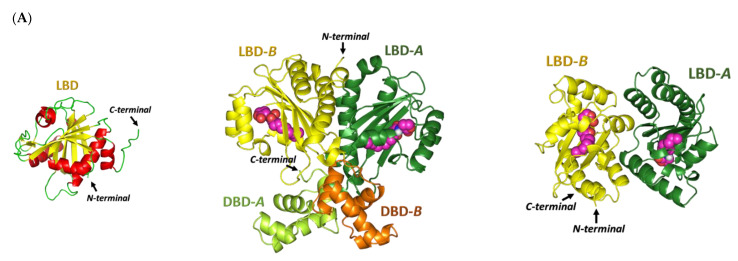
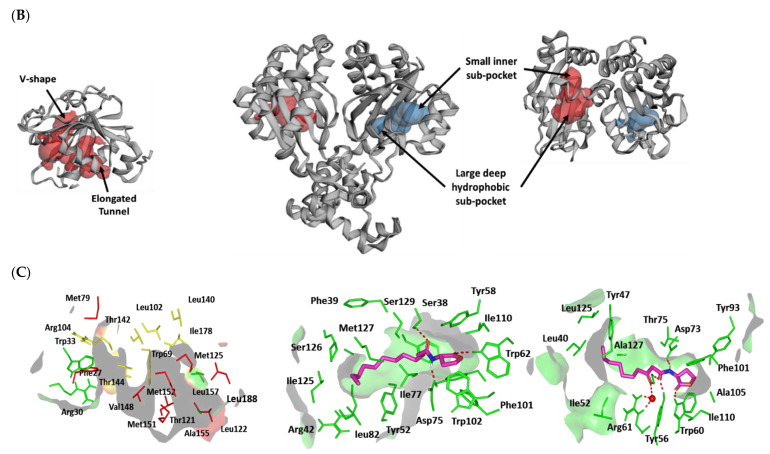
Figure 6.
Architectures of three P. aeruginosa virulence-regulating biological targets. (A) Overall 3D cartoon representations of the P. aeruginosa LasI-type AHL synthase ligand-binding domain (PDB ID: 1R05; right panel), as well as QscR (PDB ID: 6CC0; middle panel) and LasR (PDB ID: 6MVN; right panel) quorum-sensing transcription proteins. Each quorum-sensing protein is colored differently in regard to its ligand-binding domain (LBD) and/or DNA-binding domain (DBD) as light/dark green and dark/light orange for protomers A and B, respectively. Ligands are represented as magenta spheres with the exception of LasI AHL synthase, where no ligand was co-crystallized with the protein. (B) Calculated putative pockets of the binding site topology at the three P. aeruginosa biological targets via the web-based Computed Atlas of Surface Topography of proteins (CASTp) server (http://sts.bioe.uic.edu/castp/index.html; accessed on 30 October 2021) using 1.4 Å radius probe, visualized as surface 3D representation, and colored in different colors (blue and red) for each target protomer. Estimated pocket area and volume were analytically calculated using Richard’s solvent-accessible surface model. (C) Surface representation of the binding site with an overlay of whichever co-crystallized ligand (magenta sticks) of its respective bacterial target is present: C12-HSL (6CC0) and 3-O-C10-HSL (6MVN). Hydrogen bonding is shown as dashed lines (red), while residues (green lines), only amino acids located within 5 Å radius distance from the bound ligand, are displayed and labeled in sequence numbers. Crystallized water molecule bridging the ligand/residue interactions is shown as red sphere.