Abstract
The discovery of immune checkpoint inhibitors (ICIs) has revolutionized the care of cancer patients. However, the response to ICI therapy exhibits substantial interindividual variability. Efforts have been directed to identify biomarkers that predict the clinical response to ICIs. In recent years, the gut microbiome has emerged as a critical player that influences the efficacy of immunotherapy. An increasing number of studies have suggested that the baseline composition of a patient's gut microbiota and its dysbiosis are correlated with the outcome of cancer immunotherapy. This review tackles the rapidly growing body of evidence evaluating the relationship between the gut microbiome and the response to ICI therapy. Additionally, this review highlights the impact of antibiotic-induced dysbiosis on ICI efficacy and discusses the possible therapeutic interventions to optimize the gut microbiota composition to augment immunotherapy efficacy.
Keywords: immune checkpoint inhibitors, microbiome, cancer treatment, immunotherapy response, dysbiosis
INTRODUCTION
Immune checkpoint inhibitors (ICIs) that target programmed cell death-1 (PD-1), programmed cell death-ligand 1 (PDL-1), or cytotoxic T lymphocyte–associated antigen 4 (CTLA-4) have revolutionized the treatment of several malignancies owing to their higher efficacy and lower toxicity as compared with traditional cytotoxic drugs. They are now being used in the standard care of several malignancies.[1,2] These drugs are monoclonal antibodies that target specific proteins on cells, namely PD-1, its ligand (PDL-1), or CTLA-4, that are involved in inhibitory regulation of immune responses.[3] This in turn allows immune cells to better target tumor cells presenting non–self-antigens, thus overcoming tumor immune subversion, which is often seen in malignancies. Despite these advances, the clinical efficacy of ICIs is highly variable among cancer patients. A considerable proportion of patients show primary resistance to ICIs, and others show disease progression or relapse due to secondary resistance after an initial response.[4–6] Several of these resistance mechanisms have been described, and they can be classified as intrinsic (related to characteristics of the cancer cells) or extrinsic (related to interactions among the ICI, T cells, macrophages, and the enteric microbiome, among other elements). Initially, PDL-1 and PD-1 receptor expression were the biomarkers used to predict a response to immunotherapy. More recently, additional predictive biomarkers have included DNA mismatch repair, microsatellite instability, and tumor mutational burden.[7] Most of the evidence has focused on tumor-related characteristics. However, another important determinant of the response to ICIs is the host immune system. Accordingly, factors that modulate the immune system, such as the gut microbiome, could also affect ICI efficacy.[8]
The microbiota that resides in the gastrointestinal tract provides essential health benefits to its host, particularly by regulating immune homeostasis. Moreover, a growing body of evidence suggests that alteration of this gut microbial community can cause immune dysregulation.[9] Recently, multiple clinical studies have demonstrated that the gut microbiome is an important regulator of systemic immune reactions and is involved in the response to ICI immunotherapy.[10–13] The first study linking the gut microbiome composition to ICI efficacy was a preclinical study using murine models published in 2015 that revealed a role for Bifidobacterium in enhancing antitumor immunity and augmenting the effect of ICIs in vivo.[14] Interestingly, such findings paved the way for numerous clinical studies published since 2017, which have established the association between the gut microbiome and response to ICIs in cancer patients. In this review, we summarize currently available data addressing the impact of the baseline gut microbiome, and its alteration with antibiotics, on ICI efficacy, and discuss the therapeutic implications of these findings in the context of cancer immunotherapy using ICIs.
SEARCH STRATEGY
References for this review were identified by using PubMed, Medline, and Embase with the search terms: immune checkpoint inhibitors, immunotherapy, cancer, responders, nonresponders, microbiota, gut microbiome, antibiotics, antimicrobials, immune-related adverse events, fecal microbiota transplant, and probiotics. We included only full text publications written in English. No time restriction was applied. Articles were initially selected by the title and the abstract. The full text was subsequently analyzed. The reference list of the included articles was also screened by FAA and RZ for relevant papers.
GUT MICROBIOTA AND RESPONSE TO IMMUNE CHECKPOINT INHIBITORS (ICIs)
The gut microbiome is a dynamic ecosystem that plays an essential role in maintaining immune homeostasis by tailoring the local and the systemic immune system.[10] Recently, many studies have suggested a role for the microbiome in the response of tumors to ICIs. Although the exact mechanisms have not been fully elucidated, current hypotheses suggest that the gut microbiome produces several molecules that affect the growth of cancer cells and modulate anticancer immunity. These molecules act as messengers that enter the circulation and signal to enhance the systemic immune cell responsiveness.[15] Such products include 1) bacterial toxins that induce immunogenic and necrotic cell death, 2) pathogen-associated molecular patterns (PAMPS) that activate Toll-like receptors and stimulate innate immunity, and 3) bacterially derived metabolites, the best known of which are the short-chain fatty acids (SCFAs) like butyrate, which can trigger cancer cell apoptosis by activating dendritic cells and cytokine production.[16–21] Several studies have also suggested possible mechanisms by which specific bacterial species of the gut microbiota exert their effect on the immune system and thus affect the response to ICIs (Table 1).
Table 1.
Responder bacteria associated with a positive response to immunotherapy and the proposed mechanisms by which they modulate the anticancer efficacy of ICI therapy
|
Main Responder Bacteria
|
References
|
Checkpoints Targeted
|
Proposed Mechanism
|
| Akkermansia | |||
| A. muciniphila | Routy et al, 2018 [55] | CTLA-4, PD-1 | Stimulate secretion of cytokines by MHC Class II restricted CD4+ T cells and DCs in the peripheral blood |
| A. muciniphila | Wind et al, 2020[47] | ||
| A. muciniphila | Zheng et al, 2019[59] | ||
| A. muciniphila | Salgia et al, 2020[46] | ||
| A. muciniphila | Botticelli et al, 2018[50] | ||
| Bacteroides | |||
| B. thetaiotaomicron, B. fragilis | Vétizou et al, 2015[48] | CTLA-4, PD-1 | Induce TH1 immune responses in tumor-draining lymph nodes and maturation DCs |
| B. thetaiotaomicron, B. caccae | Frankel et al, 2017[41] | ||
| B. plebeius | Botticelli et al, 2018[50] | ||
| B. eggerthii, B. thetaiotaomicron, B. massiliensis | Wind et al, 2020[47] | ||
| B. eggerthii | Salgia et al, 2020[46] | ||
| Bifidobacterium | |||
| B. breve, B. longum | Sivan et al, 2015[14] | CTLA-4, PD-1 | Increase accumulation of antigen-specific CD8+ TILs and MHC Class II DCs |
| B. longum | Botticelli et al, 2018[50] | ||
| B. longum | Matson et al, 2018[51] | ||
| B. longum | Jin et al, 2019[52] | ||
| B. pseudolongum | Mager et al, 2020[35] | ||
| B. adolescentis | Salgia et al, 2020[46] | ||
| Ruminococcaceae | |||
| Unspecified | Routy et al, 2018[55] | PD-1 | Increase CD8+ TILs and increase levels of CD4+ and CD8+ T cells in the peripheral blood |
| Unspecified | Gopalakrishnan et al, 2018[43] | ||
| R. obeum, R. bromii | Zheng et al, 2019[59] | ||
| Ruminococcaceae UCG 13 | Hakozaki et al, 2020[56] | ||
| Faecalibacterium | |||
| F. prausnitzii | Frankel et al, 2017[41] | CTLA-4, PD-1 | |
| F. prausnitzii L2-6 | Chaput et al, 2017[44] | ||
| Unspecified | Gopalakrishnan et al, 2018[43] | ||
| F. prausnitzii | Botticelli et al, 2018[50] | ||
ICI: immune checkpoint inhibitor; DC: dendritic cell; MHC: major histocompatibility complex; TH1: T-helper 1; TIL: tumor-infiltrating lymphocyte
The key to understanding the role of baseline gut microbiome and its disruption in human diseases and response to treatment is the stability of the microbiota over time. Sequencing studies have revealed that each individual harbors a unique collection of gut microbial species. Several studies have also shown remarkable long-term within-individual stability in the gut microbial abundance over years.[22–25] For example, by using 16S rRNA and whole genome sequencing techniques to characterize bacterial strain composition in the fecal microbiota, Faith et al[26] showed that on average 60% of the microbial strains harbored in each adult's intestine were retained over the course of 5 years. Moreover, a gut microbial composition with higher baseline diversity was more stable over time,[23,25] and members of the Bacteroidetes, Bifidobacterium, and Actinobacteria were more stable components of the microbiota than members of other phyla.[25,26] On the other hand, several studies have shown that the gut microbiota varied by various factors such as age, diet, geographic location, smoking, exercise, infections, and antibiotic administration.[27–31] This raises an important question: If the gut microbiome can be influenced by so many factors, how can it be used as a biomarker to predict response to ICIs? The study of metabolites as common outputs of bacterial metabolic function introduces the notion of a functional classification of the gut microbiome rather than a taxonomic one. It is plausible that different taxonomic groups of bacteria have evolved to perform similar metabolic functions, and thus can be classified by the type of metabolites they produce. Despite the variation in microbiota composition among individuals and over time, the gut microbiome metabolic pathways are what remain stable and mediate their interaction with the immune system to modify the response to ICIs. To date, most of the studies have focused on the baseline microbiota composition as a predictive marker of response to ICIs. Few studies have tried to establish the role of specific bacterial metabolites such as SCFAs, glycerophospholipids, and inosine. For example, Nomura et al[32] examined fecal and plasma levels of SCFAs in patients with solid tumors treated with anti–PD-1 in a prospective cohort study. They found that high concentrations of SCFAs were associated with longer progression-free survival (PFS). On the contrary, Coutzac et al[33] demonstrated that anti-CTL4 therapy efficacy was negatively influenced by systemic SCFAs. Xu et al[34] showed that better efficacy of anti–PD-1 treatment was associated with the glycerophospholipid pathway. Mager et al[35] found that mice administered with inosine showed improved antitumor effects from anti-CTL4 therapy. However, the exact mechanisms by which these metabolites modify antitumor immunity are still under investigation. A possible mechanism is related to the accumulation of CD8+ T cells in the tumor microenvironment and increased production of IFN-γ in response to SCFA.[36–38] Further mechanistic studies are needed to define the immunomodulatory role of these metabolites. Such level of understanding of the molecular details of the immune system–microbiota interaction would open a new area in this field, resulting in numerous possibilities for the prognostic and therapeutic use of the gut microbiome.[39,40]
Responders vs Nonresponders
To evaluate the effect of the gut microbiome on ICI response, it is important to determine the composition of the individual's gut flora. Sequencing technologies such as metagenomic shotgun sequencing and 16S RNA gene sequencing have been widely used for this purpose. Subsequently, patients treated with ICI therapy are typically classified as “responders” (R) or “nonresponders” (NR) based on the RECIST (Response Evaluation Criteria in Solid Tumors) criteria in most cases.[41,42] As a result, multiple correlations have been drawn that link particular bacterial species to the observed clinical responses. Most studies thus far have been conducted in patients with melanoma, although more recent ones have included patients with other solid tumors such as non–small cell lung cancer (NSCLC) and renal cell carcinoma (RCC).
Preclinical studies have been largely suggestive of the role of certain microbes in the clinical response to ICIs. Some of these correlations were later re-established in human studies. For example, a study of 43 melanoma patients found Faecalibacterium to be correlated with a positive response as demonstrated by prolonged PFS. In addition to responding better to anti–PD-1 therapy, the study revealed that transplanting fecal microbiota rich in Faecalibacterium from R patients to mice resulted in significant reduction in tumor growth, as well as an increase in the number of CD45+ immune and CD8+ T cells in the gut, as compared with mice with fecal microbiota transplant (FMT) from NR patients.[43] Other studies have also confirmed this positive correlation.[44,45]
Bacteroides species, particularly B. fragilis and B. thetaiotaomicron, have also been correlated with a positive response.[45–50] This was demonstrated in a murine study, whereby germ-free or antibiotic-treated mice did not respond to CTLA-4 blockade until gavage with B. fragilis, immunization with B. fragilis polysaccharides, or adoptive transfer of B. fragilis–specific T cells. The immunostimulatory role of Bacteroides was further demonstrated when treatment with CTLA-4–blocking antibodies in mice treated with FMT from R patients favored the outgrowth of B. fragilis.[48] As in murine studies, a study on melanoma patients also determined a positive correlation between Bacteroides species, namely B. massiliensis, and Streptococcus parasanguinis and outcome. Patients with large concentrations of these two bacteria had prolonged PFS and overall survival (OS), respectively. However, patients with large numbers of Peptostreptococcaceae had a shorter duration of both PFS and OS.[47]
Bifidobacterium is another important genus that has been consistently shown to be correlated with a positive response, as defined by the RECIST criteria.[51,52] This was established in early studies, whereby Bifidobacterium-treated mice displayed significantly improved tumor control in comparison with their non–Bifidobacterium-treated counterparts. In one study, oral administration of Bifidobacterium alone improved tumor control to the same degree as PD-L1 therapy blockade.[14] Another study conducted on a murine model of colorectal cancer also confirmed a significant role for Bifidobacterium pseudolongum. In this study, monocolonization of mice with either Lactobacillus johnsonii, Olsenella species, or B. pseudolongum significantly enhanced CTLA-4 blockade activity. B. pseudolongum had the most robust ICI response, owing to its correlation with an elevated level of inosine, a metabolite that promotes Th1 cell activation through adenosine 2A (A2A) receptor signaling.[35]
Correlation studies conducted on human subjects have also identified numerous bacterial species that impact the response to ICIs (Table 1). Bifidobacterium was again noted to favor a positive response, particularly B. longus and B. adolescentis in NSCLC and melanoma studies, respectively. These two studies also noted other bacteria correlated with R patients, including the previously noted Lactobacillus and Enterococcus faecium. Other R species included Prevotella copri, Alistipes putredinis, Parabacteroides, Methanobacteriaceae, Clostridium, Collinsella aerofaciens, Klebsiella pneumoniae, Veillonella parvula, Parabacteroides merdae, and Syntrophococcus. Importantly, both studies noted Ruminococcus to be associated with the NR group, particularly R. obeum. Other NR species included Sutterella, Bilophila, and Roseburia intestinalis.[48,51–54] Intriguingly, some studies have found Ruminococcaceae to have a general positive prognostic significance, which raises the question as to the cause of this difference.[43,55,56]
Akkermansia muciniphila was also consistently found to be associated with favorable clinical outcomes and improved PFS in NSCLC and RCC studies conducted by Routy et al[55] and Derosa et al.[57] A positive correlation was also noted in a study on melanoma patients.[51] In addition, a study comparing a control group with NSCLC patients found that A. muciniphila was more abundant in the gut of control participants.[49]
Chronology has been an interesting aspect to examine as well. A study on metastatic RCC included temporal profiling of the microbiome in patients treated with ICIs. In addition to identifying A. muciniphila, B. adolescentis, Barnesiella intestinihominis, Odoribacter splanchnicus, and Bacteroides eggerthii as correlated with clinical benefits, the study revealed a general increase in the relative abundance of A. muciniphila in patients deriving those benefits, with a relative decrease in abundance in those not deriving benefit. Other studies differ in reporting temporal changes, whereby some found no significant change in the microbiome composition over the course of ICI therapy, whereas others determined that microbial diversity changes in the first few weeks of treatment could be an early predictor of response.[44,46,58,59] This raises hope for the potential utility of microbiome modulation in improving outcomes of patients receiving ICI treatment.
The difference in results of all these studies is noteworthy. As detailed above, different studies have identified different responder bacteria. Although for the most part congruent, some inconsistencies were noted with certain bacteria such as B. eggerthii and Ruminococcaceae—among others—whereby different studies identified them as R or NR bacteria.[47] No clear explanation has yet been provided for these variations. The type of ICI used may be a possible explanation, as not all studies encompassed both PD-1/PD-L1 and CTLA-4 blockade. In the case of Ruminococcaceae, all studies have included only PD-1/PD-L1 inhibition. Despite most studies identifying these bacteria as R, a study by Jin et al[52] found them to be associated with the NR group, thus disputing the theory that specific R bacteria are associated with the subtype of the ICI at hand. Another possibility is that geographic location or cancer type may play a role as well. However, a few studies that have examined these variables have noted that the diversity of the microbiome per se is the main factor affecting response, rather than geographic locations.[47] In addition, multiple bacteria have been independently shown to be responders in studies conducted in vastly separate countries such as China, The Netherlands, Italy, and France.[47,49,55,59] Similarly, the same bacteria were found to be R bacteria in patients with different types of malignancy, such as the case of A. muciniphila in NSCLC, RCC, and melanoma.[47,55] It is true that currently available studies have not sufficiently accounted for confounding variables such as age or body mass index (BMI) which may in part play a role in the minor inconsistencies of some study results.[47] What remains most consistent, however, is the correlation between the diversity of the microbiome and favorable anticancer efficacy of ICIs. Abundance of R species is also favorable, but more studies are needed to identify implicated species and the means of interpreting their presence.[49,52] Although the exact mechanisms have not yet been fully elucidated, it may be related to the intricate cross talk between microbes through signaling molecules such as SCFAs.[49,52]
The role of the gut microbiome in human health extends further to influence the tumor microenvironment. A new trend to study how the intratumor microbial ecosystem—particularly in tumor locations that the gut microbes can reach—affects clinical outcome has emerged in recent years. A study conducted by Riquelme et al[60] examined the role of intratumor microbiome on long-term survival of pancreatic adenocarcinoma patients. They analyzed the tumor microbiome composition of 68 patients who underwent surgical resection of pancreatic adenocarcinoma. They found that greater intratumor microbial diversity (defined by the number of species) and higher abundance of Pseudoxanthomonas, Saccharopolyspora, Streptomyces, and Bacillus clausii were associated with better survival outcome. They also found a strong correlation between CD8+ and granzyme B tissue densities with the intratumor microbial diversity, which suggested that the tumor microbiome might influence the degree of CD8+ T-cell infiltration and activation to affect antitumor immune response. Interestingly, they demonstrated that the intratumor microbiome cross-talks with the gut microbiome. By performing FMT in mice previously treated with antibiotics and challenged with orthotopic implantation of syngeneic cancer lines, they were able to detect human donor bacteria in the murine tumor microbiome post FMT, whereas it remained absent from mice who did not receive FMT, suggesting that potentially the gut microbiome has the capacity to translocate to pancreatic tumors and this colonization can modify the tumor microbiome and antitumor immunity. However, studies of this topic are limited and the impact of tumor microbiome on prognosis is still not clear.
Manipulation of the Gut Microbiome Impacts ICI Efficacy
Antibiotic-induced dysbiosis and the therapeutic effect of probiotics
Intestinal dysbiosis is defined as the imbalance in the baseline structure and function of the gut microbiota. This is typically represented by major shifts in microbial composition, resulting in decreased overall diversity, decreased abundance of anti-inflammatory species such as Faecalibacterium prausnitzii and increased abundance of different Enterobacteriaceae species. As such, changes in the abundance of particular species can be considered as markers of dysbiosis.[61] Dysbiosis can be induced by exposure to various environmental factors such as antibiotics, diet, and infections. It has been associated with the pathogenesis of multiple intestinal diseases such as inflammatory bowel disease and colorectal cancer.[12,62–65] Several studies have demonstrated the important role played by antibiotics in intestinal dysbiosis. It has been shown that antibiotics restrict the microbial diversity in the gastrointestinal tract and displace certain bacterial taxa.[66–68] This alteration of taxonomic composition appears shortly after the administration of antibiotics and can persist for long periods.[68–71] A study of healthy human volunteers before, during, and after a 5-day course of oral ciprofloxacin demonstrated decreased abundance of about a third of the bacterial taxa in the gut during treatment. Although the microbiota largely returned to its pretreatment composition in about 4 weeks after treatment, several taxa failed to recover within 6 months.[68] Another study of the human fecal microbiota showed that a 7-day course of clindamycin reduced the diversity of Bacteroides, which did not return to the pretreatment composition for up to 2 years after treatment.[69] These studies suggest that although antibiotic-induced dysbiosis can be reversible, it can lead to persistent long-term impacts on the gut microbiome. The altered composition of the gut microbiome affects the mutual relationship between the microbiome and the host immune system.[10,72] Antibiotics are also shown to alter gene expression and metabolite production by the gut microbiome.[73,74] The administration of antibiotics can reduce the expression of antimicrobial peptides by intestinal cells, downregulate genes encoding MHC class I and II, and reduce the frequency of CD4+ T cells expressing IFN-γ and interleukin-17. Antibiotics also impact host immunity by reducing the production of bacterial metabolites like SCFAs, which have been implicated in regulating many aspects of intestinal immunity.[10,75] Interestingly, the impact of antibiotics on the gut microbiome exhibits significant interindividual variability.[68] Both drug-related factors (such as antibiotic class, timing of exposure, and route of administration) and host-related factors (such as age and microbiota composition) influence the perturbations of the gut microbiome caused by antibiotics. For example, oral vancomycin is a powerful modulator of the microbiota but not intravenous vancomycin. The microbiota of infants has been shown to be more affected by antibiotics than adults.[76] The initial state of the gut microbiota appears to influence its reshaping by antibiotics in adults. In a study of 18 healthy volunteers, a 7-day regimen of cefprozil led to similar qualitative alterations of microbiota in most subjects, but to drastically different alterations in a subset of individuals with a predominant Bacteroides enterotype.[77]
On the other hand, probiotics have demonstrated a beneficial role on human gastrointestinal diseases through influencing the intestinal microbiota.[78] They are suggested to restore microbial balance and recover the intestinal homeostasis, thus serving as a therapeutic option in several gastrointestinal disorders.[79,80] The mechanisms by which probiotics exert their therapeutic effects include: improvement of cell barrier function; antagonist activity against pathogenic bacteria; modulation of intestinal cytokine signaling; and exhibition of anti-inflammatory properties.[81–83] Probiotics' anti-inflammatory effect is mediated by the downregulation of proinflammatory cytokines like TNF-α and attenuation of the NF-kB signaling pathway.[79] Moreover, they can induce regulatory T cells, which inhibit the effector T cells that would cause inflammation.[83] Certain probiotics have been shown to reduce the rate of infection with Clostridium difficile and the incidence of antibiotic-associated diarrhea.[84–87] Accordingly, the association between the gut microbiome and the clinical response to ICIs makes it logical to hypothesize that the manipulation of the gut microbiome by antibiotics or probiotics would influence ICI efficacy.
Antibiotic-related dysbiosis impact on ICI efficacy
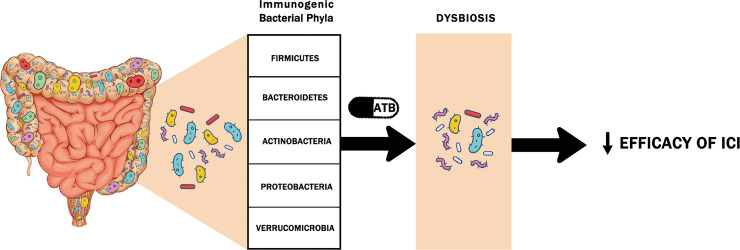
Antibiotic-related dysbiosis can eliminate the most immunogenic bacteria required to engage the immune system in the response to ICIs (Figure 1).[57] Moreover, by downregulating the expression of MHC class I and II, which are necessary for antigen presentation, they help tumor cells avoid immune recognition and thus decrease the efficacy of ICI therapy. To date, multiple clinical studies have attempted to determine the impact of antibiotics on the clinical efficacy of ICIs (Table 2).[55,57,88–98] In most of these studies, antibiotics were associated with reduced ICI efficacy, and both univariant and multivariant analyses confirmed that antibiotics received during a critical timeframe in patients receiving ICIs are independently associated with worse clinical outcomes, suggesting that an intact gut microbiome is needed to reinforce the immune system during immunotherapy. Interestingly, several meta-analyses have shown through subgroup analysis that this association is independent of cancer type, age, sample size, therapeutic strategy, ICI type, and timing of antibiotics.
Figure 1.
The five immunogenic bacterial phyla of the gut microbiome implicated in the response to immune checkpoint inhibitors (ICIs). It is speculated that antibiotic (ATB) use induces a dysbiosis that alters the concentrations of these immunogenic bacteria required to engage the immune system in the response to ICIs, thus reducing their efficacy.
Table 2.
Summary of available studies addressing the impact of antibiotics on patients with cancer treated with ICI therapy
|
Study
|
Cancer Type
|
Pts,
n |
ICI
|
Antibiotic Timing With Respect to ICI Initiation
|
Outcomes
|
Statistics,
p
or
HR (95% CI) |
| Kaderbahi et al, 2017[89] | NSCLC | 74 | Anti–PD-1 | Within 3 mo before ICI initiation | No change in PFS No change in RR | p = 0.72 p = 0.75 |
| Hakozaki et al, 2019[88] | NSCLC | 90 | Anti–PD-1 | Within 1 mo after ICI initiation | No change in OS | 2.0 (0.7–5.8) |
| Schett et al, 2020[90] | NSCLC | 218 | Anti–PDL-1 | Within 2 mo before ICI initiation | PFS (1.4 vs 5.5 mo) OS (1.8 vs 15.4 mo) PD (72.7 vs 43.9%) | 2.2 (1.5–3.3) 2.6 (1.7–4.0) p = 0.02 |
| Chalabi et al, 2020[91] | NSCLC | 757 | Anti–PDL-1 | Within 1 mo before and after ICI initiation | OS (8.5 vs 14.1 mo) | 1.3 (1.1–1.6) |
| Ruiz-Patino et al, 2020[92] | NSCLC | 140 | Anti–PDL-1 with chemotherapy, anti–PDL-1 alone | Within 1 mo before ICI initiation During treatment Anytime within treatment period | OS (20.3 vs 40.6 mo) OS (24.7 vs 40.6 mo) No change in PFS No change in RR | p < 0.05 p < 0.05 p > 0.05 p > 0.05 |
| Lalani et al, 2020[93] | RCC | 146 | Anti–PD-1, anti–PDL-1 alone or with chemotherapy | Within 1 mo before or after ICI initiation | RR (12.9 vs 34.8%) PFS (2.6 vs 8.1 mo) | p = 0.026 2.0 (1.2–3.2) |
| Elkrief et al, 2019[94] | Melanoma | 74 | CTLA-4 with chemotherapy, CTLA-4 alone, anti–PD-1 | Within 1 mo before ICI initiation | RR (0 vs 34%) PFS (2.4 vs 7.3 mo) | p = 0.01 p = 0.01 |
| Derosa et al, 2018[57] | NSCLC RCC | 239 121 | Anti–PD-1, anti–PD-1 + CTLA-4 | Within 1 mo before ICI initiation | NSCLC: PFS (1.9 vs 3.8 mo) OS (7.9 vs 24.6 mo) RCC: PD (75 vs 22%) PFS (1.9 vs 7.4 mo) OS (17.3 vs 30.6 mo) | 1.5 (1.0–2.2) 4.4 (2.6–7.7) p < 0.01 3.1 (1.4–6.9) 3.5 (1.1–10.8) |
| Routy et al, 2018[55] | NSCLC RCC UC | 140 67 42 | Anti–PD-1, anti–PDL-1 | Within 2 mo before or 1 mo after ICI initiation | PFS (3.5 vs 4.1 mo) OS (11.5 vs 20.6 mo) | p = 0.017 p < 0.001 |
| Tinsley et al, 2020[95] | NSCLC RCC Melanoma | 291 | Anti-PDL-1, CTLA-4 | Within 2 wk before or 6 wk after ICI initiation | PFS (3.1 vs 6.3 mo) OS (10.4 vs 21.7 mo) | p = 0.003, HR 1.6 p = 0.002, HR 1.7 |
| Ahmed et al, 2018[96] | Multiple | 60 | ICI with chemotherapy, anti–PD-1 or anti–PDL-1 alone | Within 2 wk before and/or after ICI initiation | RR (29.4 vs 62.8%) PFS (0 vs 22.5 wk) OS (24 vs 89 mo) | 1.9 (1.2–3.0) 1.6 (0.8–3.0) 2.9 (1.1–8.1) |
| Pinato et al, 2019[97] | Multiple | 196 | Anti–PD-1 Anti–PDL-1 | Within 1 mo before ICI initiation Anytime within treatment period | OS (2 vs 26 mo) PD (81 vs 44%) No change in OS | 7.4 (4.2–12.9) p < 0.001 p = 0.76 |
| Iglesias-Santamaría, 2020[98] | Multiple | 102 | Anti–PDL-1, CTLA-4 | Within 4 wk before or after ICI initiation anytime within treatment period | No change in PFS No change in OS PFS with ↑ AE* (3.1 vs 8.2 mo) OS with ↑ AE* (9.4 vs 17.8 mo) | 1.4 (0.9–2.3) 1.5 (0.9–2.7) 2.3 (1.2–4.6) 2.3 (1.2–4.9) |
ICI: immune checkpoint inhibitor; Pts: patients; HR: hazard ratio; NSCLC: non–small cell lung cancer; PFS: progression-free survival; RR: response rate; OS: overall survival; PD: primary progressive disease; RCC: renal cell carcinoma; UC: urothelial carcinoma; AE, antibiotic exposure; ↑ indicates increase.
AE defined as number of days of antibiotic use divided by the number of days of ICI use.
To further determine which type of antibiotics most significantly affected ICI efficacy, Ahmed et al[96] divided the patients into narrow-spectrum antibiotics recipients and broad-spectrum antibiotics recipients. Narrow-spectrum antibiotics were defined as antibiotics only covering gram-positive bacteria, whereas broad-spectrum antibiotics included antibiotics that would cover gram-positive and gram-negative with or without anaerobic bacteria. It was found that narrow-spectrum antibiotics did not affect the response rate (RR) to ICIs. In contrast, patients who received broad-spectrum antibiotics had a lower RR (25% vs 61%) and took longer time to respond to immunotherapy. These findings suggested that broad-spectrum antibiotics might have impoverished the microbes involved in stimulatory immune responses, creating a habitat for the microbes that induce suppressive immune responses, reducing ICI efficacy.
The study by Tinsley et al[95] was the first to shed light on the importance of antibiotic duration and cumulative antibiotic use. Recipients of antibiotics were categorized as either having had a single course of antibiotics, or cumulative courses of antibiotics, defined as concurrent or successive antibiotics for more than 7 days. The latter had significantly lower PFS and OS. Similar results were reported by Iglesias-Santamaría[98] whereby patients with higher “antibiotic exposure,” defined as % “days of antibiotic/days of ICI,” had significantly reduced clinical outcomes.
Derosa et al[57] examined the impact of antibiotic timing with respect to ICI initiation on the prognostic role of antibiotic administration. After finding that antibiotics administered within 30 days of starting ICIs were associated with decreased OS and PFS, they examined the effect of antibiotics within 60 days of ICI initiation. Although they were still associated with a worse clinical outcome, the effect was not as pronounced as within 30 days before ICI. These findings can be justified by evidence showing subsequent recovery of the gut microbiome within 1–3 months after antibiotic discontinuation.[99] This implies that there is a specific timeframe during which antibiotics will have the greatest impact on ICI efficacy. However, it remains difficult to define this timeframe considering the differences in the timing of antibiotic administration between the different studies. Ruiz-Patiño et al[92] also investigated whether antibiotic timing plays a role, to find if the effect of antibiotics would differ between patients who received them before ICI initiation and those who received them concomitantly with ICIs. In both groups, antibiotic use was associated with reduced OS with no significant difference observed between them. In contrast, Schett et al[90] showed that antibiotics had a negative impact on OS only when administered 2 months prior to ICI initiation, but not when administered during treatment or after ICI discontinuation. Similar results were also reported in the prospective study of Pinato et al.[97]
While multiple studies have concluded that, in general, antibiotic-induced dysbiosis can potentially dampen the antitumor effects of ICIs, some studies have produced evidence that is at odds with such findings. In a retrospective study by Kaderbhai et al,[89] investigators assessed the treatment outcomes of patients with NSCLC who were receiving anti–PD-1 immunotherapy with respect to antibiotic usage within the 3 months prior to the first dose; the study demonstrated a lack of association between response to ICI and antibiotic use. However, these results might have been affected by the long cutoff of 3 months, considering that the gut microbiome repopulation can occur within 4 weeks after antibiotic discontinuation. Another study by Hakozaki et al[88] on NSCLC patients also showed no effect of antibiotics administered within 1 month after initiation of ICI. Taking it a step further, a study by Metges et al[100] examined the outcomes of antibiotic use prior to and during treatment with ICIs in a cohort of 325 patients with advanced NSCLC and deemed that a survival advantage was appreciated in patients who had received antibiotics up to 60 days before or during immunotherapy (median survival, 16.2 months vs 11.5 months). Equivalently, the use of antibiotics during immunotherapy correlated with increased survival in a study by Masini et al.[101] In a similar vein, one study by Vétizou et al[48] that analyzed the factors that affect CTLA-4 blockade in patients with melanoma concluded that vancomycin could potentially augment the antitumor effect observed with anti–CTLA-4 treatment; this was attributed to a potentially induced favorable representation of certain components of the microbiota such as Bacteroidales as opposed to Clostridiales.
In summary, it has been well established that altering the gut microbiome can potentially impair the response to ICIs. However, if gut microbiome is to be used as a biomarker to predict the response to ICIs, we need to better understand in depth how it is affected by antibiotics. The above-mentioned studies need to be interpreted in the context of their study design. First, they are all retrospective observational studies, and thus their results reflect a general prognostic association and not causation, and they can be affected by selection bias and unmeasured confounders. The conflicting findings among the different studies are difficult to explain and may result from the inherent limitations of retrospective analysis such as small population size, variation in patient characteristics between groups, and different definitions of studied variables such as the antibiotic use window and antibacterial spectrum. Additional studies are needed to answer controversial questions regarding the effect of several variables that were not taken into account in most of the aforementioned studies, such as the duration of antibiotic treatment, the class of the antibiotic and its spectrum, the route of administration (intravenous versus oral), the indication for administration (infection treatment vs prophylaxis), and the severity of the infection. It is challenging to infer a causal link between the use of antibiotics and decreased response to ICIs. A confounding factor could be that frail patients who are less likely to benefit from cancer treatment are more likely to acquire infections that need antibiotic treatment. Moreover, it may be possible that the infections themselves have an immunosuppressive effect that alters the response to ICIs. Therefore, future studies are warranted to focus on the experimental designs to control for confounding factors and further delineate the mechanistic basis of the effects of antibiotics on ICI therapy.
Use of probiotics to modify the microbiome to a pattern favorable for ICI therapy
Due to the beneficial effect of probiotics on dysbiosis-related intestinal disorders, several studies investigated the role of probiotics in improving efficacy of ICI. Studies using mice models demonstrated that the efficacy of PDL-1 inhibitors was ameliorated with supplementation of probiotics containing Bifidobacterium species by augmenting the activity of tumor-specific CD8 T cells.[14] Similarly, in addition to a noteworthy association between the presence of A. muciniphila and favorable clinical outcomes, preclinical studies have demonstrated that supplementation with A. muciniphila improved PD-1 efficacy in mice models.[55] A study by Vétizou et al[48] conducted on mice models deemed that the antitumor efficacy of CTLA-4 antibodies improved after oral administration of B. fragilis with B. thetaiotaomicron or Burkholderia cepacia. The enhanced efficacy is associated with increased dendritic cell maturation and Th-1 predominant response.
A clinical single-center retrospective study was performed by Tomita et al[102] to investigate the role of probiotic treatment on PFS and OS in patients with advanced NSCLC treated with ICI. The study included 118 patients, 39% of whom received probiotics within 6 months of ICI therapy or during treatment. Probiotics were prescribed for alleviation of constipation or diarrhea. To minimize bias arising from participants' baseline characteristics, the authors conducted univariate analysis and survival analysis. They adjusted for several factors including age, sex, smoking history, initial staging, first- or second-line ICI therapy, monotherapy or combination therapies, use of antibiotics, and others. The authors were able to identify the use of probiotics as an independent prognostic factor associated with improved PFS and OS in patients with advanced NSCLC. This study also showed that patients who received antibiotics followed by probiotics had improved survival outcomes when compared with patients who received antibiotics alone. Another study by Takada et al[103] showed that probiotics administration improves PFS in patients with advanced or recurrent NSCLC treated with a PD-1 inhibitor. The study retrospectively analyzed data of 294 patients with NSCLC from multiple centers across Japan and used inverse probability of treatment weighting. This study revealed that the use of probiotics is an independent prognostic factor associated with increased PFS but not OS. Overall, the current clinical evidence is still limited. In light of the available promising data, future randomized clinical trials are required to investigate the role of probiotics in modulating response to immunotherapy and assess its therapeutic potential.
Fecal microbiota transplant
In recent years, increased effort uncovered the rising role of FMT as a method of microbiota modulation with therapeutic implications for ICI therapy. A study by Baruch et al[104] highlighted the promising role of FMT in improving therapy outcome. The authors performed an open-label single-armed randomized clinical trial on patients with metastatic melanoma resistant to at least one line of PD-1 inhibitors. Patients initially received oral antibiotics to deplete their baseline intestinal microbiota. Then, they received FMT from metastatic melanoma patients who were previously treated with PD-1 inhibitors and achieved at least 1 year of remission. Three patients had response to treatment, including one patient with complete remission. Davar et al[105] similarly conducted an open-label single-armed study that highlighted the improved clinical benefit in PD-1 refractory melanoma patients after responder-derived FMT, with increase in the abundance of taxa associated with a positive response.
The mechanisms through which a subsequent response to anti–PD-1 therapy was obtained following microbiome manipulation via FMT are interconnected to the remodeling of the gut microbiome–host immunity interface. Brauch et al[104] readily revealed that participants had an upregulated expression of genes correlated with the innate immune system and the antigen presentation machinery. Moreover, all participants had augmentation of T-cell activation. This pointed out the novel possibility of alternative mechanisms independent of PD-1 signaling that could further contribute to the enhanced antitumor activity. This encompasses a myriad of possible mechanisms that pertain to a noted enhanced signaling through antitumoral immune pathways, that is, increased IFN-γ signaling, escalated CD8+ cellular infiltration into the tumoral microenvironment, production of immunomodulatory metabolites by the microbiome constituents, and exclusion of potentially immunosuppressive microbes via microbiome reconstitution.[106]
Several randomized controlled trials are currently in progress to further investigate the role of FMT as an intervention to enhance the response to immunotherapy. However, to confirm its therapeutic efficacy, future research should focus on conducting double-blinded studies to account for bias or confounding variables that might be affecting the results.
CONCLUSION
In recent years, the intricate interplay between the gut microbiome and overall health has become an area of increasing interest. Growing evidence introduces the notion of relying on microbiome characterization as a prognostic marker capable of forecasting a patient's clinical response or resistance to ICIs. Several studies set out to determine the bacterial composition that influences the clinical response to ICIs, culminating in promising results that linked the presence of certain taxa such as Akkermansia, Ruminococcaceae, Faecalibaterium, Bacteroides, and Bifidobacterium to positive outcomes including reduction in tumor growth and increase in prolonged PFS and OS in some studies. These results opened doors for further examination as to how the manipulation of the microbiome could potentially affect ICI efficacy. Antibiotic-related dysbiosis has generally been linked to reduced ICI efficacy in most studies. On the other hand, supplementation with probiotics such as Bifidobacterium, which restores the gut dysbiosis, has emerged as a potential method through which augmentation of response to ICIs is possible. Additionally, alteration of the microbiome via FMT is being investigated in multiple randomized clinical trials to further elucidate its role as a therapeutic intervention able to shift outcomes in patients receiving ICIs.
Figure 3 provides a brief graphic summary of this review. We examined the most up-to-date evidence establishing the association between the gut microbiome and response to ICIs, the factors that influence the composition of the gut microbiome, possible therapeutic interventions to modulate the microbiome (Figure 2), and we presented some key unanswered questions to be addressed in future research (Figure 3). Complexity arises from the presence of several uncertainties on the matter, such as the factors impacting the gut microbiome composition, the modulatory effects of antibiotics on ICIs, the exact nature of “favorable” versus “non-favorable” bacterial species, the role of bacterial metabolites, and the precise influence of antibiotics and probiotics on ICIs response. More studies are warranted to better characterize the exact nature of the microbiome-ICI interaction, to propose novel and reliable biomarkers that enable prediction of future response or resistance to ICIs. It is important that future studies focus on standardization of sampling, sequencing techniques, data analysis, and study design in order to minimize the inconsistencies amongst the present data.
Figure 3.
Graphic summary of the emerging role of the gut microbiome in the cancer response to immune checkpoint inhibitors (ICIs). The baseline gut microbiome and its alteration with antibiotics have been shown to affect ICI efficacy by tailoring the immune system. Probiotics and fecal microbiota transplant (FMT) provide promising therapeutic interventions to enhance the response to ICIs.
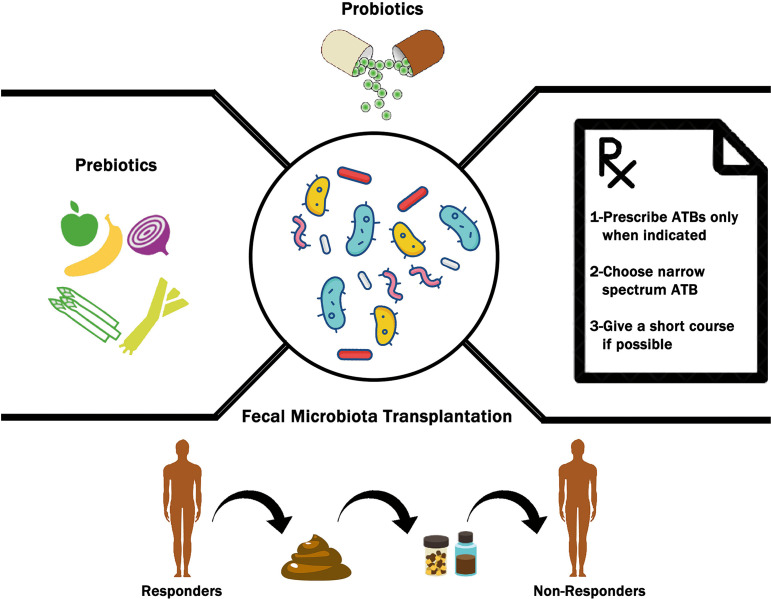
Figure 2.
The possible therapeutic interventions to optimize unfavorable gut microbiome into a favorable composition in order to improve the clinical outcomes of immune checkpoint inhibitors (ICIs). These interventions range from complex microbiome transfers in the form of fecal microbiota transplant (FMT), to delivery of microbes by probiotics. Additional interventions include prebiotic use to favor the growth of beneficial gut microbiome and following specific considerations when prescribing antibiotics (ATBs) in patients treated with ICIs.
Acknowledgment
We thank Amani Araji (Beirut Arab University, Beirut, Lebanon) for her assistance in the creation of the illustrations.
Author Interview
To hear more about this article, visit the JIPO YouTube Channel at https://m.youtube.com/watch?v=JQMvpKMOw4E.
Funding Statement
Source of Support: Dr. Yeung receives support from Bristol-Myer Squibb, Inc. and DepoMed, Inc. The other authors have nothing to disclose.
Footnotes
Conflict of Interest: Dr. Yeung is a member of an expert panel for Celgene, Inc. The other authors have nothing to disclose.
References
- 1.Elias R, Morales J, Rehman Y, Khurshid H. Immune checkpoint inhibitors in older adults. Curr Oncol Rep . 2016;18:47. doi: 10.1007/s11912-016-0534-9. [DOI] [PubMed] [Google Scholar]
- 2.Marin-Acevedo JA, Soyano AE, Dholaria B, et al. Cancer immunotherapy beyond immune checkpoint inhibitors. J Hematol Oncol . 2018;11:8. doi: 10.1186/s13045-017-0552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saman H, Uddin S, Raza S, et al. Understanding checkpoint inhibitors in cancer therapy, mechanisms of action, resistance and future challenges. Clin Oncol Res . 2020. pp. 1–13. [DOI]
- 4.Seto T, Sam D, Pan M. Mechanisms of primary and secondary resistance to immune checkpoint inhibitors in cancer. Med Sci (Basel) . 2019;7:14. doi: 10.3390/medsci7020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujiwara Y, Mittra A, Naqash AR, Takebe N. A review of mechanisms of resistance to immune checkpoint inhibitors and potential strategies for therapy. Cancer Drug Resist . 2020;3:252–275. doi: 10.20517/cdr.2020.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haslam A, Prasad V. Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw Open . 2019;2:e192535–e192535. doi: 10.1001/jamanetworkopen.2019.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darabi S, Braxton DR, Eisenberg BL, Demeure MJ. Predictive biomarkers for immunotherapy response beyond PD-1/PD-L1. Oncology (Williston Park) . 2020;34:321–327. doi: 10.46883/ONC.2020.3408.0321. [DOI] [PubMed] [Google Scholar]
- 8.Prelaj A, Tay R, Ferrara R, et al. Predictive biomarkers of response for immune checkpoint inhibitors in non-small-cell lung cancer. Eur J Cancer . 2019;106:144–159. doi: 10.1016/j.ejca.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Wu H-J, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes . 2012;3:4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ubeda C, Pamer EG. Antibiotics, microbiota, and immune defense. Trends Immunol . 2012;33:459–466. doi: 10.1016/j.it.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Deng Y, Chu Q, Zhang P. Gut microbiome and cancer immunotherapy. Cancer Lett . 2019;447:41–47. doi: 10.1016/j.canlet.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 12.Shui L, Yang X, Li J, et al. Gut microbiome as a potential factor for modulating resistance to cancer immunotherapy. Front Immunol . 2020;10:2989. doi: 10.3389/fimmu.2019.02989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Ma R, Liu F, et al. Modulation of gut microbiota: a novel paradigm of enhancing the efficacy of programmed death-1 and programmed death ligand-1 blockade therapy. Front Immunol . 2018;9:374. doi: 10.3389/fimmu.2018.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science . 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorjifard S, Goldszmid RS. Microbiota-myeloid cell crosstalk beyond the gut. J Leukoc Biol . 2016;100:865–879. doi: 10.1189/jlb.3RI0516-222R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iraporda C, Errea A, Romanin DE, et al. Lactate and short chain fatty acids produced by microbial fermentation downregulate proinflammatory responses in intestinal epithelial cells and myeloid cells. Immunobiology . 2015;220:1161–1169. doi: 10.1016/j.imbio.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Malla S, Niraula NP, Singh B, et al. Limitations in doxorubicin production from Streptomyces peucetius. Microbiol Res. 2010;165:427–435. doi: 10.1016/j.micres.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Paulos CM, Wrzesinski C, Kaiser A, et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J Clin Invest . 2007;117:2197–2204. doi: 10.1172/JCI32205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science . 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature . 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurav A, Sivaprakasam S, Bhutia YD, et al. Slc5a8, a Na+-coupled high-affinity transporter for short-chain fatty acids, is a conditional tumour suppressor in colon that protects against colitis and colon cancer under low-fibre dietary conditions. Biochem J . 2015;469:267–278. doi: 10.1042/BJ20150242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garud NR, Good BH, Hallatschek O, Pollard KS. Evolutionary dynamics of bacteria in the gut microbiome within and across hosts. PLoS Biol . 2019;17:e3000102. doi: 10.1371/journal.pbio.3000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frost F, Kacprowski T, Rühlemann M, et al. Long-term instability of the intestinal microbiome is associated with metabolic liver disease, low microbiota diversity, diabetes mellitus and impaired exocrine pancreatic function. Gut . 2021;70:522–530. doi: 10.1136/gutjnl-2020-322753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehta RS, Abu-Ali GS, Drew DA, et al. Stability of the human faecal microbiome in a cohort of adult men. Nat Microbiol . 2018;3:347–355. doi: 10.1038/s41564-017-0096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Wang D, Garmaeva S, et al. The long-term genetic stability and individual specificity of the human gut microbiome. Cell . 2021;184:2302–2315.e12. doi: 10.1016/j.cell.2021.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science . 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasan N, Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ . 2019;7:e7502. doi: 10.7717/peerj.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monda V, Villano I, Messina A, et al. Exercise modifies the gut microbiota with positive health effects. Oxid Med Cell Longev . 2017;2017:3831972. doi: 10.1155/2017/3831972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodríguez JM, Murphy K, Stanton C, et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis . 2015;26:26050. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biedermann L, Zeitz J, Mwinyi J, et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One . 2013;8:e59260. doi: 10.1371/journal.pone.0059260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyakht AV, Kostryukova ES, Popenko AS, et al. Human gut microbiota community structures in urban and rural populations in Russia. Nat Commun . 2013;4:2469. doi: 10.1038/ncomms3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nomura M, Nagatomo R, Doi K, et al. Association of short-chain fatty acids in the gut microbiome with clinical response to treatment with nivolumab or pembrolizumab in patients with solid cancer tumors. JAMA Netw Open . 2020;3:e202895. doi: 10.1001/jamanetworkopen.2020.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coutzac C, Jouniaux J-M, Paci A, et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat Commun . 2020;11:2168. doi: 10.1038/s41467-020-16079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X, Lv J, Guo F, et al. Gut microbiome influences the efficacy of PD-1 antibody immunotherapy on MSS-type colorectal cancer via metabolic pathway. Front Microbiol . 2020;11:814. doi: 10.3389/fmicb.2020.00814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mager LF, Burkhard R, Pett N, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science . 2020;369:1481–1489. doi: 10.1126/science.abc3421. [DOI] [PubMed] [Google Scholar]
- 36.Danne C, Sokol H. Butyrate, a new microbiota-dependent player in CD8+ T cells immunity and cancer therapy? Cell Rep Med . 2021;2:100328. doi: 10.1016/j.xcrm.2021.100328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kespohl M, Vachharajani N, Luu M, et al. The microbial metabolite butyrate induces expression of Th1-associated factors in CD4+ T cells. Front Immunol . 2017;8:1036. doi: 10.3389/fimmu.2017.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachem A, Makhlouf C, Binger KJ, et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity . 2019;51:285–297.e5. doi: 10.1016/j.immuni.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Thaiss CA, Elinav E. Exploring new horizons in microbiome research. Cell Host Microbe . 2014;15:662–667. doi: 10.1016/j.chom.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 40.Levy M, Thaiss CA, Elinav E. Metabolites: messengers between the microbiota and the immune system. Genes Dev . 2016;30:1589–1597. doi: 10.1101/gad.284091.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frankel AE, Coughlin LA, Kim J, et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia . 2017;19:848. doi: 10.1016/j.neo.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierrard J, Seront E. Impact of the gut microbiome on immune checkpoint inhibitor efficacy—a systematic review. Curr Oncol . 2019;26:395. doi: 10.3747/co.26.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science . 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol . 2017;28:1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 45.Frankel AE, Deshmukh S, Reddy A, et al. Cancer immune checkpoint inhibitor therapy and the gut microbiota. Integr Cancer Ther . 2019;18:1534735419846379. doi: 10.1177/1534735419846379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salgia NJ, Bergerot PG, Maia MC, et al. Stool microbiome profiling of patients with metastatic renal cell carcinoma receiving anti-PD-1 immune checkpoint inhibitors. Eur Urol . 2020;78:498–502. doi: 10.1016/j.eururo.2020.07.011. [DOI] [PubMed] [Google Scholar]
- 47.Wind TT, Gacesa R, Vich Vila A, et al. Gut microbial species and metabolic pathways associated with response to treatment with immune checkpoint inhibitors in metastatic melanoma. Melanoma Res . 2020;30:235–246. doi: 10.1097/CMR.0000000000000656. [DOI] [PubMed] [Google Scholar]
- 48.Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science . 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vernocchi P, Gili T, Conte F, et al. Network analysis of gut microbiome and metabolome to discover microbiota-linked biomarkers in patients affected by non-small cell lung cancer. Int J Mol Sci . 2020;21:8730. doi: 10.3390/ijms21228730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Botticelli A, Putignani L, Zizzari I, et al. Changes of microbiome profile during nivolumab treatment in NSCLC patients. J Clin Oncol . 2018;36(15 suppl):e15020–e15020. [Google Scholar]
- 51.Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science . 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin Y, Dong H, Xia L, et al. The diversity of gut microbiome is associated with favorable responses to anti-programmed death 1 immunotherapy in Chinese patients with NSCLC. J Thorac Oncol . 2019;14:1378–1389. doi: 10.1016/j.jtho.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 53.Song P, Yang D, Wang H, et al. Relationship between intestinal flora structure and metabolite analysis and immunotherapy efficacy in Chinese NSCLC patients. Thorac Cancer . 2020;11:1621–1632. doi: 10.1111/1759-7714.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katayama Y, Yamada T, Shimamoto T, et al. The role of the gut microbiome on the efficacy of immune checkpoint inhibitors in Japanese responder patients with advanced non-small cell lung cancer. Transl Lung Cancer Res . 2019;8:847–853. doi: 10.21037/tlcr.2019.10.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science . 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 56.Hakozaki T, Richard C, Elkrief A, et al. The gut microbiome associates with immune checkpoint inhibition outcomes in patients with advanced non-small cell lung cancer. Cancer Immunol Res . 2020;8:1243–1250. doi: 10.1158/2326-6066.CIR-20-0196. [DOI] [PubMed] [Google Scholar]
- 57.Derosa L, Hellmann MD, Spaziano M, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol . 2018;29:1437–1444. doi: 10.1093/annonc/mdy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Agarwal A, Modliszewski J, Davey L, et al. Investigating the role of the gastrointestinal microbiome in response to immune checkpoint inhibitors (ICIs) among patients (pts) with metastatic renal cell carcinoma (mRCC) J Clin Oncol . 2020;38(6 suppl):730. [Google Scholar]
- 59.Zheng Y, Wang T, Tu X, et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J Immunother Cancer . 2019;7:193. doi: 10.1186/s40425-019-0650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riquelme E, Zhang Y, Zhang L, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell . 2019;178:795–806.e12. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mahnic A, Breskvar M, Dzeroski S, et al. Distinct types of gut microbiota dysbiosis in hospitalized gastroenterological patients are disease non-related and characterized with the predominance of either Enterobacteriaceae or Enterococcus. Front Microbiol . 2020;11:120. doi: 10.3389/fmicb.2020.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zareef R, Younis N, Mahfouz R. Inflammatory bowel disease: a key role for microbiota? Meta Gene . 2020;25:100713. [Google Scholar]
- 63.Carding S, Verbeke K, Vipond DT, et al. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis . 2015;26:26191. doi: 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu S, Rhee K-J, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med . 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Angelakis E, Armougom F, Million M, Raoult D. The relationship between gut microbiota and weight gain in humans. Future Microbiol . 2012;7:91–109. doi: 10.2217/fmb.11.142. [DOI] [PubMed] [Google Scholar]
- 66.Lange K, Buerger M, Stallmach A, Bruns T. Effects of antibiotics on gut microbiota. Dig Dis . 2016;34:260–268. doi: 10.1159/000443360. [DOI] [PubMed] [Google Scholar]
- 67.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A . 2011;108(suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol . 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J . 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 70.Heinsen F-A, Knecht H, Neulinger SC, et al. Dynamic changes of the luminal and mucosa-associated gut microbiota during and after antibiotic therapy with paromomycin. Gut Microbes . 2015;6:243–254. doi: 10.1080/19490976.2015.1062959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Looft T, Johnson TA, Allen HK, et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci U S A . 2012;109:1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dufour V, Millon L, Faucher J-F, et al. Effects of a short-course of amoxicillin/clavulanic acid on systemic and mucosal immunity in healthy adult humans. Int Immunopharmacol . 2005;5:917–928. doi: 10.1016/j.intimp.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 73.Cerutti A, Rescigno M. The biology of intestinal immunoglobulin A responses. Immunity . 2008;28:740–750. doi: 10.1016/j.immuni.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wlodarska M, Willing B, Keeney KM, et al. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun . 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host–microbiota mutualism. Nat Rev Microbiol . 2011;9:233–243. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 76.Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut . 2016;65:1906–1915. doi: 10.1136/gutjnl-2016-312297. [DOI] [PubMed] [Google Scholar]
- 77.Raymond F, Ouameur AA, Déraspe M, et al. The initial state of the human gut microbiome determines its reshaping by antibiotics. ISME J . 2016;10:707–720. doi: 10.1038/ismej.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Simone C. The unregulated probiotic market. Clin Gastroenterol Hepatol . 2019;17:809–817. doi: 10.1016/j.cgh.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 79.Solga SF, Diehl AM. Non-alcoholic fatty liver disease: lumen-liver interactions and possible role for probiotics. J Hepatol . 2003;38:681–687. doi: 10.1016/s0168-8278(03)00097-7. [DOI] [PubMed] [Google Scholar]
- 80.Sanders ME, Heimbach JT, Pot B, et al. Health claims substantiation for probiotic and prebiotic products. Gut Microbes . 2011;2:127–133. doi: 10.4161/gmic.2.3.16174. [DOI] [PubMed] [Google Scholar]
- 81.Jones JL, Foxx-Orenstein AE. The role of probiotics in inflammatory bowel disease. Dig Dis Sci . 2007;52:607–611. doi: 10.1007/s10620-006-9225-y. [DOI] [PubMed] [Google Scholar]
- 82.Gionchetti P, Rizzello F, Campieri M. Probiotics in gastroenterology. Curr Opin Gastroenterol . 2002;18:235–239. doi: 10.1097/00001574-200203000-00014. [DOI] [PubMed] [Google Scholar]
- 83.Boirivant M, Strober W. The mechanism of action of probiotics. Curr Opin Gastroenterol . 2007;23:679–692. doi: 10.1097/MOG.0b013e3282f0cffc. [DOI] [PubMed] [Google Scholar]
- 84.Pozzoni P, Riva A, Bellatorre AG, et al. Saccharomyces boulardii for the prevention of antibiotic-associated diarrhea in adult hospitalized patients: a single-center, randomized, double-blind, placebo-controlled trial. Am J Gastroenterol . 2012;107:922–931. doi: 10.1038/ajg.2012.56. [DOI] [PubMed] [Google Scholar]
- 85.Sampalis J, Psaradellis E, Rampakakis E. Efficacy of BIO K+ CL1285 in the reduction of antibiotic-associated diarrhea—a placebo controlled double-blind randomized, multi-center study. Arch Med Sci . 2010;6:56–64. doi: 10.5114/aoms.2010.13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Song HJ, Kim J-Y, Jung S-A, et al. Effect of probiotic Lactobacillus (Lacidofil® cap) for the prevention of antibiotic-associated diarrhea: a prospective, randomized, double-blind, multicenter study. J Korean Med Sci . 2010;25:1784–1791. doi: 10.3346/jkms.2010.25.12.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McFarland LV. Meta-analysis of probiotics for the prevention of antibiotic associated diarrhea and the treatment of Clostridium difficile disease. Am J Gastroenterol . 2006;101:812–822. doi: 10.1111/j.1572-0241.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 88.Hakozaki T, Okuma Y, Omori M, Hosomi Y. Impact of prior antibiotic use on the efficacy of nivolumab for non-small cell lung cancer. Oncol Lett . 2019;17:2946–2952. doi: 10.3892/ol.2019.9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaderbhai C, Richard C, Fumet JD, et al. Antibiotic use does not appear to influence response to nivolumab. Anticancer Res . 2017;37:3195–3200. doi: 10.21873/anticanres.11680. [DOI] [PubMed] [Google Scholar]
- 90.Schett A, Rothschild SI, Curioni-Fontecedro A, et al. Predictive impact of antibiotics in patients with advanced non small-cell lung cancer receiving immune checkpoint inhibitors: antibiotics immune checkpoint inhibitors in advanced NSCLC. Cancer Chemother Pharmacol . 2020;85:121–131. doi: 10.1007/s00280-019-03993-1. [DOI] [PubMed] [Google Scholar]
- 91.Chalabi M, Cardona A, Nagarkar DR, et al. Efficacy of chemotherapy and atezolizumab in patients with non-small-cell lung cancer receiving antibiotics and proton pump inhibitors: pooled post hoc analyses of the OAK and POPLAR trials. Ann Oncol . 2020;31:525–531. doi: 10.1016/j.annonc.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 92.Ruiz-Patiño A, Barrón F, Cardona AF, et al. Antibiotics impair immune checkpoint inhibitor effectiveness in Hispanic patients with non-small cell lung cancer (AB-CLICaP) Thorac Cancer . 2020;11:2552–2560. doi: 10.1111/1759-7714.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lalani A-KA, Xie W, Braun DA, et al. Effect of antibiotic use on outcomes with systemic therapies in metastatic renal cell carcinoma. Eur Urol Oncol . 2020;3:372–381. doi: 10.1016/j.euo.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Elkrief A, El Raichani L, Richard C, et al. Antibiotics are associated with decreased progression-free survival of advanced melanoma patients treated with immune checkpoint inhibitors. Oncoimmunology . 2019;8:e1568812. doi: 10.1080/2162402X.2019.1568812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tinsley N, Zhou C, Tan G, et al. Cumulative antibiotic use significantly decreases efficacy of checkpoint inhibitors in patients with advanced cancer. Oncologist . 2020;25:55–63. doi: 10.1634/theoncologist.2019-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ahmed J, Kumar A, Parikh K, et al. Use of broad-spectrum antibiotics impacts outcome in patients treated with immune checkpoint inhibitors. Oncoimmunology . 2018;7:e1507670. doi: 10.1080/2162402X.2018.1507670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pinato DJ, Howlett S, Ottaviani D, et al. Association of prior antibiotic treatment with survival and response to immune checkpoint inhibitor therapy in patients with cancer. JAMA Oncol . 2019;5:1774–1778. doi: 10.1001/jamaoncol.2019.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Iglesias-Santamaría A. Impact of antibiotic use and other concomitant medications on the efficacy of immune checkpoint inhibitors in patients with advanced cancer. Clin Transl Oncol . 2020;22:1481–1490. doi: 10.1007/s12094-019-02282-w. [DOI] [PubMed] [Google Scholar]
- 99.Raymond F, Déraspe M, Boissinot M, et al. Partial recovery of microbiomes after antibiotic treatment. Gut Microbes . 2016;7:428–434. doi: 10.1080/19490976.2016.1216747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Metges J-P, Michaud E, Deniel Lagadec D, et al. Impact of anti-infectious and corticosteroids on immunotherapy: nivolumab and pembrolizumab follow-up in a French study. Ann Oncol . 2018. 29:viii431.
- 101.Masini C, Berselli A, Romagnani A, et al. Results of an Italian CORE-IMMUNO study: safety and clinical-related biomarkers as predictors of immunotherapy (IT) benefit in real-world treatment of various advanced tumors (ATs) J Clin Oncol . 2019;37(15 suppl):e14156–e14156. [Google Scholar]
- 102.Tomita Y, Ikeda T, Sakata S, et al. Association of probiotic Clostridium butyricum therapy with survival and response to immune checkpoint blockade in patients with lung cancer. Cancer Immunol Res . 2020;8:1236–1242. doi: 10.1158/2326-6066.CIR-20-0051. [DOI] [PubMed] [Google Scholar]
- 103.Takada K, Shimokawa M, Takamori S, et al. Clinical impact of probiotics on the efficacy of anti-PD-1 monotherapy in patients with nonsmall cell lung cancer: a multicenter retrospective survival analysis study with inverse probability of treatment weighting. Int J Cancer . 2021;149:473–482. doi: 10.1002/ijc.33557. [DOI] [PubMed] [Google Scholar]
- 104.Baruch EN, Youngster I, Ben-Betzalel G, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science . 2021;371:602–609. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 105.Davar D, Dzutsev AK, McCulloch JA, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science . 2021;371:595–602. doi: 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rebeck ON, Dantas G, Schwartz DJ. Improving ICI outcomes with a little help from my microbial friends. Cell Host Microbe . 2021;29:155–157. doi: 10.1016/j.chom.2021.01.012. [DOI] [PubMed] [Google Scholar]