.National Research Council. Nutrient requirements of dairy cattle, 6th revised ed., update. Washington, D.C: National Academy Press; 1989. [Google Scholar]
- 29.Ng T K, Zeikus J G. Differential metabolism of cellobiose and glucose by Clostridium thermocellum and Clostridium thermohydrosulfuricum. J Bacteriol. 1982;150:1391–1399. doi: 10.1128/jb.150.3.1391-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niwa T, Inoue S, Tsuruoka T, Koaze Y, Niida T. “Nojirimycin” as a potent inhibitor of glucosidase. Agric Biol Chem. 1970;34:966–968. [Google Scholar]
- 31.Nochur S V, Jacobson G R, Roberts M F, Demain A L. Mode of sugar phosphorylation in Clostridium thermocellum. Appl Biochem Biotechnol. 1992;33:33–41. [Google Scholar]
- 32.Ogimoto K, Imai S. Atlas of rumen microbiology. Tokyo, Japan: Japan Scientific Societies Press; 1981. [Google Scholar]
- 33.Ohmiya K, Shimizu S. β-Glucosidase from Ruminococcus albus. Methods Enzymol. 1988;160:408–414. [Google Scholar]
- 34.Parker L L, Hall B G. Characterization and nucleotide sequence of the cryptic cel operon of Escherichia coli K12. Genetics. 1990;124:455–471. doi: 10.1093/genetics/124.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettipher G L, Latham M J. Production of enzymes degrading plant cell walls and fermentation of cellobiose by Ruminococcus flavefaciens in batch and continuous culture. J Gen Microbiol. 1979;110:29–38. [Google Scholar]
- 36.Postma P W, Lengeler J W. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1985;49:232–269. doi: 10.1128/mr.49.3.232-269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinehart C A, Hubbard J S. Energy coupling in the active transport of proline and glutamate by the photosynthetic halophile Ectothiorhodospira halophila. J Bacteriol. 1976;127:1255–1264. doi: 10.1128/jb.127.3.1255-1264.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell J B. Fermentation of cellodextrins by cellulolytic and noncellulolytic rumen bacteria. Appl Environ Microbiol. 1985;49:572–576. doi: 10.1128/aem.49.3.572-576.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell J B, Baldwin R L. Substrate preferences in rumen bacteria: evidence of catabolite regulatory mechanisms. Appl Environ Microbiol. 1978;36:319–329. doi: 10.1128/aem.36.2.319-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell J B, Dombrowski D B. Effect of pH on the efficiency of growth by pure cultures of rumen bacteria in continuous culture. Appl Environ Microbiol. 1980;39:604–610. doi: 10.1128/aem.39.3.604-610.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell J B, Wallace R J. Energy yielding and consuming reactions. In: Hobson P N, editor. The rumen microbial ecosystem. London, United Kingdom: Elsevier Applied Science; 1988. pp. 185–215. [Google Scholar]
- 42.Sepúlveda F V, Robinson J W L. Harmaline, a potent inhibitor of sodium-dependent transport. Biochim Biophys Acta. 1974;373:527–531. doi: 10.1016/0005-2736(74)90035-2. [DOI] [PubMed] [Google Scholar]
- 43.Snedecor G W, Cochran W G. Statistical methods. 6th ed. Ames: Iowa State University Press; 1967. [Google Scholar]
- 44.Stewart C S, Bryant M P. The rumen bacteria. In: Hobson P N, editor. The rumen microbial ecosystem. London, United Kingdom: Elsevier Applied Science; 1988. pp. 21–75. [Google Scholar]
- 45.Strobel H J, Cardwell F C, Dawson K A. Carbohydrate transport by the anaerobic thermophile Clostridium thermocellum LQRI. Appl Environ Microbiol. 1995;61:4012–4015. doi: 10.1128/aem.61.11.4012-4015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strobel H J, Russell J B. Non-proton-motive-force-dependent sodium efflux from the ruminal bacterium Streptococcus bovis: bound versus free pools. Appl Environ Microbiol. 1989;55:2664–2668. doi: 10.1128/aem.55.10.2664-2668.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thurston B, Dawson K A, Strobel H J. Cellobiose versus glucose utilization by the ruminal bacterium Ruminococcus albus. Appl Environ Microbiol. 1993;59:2631–2637. doi: 10.1128/aem.59.8.2631-2637.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Soest P J. Use of detergents in the analysis of fibrous feeds. II. A rapid method for the determination of fiber and lignin. J Assoc Off Anal Chem. 1963;46:829–835. [Google Scholar]
- 49.Van Soest P J, Wine R H. Use of detergents in the analysis of fibrous feeds. IV. Determination of plant cell-wall constituents. J Assoc Off Anal Chem. 1967;46:829–835. [Google Scholar]
- 50.Wells J E, Russell J B, Shi Y, Weimer P J. Cellodextrin efflux by the cellulolytic ruminal bacterium Fibrobacter succinogenes and its potential role in the growth of nonadherent bacteria. Appl Environ Microbiol. 1995;61:1757–1762. doi: 10.1128/aem.61.5.1757-1762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams A G, Withers S E, Strachan N H. Postprandial variations in the activity of polysaccharide-degrading enzymes in microbial populations from the digesta solids and liquor fractions of rumen contents. J Appl Bacteriol. 1989;66:15–26. doi: 10.1111/j.1365-2672.1989.tb02449.x. [DOI] [PubMed] [Google Scholar]
- 52.Wulf-Strobel C R, Wilson D B. Cloning, sequencing, and characterization of membrane-associated Prevotella ruminicola B14 β-glucosidase with cellodextrinase and cyanoglycosidase activities. J Bacteriol. 1995;177:5884–5890. doi: 10.1128/jb.177.20.5884-5890.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang C-M J, Russell J B. Resistance of proline-containing peptides to ruminal degradation in vitro. Appl Environ Microbiol. 1992;58:3954–3958. doi: 10.1128/aem.58.12.3954-3958.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]