Abstract
The global transcriptional response of Escherichia coli to styrene and potential influence of exposure source was determined by performing RNA sequencing (RNA-seq) analysis on both styrene-producing and styrene-exposed cells. In both cases, styrene exposure appears to cause both cell envelope and DNA damage, to which cells respond by down-regulating key genes/pathways involved in DNA replication, protein production, and cell wall biogenesis. Among the most significantly up-regulated genes were those involved with phage shock protein response (e.g. pspABCDE/G), general stress regulators (e.g. marA, rpoH), and membrane-altering genes (notably, bhsA, ompR, ldtC), whereas efflux transporters were, surprisingly, unaffected. Subsequent studies with styrene addition demonstrate how strains lacking ompR [involved in controlling outer membrane (OM) composition/osmoregulation] or any of tolQ, tolA, or tolR (involved in OM constriction) each displayed over 40% reduced growth relative to wild-type. Conversely, despite reducing basal fitness, overexpression of plsX (involved in phospholipid biosynthesis) led to 70% greater growth when styrene exposed. These collective differences point to the likely importance of OM properties in controlling native styrene tolerance. Overall, the collective behaviours suggest that, regardless of source, prolonged exposure to inhibitory styrene levels causes cells to shift from‘growth mode’ to ‘survival mode’, redistributing cellular resources to fuel native tolerance mechanisms.
Keywords: Styrene, Toxicity, RNA sequencing
Introduction
Through metabolic engineering and synthetic biology strategies, important progress continues to be made towards the microbial production of value-added chemicals and biofuels from renewable substrates (Matsumoto et al., 2017). One common and persistent challenge, however, remains the cytotoxicity associated with the accumulation of many target end-products of interest. As one example, many recent studies have focused on demonstrating and improving the microbial biosynthesis of a diversity of aromatic chemicals, as recently reviewed (Machas et al., 2019; Noda & Kondo, 2017; Thompson et al., 2015). Aromatics serve a range of industrial uses, including as pharmaceutical precursors, plastic monomers, and bioenergy compounds. However, with highly lipophilic and solvent-like properties, cytotoxicity has commonly been reported due to the propensity of many aromatics to accumulate in the cell membrane, upon which they both disrupt membrane integrity and inhibit membrane protein function (Antunes-Madeira & Madeira, 1989; Jarboe et al., 2010; Scott & Finnerty, 1976; Sikkema et al., 1995). Accordingly, end-product toxicity remains one of the key limitations facing the production of most aromatic biochemicals (Li et al., 2005; McKenna & Nielsen, 2011; Vargas-Tah et al., 2015; Yoon et al., 2007).
In light of the above limitations, there remains a pressing need to develop genetic mechanisms for increasing tolerance towards inhibitory bioproducts, including aromatic biochemicals. To design truly effective tolerance engineering strategies, it is useful to first understand (i) the nature of cellular damage caused by the compound and (ii) how cells naturally respond to this damage. One may begin to gain such collective insights by characterizing the cells’ global transcriptomic response following exposure to the compound of interest. Such approaches have been employed, for example, to investigate how Escherichia coli behaves when exposed to acetate (Arnold et al., 2001), iso-butanol (Brynildsen & Liao, 2009), ethanol (Horinouchi et al., 2010), free fatty acids (Lennen et al., 2011), and 1,4-butanediol (Rau et al., 2016), among other compounds (Pomposiello et al., 2001; Visvalingam et al., 2013; Zheng et al., 2001). Meanwhile, with specific respect to aromatics, E. coli’s transcriptional response has also been characterized with respect to toluene (Yung et al., 2016), p-hydroxybenzoic acid (Van Dyk et al., 2004), and cinnamaldehyde (Lin et al., 2017; Visvalingam et al., 2013). Furthermore, aromatic induced transcriptional responses have also been reported for 2-phenylethanol in Saccharomyces cerevisiae (Jin et al., 2018) and toluene in Pseudomonas putida (Molina-Santiago et al., 2017).
Contributing to this growing knowledge base, the present study aimed to characterize the transcriptomic response of E. coli to both exogenously added and internally produced styrene via RNA sequencing (RNA-seq) analysis. Styrene is a bio-monomer compound that can be synthesized via an engineered pathway, but is quite toxic (complete inhibition of E. coli growth above ∼250 mg/l) (McKenna & Nielsen, 2011). Interestingly, meanwhile, past studies suggest that differences exist with respect to how E. coli responds to styrene when it is directly added to cultures versus produced by cells (Lian et al., 2016). Specifically, Lian et al. found that while loss of membrane integrity (quantified as membrane leakage) occurred in <10% of cells exposed to styrene via its external addition, >50% of styrene-producing cells showed damaged membranes. Inspired in part by this observation, another key objective of this study was to explore and characterize potential differences in E. coli’s transcriptional response to styrene across two distinct exposure modes: extracellular addition and intracellular production. As will be demonstrated, the collective data suggest that styrene damages cells at multiple levels, causing them to shift from ‘growth mode’ to ‘survival mode’. Down-regulation of multiple growth-essential processes is met by the up-regulation of several stress response systems, notably including several involved in modifying the cell envelope. With an improved and comprehensive understanding of the mechanisms involved in styrene stress and possible roles played by native tolerance strategies, the outcomes of this study may ultimately facilitate the future development of genetic strategies aimed at enhancing tolerance towards styrene and/or other aromatics and, in turn, improve their bioproduction.
Materials and Methods
Strains and Cultivation Conditions
A total of three conditions representing different styrene exposure modes were investigated in this study: styrene production (P), styrene addition (A), and a no styrene (production or addition) control (C). E. coli NST74 (ATCC 31884; a phenylalanine-overproducing strain) (Tribe, 1987) carrying pTrcColaK-PAL2 and pTrc99A-FDC1 (expressing PAL2 from Arabidopsis thaliana and FDC1 from S. cerevisiae, respectively, to collectively convert endogenous phenylalanine to styrene) was used as the styrene producing strain in P. E. coli NST74 carrying the empty vectors pTrcColaK and pTrc99A was used as a non-producing strain in A and C. In all cases, seed cultures were grown in 3 ml LB broth supplemented with 100 µg/ml ampicillin and 30 µg/ml kanamycin at 32°C while shaking at 200 rpm for 12–16 hr. Seed cultures were then used to inoculate 50 ml of pH 6.8 MM1 media [a phosphate-limited minimal media described by Machas et al. (2016)] supplemented with 1.5% (wt/vol) glucose in 100 ml Teflon-capped corning bottles (sealed bottles were used to prevent styrene loss via evaporation) at an initial OD600 of ∼0.01. Prior to sealing, the headspace of each culture was exchanged with pure O2 gas to ensure that aerobic conditions were maintained throughout the experiment. Once sealed, cultures were incubated at 32°C while shaking at 200 rpm. Upon reaching OD600 ∼1.0, all cultures were briefly opened and induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) at a final concentration of 0.2 mM, after which the headspace was again flushed with pure O2 before resealing. Culturing continued as described for a total of 27 hr. In the case of exogenous styrene addition, styrene (99%, stabilized with 10–15 ppm 4-tert-butyl-catechol; Alfa Aesar, Tewksbury, MA, USA) was added to the culture according to the following schedule (values represent final concentrations after each addition): 0 mg/l at 0 hr, 25 mg/l at 13 hr, 65 mg/l at 23 hr, 165 mg/l at 25 hr. The gradual increase in styrene concentration was designed to mimic its experimental accumulation profile in styrene producing cultures, as characterized via preliminary experiments.
RNA-seq Analysis
At 27 hr post-inoculation, cells from four independent biological replicates were harvested for RNA extraction using the RNeasy Mini Kit (Qiagen, Germantown, MD, USA) according to vendor protocols. Total RNA from two independent biological replicates was pooled at equimolar concentrations to create a single sample, after which two samples were sequenced for each condition (Schurch et al., 2016). RNA degradation and contamination were monitored on 1% agarose gels. After rRNA depletion using a RiboZero kit (Illumina, San Diego, CA), random hexamer priming was used to generate cDNA and library preparation was performed using a Nextera library prep kit (Illumina) according to manufacturer instructions. Paired end sequencing (2 × 150) was performed using an Illumina NextSeq at the DNASU Sequencing Core at Arizona State University. Reads had adapters removed and were quality trimmed using the default settings of Trim Galore (https://github.com/FelixKrueger/TrimGalore) prior to being mapped to the E. coli MG1655 genome (including the PAL2 and FDC1 genes) using STAR (Dobin et al., 2013). Read counts were assigned to transcript features using featureCounts (Liao et al., 2014). Differential gene expression analysis was performed using DESeq2 (Love et al., 2014) using the default median of ratios method for normalization (Anders & Huber, 2010). Differences in transcript levels were determined and are reported as log2-fold change (L2FC), with positive and negative values indicating up- and down-regulated expression of genes relative to the unexposed control, respectively. Genes displaying significant differential expression were determined as those maintaining a false discovery rate (FDR) adjusted p-value < .1 when compared to the unexposed control (as calculated by L2FC) using the Benjamini–Hochberg adjustment method (Benjamini & Hochberg, 1995).
Gene Ontology and KEGG Pathway Analyses
Gene ontology (GO) term analysis was performed using GeneSCF (Subhash & Kanduri, 2016) employing all three databases for GO analysis (i.e. biological process, molecular function, cellular components) and the E. coli organism database. For KEGG pathway analysis and identification of enriched pathways, KOBAS 3.0 (KEGG Orthology Based Annotation System) was utilized (Ai & Kong, 2018; Xie et al., 2011). Significantly over-represented GO terms and KEGG pathways were reported for only differentially expressed (DE) genes if, when compared to all genes in the E. coli K-12 genome, a p-value < .05 cut-off was met.
Assaying Differences in Styrene Sensitivity Amongst E. coli Single Gene Deletion Mutants
E. coli strains containing deletions cassettes for each single gene of interest (GOI; e.g. GOI::FRT-kanR-FRT) were obtained from the Coli Genetic Stock Center (CGSC; New Haven, CT) whereas plasmids used to overexpress each single GOI were obtained from the ASKA plasmid collection (Kitagawa et al., 2005) and transformed into wild-type E. coli BW25113 (all genes and associated collection designations are summarized in Supplementary Table S1). All single gene deletion mutants and overexpressing strains were tested with respect to their relative sensitivity to exogenous styrene, as compared with E. coli BW25113 and BW25113 carrying the pCA24N control plasmid, respectively. Seed cultures were prepared as above and used to inoculate 20 ml of pH 6.8 M9 media with 1.5% (wt/vol) glucose containing either 0 or 100 mg/l styrene. For overexpressing strains, both 10 and 100 µM IPTG were tested for induction, in both cases with their addition occurring at inoculation. Cultures were grown in foil-capped, sealed glass vials for 6 hr at 37°C while mixing at 200 rpm, after which cell growth was subsequently determined by measuring OD600. Further characterization of the growth of E. coli BW25113 ΔompR and BW25113 individually overexpressing ompF, plsX, and tolA was also subsequently performed across a wider range of styrene concentrations to more carefully assess differences in relative behaviours. Seed cultures (0.4 ml, prepared as above) were used to inoculate media containing either 0, 75, 125, or 175 mg/l styrene. Cultures were again grown in foil-capped, sealed glass vials for 6 hr at 37°C while mixing at 200 rpm, after which cell growth was subsequently determined by measuring OD600.
Results and Discussion
Determining E. coli’s Global Transcriptional Response to Styrene Exposure
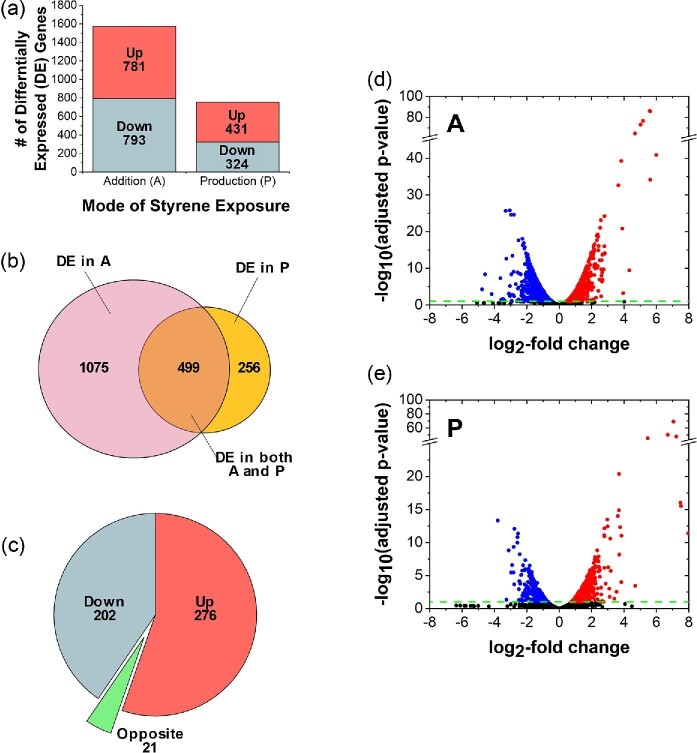
The transcriptional response of E. coli to styrene was determined across two different exposure modes—styrene addition (A) and styrene production (P)—along with a control (C) containing no styrene. To promote similar culture environments between the two exposure modes, the schedule of styrene addition in A was designed to closely mimic its accumulation profile during P, as determined via preliminary experiments and illustrated in Supplementary Fig. S1. In the case of P, a total of 135 ± 24 mg/l styrene had accumulated by 27 hr. At this point, the OD600 values of A and P were 2.00 ± 0.09 and 2.12 ± 0.12, respectively; suggesting both styrene-exposed cultures had reached a similar growth stage. At 27 hr, cells were harvested from each culture and total RNA was extracted for sequencing and analysis. A sufficient level of sequencing depth (i.e. >10 M non-rRNA fragments) was attained to perform differential gene expression analysis (Haas et al., 2012), with 11.4 and 17.2, 14.7 and 14.0, and 12.5 and 14.8 M mapped paired-end reads for A, P, and C, respectively. Differential expression analysis was therefore performed to characterize changes in E. coli’s transcriptome as a result of styrene exposure in A and P, in each case as reported relative to C (note: hereafter, ‘A’ and ‘P’ refer to A vs. C and P vs. C, respectively). In total, 1,574 and 755 DE genes were identified for A and P, respectively (Fig. 1a; see Supplementary Material for complete data), while 793 and 629 of these showed more significant levels of differential expression when considering L2FC > 1 or < −1 as an additional, more stringent cut-off. For comparison, in response to exogenous toluene exposure at 200 mg/l, a total of 641 DE genes were identified in E. coli with L2FC > 1 or < −1 and p-value < .05 (Yung et al., 2016). Fig. 1d and e further illustrate the relationship between FDR adjusted p-value and L2FC for both A and P across each entire dataset. Among DE genes identified in response to styrene, approximately 50% were up-regulated in A, compared to 56% in P (Fig. 1a). Meanwhile, a total of 499 genes were DE under both exposure modes, compared to 1 075 and 256 genes that were uniquely DE in A and P alone, respectively (Fig. 1b; Hulsen et al., 2008). Since the majority (i.e. 66%) of DE genes in P significantly overlap with those in A, this suggests that the overall response is predominantly conserved between the two different exposure modes. Among the 499 commonly DE genes, 276 were up-regulated and 202 were down-regulated under both A and P (Fig. 1c). Interestingly, 21 genes were DE in opposite directions for P versus A (Fig. 1c; for a complete list see Supplementary Table S2). Overall, while this unique subset of genes is somewhat broadly distributed, the presence of several involved in regulating the cell envelope (i.e. mepS, ompT, dacC) and general stress response (i.e. cspG, elaB, hdeA) could suggest that differences (however small) might exist with respect to how E. coli experiences and responds to styrene when exposure occurs internally versus externally. More detailed characterizations are needed to fully explore this prospect.
Fig. 1.
(a) Total differentially expressed (DE) genes identified for styrene addition (A) and production (P), both relative to a no styrene control (C). Upper (red) and lower (blue) bars, along with associated inset values, show total DE genes up-regulated or down-regulated, respectively. (b) A Venn diagram (created with BioVenn; Hulsen et al., 2008) comparing DE genes unique to A (maroon, left) versus unique to P (gold, right), as well as DE genes common to both conditions (orange, middle). (c) For DE genes common to both A and P, the number of genes down-regulated under both conditions (blue), up-regulated under both conditions (red), and oppositely DE (green) are compared. Note: for (a)–(c), only DE genes with FDR adjusted p-value < .1 are included. Volcano plots of all DE genes (down-regulated, blue; up-regulated, red; no significant differential expression, black) identified in the case of (D) styrene addition (A) or (E) styrene production (P), with a significance threshold of FDR adjusted p-value < .1 (green dotted line).
Despite similarities in overall behaviour (note: the Pearson correlation coefficient was calculated as 0.64, suggesting a moderate to strong correlation), expression patterns of many individual genes varied more significantly between A and P, as summarized in Fig. 2. For example, the most highly up-regulated genes (i.e. the phage shock protein operon pspABCDE and pspG) showed much higher DE in P than in A (e.g. up to ∼10-fold greater in the case of pspG). Other genes followed a similar trend of increased differential expression in P (e.g. trpE, marR, ymgA, ymdF, fadM, ycgR), whereas others still were more significantly DE in A (e.g. ldtC, ibpB). Overall, however, while it should be appreciated that styrene levels in the extracellular environment were closely matched between A and P, it unfortunately remains impossible to tightly control for small differences in intracellular styrene levels which may have also contributed to differences in E. coli’s responses between the two cases. That said, these levels are expected to be close since, as revealed in past characterizations, the maximum inhibitory concentration of styrene against E. coli is at least very similar between the two cases (both ∼250–260 mg/l) (McKenna & Nielsen, 2011).
Fig. 2.
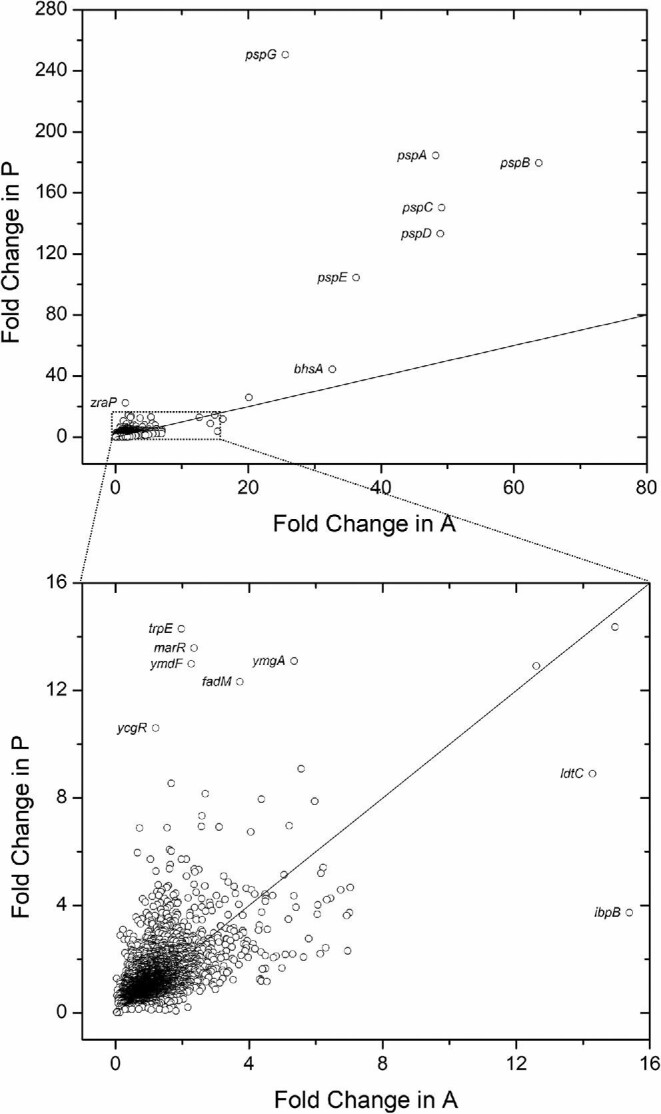
Fold change of 3 986 E. coli genes for styrene production (P) versus styrene addition (A), both relative to the no styrene control (C). Each circle represents a different gene while the solid line represents y = x. The lower graph represents an inset of the complete dataset, the region of which is depicted via a dashed box in the upper graph.
Characterizing E. coli’s Styrene Response Via Gene Ontology and KEGG Pathway Analysis
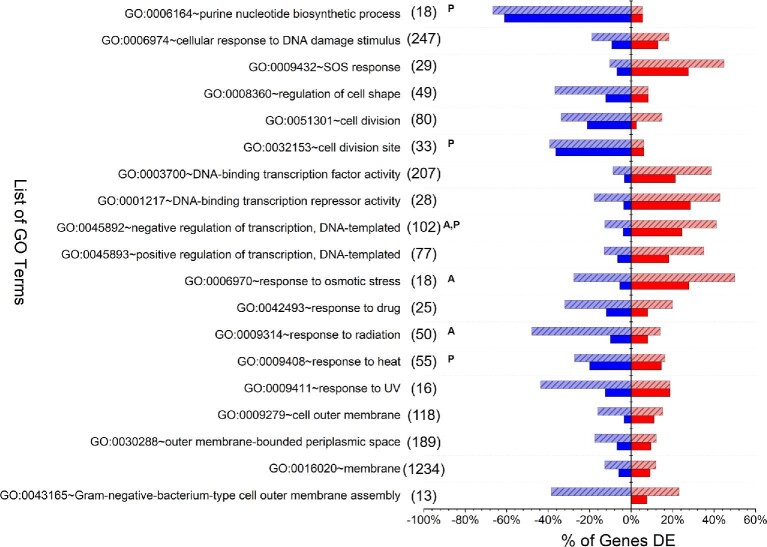
Beyond characterizing differential expression behaviours of individual genes, identification of toxicity and tolerance mechanisms may also be accomplished by examining up- and down-regulated gene families. Accordingly, the function of all DE genes was next further analyzed based on GO terms, revealing that, in total, 23 and 47 GO terms were over-represented (p-value < .05) in A and P, respectively. A total of 15 GO terms were over represented in both A and P, including many from very broad categories (e.g. ‘GO:0005829∼cytosol’ which contains 1018 genes). Fig. 3 summarizes a collection of GO terms of particular interest, along with the percentage of genes in each GO term significantly up- or down-regulated in A and/or P. Notably included among over-represented GO terms are those related to DNA synthesis and cell division (largely down-regulated), as well as responses to DNA damage (e.g. SOS response) and various stresses (mostly up-regulated).
Fig. 3.
Comparison of DE genes identified for styrene addition (A) and styrene production (P) across a selection of GO terms of notable significance and/or particular interest to this study. For each GO term, the total number of associated E. coli genes is listed in parenthesis. The y-axis indicates the percentage of total genes in each GO term that were significantly down-regulated (lower, blue) or up-regulated (upper, red) for A (light red or light blue with stripes) and P (solid red or solid blue). Superscripts ‘A’ and/or ‘P’ indicate that the entire GO term was significantly over-represented (p-value < .05) for that condition, as compared to the E. coli genome.
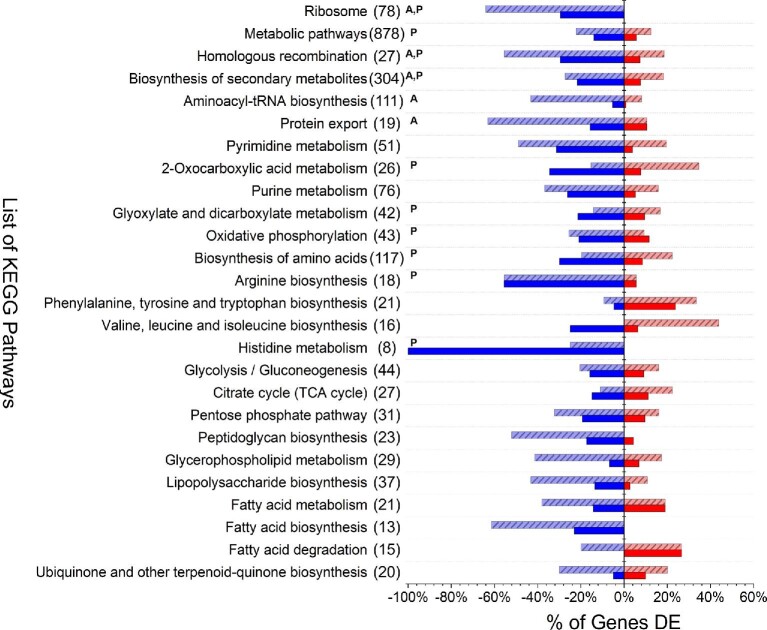
KEGG pathway analysis was next also performed on all DE genes, revealing that 5 and 14 pathways (among 138 total, Sajed et al., 2016) were over-represented (p-value < .05) in A and P, respectively. Of these, three pathways were over-represented in both conditions: ‘ribosome’ (eco03010), ‘biosynthesis of secondary metabolites’ (eco01110), and ‘homologous recombination’ (eco03440). Fig. 4 summarizes a collection of KEGG pathways of particular interest, along with the percentage of genes in each pathway significantly up- or down-regulated in A and/or P. Most notably, included among over-represented KEGG pathways are those related to amino acid and protein production, as well as the biosynthesis of membrane and cell wall components (all predominantly down-regulated). Among notable similarities between A and P was the down-regulation of the majority of genes (12 and 11 of 17 total, respectively) involved in converting α-ketoglutarate to glutamate, proline, arginine, and/or glutamine. The shutdown of pathways responsible for consuming α-ketoglutarate perhaps represents a strategy for increasing energy generation by promoting the availability of precursors for and flux through the TCA cycle.
Fig. 4.
Comparison of DE genes identified for styrene addition (A) and styrene production (P) across a selection of KEGG pathways of notable significance and/or particular interest to this study. For each KEGG pathway, the total number of associated E. coli genes is listed in parenthesis. The y-axis indicates the percentage of total genes in each KEGG pathway that were significantly down-regulated (lower, blue) or up-regulated (upper, red) for A (light red or blue with stripes) and P (solid dark red or blue). Superscript ‘A’ and/or ‘P’ indicates that the entire KEGG pathway was significantly over-represented (p-value < .05) for that condition, as compared to the E. coli genome.
Interpreting E. coli’s Transcriptional Response to Styrene
At a high level, the GO term and KEGG pathway analysis results suggest that styrene exposure triggers a widespread cellular response which involves the activation of stress response systems shutting down several growth essential processes; including DNA synthesis and protein production to cell wall biosynthesis and cell division. To provide a detailed understanding of how E. coli is impacted by and responds to styrene, a subset of the most statistically significant and/or unique outcomes across the datasets were next selected for further analysis and discussion, organized according to major cellular functions, processes, or roles. Taken together, one emergent pattern to be gleaned from the overall data is that styrene exposure, which appears to cause cell damage via multiple mechanisms, triggers the shutdown of cell growth in a ‘bottom up’ manner and, in the process, promotes a shift in cell behaviour from ‘growth mode’ to ‘survival mode’. This general behaviour, which has also been described with respect to stress-induced changes in the proteome (Guo & Gross, 2014), is likely used as a strategy that allows cells to focus on deploying mechanisms to counter stresses while waiting for the return of suitable conditions for resuming growth.
DNA replication, damage, and repair
In both A and P, numerous genes associated with the GO term ‘DNA replication’ (GO:0006260) were significantly repressed, including, for example, both dnaX (L2FC = −1.94 for A) and dnaA (L2FC = −1.48 and −0.93 for A and P, respectively). Pathways responsible for supplying nucleotide precursors (e.g. purine and pyrimidine biosynthesis) were also repressed in both A and P. Considering their energy intensive nature, down-regulation of these pathways may represent a strategy for conserving and reallocating resources towards other functions of more immediate importance to cell survival. In addition to inhibiting DNA replication, styrene exposure also appears to elicit DNA damage in a manner similar to exposure to UV radiation or DNA-arresting chemicals (Janion, 2008; Lou et al., 2012); conditions known to trigger the SOS response for DNA repair (Dörr et al., 2009; Lou et al., 2012). From the GO term ‘SOS response’ (GO:0009432), 13 and 8 of 29 total genes were up-regulated in A and P, respectively. Liang et al. also observed significant up-regulation of many SOS response system genes in a styrene-tolerant E. coli mutant (Liang et al., 2019). Here, up-regulated genes included dinB and umuDC, encoding DNA polymerases Pol IV and V, respectively. In addition to being low-fidelity, Pol IV and V enable DNA replication through damage and lesions that Pol III cannot (Finkel, 2006). Therefore, up-regulation of Pol IV and V might suggest an attempt to improve survivability by incorporating beneficial mutations. Lastly, in over 70 separate transcriptomic studies investigating E. coli’s response to compounds including as iso-butanol, salicylate, and various acids, dps (encoding a DNA-binding protein that protects DNA from breakage, Calhoun & Kwon, 2011) has been identified as a significantly DE gene (Alekshun & Levy, 1999; Erickson et al., 2017). Here, dps was also highly up-regulated in P (L2FC = 1.95). Whereas styrene has previously been shown to damage the membrane in E. coli (Lian et al., 2016), the observation that it also potentially acts as a DNA-damaging agent is a new finding. This apparent duality of styrene toxicity makes it similar to p-coumaric acid which, in addition to increasing membrane permeability, binds/damages DNA by intercalating into the double helix (Lou et al., 2012).
Cell division
Of the 33 total genes associated with the GO term ‘cell division site’ (GO:0032153), 15 and 14 were DE for A and P, respectively; 13 and 12 of which were down-regulated. Several vital components of the ‘divisome’ (Du & Lutkenhaus, 2017) belong to the dcw cluster, numerous of which were significantly down-regulated in both A (i.e. murE, mraY, ftsW, murG, murC, ddlB, ftsQ) and P (i.e. ftsW, murG, murC, ddlB, ftsQ, ftsA, ftsZ lpxC). Meanwhile, in addition to promoting DNA repair, the SOS response also influences cell division, notably via induction of the inhibitor protein encoded by sulA (L2FC = 0.86 and 1.90 in A and P, respectively) (Fonville et al., 2010). Concurrently, additional genes involved in synchronizing cell envelope division, controlling elongation machinery, and mediating cell wall synthesis were also significantly down-regulated in both A and P, including those comprising the Tol system (tolQ, tolA, tolR) which controls initiation of outer membrane (OM) constriction (Gray et al., 2015). Damage to the Tol system has been shown to lead to decreased OM integrity and periplasmic leakage, and thus, increased sensitivity to drugs and other stresses (Gray et al., 2015). Since the processes of initiating OM constriction have been reported to be particularly vulnerable to damage in E. coli (Egan, 2018), down-regulation of the Tol system could signify that E. coli is invoking a conservative approach towards limiting its susceptibility to stress-induced damage during styrene exposure.
Global stress response
Unsurprisingly, styrene exposure caused extensive transcriptome remodelling with respect to several stress response systems. Expression of rpoH (encoding the heat shock responsive sigma factor, σ32), for example, was significantly increased in A (L2FC = 1.48). Meanwhile, two of the most significantly up-regulated genes in both A and P were ibpA and ibpB (L2FC: A = 2.27 and 3.94; P = 1.08 and 1.90, respectively), both encoding small heat shock chaperones belonging to the σ32 regulon (Kitagawa et al., 2002). Increased expression of rpoH and other genes in its regulon have been reported in response to other chemical stresses, including for both ethanol (VanBogelen et al., 1987) and n-butanol (Rutherford et al., 2010). Up-regulation of yibA was also observed in A and P (L2FC = 1.83 and 1.60, respectively). E. coli strains lacking yibA display increased sensitivity to nalidixic acid, tetracycline, and mitomycin (Han et al., 2010), as well as UV and X radiation (Sargentini et al., 2016); overexpression of yibA, however, has been shown to increase E. coli’s n-butanol tolerance by ∼13% (Reyes et al., 2011). Part of the marRAB locus, marA encodes a dual transcriptional regulator that modulates expression of several genes involved in resistance to multiple antibiotics and other inhibitory compounds/conditions (Randall & Woodward, 2002) (including salicylate, Cohen et al., 1993), and was highly up-regulated in both A and P (L2FC = 1.08 and 2.52, respectively). While this is consistent with previous characterizations of E. coli’s response to other inhibitory conditions, where marA expression was almost always up-regulated (93% of the time), including when exposed to phenolic compounds (Alekshun, Levy, 1999; Erickson et al., 2017), it differs from the results of Liang et al. (2019), who found marA to be down-regulated in a styrene-tolerant mutant isolate. Although marA expression was up-regulated, several genes that it regulates—and which typically play key tolerance roles—were not in either A or P; including soxS as well as acrAB and tolC (encoding the AcrAB-TolC multi-drug RND efflux transporter). This discrepant behaviour may due to the fact that the MarA regulon is known to be governed in a concentration dependent manner (Garcia-Bernardo & Dunlop, 2013; Martin et al., 2008).
Membrane stress response
Styrene and other aromatics are commonly reported to cause membrane damage and stress (Antunes-Madeira & Madeira, 1989; Lian et al., 2016; Sikkema et al., 1994). Therefore, as expected, several stress responses associated with membrane-related damage were activated in both A and P. In E. coli, five main envelope stress response systems have been identified, including the Psp, Cpx, Bae, Rcs, and σE signalling pathways, which collectively work to restore the cell envelope upon damage, as well as maintain its stability and integrity (Rowley et al., 2006). Up-regulation of the Psp and σE signalling pathways was observed in both A and P. The Psp (phage shock protein) system (comprised of pspABCDE operon along with pspF and pspG), for example, has been shown to respond to multiple stresses, including exposure to ethanol, methanol, and other hydrophobic solvents (e.g. n-hexane, cyclohexane) (Flores-Kim & Darwin, 2016; Manganelli & Gennaro, 2017). Here, individual components of the Psp system were among the most highly up-regulated genes (L2FC = 4.68–5.99 in A, 6.70–7.99 in P; Fig. 2). Strains lacking a functional Psp system have been reported to display difficulties in maintaining proton motive force when exposed to different stresses (e.g. heat, osmotic shock, ethanol); suggesting that it plays a key role in maintaining inner membrane stability and rigidity under stress (Flores-Kim & Darwin, 2016; Manganelli & Gennaro, 2017). Meanwhile, up-regulated in both A and P (L2FC = 0.63 and 2.05, respectively), activation of σE (encoded by rpoE) has been found to improve OM stability by activating expression of genes responsible for re-folding membrane proteins, reducing expression of new OM proteins, and rapidly modifying the cell envelope upon sensing stress (Mitchell & Silhavy, 2019).
Cell envelope modification
Microbes employ a variety of mechanisms to counter chemical-induced stress/damage to the cell envelope as well as increase chemical tolerance, including by modifying the structure and composition of the cell envelope (Bui le et al., 2015; Heipieper et al., 2003; Junker & Ramos, 1999; Sandoval & Papoutsakis, 2016; Tan et al., 2016; Tan, Khakbaz, et al., 2017). One important mechanism involves maintaining proper cross-linking of the peptidoglycan (PG) layer; including both intermolecular cross-linking as well as cross-linking of the PG to the OM lipoprotein (Lpp) via LdtC. Here, significant up-regulation of ldtC was observed in both A and P (L2FC = 3.84 and 3.15, respectively). Stabilization of the OM to prevent damage has been reported to maintain or increase cell wall stability in response to penicillin exposure (Braun & Rehn, 1969; Braun & Wolff, 1975; Surmann et al., 2016). The composition and structure of the OM has also been reported as a strong determinant of tolerance, especially in the case of hydrophobic solvents (Glebes et al., 2014; Lennen & Pfleger, 2013; Sherkhanov et al., 2014; Tan et al., 2016). Two small RNAs (sRNAs), omrA (L2FC = 4.33 and 4.69 in A and P, respectively) and omrB (L2FC = 3.90 and 3.84) play an important role to this end. Activation of omrA and omrB has also been reported following exposure to butanol, furfural, geraniol, and succinic acid (Rau et al., 2015). Expression of ompF, meanwhile, which belongs to the General Bacterial Porin family and facilitates diffusion of various small (<600 Da; note: styrene is ∼104 Da) molecules (e.g. ions, antibiotics, small proteins) across the OM (Nikaido, 1989; Pichler & Emmerstorfer-Augustin, 2018). Strains lacking ompF have previously been shown to display improved tolerance to externally-added short-chain fatty acids (Rodriguez-Moya & Gonzalez, 2015), as well as improved membrane integrity and increased production of fatty acids (Tan, Black, et al., 2017). Expression of ompF was also previously found to be repressed in a series of isolated mutants displaying enhanced tolerance to various hydrophobic solvents (e.g. cyclohexane, xylene) (Aono & Kobayashi, 1997). Here, ompF was significantly down-regulated in both A and P (L2FC = −1.89 and −2.55, respectively), perhaps suggesting an effort to prevent styrene (added, or produced and excreted) from (re)entering the cell. Lastly, bhsA (encoding an OM protein) was also among the most highly up-regulated genes in both A and P (L2FC = 5.03 and 5.47, respectively). Induction of bhsA expression has previously been observed in response to various stresses (e.g. hydrogen peroxide (Zheng et al., 2001), cadmium (Egler et al., 2005), and salicylate, (Pomposiello et al., 2001)). Over-expression of bhsA, meanwhile, has been shown to significantly alter cell surface hydrophobicity (direction dependent on strain), resulting in increased tolerance to and production of octanoic acid (Chen et al., 2018).
Efflux transporters
One surprising observation of this study was the fact that multi-drug resistant (MDR) efflux transporters were largely unrepresented amongst significantly DE genes in both A and P. As discussed above, this included the AcrAB-TolC RND efflux pump, which was previously implicated as important for E. coli growth in the presence of styrene, with removal of this transporter (by deletion of acrB in NST74) resulting in inhibition of both growth and styrene production (Mingardon et al., 2015). In addition to RND efflux pumps, E. coli also encodes numerous other MDR transporters from different families; however, most were not significantly responsive to styrene exposure, including in both A and P. Overall, only expression of mdlA, emrE, emrY, fsr, ynfM, ydiM, and ydeA was increased for A, along with mdlA, mdtK, mdtG, mdtH, ynfM, yebQ, yghB, yqjA, and ydeA for P. Lack of representation of MDR efflux transporters may, however, be an artefact of time-dependent behaviour. For example, using Pseudomonas putida KT2440 and toluene, Molina-Santiago et al. demonstrated that transcriptional changes in efflux pump expression after short-term (1 hr) exposure were much greater than those measured after the long-term (several hours) exposure (Molina-Santiago et al., 2017). In the present study, only long-term (i.e. 27 hr) styrene exposure was investigated and it is likely that acute responses would differ, both in terms of the nature of DE genes and their relative expression levels. Further investigation is necessary to characterize E. coli’s acute response and/or dynamic behaviours occurring immediately after styrene exposure.
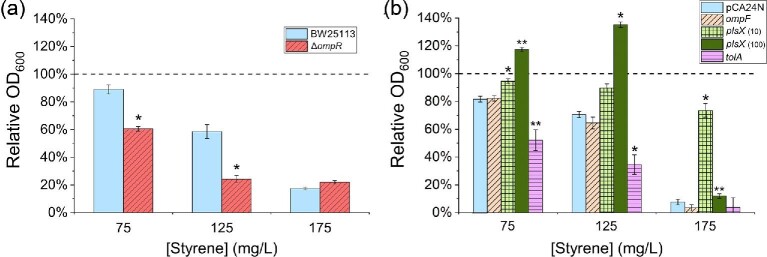
Comparing Differences in Styrene Sensitivity Among Single Gene Deletion Mutants and Overexpressing Strains
A subset of DE genes was lastly selected for further functional investigation. This not only included those most highly DE genes in A and/or P, but also those showing lower differential expression which are known or hypothesized to be important in affecting solvent tolerance and/or membrane integrity. In this case, all genes were characterized with respect to the impacts of both their deletion (where non-essential) and over-expression on growth in the presence of 100 mg/l exogenous styrene. As seen in Fig. 5a, when compared to the wild type, for many of the selected genes, their sole deletion resulted in only a modest change in relative growth due to the presence of styrene. This outcome is not altogether surprising, however, since solvent tolerance is generally considered to be a multigenic phenotype (Alper et al., 2006; Alper & Stephanopoulos, 2007). That said, styrene sensitivity was significantly increased for several mutants, most notably including ΔompR, ΔtolQ, ΔtolA, and ΔtolR. Together with EnvZ, OmpR makes up a signal transduction system (L2FC: ompR = 0.80 and 0.86 for A and P, respectively; envZ was not significantly DE) which acts as an important regulator of OM composition and is involved in osmoregulation (Wang et al., 2012). In response to environmental changes, OmpR has been shown to activate the acid and osmotic stress responses in both E. coli and Salmonella typhirium (Chakraborty et al., 2017) and control expression of the aforementioned sRNAs omrA and omrB (Guillier & Gottesman, 2006) as well as the OM porin ompF (Pratt et al., 1996). As it acts as a positive activator (Brosse et al., 2016), deletion of ompR has been shown to block expression of ompF (Mizuno & Mizushima, 1987). It is further noted, however, that micF, which encodes a regulatory sRNA that represses ompF expression (Andersen et al., 1989), was also significantly DE in the presence of styrene (L2FC = 2.02 and 2.75 for A and P, respectively), which perhaps explains the observed down-regulation of ompF even as ompR was up-regulated. Thus, it is possible that, in the case of styrene, the role of OmpR in meditating native tolerance is more significantly associated with its influence on modulating changes in OM composition/structure than in preventing styrene import. This is further supported in Fig. 5a, where the ompF mutant showed no significant change in styrene tolerance relative to wild type. Meanwhile, as discussed above, TolQ, TolR, and TolA are all prominently involved in cell division processes. It is further noted, however, that deletion of any of tolQ, tolA, or tolR also significantly reduced E. coli’s baseline fitness, with styrene only further compounding this disadvantageous behaviour.
Fig. 5.
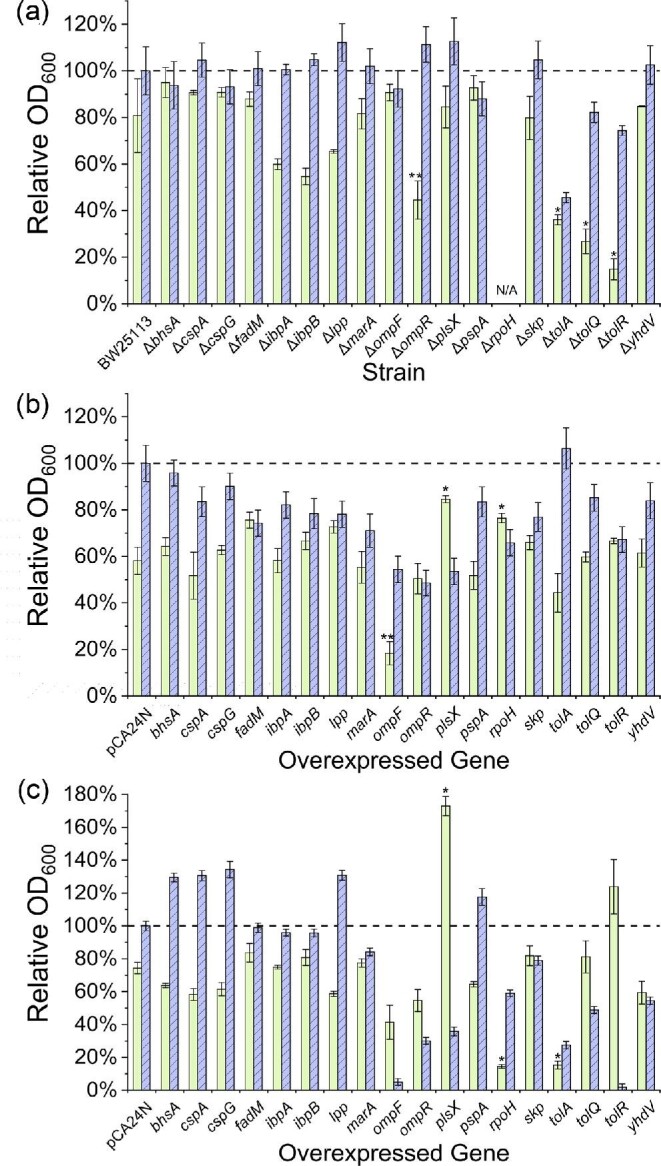
(a) Relative growth of single gene deletion mutants of interest comparing the final OD600 of deletion mutants following 6 hr of exposure to 100 mg/l styrene relative to no styrene control (green, solid) as well as the final OD600 of deletion mutants and wild-type E. coli BW25113 both with no styrene added (blue, striped). Error bars reported at one standard deviation (n = 4 for single gene deletion mutants, n = 8 for BW25113). (b) Relative growth of single gene overexpression strains comparing the final OD600 of strains induced with 10 µM IPTG following 6 hr of exposure to 100 mg/l styrene relative to no styrene control (green, solid) as well as the final OD600 of the single gene overexpression strains and wild-type E. coli BW25113 pCA24N control with no styrene added (blue, striped). Error bars reported at one standard deviation (n = 3). (c) Same conditions as (b) except using 100 µM IPTG for induction. Dashed lines indicate relative growth of 100% (i.e. no growth difference in the presence versus absence of styrene or no growth difference between strain of interest and the respective control both with no styrene). * indicates p < .001 and ** indicates p < .005 for two-tailed Student's t-test when compared to the respective control strain (BW25113 or BW25113 pCA24N).
With respect to the overexpression of single genes of interest, styrene sensitivity was characterized at two induction levels (10 and 100 µM IPTG) in order capture potentially unique titration effects. As seen in Fig. 5b and c, overexpression of several genes (e.g. ompF, ompR, plsX) resulted in significantly reduced fitness, even in the absence of styrene (and especially when induced with 100 µM IPTG). Meanwhile, sensitivity to styrene was found to increase for several overexpressed genes, as was most prominently observed in the cases of ompF and tolA. Interestingly, overexpression of plsX (a phosphotransacetylase involved in the biosynthesis of membrane phospholipids (Lu et al., 2006; Röttig & Steinbüchel, 2013; Yoshimura et al., 2007); L2FC = −2.21 for A, not DE for P) resulted in improved growth (compared to the control strain) in the presence of styrene under both induction conditions, with the strain even reaching a higher final OD600 with 100 µM IPTG when exposed to styrene versus the no styrene control (0.36 ± 0.01 vs. 0.21 ± 0.02). Previously, plsX was implicated to have an effect on tolerance to isobutanol (Minty et al., 2011) and isoprenol (Babel & Krömer, 2020) in E. coli, whereas overexpression of plsX in Clostridium acetobutylicum improved tolerance to butanol, butyrate and acetate while enhancing levels of saturated fatty acids in the membrane (Alsaker et al., 2010). Lastly, rpoH overexpression interestingly led to both significantly improved and reduced relative growth (compared to the control) at low and high induction levels, respectively. The variable influence of this regulator is perhaps unsurprising given the large number of genes whose expression it controls and its previously implicated role controlling E. coli’s solvent tolerance; though these results provide evidence that the relative level of rpoH expression may be a strong determinant of any tolerance phenotype.
Finally, considering their prominent behaviours displayed in the preliminary screen (Fig. 5), differences in the styrene sensitivity of the ΔompR mutant as well as strains individually overexpressing ompF, plsX, and tolA were further characterized across a wider range of styrene concentrations (0–175 mg/l). As seen in Fig. 6a, the relative growth of the ompR mutant was significantly lower (∼33–40%) when exposed to 75 or 125 mg/l styrene, however, by 175 mg/l styrene, both strains grew equally poor (final OD600 = 0.077 ± 0.004 and 0.069 ± 0.004 for BW25113 and BW25113 ompR, respectively). This suggests that, although it may be important for improving tolerance at moderate concentrations, OmpR alone is not responsible for dictating the E. coli’s maximum inhibitory styrene threshold (note: as a cursory test of the broader importance of ompR with respect to general aromatic tolerance, similar differences in sensitivity of the ompR mutant were also analogously observed for 2-phenylethanol; Supplementary Fig. S2). Meanwhile, as seen in Fig. 6b, overexpression of ompF had no discernable impact on styrene tolerance. Strains overexpressing tolA showed increased sensitivity to styrene relative to the control, further suggesting that (a) tolA and the Tol-Pal system in general play an important role in styrene tolerance and (b) expression levels of the associated genes are an important consideration to this end. Lastly, although overexpression of plsX resulted in improved growth in the presence of styrene, this behaviour was highly dependent upon the induction strength used. In particular, at high induction (100 µM IPTG), plsX overexpression led to improved tolerance at moderate styrene levels, with ∼17 and 35% increases in final OD600 being achieved at 75 and 125 mg/l styrene, respectively (both relative to the no styrene control). As with the deletion of ompR, however, no difference in the maximum inhibitory styrene threshold was observed. In contrast, low induction (10 µM IPTG) of plsX overexpression showed no major improvements in relative growth at moderate styrene levels, however, significant growth was instead uniquely maintained at higher styrene concentrations (i.e. 74 ± 5% relative growth compared to no styrene control at 175 mg/l styrene). This suggests that plsX is a promising target for tolerance engineering strategies, with optimization of its expression level clearly being an important facet of any future design. Future work will focus on characterizing role of these genes during styrene production to determine if and to what extent their overall importance is likewise conserved.
Fig. 6.
(a) Relative growth of wild-type E. coli BW25113 (control; solid, light blue) and the BW25113 ΔompR mutant (red, striped). (b) Relative growth of BW25113 harbouring pCA24N plasmid (control; solid, light blue) BW25113 harbouring appropriate plasmids used to overexpress ompF (10 µM IPTG; diagonal striped, beige), plsX (10 and 100 µM IPTG; light green, checkered and dark green, dotted, respectively) and tolA (10 µM IPTG; horizontal striped, pink). For (a) and (b), OD600 were measured following 6 hr of exposure to 75, 125, or 175 mg/l styrene, each relative to a no styrene control. Error bars reported at one standard deviation (n = 3). Dashed lines indicate relative growth of 100% (i.e. no growth difference in the presence versus absence of styrene). * indicates p < .001 and ** indicates p < .005 for two-tailed Student's t-test when compared to the respective control strain (BW25113 or BW25113 pCA24N) at the equivalent concentrations of styrene.
Conclusion
E. coli’s transcriptional response was analyzed following prolonged exposure to styrene, which was either exogenously added or endogenously produced. Most behaviours were conserved between the two exposure modes, including prominent responses such as the up-regulation of phage shock response, DNA damage response and cell envelope-altering genes as well as down-regulation of ribosomal and nucleotide biosynthesis genes. The collective behaviours observed support an inhibition-response model wherein styrene first causes both cell envelope and DNA damage, after which E. coli responds by reducing the activity of DNA biosynthesis/repair, amino acid biosynthesis, protein production, and cell wall biogenesis, while also disrupting normal cell division. At the same time, key resources are instead shifted towards supporting a range of tolerance mechanisms, chiefly including a multi-level strategy focused on strengthening the cell membrane through structural and significant remodelling of the cell membrane; key processes at least partially influenced by ompR, the Tol-Pal system, and plsX. Overall, the collective results suggest that, in response to inhibitory levels of styrene, E. coli shifts its collective behaviours from a ‘growth mode’ towards ‘survival mode’, likely as part of a bet-hedging strategy where current fitness is traded for the prospect of future benefits. Finally, with their specific abilities to influence the properties of the OM, present findings suggest that ompR, tolQRA, and plsX may each represent promising targets for future tolerance engineering efforts.
Supplementary Material
Contributor Information
Michael Machas, Chemical Engineering, School for Engineering of Matter, Transport, and Energy, Arizona State University, Tempe, AZ 85287-6106, USA.
Gavin Kurgan, School of Life Sciences, Arizona State University, Tempe, AZ 85287-6106, USA.
Omar A Abed, Chemical Engineering, School for Engineering of Matter, Transport, and Energy, Arizona State University, Tempe, AZ 85287-6106, USA.
Alyssa Shapiro, Chemical Engineering, University of Michigan, Ann Arbor, MI 48109, USA.
Xuan Wang, School of Life Sciences, Arizona State University, Tempe, AZ 85287-6106, USA.
David Nielsen, Chemical Engineering, School for Engineering of Matter, Transport, and Energy, Arizona State University, Tempe, AZ 85287-6106, USA.
Funding
This work was conducted with support from the National Science Foundation (CBET-1511637, CBET-1903497). RNA-seq analysis was supported by Illumina and Genomic Core of ASU.
Conflict of Interest
The authors declare no conflict of interest.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
References
- Ai C., Kong L. (2018). CGPS: A machine learning-based approach integrating multiple gene set analysis tools for better prioritization of biologically relevant pathways. Journal of Genetics and Genomics 45, 489–504. 10.1016/j.jgg.2018.08.002. [DOI] [PubMed] [Google Scholar]
- Alekshun M. N., Levy S. B. (1999). The mar regulon: Multiple resistance to antibiotics and other toxic chemicals. Trends in Microbiology, 7, 410–413. [DOI] [PubMed] [Google Scholar]
- Alper H., Moxley J., Nevoigt E., Fink G. R., Stephanopoulos G. (2006). Engineering yeast transcription machinery for improved ethanol tolerance and production. Science, 314, 1565–1568. 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- Alper H., Stephanopoulos G. (2007). Global transcription machinery engineering: A new approach 548 for improving cellular phenotype. Metabolic Engineering, 9, 258–267. 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Alsaker K. V., Paredes C., Papoutsakis E. T. (2010). Metabolite stress and tolerance in the production of biofuels and chemicals: Gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnology and Bioengineering, 105, 1131–1147. 10.1002/bit.22628. [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biology, 11, R106. 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J., Forst S. A., Zhao K., Inouye M., Delihas N. (1989). The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. Journal of Biological Chemistry, 264, 17961–17970. [PubMed] [Google Scholar]
- Antunes-Madeira M. C., Madeira V. M. (1989). Membrane fluidity as affected by the insecticide lindane. Biochimica et Biophysica Acta (BBA)—Biomembranes, 982, 161–166. [DOI] [PubMed] [Google Scholar]
- Aono R., Kobayashi H. (1997). Cell surface properties of organic solvent-tolerant mutants of Escherichia coli K-12. Applied and Environmental Microbiology, 63, 3637–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold C. N., McElhanon J., Lee A., Leonhart R., Siegele D. A. (2001). Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. Journal of Bacteriology, 183, 2178–2186. 10.1128/JB.183.7.2178-2186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babel H., Krömer J. O. (2020). Evolutionary engineering of E. coli MG1655 for tolerance against isoprenol. Biotechnology for Biofuels, 13, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Braun V., Rehn K. (1969). Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. European Journal of Biochemistry FEBS, 10, 426–438. [DOI] [PubMed] [Google Scholar]
- Braun V., Wolff H. (1975). Attachment of lipoprotein to murein (peptidoglycan) of Escherichia coli in the presence and absence of penicillin FL 1060. Journal of Bacteriology, 123, 888–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosse A., Korobeinikova A., Gottesman S., Guillier M. (2016). Unexpected properties of sRNA promoters allow feedback control via regulation of a two-component system. Nucleic Acids Res., 44, 9650–9666. 10.1093/nar/gkw642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsen M. P., Liao J. C. (2009). An integrated network approach identifies the isobutanol response network of Escherichia coli. Molecular Systems Biology, 5, 277. 10.1038/msb.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui le M., Lee J. Y., Geraldi A., Rahman Z., Lee J. H., Kim S. C. (2015). Improved n-butanol tolerance in Escherichia coli by controlling membrane related functions. Journal of Biotechnology, 204, 33–44. 10.1016/j.jbiotec.2015.03.025. [DOI] [PubMed] [Google Scholar]
- Calhoun L. N., Kwon Y. M. (2011). Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: A review. Journal of Applied Microbiology 110, 375–386. 10.1111/j.1365-2672.2010.04890.x. [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Winardhi R. S., Morgan L. K., Yan J., Kenney L. J. (2017). Non-canonical activation of OmpR drives acid and osmotic stress responses in single bacterial cells. Nature Communications, 8, 1587. 10.1038/s41467-017-02030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Reinhardt M., Neris N., Kerns L., Mansell T. J., Jarboe L. R. (2018). Lessons in membrane engineering for octanoic acid production from environmental Escherichia coli Isolates. Applied and Environmental Microbiology, 19, 84. 10.1128/AEM.01285-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. P., Levy S. B., Foulds J., Rosner J. L. (1993). Salicylate induction of antibiotic resistance in Escherichia coli: Activation of the mar operon and a mar-independent pathway. Journal of Bacteriology, 175, 7856–7862. 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T., Lewis K., Vulić M. (2009). SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genetics, 5, e1000760. 10.1371/journal.pgen.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S., Lutkenhaus J. (2017). Assembly and activation of the Escherichia coli divisome. Molecular Microbiology, 105, 177–187. 10.1111/mmi.13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan A. J. F. (2018). Bacterial outer membrane constriction. Molecular Microbiology, 107, 676–687. 10.1111/mmi.13908. [DOI] [PubMed] [Google Scholar]
- Egler M., Grosse C., Grass G., Nies D. H. (2005). Role of the extracytoplasmic function protein family sigma factor RpoE in metal resistance of Escherichia coli. Journal of Bacteriology, 187, 2297–2307. 10.1128/JB.187.7.2297-2307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson K. E., Winkler J. D., Nguyen D. T., Gill R. T., Chatterjee A. (2017). The tolerome: A database of transcriptome-level contributions to diverse Escherichia coli resistance and tolerance phenotypes. ACS Synthetic Biology, 6, 2302–2315. 10.1021/acssynbio.7b00235. [DOI] [PubMed] [Google Scholar]
- Finkel S. E. (2006). Long-term survival during stationary phase: Evolution and the GASP phenotype. Nature Reviews Microbiology, 4, 113–120. 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- Flores-Kim J., Darwin A. J. (2016). The phage shock protein response. Annual Review of Microbiology, 70, 83–101. 10.1146/annurev-micro-102215-095359. [DOI] [PubMed] [Google Scholar]
- Fonville N. C., Bates D., Hastings P. J., Hanawalt P. C., Rosenberg S. M. (2010). Role of RecA and the SOS response in thymineless death in Escherichia coli. PLoS Genetics, 6, e1000865. 10.1371/journal.pgen.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bernardo J., Dunlop M. J. (2013). Tunable stochastic pulsing in the Escherichia coli multiple antibiotic resistance network from interlinked positive and negative feedback loops. PLoS Computational Biology, 9, e1003229. 10.1371/journal.pcbi.1003229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glebes T. Y., Sandoval N. R., Reeder P. J., Schilling K. D., Zhang M., Gill R. T. (2014). Genome-wide 622 mapping of furfural tolerance genes in Escherichia coli. PLoS One, 9, e87540. 10.1371/journal.pone.0087540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray A. N., Egan A. J., Van't Veer I. L., Verheul J., Colavin A., Koumoutsi A., Biboy J., Altelaar A. F., Damen M. J., Huang K. C., Simorre J. P., Breukink E., den Blaauwen T., Typas A., Gross C. A., Vollmer W. (2015). Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. Elife, 4, e07118. 10.7554/eLife.07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M., Gottesman S. (2006). Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Molecular Microbiology, 59, 231–247. 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- Guo M. S., Gross C. A. (2014). Stress-induced remodeling of the bacterial proteome. Current Biology, 24, R424–R434. 10.1016/j.cub.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Chin M., Nusbaum C., Birren B. W., Livny J. (2012). How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes? BMC Genomics [Electronic Resource], 13, 734. 10.1186/1471-2164-13-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Dorsey-Oresto A., Malik M., Wang J. Y., Drlica K., Zhao X., Lu T. (2010). Escherichia coli genes that reduce the lethal effects of stress. BMC Microbiology [Electronic Resource], 10, 35. 10.1186/1471-2180-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heipieper H. J., Meinhardt F., Segura A. (2003). The cis–trans isomerase of unsaturated fatty acids in Pseudomonas and Vibrio: Biochemistry, molecular biology and physiological function of a unique stress adaptive mechanism. FEMS Microbiology Letters, 229, 1–7. [DOI] [PubMed] [Google Scholar]
- Horinouchi T., Tamaoka K., Furusawa C., Ono N., Suzuki S., Hirasawa T., Yomo T., Shimizu H. (2010). Transcriptome analysis of parallel-evolved Escherichia coli strains under ethanol stress. BMC Genomics [Electronic Resource], 11, 579. 10.1186/1471-2164-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T., de Vlieg J., Alkema W. (2008). BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics [Electronic Resource], 9, 488. 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janion C. (2008). Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. International Journal of Biological Sciences, 4, 338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarboe L. R., Zhang X., Wang X., Moore J. C., Shanmugam K. T., Ingram L. O. (2010). Metabolic engineering for production of biorenewable fuels and chemicals: Contributions of synthetic biology. Journal of Biomedicine and Biotechnology, 2010, 1. 10.1155/2010/761042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D., Gu B., Xiong D., Huang G., Huang X., Liu L., Xiao J. (2018). A transcriptomic analysis of Saccharomyces cerevisiae under the stress of 2-phenylethanol. Current Microbiology, 75, 1068–1076. 10.1007/s00284-018-1488-y. [DOI] [PubMed] [Google Scholar]
- Junker F., Ramos J. L. (1999). Involvement of the cis/trans isomerase Cti in solvent resistance of Pseudomonas putida DOT-T1E. Journal of Bacteriology, 181, 5693–5700/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H. (2005). Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Research, 12, 291–299. 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Kitagawa M., Miyakawa M., Matsumura Y., Tsuchido T. (2002). Escherichia coli small heat shock proteins, IbpA and IbpB, protect enzymes from inactivation by heat and oxidants. European Journal of Biochemistry FEBS, 269, 2907–2917. [DOI] [PubMed] [Google Scholar]
- Lennen R. M., Kruziki M. A., Kumar K., Zinkel R. A., Burnum K. E., Lipton M. S., Hoover S. W., Ranatunga D. R., Wittkopp T. M., Marner W. D., Pfleger B. F. (2011). Membrane stresses induced by overproduction of free fatty acids in Escherichia coli. Applied and Environmental Microbiology, 77, 8114–8128. 10.1128/AEM.05421-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennen R. M., Pfleger B. F. (2013). Modulating membrane composition alters free fatty acid tolerance in Escherichia coli. PLoS One, 8, e54031. 10.1371/journal.pone.0054031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Xie D., Frost J. W. (2005). Benzene-free synthesis of catechol: Interfacing microbial and chemical catalysis. Journal of the American Chemical Society, 127, 2874–2882. 10.1021/ja045148n. [DOI] [PubMed] [Google Scholar]
- Lian J., McKenna R., Rover M. R., Nielsen D. R., Wen Z., Jarboe L. R. (2016). Production of biorenewable styrene: Utilization of biomass-derived sugars and insights into toxicity. Journal of Industrial Microbiology & Biotechnology, 43, 595–604. 10.1007/s10295-016-1734-x. [DOI] [PubMed] [Google Scholar]
- Liang L., Liu R., Foster K. E. O., AlakshChoudhury, Cook S., Cameron J., Srubar W. V. 3rd, Gill R. T. (2019). Genome engineering of E. coli for improved styrene production and polymerization. Metabolic Engineering. 10.1016/j.ymben.2019.09.007. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W. (2014). featureCounts: An efficient general purpose program for 679 assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lin S., Liang R., Zhang T., Yuan Y., Shen S., Ye H. (2017). Microarray analysis of the transcriptome of the Escherichia coli (E. coli) regulated by cinnamaldehyde (CMA). 28, 500–515. 10.1080/09540105.2017.1300875. [DOI] [Google Scholar]
- Lou Z., Wang H., Rao S., Sun J., Ma C., Li J. (2012). p-Coumaric acid kills bacteria through dual damage mechanisms. Food Control, 25, 550–554. [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. J., Zhang Y. M., Grimes K. D., Qi J., Lee R. E., Rock C. O. (2006). Acyl-phosphates initiate membrane 689 phospholipid synthesis in Gram-positive pathogens. Molecular Cell, 23, 765–772. 10.1016/j.molcel.2006.06.030. [DOI] [PubMed] [Google Scholar]
- Machas M., Kurgan G., Jha A. K., Flores A., Schneider A., Coyle S., Varman A. M., Wang X., Nielsen D. R. (2019). Emerging tools, enabling technologies, and future opportunities for the bioproduction of aromatic chemicals. Journal of Chemical Technology and Biotechnology, 94, 38–52. 10.1002/jctb.5762. [DOI] [Google Scholar]
- Machas M., McKenna R., Nielsen D. R. (2016). Expanding upon styrene biosynthesis to engineer a novel route to 2-phenylethanol. Biotechnology Journal 10.1002/biot.201700310. [DOI] [PubMed] [Google Scholar]
- Manganelli R., Gennaro M. L. (2017). Protecting from envelope stress: Variations on the Phage-Shock-Protein Theme. Trends in Microbiology, 25, 205–216. 10.1016/j.tim.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. G., Bartlett E. S., Rosner J. L., Wall M. E. (2008). Activation of the Escherichia coli marA/soxS/rob regulon in response to transcriptional activator concentration. Journal of Molecular Biology, 380, 278–284. 10.1016/j.jmb.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T., Tanaka T., Kondo A. (2017). Engineering metabolic pathways in Escherichia coli for constructing a “microbial chassis” for biochemical production. Bioresource Technology, 245, 1362–1368. [DOI] [PubMed] [Google Scholar]
- McKenna R., Nielsen D. R. (2011). Styrene biosynthesis from glucose by engineered E. coli. Metabolic Engineering, 13, 544–554. 10.1016/j.ymben.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Mingardon F., Clement C., Hirano K., Nhan M., Luning E. G., Chanal A., Mukhopadhyay A. (2015). Improving olefin tolerance and production in E. coli using native and evolved AcrB. Biotechnology and Bioengineering, 112, 879–888. 10.1002/bit.25511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty J. J., Lesnefsky A. A., Lin F., Chen Y., Zaroff T. A., Veloso A. B., Xie B., McConnell C. A., Ward R. J., Schwartz D. R., Rouillard J. M., Gao Y., Gulari E., Lin X. N. (2011). Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microbial Cell Factories, 10, 18. 10.1186/1475-2859-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. M., Silhavy T. J. (2019). Envelope stress responses: Balancing damage repair and toxicity. Nature Reviews Microbiology, 17, 417–428. 10.1038/s41579-019-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Mizushima S. (1987). Isolation and characterization of deletion mutants of ompR and envZ, regulatory genes for expression of the outer membrane proteins OmpC and OmpF in Escherichia coli. Journal of Biochemistry, 101, 387–396. 10.1093/oxfordjournals.jbchem.a121923. [DOI] [PubMed] [Google Scholar]
- Molina-Santiago C., Udaondo Z., Gómez-Lozano M., Molin S., Ramos J. L. (2017). Global transcriptional response of solvent-sensitive and solvent-tolerant Pseudomonas putida strains exposed to toluene. Environmental Microbiology, 19, 645–658. 10.1111/1462-2920.13585. [DOI] [PubMed] [Google Scholar]
- Nikaido H. (1989). Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrobial Agents and Chemotherapy, 33, 1831–1836. 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda S., Kondo A. (2017). Recent advances in microbial production of aromatic chemicals and derivatives. Trends in Biotechnology, 35, 785–796. 10.1016/j.tibtech.2017.05.006. [DOI] [PubMed] [Google Scholar]
- Pichler H., Emmerstorfer-Augustin A. (2018). Modification of membrane lipid compositions in single-727 celled organisms—From basics to applications. Methods (San Diego, CA), 147, 50–65. 10.1016/j.ymeth.2018.06.009. [DOI] [PubMed] [Google Scholar]
- Pomposiello P. J., Bennik M. H., Demple B. (2001). Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. Journal of Bacteriology, 183, 3890–3902. 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt L. A., Hsing W., Gibson K. E., Silhavy T. J. (1996). From acids to osmZ: Multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Molecular Microbiology, 20, 911–917. [DOI] [PubMed] [Google Scholar]
- Randall L. P., Woodward M. J. (2002). The multiple antibiotic resistance (mar) locus and its significance. Research in Veterinary Science, 72, 87–93. 10.1053/rvsc.2001.0537. [DOI] [PubMed] [Google Scholar]
- Rau M. H., Bojanovič K., Nielsen A. T., Long K. S. (2015). Differential expression of small RNAs under chemical stress and fed-batch fermentation in E. coli. BMC Genomics [Electronic Resource], 16, 1051. 10.1186/s12864-015-2231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau M. H., Calero P., Lennen R. M., Long K. S., Nielsen A. T. (2016). Genome-wide Escherichia coli stress response and improved tolerance towards industrially relevant chemicals. Microbial Cell Factories, 15, 176. 10.1186/s12934-016-0577-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes L. H., Almario M. P., Kao K. C. (2011). Genomic library screens for genes involved in n-butanol tolerance in Escherichia coli. PLoS One, 6, e17678. 10.1371/journal.pone.0017678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Moya M., Gonzalez R. (2015). Proteomic analysis of the response of Escherichia coli to short-chain fatty acids. Journal of Proteomics, 122, 86–99. 10.1016/j.jprot.2015.03.033. [DOI] [PubMed] [Google Scholar]
- Röttig A., Steinbüchel A. (2013). Acyltransferases in bacteria. Microbiology and Molecular Biology Reviews, 77, 277–321. 10.1128/MMBR.00010-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley G., Spector M., Kormanec J., Roberts M. (2006). Pushing the envelope: Extracytoplasmic 749 stress responses in bacterial pathogens. Nature Reviews Microbiology, 4, 383–394. 10.1038/nrmicro1394. [DOI] [PubMed] [Google Scholar]
- Rutherford B. J., Dahl R. H., Price R. E., Szmidt H. L., Benke P. I., Mukhopadhyay A., Keasling J. D. (2010). Functional genomic study of exogenous n-butanol stress in Escherichia coli. Applied and Environmental Microbiology, 76, 1935–1945. 10.1128/AEM.02323-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajed T., Marcu A., Ramirez M., Pon A., Guo A. C., Knox C., Wilson M., Grant J. R., Djoumbou Y., Wishart D. S. (2016). ECMDB 2.0: A richer resource for understanding the biochemistry of E. coli. Nucleic Acids Research, 44, D495–501. 10.1093/nar/gkv1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval N. R., Papoutsakis E. T. (2016). Engineering membrane and cell-wall programs for tolerance to toxic chemicals: Beyond solo genes. Current Opinion in Microbiology, 33, 56–66. 10.1016/j.mib.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargentini N. J., Gularte N. P., Hudman D. A. (2016). Screen for genes involved in radiation survival of Escherichia coli and construction of a reference database. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis, 793–794, 1–14. 10.1016/j.mrfmmm.2016.10.001. [DOI] [PubMed] [Google Scholar]
- Schurch N. J., Schofield P., Gierliński M., Cole C., Sherstnev A., Singh V., Wrobel N., Gharbi K., Simpson G. G., Owen-Hughes T., Blaxter M., Barton G. J. (2016). How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA, 22, 839–851. 10.1261/rna.053959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C. C., Finnerty W. R. (1976). Characterization of intracytoplasmic hydrocarbon inclusions from the hydrocarbon-oxidizing Acinetobacter species HO1-N. Journal of Bacteriology, 127, 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherkhanov S., Korman T. P., Bowie J. U. (2014). Improving the tolerance of Escherichia coli to medium-chain fatty acid production. Metabolic Engineering, 25, 1–7. 10.1016/j.ymben.2014.06.003. [DOI] [PubMed] [Google Scholar]
- Sikkema J., de Bont J. A., Poolman B. (1994). Interactions of cyclic hydrocarbons with biological membranes. Journal of Biological Chemistry, 269, 8022–8028. [PubMed] [Google Scholar]
- Sikkema J., de Bont J. A., Poolman B. (1995). Mechanisms of membrane toxicity of hydrocarbons. Microbiological Reviews, 59, 201–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subhash S., Kanduri C. (2016). GeneSCF: A real-time based functional enrichment tool with support for multiple organisms. BMC Bioinformatics, 17, 365. 10.1186/s12859-016-1250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmann K., Ćudić E., Hammer E., Hunke S. (2016). Molecular and proteome analyses highlight the importance of the Cpx envelope stress system for acid stress and cell wall stability in Escherichia coli. Microbiologyopen, 5, 582–596. 10.1002/mbo3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z., Black W., Yoon J. M., Shanks J. V., Jarboe L. R. (2017). Improving Escherichia coli membrane integrity and fatty acid production by expression tuning of FadL and OmpF. Microbial Cell Factories, 16, 38. 10.1186/s12934-017-0650-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z., Khakbaz P., Chen Y., Lombardo J., Yoon J. M., Shanks J. V., Klauda J. B., Jarboe L. R. (2017). Engineering Escherichia coli membrane phospholipid head distribution improves 785 tolerance and production of biorenewables. Metabolic Engineering, 44, 1–12. 10.1016/j.ymben.2017.08.006. [DOI] [PubMed] [Google Scholar]
- Tan Z., Yoon J. M., Nielsen D. R., Shanks J. V., Jarboe L. R. (2016). Membrane engineering via trans unsaturated fatty acids production improves Escherichia coli robustness and production of biorenewables. Metabolic Engineering, 35, 105–113. 10.1016/j.ymben.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Thompson B., Machas M., Nielsen D. R. (2015). Creating pathways towards aromatic building blocks 791 and fine chemicals. Current Opinion in Biotechnology, 36, 1–7. 10.1016/j.copbio.2015.07.004. [DOI] [PubMed] [Google Scholar]
- Tribe D. E. (1987). Novel microorganism and method. U.S. Patent No.4,681,852.
- VanBogelen R. A., Acton M. A., Neidhardt F. C. (1987). Induction of the heat shock regulon does not produce thermotolerance in Escherichia coli. Genes & Development, 1, 525–531. [DOI] [PubMed] [Google Scholar]
- Van Dyk T. K., Templeton L. J., Cantera K. A., Sharpe P. L., Sariaslani F. S. (2004). Characterization of the Escherichia coli AaeAB efflux pump: A metabolic relief valve? Journal of Bacteriology, 186, 7196–7204. 10.1128/JB.186.21.7196-7204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Tah A., Martinez L. M., Hernandez-Chavez G., Rocha M., Martinez A., Bolivar F., Gosset G. (2015). Production of cinnamic and p-hydroxycinnamic acid from sugar mixtures with engineered Escherichia coli. Microbial Cell Factories, 14, 6. 10.1186/s12934-014-0185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvalingam J., Hernandez-Doria J. D., Holley R. A. (2013). Examination of the genome-wide transcriptional response of Escherichia coli O157:H7 to cinnamaldehyde exposure. Applied and Environmental Microbiology, 79, 942–950. 10.1128/AEM.02767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. C., Morgan L. K., Godakumbura P., Kenney L. J., Anand G. S. (2012). The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO Journal, 31, 2648–2659. 10.1038/emboj.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., Kong L., Gao G., Li C. Y., Wei L. (2011). KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research, 39, W316–W322. 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S. H., Lee E. G., Das A., Lee S. H., Li C., Ryu H. K., Choi M. S., Seo W. T., Kim S. W. (2007). Enhanced vanillin production from recombinant E. coli using NTG mutagenesis and adsorbent resin. Biotechnology Progress, 23, 1143–1148. 10.1021/bp070153r. [DOI] [PubMed] [Google Scholar]
- Yoshimura M., Oshima T., Ogasawara N. (2007). Involvement of the YneS/YgiH and PlsX proteins in phospholipid biosynthesis in both Bacillus subtilis and Escherichia coli. BMC Microbiology [Electronic Resource], 7, 69. 10.1186/1471-2180-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung P. Y., Grasso L. L., Mohidin A. F., Acerbi E., Hinks J., Seviour T., Marsili E., Lauro F. M. (2016). Global transcriptomic responses of Escherichia coli K-12 to volatile organic compounds. Scientific Reports, 6, 19899. 10.1038/srep19899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Wang X., Templeton L. J., Smulski D. R., LaRossa R. A., Storz G. (2001). DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. Journal of Bacteriology, 183, 4562–4570. 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.