Abstract
Simple Summary
When two different species or subspecies of animals have progeny, we speak about hybrid organisms, which present a mixture of the genetic characteristics of their parents. This phenomenon occurs in nature in a common way, although most of the time the hybrids between species are sterile. In this work, the venom characteristics of hybrids from two species of rattlesnakes were studied: Crotalus aquilus (father) and Crotalus polystictus (mother), both endemic to central Mexico. Scale numbers (phenetic analysis) and venom protein were compared between hybrids (females and males), biological parents, and adult individuals of the two species. The presence and activity of the main types of enzymes in these venoms were analyzed, and the lethal dose was determined in mice. Through the phenetic analysis, it was observed that the hybrids were more similar to C. polystictus (mother), the presence of proteins and enzymatic activity resulted in a combination of the two species, but the lethality of the venom was greater in the hybrids. These results allow us to learn more about the way in which the hybridization phenomenon influences the characteristics of rattlesnake venom. Some of the applications of this knowledge could be used to develop more effective antidotes.
Abstract
Hybridization is defined as the interbreeding of individuals from two populations distinguishable by one or more heritable characteristics. Snake hybridization represents an interesting opportunity to analyze variability and how genetics affect the venom components between parents and hybrids. Snake venoms exhibit a high degree of variability related to biological and biogeographical factors. The aim of this work is to analyze the protein patterns and enzymatic activity of some of the main hemotoxic enzymes in snake venoms, such as serine proteases (trypsin-like, chymotrypsin-like, and elastase-like), metalloproteases, hyaluronidases, and phospholipase A2. The lethal dose of 50 (LD50) of venom from the Crotalus aquilus (Cabf) and Crotalus polystictus (Cpbm) parents and their hybrids in captivity was determined, and phenetic analysis is also conducted, which showed a high similarity between the hybrids and C. polystictus. The protein banding patterns and enzymatic activity analyze by zymography resulted in a combination of proteins from the parental venoms in the hybrids, with variability among them. In some cases, the enzymatic activity is higher in the hybrids with a lower LD50 than in the parents, indicating higher toxicity. These data show the variability among snake venoms and suggest that hybridization is an important factor in changes in protein concentration, peptide variability, and enzymatic activity that affect toxicity and lethality.
Keywords: rattlesnake, venom enzymes, proteases, hyaluronidases, phospholipase A2, hybridization
1. Introduction
Snake venoms are a mixture of proteins, peptides, and toxins that vary extensively between and within snake species [1]. This variability is widely reported at different taxonomic levels, from the family level down to variations between organisms of the same species. They are also related to geographical location or type of feeding and to other variables, such as age-dependent changes, sexual dimorphism, and factors associated with feeding type, which can affect the degree of the venom’s lethality; however, the mechanisms that cause variation in venoms remain mostly unknown [2,3]. Nevertheless, the processes of gene duplication and positive selection appear to be the predominant mechanisms for generating diversity in snake venoms [1]. The variation in the composition of venoms is of particular interest in terms of their mechanisms of action [4,5], as well as their medical implications [6] and consequences for the efficacy of snakebite treatments [1]. Another phenomenon associated with gene variation is hybridism [7,8], which can be defined as the interbreeding of individuals from two populations, or groups of populations, that are distinguishable by one or more heritable characteristics, or the incorporation of genes from differentiated populations into another. This phenomenon has been reported in several organisms as an adaptive variable [8,9], usually associated with geographical areas where populations of nearby species are found [10] and could be related to the mechanism that contributes to speciation [11,12,13]. In the case of snakes, hybridism is also associated with variability in venom composition [2,3]. Hybridization in snakes has been reported based on analyses of morphological and genotype features [11]. In rattlesnakes, hybridization was first documented by Bailey [14], who identified a hybrid case between Crotalus horridus and Sistrurus catenatus by conducting a thorough examination of scalation, body proportions, and color patterns. There are just a few works reporting the biological and enzymatic activities of the venoms in hybrids from C. atrox and C. scutulatus [15], Bothrops erythromelas and B. neuwiedi [11], C. s. scutulatus and C. oreganus helleri [16], C. viridis and C. s. scutulatus [3], and Protobothrops flavoviridis and P. elegans [17].
During 2006 and 2007, seven hybrid organisms were born from two hybridization events that occurred between a captive individual of Crotalus aquilus, the biological father (Cabf), and Crotalus polystictus, the biological mother (Cpbm), at the Animal Resource Facility (Autonomus University of Querétaro, México). Both species are endemic to central Mexico [18,19,20]. These rattlesnakes are part of the Viperidae family [21,22], with hemotoxic venoms that act directly on the cells and hemostatic system, causing extensive local lesions, hemorrhages, edema, tissue myonecrosis, and a variety of alterations in blood coagulation [23,24]. Such damage is the result of the activity of several components of the venom, including metalloproteases, serine proteases, hyaluronidases, phospholipase A2, L-amino oxidases, and acetylcholinesterase, as well as non-enzymatic proteins such as type C lectin proteins and other minor components [25,26].
In this work, the main hemotoxic enzymatic activity types (serine proteases, metalloproteases, hyaluronidases, and phospholipase A2) were compared, as well as the LD50 in mice, and the differences between venom patterns were obtained by reverse-phase HPLC, MALDI-TOF-MS. On the contrary, a phenetic analysis, based on the number and morphological characteristics of the scales, was also determined to establish the similarity between the parental species and hybrids.
2. Materials and Methods
2.1. Phenetic Analysis
Captive male Crotalus aquilus and female Crotalus polystictus rattlesnakes and their hybrids born in 2006 and 2007 were housed and cared for in the Animal Resource Facility (Autonomous University of Querétaro, Querétaro, México). Phenetic analysis was performed comparing the number of dorsal spotting and tail band patterns, and the characteristics of the number of scales around the rattle, subcaudal, ventral, supralabial, infralabial, dorsal, and intersupraocular were also counted from the following juvenile hybrids (250–325 mm long) of the first hybridization (four years old) in 2006: Male hybrid 1 (MH1); female hybrid 2 (FH2); female hybrid 3 (FH3); male hybrid 4 (MH4); juvenile hybrids of the second hybridization (three years old) in 2007: Female hybrid 5 (FH5), male hybrid 6 (MH6), and male hybrid 7 (MH7); three males of C. aquilus: Two juveniles, MCa8 and MCa9 (250–325 mm long), and one adult (>450 mm long) that was the biological father (Cabf); and two C. polystictus adults (>450 mm long), a male (MCp10) and the biological mother (Cpbm). A second analysis was performed using the characteristic of the number of scales plus the characteristic of the protein bands, using the two protein bands of Cabf (26 kDa) and Cpbm (31 kDa) from the SDS-PAGE analysis. Each characteristic was taken as an operational taxonomic unit (OTU). These data were used to form a similarity matrix, analyzed by the taxonomic similarity coefficient to clustering similar groups using the unweighted pair group method with the arithmetic mean (UPGMA) method in the NTSYSpc program version 2.2 [27].
2.2. Venom Harvest
The venom used for the determination of serine proteases (trypsin- and chymotrypsin-like), metalloproteases (zymography), and the LD50 was obtained by manual extraction (milking) of 11 rattlesnakes, seven of which were from the four-year-old hybrids of the first hybridization (MH1, FH2, FH3, and MH4) and the three-year-old hybrids; the following three of the second hybridization event (three years old): FH5, MH6, and MH7; the following two male adults of Crotalus aquilus: MCa8 and the biological father Cabf; and two adults of C. polystictus, MCp10 and the biological mother Cpbm. The venom was immediately stored on ice, lyophilized, and then kept at −70 °C until use, only during the first three days after milking. In 2017, Cabf, Cpbm, and two hybrids (FH2 and FH5) died, making it necessary to determine the enzymatic activity of elastases, metalloproteases, phospholipase A2, and hyaluronidases from the venom obtained from five adult hybrids, namely, three 11-year-old hybrids (MH1, FH3, and MH4) and two 10-year-old hybrids (MH6 and MH7), as well as the pooled venom of four male adults of Crotalus aquilus, MCa-PV, and pooled venom of four female adults of Crotalus polystictus, FCp-PV.
2.3. Polyacrylamide Gel Electrophoresis
The lyophilized venom was diluted in tridistillated water and the protein concentration was determined [28] using as protein standard BSA (Sigma-Aldrich, catalog number 05470, St. Louis, MO, USA) at different concentrations. SDS-PAGE using 20 µg of venom protein was analyzed under reducing (4% of β-mercaptoethanol and 5 min at boiling water temperature) and non-reducing conditions [29], using a 10% or 12% polyacrylamide gel in a Mini Protean II unit (Bio-Rad Hercules, CA, USA) or a Hoefer MiniVE system (Hoefer, Holliston, MA, USA). The protein bands were stained with Coomassie Blue G250 dye (Bio-Rad, catalog number 1610406, CA, USA), unstained with a 40% and 10% methanol–acetic acid solution, respectively, and imaged on an HP Scanjet 4570c scanner.
2.4. Zymography Assays
2.4.1. Serine Proteases Zymography
Proteolytic activity measured by zymography was detected based on the methodology reported by Ohlsson et al. [30] and Vinokurov et al. [31]. For trypsin and chymotrypsin zymography, after electrophoresis of 50 and 100 µg of venom protein, respectively, the gel was washed with 0.1 M Tris-HCl pH 8 buffer and then a cellulose membrane (Bio-Rad, catalog number 1620112, CA, USA) previously embedded in the corresponding substrate, BApNA (Sigma-Aldrich, catalog number B4875, St. Louis, MO, USA) or SAAPFpNA (Sigma-Aldrich, catalog number S7388, St. Louis, MO, USA), respectively. Both substrates were previously prepared in dimethylformamide and diluted 1:20 (v/v) in 0.1 M Tris-HCl to obtain a final concentration of 1 mM. The membranes were placed on top of the electrophoresis gel and incubated at 37 °C for 2–2.5 h or until bands with yellow coloration appeared. The cellulose membrane was serially washed for 5 min with sodium nitrite (NaNO2) 0.1% in HCl 1 M, ammonium sulfate (NH4SO3NH2) 0.5% in HCl 1 M, and 0.05% N-(1-naftil) ethylenediamine dihydrochloride (C12H14N2 2HCl) in 47.5% ethanol. After incubation, the gels were stained with Coomassie Blue G250 dye and unstained with 10% acetic acid.
2.4.2. Metalloprotease Zymography
An analysis of 20 µg of the venom protein samples was conducted in a 10% or 12% SDS-PAGE gel co-polymerized with 0.12% gelatin (Sigma-Aldrich, catalog number G2500, St. Louis, MO, USA) as the substrate [32]. After electrophoresis, the gel was washed in a 2.5% (v/v) Triton X100 solution for 1 h and later for 30 min in a 0.05 M Tris-HCl pH 7.4 buffer, then finally incubated for 2–3 h in a 0.2 M NaCl, 0.005 M CaCl2, 0.002% (v/v) Triton X-100, 0.001 M cysteine, and 0.05 M Tris-HCl pH 8 buffer. After incubation, the gel was stained with Coomassie Blue G250 dye and then unstained with 40% methanol–10% acetic acid.
2.4.3. PLA2 Zymography
Twenty micrograms of the venom protein samples were run in 10% SDS-PAGE gel under non-reducing conditions; then, the gel was washed for 1 h with 2.0% (v/v) Triton X-100 and 0.5 M Tris-HCl pH 7.4 buffer. The gel was subsequently washed for 30 min in a buffer with 140 mM NaCl, 2.5 mM CaCl2, and 50 mM Tris-HCl pH 7.4 buffer and then incubated for 2.5 h at 37 °C on a 1% agarose (Sigma-Aldrich, catalog number A9539, St. Louis, MO, USA) gel co-polymerized with a solution containing 50 mM Tris-HCl (pH 7.4), 140 mM NaCl, 2.5 mM CaCl2, and 2% egg yolk. Light zones indicate the presence of PLA2 [33].
2.4.4. Hyaluronidases Zymography
A 10% SDS-PAGE gel was co-polymerized with 1 mg/mL of hyaluronic acid (Sigma-Aldric, catalog number H7630, St. Louis, MO, USA), and then 100 µg of the venom protein samples was analyzed via non-reducing electrophoresis. The gel was washed for 30 min with 0.2 M sodium acetate (pH 6), 0.15 M NaCl, and 2.5% (v/v) X-100 Triton buffer solution and subsequently incubated for 2.5–3 h at 37 °C in the same buffer solution without Triton. The gel was stained with alcian blue dye (Sigma-Aldrich, catalog number A5268, St. Louis, MO, USA) at 0.5% (w/v) and subsequently de-stained with a solution containing 40% methanol and 10% acetic acid [34].
2.5. Enzymatic Assays
2.5.1. Serine Proteases
The proteolytic activity was determined according to the method reported by Erlanger et al. [35]. Briefly, 20 μL of Trypsin and chymotrypsin-like substrates BApNA or SAAPFpNA were used at 0.01 M in dimethyl sulfoxide (DMSO), and then 20 μL of the venom sample was added and carried to a final volume of 240 μL in 0.1 M Tris-HCl buffer pH 8. The reaction was incubated for 2 h at 37 °C. For the elastase-like activity, 10 μL of N-methoxysuccinyl-Ala-Ala-Pro-Val p-nitroanilide (Sigma-Aldrich, catalog number M4765, St. Louis, MO, USA) at 0.01 M in DMSO was used and carried to a final volume of 120 μL in 0.1 M Tris-HCl buffer pH 5. Trypsin-like activity was determined using 20 μL of venom sample from a mix of 1 μL of crude venom diluted in 99 μL of distilled water. Chymotrypsin-like activity was determined using 20 μL of the venom sample from a mix of 1 μL of crude venom diluted in 999 μL of distilled water. For elastase-like activity, 50 μg of the protein venom sample was used. The enzymatic activity was expressed as specific activity, calculating the amount of protein in each sample according to the Bradford method. The absorbance of liberated p-nitro aniline of the mixture was measured in a spectrophotometer (Benchmark Plus, Bio-Rad, USA) at 405 nm and 37 °C in a 96-well plate. A unit of proteolytic activity is defined as an increase of 0.01 absorbance at 405 nm per min, while specific peptidase activity is expressed as units of proteolytic activity per microgram of protein venom. The experiments were carried out in triplicate. In each case, bovine trypsin (Sigma-Aldrich, catalog number T1426, St. Louis, MO, USA), bovine chymotrypsin (Sigma-Aldrich, catalog number C4129, St. Louis, MO, USA), and porcine elastase (Sigma-Aldrich, catalog number E1250, St. Louis, MO, USA) were used as positive controls.
2.5.2. Metalloprotease Proteolytic Activity
Proteolytic activity was assayed according to Gutiérrez et al. [36] and Ponce et al. [37] with modifications. Briefly, total enzymatic activity was measured in the first set of tubes where 100 µg of venom protein and 500 µL of 2% (w/v) casein (Sigma-Aldrich, catalog number C3400, St. Louis, MO, USA) were diluted in 0.1 mM Tris–HCl (pH 7.4) and 0.15 M NaCl buffer and carried to a final volume of 600 µL. After 2.5 h of incubation at 37 °C, the reaction was stopped by adding 500 µL of 5% (w/v) trichloroacetic acid (TCA) at 37 °C for 30 min at room temperature. The mixture was centrifuged at 12,000× g for 10 min, and casein hydrolysis was measured from 200 µL of the supernatant at 280 nm. In the second set of tubes, venom samples were incubated with 50 mM EDTA (Sigma-Aldrich, catalog number E9884, St. Louis, MO, USA) for 30 min before casein was added and the same procedure was performed (EDTA inhibits activity of the metalloproteases). The total activity minus the activity in the presence of EDTA represents the activity of metalloproteases. Water instead of venom was used as a negative control and it was subtracted from the venom samples. The specific activity is reported as activity units (AUs) per microgram of AU)/venom protein, where one activity unit corresponded to an increase in absorbance of 0.01 at 280 nm.
2.5.3. Phospholipase A2 (PLA2) Activity
The activity of phospholipase A2 of 100 ng of venom protein was analyzed by a colorimetric assay using an sPLA2 Assay Kit (Cayman Chemical, catalogue number 765,001, Ann Arbor, MI, USA). The assay used diheptanoyl phosphatidylcholine analog as the substrate. After hydrolysis of the substrate at the SN-2 position of the thioester bond by the phospholipases, the free thiols generated were detected using 5,5′-dithio-bis-(2-nitrobenzoic acid) (DTNB). The absorbance increase was measured using a spectrophotometer (Benchmark Plus, Bio-Rad, USA) at 414 nm each min for 10 min. The bee venom PLA2 (Cayman Chemical, catalogue number 765,016, Ann Arbor, MI, USA) was used as the positive control. The PLA2-specific activity is expressed as hydrolyzed substrate per minute per microgram of protein. Water instead of venom was used as a negative control and was subtracted from the venom samples.
2.5.4. Hyaluronidase Activity
Concentrations of 20–200 μg of venom protein were used. A sample of 100 μL of buffer and 100 μL of 1 mg/mL of hyaluronic acid (Sigma-Aldrich, catalog number H7630, St. Louis, MO, USA) as substrate were added to a final volume of 250 μL (buffer volume varies depending on the volume of each sample concentration to be used). After mixing, it was incubated for 15 min at 37 °C, and subsequently, 1 mL of 2.5% hexadecyltrimethylammonium bromide in 2% NaOH was added and allowed to stand for 30 min, which was read at a wavelength of 400 nm in a plate reader (Benchmark Plus, Bio-Rad, USA) [38].
2.6. RP-HPLC
The analysis was conducted using 200 μg of the adult pooled venoms of MCa-PV and FCp-PV and the individual hybrid samples MH1, MH4, MH6, MH7, and FH3 using a C18 column (150 × 4.6 mm; particle size: 3.5 μm) equilibrated with solution A (0.1% trifluoroacetic acid TFA in H2O) using an Agilent 1200 chromatograph (Agilent 1200, Agilent Technologies, CA, USA). Elution was performed at 1 mL/min using a gradient toward solution B (acetonitrile ACN and 0.1% of TFA) as follows: 0% B by 5 min, 0–60% B over 60 min. Absorbance was monitored at 280 nm.
2.7. MALDI-TOF-MS
The adult pooled venom samples of MCa-PV and FCp-PV and the hybrids (MH1, MH4, MH6, MH7, and FH3) were analyzed using 70 µg diluted in 3 µL of sinapinic acid matrix (10 mg/mL in ddH2O), spotted onto MALDI target plates and analyzed using an Ultraflex-TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). The mass analyzer was operated in reflector mode using a 20 kV accelerating voltage to measure the ion abundance. The software Flexcontrol (Bruker Daltonics, Bremen, Germany) was used for data analysis.
2.8. Acute Toxicological Assay
The LD50 of the pooled venoms of four male hybrids (MH-PV; MH1, MH4, MH6, and MH7), three female hybrids (FH-PV; FH2, FH3, and FH5), Cabf, and Cpbm was measured in groups of four mice of the CD-1 strain (18–20 g) by intraperitoneal injection of 0.5 mL of venom doses (0.5, 0.75, 1, 1.5, 2, and 3 μg of venom protein per milliliter of saline water), and a control group injected with isotonic saline solution was included. The venom was kept at −70 °C and was used only during the first three days after milking. The animals were kept under controlled temperature, humidity, and 24 h light-dark cycle conditions. The LD50 was calculated by linear regression analysis of the log10 of the doses versus the percentage of survival within the first 24 h of venom injection. The protocol was approved by the Bioethics Committee of the Natural Sciences Faculty of the Autonomus University of Querétaro, México (01FCN2014). At all times, the ethical guidelines of the Norma Oficial Mexicana NOM-062-ZOO [39] were observed.
3. Results and Discussion
3.1. Phenetic Analysis
C. polystictus appears in the triseriatus group, a small montane rattlesnake species [40], where C. aquilus is included. In analyses (by BEAST) based on gene sequences of mitochondrial DNA, it appears as a clade sister to the C. durissus group; however, analysis by BI (concatenated Bayesian) groups it as a sister of the C. cerastes group and as a sister group of the C. triseriatus group in ML (maximum likelihood) analysis conducted by Reyes-Velazco et al. [41] and by Alencar et al. [22]. This variation indicates that more analyses are needed to establish their phylogenetic relation.
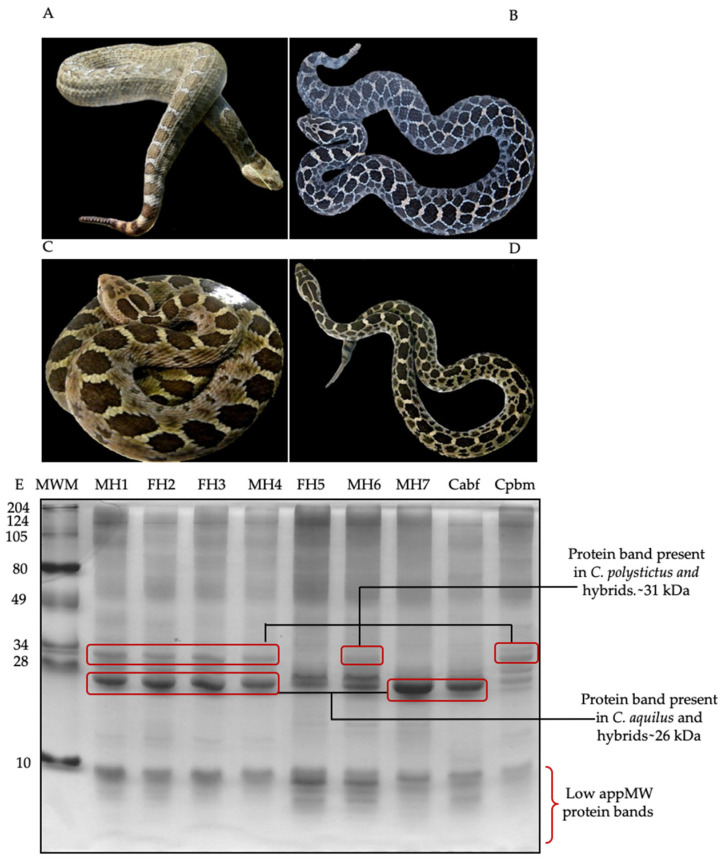
To study the phenetic relationship between C. aquilus, C. polystictus, and the hybrids, the characteristics of the number of scales and two protein bands were used. With respect to scales, the hybrids showed a combination of numbers and types from both parental species, according to data reported by Meik et al. [20] for C. aquilus, and by McCranie [18] for C. polystictus (Table 1). The hybrids presented a mixture of dorsal coloring patterns from the parents, with similarities to one or the other parent. MH1 was similar to Cabf, whereas MH6 was similar to Cpbm (Figure 1A–D).
Table 1.
Comparison between number of scales for C. aquilus, C. polystictus, Cabf, Cpbm, and the hybrids.
| Scales | MCa+ | FCa+ | Cabf | MCp * | FCp * | Cpbm | MHs | FHs |
|---|---|---|---|---|---|---|---|---|
| Dorsal spotting pattern | 24–35 | 26–32 | 27–30 | 30–47 | 30–47 | ND | 32–39 | 31–34 |
| Tail band pattern | 2–4 | 4–6 | 4 | ND | ND | 5 | 4–7 | 3–5 |
| Scales around the rattle | 8–10 | 8–10 | 10 | ND | ND | 12 | 10–12 | 10–12 |
| Subcaudal | 18–24 | 23–28 | 25–26 | 24–29 | 17–25 | 20 | 24–27 | 19–21 |
| Ventral | 141–150 | 136–144 | 143–152 | 161–177 | 167–187 | 165 | 155–158 | 157–164 |
| Supralabial | 11–13 | 10–13 | 11–12 | 12–15 | 12–15 | 14 | 13–14 | 12–14 |
| Infralabial | 11–12 | 10–12 | 11–12 | 11–16 | 11–16 | 14 | 11–14 | 12–14 |
| Interrectal | 20–25 | 20–26 | ND | ND | ND | ND | ND | ND |
| Middle body scales | 21–24 | 21–23 | 23 | 25–28 | 25–28 | 25 | 25–26 | 25 |
| Intersupraocular | 3 | 2–4 | ND | ND | ND | 3 | ND | ND |
Data of the number of scales of C. aquilus: Males (MCa+) and females (FCa+) [15] and the C. aquilus biological father (Cabf); C. polystictus: Males (MCp *) and females (FCp *) [18], the C. polystictus biological mother (Cpbm), four male hybrids (MHs), and three female hybrids (FHs). * Not determined (ND).
Figure 1.
Dorsal banding pattern and protein profile of 20 µg of venom in a 12% SDS-PAGE. (A) C. aquilus (Cabf), (B) C. polystictus (Cpbm), (C) male hybrid 1 (MH1), (D) male hybrid 6 (MH6), (E) protein pattern. Hybrids of the first hybridization event (four years old): MH1, FH2, FH3, and MH4; hybrids of the second hybridization event (three years old): FH5, MH6, and MH7; C. aquilus biological father (Caqbf); C. polystictus biological mother (Cpbm). Molecular weight markers (MWMs) are indicated on the left side. M, male; F, female.
With respect to the venom electrophoretic profiles, some differences in the protein banding patterns were observed. Cpbm presented four protein bands around 28–34 kDa, while Cabf presented only two. The protein pattern between the hybrids MH1, FH2, FH3, MH4, and MH6 and Cpbm was similar, showing a protein band of an apparent molecular weight of 31 kDa (Figure 1E). On the contrary, the protein pattern of FH5 and MH7 was more similar to that of Cabf, presenting a protein band with an apparent molecular weight of ~26 kDa, also present in the other hybrid venoms but not in Cpbm. The hybrids had a combination of protein bands from both parental species, including ~31 and ~26 kDa bands, which were present in most of the hybrids (MH1, FH2, FH3, MH4, and MH6) and apparently not shared among the biological parents. FH5, MH6, and MH7 (second hybridization event), unlike the other hybrids, showed a greater number of low molecular weight bands—less than 7 kDa.
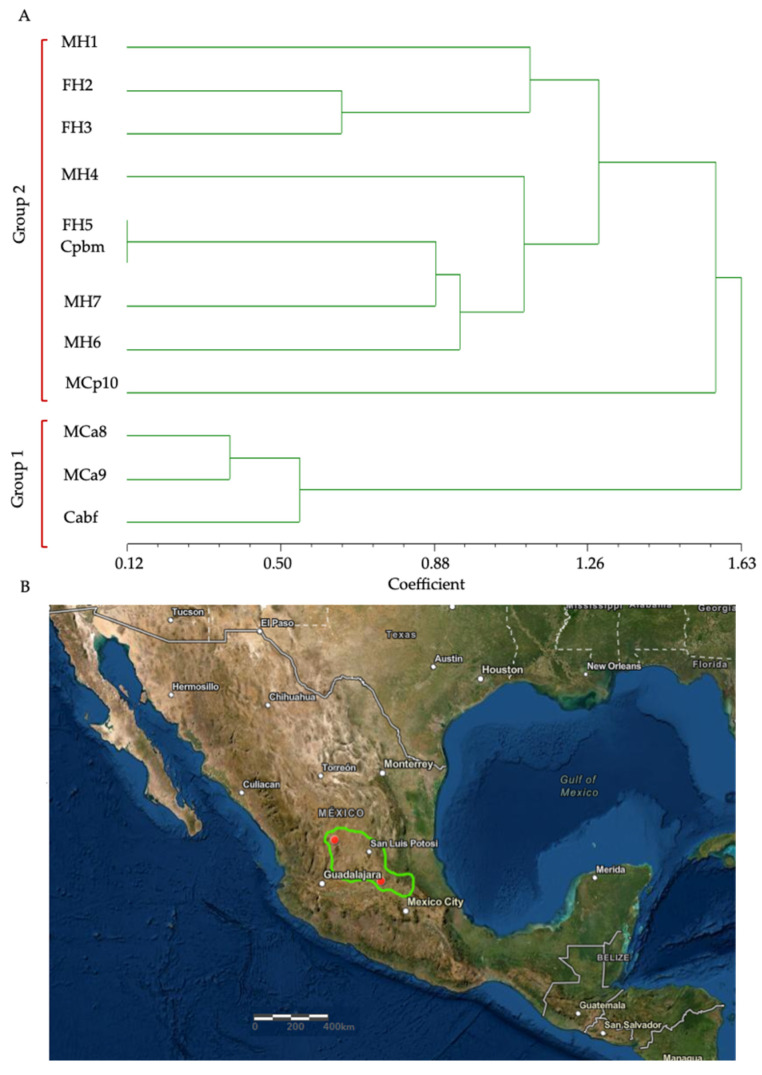
In the phenogram obtained by using the characteristics of the number of scales and two protein bands of ~26 and ~31 kDa specific for the parental Cabf and Cpbm venoms, two main groups were identified. The first was conformed to by all of the hybrids and the C. polystictus adults (MCp10 and Cpbm). The second group was formed by the C. aquilus adults (MCa8, MCa9, and Cabf). FH5 was more similar to the biological mother (Cpbm) than to C. aquilus (Figure 2A). However, it is important to observe that even though MH7 and Cabf were clearly similar in their protein patterns, they were divided into different groups in the cluster analysis. This could be due to the cluster analysis being conducted using only the following two differential protein bands: Cabf (26 kDa) and Cpbm (31 kDa), which were not shared between them. All of the other protein bands were not included in the analysis due to being shared between the parental venoms.
Figure 2.
Phenogram of C. aquilus, C. polystictus, and hybrid rattlesnakes. The phenetic diagram was constructed using the UPGMA method [27] based on the number of scales and protein bands. (A) Hybrids of the first hybridization event (four years old): MH1, FH2, FH3, and MH4; hybrids of the second hybridization event (three years old): FH5, MH6, and MH7; males of C. aquilus (MCa8 and MCa9) and C. aquilus biological father (Cabf); C. polystictus male (MCp10) and C. polystictus biological mother (Cpbm). M, male; F, female. (B) possible sympatric distribution of C. aquilus and C. Polystictus in central Mexico.
The phylogenetic analyses using the morphological characteristics (scales and gene sequences) [41,42] showed results indicative of high similarity between C. aquilus and C. polystictus, and the hybridization events indicate that the reproductive isolation between C. aquilus and C. polystictus is not complete [43] and that they could be closely related. In addition, their distribution converged in central Mexico, where, in some areas, C. aquilus and C. polystictus are sympatrically distributed [18,19], which could represent a possible natural hybridization zone (Figure 2B).
3.2. Enzymatic Activity and Zymography
3.2.1. Snake Venom Serine Proteases (SVSPs)
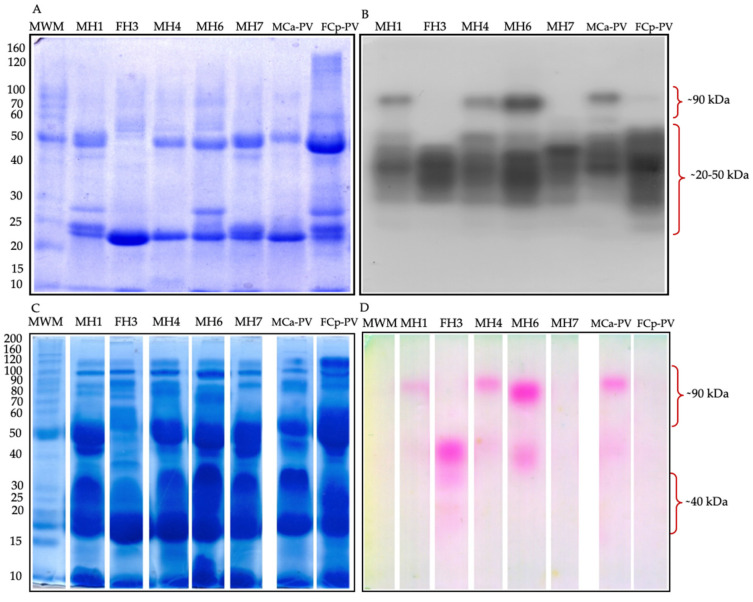
The zymography for both serine protease types showed bands in the range of ~20 to ~90 kDa for the hybrids and were similar to the previously reported protein pattern banding of MCa-PV and FCp-PV [44]. In trypsin-like zymography, the hybrids MH1, MH4, and MH6 showed a similar band pattern to Ca, presenting a protease band of ~90 kDa, while the hybrids FH3 and MH7 and FCp-PV lacked this protease band (Figure 3A,B). A high molecular weight chymotrypsin-like activity was detected by zymography with a band of ~90 kDa that was present only in the hybrids MH1 and MH4 and in MCa-PV. FH3 showed a protease of ~35 kDa, while MH6 presented a protease of ~85 kDa, both of which were not detected in MCa-PV, FCp-PV, or the other hybrids. MH7 and Cp showed no chymotrypsin-like activity by zymography (Figure 3C,D). For both types of zymography, the hybrids MH1 and MH4 shared a protease band of ~90 kDa, not shown in C. polystictus, perhaps inherited from C. aquilus.
Figure 3.
Venom serine protease-like zymography. (A) 50 µg of venom protein in a 10% SDS-PAGE gel. (B) Trypsin-like activity was determined using BApNA as the substrate and incubated for 2.5 h. (C) 100 µg of venom protein in a 10% SDS-PAGE gel. (D) Chymotrypsin-like proteolytic activity determined using SAAFpNA as the substrate and incubated for 2.5 h. Hybrids of the first hybridization event (11 years old): MH1, FH3, and MH4; hybrids of the second hybridization event (10 years old): MH6 and MH7; pooled venom of four male organisms of C. aquilus (MCa-PV), pooled venom of four female organisms of C. polystictus (FCp-PV). MWM, molecular weight marker.
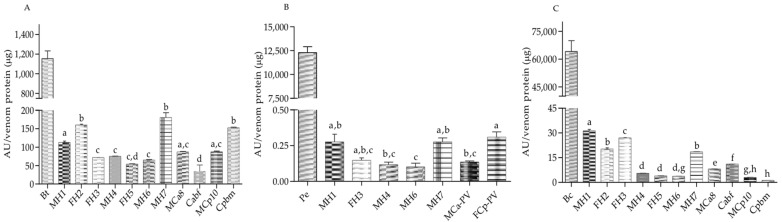
For trypsin-like enzymatic activity, both MCa8 and MCp10 (excluding Cabf with lower activity) showed similar activities. Cpbm showed the highest trypsin-like activity between the parental species. In terms of the chymotrypsin-like activity, the organisms of C. aquilus showed the highest activity compared to the C. polystictus organisms. The hybrids FH2 and MH7 showed the highest trypsin-like enzymatic activity, whereas, for chymotrypsin-like activity, this was presented by MH1. For the parental species, the highest value of elastase-like activity corresponded to FCp-PV, while both the enzymatic activities between the hybrids MCa-PV and FCp-PV were similar. The highest values of enzymatic activity between the serine proteases corresponded to trypsin-like proteases (Figure 4).
Figure 4.
Specific enzymatic activity of serine proteases. (A) Trypsin-like specific activity on BApNA substrate; (B) chymotrypsin-like specific activity on SAAPFpNA substrate; (C) elastase activity of 5 µg of venom protein. Hybrids of the first hybridization event (four years old): MH1, MH2, FH3, and MH4; hybrids of the second hybridization event (three years old): FH5, MH6, and MH7; males of C. aquilus: MCa8 and Cabf; organism of C. polystictus: A male, MCp10, and a female, Cpbm, used for the determination of trypsin- and chymotrypsin-like activities. Hybrids of the first hybridization event (11 years old): MH1, FH3, and MH4; hybrids of the second hybridization event (10 years old): MH6 and MH7; and pooled venom of C. aquilus (MCa-PV) and of C. polystictus (FCp-PV) were used for the determination of elastase-like activity. Bt (bovine trypsin), Bc (bovine chymotrypsin), and Pe (porcine elastase) were used as positive controls. Lowercase letters indicate significant statistical differences (Tukey; p < 0.05).
The proteolytic activity in the solution, as well as the zymography, showed high variability between the hybrids and between the adults. However, in terms of the enzymatic values of the trypsin-, chymotrypsin-, and elastase-like activities, they were similar or higher for some hybrids’ venoms compared to their biological parents’ venoms. Hybrids from Bothrops erythromelas and Bothrops neuwiedi showed a high variability in their enzymatic activity, particularly in neonates, where an increase in trypsin-like activity was related to an increase in body growth, with similar activity to the parents when the hybrids were 12 months old [11]. Here, we observed that the three- and four-year-old hybrids showed similar values of trypsin- and chymotrypsin-like proteases and, in some cases, higher enzymatic activity than their parents.
3.2.2. Gelatinolytic Activity
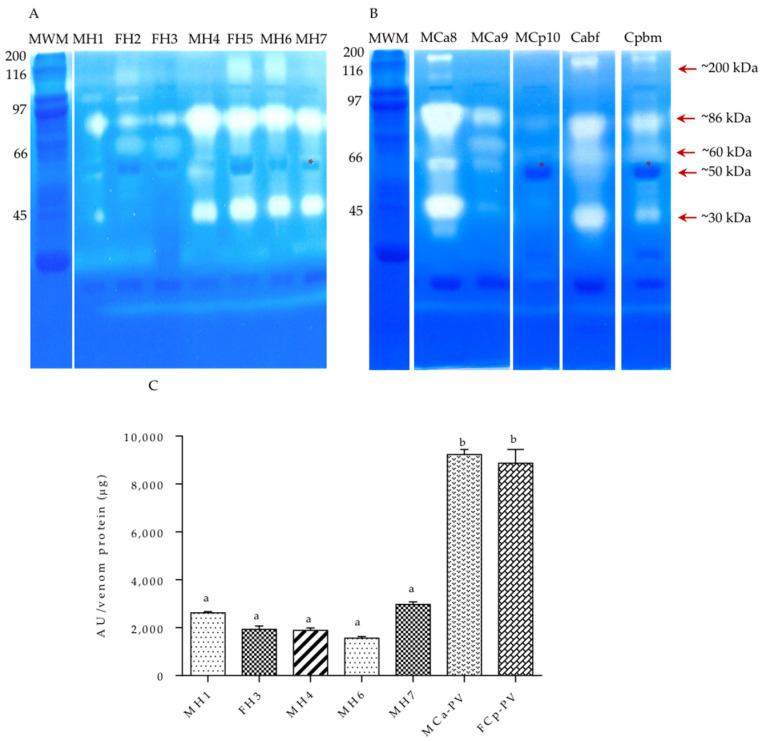
After SDS-PAGE, the snake venom metalloproteases (SVMPs) were identified using gelatin as the substrate, copolymerized with polyacrylamide, and incubated for 24 h (Figure 5A,B). A similar proteolytic banding pattern was observed with proteases of ~200, ~86, ~60, ~50, and ~30 kDa between the hybrids and adults. These data are similar to those reported for C. aquilus and C. polystictus by this work group [44]. The hybrids MH1, FH2, FH5, MH6, and MH7 and Cpbm, MCa8, and MCa9 presented a band of ~86 kDa MW, not observed in Cabf. The hybrids MH1, FH2, and FH3 and the C. polystictus adults did not present the ~31 kDa protease present in the other organisms. Moreover, a band of ~50 kDa was observed in all of the organisms, but only the hybrids and C. aquilus individuals showed proteolytic activity. The highest metalloprotease activity was present in MCa-PV and FCp-PV venoms and the other hybrids, which showed similar values of enzymatic activity (Figure 5C).
Figure 5.
Venom metalloproteinase activity. (A) Zymography in SDS-PAGE co-polymerized with gelatin using 20 µg of protein venom under no reducing conditions. (B) Enzymatic activity of 100 µg of venom protein. Hybrids of the first hybridization event (four years old): MH1, MH2, FH3, and MH4; hybrids of the second hybridization event (three years old): FH5, MH6, and MH7; males of C. aquilus: MCa8, MCa9, and Cabf; organism of C. polystictus: A male, MCp10, and a female, Cpbm, used for zymography analysis. (C) Hybrids of the first hybridization event (11 years old): MH1, FH3, and MH4; hybrids of the second hybridization event (10 years old): MH6 and MH7; and pooled venom of venoms MCa-PV and FCp-PV were used for the determination of metalloproteinase activity. MWM, molecular weight marker. Red asterisk shows a protein shared between C. polystictus and the hybrids. Lowercase letters indicate significant statistical difference (Tukey; p < 0.05).
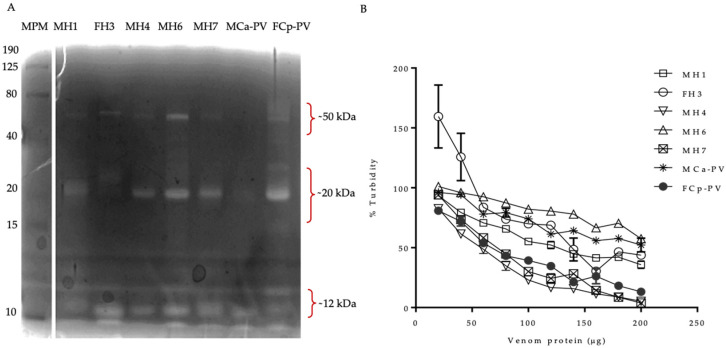
3.2.3. Snake Venom Hyaluronidases (SVHs)
The hyaluronidase activity was determined by zymography and turbidimetry using hyaluronic acid (Figure 6). The venoms of the parents showed hyaluronidases of ~12, ~25, and ~50 kDa (absent in MCa-PV), as previously reported [44], and these protein bands were found in all of the hybrid samples studied. The turbidimetric assay showed a lower activity for Ca than the hybrids MH1, MH4, and MH7 and Cp. More variability in the hyaluronidase pattern banding was observed for the ~25 to ~50 kDa bands; for example, MCa-PV did not present hyaluronidases in the range of ~50 kDa. Cp showed higher values of SVH than MCa-PV, while the other hybrids showed similar activity values to MCa-PV but lower than FCp-PV. The enzymatic values of the SVHs were higher for the MH7 and MH4 hybrids compared to the other hybrids and to MCa-PV and FCp-PV. The data were similar to those reported; however, SVH proteins of ~90 were not detected [44].
Figure 6.
Hyaluronidase activity. (A) Hyaluronidase zymography of 100 µg of venom protein. (B) Enzymatic activity of hyaluronidases using 20–200 µg of venom protein. Hybrids of the first hybridization event (11 years old): MH1, FH3, and MH4; hybrids of the second hybridization event (10 years old): MH6 and MH7; pooled venom of MCa-PV and FCp-PV. The MWMs are to the left.
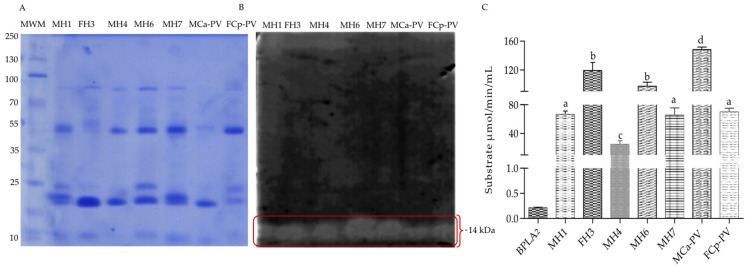
3.2.4. Snake Venom Phospholipase A2 (SVPLA2) Activity
For phospholipase A2 activity, the zymography showed a phospholipase band of ~14 kDa in all of the samples, similar to the reported data [44] (Figure 7); however, MCa-PV and the hybrid FH3 showed the highest activity. The MH6 showed higher phospholipase A2 activity than Cp, while the other hybrids showed similar/or lower values to the MCa-PV and FCp-PV species. The variability in the enzymatic activity values could indicate the presence of different isoforms of PLA2 in the analyzed venoms, which may be related to inter- or intraspecific variation in the venoms [45].
Figure 7.
Snake venom phospholipase A2 activity. (A) 10% SDS-PAGE using 100 ng of venom protein. (B) Zymography on agarose gel copolymerized with egg yolk. (C) Enzymatic activity of 100 ng of venom protein. Hybrids of the first hybridization event (11 years old): MH1, FH3, and MH4; hybrids of the second hybridization event (10 years old): MH6 and MH7; pooled venom of MCa-PV and FCp-PV. The MWMs are to the left. Lowercase letters indicate significant statistical difference (Tukey; p < 0.05).
3.3. RP-HPLC and MALDI-TOF-MS
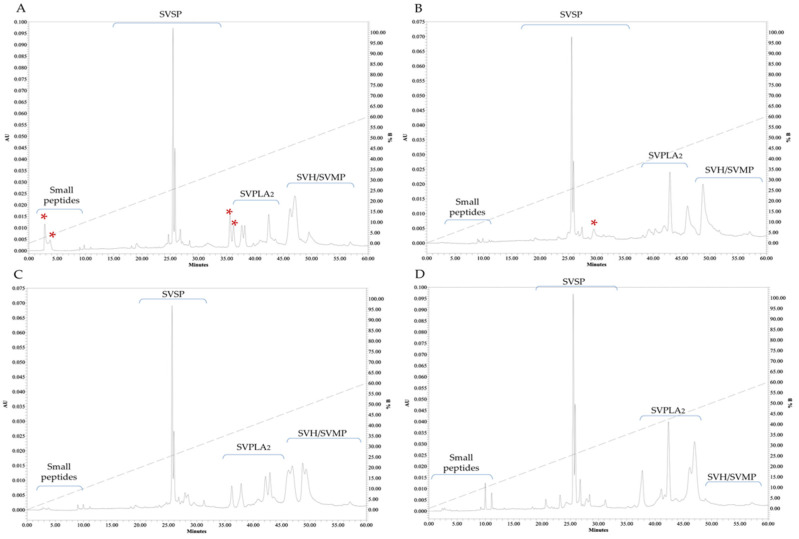
The protein fractionation by RP-HPLC showed different profiles between the venoms. Peaks between 35 and 55 min were found, where proteins such as phospholipase A2 (35–45 min) [46] and metalloproteases and hyaluronidases (≥50 min) [47] are usually eluted. Fewer peaks were found between 20 and 30 min retention times, in accordance with serine proteases (30–35 min) [48]. The last group, probably small peptides, eluted between 5 and 10 min [47]. The reported data for MCa-PV and FCp-PV showed eluted peaks at 36 min, which were not shared between them [44]. These peaks were present in the venoms of some of the hybrids, with a greater variability at 25 and 30–45 min, corresponding to the elution times of SVSP and SVPLA2; fractions were not detected after 60 min (Figure 8). However, further identification of the proteins must be confirmed by other methods.
Figure 8.
Protein fractionation of the venom by RP-HPLC. Pooled venoms of C. aquilus (A), C. polystictus (B), the male hybrids (MH1, MH4, MH6, and MH7) (MHs) (C), and a female hybrid (FH3) (D). Red asterisks show the peaks not shared between the samples. SVSP, snake venom serine protease; SVMPs, snake venom metalloprotease; SVPLA2, snake venom phospholipase; SVH, snake venom hyaluronidase. Absorbance units (AUs) at 280 nm. Enzymatic activity was not determined. The family proposed for the enzymes was based on similarity of the elution profiles with the reported data.
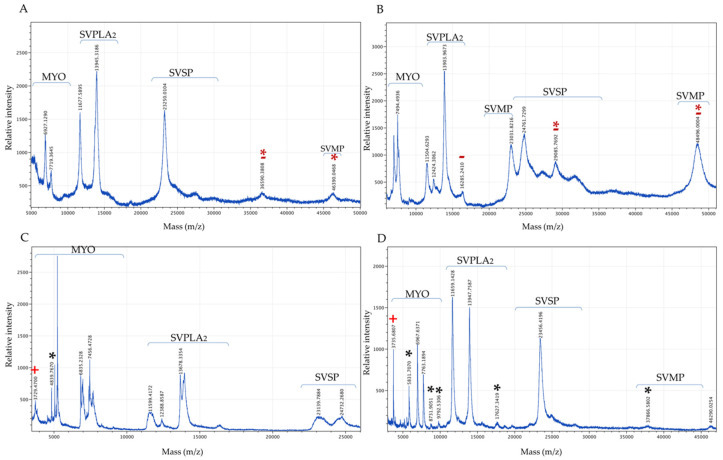
With respect to MS (Figure 9), the hybrids showed lower peak intensity for SVMP compared to the MCa-PV and FCp-PV venoms. These results are consistent with the SVMP enzymatic activity. The peaks of lower molecular weight and in the range of SVPLA2 were the most abundant in the hybrids, whereas the peaks in the range of SVSP and SVMP were the most abundant for the MCa-PV and FCp-PV venoms. The FH3 venom showed a greater abundance of peaks, followed by the MH, MCa-PV, and FCp-PV venoms. Additionally, all of the venoms presented compounds of lower molecular weight (3–11 kDa), suggesting crotamine-like myotoxins (MYO)—a small, non-enzymatic protein of approximately 4–5 kDa with a spastic paralysis effect in the hind limbs of mice and necrosis in muscle cells [49]. These MYO peaks were more abundant and variable in the hybrid organisms. Peaks not shared between the parental venoms were observed. The hybrids, either male or female, also presented peaks not shared between them or between their parents. The venom profiles indicate a differential pattern of protein between the parental and hybrid venoms (Table 2). The molecular weights of SVSP, SVMP, and SVPLA2 were similar to those observed by zymography; however, a larger number of proteases were found by zymography than by MALDI-TOF-MS, perhaps due to the long incubation time used to measure the activity.
Figure 9.
MALDI-TOF-MS analysis of snake venom. Pooled venoms of MCa-PV (A), FCp-PV (B), the male hybrids (MH1, MH4, MH6, and MH7) (MHs) (C), and a female hybrid (FH3) (D). Red asterisks indicate the peaks not shared between the MCa-PV and FCp-PV venoms; red minus signs indicate exclusive peaks for MCa-PV and FCp-PV; black asterisks indicate exclusive and unshared peaks between the male and female hybrid venoms; red plus signs indicate a peak present only in both hybrid venoms. MYO, myotoxin; SVSP, snake venom serine protease; SVPLA2, snake venom phospholipase; SVMP, snake venom metalloprotease. SVPLA2 and SVSP peaks.
Table 2.
Enzyme families observed for C. aquilus, C. polystictus, and the hybrids.
| Protein Family | Data Reported | MCa-PV | FCp-PV | MH | FH |
|---|---|---|---|---|---|
| SVMP | Reported for C. polistictus in the range of 20 and 30–40 kDa [50,51] | 23.2 kDa | 23 kDa | 23 kDa | 23.4, 37.8, and 46.2 kDa |
| SVSP | Reported for C. molossus venom between 24 and 30 kDa [49] | 27 kDa | 24.7 and 29 kDa | 24.7 kDa | NF |
| SVPLA2 | Reported in the range of 13–18 kDa [50] | 12.4, 16.2, and 13.9 kDa | 11.5, 12.4, 13.9, and 16.2 kDa | 11.5, 12.3, and 13.6 kDa | 11.6, 13.9, and 17.6 kDa |
| MYO | Reported in the range of 4–5 kDa in snake venoms and one MYO of 10 kDa in C. molossus [49] | 6.9 and 7.7 kDa | 7.4 kDa | 3.7, 4.8, 6.8, and 7.4 kDa | 3.7, 5.8, 6.9, 7.7, 8.7, and 9.7 kDa |
MCa-PV, FCp-PV, four male hybrids (MH1, MH4, MH6, and MH7) (MHs) and a female hybrid (FH3). NF, not found.
3.4. Lethal Dose-50 (LD50)
In order to calculate the LD50, concentrations of 0.5, 0.75, 1, 1.5, 2, and 3 μg of venom protein per milliliter in saline water were tested. Pooled venoms were used because of the low amount of venom from the juvenile organisms. Pool 1, which comprised venom from the female hybrids (FH-PV; FH2, FH3, and FH5), pool 2, which comprised venom from the male hybrids (MH-PV; MH1, MH4, MH6, and MH7), and the individual samples of the biological father, C. aquilus (Cabf), and the biological mother, C. polystictus (Cpbm), were tested. The FH-PV and Cabf venoms showed a lethal effect as a function of concentration, with an LD50 of 1.02 and 2.04 μg of venom protein per gram of body weight, respectively. A similar LD50 value of 2.24 μg of venom protein per gram of body weight was reported for C. aquilus [52]. For MH-PV and Cpbm, no lethal effects were detected with the doses used; however, it has been reported to have an LD50 of 3.4 μg of venom per gram of body weight [53], of 4.5 μg of venom protein per gram of body weight in neonate venom, and of 5.5 μg of venom protein per gram of body weight in adult venom of C. polystictus using non-Swiss albino mice [54]. Hybridism has been shown to be associated with variability in venom composition [2,11], and in previous reports, high variability in enzymatic activity has been observed [11]. In this study, the female hybrids showed the lowest LD50, meaning that they have a more toxic venom than their parents. This could be associated with the high variability of enzymes observed as a result of genetic variability due to the toxic effect of the venom being a result of the combination or synergism of the different enzymes [3].
With the exception of metalloprotease activity, some hybrids showed similar or higher enzymatic values (MH1, FH2, FH3, and MH7) with respect to the adults (Cabf, Cpbm, MCa-PV, and FCp-PV). When comparing the venoms of the juvenile hybrids (trypsin- and chymotrypsin-like activities) or the adult hybrids (elastase-like, SVPLA2, and SVHA activities) versus the parents, it was of particular interest that FH3, included in the pooled venom with the lowest LD50, showed similar or higher values in both SVSP (trypsin-, chymotrypsin-, and elastase-like protease) and PLA2 activities compared to the other organisms. However, despite male hybrids in some cases showing higher values of SVSP (trypsin-like) or similar PLA2 activities than some of the female hybrids, none of the male hybrids showed high enzymatic activity. For example, MH1 and MH7 showed similar or higher SVSP (trypsin-, chymotrypsin-, and elastase-like) proteolytic activity compared to the female hybrids and Cabf and Cpbm, but they showed lower PLA2 activity than MCa-PV and FH3 (which presented lethality under the used doses). The PLA2 activity for the MH1, MH4, and MH7 hybrids was similar to that of C. polystictus (which did not present lethality under the used doses). Neither parental species showed both high SVSP and PLA2 activities compared to the female hybrids. Moreover, it was observed that the male hybrid MH6 showed an SVPLA2 activity similar to the female hybrid FH3, but lower SVSP (chymotrypsin-like) and trypsin-like activities than the female hybrid FH2. For C. molossus nigrescens venom, it has been reported that at least two SVSPs are the main compounds associated with its lethality [49]. Additionally, PLA2 is one of the main components of snake venom, which helps immobilization and rapid death of prey and is associated with myotoxic, cytolytic, neurotoxic, and edema formation [44,51]. Refs. [49,55] Perhaps high values of both families of enzymes could be related to the lethality of the female hybrids that showed high SVPLA2 and SVSP activity values. These results suggest an important role for SVPLA2, SVSP, and MYO in the lethality of hybrids.
In other species, such as Crotalus viridis or Crotalus horridus, whose venoms are usually hemotoxic, neurotoxins have also been found and explained, based on their hybridization with Crotalus species, which are phylogenetically related, with the presence of neurotoxins in their venoms. The presence of neurotoxins in hybrids is assumed to be an adaptive advantage and a source of intraspecific variation among snake venom; however, it has been reported that in the hybridization of C. s. scutulatus and C. viridis, the Mojave toxin (PLA2) is not necessarily a strong selective advantage source for snake venom due to the fact that the toxins acquired through this phenomenon are confined to specific areas or hybridization zones where the two species are in near contact and the toxins acquired do not spread outside of the hybridization boundaries into the gene pool of the parental populations. However, the authors cannot exclude the possibility of a different outcome under different selective regimes, since they cannot reject the possibility that, given sufficient time, a degree of introgression of Mojave toxin genes may occur [3]. Moreover, ontogenetic changes are related to a low LD50, as some species of rattlesnakes as neonates could have more lethal venom than adults, which tends to show higher SVMP enzymatic activity, as observed in studies in adults and neonates of C. polystictus. However, no significant differences between the juveniles’ (PLA2, SVMP, and kallikrein-like) and adults’ enzymatic activity were observed [54]. Santoro et al. [11] reported ontogenetic changes (lower amidolytic and proteolytic activities) in the neonate hybrids of Bothrops erythromelas and Bothrops neuwiedi when they were three months old, with no differences to their parents when they were 12 months old. The analyses in this study showed juvenile hybrids of three and four years old with low SVMP activity and more protein, peptide, and enzyme variability in the protein families of their venoms than the parental species. This variation could include several isoforms that may present a distinct functional activity and could represent an advantage in the functionality, improving the capture of prey, digestion, or the defensive efficacy of the venom [55,56,57] and could have adaptive implications, facilitating the response to changing environmental pressures [58,59].
4. Conclusions
The phenetic analysis indicated a higher similarity between the hybrids and C. polystictus than with C. aquilus; however, the lethality of their venom was different, with the venom of the juvenile female hybrids being more lethal than that of their corresponding parents. Snake venom is a very complex mixture of different compounds, and its lethality is mainly the result of a combination of enzymes and toxins. The high values in SVSP and PLA2 and the higher variability of peptides of low molecular weight (maybe crotamine-like peptides) in the female hybrids could be related to their toxicity. The phenomenon of snake hybridization represents a way to increase the variability of snake venoms and an opportunity to observe changes in some specific protein families, which could help us to understand the mechanism of variability itself and in the development of more effective antidotes.
Acknowledgments
The authors thank Oscar Antonio Rayas Estrada, Ulises Padilla Garcia, Ricardo Daniel Valencia García, Sergio Hugo Nieves Morán, Oscar Daniel Robles Pérez, Yazmhinne Abril Her-nández Romero, Ana Valeria Romero Juarez, Carlos Eduardo Ramírez Alvarado, Cruz Daniel Juárez Acosta, and Daniel de los Cobos Landeros for the venom harvest; Norma Hernández Camacho for access to the Animal Resource Facility; and Luis Fernando Díaz-Peña for his technical support.
Author Contributions
Conceptualization, O.R.-P., A.B.-L. and T.G.-G.; investigation, O.R.-P.; methodology, O.R.-P., E.M.-O., M.S.C.-P., J.L.C.-G., J.A.G.-A. and P.H.-P.; formal analysis, O.R.-P. and J.L.C.-G., J.A.G.-A., P.H.-P., A.B.-L. and T.G.-G.; resources, A.B.-L. and T.G.-G.; data curation, O.R.-P., J.L.C.-G., J.A.G.-A., C.S.-G., P.H.-P., A.B.-L. and T.G.-G.; writing—original draft preparation, O.R.-P.; writing—review and editing, O.R.-P., A.B.-L. and T.G.-G.; supervision, A.B.-L. and T.G.-G.; project administration, A.B.-L. and T.G.-G.; funding acquisition, A.B.-L. and T.G.-G. All authors have read and agreed to the published version of the manuscript.
Funding
We thank to CONACyT for O.R.-P. PhD fellowship and for the financial support by FOPER-UAQ 2017.
Institutional Review Board Statement
This study was approved by the Committee of Bioethics of the Faculty of Natural Sciences, Autonomous University of Queretaro, and the in vivo studies were performed according to the guidelines of the Declaration of Helsinki and the NOM 062-ZOO (1999).
Informed Consent Statement
Not applicable.
Data Availability Statement
This manuscript was read and approved by all of the named authors, and there were no other persons who satisfied the criteria for authorship but are not listed. The order of authors listed in the manuscript was approved by all authors. The authors gave due consideration to the protection of intellectual property associated with this work, and there are no impediments to publication, including the timing of publication, with respect to intellectual property. The authors followed the regulations of their institutions concerning intellectual property. Any aspect of the work covered in this manuscript that involved experimental animals was conducted with the ethical approval of all relevant bodies, and such approvals are acknowledged within the manuscript. The corresponding author is the sole contact for the editorial process (including the editorial manager and direct communications with the office). They are responsible for communicating with the other authors about progress, submissions of revisions, and final approval of proofs. Current, correct email addresses from all authors have been provided, accessible by the corresponding author, and configured to accept emails.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Casewell N.R., Jackson T.N.W., Laustsen A.H., Sunagar K. Causes and Consequences of Snake Venom Variation. Trends Pharmacol. Sci. 2020;41:570–581. doi: 10.1016/j.tips.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chippaux J.P., Williams V., White J. Snake Venom Variability: Methods of Study, Results and Interpretation. Toxicon. 1991;29:1279–1303. doi: 10.1016/0041-0101(91)90116-9. [DOI] [PubMed] [Google Scholar]
- 3.Zancolli G., Baker T.G., Barlow A., Bradley R.K., Calvete J.J., Carter K.C., de Jager K., Owens J.B., Price J.F., Sanz L., et al. Is Hybridization a Source of Adaptive Venom Variation in Rattlesnakes? A Test, Using a Crotalus Scutulatus × Viridis Hybrid Zone in Southwestern New Mexico. Toxins. 2016;8:188. doi: 10.3390/toxins8060188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch V.J. Inventing an Arsenal: Adaptive Evolution and Neofunctionalization of Snake Venom Phospholipase A2 Genes. BMC Evol. Biol. 2007;7:2. doi: 10.1186/1471-2148-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatakeyama D.M., Tasima L.J., Bravo-Tobar C.A., Serino-Silva C., Tashima A.K., Rodrigues C.F.B., da Silva Aguiar W., da Costa Galizio N., de Lima E.O.V., Kavazoi V.K., et al. Venom Complexity of Bothrops Atrox (Common Lancehead) Siblings. J. Venom. Anim. Toxins Incl. Trop. Dis. 2020;26:e20200018. doi: 10.1590/1678-9199-jvatitd-2020-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massey D.J., Calvete J.J., Sánchez E.E., Sanz L., Richards K., Curtis R., Boesen K. Venom Variability and Envenoming Severity Outcomes of the Crotalus Scutulatus Scutulatus (Mojave Rattlesnake) from Southern Arizona. J. Proteom. 2012;75:2576–2587. doi: 10.1016/j.jprot.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 7.Kindler C., Chèvre M., Ursenbacher S., Böhme W., Hille A., Jablonski D., Vamberger M., Fritz U. Hybridization Patterns in Two Contact Zones of Grass Snakes Reveal a New Central European Snake Species. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-07847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zinenko O., Sovic M., Joger U., Gibbs H.L. Hybrid Origin of European Vipers (Vipera Magnifica and Vipera Orlovi) from the Caucasus Determined Using Genomic Scale DNA Markers. BMC Evol. Biol. 2016;16:1–13. doi: 10.1186/s12862-016-0647-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewontin R.C., Birch L.C. Hybridization as a Source of Variation for Adaptation to New Environments. Evolution. 1966;20:315. doi: 10.1111/j.1558-5646.1966.tb03369.x. [DOI] [PubMed] [Google Scholar]
- 10.Harrison R.G., Larson E.L. Hybridization, Introgression, and the Nature of Species Boundaries. J. Hered. 2014;105:795–809. doi: 10.1093/jhered/esu033. [DOI] [PubMed] [Google Scholar]
- 11.Santoro M.L., do Carmo T., Cunha B.H.L., Alves A.F., Zelanis A., Serrano S.M.D.T., Grego K.F., Sant’Anna S.S., Barbaro K.C., Fernandes W. Ontogenetic Variation in Biological Activities of Venoms from Hybrids between Bothrops Erythromelas and Bothrops Neuwiedi Snakes. PLoS ONE. 2015;10:e0145516. doi: 10.1371/journal.pone.0145516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowling T.E., Secor C.L. The role of hybridization and introgression in the diversification of animals. Annu. Rev. Ecol. Syst. 1997;28:593–619. doi: 10.1146/annurev.ecolsys.28.1.593. [DOI] [Google Scholar]
- 13.Bullini L. Origin and Evolution of Animal Hybrid Species. Trends Ecol. Evol. 1994;9:422–426. doi: 10.1016/0169-5347(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 14.Bailey R.M. An Intergeneric Hybrid Rattlesnake. Am. Nat. 2015;76:376–385. doi: 10.1086/281054. [DOI] [Google Scholar]
- 15.Aird S.D., Thirkhill L.J., Seebart C.S., Kaiser I.I. Venoms and Morphology of Western Diamondback/Mojave Rattlesnake Hybrids. J. Herpetol. 1989;23:131. doi: 10.2307/1564018. [DOI] [Google Scholar]
- 16.Smith C.F., Mackessy S.P. The Effects of Hybridization on Divergent Venom Phenotypes: Characterization of Venom from Crotalus Scutulatus Scutulatus × Crotalus Oreganus Helleri Hybrids. Toxicon. 2016;120:110–123. doi: 10.1016/j.toxicon.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Aird S.D., Aggarwal S., Villar-Briones A., Tin M.M.-Y., Terada K., Mikheyev A.S. Snake Venoms Are Integrated Systems, but Abundant Venom Proteins Evolve More Rapidly. BMC Genom. 2015;16:647. doi: 10.1186/s12864-015-1832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCranie J.R. Crotalus Polystictus. Soc. Study Amphib. Reptiles. 1976;180:1–2. [Google Scholar]
- 19.Campbell J.A., Lamar W.W. The Venomous Reptiles of Latin America. Q. Rev. Biol. 1989;65:516–517. doi: 10.1086/417006. [DOI] [Google Scholar]
- 20.Meik J., Mociño-Deloya E., Setser K. New Distribution Records for the Queretero Dusky Rattlesnake Crotalus Aquilus (Viperidae), with Comments on Morphology and Habitat Use. West. N. Am. Nat. 2007;67:601–604. doi: 10.3398/1527-0904(2007)67[601:NDRFTQ]2.0.CO;2. [DOI] [Google Scholar]
- 21.Oukkache N., Lalaoui M., Ghalim N. General Characterization of Venom from the Moroccan Snakes Macrovipera Mauritanica and Cerastes Cerastes. J. Venom. Anim. Toxins Incl. Trop. Dis. 2012;18:411–420. doi: 10.1590/S1678-91992012000400009. [DOI] [Google Scholar]
- 22.Alencar L.R.V., Quental T.B., Grazziotin F.G., Alfaro M.L., Martins M., Venzon M., Zaher H. Diversification in Vipers: Phylogenetic Relationships, Time of Divergence and Shifts in Speciation Rates. Mol. Phylogenetics Evol. 2016;105:50–62. doi: 10.1016/j.ympev.2016.07.029. [DOI] [PubMed] [Google Scholar]
- 23.Maruñak S.L., Ruíz de Torrent R.M., Teibler G.P., Gay C.C., Leiva L., Acosta De Pérez O. Acción Del Veneno de Bothrops Jararacussu de Argentina Sobre La Coagulación Sanguínea. Volume 8 Universidad de Buenos Aires, Facultad de Ciencias Veterinarias; Corrientes, Argentina: 2005. [Google Scholar]
- 24.Slagboom J., Kool J., Harrison R.A., Casewell N.R. Haemotoxic Snake Venoms: Their Functional Activity, Impact on Snakebite Victims and Pharmaceutical Promise. Br. J. Haematol. 2017;177:947–959. doi: 10.1111/bjh.14591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvete J.J. Proteomic Tools against the Neglected Pathology of Snake Bite Envenoming. Expert Rev. Proteom. 2011;8:739–758. doi: 10.1586/epr.11.61. [DOI] [PubMed] [Google Scholar]
- 26.Xiong S., Huang C. Synergistic Strategies of Predominant Toxins in Snake Venoms. Toxicol. Lett. 2018;287:142–154. doi: 10.1016/j.toxlet.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Rohlf F. NTSYS-Pc: Microcomputer Programs for Numerical Taxonomy and Multivariate Analysis. Am. Stat. 1987;41:330. doi: 10.2307/2684761. [DOI] [Google Scholar]
- 28.Bradford M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Ohlsson B.G., Weström B.R., Karlsson B.W. Enzymoblotting: A Method for Localizing Proteinases and Their Zymogens Using Para-Nitroanilide Substrates after Agarose Gel Electrophoresis and Transfer to Nitrocellulose. Anal. Biochem. 1986;152:239–244. doi: 10.1016/0003-2697(86)90404-5. [DOI] [PubMed] [Google Scholar]
- 31.Vinokurov K.S., Oppert B., Elpidina E.N. Notes & Tips an Overlay Technique for Postelectrophoretic Analysis of Proteinase Spectra in Complex Mixtures Using p-Nitroanilide Substrates. Anal. Biochem. 2005;337:164–166. doi: 10.1016/j.ab.2004.10.043. [DOI] [PubMed] [Google Scholar]
- 32.Rice K., Peralta R., Bast D., De Azavedo J., McGavin M.J. Description of Staphylococcus Serine Protease (Ssp) Operon in Staphylococcus Aureus and Nonpolar Inactivation of SspA-Encoded Serine Protease. Infect. Immun. 2001;69:159–169. doi: 10.1128/IAI.69.1.159-169.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossignol G., Merieau A., Guerillon J., Veron W., Lesouhaitier O., Feuilloley M.G.J., Orange N. Involvement of a Phospholipase C in the Hemolytic Activity of a Clinical Strain of Pseudomonas Fluorescens. BMC Microbiol. 2008;8:189. doi: 10.1186/1471-2180-8-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guntenhöner M.W., Pogrel M.A., Stern R. A Substrate-Gel Assay for Hyaluronidase Activity. Matrix. 1992;12:388–396. doi: 10.1016/S0934-8832(11)80035-1. [DOI] [PubMed] [Google Scholar]
- 35.Erlanger B.F., Kokowsky N., Cohen W. The Preparation and Properties of Two New Chromogenic Substrates of Trypsin. Arch. Biochem. Biophys. 1961;95:271–278. doi: 10.1016/0003-9861(61)90145-X. [DOI] [PubMed] [Google Scholar]
- 36.Gutiérrez J.M., Sanz L., Escolano J., Fernández J., Lomonte B., Angulo Y., Rucavado A., Warrell D.A., Calvete J.J. Snake Venomics of the Lesser Antillean Pit Vipers Bothrops Caribbaeus and Bothrops Lanceolatus: Correlation with Toxicological Activities and Immunoreactivity of a Heterologous Antivenom. J. Proteome Res. 2008;7:4396–4408. doi: 10.1021/pr8003826. [DOI] [PubMed] [Google Scholar]
- 37.Ponce-Soto L.A., Bonfim V.L., Novello J.C., Navarro Oviedo R., Yarlequé Chocas A., Marangoni S. Isolation and Characterization of a Serine Protease, Ba III-4, from Peruvian Bothrops Atrox Venom. Protein J. 2007;26:387–394. doi: 10.1007/s10930-007-9078-z. [DOI] [PubMed] [Google Scholar]
- 38.Di Ferrante N. Turbidimetric Measurement of Acid Mucopolysaccharides and Hyaluronidase Activity. J. Biol. Chem. 1956;220:303–306. doi: 10.1016/S0021-9258(18)65354-2. [DOI] [PubMed] [Google Scholar]
- 39.NOM-062-ZOO Norma Oficial Mexicana Especificaciones Técnicas Para La Producción, Cuidado y Uso de Los Animales de Laboratorio (NOM-062-ZOO 1999) [(accessed on 20 February 2022)];D. Of. Fed. 2001 :107–165. Available online: http://www.anmm.org.mx/bgmm/1864_2007/2002-138-3-295-298.pdf. [Google Scholar]
- 40.Klauber L.M. Rattlesnakes: Their Habits, Life Histories, and Influence on Mankind. Zoological Society of San Diego, University of California Press; Berkeley, CA, USA: 1972. [Google Scholar]
- 41.Reyes-Velasco J., Meik J.M., Smith E.N., Castoe T.A. Phylogenetic Relationships of the Enigmatic Longtailed Rattlesnakes (Crotalus Ericsmithi, C. Lannomi, and C. Stejnegeri) Mol. Phylogenetics Evol. 2013;69:524–534. doi: 10.1016/j.ympev.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 42.Blair C., Sánchez-Ramírez S. Diversity-Dependent Cladogenesis throughout Western Mexico: Evolutionary Biogeography of Rattlesnakes (Viperidae: Crotalinae: Crotalus and Sistrurus) Mol. Phylogenetics Evol. 2016;97:145–154. doi: 10.1016/j.ympev.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 43.Mallet J. Hybridization as an Invasion of the Genome. Trends Ecol. Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 44.Roldán-Padrón O., Castro-Guillén J.L., García-Arredondo J.A., Cruz-Pérez M.S., Díaz-Peña L.F., Saldaña C., Blanco-Labra A., García-Gasca T. Snake Venom Hemotoxic Enzymes: Biochemical Comparison between Crotalus Species from Central Mexico. Molecules. 2019;24:1489. doi: 10.3390/molecules24081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Casewell N.R., Wagstaff S.C., Wuster W., Cook D.A.N., Bolton F.M.S., King S.I., Pla D., Sanz L., Calvete J.J., Harrison R.A. Medically Important Differences in Snake Venom Composition Are Dictated by Distinct Postgenomic Mechanisms. Proc. Natl. Acad. Sci. USA. 2014;111:9205–9210. doi: 10.1073/pnas.1405484111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marangoni F.A., Ponce-Soto L.A., Marangoni S., Landucci E.C.T. Unmasking Snake Venom of Bothrops Leucurus: Purification and Pharmacological and Structural Characterization of New PLA2 Bleu TX-III. Biomed. Res. Int. 2013;2013:941467. doi: 10.1155/2013/941467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lomonte B., Calvete J.J. Strategies in ‘Snake Venomics’ Aiming at an Integrative View of Compositional, Functional, and Immunological Characteristics of Venoms. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017;23:26. doi: 10.1186/s40409-017-0117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menaldo D.L., Bernardes C.P., Santos-Filho N.A., Moura L.D.A., Fuly A.L., Arantes E.C., Sampaio S.V. Biochemical Characterization and Comparative Analysis of Two Distinct Serine Proteases from Bothrops Pirajai Snake Venom. Biochimie. 2012;94:2545–2558. doi: 10.1016/j.biochi.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Borja M., Neri-Castro E., Pérez-Morales R., Strickland J., Ponce-López R., Parkinson C., Espinosa-Fematt J., Sáenz-Mata J., Flores-Martínez E., Alagón A., et al. Ontogenetic Change in the Venom of Mexican Black-Tailed Rattlesnakes (Crotalus Molossus Nigrescens) Toxins. 2018;10:501. doi: 10.3390/toxins10120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X., Manjunatha Kini R., Doley R. Handbook of Venoms and Toxins of Reptiles. CRC Press; Boca Raton, FL, USA: 2009. Snake Venom Phospholipase A2 Enzymes; pp. 173–205. [Google Scholar]
- 51.Rael E.D., Rivas J.Z., Chen T., Maddux N., Huizar E., Lieb C.S. Differences in Fibrinolysis and Complement Inactivation by Venom from Different Northern Blacktailed Rattlesnakes (Crotalus Molossus Molossus) Toxicon. 1997;35:505–513. doi: 10.1016/S0041-0101(96)00139-0. [DOI] [PubMed] [Google Scholar]
- 52.Rivas E., Neri-Castro E., Bénard-Valle M., Hernánez-Dávila A.I., Zamudio F., Alagón A. General Characterization of the Venoms from Two Species of Rattlesnakes and an Intergrade Population (C. Lepidus × Aquilus) from Aguascalientes and Zacatecas, Mexico. Toxicon. 2017;138:191–195. doi: 10.1016/j.toxicon.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Mackessy S. Venom Composition in Rattlesnakes: Trends and Biological Significance. Biol. Ratt. 2008;495:510. [Google Scholar]
- 54.Mackessy S.P., Leroy J., Mociño-Deloya E., Setser K., Bryson R.W., Saviola A.J. Venom Ontogeny in the Mexican Lance-Headed Rattlesnake (Crotalus Polystictus) Toxins. 2018;10:271. doi: 10.3390/toxins10070271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amazonas D.R., Freitas-de-Sousa L.A., Orefice D.P., Sousa L.F.D., Martinez M.G., Mourão R.H., Chalkidis H.M., Camargo P.B., Moura-da-Silva A.M. Evidence for Snake Venom Plasticity in a Long-Term Study with Individual Captive Bothrops Atrox. Toxins. 2019;11:294. doi: 10.3390/toxins11050294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Casewell N.R., Wüster W., Vonk F.J., Harrison R.A., Fry B.G. Complex Cocktails: The Evolutionary Novelty of Venoms. Trends Ecol. Evol. 2013;28:219–229. doi: 10.1016/j.tree.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 57.Calvete J.J. Venomics: Integrative Venom Proteomics and Beyond. Biochem. J. 2017;474:611–634. doi: 10.1042/BCJ20160577. [DOI] [PubMed] [Google Scholar]
- 58.Margres M.J., Wray K.P., Hassinger A.T.B., Ward M.J., McGivern J.J., Moriarty Lemmon E., Lemmon A.R., Rokyta D.R. Quantity, Not Quality: Rapid Adaptation in a Polygenic Trait Proceeded Exclusively through Expression Differentiation. Mol. Biol. Evol. 2017;34:3099–3110. doi: 10.1093/molbev/msx231. [DOI] [PubMed] [Google Scholar]
- 59.Amazonas D.R., Portes-Junior J.A., Nishiyama-Jr M.Y., Nicolau C.A., Chalkidis H.M., Mourão R.H.V., Grazziotin F.G., Rokyta D.R., Gibbs H.L., Valente R.H., et al. Molecular Mechanisms Underlying Intraspecific Variation in Snake Venom. J. Proteom. 2018;181:60–72. doi: 10.1016/j.jprot.2018.03.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This manuscript was read and approved by all of the named authors, and there were no other persons who satisfied the criteria for authorship but are not listed. The order of authors listed in the manuscript was approved by all authors. The authors gave due consideration to the protection of intellectual property associated with this work, and there are no impediments to publication, including the timing of publication, with respect to intellectual property. The authors followed the regulations of their institutions concerning intellectual property. Any aspect of the work covered in this manuscript that involved experimental animals was conducted with the ethical approval of all relevant bodies, and such approvals are acknowledged within the manuscript. The corresponding author is the sole contact for the editorial process (including the editorial manager and direct communications with the office). They are responsible for communicating with the other authors about progress, submissions of revisions, and final approval of proofs. Current, correct email addresses from all authors have been provided, accessible by the corresponding author, and configured to accept emails.