Abstract
A 22-year-old man, after a hematopoietic stem cell transplant, suffered long-term pneumonia caused by blaKPC-2-positive K. pneumoniae and blaKPC-33-positive K. pneumoniae alternately and finally achieved pathogenic clearance and improvement of clinical infectious conditions after using ceftazidime–avibactam in combination with imipenem as salvage therapy. This case provides a reference for treating infection caused by K. pneumoniae with a KPC variant in countries lacking new antimicrobial agents.
Keywords: KPC-2, KPC-33, ceftazidime–avibactam, imipenem
1. Introduction
With the widespread use of ceftazidime–avibactam, KPC-variant-producing Klebsiella pneumoniae resistant to ceftazidime–avibactam continues to be identified in clinics worldwide [1,2]. NCBI data show that more KPC variants originating from blaKPC-2 or blaKPC-3 were reported in 2020 than in the previous seventeen years combined [3]. A variety of factors tend to mislead clinical anti-infective therapy, including the fact that conventional carbapenemase assays are often falsely negative when detecting KPC variants [4], that strains are often resistant to ceftazidime–avibactam but regain susceptibility to carbapenems such as imipenem, as well as the fact that ESBL phenotypic tests are often positive. There are no uniform recommendations for treating infections caused by KPC-variant-producing strains, especially in countries where new effective antimicrobial agents such as meropenem–vaborbactam are unavailable [4]. Taken together, this requires vigilance against the emergence of KPC variants during therapy which would result in therapy failure.
Here, we report a case of long-term pneumonia caused by alternating blaKPC-2-positive K. pneumoniae and blaKPC-33-positive K. pneumoniae in a patient with a severe hematological disease who achieved eventual pathogenic clearance and improvement of clinical infectious conditions after using ceftazidime–avibactam in combination with imipenem as salvage therapy.
2. Case Presentation
A 22-year-old man was diagnosed with non-Hodgkin lymphoma in December 2020 at a local hospital and underwent a hematopoietic stem cell transplant in July 2021. The patient developed fever, cough, and sputum one week after transplant, while carbapenem-resistant K. pneumoniae (CRKP), carrying blaKPC-2, were isolated from blood, sputum, and rectal swabs, respectively. After being diagnosed with bloodstream infection and pneumonia, he was given anti-infective treatment with meropenem (1 g q8h), polymyxin B (100 mu q12h), tigecycline (100 mg q12h), ceftazidime–avibactam (2.5 g q8h), and other antimicrobial agents successively because of no improvement in clinical symptoms. The patient’s cough and sputum symptoms still did not improve. A lung computed tomography (CT) plain scan showed multiple nodules and patchy shadows in both lungs, which was consistent with the manifestation of pneumonia, so he was admitted to Huashan Hospital affiliated with Fudan University to treat the fever with a cough and sputum in October 2021 (defined as day 1).
After admission, the patient continued anti-infective therapy with ceftazidime–avibactam (2.5 g, q8h, ivgtt) (day 1–day 7). During this time, Acinetobacter spp. was isolated from blood culture on day 4. However, since the Acinetobacter spp. strain was susceptible to ceftazidime–avibactam (MIC = 4 mg/L), the anti-infective regimen was not changed. However, he still had a cough and sputum, and CT showed a slight improvement in lung inflammation compared to the previous CT scan. On day 6, ceftazidime–avibactam-resistant CRKP (named K. pneumoniae HS01) was isolated from a sputum specimen, confirmed using the MALDI-TOF/MS system (bioMérieux, France), which was susceptible to imipenem and tigecycline (Table 1). While the five carbapenemases (KPC, NDM, VIM, IMP, and OXA-48-like) were not detected using the immunochromatographic NG-test Carba 5 assay (NG Biotech, France) in K. pneumoniae HS01, the blaKPC-33 gene was confirmed via polymerase chain reaction and sequencing. On day 8, the anti-infective regimen was switched to imipenem–cilastatin sodium (1 g, q8h, ivgtt) combined with tigecycline (100 mg, q12h, ivgtt) based on the result of antimicrobial susceptibility testing. After one week, the patient’s cough and sputum symptoms improved compared with before.
Table 1.
Antimicrobial susceptibility and resistance genes of K. pneumoniae HS01 and K. pneumoniae HS02.
| Antimicrobial Agents | MICs (mg/L) and Resistance Genes | |||
|---|---|---|---|---|
| K. pneumoniae HS 01 | K. pneumoniae HS 02 | |||
| Amikacin | >128 | R | >128 | R |
| Ceftazidime | >32 | R | >32 | R |
| Cefepime | >128 | R | >128 | R |
| Aztreonam | >128 | R | >128 | R |
| Meropenem | 4 | R | >64 | R |
| Imipenem | 0.5 | S | 64 | R |
| Piperacillin–Tazobactam | >256 | R | >256 | R |
| Cefoperazone–Sulbactam | >128 | R | >128 | R |
| Ceftazidime–Avibactam | >64 | R | 4 | S |
| Aztreonam–Avibactam | 4 | S | 2 | S |
| Meropenem–Vaborbactam | 4 | S | 16 | R |
| Imipenem–Relebactam | 0.25 | S | 1 | S |
| Cefiderocol | 8 | I | 2 | S |
| Levofloxacin | >16 | R | >16 | R |
| Ciprofloxacin | >8 | R | >8 | R |
| Tigecycline | 2 | S | 2 | S |
| Polymyxin B | >16 | R | >16 | R |
| NG-test Carba 5 | Negative | Positive | ||
| β-lactams | blaCTX-M-65, blaLAP-2, blaSHV-12, blaTEM-1B, blaKPC-33 | blaCTX-M-65, blaLAP-2, blaSHV-12, blaTEM-1B, blaKPC-2 | ||
| Quinolone | qnrS1 | qnrS1 | ||
| Aminoglycoside | addA2 | addA2 | ||
| Fosfomycin | fosA | fosA | ||
| Phenicol | catA2 | catA2 | ||
| Trimethoprim | dfrA14 | dfrA14 | ||
R, resistant; S, susceptible.
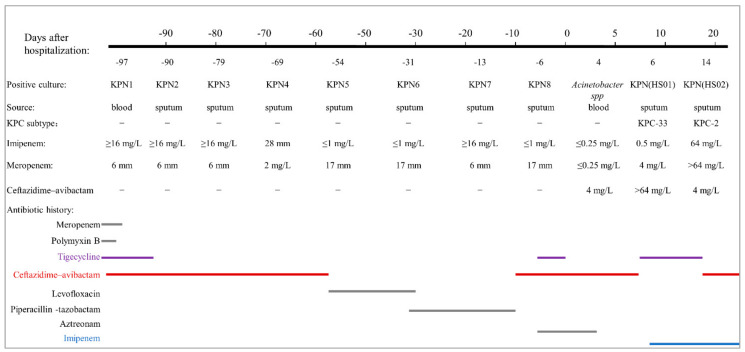
However, on day 14, the ceftazidime–avibactam-susceptible CRKP (K. pneumoniae HS02) was isolated from the sputum specimen, which was susceptible to ceftazidime–avibactam and tigecycline but resistant to imipenem and meropenem (Table 1). KPC was detected using NG-test Carba 5, and PCR and DNA sequencing confirmed blaKPC-2. According to different treatment plans, the K. pneumoniae may switch its corresponding dominant subtype (blaKPC-2-positive or blaKPC-33-positive isolate) as needed, resulting in treatment failure. The antimicrobial regimen was changed again to achieve the simultaneous treatment and prevention of dominant clone switching. Ceftazidime–avibactam (2.5 g q8h, ivgtt, to treat the infection caused by blaKPC-2-positive K. pneumoniae) combined with imipenem (1 g q8h, ivgtt, to prevent switching to blaKPC-33-positive K. pneumoniae) was started. The patient then showed significant improvement in cough and sputum symptoms. On days 21 and 23, CT showed an attractive shadow compared with the previous one, and the sputum culture for K. pneumoniae was negative. On day 23 after admission, the patient ended this phase of anti-infective treatment and recovered (Figure 1).
Figure 1.
Antibiotic history and the results of microbiology cultures. (The doses of antibiotics are as follows: meropenem, 1 g every 8 h, day 100 to day 94; polymyxin B, 1 million units every 12 h, day 99 to day 97; tigecycline, 100 mg every 12 h, day 99 to day 91, day 4 to day 1, day 7 to day 14; ceftazidime–avibactam, 2.5 g every 8 h, day 97 to day 56, day 9 to day 7, day 15 to day 25; levofloxacin, day 56 to day 32; piperacillin–tazobactam, 4.5 g every 8 h, day 32 to day 9; aztreonam 2 g every 8 h, day 7 to day 1; imipenem 1 g every 8 h, day 8 to day 25).
Eight K. pneumoniae strains (KPN1~KPN8) were isolated from the patient in the local hospital. Though genomic information on these strains was not available, we can infer that KPN 1, KPN 2, KPN 3, and KPN 7 carry blaKPC-2, and KPN 4, KPN 5, KPN 6, and KPN 8 carry blaKPN-33 based on results of antimicrobial susceptibility testing. Whole-genome sequencing analysis revealed that K. pneumoniae HS01 and K. pneumoniae HS02 belonged to ST 11 and carried the same antibiotics resistance genes (Table 1). However, K. pneumoniae HS01 carried blaKPC-33, while K. pneumoniae HS02 carried blaKPC-2. These results suggest that blaKPC-33-carrying K. pneumoniae evolved from blaKPC-2-carrying K. pneumoniae.
3. Discussion
Carbapenem-resistant Enterobacterales (CREs) have been ranked as urgent critical threats by WHO due to their high resistance rate and high mortality rate [5]. Clinical studies showed that the mortality rate of CRKP bloodstream infections has reached over 50%, which is 2–3 times higher than that of carbapenem-susceptible K. pneumoniae [6]. Ceftazidime–avibactam is favored as a first-line anti-infective agent for treating CRKP infections because it can inhibit the activity of class A, C, and some class D carbapenemases [7]. Unfortunately, the rapid emergence of KPC variants, which undergo amino acid substitutions or insertions at key sites compared to KPC-2 or KPC-3, rendering strains resistant to ceftazidime–avibactam, has posed a new challenge in healthcare facilities [2].
Of note is that patients treated with ceftazidime–avibactam against infection for about two weeks have a significant risk factor for the emergence of the KPC variant [1,2]. The reason for the resistant strains’ emergence after using ceftazidime–avibactam is unclear, although it is speculated that it may be related to the inadequate dose of avibactam. Thus, it is essential to send specimens for microbiological culture and antimicrobial susceptibility testing on time and several times when using ceftazidime–avibactam as an anti-infective therapy to keep track of the dynamic changes in KPC in patients. The treatment of KPC-variant-producing K. pneumoniae infection is currently unclear. Data in vitro show the excellent antibacterial activity of meropenem–vaborbactam against KPC-variant-producing K. pneumoniae [8]. Sporadic case reports have demonstrated the success of meropenem–vaborbactam in treating blaKPC-31-positive K. pneumoniae infections [9]. Unfortunately, meropenem–vaborbactam is currently only available in a few countries. At the same time, new subtypes of KPC have been detected in several regions and countries, and effective anti-infective treatment options are urgently needed [1,2]. It is noteworthy that tigecycline had excellent activity against blaKPC-33 carrying K. pneumoniae in vitro but had little effect in vivo. This may be related to the dose and duration of treatment in this case. There are few reports on the use of tigecycline in treating KPC-variant-producing K pneumoniae infections, and further validation is still needed.
Interestingly, most KPC-variant-producing K. pneumoniae regained susceptibility to meropenem or imipenem, suggesting a potential therapeutic role for these antibiotics. Of the nine cases of KPC-variant-producing K. pneumoniae infections treated with carbapenems or in combination with other antimicrobial agents, five diseases were successfully cured [2,10,11,12,13,14]. However, in the present case, after the discontinuation of ceftazidime–avibactam and use of imipenem in combination with tigecycline for KPC-33-producing K. pneumoniae (imipenem MIC = 0.5 mg/L), KPC-2-carrying K. pneumoniae quickly reverted to the dominant clone, failing to use imipenem as an alternative treatment regimen. Not coincidentally, a report by Shi et al. [2] similarly showed the failure of imipenem as salvage therapy for KPC-33-producing K. pneumoniae when the KPC-2 clone reappeared in the patient after treatment with imipenem, which may be related to the presence of both KPC-2 as well as KPC-33 clones in the patient. However, KPC variants are often ignored because most carbapenemase phenotypic methods fail to detect KPC variants. Ding et al. [4] reported that KPC variants were not detected using two carbapenemase phenotypic methods (the modified carbapenem inactivation method and the 3-aminophenyl boronic and EDTA method), including KPC-33, KPC-35, KPC71, KPC-76, KPC-78, and KPC-79. Furthermore, clinical laboratories encountering strains that have specific resistance phenotypes (for example, a strain harboring blaKPC-33 was resistant to ceftazidime–avibactam but susceptible to imipenem) and that are carbapenemase-negative when using carbapenemase phenotypic methods should further define the resistance mechanism by using sequencing to identify possible genetic subtypes. The combination regimen of ceftazidime–avibactam and imipenem was ultimately successful in this case. The combination of ceftazidime–avibactam and imipenem was able to kill both blaKPC-2-positive K. pneumoniae and blaKPC-33-positive K. pneumoniae, blocking the possibility of repeated substitution between blaKPC-2 and blaKPC-33. In vitro data similarly showed that the combination of ceftazidime–avibactam and imipenem effectively prevented the emergence of a KPC-variant-resistant subgroup of KPC-3-producing K. pneumoniae [15]. However, whether the combination of ceftazidime–avibactam and carbapenems is effective in preventing the emergence of KPC-variant-resistant strains in vivo has not been conclusively established, and the successful combination of ceftazidime–avibactam and imipenem, in this case, may provide a reference for countries where new antimicrobial agents such as meropenem–vaborbactam are not yet available.
The emergence of carbapenemase-producing K. pneumoniae has created a “superstorm” in clinical anti-infective therapy worldwide due to its widespread resistance profile. The emergence of KPC variants has undoubtedly added to this, especially in countries with no new effective antibacterial agents. Because routine laboratory testing methods often produce false-negative results when detecting new KPC variants, they may be considered carbapenemase-negative K. pneumoniae. During the evolution from blaKPC-2 to blaKPC-33 and other new subtypes, if clinicians abandon ceftazidime–avibactam based on the results of antimicrobial susceptibility testing and choose other antimicrobial agents, this may delay anti-infective therapy. Therefore, innovative clinical thinking is needed to treat infections caused by these emerging strains. In this case, although the strain was resistant to ceftazidime–avibactam, ceftazidime–avibactam was still used in order to prevent the next-step mutation according to the evolutionary characteristics of the bacteria. Finally, the combination of ceftazidime–avibactam and imipenem successfully eliminated the bacteria and restored the patient’s health.
Acknowledgments
We thank Professor Zou Mingxiang, from the department of laboratory medicine of Xiangya Hospital, Central South University, for assisting in reviewing the patient’s medical history information.
Author Contributions
Methodology, S.S. and R.H.; software, D.Y. and Y.G.; writing—original draft preparation, L.D.; writing—review and editing, F.H. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study protocol was approved by the Institutional Review Board of Huashan Hospital, Fudan University (No. 2018-408).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used during the current study are available from the corresponding author on reasonable request. The bacterial genome sequences have been uploaded to NCBI with the following accession numbers: K. pneumoniae HS 01, JAKWJN000000000, K. pneumoniae HS 02, JAKWJM000000000.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant no. 82172311, 81902100, and 32141002), China Antimicrobial Surveillance Network (2020QD049), and the Shanghai Public Health System Construction Three-year Action Plan (GWV-10.2-XD02). The funders had no role in study design, data collection, analysis, publishing decisions, or manuscript preparation.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Venditti C., Butera O., Meledandri M., Balice M.P., Cocciolillo G.C., Fontana C., D’Arezzo S., De Giuli C., Antonini M., Capone A., et al. Molecular analysis of clinical isolates of ceftazidime-avibactam-resistant Klebsiella pneumoniae. Clin. Microbiol. Infect. 2021;27:1040.e1–1040.e6. doi: 10.1016/j.cmi.2021.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Shi Q., Yin D., Han R., Guo Y., Zheng Y., Wu S., Yang Y., Li S., Zhang R., Hu F. Emergence and Recovery of Ceftazidime-avibactam Resistance in blaKPC-33-Harboring Klebsiella pneumoniae Sequence Type 11 Isolates in China. Clin. Infect. Dis. 2020;71:S436–S439. doi: 10.1093/cid/ciaa1521. [DOI] [PubMed] [Google Scholar]
- 3.Lebreton F., Corey B.W., McElheny C.L., Iovleva A., Preston L., Margulieux K.R., Cybulski R.J., Mc Gann P., Doi Y., Bennett J.W. Characterization of KPC-82, a KPC-2 Variant Conferring Resistance to Ceftazidime-Avibactam in a Carbapenem-Nonsusceptible Clinical Isolate of Citrobacter koseri. Antimicrob. Agents Chemother. 2021;65:e0015021. doi: 10.1128/AAC.00150-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding L., Shi Q., Han R., Yin D., Wu S., Yang Y., Guo Y., Zhu D., Hu F. Comparison of Four Carbapenemase Detection Methods for blaKPC-2 Variants. Microbiol. Spectr. 2021;9:e0095421. doi: 10.1128/Spectrum.00954-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lasko M.J., Nicolau D.P. Carbapenem-Resistant Enterobacterales: Considerations for Treatment in the Era of New Antimicrobials and Evolving Enzymology. Curr. Infect. Dis. Rep. 2020;22:6. doi: 10.1007/s11908-020-0716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-David D., Kordevani R., Keller N., Tal I., Marzel A., Gal-Mor O., Maor Y., Rahav G. Outcome of carbapenem resistant Klebsiella pneumoniae bloodstream infections. Clin. Microbiol. Infect. 2012;18:54–60. doi: 10.1111/j.1469-0691.2011.03478.x. [DOI] [PubMed] [Google Scholar]
- 7.Yin D., Wu S., Yang Y., Shi Q., Dong D., Zhu D., Hu F. Results from the China Antimicrobial Surveillance Network (CHINET) in 2017 of the In Vitro Activities of Ceftazidime-Avibactam and Ceftolozane-Tazobactam against Clinical Isolates of Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019;63:e02431-18. doi: 10.1128/AAC.02431-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hackel M.A., Lomovskaya O., Dudley M.N., Karlowsky J.A., Sahm D.F. In Vitro Activity of Meropenem-Vaborbactam against Clinical Isolates of KPC-Positive Enterobacteriaceae. Antimicrob. Agents Chemother. 2018;62:e01904-17. doi: 10.1128/AAC.01904-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiseo G., Falcone M., Leonildi A., Giordano C., Barnini S., Arcari G., Carattoli A., Menichetti F. Meropenem-Vaborbactam as Salvage Therapy for Ceftazidime-Avibactam-, Cefiderocol-Resistant ST-512 Klebsiella pneumoniae–Producing KPC-31, a D179Y Variant of KPC-3. Open Forum Infect. Dis. 2021;8:ofab141. doi: 10.1093/ofid/ofab141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giddins M.J., Macesic N., Annavajhala M.K., Stump S., Khan S., McConville T.H., Mehta M., Gomez-Simmonds A., Uhlemann A.-C. Successive Emergence of Ceftazidime-Avibactam Resistance through Distinct Genomic Adaptations in blaKPC-2-Harboring Klebsiella pneumoniae Sequence Type 307 Isolates. Antimicrob. Agents Chemother. 2018;62:2000022. doi: 10.1128/AAC.02101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shields R.K., Potoski B.A., Haidar G., Hao B., Doi Y., Chen L., Press E.G., Kreiswirth B.N., Clancy C.J., Nguyen M.H. Clinical Outcomes, Drug Toxicity, and Emergence of Ceftazidime-Avibactam Resistance Among Patients Treated for Carbapenem-Resistant Enterobacteriaceae Infections: Table. Clin. Infect. Dis. 2016;63:1615–1618. doi: 10.1093/cid/ciw636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shields R.K., Chen L., Cheng S., Chavda K.D., Press E.G., Snyder A., Pandey R., Doi Y., Kreiswirth B.N., Nguyen M.H., et al. Emergence of Ceftazidime-Avibactam Resistance Due to Plasmid-Borne blaKPC-3 Mutations during Treatment of Carbapenem-Resistant Klebsiella pneumoniae Infections. Antimicrob. Agents Chemother. 2017;61:e02097-16. doi: 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaibani P., Campoli C., Lewis R.E., Volpe S.L., Scaltriti E., Giannella M., Pongolini S., Berlingeri A., Cristini F., Bartoletti M., et al. In vivo evolution of resistant subpopulations of KPC-producing Klebsiella pneumoniae during ceftazidime/avibactam treatment. J. Antimicrob. Chemother. 2018;73:1525–1529. doi: 10.1093/jac/dky082. [DOI] [PubMed] [Google Scholar]
- 14.Cano Á., Guzmán-Puche J., García-Gutiérrez M., Castón J.J., Gracia-Ahufinger I., Perez-Nadales E., Recio M., Natera A.M., Marfil-Pérez E., Martínez-Martínez L., et al. Use of carbapenems in the combined treatment of emerging ceftazidime/avibactam-resistant and carbapenem-susceptible KPC-producing Klebsiella pneumoniae infections: Report of a case and review of the literature. J. Glob. Antimicrob. Resist. 2020;22:9–12. doi: 10.1016/j.jgar.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Göttig S., Frank D., Mungo E., Nolte A., Hogardt M., Besier S., Wichelhaus T.A. Emergence of ceftazidime/avibactam resistance in KPC-3-producing Klebsiella pneumoniae in vivo. J. Antimicrob. Chemother. 2019;74:3211–3216. doi: 10.1093/jac/dkz330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used during the current study are available from the corresponding author on reasonable request. The bacterial genome sequences have been uploaded to NCBI with the following accession numbers: K. pneumoniae HS 01, JAKWJN000000000, K. pneumoniae HS 02, JAKWJM000000000.