Abstract
The genes man26a and man2A from Cellulomonas fimi encode mannanase 26A (Man26A) and β-mannosidase 2A (Man2A), respectively. Mature Man26A is a secreted, modular protein of 951 amino acids, comprising a catalytic module in family 26 of glycosyl hydrolases, an S-layer homology module, and two modules of unknown function. Exposure of Man26A produced by Escherichia coli to C. fimi protease generates active fragments of the enzyme that correspond to polypeptides with mannanase activity produced by C. fimi during growth on mannans, indicating that it may be the only mannanase produced by the organism. A significant fraction of the Man26A produced by C. fimi remains cell associated. Man2A is an intracellular enzyme comprising a catalytic module in a subfamily of family 2 of the glycosyl hydrolases that at present contains only mammalian β-mannosidases.
Members of the genus Cellulomonas are common in environments rich in decaying plant material. They degrade cellulose, xylans, glucans, mannans, chitin, and starch. Extracellular enzymes hydrolyzing some of these polysaccharides have been described from a number of strains. As with other polysaccharide-degrading microorganisms, purification of an enzyme is complicated by the production of complex systems of enzymes with similar activities. Consequently, cloning of the genes encoding the enzymes and expression of the genes in heterologous hosts are used extensively to obtain individual enzymes (4). Several cellulases and xylanases from Cellulomonas fimi were characterized in this manner (7, 10, 12, 39, 40, 46, 54, 64), but mannan-degrading enzymes have not been described.
Mannans are linear polymers of mannose, or of mannose and glucose, and are linked β-1,4. The principal hemicelluloses in soft woods, accounting for up to 25% of the dry weight, are O-acetylgalactoglucomannans in which the backbone comprises mannose and glucose in the ratio 3:1; the glucose residues may be distributed randomly (49, 61). Galactose monomers are linked α-1,6 to some of the mannose residues (61); some 2- and 3-hydroxyls of the mannose residues and, to a lesser extent, of the glucose residues, are acetylated (49). In some green algae, the crystalline structural component of the cell wall is mannan, not cellulose. Mannans function as storage carbohydrates in the bulbs and endosperm of some plants: ivory nut mannan (INM), from Phytelphus macrocarpa, is an insoluble crystalline material comprising only mannose, with a backbone conformation very similar to that of cellulose; the backbone of locust bean gum (LBG), from Ceratonia siliqua, also comprises only mannose, with galactose monomers linked to it randomly by α-1,6 bonds. The hydrolysis of O-acetylgalactomannan into its monomeric components requires endo-β-mannanase, β-mannosidase, β-glucosidase, α-galactosidase, and acetyl esterase (49). The hydrolysis of INM requires only endo-β-mannanase and β-mannosidase; hydrolysis of LBG requires α-galactosidase in addition to these two enzymes.
β-Mannanases (mannan endo-1,4-β-mannosidase; EC 3.2.1.78) are produced by plants, fungi, and bacteria. They catalyze the random hydrolysis of β-1,4 mannosidic linkages within the backbones of mannans, galactomannans, and glucomannans. Well-characterized enzymes include Man1 from Trichoderma reesei (25, 56), ManA from Streptomyces lividans (2), ManA from Pseudomonas fluorescens subsp. cellulosa (8, 9), and an enzyme from Aspergillus niger (38). The amino acid sequences of several enzymes, deduced from the sequences of the genes encoding them (2, 9, 41, 42, 55), place them in families 5 and 26 of the glycosyl hydrolases (26–28), both of which are in clan GH-A of retaining enzymes (8, 29). The crystal structure of a mannanase from family 5 has been determined (31). A particular enzyme may comprise one or more modules (9, 21, 42, 55).
β-Mannosidases (β-1,4-mannoside mannohydrolase; EC 3.2.1.25) catalyze the removal of β-d-mannose residues from the nonreducing ends of oligosaccharides. Some eucaryotic β-mannosidases, in family 2 of glycosyl hydrolases, process the N-linked oligosaccharides of glycoproteins (1, 11, 15, 35). A. niger, Aspergillus awamori, and T. reesei fungi secrete β-mannosidases that act preferentially on shorter manno-oligosaccharides (43). An enzyme from the archaeon Pyrococcus furiosus, the only procaryotic β-mannosidase sequenced to date, is in family 1 of the glycosyl hydrolases (3).
α-Galactosidases (melibiase or α-d-galactoside galactohydrolase; EC 3.2.1.22) remove α-1,6-linked galactose residues from galactomannan polymers. They are in families 27 and 36 of the glycosyl hydrolases (28). Bacterial enzymes are intracellular (20, 65) or extracellular (37, 38, 58).
This study describes the cloning and sequencing of two genes from C. fimi that encode a β-mannanase and a β-mannosidase, their expression in Escherichia coli, and the preliminary characterization of the encoded enzymes. The enzymes belong to families 26 and 2, respectively, of the glycosyl hydrolases. The β-mannanase has an unusual modular structure.
MATERIALS AND METHODS
Materials.
Restriction endonucleases were from New England Biolabs (Mississauga, Ontario, Canada), Gibco-BRL (Burlington, Ontario, Canada), or Boehringer Mannheim (Laval, Quebec, Canada). T4 DNA ligase was from Gibco-BRL. Vent DNA polymerase was from New England Biolabs. PWO and HiFi DNA polymerases were from Boehringer Mannheim. Isopropyl-β-d-thiogalactopyranoside (IPTG), 4-methylumbelliferyl-β-d-mannoside (MUβMan), 4-methylumbelliferyl-α-D-galactoside (MUαGal), 5-bromo-4-chloro-3-indolyl-β-d-glucoside (X-Gluc), p-nitrophenyl glycosides (other than that of mannobiose), LBG (purity not specified), and carboxymethyl cellulose (CM-cellulose; sodium salt, low viscosity; nominal degree of polymerization, 400; nominal degree of substitution, 0.7) were from Sigma (St. Louis, Mo.). INM (purity, >98%) and azo-carob galactomannan (ACG; low viscosity, purity, >98%) were from Megazyme (North Rocks, New South Wales, Australia). p-Nitrophenyl-β-mannobioside (PNPM2) was synthesized by enzymatic transglycosylation (unpublished data). Birchwood xylan was from Carl Roth KG (Karlsruhe, Germany). Zeocin was from Invitrogen (San Diego, Calif.). Mouse anti-His6 serum was from Dianova (Hamburg, Germany). Rabbit anti-mouse immunoglobulin G serum conjugated to horseradish peroxidase was from Dako Diagnostics (Carpinteria, Calif.).
Bacterial strains, plasmids, and bacteriophages.
The E. coli strains used were JM101 (51), DH5α (51), BL21(DE3) (24), and XLOLR (Stratagene). The C. fimi strain used was ATCC 484. The plasmids used were pZErO (Invitrogen), pBluescript SK and KS (Stratagene) and pET27b and pET28a (Novagen). The bacteriophages used were λZAPII and ExAssist (Stratagene). Bacterial stocks were maintained at −70°C in Luria-Bertani (LB) medium containing 15% glycerol. Plasmid DNA was stored in water at −20°C. Bacteriophage lambda was stored at 4°C or −70°C in TYP medium.
Media and growth conditions.
E. coli strains were grown routinely in LB medium (51) at 37°C or in TYP medium (51) at room temperature for protein production. Media were supplemented with 50 μg of kanamycin or 100 μg of ampicillin ml−1, depending on the plasmid-encoded resistance. Strains carrying pZErO and derivatives thereof were grown in low-salt LB (51) supplemented with 50 μg of Zeocin ml−1. C. fimi was grown at 30°C in basal salts medium (57) supplemented with 0.2% (wt/vol) carbon source and 50 μg of kanamycin ml−1. Liquid cultures were shaken at 220 rpm. Solid media contained 1.5% agar (Difco).
DNA manipulations.
All oligodeoxynucleotide primers were synthesized by the Nucleic Acid and Protein Service (NAPS) Unit of the University of British Columbia by using an Applied Biosystems model 380A DNA synthesizer and were purified by extraction with n-butanol (53). DNA was sequenced by the NAPS Unit by using the AmpliTaq dye termination cycle sequencing protocol with the addition of 5% dimethyl sulfoxide (DMSO) and an Applied Biosystems model 377 sequencer. C. fimi DNA was amplified by PCR. Reaction mixtures contained 10 to 100 ng of template, 40 pmol of each primer, the recommended polymerase buffer, 10% DMSO, 0.2 mM 2′-deoxynucleoside 5′-triphosphates, and 1 U of Vent, PWO, or HiFi polymerase in a final volume of 100 μl. Incubation was for 30 cycles of 94°C for 30 s, 64 to 67°C for 30 s, and 72°C for 45 to 90 s, depending on the reaction. DNA was manipulated routinely as described previously (51). Restriction endonucleases were used as recommended by the suppliers. DNA fragments were separated by agarose gel electrophoresis (51); they were then recovered from the gels and purified by using the Qiaex II Gel Extraction Kit (Qiagen, Santa Clarita, Calif.). Ligation reaction mixtures contained 100 to 500 ng of total DNA at an insert-to-vector molar ratio of 10, 1 U of T4 DNA ligase, and the recommended buffer in a final volume of 100 μl. Mixtures were incubated at 23°C for 2 h and then desalted by butanol precipitation (62). E. coli cells were transformed either by electroporation (GenePulser; Bio-Rad) or by heat shock of cells prepared by the quick chemical competent cell protocol in TSS buffer (51).
Lambda library screening.
The agar in petri dishes (150 mm in diameter) containing 30 ml of NZY medium (51) was overlaid with 10 ml of NZY medium containing 0.4% ACG and 1.5% agar. After solidification, the plates were dried for several hours before they were overlaid with a mixture of E. coli cells and an appropriate dilution of a C. fimi λ ZAPII library (39, 40) in 3.5 ml of NZY medium with 0.1 to 0.5 mM IPTG and 0.7% agar. Mannanase-positive clones produced haloes of hydrolysis after incubation at 37°C for 16 to 24 h (9). After secondary and tertiary screening, mannanase-positive phagemids (pBluescript SK) containing genomic DNA inserts were excised in vivo from lambda DNA, recircularized, and transduced into E. coli XLOLR cells (54). The excised form of the entire genomic λ ZAPII library was screened for E. coli clones producing β-mannosidase by plating appropriate dilutions on NZY plates and then, after incubation at 37°C overnight, replicating the colonies onto NZY plates containing 0.5 mM MUβMan and 0.3 mM IPTG. Positive clones fluoresced under UV light (63).
Detection of enzyme activity.
Mannanase-positive E. coli clones were detected by plating on LB containing 0.2% ACG (9). Zones of hydrolysis were visualized after incubation at 37°C for 12 to 16 h. β-Mannanase or α-galactosidase activity was detected on plates containing 0.1 to 0.5 mM MUβMan or MUαGal, respectively. After incubation, the hydrolysis product was detected under UV light. Mannanases in culture supernatants or cell extracts were screened by zymograms by using nonreducing sodium dodecyl sulfate (SDS)-polyacrylamide gels incorporating 0.5% ACG (9). β-Mannosidase, α-galactosidase, and β-glucosidase in extracts were screened by incubating nonreducing SDS-polyacrylamide gels after electrophoresis in 1 mM MUβMan, 1 mM MUαGal, or 0.5 mM X-Gluc, respectively (33, 63). Mannanase activity was screened during purification by spotting 10- to 20-μl samples onto plates containing 2.0% agar and 0.5% LBG in 50 mM phosphate buffer (pH 7.0) (56); after incubation for 12 to 16 h at 37°C, hydrolysis was detected by staining the plates with Congo red (60).
Location of mannanase activity in C. fimi.
A liquid culture of C. fimi was grown for 6 days in minimal medium containing 0.2% LBG. One hour before harvesting, chloramphenicol was added (40 μg ml−1) to stop further protein synthesis. Mannanase activity was measured in samples of the whole culture, the culture supernatant, and the cells. The cells were recovered by centrifugation, washed three times with 50 mM phosphate buffer (pH 7.0), and resuspended to the starting volume in the same buffer. Activities were determined by incubating 210-μl samples of each preparation with 75 μl of 2% ACG, 5 μl of 10% sodium azide, and 6 μl of chloramphenicol (20 mg ml−1) for 16 h at 37°C. Reactions were stopped by adding 750 μl of 95% ethyl alcohol. The precipitated ACG was removed by centrifugation for 2 min at 13,000 rpm. The release of soluble azo-manno-oligosaccharides was determined from the A590 of the supernatant. The results were normalized to the activity in the whole culture being 100%.
Production and purification of enzymes.
Cloned genes were expressed from pET vectors in E. coli BL21(DE3) cells. Cultures, shaken at room temperature, were induced with 0.2 to 0.4 mM IPTG in mid-exponential phase and then incubated for a further 24 to 36 h. The cultures were centrifuged at 5,000 rpm and 4°C for 10 min. Mannanase or mannosidase was purified from the cells as follows. The cells were resuspended in binding buffer (5 mM imidazole, 500 mM NaCl, 20 mM Tris-HCl; pH 7.9) and ruptured by passage three times through a French pressure cell. Debris and unbroken cells were removed by centrifugation at 15,000 rpm and 4°C for 30 min. The supernatant was passed through a 5- to 10-ml His-Bind metal chelating affinity column at a flow rate of 0.5 ml min−1. Mannanase was also purified from the supernatant of the E. coli culture. The supernatant was concentrated to 50 ml and exchanged into binding buffer by using an Ultrasette tangential flow concentrator with a 1-kDa cutoff (Filtron). The concentrated solution was passed through a 15-ml His-Bind column at a flow rate of 0.5 ml min−1. In all cases, the His-Bind columns were washed with 3 to 6 volumes of binding buffer at a flow rate of 1 ml min−1, after which adsorbed proteins were eluted by stepwise increases in the concentration of imidazole in the buffer from 5 to 500 mM. The protein content was screened by measuring the A280. Fractions of 5 ml were collected. Protein-containing fractions were screened for enzyme activity. The enzymes were eluted, usually with 50 to 120 mM imidazole. Enzyme-containing fractions were pooled and then concentrated, and the buffer was exchanged by diafiltration through an Amicon PM10 membrane. Purity was evaluated by SDS-polyacrylamide gel electrophoresis (PAGE) (34). β-Mannosidase was partially purified from cells of C. fimi as follows. C. fimi was grown overnight at 30°C in 2 liters of basal salts medium with 0.2% glucose. The cells were recovered by centrifugation for 10 min at 5,000 rpm and 4°C, resuspended in 50 ml of buffer, and ruptured by four passages through a French pressure cell. Debris and unbroken cells were removed by centrifugation at 4,000 rpm and 4°C for 1 h. The buffer in the supernatant was changed to 20 mM Tris-HCl (pH 5.8) by diafiltration. The extract was passed through an EconoQ (Bio-Rad) column. Adsorbed proteins were eluted with 20 column volumes of a linear gradient of 0 to 700 mM NaCl in the same buffer. Fractions of 5 ml were collected and screened for activity. The fractions with the highest activity were pooled and concentrated as described above.
Analysis of proteins.
The concentrations of proteins were determined by measuring the A280 (47). N-terminal amino acid sequences of proteins or fragments thereof were determined after separation by SDS-PAGE, electroblotting to polyvinylidene membranes, and excision of the appropriate bands after visualization by staining with Coomassie blue. Automated Edman sequencing was done by the NAPS Unit at the University of British Columbia or at the Protein Microchemistry Facility, University of Victoria, British Columbia, Canada, by using Applied Biosystems 476A gas-phase sequenators. The molecular weights of proteins were determined by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Protein samples (0.5 to 3.0 mg ml−1) were desalted by depositing 5- to 10-μl drops onto dialysis membranes (Millipore MF; 0.025-μm pore size) floating on distilled water. After 12 to 16 h, 1 μl of dialyzed sample was transferred to the sample holder and dried for 5 min. The sample was overlaid with 1 μl of matrix (supersaturated sinapinic acid solution in 70% acetonitrile and 0.1% trifluoroacetate) and dried for 5 min (32). Spectra were obtained with a Mass Phoresis instrument (Ciphergen, Inc.).
RESULTS
Galactomannan-degrading enzymes produced by C. fimi.
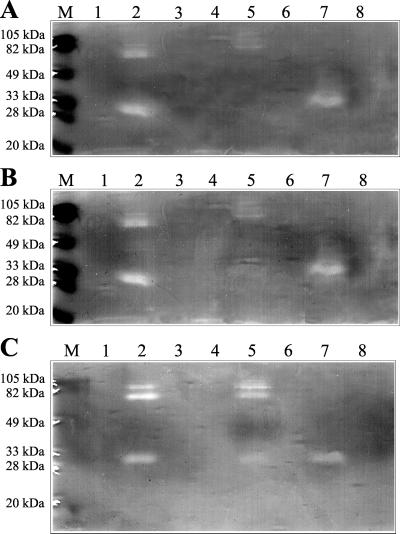
After growth of C. fimi for 6 days in minimal medium with LBG as a carbon source, most of the mannanase activity was in the culture supernatant; there was a low level of activity in a cell extract. Supernatants from cultures grown with different carbon sources were screened for mannanases by zymograms with ACG as substrate. There were three proteins with endomannanase activity when LBG was the carbon source: two major proteins of 75 and 30 kDa and a minor protein of 100 kDa (Fig. 1). The N-terminal sequence of the mannanase of 75 kDa was APADET. The production of all three proteins was reduced when glucose and LBG were added together as carbon sources. Only the 30-kDa protein was produced with CM-cellulose as a carbon source. Mannanases were not produced with mannose, galactose, xylan, or glycerol as a carbon source.
FIG. 1.
Mannanases from C. fimi analyzed by nonreducing SDS-PAGE using zymograms of culture supernatants. The cultures were grown in minimal medium with no addition (lane 1) or supplemented (0.2%) with LBG (lane 2), mannose (lane 3), glycerol (lane 4), glucose and LBG (lane 5), xylan (lane 6), CM-cellulose (lane 7), or galactose (lane 8). Prestained molecular weight standards are shown in lane M. The cultures were incubated for 2 days (A), 6 days (B), and 11 days (C).
β-Mannosidase and α-galactosidase activities were present only in the cell extract; zymograms revealed single proteins of 60 and 120 kDa, respectively. The mannosidase comigrated with a β-glucosidase (63), so it was partially purified. Its N-terminal amino acid sequence was MITQDLYD. Zymograms of cell extracts showed that β-mannosidase was produced only in the presence of mannan or mannose.
Screening of a genomic library of C. fimi DNA for mannanase genes and sequencing of a mannanase gene.
A library of C. fimi genomic DNA was prepared previously in λ ZAPII (39). The library was screened for mannanase-positive clones on ACG plates with IPTG. Eight positive plaques were obtained. Phagemids (pBluescript with C. fimi DNA inserts) were excised from the clones and transferred to E. coli XLOLR. The strains were designated CMan1 to CMan8; all of them accumulated in the culture medium at least two proteins with mannanase activity and of similar sizes, the largest being approximately 100 kDa. The clones producing the most activity, CMan2 and CMan4, were analyzed further. They carried plasmids, pCMan2 and pCMan4, with inserts of 4.3 and 6.3 kbp, respectively. Restriction mapping and partial sequencing showed that the inserts had a DNA sequence in common. Compared with that in pCMan2, the insert in pCMan4 had an extra 65 bp at the 5′ end, after which the sequences were the same. As it appeared that the two inserts encoded the same mannanase, the insert in pCMan2 was sequenced in its entirety.
A putative translational start codon was not present in the sequence of the insert in pCMan2. The similarity of the mannanase profiles in strains CMan1 to CMan8 led to the library being screened for further mannanase-positive clones. Five further clones were treated as described above and screened by zymograms. CMan30 produced two mannanases similar in size to those produced by C. fimi. It carried a plasmid with an insert of 3.0 kbp, the restriction pattern for which showed that it also contained the sequence common to pCMan2 and pCMan4. Sequencing of the 5′ end of the insert in pCMan30 revealed an ATG start codon 540 bp downstream of its 5′ end and 6 and 72 bp, respectively, upstream of the 5′ ends of the inserts in pCMan4 and pCMan2, thus completing the sequence of a putative mannanase gene (GenBank accession number AF126471).
The open reading frame covered by the inserts in pCMan2, pCMan4, and pCMan30 was 3,033 bp long, encoding a polypeptide of 1,011 amino acids. The molecular weight of the polypeptide, calculated from the deduced amino acid sequence, was 107,033. The N-terminal amino acid sequence was characteristic of a signal peptide, with the cleavage site predicted to be between Ala40 and Ala41 in the sequence 37PAPA↓APV43 (44). This finding agreed with the consensus leader peptide cleavage sequence A/VXA↓A derived from other extracellular enzymes from C. fimi (12, 39, 40, 46, 54, 64). Cleavage at this point would give a mature polypeptide of about 103 kDa, similar to that of the largest polypeptide with mannanase activity (∼100 kDa) detected in supernatants from C. fimi cultures. Amino acids 50 to 55 in the deduced sequence were APADET, matching the N-terminal sequence of the mannanase of 75 kDa in the supernatants. Ala50 in the Man26A sequence is designated residue 1 in the mature polypeptide, which is 961 amino acids long.
Analysis of the deduced amino acid sequence of the mannanase.
Alignment of the deduced amino acid sequence of the mannanase with those of proteins in the database revealed a modular structure. The N-terminal part of the polypeptide shared sequence identity with the catalytic modules of enzymes in family 26 of the retaining glycosyl hydrolases, which contains mannanases and glucanases. Therefore, in keeping with the recently proposed nomenclature for enzymes hydrolyzing the polysaccharides in the cell walls of plants (30), the enzyme from C. fimi is designated Man26A. Man26A shares the greatest identity with mannanase ManA from P. fluorescens subsp. cellulosa, a single module polypeptide (9): amino acids 1 to 398 of Man26A are 40% identical to amino acids 3 to 383 of mature ManA (Fig. 2). Thus, the catalytic module of Man26A comprises at least residues 1 to 398, based on the alignment with ManA. Glu174 and Glu282 in mature ManA are the catalytic carboxylic amino acids (8). These residues are strictly conserved in the enzymes of family 26; the corresponding residues in mature Man26A are Glu176 and Glu283 (Fig. 2).
FIG. 2.
Alignment of the first 398 amino acids from C. fimi Man26A (Cf Man26A; GenBank AF126471) with the entire sequence of ManA from P. fluorescens subsp. cellulosa (Pf ManA; GenBank X82179). Asterisks indicate identical residues. The catalytic glutamates are in boldface.
Amino acids 637 to 830 of mature Man26A share 23 to 30% sequence identity with S-layer homology (SLH) domains, which typically comprise three repeats of about 60 amino acids each (45). There are three repeats of about 60 amino acids each that are 20% identical in this part of Man26A, covering residues 647 to 826 (Fig. 3). None of the extracellular proteins from C. fimi described previously contain SLH modules. Such modules usually anchor the proteins containing them to the bacterial cell wall. They are present in some xylanases and pullulanases and in cellulosome-anchoring proteins (19, 36, 50).
FIG. 3.
Alignment of the SLH domain repeats of Man26A with the SLH repeats from bacterial proteins. The numbers indicate the positions of the first amino acid residue of each sequence in the different proteins. Cf, C. fimi Man26A; Ta, S-layer protein from Thermus aquaticus (GenBank A47024); Ba, S-layer protein from Bacillus anthracis (GenBank X99724); Ct, cellulosome-anchoring protein from Clostridium thermocellum (GenBank X67506); Tm, outer membrane protein from Thermotoga maritima (GenBank X68276). Asterisks indicate residues that are identical or conserved in all sequences.
Amino acids 450 to 636 and 831 to 961 of Man26A do not share sequence identity with any proteins in the database. They may comprise functional modules.
Such a multimodule structure is common in the extracellular enzymes produced by C. fimi (10).
Production of Man26A in E. coli.
The man26A gene was subcloned into the expression vector pET27b as a fusion encoding Man26A with a C-terminal H6 tag. The plasmid, pET27Man26A, was transformed into E. coli BL21(DE3). This strain yielded about 70 mg of active Man26A liter−1 after purification (see Materials and Methods), with ca. 10 mg recovered from the culture supernatant and ca. 60 mg from the cell extract.
Properties of Man26A.
The Man26A produced by E. coli was ∼100 kDa (Fig. 4); its N-terminal amino acid sequence was APAPAAP, corresponding to residues −14 to −8 relative to the N terminus of the enzyme produced by C. fimi. This means either that E. coli and C. fimi use different leader peptide processing sites in proMan26A or that they use the same site and C. fimi removes further amino acids from the N terminus. Man26A hydrolyzed PNPM2 very slowly; prolonged incubation was required to detect activity. The hydrolysis of LBG was assayed at concentrations from 0.01 to 4.5 mg ml−1. Higher concentrations were not tested because of the viscosity. An apparent kcat/Km of 1,150 ml mg−1 min−1 was calculated from the initial slope of a plot of v versus [S]. The pH and temperature optima were 5.5 and 42°C, respectively. The enzyme was stable for >2 h at this temperature.
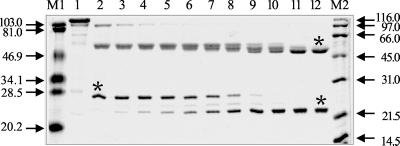
FIG. 4.
Digestion of Man26A, produced in E. coli, with protease from C. fimi. Man26A was digested with protease (1 μg of protease and 60 μg of Man26A) at 37°C. Samples were removed at intervals and boiled in loading buffer for 2 min to stop digestion. Digestion times were 0, 15, 30, 45, 60, and 90 min (lanes 1 to 6) and 2, 3, 5, 7, 16, and 24 h (lanes 7 to 12). Molecular weight standards are in lanes M1 and M2. The N termini of the polypeptides marked with an asterisk were sequenced.
Proteolysis of Man26A.
C. fimi secretes a protease(s) that releases discrete fragments, corresponding to individual modules, from other polysaccharide hydrolases secreted by the organism (22, 23, 52). When Man26A produced in E. coli was digested with the protease (23), several discrete fragments were released, two of which were resistant to further hydrolysis (Fig. 4). The largest stable fragment, which arose by hydrolysis of a fragment of 55 to 60 kDa, had the N-terminal amino acid sequence AGALP and a mass of 50 kDa, making it about 460 amino acids long. Therefore, it comprised the catalytic module plus 50 to 60 amino acids at the C terminus. The sequence AGALP starts nine residues after the N terminus of Man26A produced by E. coli and six residues before the N terminus of the mature protein produced by C. fimi. The other stable fragment had the N-terminal amino acid sequence AHPGVE, corresponding to residues 637 to 642 of the mature protein, just before the first SLH repeat starting at residue 647; its mass was 21 kDa, making it about 190 amino acids long. This stable fragment arose by trimming of the N and C termini of a fragment of about 28 kDa with the N-terminal amino acid sequence VNSAE, corresponding to residues 618 to 622 of the mature protein.
The nature of these fragments supported the modular structure proposed for Man26A. As with other polysaccharide hydrolases produced by C. fimi, the initial sites of proteolysis were between the modules. Besides cutting Man26A from E. coli between the modules, the protease from C. fimi also trimmed the N terminus of the catalytic module.
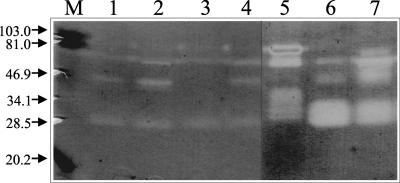
When supernatants from cultures of C. fimi grown with INM, LBG, CM-cellulose, or LBG and CM-cellulose were screened by zymogram after nondenaturing gel electrophoresis, the patterns of active bands were identical except for that for the culture grown with CM-cellulose. Furthermore, the same patterns were obtained for Man26A produced in E. coli and digested either with C. fimi protease or with the supernatant from the culture of C. fimi grown with LBG (Fig. 5). This suggests that Man26A is the only mannanase produced by C. fimi and that all of the multiple bands seen on the zymograms (Fig. 1) arise by proteolysis of this 100-kDa protein in the cultures.
FIG. 5.
Origin of the multiple mannanases from C. fimi analyzed by using nonreducing SDS-PAGE zymograms. Cultures of C. fimi were incubated for 11 days at 30°C in minimal medium supplemented (0.2%) with INM (lane 1), LBG (lane 2), CM-cellulose (lane 3), or LBG and CM-cellulose (lane 4). Samples of the culture supernatants were analyzed. Man26A (0.05 μg) produced in E. coli was analyzed without pretreatment (lane 5), after digestion (1 h, 37°C) with protease from C. fimi (lane 6), or with the supernatant from a culture of C. fimi grown with LBG (lane 7).
Location of mannanase activity.
After growth of a culture of C. fimi for 6 days in the presence of LBG, the relative levels of mannanase activity in the whole culture, associated with the cells, and in the culture supernatant were 100, 80, and 55%, respectively. Thus, a significant fraction of the activity appeared to be cell associated, possibly through binding to the surface by the SLH domain. The discrepancy between the activity in the culture and the sum of the activities associated with the cells and in the culture supernatant could have been caused by residual LBG in the culture interfering with hydrolysis of the test substrate, ACG. Any residual LBG would have been separated from the cells by centrifugation. Man26A produced in E. coli did not bind to a peptidoglycan fraction prepared from C. fimi.
Isolation and sequencing of a mannosidase gene.
Mannosidase-positive plaques were not obtained by direct screening of the λ ZAPII library with MUβMan. Therefore, the library was rescreened in its excised form as E. coli colonies carrying pBluescript phagemids with inserts of C. fimi DNA. Two positive clones were obtained. The plasmids they carried were designated pCManI and pCManII and contained fragments of C. fimi DNA of 2.6 and 6.5 kbp, respectively. Restriction mapping and partial sequencing showed them to have a DNA fragment in common. The 5′ end of the insert in pCManI was 150 bp downstream of the 5′ end of that in pCManII. Therefore, the insert in pCManI was sequenced. It contained an open reading frame of 2,526 bases (GenBank accession number AF126472), encoding a protein of 842 amino acids with a calculated molecular weight of 94,960.
Analysis of the deduced amino acid sequence of the mannosidase.
The N-terminal amino acid sequence deduced for the mannosidase matched that for the enzyme partially purified from C. fimi. Signature consensus sequences are defined for each family of glycosyl hydrolases to reduce the ambiguity in the classification of newly reported enzymes. The mammalian mannosidases are in family 2 of the glycosyl hydrolases, but their alignments with the consensus sequences for this family suggest that they form a subfamily within it. Alignment of its amino acid sequence placed the enzyme from C. fimi in this subfamily (Fig. 6); it was designated Man2A (30). At present, it is the only procaryotic enzyme in the subfamily. The conserved catalytic carboxyls in family 2 are glutamates. In Man2A, Glu429 and Glu519 are predicted to be the acid-base catalyst and the nucleophile, respectively (Fig. 6). Unlike Man26A, Man2A comprises only a catalytic module.
FIG. 6.
Alignment of the β-mannosidase subfamily in family 2 of the glycosyl hydrolases. Goat Man, Capra hicus β-mannosidase (GenBank U46067); Bovine Man, Bos taurus β-mannosidase (GenBank U17432); Human Man, Homo sapiens β-mannosidase (GenBank U60337); C. fimi Man, C. fimi β-mannosidase (GenBank AF126472). Asterisks indicate identical residues. The putative catalytic glutamates are in boldface.
Production of Man2A in E. coli.
The man2A gene was subcloned into the expression vector pET28A(+), yielding the plasmid pET28Man. The protein encoded by this plasmid carried a C-terminal H6 tag for purification. Cell extracts from E. coli BL21(D3) carrying pET28Man yielded up to 300 mg of purified Man2A liter−1 of culture. The purity was estimated to be >95% by SDS-PAGE. The N-terminal amino acid sequence of the enzyme was MITQDLYDG, matching that of the enzyme produced in C. fimi.
Properties of Man2A.
Man2A hydrolyzed p-nitrophenyl-β-mannoside (PNPM) readily. It had low activity on p-nitrophenyl-β-galactoside. It did not hydrolyze p-nitrophenyl derivatives of α-mannose, β-N-acetylglucosamine, β-glucose, β-xylose, β-cellobiose, or β-gentiobiose. The optimum pH for the hydrolysis of PNPM was 7.0. Although the rate of hydrolysis of PNPM was fastest at 55°C, the enzyme was unstable at this temperature. The half-life of the enzyme was 27 h at 37°C. Man2A was inhibited by PNPM at concentrations >400 μM (data not shown), so the values of 167 s−1 for kcat, 0.3 mM for Km, and 500 s−1 · mM−1 for kcat/Km were only estimates.
Size exclusion chromatography of Man2A gave a mass of 103 kDa, indicating that the native enzyme was a monomer.
DISCUSSION
All of the extracellular glycosyl hydrolases produced by C. fimi, including Man26A, are modular proteins, comprising a catalytic module and at least one other module. Man26A is the only one with an SLH module. It appears to contain two other modules, comprising amino acids 450 to 636 and 831 to 961, that are unrelated to modules in the database and are of unknown function. The mannanases produced by other microorganisms vary in complexity; they range in length from 306 (2) to 1,332 (42) amino acids. Mannanases from S. lividans and P. fluorescens subsp. cellulosa comprise catalytic modules only, from families 5 and 26, respectively, of the glycosyl hydrolases (2, 9). A mannanase from Agaricus bispora comprises a catalytic module from family 5 connected to an N-terminal cellulose-binding module from family II by a proline-rich linker (66). Thermophilic Caldocellulosiruptor spp. produce a number of complex enzymes with two catalytic modules, at the N and C termini, connected by one or two cellulose-binding modules from family III; in some of them, one of the catalytic modules is a mannanase from family 5 (42) or family 26 (21). The activity of Man26A against LBG is comparable to those of other mannanases (2, 8, 14, 56), but exact comparisons are difficult because of the viscosity of the substrate.
SLH modules are present in other polysaccharidases, including xylanases, pullulanases, lichenases, and endoglucanases (6), where they may be involved in attaching the enzymes to the cell surfaces. In growing cells of Clostridium thermocellum, cellulosomes are attached to the cell envelope through SLH modules (5). Although some SLH modules bind to peptidoglycan, others appear to bind to secondary cell wall polysaccharides rather than to peptidoglycan (16, 50). It is possible, given their diverse sequences, that not all SLH modules recognize and interact with the same components of the cell surface; some interact with other SLH modules (36). Man26A produced by C. fimi appears to be transiently cell associated but not to bind to peptidoglycan. Further analysis is required to define the exact role(s) of the SLH module in the functioning and location of the enzyme.
Man26A appears to be processed further by C. fimi after removal of the leader peptide. The N terminus of the mature enzyme from C. fimi is 13 amino acids downstream of the site processed by E. coli. The N-terminal amino acid sequences of some enzymes secreted by C. fimi are the same when the enzymes are produced by E. coli or C. fimi (7, 12, 39, 40, 46, 64), but, like Man26A, cellobiohydrolase Cel48A (formerly CbhB; see reference 30) also appears to be processed further by C. fimi after removal of the leader peptide (54). The relevant sequences are as follows: Cel48A, PAIA↓1AAGAGQPATVTVPAASPVRA↓2AVDGE; Man26A, PAQS↓1APAPAAPVAGALPT↓2APADE; ↓1 and ↓2 indicate the sites of hydrolysis producing the N termini of the enzymes produced by E. coli and C. fimi, respectively. T. reesei removes a further eight amino acids from the N terminus of Man1 after the processing of the leader peptide (55).
Family 2 of the glycosyl hydrolases includes enzymes from eubacteria and eucaryotes, but Man2A is the first eubacterial β-mannosidase to be assigned to the family. An interesting difference between Man2A and the mammalian enzymes in the same subfamily is the cysteine residues: the mammalian enzymes contain 13 conserved cysteines; Man2A contains only 3 of these conserved cysteines plus 2 others (Fig. 6). Unlike some other glycosidases, Man2A appears to be a monomer. Its activity is comparable to that reported for a mannosidase from Thermotoga neopolitana (14). Like Man2A, some other glycosidases, including a β-glucosidase (17) and β-galactosidases (48), are inhibited by substrate.
Detailed characterization of Man26A and Man2A, including their combined actions on manno-oligosaccharides and mannans, is in progress.
ACKNOWLEDGMENTS
We thank N. R. Gilkes, D. G. Kilburn, P. Tomme, and S. G. Withers for helpful discussions and Brad McLean for help with the preparation of the figures and tables.
This research was supported by the Natural Sciences and Engineering Research Council of Canada and the Protein Engineering Network of Centers of Excellence.
REFERENCES
- 1.Alkhayat A H, Kraemer S A, Leipprandt J R, Macek M, Kleijer W J, Friderici K H. Human β-mannosidase cDNA characterization and first identification of a mutation associated with human β-mannosidosis. Hum Mol Genet. 1998;7:75–83. doi: 10.1093/hmg/7.1.75. [DOI] [PubMed] [Google Scholar]
- 2.Arcand N, Kluepfel D, Paradis F W, Morosoli R, Sharek F. β-Mannanase of Streptomyces lividans 66: cloning and DNA sequencing of the manA gene and characterization of the enzyme. Biochem J. 1993;290:857–863. doi: 10.1042/bj2900857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer M W, Bylina E J, Swanson R V, Kelly R M. Comparison of a β-glucosidase and a β-mannosidase from the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1996;271:23749–23755. doi: 10.1074/jbc.271.39.23749. [DOI] [PubMed] [Google Scholar]
- 4.Béguin P, Gilkes N R, Kilburn D G, Miller R C, Jr, O’Neill G P, Warren R A J. Cloning of cellulase genes. Crit Rev Biotechnol. 1987;6:129–162. [Google Scholar]
- 5.Béguin P, Lemaire M. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit Rev Biochem Mol Biol. 1996;3:201–236. doi: 10.3109/10409239609106584. [DOI] [PubMed] [Google Scholar]
- 6.Beveridge T J, Pouwels P H, Sars M, Kotiranta A, Lounatma K, Kari K, Keruso E, Haapasolo M, Egelseer E M, Scocher I, Sleytr U B, Morelli L, Callegari M-L, Nomellini J F, Bingle W H, Smit J, Leibovitz E, Lemaire M, Miras I, Salamitou S, Beguin P, Ohayon H, Gounon P, Matuschek M, Sahm K, Bahl H, Grogono-Thomas R, Dworkin J, Blaser M J, Woodland R M, Newell D G, Kessel M, Koval S F. Function of S-layers. FEMS Microbiol Rev. 1997;20:99–149. doi: 10.1111/j.1574-6976.1997.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 7.Black G W, Hazlewood G P, Millward-Sadler S J, Laurie J I, Gilbert H J. A modular xylanase containing a novel non-catalytic xylan-specific binding domain. Biochem J. 1995;307:191–195. doi: 10.1042/bj3070191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolam D N, Hughes N, Virden R, Lakey J H, Hazlewood G P, Henrissat B, Braithwaite K L, Gilbert H J. Mannanase A from Pseudomonas fluorescens subsp. cellulosa is a retaining glycosyl hydrolase in which E12 and E320 are the putative catalytic residues. Biochemistry. 1996;35:16195–16204. doi: 10.1021/bi961866d. [DOI] [PubMed] [Google Scholar]
- 9.Braithwaite, K. L., G. W. Black, G. P. Hazlewood, B. R. Ali, and H. J. Gilbert. A non-modular endo-β-mannanase from Pseudomonas fluorescens subsp. cellulosa. Biochem. J. 305:1005–1010. [DOI] [PMC free article] [PubMed]
- 10.Bray M R, Creagh A L, Damude H G, Gilkes N R, Haynes C A, Jervis E, Kilburn D G, MacLeod A M, Meinke A, Miller R C, Jr, Rose D R, Shen H, Tomme P, Tull D, White A, Withers S G, Warren R A J. β-1,4-Glycanases of Cellulomonas fimi: families, mechanisms, and kinetics. ACS (Am Chem Soc) Symp Ser. 1996;655:64–84. [Google Scholar]
- 11.Chen H, Leipprandt J R, Travis C E, Sopher B L, Jones M Z, Cavanagh K T, Friderici K H. Molecular cloning and characterization of bovine β-mannosidase. J Biol Chem. 1995;270:3841–3848. doi: 10.1074/jbc.270.8.3841. [DOI] [PubMed] [Google Scholar]
- 12.Clarke J H, Davidson K, Gilbert H J, Fontes C M G A, Hazlewood G P. A modular xylanase from mesophilic Cellulomonas fimi contains the same cellulose-binding and thermostabilising domains as xylanases from thermophilic bacteria. FEMS Microbiol Lett. 1996;139:27–35. doi: 10.1111/j.1574-6968.1996.tb08175.x. [DOI] [PubMed] [Google Scholar]
- 13.Clarke J H, Laurie J I, Gilbert H J, Hazlewood G P. Multiple xylanases of Cellulomonas fimi are encoded by distinct genes. FEMS Microbiol Lett. 1991;83:305–310. doi: 10.1016/0378-1097(91)90493-t. [DOI] [PubMed] [Google Scholar]
- 14.Duffaud G D, McCutchen C M, Leduc P, Parker K N, Kelly R M. Purification and characterization of extremely thermostable β-mannanase, β-mannosidase, and α-galactosidase from the hyperthermophilic eubacterium Thermotoga neopolitana 5068. Appl Environ Microbiol. 1997;63:169–177. doi: 10.1128/aem.63.1.169-177.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duran P, Lehn P, Callebaut I, Fabrega S, Henrissat B, Mornon J-P. Active-site motifs of lysosomal acid hydrolases: invariant features of clan GH-A glycosyl hydrolases deduced from hydrophobic cluster analysis. Glycobiology. 1997;7:277–284. doi: 10.1093/glycob/7.2.277. [DOI] [PubMed] [Google Scholar]
- 16.Egelseer E M, Leitner K, Jarosch M, Hotzy C, Zayni S, Sleytr U B, Sara M. The S-layer proteins of two Bacillus stearothermophilus wild-type strains are bound via their N-terminal region to a secondary cell wall polymer of identical composition. J Bacteriol. 1998;180:1488–1495. doi: 10.1128/jb.180.6.1488-1495.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estrada P, Mata I, Dominguez J M, Castillon M P, Acebal C. Kinetic mechanism of β-glucosidase from Trichoderma reesei QM 9414. Biochim Biophys Acta. 1990;1033:298–304. doi: 10.1016/0304-4165(90)90137-l. [DOI] [PubMed] [Google Scholar]
- 18.Fanutti C, Ponyi T, Black G W, Hazlewood G P, Gilbert H J. The conserved non-catalytic 40-residue sequence in cellulases and hemicellulases from anaerobic fungi functions as a protein docking domain. J Biol Chem. 1995;270:29314–29322. doi: 10.1074/jbc.270.49.29314. [DOI] [PubMed] [Google Scholar]
- 19.Fujino T, Béguin P, Aubert J-P. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in the attachment of the cellulosome to the cell surface. J Bacteriol. 1993;175:1891–1899. doi: 10.1128/jb.175.7.1891-1899.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gherardini F, Babcock M, Salyers A. Purification and characterization of two α-galactosidases associated with the catabolism of guar gum and other β-galactosides by Bacteroides ovatus. J Bacteriol. 1985;161:500–506. doi: 10.1128/jb.161.2.500-506.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbs M D, Elinder A U, Reeves R A, Bergquist P L. Sequencing, cloning and expression of a β-1,4-mannanase gene, manA, from the extremely thermophilic anaerobic bacterium, Caldicellulosiruptor RT8B.4. FEMS Microbiol Lett. 1996;141:37–43. doi: 10.1111/j.1574-6968.1996.tb08360.x. [DOI] [PubMed] [Google Scholar]
- 22.Gilkes N R, Kilburn D G, Miller R C, Jr, Warren R A J. Structural and functional analysis of a bacterial cellulase by proteolysis. J Biol Chem. 1989;264:17802–17808. [PubMed] [Google Scholar]
- 23.Gilkes N R, Warren R A J, Miller R C, Jr, Kilburn D G. Precise excision of the cellulose-binding domain from two Cellulomonas fimi cellulases by a homologous protease and the effects on catalysis. J Biol Chem. 1988;263:10401–10407. [PubMed] [Google Scholar]
- 24.Grodber J, Dunn J J. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988;170:1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harjunpaa V, Teleman A, Siika-Aho M, Drakenberg T. Kinetic and stereochemical studies of mannooligosaccharide hydrolysis catalyzed by β-mannanases from Trichoderma reesei. Eur J Biochem. 1995;234:278–283. doi: 10.1111/j.1432-1033.1995.278_c.x. [DOI] [PubMed] [Google Scholar]
- 26.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1990;280:304–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henrissat B, Bairoch A. Updating the sequence based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon J-P, Davies G. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc Natl Acad Sci USA. 1995;92:7090–7094. doi: 10.1073/pnas.92.15.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henrissat B, Teeri T T, Warren R A J. A scheme for designating enzymes that hydrolase the polysaccharides in the cell walls of plants. FEBS Lett. 1998;425:352–354. doi: 10.1016/s0014-5793(98)00265-8. [DOI] [PubMed] [Google Scholar]
- 31.Hilge M, Gloor S M, Rypniewski W, Sauer O, Heightman T D, Zimmerman W, Winterhalter K, Piontek K. High-resolution native and complex structures of thermostable β-mannanase from Thermomonospora fusca—substrate specificity in glycosyl hydrolase family 5. Structure. 1998;6:1433–1444. doi: 10.1016/s0969-2126(98)00142-7. [DOI] [PubMed] [Google Scholar]
- 32.Kallweit U, Boernsen K O, Kresbach G M, Widmer H M. Matrix compatible buffers for analysis of proteins with matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom. 1996;10:845–849. [Google Scholar]
- 33.Kohchi C, Toh-e A. Cloning of a Candida pelliculosa β-glucosidase gene and its expression in Saccharomyces cerevisiae. Mol Gen Genet. 1986;203:89–94. doi: 10.1007/BF00330388. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Leipprandt, J. R., S. A. Kraemer, B. E. Haithcock, H. Chen, J. L. Dyme, K. H. Friderici, and M. Z. Jones. Caprine β-mannosidase: sequencing and characterization of the cDNA and identification of the molecular defect of caprine β-mannosidosis. Genomics 37:51–56. [DOI] [PubMed]
- 36.Lemaire M, Ohayon H, Gounon P, Fujino T, Béguin P. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J Bacteriol. 1995;177:2451–2459. doi: 10.1128/jb.177.9.2451-2459.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margolles-Clark E, Tenkanen M, Luonteri E, Penttila M. Three α-galactosidase genes of Trichoderma reesei cloned by expression in yeast. Eur J Biochem. 1996;240:104–111. doi: 10.1111/j.1432-1033.1996.0104h.x. [DOI] [PubMed] [Google Scholar]
- 38.McLeary B V, Taravel F R, Joseleau J-P. Characterisation of the oligosaccharides produced on hydrolysis of galactomannan with β-d-mannanase. Carbohydr Res. 1983;118:91–109. [Google Scholar]
- 39.Meinke A, Gilkes N R, Kilburn D G, Miller R C, Jr, Warren R A J. Cellulose-binding polypeptides from Cellulomonas fimi: endoglucanase D (CenD), a family A β-1,4-glucanase. J Bacteriol. 1993;175:1910–1918. doi: 10.1128/jb.175.7.1910-1918.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meinke A, Gilkes N R, Kwan E, Kilburn D G, Miller R C, Jr, Warren R A J. Cellobiohydrolase A (CbhA) from the cellulolytic bacterium Cellulomonas fimi is a β-1,4-exocellobiohydrolase analogous to Trichoderma reesei CBHII. Mol Microbiol. 1994;12:413–442. doi: 10.1111/j.1365-2958.1994.tb01030.x. [DOI] [PubMed] [Google Scholar]
- 41.Millward-Sadler S J, Hall J, Black G W, Hazlewood G P, Gilbert H J. Evidence that the Piromyces gene family encoding endo-1,4-mannanases arose through gene duplication. FEMS Microbiol Lett. 1996;141:183–188. doi: 10.1111/j.1574-6968.1996.tb08382.x. [DOI] [PubMed] [Google Scholar]
- 42.Morris D D, Reeves R A, Gibbs M D, Saul D J, Bergquist P. Correction of the the β-mannanase domain of the celC pseudogene from Caldocellulosiruptor saccharolyticus and activity of the gene product on kraft pulp. Appl Environ Microbiol. 1995;61:2262–2269. doi: 10.1128/aem.61.6.2262-2269.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neustroev K N, Krylov A S, Firsov L M, Abroskina O N, Khorlin A Y. Isolation and properties of β-mannosidase from Aspergillus awamori. Biokhimiya. 1991;56:1406–1412. [Google Scholar]
- 44.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Prot Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 45.Olabarria G, Carrascosa J, Pedro M A, Berenguer J. A conserved motif in S-layer proteins is involved in peptidoglycan binding in Thermus thermophilus. J Bacteriol. 1996;178:4765–4772. doi: 10.1128/jb.178.16.4765-4772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Neill G, Goh S H, Warren R A J, Kilburn D G, Miller R C., Jr Structure of the gene encoding the exoglucanase of Cellulomonas fimi. Gene. 1986;44:325–330. doi: 10.1016/0378-1119(86)90197-6. [DOI] [PubMed] [Google Scholar]
- 47.Pace N C, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Posci I, Taylor S A, Richardson A C, Smith B V, Price R G. Comparison of several new chromogenic galactosides as substrates for various β-d-galactosidases. Biochim Biophys Acta. 1993;1163:54–60. doi: 10.1016/0167-4838(93)90278-y. [DOI] [PubMed] [Google Scholar]
- 49.Puls J, Schuseil J. Chemistry of hemicelluloses: relationship between hemicellulose structure and enzymes required for hydrolysis. In: Coughlan M P, Hazlewood G P, editors. Hemicelluloses and hemicellulases. London, England: Portland Press; 1996. pp. 1–28. [Google Scholar]
- 50.Ries W, Hotzy C, Schocher I, Sleytr U B, Sagra M. Evidence that the N-terminal part of the S-layer protein from Bacillus stearothermophilus PV72/p2 recognizes a secondary cell wall polymer. J Bacteriol. 1997;179:3892–3898. doi: 10.1128/jb.179.12.3892-3898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 52.Sandercock L, Meinke A, Warren R A J. Degradation of cellulases in cultures of Cellulomonas fimi. FEMS Microbiol Lett. 1996;143:7–12. [Google Scholar]
- 53.Sawadago M, van Dyke M W. A rapid method for the purification of deprotected oligodeoxynucleotides. Nucleic Acids Res. 1991;19:674. doi: 10.1093/nar/19.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen H, Gilkes N R, Kilburn D G, Miller R C, Jr, Warren R A J. Cellobiohydrolase B, a second exocellobiohydrolase from the cellulolytic bacterium Cellulomonas fimi. Biochem J. 1995;311:67–74. doi: 10.1042/bj3110067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stålbrand H, Saloheimo A, Vehmaanpera J, Henrissat B, Penttila M. Cloning and expression in Saccharomyces cerevisiae of a Trichoderma reesei β-mannanase gene containing a cellulose-binding domain. Appl Environ Microbiol. 1995;61:1090–1097. doi: 10.1128/aem.61.3.1090-1097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stålbrand H, Siika-Aho M, Tenkanen M, Viikari L. Purification and characterisation of two β-mannanases from Trichoderma reesei. J Biotechnol. 1993;29:229–242. [Google Scholar]
- 57.Stewart B J, Leatherwood J M. Derepressed synthesis of cellulase by Cellulomonas. J Bacteriol. 1976;128:609–615. doi: 10.1128/jb.128.2.609-615.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Talbot G, Sygusch J. Purification and characterization of thermostable β-mannanase and α-galactosidase from Bacillus stearothermophilus. Appl Environ Microbiol. 1990;56:3505–3510. doi: 10.1128/aem.56.11.3505-3510.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamaru Y, Araki T, Amagoi H, Mori H, Morishita T. Purification and characterization of an extracellular β-1,4-mannanase from a marine bacterium, Vibrio sp. strain MA-138. Appl Environ Microbiol. 1995;61:4454–4458. doi: 10.1128/aem.61.12.4454-4458.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teather R M, Wood P J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982;43:777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tenkanen M, Makkonen M, Perttula M, Viikari L, Teleman A. Action of Trichoderma reesei on galactoglucomannan in pine kraft pulp. J Biotechnol. 1997;57:191–204. doi: 10.1016/s0168-1656(97)00099-0. [DOI] [PubMed] [Google Scholar]
- 62.Thomas M G. A procedure for second-round differential screening of cDNA libraries. BioTechniques. 1994;16:988–989. [PubMed] [Google Scholar]
- 63.Wakarchuk W W, Kilburn D G, Miller R C, Jr, Warren R A J. β-Glucosidases of Cellulomonas fimi. J Gen Microbiol. 1984;130:1385–1389. [Google Scholar]
- 64.Wong W K R, Gerhard B, Guo Z M, Kilburn D G, Warren R A J, Miller R C., Jr Characterisation and structure of an endoglucanase gene cenA of Cellulomonas fimi. Gene. 1986;44:315–324. doi: 10.1016/0378-1119(86)90196-4. [DOI] [PubMed] [Google Scholar]
- 65.Xiuzhu D, Schyns P J Y M J, Stams A J M. Degradation of galactomannan by a Clostridium butyricum strain. Antonie Leewenhoek. 1991;60:109–114. doi: 10.1007/BF00572700. [DOI] [PubMed] [Google Scholar]
- 66.Yagüe E, Mehak-Zunic M, Morgan L, Wood D A, Thurston C F. Expression of CEL2 and CEL4, two proteins from Agaricus bisporus with similarity to fungal cellobiohydrolase I and β-mannanase, respectively, is regulated by carbon source. Microbiology. 1997;143:239–244. doi: 10.1099/00221287-143-1-239. [DOI] [PubMed] [Google Scholar]