Abstract
The in situ community structure of Prochlorococcus populations in the eastern North Atlantic Ocean was examined by analysis of Prochlorococcus 16S rDNA sequences with three independent approaches: cloning and sequencing, hybridization to specific oligonucleotide probes, and denaturing gradient gel electrophoresis (DGGE). The hybridization of high-light (HL) and low-light (LL) Prochlorococcus genotype-specific probes to two depth profiles of PCR-amplified 16S rDNA sequences revealed that in these two stratified water columns, an obvious niche-partitioning of Prochlorococcus genotypes occurred. In each water column a shift from the HL to the LL genotype was observed, a transition correlating with the depth of the surface mixed layer (SML). Only the HL genotype was found in the SML in each water column, whereas the LL genotype was distributed below the SML. The range of in situ irradiance to which each genotype was subjected within these distinct niches was consistent with growth irradiance studies of cultured HL- and LL-adapted Prochlorococcus strains. DGGE analysis and the sequencing of Prochlorococcus 16S rDNA clones were in full agreement with the genotype-specific oligonucleotide probe hybridization data. These observations of a partitioning of Prochlorococcus genotypes in a stratified water column provide a genetic basis for the dim and bright Prochlorococcus populations observed in flow cytometric signatures in several oceanic provinces.
The world’s oceans are estimated to contribute around half of the global net primary productivity, with approximately one-quarter of this attributed to oligotrophic regions (10). These oligotrophic areas are dominated by the oxygenic photosynthetic picoplankton, a component of the phytoplankton whose size is between 0.2 and 2.0 μm and whose prokaryotic part is represented by the genera Synechococcus and Prochlorococcus (5, 6, 26, 48). This latter group accounts for a large fraction of the biomass in the central oceanic gyres (15, 17, 28), for example, 60% of the biomass and 30% of the particulate organic carbon in the central Pacific Ocean (4). Furthermore, Prochlorococcus appears to be more abundant and extends deeper down the water column than its Synechococcus counterpart, notably in the oligotrophic Pacific and North Atlantic Oceans (3, 48). The high depth-integrated abundance of Prochlorococcus results from high cell densities of 104 to 105 cells/ml throughout the water column, from surface water to depths of 150 to 200 m (5, 26, 43). Although it is now well recognized that Prochlorococcus plays a crucial role in global carbon cycling, little is known of the genetic structure of populations down a water column. This has implications for the accuracy of depth-integrated primary productivity measurements since specific genotypes may have widely differing photosynthetic efficiencies or nutrient acquisition capacities and so effect different rates of carbon dioxide conversion into biomass.
The presence of multiple Prochlorococcus populations is suggested by the frequent observations of bimodal red fluorescence distributions (dim and bright populations) revealed by flow cytometry (5, 28, 43) and by the increase in chlorophyll fluorescence per cell (5) and high divinyl-chlorophyll (DV chl) b/DV chl a ratio of deep Prochlorococcus populations (28, 43). Moreover, culture studies have indicated that all Prochlorococcus isolates can be defined as high-light (HL) or low-light (LL) adapted (21–23, 30). The LL-adapted strains possess a higher DV-chl b/DV-chl a ratio, their optimal level of growth irradiance is lower, and they become photoinhibited at irradiances at which the HL-adapted strains grow maximally (21, 23).
Cultured Prochlorococcus strains and field populations have been shown to be genetically diverse by restriction fragment length polymorphism (RFLP) analysis (33) and rpoC1 (9, 27) and petBD sequence comparisons (38). Furthermore, the relatedness of Prochlorococcus strains appears to be closely correlated with their depth of isolation rather than their geographical origin (9, 33, 38, 40). The co-occurrence of genetically and physiologically distinct Prochlorococcus strains was conclusively demonstrated at two sites in the North Atlantic, and interestingly, the HL- or LL-adapted physiology observed within each pair of strains was associated with only limited 16S rRNA sequence diversity (microdiversity) (23).
The combination of molecular and physiological studies of cultured Prochlorococcus strains and photosynthetic pigment analyses of field Prochlorococcus populations has demonstrated the physiological and genetic diversity of Prochlorococcus, but these studies have not yet revealed the vertical distribution of Prochlorococcus populations in situ. In order to examine the community structure of natural populations of Prochlorococcus, PCR-amplified 16S rDNA sequences from depth profiles in the eastern North Atlantic were analyzed by hybridization to genotype-specific probes, by cloning and sequencing, and by denaturing gradient gel electrophoresis (DGGE). The 16S rRNA gene is useful for the design of genotype-specific oligonucleotides since Southern hybridization analysis carried out in our own laboratory suggests that it is present as a single copy in all of the Prochlorococcus strains so far tested (40). Using these molecular methods we show that a partitioning of genetically distinct Prochlorococcus populations exists in situ, which can be correlated with physical features of the water column.
MATERIALS AND METHODS
Sampling.
Water samples were collected in July 1996 during an NERC Plankton Reactivity In the Marine Environment cruise in the eastern North Atlantic aboard the R.R.S. Discovery. Samples were obtained from discrete depths on 11 July (profile 1, 36.99°N, 19.68°W at 10, 20, 30, 40, 50, 60, 70, and 90 m) and 16 July (profile 2, 36.47°N, 19.2°W at 10, 30, 40, 50, 60, 70, 90, and 110 m), with a rosette of 25-liter Go-Flo bottles. Conductivity, temperature, barometric pressure, and chl a were measured simultaneously. Seawater (10 liters) was filtered from each depth onto 47-mm-diameter 0.45-μm-pore-size polysulfone filters (Supor-450; Gelman Sciences Inc., Ann Arbor, Mich.) in a gentle vacuum. A DNA buffer (10 mM Tris-HCl [pH 8.0], 100 mM EDTA, 0.5 M NaCl) (1 ml) was drawn through each filter before storage at −20°C prior to DNA extraction. Inorganic nutrients were analyzed by using a Technicon segmented flow colorimetric autoanalyzer as described by Woodward and Rees (46). Picophytoplankton were enumerated by using a FACSort flow cytometer (Becton Dickinson) equipped with a 15-mW argon ion laser exciting at 488 nm as described previously (37).
DNA isolation.
DNA was prepared from Prochlorococcus and Synechococcus strains (the origins of which have been reported elsewhere [40]) and from Escherichia coli by resuspending pelleted cells in a lysis buffer (0.25 M Tris-HCl [pH 8.0], 25% sucrose, 10 mg of lysozyme [Sigma] per ml) and incubated at 37°C for 1 h. Sarkosyl (1% [vol/vol]) and proteinase K (final concentration, 200 μg/ml) were then added, and the cell lysate was incubated at 65°C for 1 h. Proteins were extracted once with phenol-chloroform (25:24) and once with chloroform-isoamyl alcohol (24:1) before nucleic acids were precipitated with 1 volume of isopropanol and 0.4 volumes of 7.5 M ammonium acetate at room temperature. Nucleic acids were recovered by centrifugation, washed once with 70% ethanol, and resuspended in TE (50 mM Tris-HCl [pH 8.0], 10 mM EDTA).
Isolation of environmental DNA.
Filters were cut into small pieces and incubated in 0.7 ml of lysis solution (45 mM glucose, 23 mM Tris [pH 8.0], 59 mM EDTA) containing lysozyme at a concentration of 1 mg/ml. Cells were lysed by the addition of sodium dodecyl sulfate (SDS) to a final concentration of 1.5% (wt/vol) and incubated for 30 min at room temperature. The cell lysates were extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1) before nucleic acids were precipitated with 1 volume of isopropanol and 0.4 volume of 7.5 M ammonium acetate at room temperature. Nucleic acids were recovered by centrifugation, washed once with 70% ethanol, and resuspended in TE.
PCR amplification.
16S rDNA sequences were amplified from environmental DNA isolated from profiles 1 and 2 with the primers shown in Table 1. The OXY107F and OXY1313R primers (39) were used in dot blot hybridizations and the construction of clone libraries, and the GC clamp primer CYA359F (25) and the PRO1017R primer were used for DGGE analysis. Control DNAs for use in dot blot hybridizations were prepared by the amplification of 16S rDNA sequences from cultured Prochlorococcus and Synechococcus strains with the OXY107F and OXY1313R primers or the 27F and 1224R primers for E. coli. PCR amplification was carried out in a total reaction volume of 100 μl containing 10 ng of template DNA, 200 μM concentrations of deoxynucleoside triphosphates, 1.5 mM MgCl2, 0.2 μM concentrations of primers, 1 mg of bovine serum albumin (Sigma) per ml, and 2.5 U of Taq polymerase in a 1× enzyme buffer (GIBCO BRL, Life Technologies Ltd., Paisley, Scotland). The amplification conditions comprised steps at 95°C for 5 min, 80°C for 1 min for the addition of the Taq polymerase, and 30 cycles at 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min. DNA from each depth was amplified in duplicate, and the reaction mixtures were pooled prior to further use. Prochlorococcus 16S rDNA sequences were amplified for DGGE analysis by using the PCR conditions described above, except that the 100-μl reaction mixture contained 250 μM concentrations of deoxynucleoside triphosphates, 0.5 μM concentrations of primers, and 1.0 U of Taq polymerase.
TABLE 1.
Oligonucleotide primers and probes used in this study
| Primer or probe | Specific target DNA | Sequence (5′ to 3′) |
|---|---|---|
| PCR primers | ||
| OXY107Fa | Oxygenic phototroph biased | GGA CGG GTG AGT AAC GCT RA |
| OXY1313Ra | Oxygenic phototroph biased | CTT CAC GTA GGC GAG TTG CAG C |
| CYA359Fb | Oxygenic phototroph specific | GGG GAA TYT TCC GCA ATG GG |
| PRO1017R | Prochlorococcus, excluding MIT9303, MIT9313, MIT9302 | TCC CGA AGG CAC CCT CWA AA |
| 27F | Eubacterial | AGA GTT TGA TCM TGG CTC AG |
| 1224R | Eubacterial | CAT TGT AGC ACG TGT GTA |
| Oligonucleotide probes | ||
| S1PRO634R | HLI Prochlorococcus | GCC GAT CAG TTT CCA CTG |
| S2PRO634R | HLII Prochlorococcus | GCC TTT CAG TTT CCA CTG |
| DPRO634R | LL Prochlorococcus (except SS120) | GCC AAT CAG TTT CCA CTG |
| SARG634R | SS120 | GCC CTT CAG TTT CCA CTG |
| MIT1023R | MIT9303 | TGC GTT CCC AAA GGC ACT |
| EUB338 | Eubacteria | GCT GCC TCC CGT AGG AGT |
| PRO444Rc | Prochlorococcus, excluding MIT9303 and MIT9313 | TAT TCC TCA AGT ACC GTC ATA |
| MAR572R | Marine picophytoplankton clade (40) biased | GCC GCC TGC GGA CGC TTT |
Based on primers described previously (39).
A GC clamp consisting of a 40-nucleotide GC-rich sequence (5′-CGC CCG CCG CGC CCC GCG CCG GTC CCG CCG CCC CCG CCC-3′) is attached to the 5′ end of the primer (25). R represents A or G, W represents A or T, Y represents T or C, and M represents A or C.
Based on SAR6R (13).
Clone libraries.
Two clone libraries were constructed. For the first library, PCR-amplified oxygenic phototroph 16S rDNA sequences from each depth of profile 2 were ligated into the pCRII TA cloning vector before transformation into E. coli InvαF′ (Invitrogen Corporation, San Diego, Calif.). For each depth, 50 transformants were picked and screened on colony blots with a Prochlorococcus-specific probe (13). For the second clone library, oxygenic phototroph 16S rDNA sequences from depths of 10 and 90 m were amplified in quintuplicate reactions as described above, except that 5% acetamide (final concentration) was included in the reaction mixture. After ligation into the pCRII TA cloning vector and transformation, 300 transformants were picked and screened as described above, by using the MAR572R probe. Plasmid DNA was isolated from Prochlorococcus-positive clones with a QIAprep Miniprep kit (Qiagen Ltd., Crawley, West Sussex, Great Britain). Clones were analyzed by amplification with the Prochlorococcus-specific primers, followed by EcoRI RFLP analysis or DGGE analysis, prior to sequencing of representative RFLP or DGGE types.
DNA sequencing.
Double-stranded plasmid DNAs were sequenced bidirectionally with an ABI 373A automated sequencer (Applied Biosystems, Foster City, Calif.).
Phylogenetic analysis.
Sequences were checked for the presence of chimeric artifacts by using the Ribosomal Database Project program CHECK_CHIMERA (20) and then aligned with the alignment editor DCSE (7). The phylogenetic tree was inferred from 1,114 unambiguously aligned bases by the neighbor-joining method with TREECON software, version 1.2 (41).
Oligonucleotide probes.
Probes (20 to 30 ng) were end labeled with γ-32P by using T4 polynucleotide kinase in a 1× forward kinase buffer (GIBCO BRL) for 30 min. Unincorporated nucleotides were removed by purification through a Sephadex G-25 column (32).
Dot blot hybridization of DNA vertical profiles.
16S rDNA amplicons were purified by using the QIAquick PCR purification kit (Qiagen Ltd.) and quantified spectrophotometrically before being denatured and blotted onto nylon membranes (Zetaprobe; Bio-Rad Laboratories Ltd., Hemel Hempstead, Hertfordshire, United Kingdom). For the control DNAs, serial dilutions from 0 to 50 ng of PCR products were blotted, whereas for the natural samples 30 ng of PCR products was blotted in triplicate. Blots were prehybridized in 5 ml of Z-hyb buffer (1 mM EDTA, 0.25 M Na2HPO4 [pH 7.2], 7% [wt/vol] SDS) for 30 min at 30°C and hybridized in 5 ml of fresh Z-hyb buffer containing labeled oligonucleotides overnight at 30°C as described previously (16). Membranes were washed in 0.2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–0.1% SDS for three 15-min washes at 30°C followed by a 10-min wash at the stringency temperature. The stringency wash temperatures (temperature of dissociation [Td]) for individual probes were assessed empirically by producing wash curves at 4 to 5°C increments from 30 to 70°C as described previously (47). Wash temperatures were as follows: SARG634R, 44°C; S2PRO634R, 41°C; MIT1023R, 41°C. For the probes S1PRO634R and DPRO634R, higher stringency temperatures were employed (50 and 46°C, respectively). For EUB338 46°C was the washing temperature, on the basis of experiments described previously (16). Hybridization was quantified by using a phosphorimager and Image Quant software (Molecular Dynamics). The relative levels of hybridization of the S1PRO634R and DPRO634R genotype-specific probes to total oxygenic phototroph 16S rDNA sequences were calculated as reported previously (16) according to the following equation:
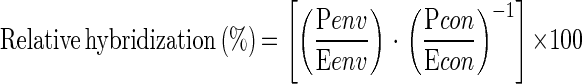 |
where Penv and Eenv represent hybridization to environmental DNA of the specific and eubacterial probes, respectively, and Pcon and Econ are the slopes of the specific (S1PRO634R or DPRO634R) and eubacterial probe-binding curves, respectively, calculated by hybridizing each probe to serially diluted homogeneous control DNAs (MED4 and NATL1 or MIT9303 and ENATL4). The relative levels of hybridization of a given specific probe to the two homogeneous control DNAs and that of the eubacterial probe were averaged.
DGGE.
DGGE was performed by using the D Gene electrophoresis system (Bio-Rad Laboratories) as described previously (24) but with the following modifications. Polyacrylamide gels (6%) were prepared in a 1× TAE buffer (40 mM Tris-HCl [pH 8.3], 20 mM acetic acid, 1 mM EDTA) containing 36% denaturant throughout. The maximum separation of Prochlorococcus 16S rDNA sequences was ascertained by electrophoresing the PCR products in a series of gels with progressively narrower denaturing gradients. Electrophoresis was carried out in 1× TAE buffer for 3 h at 200 V.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database as accession no. AF099991 (ENATL1), AF099992 (ENATL2), AF099993 (ENATL3), AF099994 (ENATL4), AF099995 (ENATL5), AF099996 (ENATL6), and AF099997 (ENATL7).
RESULTS
Temperature and nutrient profiles of the sampling stations.
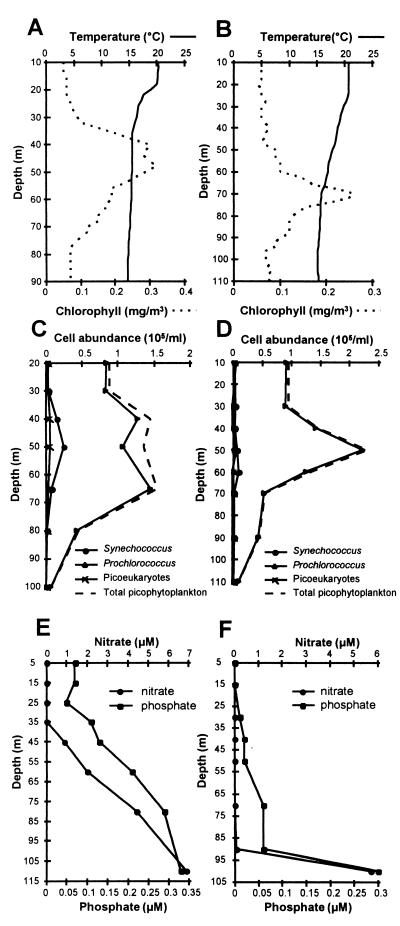
Seawater samples for the extraction of total DNAs were collected from two depth profiles at depths from 10 m to 90 to 110 m, in the northeastern Atlantic Ocean on 11 and 16 July 1996. Flow cytometry data for profiles 1 and 2 is shown in Fig. 1C and D. Prochlorococcus dominated the picophytoplankton at both stations, particularly in profile 2, where it comprised 93 to 97% of the total picophytoplankton. In profile 1 the peak abundance of Prochlorococcus was between 40 and 70 m, whereas for profile 2 the peak occurred at 50 m. For depth profile 1, a sharp thermocline was evident at around 20 m, separating the surface mixed layer (SML) at a temperature of 20°C from the water below 35 m at a temperature of 15°C (Fig. 1A). For the second depth profile taken 5 days later, the SML at a temperature of around 21°C had deepened to 30 m with the major temperature gradient extending to around 70 m (Fig. 1B). Phosphate and nitrate were depleted in the SML of profile 1 but increased in concentration below 35 m, whereas in profile 2, nutrient concentrations were very low, even at 90 m (Fig. 1E and F).
FIG. 1.
Water column properties and picophytoplankton cell abundances of two vertical profiles in the eastern North Atlantic. Temperature and fluorescence profiles (A and B), flow cytometry data of picophytoplankton cell abundance (C and D), and nutrient concentrations (E and F) for profile 1 at 36.99°N, 19.68°W (A, C, and E) and profile 2 at 36.47°N, 19.2°W (B, D, and F).
Prochlorococcus 16S rDNA clone libraries.
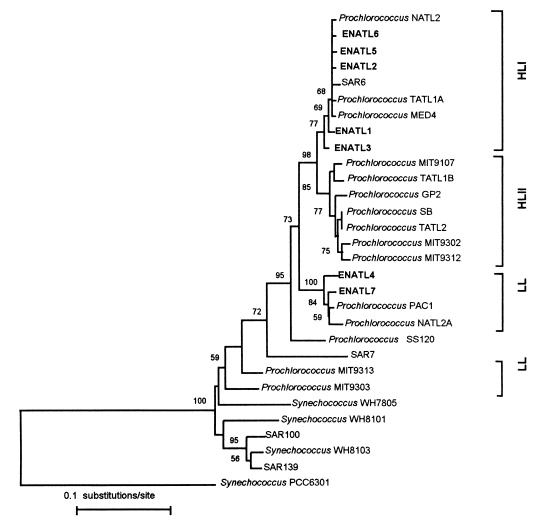
The HL-adapted clade as defined previously (40) can be further divided into two subclusters, and for the purposes of this study, we now refer to the two HL clusters as HLI and HLII and the deeply branching LL-adapted strains as LL (Fig. 2). An inspection of the available Prochlorococcus 16S rRNA sequences suggested that the presence of two EcoRI restriction sites was a good marker of the HLI sequence type. For the first clone library, a total of 91 Prochlorococcus 16S rDNA clones were retrieved from 90 m or above, and of these, 87 clones gave EcoRI restriction patterns identical to that of the HLI cluster. A clone selected at a depth of 60 m (ENATL2) was indeed found to be closely related to other members of the HLI cluster, as were two other clones at 40 m (ENATL1) and 60 m (ENATL3), which gave differing EcoRI restriction patterns (Fig. 2). The 16S rDNA clone retrieved at 110 m (ENATL4) was closely related to 16S rDNA sequences of the LL-adapted Prochlorococcus strains PAC1 and NATL2A (Fig. 2).
FIG. 2.
Phylogenetic tree showing the relationships of surface and deep Prochlorococcus environmental sequences and cultured Prochlorococcus and Synechococcus strains, inferred from 16S rRNA sequences. The tree was constructed from 1,114 unambiguously aligned bases by the neighbor-joining method. The number of bootstrap replicates supporting the branching order, of a total of 100, is shown at each node. Bootstrap values below 50 are not shown. The 16S rRNA sequence of the cyanobacterium Synechococcus sp. strain PCC6301 was used to root the tree.
The second clone library was prepared from DNA extracted from samples retrieved at 10 m and 90 m. All 39 clones retrieved at 10 m gave HLI EcoRI patterns in agreement with results from the DGGE analysis, and two selected sequences, ENATL5 and ENATL6, fell within the HLI cluster (Fig. 2). DGGE analysis of 19 clones retrieved at 90 m revealed one HL-type sequence and 18 LL-type sequences. An LL-type sequence selected, ENATL7, was closely related to ENATL4, PAC1, and NATL2A. The clones ENATL2 and ENATL6 retrieved at 60 and 10 m, respectively, were identical. The placement of the environmental sequences in either the HLI cluster or the PAC1-NATL2A cluster was also supported by phylogenetic trees constructed by using the maximum likelihood or maximum parsimony methods (data not shown).
Dot blot hybridization of DNA depth profiles.
By comparing alignments of all available Prochlorococcus 16S rDNA sequences, a variable region that allowed oligonucleotide probes to be designed to specifically target either the HLI or HLII cluster or all LL-adapted strains except SS120 was identified. This strain possessed a unique sequence in this region, giving rise to the specific oligonucleotide probe SARG634R. Since the probe specific for the LL-adapted strains also recognized MIT9303 and MIT9302, which branch more deeply within the marine picophytoplankton clade (Fig. 2), a further specific probe, MIT1023R, was designed to examine the distribution of the MIT9303 genotype alone. The specificities of all probes were checked with the CHECK_PROBE program of the Ribosomal Database Project (20) and by performing BLAST searches (1). Only environmental 16S rDNA sequences (11, 35) which are all closely related to cultured Prochlorococcus strains gave exact matches to the S1PRO634R, S2PRO634R, and DPRO634R probes, with the exception of a freshwater cyanobacterium, Gloeothece membranacea, whose 16S rRNA sequence matches that of S2PRO634R.
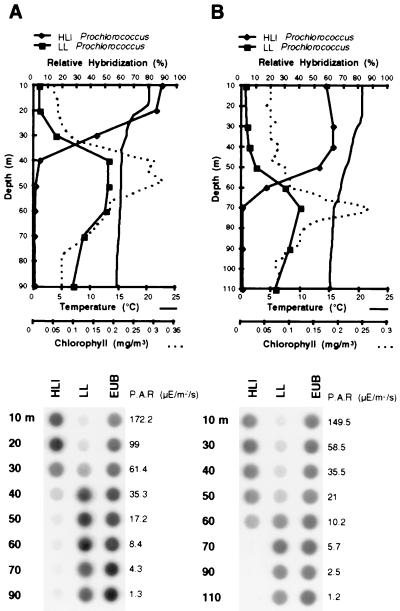
The distribution of Prochlorococcus genotypes in profiles 1 and 2 was examined by the hybridization of PCR-amplified oxygenic phototroph 16S rDNA sequences from each depth with the specific oligonucleotide probes (Table 1). Probes were always hybridized simultaneously to blots containing control DNAs for subsequent quantitation and to check the specificity of the probes. No cross-hybridization of the probes to nonhomologous sequences was observed, as shown in Fig. 3. The probes designed to specifically recognize Prochlorococcus sequences from the HLII cluster, SS120 or MIT9303, showed no hybridization to DNA amplified from either depth profile (data not shown). This suggested that these genotypes were absent or were a minor component of the Prochlorococcus population at the two sampling stations. The two probes which specifically recognized sequences from the HLI-adapted Prochlorococcus and the LL-adapted Prochlorococcus strains showed an obvious differential hybridization to both depth profiles (Fig. 4). For depth profile 1, the HLI probe hybridized significantly to DNA amplicons from the upper 30 m and gave a weak signal below 40 m, whereas the LL probe hybridized significantly only to DNA amplified from 40 m or below. Depth profile 2 showed a similar transition from the HLI to LL genotype down the water column, though the shift occurred deeper down the water column, around 50 to 60 m. For both depth profiles, the transition in distribution between the two Prochlorococcus populations appeared to occur around the midpoint of the thermocline, which was closer to the surface and steeper in profile 1. With the deepening of the mixed layer, the HLI genotype appeared to penetrate deeper into the water column, and the peak abundance of the LL genotype also occurred at a greater depth than in profile 1. The downwelling photosynthetically active radiation (PAR) measured at each depth sampled is shown to the right of the dot blots in the lower panel of Fig. 4. For each depth profile, the HLI and LL genotypes occupied distinct layers in the water column that received irradiation within the ranges from ∼10 to 170 and from ∼1 to 60 microeinsteins/m2/s, respectively.
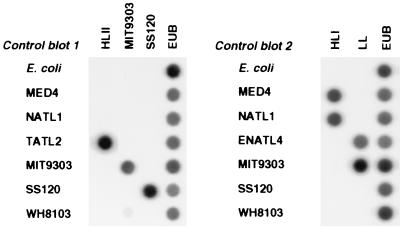
FIG. 3.
Representative dot blots showing the specificity of hybridization of each genotype-specific oligonucleotide (HLI, HLII, LL, MIT9303, and SS120) to arrays of control DNA samples (E. coli, Prochlorococcus sp. strains MED4, NATL1, TATL2, MIT9303, and SS120, Prochlorococcus 16S rDNA clone ENATL4, and Synechococcus sp. strain WH8103). EUB, eubacterial probe.
FIG. 4.
Vertical distribution of the HLI- and LL-adapted Prochlorococcus 16S rDNA genotypes in two different water columns in the eastern North Atlantic, revealed by dot blot hybridization. In the upper panels are graphs showing the differential hybridization of the HLI and LL probes to oxygenic phototroph 16S rDNA sequences PCR amplified from different depths in depth profiles 1 (A) and 2 (B), together with the temperature and fluorescence data. Dot blots corresponding to the graphs from which the relative hybridization of each genotype-specific probe was quantified are shown in the lower panels, together with the PAR measured at each depth. EUB, eubacterial probe.
DGGE analysis.
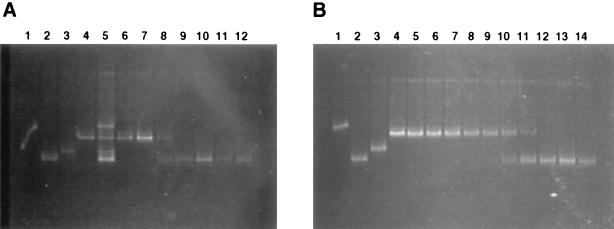
The separation of DNA amplified from each depth profile by DGGE also revealed the differential distribution of Prochlorococcus genotypes, in good agreement with observations from the dot blot hybridization analysis (Fig. 5). The transition from the HL- to LL-adapted genotype occurred at 30 m for depth profile 1 and between 50 and 60 m for depth profile 2. For both profiles, the Prochlorococcus 16S rDNA sequences dominant in surface waters comigrated with those amplified from the strains which fell into the two HL-adapted clusters (excluding MED4). The 16S rDNA sequences amplified from deeper waters from each profile comigrated with the 16S rDNA clone ENATL4 retrieved at 110 m and not with SS120, in agreement with the dot blot hybridization results and sequence data from the clone libraries of profile 2.
FIG. 5.
DGGE analysis of the distribution of Prochlorococcus 16S rDNA genotypes in depth profiles 1 (A) and 2 (B). 16S rDNA sequences amplified from cultured Prochlorococcus strains were used as controls: panel A, lane 1, MED4; lane 2, ENATL4; lane 3, SS120; lane 4, TATL2; lane 5, mixture of MED4, SS120, ENATL4, and TATL2; lane 6, 10 m; lane 7, 20 m; lane 8, 30 m; lane 9, 40 m; lane 10, 50 m; lane 11, 60 m; and lane 12, 70 m; panel B, lane 1, MED4; lane 2, ENATL4; lane 3, SS120; lane 4, NATL1; lane 5, TATL1; lane 6, TATL2; lane 7, 10 m; lane 8, 30 m; lane 9, 40 m; lane 10, 50 m; lane 11, 60 m; lane 12, 70 m; lane 13, 90 m; and lane 14, 110 m.
DISCUSSION
The high abundance of Prochlorococcus in HL surface waters down to depths at the 1% light level and the differences in chlorophyll fluorescence and pigmentation of surface and deep populations have been suspected to be due to the presence of multiple Prochlorococcus populations with different growth irradiance optima (5, 14, 28, 43). The molecular tools applied in this study have allowed us to demonstrate that genetically distinct Prochlorococcus populations are partitioned into surface and deep layers, the depths of which appear to be correlated with physical and chemical changes in the water column.
The dominance of Prochlorococcus among the picophytoplankton observed in both depth profiles is typical for this ocean province following summer stratification (44). The flow cytometry data indicated that Prochlorococcus was less abundant in the upper 30 m, in contrast to the abundance of the HL Prochlorococcus genotype revealed by the dot blot hybridization results. However, this discrepancy can be explained by the underestimation of surface Prochlorococcus cell numbers by flow cytometry due to their low chlorophyll content (48). The striking differential distribution of the two major 16S rDNA genotypes, HLI and LL, in each stratified water column is consistent with previous research, which has shown that Prochlorococcus 16S rRNA genotypes are well correlated with their light-dependent physiologies (23). Moreover, recent phylogenetic analyses of Prochlorococcus rpoC1 (9) and petBD (38) sequences from the Pacific and North Atlantic Oceans, respectively, were also suggestive of different Prochlorococcus populations adapted to high or low light levels.
Photophysiological studies of cultured Prochlorococcus strains have demonstrated that HL- and LL-adapted strains are able to grow under a range of irradiances, though they exhibit shifted growth irradiance optima (22, 23, 30, 31). The ranges of irradiance which support the growth of cultured HL- or LL-adapted Prochlorococcus strains are similar to those found in the layers occupied by the HL- and LL-adapted genotypes in situ, respectively. Thus, the relative distributions of each Prochlorococcus population within the water column which allow colonization throughout the euphotic zone are supported by culture studies.
The remarkable compartmentalization of each distinct population raises questions as to what factors govern the depth of the transition from the HL to LL population. Within the layer occupied by Prochlorococcus there are gradients of light, nutrients, temperature, and salinity. Although irradiance levels must play a fundamental role in determining the upper and lower limits of each population, the variation in the distribution of the two populations between the two depth profiles implicates additional factors. It is possible that different Prochlorococcus populations have different nutrient requirements: for example, where the surface population can grow in relatively impoverished conditions, the deep population may have a more stringent requirement for nutrients. In both depth profiles, the HL-adapted population was always confined to the nutrient-depleted waters, whereas the LL-adapted population was found within the nutricline in the first depth profile and in nutrient-depleted waters in the second depth profile. Moreover, a time series of nitrate and ammonium assimilation carried out on the same cruise at 37°N, 20°W showed f-ratios not exceeding 0.2 in the surface 30 m but increasing to 0.87 below this depth, suggesting a high proportion of new production below 30 m in this oligotrophic region (8). It is tempting to suggest that this reflects an ability of HL populations to exclusively use regenerated or reduced forms of nitrogen for growth, while LL populations largely assimilate nitrate. Analysis of Prochlorococcus vertical distributions in several ocean provinces has demonstrated, however, that the influence of nutrient concentrations on Prochlorococcus distribution and growth cannot be simply defined. Prochlorococcus populations in the Mediterranean (42) and the North Atlantic (26) appear to be nitrate limited, whereas in the oligotrophic equatorial Pacific Ocean, Prochlorococcus growth rates are high, despite low concentrations of nitrate (19).
The depth of the mixed layer appeared to strongly influence the distribution of each genotype at 37°N, 20°W, since the depth of transition from the HL- to the LL-adapted genotype deepened with the increased depth of the SML in profile 2. Thus, the HL-adapted population of Prochlorococcus may be better able to survive the turbulent mixing in this layer which must subject the cells to significant changes in irradiance. A study of the expression of the psbA genes of MED4 and SS120, in response to increased irradiance, demonstrated that psbA transcript levels increased more rapidly in the HL-adapted strain MED4 than in SS120 (12). Moreover, it has been suggested that LL-adapted Prochlorococcus cells exposed to sudden light changes would be more subject to damage to the photosystem II reaction center due to the high energy conveyed by the abundance of their antenna proteins (29). If the regulation of psbA expression is similar among other HL-adapted Prochlorococcus strains, then this study provides further evidence of their ability to cope with rapid changes in irradiance. It is possible that the ability of the HL-adapted Prochlorococcus population to respond rapidly to changes in irradiance may also allow this population to persist for longer in the water column with the onset of winter mixing.
The degree of mixing in each layer of the water column was also associated with differences in temperature, around 20°C in the mixed layer compared with 15°C below the thermocline. Hence, the temperature of each stratified layer may also have some influence on the partitioning of different Prochlorococcus populations. However, culture studies have indicated that HL- and LL-adapted Prochlorococcus strains have similar optimal growth temperatures, though the range of temperature for which growth occurs for each group may differ somewhat (21, 22).
Despite the use of oligonucleotide probes that were targeted to all known Prochlorococcus sequences, only the sequences which fell into the HLI and LL clusters were found to be dominant in both water columns studied, an observation which was also supported by DGGE analysis and by sequencing of 16S rDNA clones. In contrast, phylogenetic analysis of 16S rRNA sequences from Prochlorococcus strains isolated from the Sargasso Sea and the Gulf Stream placed the sequences within the HLII cluster or more deeply branching in the tree (23). It is possible that the different Prochlorococcus genotypes found in the eastern and western North Atlantic may indicate geographical influences on the Prochlorococcus community structure. Alternatively, Prochlorococcus strains which fall into the HLII cluster may be more abundant at different times in the seasonal cycle, changes which could best be monitored over the course of a year.
In view of the biases associated with PCR-based methods it will be important to develop in situ hybridization protocols for Prochlorococcus cells with 16S rRNA-targeted oligonucleotides, to be used alongside the methods described here. In situ hybridization has been used widely to study microbial diversity in aquatic and terrestrial environments (2) and is particularly powerful when used in conjunction with flow cytometry (36, 45). In addition, coupling [14C]NaHCO3 labeling of picoplankton populations (18) with such technology, as well as using antibodies to detect molecular markers of nutrient stress (34), will potentially allow an assessment of carbon fixation rates of individual genotypes to be correlated with their nutrient status. The molecular probes developed here will allow microbial ecologists to monitor the dynamics of specific components of the Prochlorococcus community and, when coupled to physiological measurements of single cells, will give a more complete understanding of the molecular ecology of these important marine photoautotrophs.
ACKNOWLEDGMENTS
We thank Glen Tarran and Peter Burkill for the flow cytometry data, Malcolm Woodward for the nutrient data, and Polly Machin for supplying the PRIME level-one data. We are also grateful to Gabrielle Rocap and Penny Chisholm for providing DNA from Prochlorococcus sp. strain MIT9303.
This work was supported in part by NERC grant GST/02/1081 via the PRIME thematic program and by the EU MAST III program PROMOLEC (MAS3-CT97-0128). D.J.S. is a Royal Society University Research Fellow.
Footnotes
This is contribution number 95 of the Plankton Reactivity in the Marine Environment (PRIME) program.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binder B J, Chisholm S W, Olson R J, Frankel S L, Worden A Z. Dynamics of picophytoplankton, ultraphytoplankton and bacteria in the central equatorial Pacific. Deep-Sea Res. 1996;43:907–931. [Google Scholar]
- 4.Campbell L, Nolla H A, Vaulot D. The importance of Prochlorococcus to community structure in the central North Pacific Ocean. Limnol Oceanogr. 1994;39:954–961. [Google Scholar]
- 5.Campbell L, Vaulot D. Photosynthetic picoplankton community structure in the subtropical North Pacific Ocean near Hawaii (Station ALOHA) Deep-Sea Res. 1993;40:2043–2060. [Google Scholar]
- 6.Chisholm S W, Olson R J, Zettler E R, Goericke R, Waterbury J B, Welschmeyer N A. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature. 1988;334:340–343. [Google Scholar]
- 7.Derijk P, De Wachter R. DCSE: an interactive tool for sequence alignment and secondary structure research. Comput Appl Biosci. 1993;9:735–740. doi: 10.1093/bioinformatics/9.6.735. [DOI] [PubMed] [Google Scholar]
- 8.Donald, K. M., A. P. Rees, E. M. S. Woodward, I. Joint, and G. Savidge. Uptake of carbon, nitrogen and phosphorus by phytoplankton along the 20°W meridian in the NE Atlantic between 56.5°N and 37°N. Deep-Sea Res., in press.
- 9.Ferris M J, Palenik B. Niche adaptation in ocean cyanobacteria. Nature. 1998;396:226–228. [Google Scholar]
- 10.Field C B, Behrenfeld M J, Randerson J T, Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998;281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 11.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Fernández J M, Hess W R, Houmard J, Partensky F. Expression of the psbA gene in the marine oxyphotobacteria Prochlorococcus spp. Arch Biochem Biophys. 1998;359:17–23. doi: 10.1006/abbi.1998.0862. [DOI] [PubMed] [Google Scholar]
- 13.Giovannoni S J, Rappé M S, Vergin K L, Adair N L. 16S ribosomal RNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc Natl Acad Sci USA. 1996;93:7979–7984. doi: 10.1073/pnas.93.15.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goericke R, Repeta D J. Chlorophyll a and chlorophyll b and divinyl chlorophyll a and chlorophyll b in the open subtropical North Atlantic Ocean. Mar Ecol Prog Ser. 1993;101:307–313. [Google Scholar]
- 15.Goericke R, Welschmeyer N A. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the Sargasso Sea. Deep-Sea Res. 1993;40:2283–2294. [Google Scholar]
- 16.Gordon D A, Giovannoni S J. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic Ocean and Pacific Ocean. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letelier R M, Bidigare R R, Hebel D V, Ondrusek M, Winn C D, Karl D M. Temporal variability of phytoplankton community structure based on pigment analysis. Limnol Oceanogr. 1993;38:1420–1437. [Google Scholar]
- 18.Li W K W. Primary production of prochlorophytes, cyanobacteria, and eukaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol Oceanogr. 1994;39:169–175. [Google Scholar]
- 19.Liu H B, Nolla H A, Campbell L. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat Microb Ecol. 1997;12:39–47. [Google Scholar]
- 20.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore L R. Physiological ecology of Prochlorococcus: a comparison of isolates from diverse oceanographic regimes. Ph.D. thesis. Boston: Massachusetts Institute of Technology; 1997. [Google Scholar]
- 22.Moore L R, Goericke R, Chisholm S W. Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar Ecol Prog Ser. 1995;116:259–275. [Google Scholar]
- 23.Moore L R, Rocap G, Chisholm S W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 24.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nübel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson R J, Chisholm S W, Zettler E R, Altabet M A, Dusenberry J A. Spatial and temporal distributions of prochlorophyte picoplankton in the North Atlantic Ocean. Deep-Sea Res. 1990;37:1033–1051. [Google Scholar]
- 27.Palenik B. Cyanobacterial community structure as seen from RNA polymerase gene sequence analysis. Appl Environ Microbiol. 1994;60:3212–3219. doi: 10.1128/aem.60.9.3212-3219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Partensky F, Blanchot J, Lantoine F, Neveux J, Marie D. Vertical structure of picophytoplankton at different trophic sites of the tropical northeastern Atlantic Ocean. Deep-Sea Res. 1996;43:1191–1213. [Google Scholar]
- 29.Partensky F, Hess W R, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Partensky F, Hoepffner N, Li W K W, Ulloa O, Vaulot D. Photoacclimation of Prochlorococcus sp. (Prochlorophyta) strains isolated from the North Atlantic and the Mediterranean Sea. Plant Physiol. 1993;101:285–296. doi: 10.1104/pp.101.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Partensky F, La Roche J, Wyman K, Falkowski P G. The divinyl chlorophyll a/b-protein complexes of two strains of the oxyphototrophic marine prokaryote Prochlorococcus: characterization and response to changes in growth irradiance. Photosynth Res. 1997;51:209–222. [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Scanlan D J, Hess W R, Partensky F, Newman J, Vaulot D. High degree of genetic variation in Prochlorococcus (Prochlorophyta) revealed by RFLP analysis. Eur J Phycol. 1996;31:1–9. [Google Scholar]
- 34.Scanlan D J, Silman N J, Donald K M, Wilson W H, Carr N G, Joint I, Mann N H. An immunological approach to detect phosphate stress in populations and single cells of photosynthetic picoplankton. Appl Environ Microbiol. 1997;63:2411–2420. doi: 10.1128/aem.63.6.2411-2420.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt T M, Delong E F, Pace N R. Analysis of a marine picoplankton community by 16S ribosomal RNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon N, LeBot N, Marie D, Partensky F, Vaulot D. Fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes to identify small phytoplankton by flow cytometry. Appl Environ Microbiol. 1995;61:2506–2513. doi: 10.1128/aem.61.7.2506-2513.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarran G A, Burkill P H, Woodward E M S. Phytoplankton community structure in the Arabian Sea during and after the SW monsoon, 1994. Deep-Sea Res. 1999;46:655–676. [Google Scholar]
- 38.Urbach E, Chisholm S W. Genetic diversity in Prochlorococcus populations flow cytometrically sorted from the Sargasso Sea and Gulf Stream. Limnol Oceanogr. 1998;43:1615–1630. [Google Scholar]
- 39.Urbach E, Robertson D L, Chisholm S W. Multiple evolutionary origins of prochlorophytes within the cyanobacterial radiation. Nature. 1992;355:267–270. doi: 10.1038/355267a0. [DOI] [PubMed] [Google Scholar]
- 40.Urbach E, Scanlan D J, Distel D L, Waterbury J B, Chisholm S W. Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria) J Mol Evol. 1998;46:188–201. doi: 10.1007/pl00006294. [DOI] [PubMed] [Google Scholar]
- 41.Van de Peer Y, De Wachter R. TREECON: a software package for the construction and drawing of evolutionary trees. Comput Appl Biosci. 1993;9:177–182. doi: 10.1093/bioinformatics/9.2.177. [DOI] [PubMed] [Google Scholar]
- 42.Vaulot D, Partensky F. Cell-cycle distributions of prochlorophytes in the northwestern Mediterranean Sea. Deep-Sea Res. 1992;39:727–742. [Google Scholar]
- 43.Veldhuis M J W, Kraay G W. Cell abundance and fluorescence of picoplankton in relation to growth irradiance and nitrogen availability in the Red Sea. Neth J Sea Res. 1993;31:135–145. [Google Scholar]
- 44.Veldhuis M J W, Kraay G W, Gieskes W W C. Growth and fluorescence characteristics of ultraplankton on a north-south transect in the eastern North Atlantic. Deep-Sea Res. 1993;40:609–626. [Google Scholar]
- 45.Wallner G, Fuchs B, Spring S, Beisker W, Amann R. Flow sorting of microorganisms for molecular analysis. Appl Environ Microbiol. 1997;63:4223–4231. doi: 10.1128/aem.63.11.4223-4231.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woodward, E. M. S., and A. P. Rees. Micro-nutrient and nanomolar ammonium distributions associated with the PRIME eddy. Deep-Sea Res., in press.
- 47.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zubkov M V, Sleigh M A, Tarran G A, Burkill P H, Leakey R J G. Picoplanktonic community structure on an Atlantic transect from 50 degrees N to 50 degrees S. Deep-Sea Res. 1998;45:1339–1355. [Google Scholar]