Abstract
Streptomyces strain K1-02, which was identified as a strain of Streptomyces albidoflavus, secreted at least six extracellular proteases when it was cultured on feather meal-based medium. The major keratinolytic serine proteinase was purified to homogeneity by a two-step procedure. This enzyme had a molecular weight of 18,000 and was optimally active at pH values ranging from 6 to 9.5 and at temperatures ranging from 40 to 70°C. Its sensitivity to protease inhibitors, its specificity on synthetic substrates, and its remarkably high level of NH2-terminal sequence homology with Streptomyces griseus protease B (SGPB) showed that the new enzyme, designated SAKase, was homologous to SGPB. We tested the activity of SAKase with soluble and fibrous substrates (elastin, keratin, and type I collagen) and found that it was very specific for keratinous substrates compared to SGPB and proteinase K.
Actinomycetes, particularly streptomycetes, are known to secrete multiple proteases into the culture medium (14). Some of these proteases, the serine proteases of Streptomyces griseus (1, 16, 17, 28) and Streptomyces fradiae (11, 18, 35), have been characterized structurally and enzymatically. There also have been many descriptions of isolation and partial characterization of alkaline protease activities from other members of the genus Streptomyces (2, 5, 6, 29, 39).
In these prokaryotic microorganisms, extracellular proteases are involved mainly in hydrolysis of large polypeptide substrates into smaller molecular entities which can subsequently be absorbed by the cells (8). The physiological role of extracellular proteases in differentiation of some Streptomyces species (22) has been demonstrated previously. These enzymes usually have low levels of substrate specificity and can degrade most nonstructural proteins (23, 31). Some of the excreted proteinases, the keratinases, have the ability to degrade native keratin and other insoluble proteins (2). The mechanical stability of keratin and its resistance to microbial degradation depend on tight packing of the protein chains in α-helix (α-keratin) or β-sheet (β-keratin) structures and linkage of these structures by cystine bridges. Keratinases may have a use in biotechnological valorization of keratin-containing wastes, like feathers or hair, as well as in the leather industry, in which they may have potential in the development of nonpolluting processes (26).
Previously (20), during a search for novel keratinases, we detected strong keratinolytic activities in the culture medium of a Streptomyces strain (strain K1-02) isolated from hen house soil and grown on feather meal as the sole source of carbon and energy.
In this study we identified Streptomyces strain K1-02, and then we purified and characterized the secreted keratinase. The ability of this enzyme to degrade keratin-based substrates selectively, which was greater than the ability of S. griseus protease B (SGPB) or proteinase K to degrade such substrates, and the behavior of the enzyme under various environmental conditions are discussed below.
MATERIALS AND METHODS
Microorganism and growth conditions.
A culture was grown in a basal salt medium supplemented with feather meal as described by Letourneau et al. (20). Keratinolytic enzymes were produced by a culture in a 2-liter stainless steel-glass fermentor (S.G.I. Instruments, Paris, France) that had a 1-liter working volume, was kept at 30°C, and was agitated at 500 rpm. Air was supplied at a rate of 60 liters · h−1. The fermentor was inoculated with 106 spores · ml−1, and the peak of exogenous keratinolytic activity occurred within 30 h. The culture was centrifuged at 4°C and 10,000 × g for 30 min in order to harvest the keratinase-containing supernatant.
PCR amplification of the 16S rDNA and sequence determination.
A PCR was performed in order to amplify the 16S ribosomal DNA (rDNA) of the Streptomyces strain. The primers used were direct and reverse primers 5′ AGAGTTTGATCCTGGCTCAG 3′ and 5′ GGTTACCTTGTTACGACTT 3′; this primer pair has been shown to amplify the maximum number of nucleotides in 16S rDNA from a wide variety of bacterial taxa (37). The PCR was performed as previously described (30) by using a DNA thermal cycler (model Own-E; Hybaid). Oligonucleotides were synthetized by Eurogentec (Seraing, Belgium). The DNA sequences of the PCR products were determined by using a Taq Dye Deoxy terminator cycle sequencing kit (Perkin-Elmer, Foster City, Calif.) and the protocol recommended by the supplier. Sequencing reaction products were analyzed with a model 373A automated DNA sequencer (Applied Biosystems, Foster, City, Calif.). Databases (GenBank, EMBL, etc.) were searched for sequences similar to the 16S rRNA gene sequence obtained.
Enzyme purification.
Following centrifugation of the culture, the supernatant was filtered through a 0.45-μm-pore-size membrane filter (Millipore Corp., Bedford, Mass.). The filtrate was concentrated 10-fold with a spiral cartridge concentrator (model CH2; Amicon Div., W. R. Grace and Co., Beverley, Mass.); the molecular weight cut-off value for the membrane filter was 10,000. The lyophilized retentate was dissolved in 20 mM Tris-HCl buffer (pH 8.0) and applied to a DEAE-cellulose column (5 by 40 cm; Whatman, England). The column was eluted at a rate of 5 ml · min−1 with 20 mM Tris-HCl (pH 8.0), and this was followed by step elution with 1 M NaCl in the same buffer. Fifty-milliliter fractions were collected and screened for keratinolytic activity with the keratin azure assay. Fractions that eluted with the running buffer and exhibited keratinase activity were pooled, dialyzed overnight at 4°C, lyophilized, dissolved in 20 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (pH 7.2), and placed on a carboxymethyl Accel Plus column (1.5 by 20 cm; Waters Div., Millipore Corp.). The column was eluted at a rate of 2 ml · min−1 with 20 mM MOPS buffer (pH 7.2), and this was followed by elution with a linear 0 to 0.5 M NaCl gradient in the same buffer. Two-milliliter fractions were collected and tested for activity. Active fractions that eluted with the NaCl gradient were pooled, dialyzed, and lyophilized.
Protein determination.
The protein content was determined by the Bradford method (4) by using the Bio-Rad assay reagent (Bio-Rad, Munich, Germany) and bovine serum albumin as the standard.
Electrophoretic methods. (i) Examination of purity and estimation of the molecular weight of the keratinase.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed with 12% polyacrylamide gels as described by Laemmli (19). Molecular weight markers (molecular weights, 14,000 to 170,000; Boehringer, Mannheim, Germany) were included, and the gels were silver stained.
(ii) Zymogram.
To prepare a zymogram, proteinase samples were mixed with electrophoresis sample buffer without heat denaturation prior to electrophoresis. SDS-PAGE was carried out at 4°C by using a 12% polyacrylamide gel. After electrophoresis, the gel was washed with 2.5% (vol/vol) Triton X-100 for 30 min and then with 50 mM Tris-HCl (pH 8.5) for 30 min. Gelatin (2%, wt/vol) in 50 mM Tris-HCl buffer (pH 8.5) was then poured onto the gel slab containing proteases. After 3 h of incubation at 40°C, the gel was stained with Coomassie brilliant blue R-250 and then destained. Protease bands appeared as clear zones on a blue background.
Determination of enzyme activities. (i) Assay of protease activity with insoluble keratin azure.
Protease samples were incubated with 4 mg of keratin azure (Sigma-Aldrich Chimie, St. Quentin Fallavier, France) in 1 ml of 50 mM Tris-HCl buffer (pH 8.5) at 50°C for 1 h with constant agitation at 900 rpm by using a Labnet orbital agitator (Bioblock, Illkirch, France). One unit of proteinase activity was defined as the amount of enzyme that resulted in an increase in absorbance at 595 nm (A595) of 0.01 U after reaction with keratin azure for 1 h.
(ii) Assays of protease activities with other insoluble and soluble substrates.
Proteolytic activities were also determined by using washed commercial feather meal (Point S.A., Viriat, France), type I collagen from bovine Achilles tendon (Sigma-Aldrich Chimie), soluble keratin obtained by heat treatment in dimethyl sulfoxide (DMSO) as described by Dozie et al. (10), Hammersten casein, and gelatin as substrates. Purified proteinase (1.3 μg) was incubated with 0.5% (wt/vol) substrate in 50 mM Tris-HCl buffer (pH 8.5), and the final volume was adjusted to 1 ml.
Assays were carried out at 50°C with constant agitation at 900 rpm for 30 to 120 min. The reactions were stopped by adding 5 μl of 10 M acetic acid. After centrifugation at 4°C and 10,000 × g for 10 min, 0.5 ml of each reaction mixture was added to 0.5 ml of 0.2 M sodium acetate buffer. After 1 ml of ninhydrin reagent (Sigma-Aldrich Chimie) was added, the free amino groups were measured by the procedure of Moore (24) at 570 nm.
One proteolytic unit was defined as the amount of enzyme that released 1 μmol of glycine after reaction with fibrous keratin, type I collagen, soluble keratin, or gelatin as the substrate for 1 h.
Influence of temperature and pH on enzyme activity and stability.
We determined the keratinase activities at various temperatures between 30 and 80°C in 50 mM Tris-HCl buffer (pH 8.5). Five milligrams of washed and autoclaved feather meal was suspended in 0.99 ml of buffer, and then after a temperature equilibrium was reached, 0.01 ml of purified protease (1.3 μg of protein) was added. After 30 min of incubation with constant agitation at 900 rpm, the reaction mixture was centrifuged at 10,000 × g for 10 min at 4°C, and then the A280 of the supernatant was determined by using an appropriate blank.
The thermostability of the keratinase was investigated by measuring the residual activities at 50°C with the same assay after the enzyme was incubated for 1 h at various temperatures between 30 and 80°C in 50 mM Tris-HCl buffer (pH 8.5) in the presence or absence of 2 mM CaCl2.
The optimum pH and pH stability of the keratinase were determined at 50°C by using pH values between 4 and 12; washed, autoclaved feather meal was used as the substrate. To determine the optimum pH, 5 mg of feather meal was added to 0.99 ml of a buffer containing phosphoric acid, acetic acid, boric acid, citric acid, barbital, and NaOH, and then the preparation was equilibrated at 50°C. Ten microliters of purified proteinase was added, and the preparation was incubated for 30 min with constant agitation. After centrifugation, the A280 of the supernatant was determined by using an appropriate blank.
pH stability was studied by measuring the residual activities at pH 8.5, with the same assay after the enzyme was incubated at various pH values between 4 and 12 at 25°C for 24 h.
NH2-terminal amino acid sequence.
The N-terminal sequence of the purified keratinase was analyzed at the Institut de Biologie et Chimie des Protéines (Lyon, France) by automated Edman degradation performed with a liquid phase sequence analyzer (model 473; Applied Biosystems).
Effects of proteinase inhibitors, organic solvents, detergents, reducing agents, and ionic strength on keratinase activity.
Enzyme samples containing 1.3 μg of purified keratinase in 0.9 ml of Tris-HCl buffer (pH 8.5) were incubated at room temperature for 15 min with the following inhibitors: 0.1 to 1 mM phenylmethylsulfonyl fluoride (PMSF); 1 mM p-chloromercuribenzoate; 10 mM EDTA; 1 to 10 mM 1,10-phenanthroline; 0.1 mM tosyl-l-lysylchloromethylketone; 0.1 mM tosyl-l-phenylalanylchloromethylketone (TPCK); and 2 μM pepstatin. After 15 min of incubation, 5 mg of keratin azure was added to each preparation, and the residual activity of the enzyme was measured as described above.
One-milliliter enzyme samples (1.3 μg of purified proteinase) in 50 mM Tris-HCl buffer (pH 8.5) containing DMSO (1 to 10%, vol/vol), isopropanol (1 to 15%, vol/vol), acetonitrile (10 to 50%, vol/vol), dithiothreitol (DTT) (0.1 to 0.5%, wt/vol), Triton X-100 (0.2 to 0.5%, vol/vol), SDS (0.1 to 0.5%, wt/vol) or sodium chloride (0.05 to 1 M) were incubated for 15 min at room temperature. Five milligrams of keratin azure, soluble keratin, or feather meal was added to each preparation, and the residual enzyme activity was measured as described above.
Enzyme kinetic measurements with synthetic substrates.
Most synthetic p-nitroanilide (pNA) peptides (Sigma-Aldrich Chimie) used in this study were prepared as stock solutions (concentration, 10 to 100 mg · ml−1, depending on the peptide) in DMSO for N-succinyl-p-nitroanilide derivatives (N-Suc-pNA derivatives) or in isopropanol for N-benzoyl-p-nitroanilide peptides (Bz-pNA peptides); the stock solution Bz-Arg-pNA (concentration, 10 mg · ml−1) was prepared in Tris-HCl buffer (pH 8.5). The final concentrations of these solvents in reaction mixtures never exceeded 5% (vol/vol). At least five concentrations of most of the synthetic substrates were assayed at 45°C with 0.73 × 10−7 M keratinase (molecular mass, 18 kDa, as determined by SDS-PAGE) in 50 mM Tris-HCl (pH 8.5) buffer; the only exceptions were the Suc-(Ala)2-Pro-Phe-pNA and Bz-Phe-Val-Arg-pNA reaction mixtures, in which the protease concentrations were 1.1 × 10−9 and 7.2 × 10−9 M, respectively. The hydrolysis of peptides was monitored continuously for pNA release at 410 nm for 5 min by using a Uvikon 930 spectrophotometer (Kontron Instruments, Schlieren, Switzerland). The initial velocities were then determined, and the steady-state kinetic parameters were calculated by using a Lineweaver-Burk plot and the molar absorption coefficient for pNA determined under our experimental conditions (ɛ = 8,800 liters · mol−1 · cm−1). When Bz-Pro-Phe-Arg-pNA and Suc-(Ala)2-Val-pNA were used as the substrates, the occurrence of hydrolysis products other than pNA was monitored by high-performance liquid chromatography performed with a high-performance liquid chromatography system (Kontron Instruments). One hundred-microliter portions of reaction mixtures obtained after 20, 40, 60, and 180 min of incubation of each of the two peptides with purified keratinase were loaded onto a type RP.18 Lichrospher 5μm Si 100 column (4.6 by 125 mm); Merck, Darmstadt, Germany). The samples were eluted at a rate of 1 ml · min−1 by using a linear 5 to 80% acetonitrile gradient in H2O containing 0.1% trifluoroacetic acid (TFA) at 25°C. The elution pattern was monitored at A220, and each hydrolysis product was collected; the amino acid content of each product was determined by the Waters Pico.Tag method (9) after acid hydrolysis.
RESULTS
Streptomyces strain identification.
Strain K1-02 was tentatively identified on the basis of its phenotypic and physiological characteristics (20, 38). In order to confirm the identity, a partial 16S rDNA sequence (1,266 bp) was determined. A search of databases for similar sequences pointed to the genus Streptomyces. The 16S rDNA sequence of strain K1-02 differed from the 16S rDNA sequences of Streptomyces albidoflavus (accession no. Z76676) and Streptomyces sampsonii (accession no. Z76680) by only two nucleotides (99.8% similarity). Our sequence differed from the sequences of other Streptomyces species by 18 nucleotides (Streptomyces intermedius; 98.6% similarity), 27 nucleotides (Streptomyces eurythermus; 97.9% similarity), 51 nucleotides (Streptomyces galbus; 96% similarity), 66 nucleotides (S. griseus; 94.8% similarity), and more than 51 nucleotides (38 other Streptomyces species). We concluded that strain K1-02 is a strain of Streptomyces albidoflavus.
Extracellular proteases of S. albidoflavus K1-02.
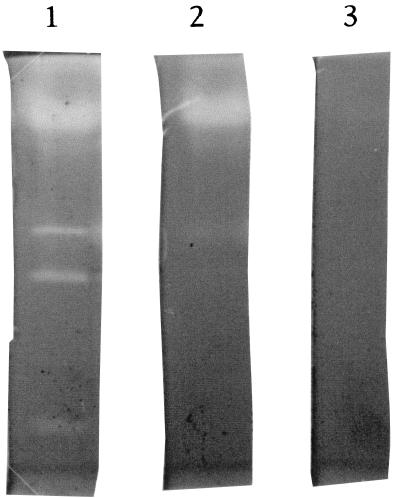
S. albidoflavus K1-02 was grown as described previously (20) on 1% feather meal basal medium. Under these conditions, at least six extracellular proteases were produced, as determined by a zymogram on gelatin (Fig. 1). Four of these six enzymes were inhibited in the presence of 10 mM EDTA, and no residual activity was observed in the presence of both 1 mM PMSF and 10 mM EDTA.
FIG. 1.
Zymogram analysis of proteases excreted by S. albidoflavus. Track 1, crude culture supernatant; track 2, supernatant treated with 10 mM EDTA; track 3, supernatant treated with 10 mM EDTA plus 1 mM PMSF.
Purification of the keratinolytic protease.
The method used to purify the enzyme from the culture medium is summarized in Table 1. The concentrated crude enzyme was first applied to a DEAE-cellulose anion-exchange column. The unbound fraction contained 48% of the total keratinolytic activity. After concentration, this sample was subjected to carboxymethyl cation-exchange chromatography, and the protein-containing keratinolytic activity peak eluted with 0.17 M NaCl. SDS-PAGE analysis of this purified peak revealed a single band (Fig. 2), indicating that the keratinase was purified to homogeneity. The overall purification factor was about 26-fold, and the final yield was 37%. The final product had a specific activity of about 17,600 U · mg−1.
TABLE 1.
Purification of the keratinase from S. albidoflavus culture medium
| Step | Total amt of protein (mg) | Total activity (U)a | Sp act (U · mg of protein−1) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Culture supernatant | 360 | 244,800 | 680 | 100 | 1.0 |
| Ultrafiltration concentrate | 250 | 202,500 | 810 | 82.7 | 1.2 |
| DEAE-cellulose chromatography | 35.3 | 117,450 | 3,321 | 48 | 4.9 |
| Carboxymethyl Accel Plus chromatography | 5.2 | 91,125 | 17,593 | 37.2 | 25.9 |
One unit of activity was defined as the amount of enzyme that resulted in an increase in A595 of 0.01 U after reaction with keratin azure for 1 h.
FIG. 2.
SDS-PAGE of purified keratinase. Lane 1, molecular mass marker proteins (α2 macroglobulin, 170 kDa; β-galactosidase, 116.4 kDa; fructose-6-phosphate kinase, 85.2 kDa; glutamate dehydrogenase, 55.6 kDa; aldolase, 39.2 kDa; triosephosphate isomerase, 26.6 kDa; trypsin inhibitor, 20.1 kDa; lysozyme, 14.3 kDa); lane 2, crude enzyme preparation; lane 3, purified keratinase.
Molecular mass of the protease.
The subunit molecular mass of the protease was estimated by comparing the electrophoretic mobility of the protease with the electrophoretic mobilities of marker proteins (Fig. 2). The apparent molecular mass was 18 kDa.
NH2-terminal amino acid sequence.
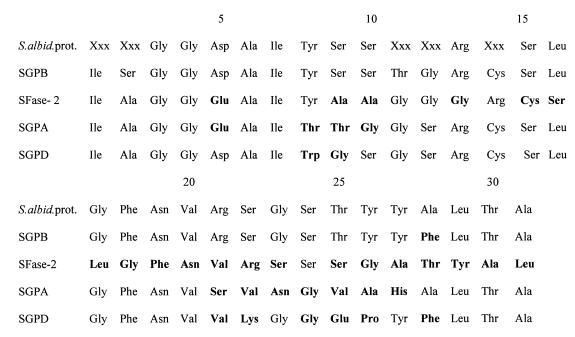
A total of 31 residues of the NH2-terminal amino acid sequence were determined (Fig. 3). The sequence obtained exhibited considerable homology with the sequences SGPB (96%) (17) and SGPA (58%) (16). The level of homology with the S. fradiae SFase-2 sequence was 23% (18).
FIG. 3.
Alignment of the N-terminal sequences of S. albidoflavus serine protease (S.albid.prot.), SGPB, SFase-2, SGPA, and SGPD. Boldface type indicates residues that are different. The data for SGPB were obtained from reference 17, the data for SFase-2 were obtained from reference 18, the data for SGPA were obtained from reference 16, and the data for SGPD were obtained from reference 33.
Effects of temperature and pH on the activity and stability of the proteinase.
The enzyme was active at a broad range of temperatures (40 to 70°C) and a broad range of pH values (pH 6 to 9.5); the optimum temperature and optimum pH were 60°C and pH 7.5, respectively. The enzyme was stable at pH 7 to 12, and more than 90% of the maximal activity was conserved at these pH values. The temperature stability of the enzyme was examined at temperatures up to 50°C in the absence of CaCl2 (80% residual activity after 1 h; measured half-life, 2 h). The temperature stability could be increased by adding 2 mM CaCl2 (which increased the half-life at 60°C 12-fold, to 72 min, compared with 6 min without CaCl2; CaCl2 did not increase the enzyme activity).
Effects of proteinase inhibitors on activity.
The effects of various synthetic and naturally occurring protease inhibitors on the proteolytic activity of S. albidoflavus keratinase (SAKase) were examined. The enzyme was completely inhibited by the serine proteinase inhibitor PMSF at a concentration of 0.1 mM and was slightly affected by a metalloproteinase inhibitor, such as 1,10-phenanthroline, at a concentration of 10 mM. None of the other specific serine proteinase inhibitors tested (tosyl-l-lysylchloromethylketone, TPCK, and pepstatin) had a significant influence on the keratinase activity.
Effects of solvents, detergents, reducing agents, and ionic strength.
Keratinase was very stable in the presence of different additives (Table 2). Reducing agents, such as β-mercaptoethanol and DTT, had no effect on proteinase activity. The nonionic detergent Triton X-100 and the anionic detergent SDS increased keratinase activity slightly; this was mainly the result of increased substrate accessibility to the enzyme. Of the chemical reagents tested, only a very high acetonitrile concentration (50%, vol/vol) and 0.5 to 1 M NaCl significantly decreased protease activity.
TABLE 2.
Effects of solvents, detergents, reducing agents, and ionic strength on the activity of purified S. albidoflavus serine proteinase
| Group | Compound | Concn (%) | Residual proteinase activity (%) |
|---|---|---|---|
| Control without additives | 100a | ||
| Detergents | Triton X-100 | 0.2b | 137 |
| 0.5 | 104 | ||
| SDS | 0.1c | 122 | |
| 0.5 | 102 | ||
| Solvents | Acetonitrile | 10b | 117 |
| 20 | 100 | ||
| 50 | 19 | ||
| Isopropanol | 1b | 96 | |
| 5 | 100 | ||
| DMSO | 1b | 91 | |
| 5 | 100 | ||
| 10 | 128 | ||
| Reducing agents | DTT | 0.1c | 96 |
| 0.5 | 103 | ||
| β-Mercaptoethanol | 0.2b | 103 | |
| 0.5 | 129 | ||
| Salt | NaCl | 0.05 | 99/92/100d |
| 0.1 | 90/82/100d | ||
| 0.5 | 76/0/100d | ||
| 1.0 | 57/0/100d |
Residual proteinase activity with keratin azure as the substrate.
Concentration (volume/volume).
Concentration (weight/volume).
Residual proteinase activity with feather meal as the substrate/residual proteinase activity with keratin azure as the substrate/residual proteinase activity with soluble keratin as the substrate.
Hydrolysis of various proteins with SAKase and other proteases.
Table 3 shows the hydrolyzing activities of SAKase, SGPB, Tritirachium album proteinase K, and α-chymotrypsin with fibrous insoluble and soluble proteins.
TABLE 3.
Enzyme activities with different soluble and insoluble substrates
| Enzyme | Sp act (U · mg−1) with:
|
Relative activities
|
|||||
|---|---|---|---|---|---|---|---|
| Fibrous keratina | Type I collagena | Soluble keratina | Gelatina | Elastin orceinb | Fibrous keratin/ type I collagen | Soluble keratin/ gelatin | |
| SAKase | 198.5 | 71.8 | 350.7 | 181.5 | 39 | 2.76 | 1.93 |
| SGPBc | 138.2 | 106.2 | 192.7 | 336.2 | 21.5 | 1.30 | 0.57 |
| Proteinase K | 167.2 | 168.6 | 244 | 375.6 | 1,400 | 0.99 | 0.65 |
| α-Chymotrypsin | 28.7 | 11.3 | 61.8 | 48.4 | 2.54 | 1.27 | |
One unit of proteolytic activity was defined as the amount of enzyme that resulted in release of 1 μmol of glycine after reaction with substrate for 1 h at pH 8.5 and 50°C.
One unit of proteolytic activity was defined as the amount of enzyme that resulted in an increase in the A578 of 0.1 U after reaction for 1 h at pH 8.5 and 50°C.
SGPB was purified from commercial pronase (Sigma) by the two-step procedure used for SAKase.
Compared with SGPB and proteinase K, SAKase had a greater ability to degrade keratin and also exhibited higher relative activities (specific activity with keratin versus specific activity with collagen and specific activity with solubilized keratin versus specific activity with gelatin). This was also true for α-chymotrypsin. The elastolytic activity of SAKase was very low compared to proteinase K activity.
Substrate specificity of purified keratinase.
P1 specificity (nomenclature of Schechter and Berger [32]) was determined with different synthetic amino acid derivatives with amino protection (Table 4). The new proteinase exhibited broad specificity with selectivity for aliphatic, hydrophobic amino acids or ionized residues, such as Arg. The nature of the amino acid at the P2 or P3 site also markedly influenced the specificity for the P1 site. For instance, proline at the P2 site had an effect on the P1 specificity [for Suc-(Ala)2-Pro-Phe-pNA and Bz-Pro-Phe-Arg-pNA]). No hydrolysis was detected with Suc-Ala-pNA, Suc-(Ala)2-pNA, Suc-Phe-pNA, and Bz-Arg-pNA. The amidase activity of the protease was markedly influenced by elongation of the peptide chain Suc-(Ala)n-pNA when n increased from two to three.
TABLE 4.
Enzyme kinetic parameters for hydrolysis of different synthetic substrates by the purified serine proteinase from S. albidoflavus
| Substrate | Km (mM) | kcat (s−1) | kcat/Km (mM−1 · s−1) |
|---|---|---|---|
| Suc-(Ala)3-pNA | 5 | 1.1 | 0.22 |
| Suc-(Ala)2-Pro-Phe-pNA | 0.615 | 505 | 821 |
| Suc-(Gly)2-Phe-pNA | 1.78 | 0.52 | 0.29 |
| Suc-(Ala)2-Val-pNA | —a | ||
| Suc-Tyr-Leu-Val-pNA | 0.392 | 0.26 | 0.67 |
| Bz-Pro-Phe-Arg-pNA | —b | ||
| Bz-Phe-Val-Arg-pNA | 2.66 | 93.7 | 35.2 |
| Bz-Tyr-ethylester | 2 | 0.187 | 0.093 |
The Ala-Val peptide bond is hydrolyzed.
The Phe-Arg peptide bond is hydrolyzed.
The proteinase exhibited esterase activity with Bz-Tyr-ethylester and had a very high proteolytic coefficient (kcat/Km) for Suc-(Ala)2-Pro-Phe-pNA, a well-known substrate for α-chymotrypsin and chymotrypsinlike proteinases. This was mainly the result of the high kcat value.
Dissolution of feather meal by the keratinase.
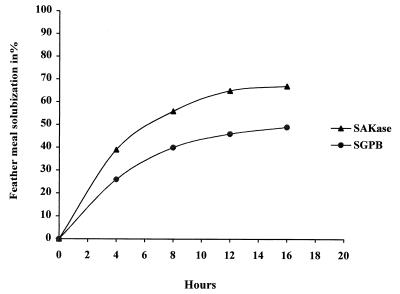
The keratinase was examined to determine its ability to solubilize feather meal. Figure 4 shows the data obtained. The S. albidoflavus protease degraded up to 67% of this fibrous substrate. In comparison, SGPB degraded only 50% of the substrate, and the rate was significantly lower. When native keratin (hair, horn) was used, only about 10% of the substrate was solubilized by our enzyme.
FIG. 4.
Feather meal hydrolysis with SAKase and SGPB. The experimental conditions were as follows: 50°C; 20 mM Tris HCl (pH 8.5); 5% (wt/vol) feather meal; constant agitation at 900 rpm; 50 keratinolytic activity units/ml was added each 4 h until the residual dry weight (measured after three washes) was constant.
DISCUSSION
The 16S rDNA of strain K1-02 differed from the previously described 16S rDNA of S. albidoflavus and S. sampsonii by only two nucleotides. It should be noted that S. sampsonii is considered a subjective synonym of S. albidoflavus (38); this synonymy has been confirmed by 16S rDNA comparisons and by studies of the 16S-23S rDNA intergenic spacer (15). Thus, strain K1-02 is most closely related to S. albidoflavus. When strain K1-02 is grown on a simple medium containing keratin-based materials, it excretes a large number of both metalloproteinases and serine proteinases, as do other Streptomyces species, such as S. fradiae and S. griseus (3, 21, 27). These late-occurring extracellular proteases, which appear after exponential growth is complete, may participate in in situ degradation of mycelium proteins during morphological differentiation (13, 22). A high yield of a pure major keratinolytic serine proteinase that exhibited 37% of the total supernatant keratinolytic activity was obtained when a simple purification scheme was used. Under the nonoptimized culture conditions, 2.6 mg of pure keratinase per liter was obtained.
Our N-terminal sequence analysis revealed a very high level of homology with the sequence of SGPB, a major component of the pronase produced by S. griseus (17), a closely related species (38); this enzyme has also been designated elastaselike enzyme III (12). The weakly alkaline, thermostable, purified enzyme had an apparent subunit molecular mass (18 kDa) that was very similar to that of SGPB (18.6 kDa) (17) or SFase-2 (19 kDa) (18), a keratinolytic enzyme of S. fradiae ATCC 14544. Protease inhibitor effects, substrate specificities, and the results of some chemical studies showed that the new keratinase may be classified as a serine proteinase belonging to the chymotrypsinlike superfamily, even if it was not inhibited by TPCK (18). The remarkable level of N-terminal sequence identity of SAKase and SGPB, together with the very similar molecular weights and differential susceptibilities to proteases inhibitors, strongly suggests that the new protease is indeed the S. albidoflavus homologue of SGPB. It is therefore likely that a small number of structurally and enzymatically closely related proteases are expressed by at least these two Streptomyces species and maybe by other species belonging to the same cluster (35).
The substrate specificities of SAKase were studied by using synthetic peptides. The purified proteinase exhibited specificity with aromatic and hydrophobic amino acid residues, such as Tyr, Phe, Ala, and Val, at the carboxyl side of the splitting point in the P1 position. SAKase is active against arginine peptide bonds, as demonstrated previously for SGPB (27). When Suc-(Ala)n-pNA is used as the substrate, a minimum length of three residues is necessary to observe peptide hydrolysis, indicating that SAKase probably has an extended active site. SAKase specificity depends mainly on secondary enzyme-substrate contacts with amino acid residues (P2, P3, etc.) more distant from the scissible bond, as illustrated by the difference between kinetic parameters observed with Suc-(Ala)2-Val-pNA and Suc-Tyr-Leu-Val-pNA. A similar observation has been made previously with other chymotrypsin-like proteinases (25). The proteolytic coefficient (kcat/Km) of SAKase with Suc-(Ala)2-Pro-Phe-pNA (821 mM−1 · s−1) is considerably higher than the proteolytic coefficients of Streptomyces pactum and S. fradiae (66 and 130 mM−1 · s−1, respectively) (2, 18) and is comparable to the proteolytic coefficient of SGPB (1,500 mM−1 · s−1) (7), for which Phe is one of the optimal P1 substrates (34).
SAKase was also tested by using fibrous substrates (keratin, collagen, and elastin) in order to compare its efficiency with the efficiencies of SGPB, proteinase K, and α-chymotrypsin. The keratinase exhibited a marked preference for keratin-based substrates. The relative activity (specific activity with keratin versus specific activity with collagen) of this enzyme was two and three times higher than the relative activities of SGPB and proteinase K (Table 3), respectively; the latter enzymes hydrolyze a broad range of insoluble proteins. The difference was even greater if elastin was used as the substrate; SAKase was 36 times less efficient than proteinase K. As fibrous substrate hydrolysis proceeds by heterogeneous phase catalysis, enzyme targeting requires the following two steps: (i) adsorption of the enzyme to the macromolecule surface by electrostatic and/or hydrophobic interactions, followed by (ii) enzyme diffusion on the surface of the substrate up to the splitting point (36). The weak ability of SAKase to hydrolyze type I collagen compared to its ability to hydrolyze fibrous keratin does not depend on a kinetic limitation linked to the initial step, enzyme adsorption to the surface of the substrate, since the enzyme behaves the same with the solubilized forms of substrates (gelatin and solubilized keratin). Thus, the observed differences between the specific activities of SAKase for fibrillar proteins such as keratin and collagen are mainly linked to differences in the primary structures of the substrates and/or in the accessibility of the enzyme to the splitting points. The hydrolytic activity of SAKase is affected when the ionic strength increases. This phenomenon is not a result of enzyme inactivation since it is not observed during hydrolysis of the soluble substrate Suc-(Ala)3-pNA and chemically solubilized feather keratin (Table 2). Therefore, the first step, protease adsorption to fibrous keratin, implies that the interaction is mainly electrostatic.
As evidence, the new enzyme is homologous to SGPB, which is excreted by S. griseus. Thus, the new keratinase, which belongs to a highly conserved protease family (33), acquired some discrete mutations that confer its ability to act during heterogeneous phase catalysis, particularly with keratin. Sidhu and Borgford (34) recently showed in a study of a broad selection of SGPB mutants that few mutations, either at the promature junction or at several sites conferring primary specificity of the mature SGPB, could lead to recombinant SGPB with modified efficiencies (kcat/Km), thermostabilities, or primary specificities. Therefore, a molecular approach to the gene coding for the SAKase should be useful for elucidating the mechanisms involved in keratinolysis in comparison with SGPB.
Even though more than 65% hydrolysis was observed with feather meal, only 10% dissolution was observed with native keratin, such as keratin azure, native feathers, or hair keratin, as described previously by other authors (2). This indicated that additional activity or treatment is needed to completely solubilize this compact fibrous protein. This point is crucial in the development of keratin-based substrate hydrolysis reactions.
ACKNOWLEDGMENTS
This work was supported by research grants from Agence de l’Environnement et de la Maîtrise de l’Energie (ADEME) and Conseil Régional du Limousin.
We are grateful to R. Faure, C. Pradoux, and C. Belzanne for technical assistance.
REFERENCES
- 1.Awad W M, Soto A R, Siegel S, Shiba W E, Bernstrom G G, Ochoa M S. Proteolytic enzymes of K-1 strain of Streptomyces griseus obtained from commercial preparation (Pronase). I. Purification of four serine endopeptidases. J Biol Chem. 1972;247:4144–4154. [PubMed] [Google Scholar]
- 2.Böckle B, Galunsky B, Muller R. Characterization of a keratinolytic serine proteinase from Streptomyces pactum DSM 40530. Appl Environ Microbiol. 1995;61:3705–3710. doi: 10.1128/aem.61.10.3705-3710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bormatova M E, Ivanova N M, Iusupova M P, Voiushina T L, Surova I A, Chestukhina G G, Stepanov V M. Proteolytic enzymes from Streptomyces fradiae: a metalloendopeptidase, subtilisin-like and trypsin-like proteinases. Biokhimiia. 1996;61:344–356. [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Chandrasekharan S, Dhar S C. Multiple proteases from Streptomyces moderatus. I. Isolation and purification of five extracellular proteases. Arch Biochem Biophys. 1987;257:395–404. doi: 10.1016/0003-9861(87)90582-0. [DOI] [PubMed] [Google Scholar]
- 6.Chauvet J, Dostal J, Archer R. Isolation of trypsin-like enzyme from Streptomyces paromomycinus by affinity adsorption through Kunitz inhibitor sepharose. Int J Pept Protein Res. 1976;8:45–55. doi: 10.1111/j.1399-3011.1976.tb02480.x. [DOI] [PubMed] [Google Scholar]
- 7.Christensen U, Ishida S, Ishii S, Mitsui Y, Iitaka Y, McClarin J, Langridge R. Interaction of Streptomyces subtilisin inhibitor with Streptomyces griseus protease A and B. Enzyme kinetic and computer simulation studies. J Biochem. 1985;98:1263–1274. doi: 10.1093/oxfordjournals.jbchem.a135393. [DOI] [PubMed] [Google Scholar]
- 8.Cohen B L. Transport and utilization of proteins by fungi. In: Payne J W, editor. Microorganisms and nitrogen sources. J. London, United Kingdom: Wiley and Sons; 1990. p. 411. [Google Scholar]
- 9.Cohen S A, Meys M, Tarvin T L. A manual of advanced techniques for amino acid analysis. Waters/Millipore publication WM02, Rev. 1. Bedford, Mass: Millipore Corp.; 1989. The Pico.Tag method; pp. 1–26. [Google Scholar]
- 10.Dozie I N S, Okeke C N, Unaeze N C. A thermostable, alkaline active keratinolytic proteinase from Chrysosporium keratinophilum. World J Microbiol Biotechnol. 1994;10:563–567. doi: 10.1007/BF00367668. [DOI] [PubMed] [Google Scholar]
- 11.Galas E, Kaluzewska M. Proteinases of Streptomyces fradiae. III. Catalytic and some physico-chemical properties of keratinolytic proteinase. Acta Microbiol Pol. 1992;41:169–177. [PubMed] [Google Scholar]
- 12.Gertler A, Trop M. The elastase-like enzyme from S. griseus (pronase) Eur J Biochem. 1971;19:90–96. doi: 10.1111/j.1432-1033.1971.tb01291.x. [DOI] [PubMed] [Google Scholar]
- 13.Ginther C L. Sporulation and the production of serine protease and cephamycin C by S. lactamdurans. Antimicrob Agents Chemother. 1979;15:522–526. doi: 10.1128/aac.15.4.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godfrey T, Reichelt J. Industrial enzymology. New York, N.Y: The Nature Press; 1983. The application of enzymes in industry; pp. 1–15. [Google Scholar]
- 15.Hain T, Ward-Rainey N, Kroppenstedt R M, Stackebrandt E, Rainey A. Discrimination of Streptomyces albidoflavus strains based on the size and number of 16S-23S ribosomal DNA intergenic spacers. Int J Syst Bacteriol. 1997;47:202–206. doi: 10.1099/00207713-47-1-202. [DOI] [PubMed] [Google Scholar]
- 16.Johnson P, Smillie L B. The amino acid sequence and predicted structure of Streptomyces griseus protease A. Can J Biochem. 1971;49:1083–1097. doi: 10.1139/o71-158. [DOI] [PubMed] [Google Scholar]
- 17.Jurasek L, Carpenter M R, Smillie L B, Getler A, Levis S, Ericsson L H. Amino acid sequence of Streptomyces griseus protease B, a major component of pronase. Biochem Biophys Res Commun. 1974;61:1095–1100. doi: 10.1016/s0006-291x(74)80396-7. [DOI] [PubMed] [Google Scholar]
- 18.Kitadokoro K, Tsuzuki H, Nakamura E, Sato T, Teraoka H. Purification, characterization, primary structure, crystallisation and preliminary crystallographic study of a serine proteinase from Streptomyces fradiae ATCC 14544. Eur J Biochem. 1994;220:55–61. doi: 10.1111/j.1432-1033.1994.tb18598.x. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Letourneau F, Soussotte V, Bressollier P, Branland P, Verneuil B. Keratinolytic activity of Streptomyces sp. S.K1-02: a new isolated strain. Lett Appl Microbiol. 1998;26:77–80. doi: 10.1046/j.1472-765x.1998.00281.x. [DOI] [PubMed] [Google Scholar]
- 21.Matsubara H, Feder J. Other bacterial, mould and yeast proteases. In: Boyer P D, editor. The enzymes. 3rd ed. Vol. 3. New York, N.Y: Academic Press; 1971. pp. 721–791. [Google Scholar]
- 22.Moncheva P A, Danova S T, Antonova S K, Ivanova I V. Physiological role of extracellular proteases and calcium ions in the processes of differentiation and antibiotic production by Streptomyces albogriseolus 444. Antibiot Khimioter. 1997;42:9–14. [PubMed] [Google Scholar]
- 23.Mono M, Togni G, Rahalison L, Frenk E. Isolation and characterization of an extracellular alkaline protease of Aspergillus fumigatus. J Med Microbiol. 1991;35:23–35. doi: 10.1099/00222615-35-1-23. [DOI] [PubMed] [Google Scholar]
- 24.Moore S. Amino acid analysis: aqueous DMSO as solvent for the ninhydrin reaction. J Biol Chem. 1968;243:6281–6283. [PubMed] [Google Scholar]
- 25.Morihara K. Comparative specificity of microbial proteinases. Adv Enzymol. 1974;41:179–243. doi: 10.1002/9780470122860.ch5. [DOI] [PubMed] [Google Scholar]
- 26.Mukohopadhyay R P, Chandra A L. Protease of a keratinolytic streptomycete to unhair goat skin. Indian J Exp Biol. 1993;31:557–558. [PubMed] [Google Scholar]
- 27.Narahashi Y, Yoda K. Studies on proteolytic enzymes (pronase) of Streptomyces griseus K-I. III. Purification and proteolytic specificity of alkaline proteinase C from pronase. J Biochem. 1973;73:831–841. doi: 10.1093/oxfordjournals.jbchem.a130146. [DOI] [PubMed] [Google Scholar]
- 28.Olafson R W, Jurasek L, Carpenter M R, Smillie L B. Amino acid sequence of Streptomyces griseus trypsin cyanogen bromide fragments and complete sequence. Biochemistry. 1975;14:1168–1177. doi: 10.1021/bi00677a011. [DOI] [PubMed] [Google Scholar]
- 29.Pokorny M, Vitale V, Turk V J, Tenko M, Zuvanic J. Streptomyces rimosus extracellular proteases. I. Characterization and evaluation of various crude preparations. Eur J Appl Microbiol Technol. 1979;8:81–90. [Google Scholar]
- 30.Quéré F, Deschamps A, Urdaci M. DNA probe and PCR specific reaction for Lactobacillus plantarum. J Appl Microbiol. 1997;82:783–790. doi: 10.1046/j.1365-2672.1997.00157.x. [DOI] [PubMed] [Google Scholar]
- 31.Reichard U, Buttner S, Eiffert H, Staid H, Ruchel R. Purification and characterization of an extracellular serine protease from Aspergillus fumigatus and its detection in tissue. J Med Microbiol. 1990;33:243–251. doi: 10.1099/00222615-33-4-243. [DOI] [PubMed] [Google Scholar]
- 32.Schechter I, Berger A. On the size of active site in proteinase. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 33.Sidhu S S, Kalmar G B, Willis L G, Borgford J. Protease evolution in Streptomyces griseus. J Biol Chem. 1995;270:7594–7600. doi: 10.1074/jbc.270.13.7594. [DOI] [PubMed] [Google Scholar]
- 34.Sidhu S S, Borgford T J. Selection of Streptomyces griseus protease B mutants with desired alterations in primary specificity using a library screening strategy. J Mol Biol. 1996;257:233–245. doi: 10.1006/jmbi.1996.0159. [DOI] [PubMed] [Google Scholar]
- 35.Sinha U, Wolz S A, Pushkaraj J L. Two new extracellular serine proteinases from Streptomyces fradiae. Int J Biochem. 1991;23:979–984. doi: 10.1016/0020-711x(91)90133-8. [DOI] [PubMed] [Google Scholar]
- 36.Wallach J M, Hornebeck W. Mechanisms involved in elastin-elastase interactions. In: Robert L, Hornebeck W, editors. Elastin and elastase. II. Boca Raton, Fla: CRC Press; 1989. pp. 3–5. [Google Scholar]
- 37.Weisburg W G, Barns M, Pelletier D A, Lane D J. Ribosomal DNA amplification for phylogenic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams S, Goodfellow T M, Alderson G. Genus Streptomyces, Waksman and Henrici 1943, 339AL. In: Williams S T, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 4. Baltimore, Md: Williams and Wilkins; 1989. pp. 2452–2492. [Google Scholar]
- 39.Yum D Y, Chung H C, Bai D H, Oh D H, Yu J H. Purification and characterization of alkaline serine protease from an alkalophilic Streptomyces sp. Biosci Biotechnol Biochem. 1994;58:470–474. doi: 10.1271/bbb.58.470. [DOI] [PubMed] [Google Scholar]