Abstract
We have been working to develop an enzymatic assay for the alcohol 2-methyl-3-buten-2-ol (232-MB), which is produced and emitted by certain pines. To this end we have isolated the soil bacterium Pseudomonas putida MB-1, which uses 232-MB as a sole carbon source. Strain MB-1 contains inducible 3-methyl-2-buten-1-ol (321-MB) and 3-methyl-2-buten-1-al dehydrogenases, suggesting that 232-MB is metabolized by isomerization to 321-MB followed by oxidation. 321-MB dehydrogenase was purified to near-homogeneity and found to be a tetramer (151 kDa) with a subunit mass of 37,700 Da. It catalyzes NAD+-dependent, reversible oxidation of 321-MB to 3-methyl-2-buten-1-al. The optimum pH for the oxidation reaction was 10.0, while that for the reduction reaction was 5.4. 321-MB dehydrogenase oxidized a wide variety of aliphatic and aromatic alcohols but exhibited the highest catalytic specificity with allylic or benzylic substrates, including 321-MB, 3-chloro-2-buten-1-ol, and 3-aminobenzyl alcohol. The N-terminal sequence of the enzyme contained a region of 64% identity with the TOL plasmid-encoded benzyl alcohol dehydrogenase of P. putida. The latter enzyme and the chromosomally encoded benzyl alcohol dehydrogenase of Acinetobacter calcoaceticus were also found to catalyze 321-MB oxidation. These findings suggest that 321-MB dehydrogenase and other bacterial benzyl alcohol dehydrogenases are broad-specificity allylic and benzylic alcohol dehydrogenases that, in conjunction with a 232-MB isomerase, might be useful in an enzyme-linked assay for 232-MB.
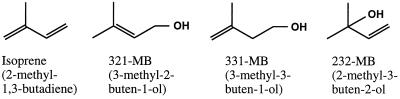
The biosphere releases a complex mixture of volatile organic compounds (VOCs) into the atmosphere (9). Some of these VOCs, such as isoprene (Fig. 1), have sufficient chemical reactivity and are emitted in such large quantities that their release can significantly alter the chemistry of the atmosphere (11). Recently, the emission of another reactive VOC, 2-methyl-3-buten-2-ol (232-MB), was described by Goldan et al. (14), who determined that 232-MB was the major nonmethane VOC emitted by a Colorado pine forest. Isoprene and 232-MB are structurally-related members of the C5 hemiterpene family of isoprenoids (3) (Fig. 1).
FIG. 1.
Naturally occurring hemiterpenes.
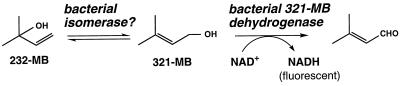
We have begun to investigate the biogenesis of 232-MB in pines such as Ponderosa and lodgepole pines. This C5 alcohol is produced in pine needles in a light-dependent manner similar to that seen for isoprene formation in other plants (16). Such studies will require a suitable analytical method for assaying 232-MB formation in pine needle extracts. Gas chromatographic methods are not well suited for this highly water- soluble alcohol, and high-pressure liquid chromatographic (HPLC) methods lack sensitivity. Consequently, we have considered a hypothetical enzyme-linked assay for 232-MB which exploits the fluorescence of NADH (Fig. 2). In this scheme 232-MB would be isomerized to 3-methyl-2-buten-1-ol (321-MB), followed by oxidation of 321-MB to 3-methyl-2-buten-1-al with concomitant reduction of NAD+ to NADH (20). However, the enzymes required for such an assay have apparently never been described.
FIG. 2.
A proposed enzyme-linked assay for 232-MB. In this assay the 232-MB would be isomerized to 321-MB by a putative isomerase. 321-MB would subsequently be oxidized to 3-methyl-2-buten-1-al by a 321-MB dehydrogenase with the concomitant reduction of NAD+ to NADH. The production of NADH would then be monitored by photometric or fluorometric means.
Towards this end we have investigated the possibility that bacteria associated with 232-MB-emitting pines might be capable of assimilating 232-MB as a sole carbon source and thus might contain enzymes that would be used for the assay diagrammed in Fig. 2. Here we describe the successful isolation of a soil bacterium that can metabolize 232-MB, and we demonstrate that it contains an inducible allylic alcohol dehydrogenase that has a high affinity for 321-MB. The properties of this allylic alcohol dehydrogenase and its relationship to other bacterial benzyl alcohol dehydrogenases are also presented.
MATERIALS AND METHODS
Materials.
Growth medium components were obtained from Difco. 232-MB and 321-MB, as well as several other substrates and substrate analogs, were supplied by Aldrich or Acros Organics. Sigma Chemical Co. supplied the DEAE anion-exchange resin, protein standards for gel filtration chromatography, coenzymes, and other reagents. The phenyl-Sepharose hydrophobic interaction resin, the Superdex fast protein liquid chromatography (FPLC) gel filtration column, and all other FPLC columns and equipment were purchased from Pharmacia (Uppsala, Sweden). Equipment and supplies for gel electrophoresis, including molecular weight markers and buffers, were provided by NOVEX. Bio-Rad Laboratories supplied the Bio-Sil SEC 250 HPLC gel filtration column.
Bacterial isolation and identification.
Bacteria capable of growth on 232-MB as a sole carbon source were isolated by enrichment culture from soil sampled under Austrian pine trees (Pinus nigra) growing locally. Soil particles were suspended by vortexing in mineral salts medium H (10), and dilutions of this suspension were used to inoculate agar plates of H medium supplemented with 100 μg of cycloheximide/ml to inhibit fungal growth; 500 μl of a 16% (vol/vol) aqueous solution of 232-MB was placed on the lid of each plate to provide a volatile carbon source, and the plates were sealed with Parafilm and incubated at room temperature. After several days colonies were picked and transferred to liquid H medium with 0.16% (vol/vol) 232-MB as a sole carbon source; positive cultures were restreaked for single colony isolates on H agar with 232-MB as above. Similar methods were used in an attempt to isolate 232-MB-utilizing bacteria from P. nigra needles; in this case individual needles were vortexed in H medium salts with 0.1% (wt/vol) Tween 20, and this rinse solution was used to inoculate plates.
One of the isolates from a soil enrichment was identified as Pseudomonas putida MB-1 by the following tests (28). MB-1 was gram negative and oxidase positive, grew at 4°C but not at 41°C, failed to hydrolyze gelatin, utilized l-arginine as a sole nitrogen source, and produced yellow pigment on King B agar that fluoresced under UV light. It was assigned to biotype B because of its ability to assimilate d-galactose, glucuronate, trigonelline, and saccharate but not nicotinate or benzylamine (28). For growth tests with isoprenoid alcohols and benzyl alcohol, cells were streaked on H agar plates and each potential carbon source was tested both by placing a 10-μl drop on the agar surface and, separately, by placing 25 μl on the lid, and then sealing the plates with Parafilm and incubating them at room temperature. Growth was scored at 3 and 7 days. P. putida MB-1 was maintained frozen in 7% (vol/vol) dimethyl sulfoxide (DMSO) at −70°C.
Acinetobacter calcoaceticus NCIB 8250 was obtained from the American Type Culture Collection as Acinetobacter sp. strain 11171. It was maintained on H agar plates with benzyl alcohol as a carbon source. P. putida PaW1 (also known as strain mt-2) containing TOL plasmid pWW0 (32) was provided by Peter Williams (University of Wales, Bangor, United Kingdom). It was maintained on H agar with 20 mM m-toluate as a carbon source. The presence of the TOL plasmid was determined by an alkaline lysis method (4). The same method was used with strain MB-1, including performance of the lysozyme step at 4, 22, and 37°C.
Growth of bacteria.
P. putida MB-1 was cultured in Hcy medium containing 0.5% (vol/vol) 232-MB as a carbon source. Medium Hcy contains H salts, 0.1% (wt/vol) casein digest, and 0.1% (wt/vol) yeast extract. Cells were grown at 25°C with moderate shaking (180 rpm) for 36 h to early stationary phase. Then the cells were harvested by centrifugation at 10,000 × g, washed three times in 50 mM KH2PO4 (pH 7.0), and then weighed and stored at −20°C. A. calcoaceticus NCIB 8250 and P. putida PaW1 were grown, harvested, and washed in the same way, except that the growth medium was Hcy containing 10 mM benzyl alcohol.
Standard enzyme assays.
The standard assay for the oxidation reaction of 321-MB dehydrogenase was carried out at 22°C and contained 100 mM Bis-Tris propane, 200 mM hydrazine (pH 9.4), 667 μM 321-MB (from a DMSO solution), and 1.3 mM NAD+. This assay was not conducted at the optimal pH (discussed below) in order to maximize the stability of the enzyme. The reaction was initiated by the addition of enzyme. Hydrazine was included in the reaction mixture in order to remove the free form of the aldehyde product as described by Shaw and Harayama (25); this resulted in more-linear initial rates of reaction. The 3-methyl-2-buten-1-al dehydrogenase assay was conducted at 25°C in 1 ml of 100 mM Bis-Tris propane (pH 9.4) containing 1.0 mM NAD+ and 500 μM 3-methyl-2-buten-1-al (from a DMSO solution). In some experiments the aldehyde dehydrogenase was assayed with benzaldehyde (500 μM) as a substrate. For all assays the reduction of NAD+ or oxidation of NADH was monitored at 340 nm with a double-beam UV-visible recording spectrophotometer (Shimadzu Scientific Instruments). The molar extinction coefficient for NADH was taken to be 6,220 M−1 cm−1, and one unit of enzyme was defined as the amount catalyzing the reduction of 1 μmol of NAD+ per min.
Inducibility.
The inducibility of 321-MB dehydrogenase was determined by measuring the enzyme activity in permeabilized cells which had been cultivated in Hcy medium with various carbon sources present at concentrations of 5 to 50 mM. Each sample was inoculated with a subculture of MB-1 which had been grown on Hcy medium for 16 to 18 h. The inoculated cultures were then grown with moderate shaking for 36 to 40 h at 25°C, and the cell density was determined (A600). The samples were harvested by centrifugation at 10,000 × g for 10 min and washed twice in 10 ml of 50 mM KH2PO4 (pH 7.0) containing 2 mM dithiothreitol (DTT) (buffer A), and then a weighed amount of each cell pellet was resuspended in 1 ml of ice-cold permeabilization buffer (100 mM KH2PO4 [pH 7.6] containing 0.05% [vol/vol] Triton X-100) per 100 mg of cells (22). The suspension was then mixed by vortexing and frozen in liquid nitrogen. The frozen suspensions were immediately thawed by swirling in a 30°C water bath, and the activity was measured by the standard assay. Reported activities were normalized for cell density in the assay based on the A600 of each sample cuvette.
Purification of 321-MB dehydrogenase.
Frozen pelleted cells (≅30 g [wet weight]) were resuspended in 1 to 2 volumes of buffer A. DNase (a few crystals) was added to the resuspension, and the mixture was homogenized. The cells were then broken by two passes through a precooled French pressure cell (American Instrument Co.) at a pressure of 14,000 lb/in2. The homogenate was centrifuged at 42,000 × g for 60 min at 4°C. The supernatant was filtered through a 0.2-μm-pore-size MCE syringe filter (Fisher) and used as the starting material for the purification of 321-MB dehydrogenase. Unless otherwise stated, all chromatography was performed on an FPLC system (Pharmacia) with precooled buffers in a cold room at 4°C.
The filtered extract (≅50 ml) was applied at a flow rate of 1.0 ml/min to a DEAE-Sephacel anion-exchange column (16 mm by 20 cm) preequilibrated in buffer A. The column was washed with 250 ml of buffer A before 321-MB dehydrogenase activity was eluted from the column by a linear gradient of 0 to 0.5 M KCl in buffer A. The gradient volume was 500 ml, and 321-MB dehydrogenase began to elute after a volume of approximately 210 ml. Fractions containing at least 20% of peak activity were pooled for further purification. The pooled fractions (≅50 ml) were applied to a phenyl-Sepharose 6 Fast Flow (low sub) column (10 mm by 3 cm) preequilibrated in buffer A. The column was washed with 50 ml of buffer A, and the enzyme was eluted by a linear gradient of 0 to 50% (vol/vol) ethylene glycol in buffer A. The total gradient volume was 30 ml, and 321-MB dehydrogenase began to elute after ≅20 ml. Ten milliliters of 60% ethylene glycol in buffer A was used to elute the remaining enzyme from the column. (In later experiments a gradient of 0 to 65% [vol/vol] ethylene glycol in buffer A was used to completely elute the enzyme.) The pooled fractions (≅20 ml) were concentrated by ultrafiltration (Amicon) under nitrogen by using a Diaflo (Amicon) membrane with a molecular mass cutoff of 10 kDa.
The concentrated enzyme (≅5 ml) was loaded at a flow rate of 0.5 ml/min onto a Superdex 200 (Pharmacia) FPLC gel filtration column (16 mm by 60 cm) preequilibrated in 50 mM KH2PO4 (pH 7.0) containing 100 mM NaCl and 2 mM DTT (buffer B). The column was then washed in buffer B at a flow rate of 1.0 ml/min. The A280 of the effluent was continually monitored by a flowthrough UV-absorbance detector (Pharmacia), and only one significant peak was observed. This peak had a retention time of 66 min and contained 321-MB dehydrogenase activity. Fractions containing at least 20% of peak activity were pooled and then concentrated for storage. The pooled fractions (≅8 ml) were concentrated to 1 ml, and the buffer was changed to buffer A in 30% glycerol, with a Centricon-10 microconcentrator (Amicon). The concentrated enzyme was then frozen in liquid nitrogen and stored at −70°C.
Protein determinations.
Protein concentrations were determined according to the method of Bradford (5), with bovine serum albumin as a standard. The A595 of 5-μl samples mixed with 200 μl of reagent was measured in 96-well microplates with a Kinetic Microplate Reader (Molecular Devices).
SDS-PAGE.
The purity of 321-MB dehydrogenase and its subunit mass were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by comparison to the migration of Mark 12 (NOVEX) molecular weight protein standards. Samples were run on a NOVEX X-cell II Mini-Cell electrophoresis system with NuPAGE precast gradient minigels (4 to 12% Bis-Tris) and NuPAGE morpholineethanesulfonic acid (MES) running buffer (50 mM MES, 50 mM Tris base, 3.5 mM SDS, and 1 mM EDTA). Gels were stained for 20 min with a solution of 0.1% (wt/vol) Coomassie blue R in 40% (vol/vol) ethanol–10% (vol/vol) acetic acid. Destaining was conducted for 10 min in 40% (vol/vol) ethanol–10% (vol/vol) acetic acid. If required, gels were destained for an additional 10 min in 90% (vol/vol) ethanol–5% (vol/vol) acetic acid. Stained gels were stored in zip-lock bags containing a solution of 50% (vol/vol) ethanol.
Native molecular mass.
The final purity of the enzyme and its native molecular mass were monitored by analytical gel filtration carried out on a Waters HPLC system (Millipore Corp.) using a Bio-Sil SEC 250 column (Bio-Rad). Buffer B was used as the mobile phase at a flow rate of 1.0 ml/min. Fifty microliters of each standard protein (2 to 10 mg/ml) or 50 μl of 321-MB dehydrogenase (0.8 mg/ml) was injected onto the column. Standards included β-amylase (200 kDa), yeast alcohol dehydrogenase (150 kDa), albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa). The absorbance of the effluent was continually monitored by a flowthrough UV-visible absorbance detector (Waters), and the retention times were calculated by using Millennium 2010 Chromatography Manager software (Millipore).
pH optima.
The pH optima for the oxidation and reduction reactions of 321-MB dehydrogenase were determined as in the standard assay except that the enzyme was incubated with the reaction mixture for 2 min and the reaction was initiated by the addition of NAD+. The following buffer systems were used for the oxidation reaction: 100 mM Bis-Tris–NaOH and 200 mM hydrazine for pH 6.0 to 7.0; 100 mM Bis-Tris propane–NaOH and 200 mM hydrazine for pH 7.1 to 9.6; and 100 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS)–NaOH and 200 mM hydrazine for pH 9.7 to 11.2. The buffers for the reduction reaction were 100 mM citric acid–NaOH for pH 3.0 to 6.6 and 100 mM Bis-Tris propane–NaOH for pH 6.7 to 9.4. In the case of the oxidation reaction, initial velocities were measured at least twice at three different nonsaturating concentrations, and for the reduction reaction, initial velocities were measured at least twice at a substrate concentration of 150 μM.
Kinetic parameters and determination of substrate specificity.
The apparent Km and Vmax for various substrates of 321-MB dehydrogenase were determined under assay conditions different from those used during the standard assay. Hydrazine, used in the standard assay during purification to trap the aldehyde product, was eliminated in order to avoid complications in the interpretation of the initial-rate data. The buffer used (glycine–NaOH [pH 9.4]) was chosen so that the results could be compared to the kinetic data obtained with the related TOL plasmid-encoded benzyl alcohol dehydrogenase (26, 27). The kinetic parameters were determined at 22°C in 1-ml reaction mixtures containing 100 mM glycine–NaOH (pH 9.4), 1.3 mM NAD+, and varying amounts of substrate alcohols added from stock solutions prepared in DMSO and diluted with water so that the final DMSO concentration in the assay was less than 1% (vol/vol). For the determination of the Km for NAD+, the same assay conditions were used with either 400 μM 321-MB or 70 μM benzyl alcohol as a substrate; these concentrations are approximately 10 times the Km values for these alcohols.
Determination of kinetic constants first involved analysis by Lineweaver-Burk plots over a range of substrate concentrations (typically 6 to 8) distributed above and below the apparent Km (30). With most alcohols, significant substrate inhibition was seen, so data were collected at low substrate concentrations. Initial velocities were typically measured 3 to 4 times at each substrate concentration, and the data were then fit with enzyme kinetics software (Enzfitter; Sigma Chemical Co.) by using the Marquandt algorithm. Substrate specificity was calculated as (apparent Vmax)/(apparent Km).
Measurement of enzyme inhibition.
The effects of various metal ion chelators and thiol-blocking reagents were determined at 22°C by the standard assay. Enzyme and inhibitor were incubated for 5 min in 50 mM KH2PO4 (pH 7.0); then the mixture was brought to standard assay conditions and the reaction was initiated by the addition of NAD+. The enzyme was incubated with the inhibitor at a neutral pH in order to minimize enzyme denaturation, and the alcohol and NAD+ substrates were omitted from the incubation mixture in an effort to prevent substrate protection.
N-terminal sequencing.
The N-terminal sequence of 321-MB dehydrogenase was determined by Macromolecular Resources (Fort Collins, Colo.) by using automated Edman degradation. 321-MB dehydrogenase was prepared for analysis by blotting the sample from a denaturing gel onto a polyvinylidene difluoride (PVDF) membrane. Five micrograms of enzyme from a phenyl-Sepharose peak fraction was electrophoresed under denaturing conditions as described above and then electroblotted onto the PVDF membrane by using the X-Cell II Mini-cell system with a blot module (NOVEX). Electroblotting was conducted at 25 V for 2 h in transfer buffer (12 mM Tris base–96 mM glycine) containing 20% (vol/vol) methanol. After transfer the membrane was stained with 0.1% Coomassie blue R in 40% (vol/vol) ethanol, destained for 10 to 20 min in 40% (vol/vol) ethanol, and then submitted for N-terminal sequencing.
RESULTS
Soil bacteria can utilize 232-MB as a sole carbon source.
As described above, we were interested in determining if soil bacteria can utilize 232-MB as a sole carbon source so that such cells might yield enzymes useful in an enzyme-linked assay for 232-MB. We determined that 232-MB enrichment cultures using soil from under a pine (P. nigra) as an inoculum contained several different bacteria capable of growth on this alcohol (typically there were hundreds of 232-MB+ colonies per gram of soil). Similar bacteria were not recovered from pine needle washings. One of the soil isolates, strain MB-1, was purified and identified as a P. putida biotype B by the methods described in Materials and Methods. This isolate could assimilate 232-MB but not the related hemiterpene alcohols 321-MB and 331-MB (Fig. 1) or the monoterpene alcohols geraniol, citronellol, linalool, (S)- or (R)-perillyl alcohol, and limonene. In addition, benzyl alcohol did not serve as a carbon source.
P. putida MB-1 contains an inducible ADH active with 321-MB.
In initial experiments it was established that cell extracts from strain MB-1 grown on 232-MB contained an alcohol dehydrogenase (ADH) activity dependent on 321-MB and NAD+; the same extracts also contained benzyl alcohol dehydrogenase activity. In order to determine whether 321-MB dehydrogenase is an inducible enzyme, cells were grown on a variety of carbon sources, permeabilized, and then assayed for ADH activity with both 321-MB and benzyl alcohol. These two alcohols were tested as substrates to determine if potential inducers would induce both ADH activities. Hcy medium with no additional carbon source was used as a control to establish the extent of growth on the carbon source versus growth on the casein and yeast extract components of the medium. The results are summarized in Table 1. Cells grown in the presence of 232-MB and, to a lesser extent, 321-MB and butanol contained 321-MB dehydrogenase activity. No 321-MB dehydrogenase activity was detected in cells grown in the presence of nontoxic levels of benzyl alcohol (5 mM) or 2-methyl-2-butanol, a structural analog of 232-MB that lacks the allylic double bond, or in cells grown on glucose or ethanol. The same pattern of induction was seen for benzyl alcohol dehydrogenase activity in these cells (Table 1). We expected that 232-MB catabolism might involve enzymes of the methylcrotonyl-coenzyme A (CoA) pathway, in a manner analogous to that for l-leucine degradation. However, growth on l-leucine, a known inducer of the methylcrotonyl-CoA pathway (21), also failed to induce 321-MB dehydrogenase activity.
TABLE 1.
Induction of 321-MB and benzyl alcohol dehydrogenase activities in P. putida MB-1
| Carbon source | Cell growth (A600) | Activitya of:
|
|
|---|---|---|---|
| 321-MB dehydrogenase (nmol min−1A600−1) | Benzyl alcohol dehydrogenase (nmol min−1A600−1) | ||
| Hcy | 1.58 | <0.5b | <0.5b |
| Hcy + 5 mM 232-MB | 2.46 | 61 | 58 |
| Hcy + 50 mM 232-MB | 4.41 | 85 | 93 |
| Hcy + 5 mM 321-MB | 2.39 | 10 | 9 |
| Hcy + 5 mM 2-methyl-2-butanol | 1.44 | <0.5 | <0.5 |
| Hcy + 23 mM l-leucine | 6.58 | <0.5 | <0.5 |
| Hcy + 5 mM benzyl alcohol | 1.58 | <0.5 | <0.5 |
| Hcy + 50 mM glucose | 8.49 | <0.5 | <0.5 |
| Hcy + 50 mM ethanol | 4.66 | <0.5 | <0.5 |
| Hcy + 5 mM 1-butanol | 2.41 | 13 | 16 |
321-MB and benzyl alcohol dehydrogenase activities were measured with permeabilized cells grown on Hcy medium with various carbon sources as described in Materials and Methods. Cell growth was assessed by the A600 of duplicate cultures, and dehydrogenase activities in permeabilized cells were measured at least twice and normalized for cell density based on the A600 of the reaction mixtures; the averages of these values are presented in the table. Data are results of a typical experiment repeated two times.
The detection limit in these assays was about 0.5 nmol min−1 A600−1.
It is known that many catabolic pathways in pseudomonads are encoded by plasmid genes (23). For example, a benzyl alcohol dehydrogenase is encoded by the TOL plasmid in P. putida (26), and we wondered if the 321-MB and benzyl alcohol dehydrogenase activities in strain MB-1 could be due to expression from a TOL-type plasmid. Using the method of Birnboim and Doly for plasmid analysis (4), we could not detect any plasmids in strain MB-1, even though by the same method the TOL plasmid pWW0 of P. putida PaW1 was readily detected.
Copurification of P. putida 321-MB and benzyl alcohol dehydrogenase activities.
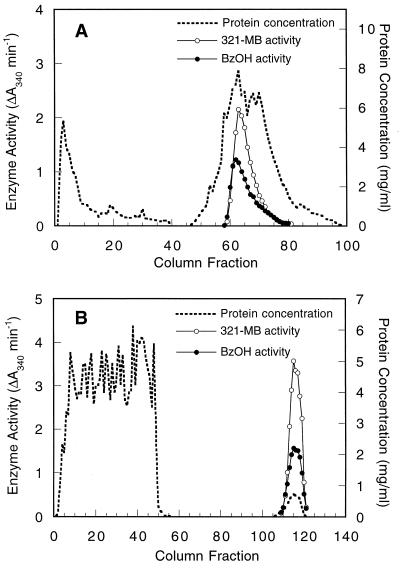
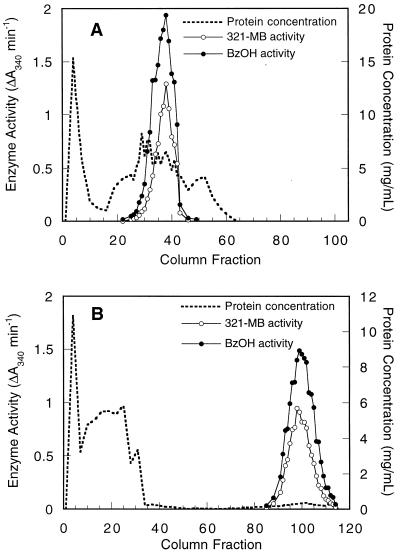
321-MB dehydrogenase was purified by adapting the methods of Chalmers et al. (8), and at each step the activity of benzyl alcohol dehydrogenase was also tested. Cell extracts were first resolved by anion-exchange chromatography on DEAE-Sephacel, and both 321-MB and benzyl alcohol dehydrogenase activities were copurified (Fig. 3A). When these fractions were pooled and chromatographed on a hydrophobic interaction column, all of the 321-MB and benzyl alcohol dehydrogenase activities were adsorbed and then coeluted with a gradient of ethylene glycol (Fig. 3B). These activities also copurified on the Superdex gel filtration column (data not shown). These results suggest that the same enzyme is responsible for the oxidation of both an allylic and an aromatic alcohol.
FIG. 3.
Coelution of P. putida MB-1 321-MB and benzyl alcohol dehydrogenase activities during DEAE-Sephacel chromatography (A) and phenyl-Sepharose chromatography (B). These chromatographic steps for the purification of 321-MB dehydrogenase are described in Materials and Methods. Dehydrogenase activities were eluted from the DEAE-Sephacel column by a linear gradient of 0 to 0.5 M KCl and from the phenyl-Sepharose column by a linear gradient of 0 to 65% ethylene glycol. The concentration of eluted protein was determined by the method of Bradford. Dehydrogenase activities for 321-MB and benzyl alcohol (BzOH) were quantified by measuring the increase in A340 under the standard assay conditions.
The results from a typical purification are summarized in Table 2. 321-MB dehydrogenase was purified 165-fold based on an increase in specific activity; similar results were obtained when benzyl alcohol was used as the substrate (data not shown). Approximately 4 to 5 mg of the purified enzyme were obtained from 30 g (wet weight) of cells. These results are similar to those of the chromosomally encoded benzyl alcohol dehydrogenase from A. calcoaceticus and the plasmid-encoded benzyl alcohol dehydrogenase from P. putida PaW1, which were purified 159- and 181-fold, respectively (18, 25). However, the specific activity of the purified enzyme with 321-MB as a substrate was 5- to 10-fold lower than published values for either of these other ADHs with benzyl alcohol as a substrate; the maximal specific activity was 38.8 μmol min−1 mg of protein−1 with Bis-Tris propane–hydrazine (pH 9.4) as the buffer and 19.3 μmol min−1 mg of protein−1 with glycine (pH 9.4) as the buffer. Purified preparations of 321-MB dehydrogenase in 50 to 65% (vol/vol) ethylene glycol were stable for several months when frozen in liquid nitrogen and stored at −70°C.
TABLE 2.
Purification of 321-MB dehydrogenase from P. putida MB-1a
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cell extract | 2,608 | 594 | 0.23 | 100 | 1 |
| DEAE-Sephacel | 290 | 398 | 1.37 | 67 | 6 |
| Phenyl-Sepharose | 8.7 | 265 | 30.6 | 45 | 134 |
| Superdex 200 | 4.7 | 179 | 37.7 | 30 | 165 |
Experimental details are described in Materials and Methods.
Subunit mass and molecular weight.
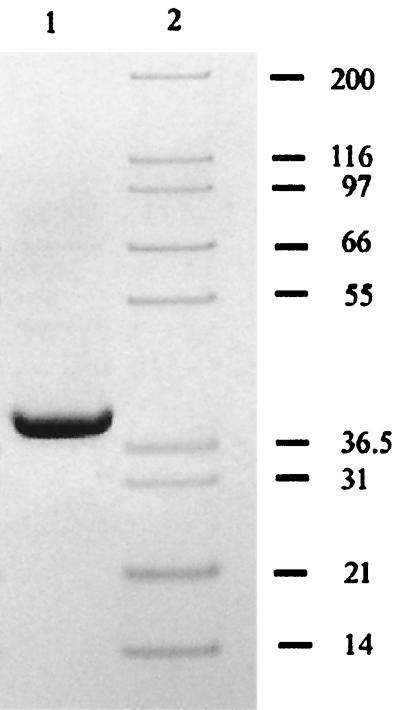
The results of SDS-PAGE analysis of samples from various phases in the purification process revealed the enrichment of a polypeptide with an approximate molecular mass of 38 kDa. Figure 4 shows that SDS-PAGE analysis of purified 321-MB dehydrogenase revealed only this single polypeptide; at a higher loading of protein, some minor bands could be detected, so the purified enzyme is approximately 95% pure. Known benzyl alcohol dehydrogenases are multimers with subunits that vary in size between 38 and 42 kDa (13, 18, 25). When the purified enzyme was subjected to analytical gel filtration chromatography, a single peak was observed in the chromatogram, and fractions from this peak, collected as they eluted from the column, were found to contain 321-MB dehydrogenase activity (data not shown). Estimates of the native molecular mass of the enzyme obtained from standards run on this column varied between 138 and 142 kDa. However, 321-MB dehydrogenase consistently had a retention time greater than or equal to that of yeast alcohol dehydrogenase, a known tetramer with a molecular weight of 150 kDa (6). Therefore, 321-MB dehydrogenase is believed to be a tetramer of identical 37.7-kDa subunits with a native molecular mass of approximately 151 kDa.
FIG. 4.
321-MB dehydrogenase purity and subunit mass determination by SDS-PAGE. The gel shown here was run and Coomassie stained as described in Materials and Methods. Lanes: 1, purified 321-MB dehydrogenase (5 μg); 2, molecular size markers (in kilodaltons).
Detection of a 3-methyl-2-buten-1-al dehydrogenase.
Extracts of P. putida MB-1 contained an aldehyde dehydrogenase that was capable of oxidizing 3-methyl-2-buten-1-al. This activity was also induced by growth on 232-MB but not by growth on glucose (data not shown). 3-Methyl-2-buten-1-al dehydrogenase was found to copurify with 321-MB dehydrogenase on the DEAE-Sephacel column but was not present in the fractions eluted from the phenyl-Sepharose column. On both of these columns the peak of 3-methyl-2-buten-1-al dehydrogenase activity coeluted with benzaldehyde dehydrogenase activity, suggesting that these activities may be the result of a single enzyme; however, no further work has been done to purify and characterize this enzyme.
Effects of pH on enzyme activity.
By using 321-MB as a substrate, the optimum pH for the oxidation reaction catalyzed by 321-MB dehydrogenase was determined to be 10.0. The reduction reaction of 321-MB dehydrogenase had an optimal pH of 5.4. These optimum pH values are similar to those for the oxidation and reduction reactions of the TOL-encoded benzyl alcohol dehydrogenase from P. putida; this enzyme was found to have an optimal pH of 9.4 for the oxidation reaction and 5.7 for the reduction reaction (25). Additionally, the maximum velocity of the reduction reaction at the optimal pH was found to be greater than that of the oxidation reaction. However, it is not uncommon for ADHs to exhibit a reductase activity that exceeds the dehydrogenase activity (18).
We noticed that the enzyme was slowly inactivated when preincubated at pH 10 at 25°C, even in the presence of DTT, while it was much more stable at pH 9.4. For this reason, routine assays of the enzyme were conducted at a pH of 9.4.
Kinetic properties and substrate specificity.
The kinetic parameters of the 321-MB dehydrogenase were first determined with 321-MB, benzyl alcohol, and NAD+. For purposes of comparison to the kinetic data of Shaw et al. (26) on the TOL plasmid-encoded benzyl alcohol dehydrogenase from P. putida, the analyses were conducted in glycine-NaOH buffer (pH 9.4). Hydrazine, present in the standard assay to help relieve aldehyde product inhibition, was omitted in kinetic assays in order to avoid complications in analysis of the results. With 400 μM 321-MB or 70 μM benzyl alcohol (10 times their Kms) as a substrate, Kms for NAD+ were 64.0 ± 5.9 or 118 ± 12.4 μM, respectively. The enzyme exhibited less than 5% of its activity when NADP+ was used as the cofactor instead of NAD+. The Kms for 321-MB and benzyl alcohol (at 1.3 mM NAD+) were 39.6 ± 3.5 and 7 ± 1.3 μM, respectively.
Purified 321-MB dehydrogenase was found to oxidize a wide variety of allylic and aromatic alcohols. To a lesser extent, it was also active with some aliphatic alcohols. These results are summarized in Table 3; the structures of the tested substrates for the enzyme are also shown. The highest enzyme specificity (apparent Vmax/apparent Km) was obtained with the allylic alcohols, 3-chloro-2-buten-1-ol, (R)-(+)-perillyl alcohol, and 321-MB, and with the aromatic alcohols, benzyl and 3-aminobenzyl alcohol. By comparing allylic, nonallylic, and aliphatic substrates, it can be seen that analogs of 321-MB lacking the allylic double bond, such as 3-methyl-1-butanol and 3-methyl-3-buten-1-ol, exhibited specificity constants 39- and 96-fold lower, respectively. These decreases were due to combinations of large increases in the Km and reductions in the Vmax (Table 3), suggesting that the allylic double bond in 321-MB serves to enhance both substrate binding and a catalytic event. The chloro analog of 321-MB, 3-chloro-2-buten-1-ol, also an allylic alcohol, was the most effective allylic substrate, whereas the aliphatic 4-carbon alcohol n-butanol was a very poor substrate, with a Km of 1,100 μM compared to 40 μM for 321-MB. Of the allylic alcohols tested, 3-chloro-2-buten-1-ol and (R)-(+)-perillyl alcohol had the lowest Kms (7 to 12 μM); perhaps these allylic alcohols show features of substrate binding in the active site that mimic the binding of the aromatic alcohols, benzyl and 3-aminobenzyl alcohol, which had similarly low Kms (7 to 11 μM [Table 3]). The purified enzyme exhibited the highest Vmax and enzyme specificity with 3-aminobenzyl alcohol as a substrate; similar results have been obtained with other bacterial benzyl alcohol dehydrogenases, where the enhanced turnover number (kcat) is attributed to the electron-donating effect of the p-amino group (19, 27). As with benzyl alcohol dehydrogenase from A. calcoaceticus (19), phenylethanol was not a substrate for the enzyme (data not shown), illustrating the importance of the presence of an aromatic group on the α-carbon of the alcohol substrate. The purified 321-MB dehydrogenase did not oxidize 232-MB, indicating that the enzyme does not catalyze isomerization of 232-MB to 321-MB. 232-MB was also a very poor inhibitor of the enzymatic oxidation of 321-MB, suggesting that the enzyme has a low binding affinity for this tertiary alcohol.
TABLE 3.
Kinetic parameters for 321-MB dehydrogenase with various substratesa
| Substrate | Structure | Apparent Vmax (U mg−1) | Apparent Km (μM) | Specificity, Vmax/Km (U mg−1 μM−1) |
|---|---|---|---|---|
| Allylic and nonallylic 3-Chloro-2-buten-1-ol | 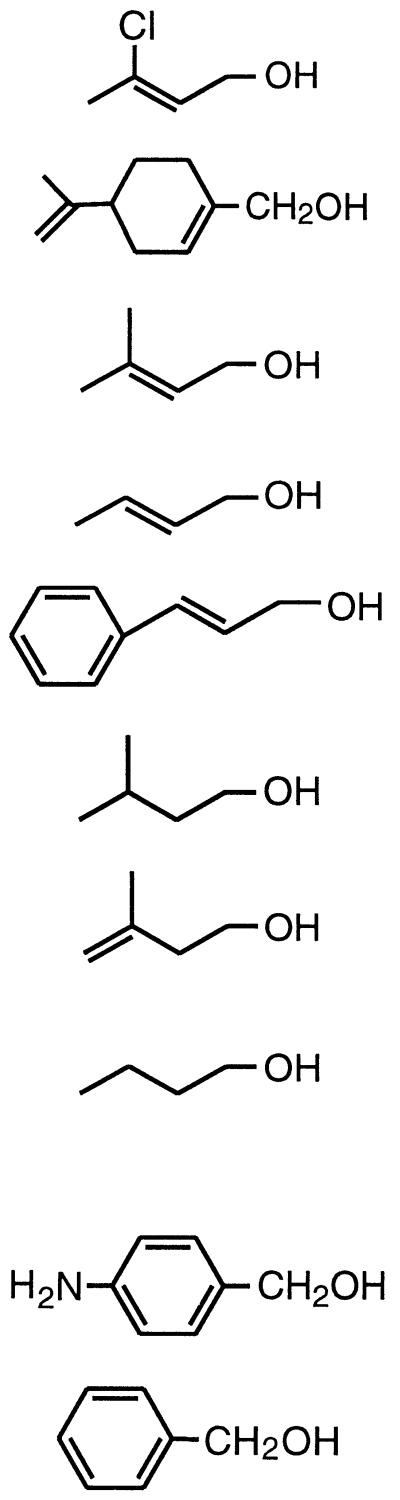 |
114 | 7 | 16 |
| (R)-(+)-Perillyl alcohol | 106 | 12 | 8.8 | |
| 321-MB | 308 | 40 | 7.7 | |
| 2-Buten-1-ol | 202 | 44 | 4.6 | |
| Cinnamyl alcoholb | 132 | 36 | 3.7 | |
| 3-Methyl-1-butanol | 57 | 270 | 0.2 | |
| 3-Methyl-3-buten-1-ol | 128 | 1,700 | 0.08 | |
| 1-Butanol | 53 | 1,100 | 0.05 | |
| Aromatic 3-Aminobenzyl alcohol | 418 | 11 | 38 | |
| Benzyl alcohol | 155 | 7 | 22 | |
Values were determined as described in Materials and Methods.
Cinnamyl alcohol, which contains an aromatic ring, is listed with the allylic substrates because it contains an allylic alcohol moiety.
Effects of metal ion chelators and thiol-blocking reagents on the activities of 321-MB dehydrogenase.
We wanted to determine if 321-MB dehydrogenase is a member of the Zn2+-dependent family of ADHs with essential cysteine residues in the active site. The enzyme was relatively insensitive to the presence of metal ion chelators in the reaction mixture. With 321-MB as a substrate, the enzyme was uninhibited in the presence of 100 mM 2,2-bipyridyl or 10 mM EDTA but exhibited a 50% reduction in activity when assayed in the presence of 100 mM EDTA. Thiol-blocking reagents such as iodoacetate and N-ethylmaleimide inhibited 321-MB dehydrogenase at moderate concentrations: a 20 or 50% reduction in activity in the presence of 2.5 mM N-ethylmaleimide or iodoacetate, respectively, and a 100 or 80% loss of activity with 10 mM N-ethylmaleimide or iodoacetate, respectively. Similar results were seen when benzyl alcohol was used as a substrate. The addition of excess DTT restored activity to the enzyme inhibited with N-ethylmaleimide but not with iodoacetate.
N-terminal sequence analysis.
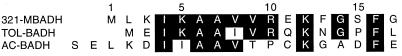
321-MB dehydrogenase was not N-terminally blocked, and the first 17 amino acid residues were unambiguously identified by Edman degradation (see below). This sequence was analyzed by the BLAST and gapped-BLAST database search programs (1), and the search revealed only one homologous enzyme, the TOL plasmid-encoded benzyl alcohol dehydrogenase of P. putida (TOL-BADH). A search of the literature revealed that the N-terminal sequence of TOL-BADH had previously been aligned with the A. calcoaceticus benzyl alcohol dehydrogenase (AC-BADH) and other homologous ADHs (7). The N-terminal sequences of these two bacterial benzyl alcohol dehydrogenases were aligned with the 321-MB alcohol dehydrogenase (321-MBADH), and the results are shown in Fig. 5. When TOL-BADH and AC-BADH are aligned with residues 4 to 16 of 321-MBADH, there are nine identical residues in the TOL-BADH sequence (69% identity) and six identical residues in the AC-BADH sequence (46% identity). These results suggest a significant degree of structural similarity among these bacterial enzymes in the N-terminal region.
FIG. 5.
Alignment of N-terminal sequences of 321-MBADH, TOL-BADH, and AC-BADH. Identical residues are highlighted on a black background.
Do known benzyl alcohol dehydrogenases oxidize 321-MB?
Since 321-MB dehydrogenase is capable of oxidizing both 321-MB and benzyl alcohol, it is conceivable that known bacterial benzyl alcohol dehydrogenases are also capable of oxidizing a nonaromatic, allylic substrate like 321-MB, although to date these enzymes have not been shown to oxidize aliphatic alcohols (13). To test this possibility, the TOL-BADH from P. putida PaW1 and the chromosomal AC-BADH were purified >100-fold from cells grown with benzyl alcohol by the same procedures used with 321-MB dehydrogenase through the phenyl-Sepharose chromatography step. In extracts from PaW1, activities for 321-MB and benzyl alcohol copurified during both DEAE-Sephacel anion-exchange chromatography (Fig. 6A) and phenyl-Sepharose hydrophobic interaction chromatography (Fig. 6B). The relative activity of benzyl alcohol dehydrogenase was consistently higher in each peak than the 321-MB dehydrogenase activity, the reverse of the results seen during chromatography of P. putida MB-1 dehydrogenases, where higher rates were seen with 321-MB (Fig. 3). It is noteworthy that in subsequent kinetic assays, where effects of substrate inhibition were minimized, the partially purified TOL-BADH exhibited similar Vmax values for benzyl alcohol and 321-MB. Km values for the two substrates were also similar: 172 μM for 321-MB and 208 μM for benzyl alcohol. The Km for benzyl alcohol is comparable to that reported previously for this enzyme (26).
FIG. 6.
Coelution of TOL-BADH with 321-MB dehydrogenase activity during DEAE-Sephacel chromatography (A) and phenyl-Sepharose chromatography (B). Conditions are the same as those described for Fig. 3.
Similar results were also observed with extracts from A. calcoaceticus grown with benzyl alcohol as the primary carbon source. Again, activities for 321-MB and benzyl alcohol copurified during both DEAE-Sephacel anion-exchange chromatography and phenyl-Sepharose chromatography (data not shown). In this case the partially purified AC-BADH showed a lower Km for benzyl alcohol than for 321-MB, 7 and 81 μM, respectively, but showed similar Vmax values with the two substrates, 36 and 31 U/mg of protein, respectively.
Although these enzymes were not purified to homogeneity, these results clearly suggest that other bacterial benzyl alcohol dehydrogenases are capable of oxidizing 321-MB. Recent results with TOL-BADH and AC-BADH expressed in and purified from Escherichia coli are consistent with this view (8a). We also noted that neither P. putida PaW1 (TOL plasmid) nor A. calcoaceticus grew with 232-MB or 321-MB as a sole carbon source, so it is likely that the presence of 321-MB dehydrogenase activity in these bacteria is a result of the broad specificity of their respective benzyl alcohol dehydrogenases.
DISCUSSION
The main goal of this work was to characterize microbial enzymes that might be used in a coupled photometric assay for 232-MB, a VOC produced in pine needles. As detailed in the Introduction, quantitation of the highly water-soluble 232-MB is an analytical challenge. We reasoned that if we could identify and purify substantial amounts of putative 232-MB isomerase and 321-MB dehydrogenase, and if these enzymes were sufficiently good catalysts, we could construct an enzyme-linked assay for 232-MB analysis (Fig. 2). The work presented here describes significant progress towards this goal.
We first determined that soil associated with pine trees contains bacteria that metabolize 232-MB. Presumably, release of 232-MB by pines with deposition to soil or release of the alcohol from leaf litter to the soil might provide a carbon source for opportunistic bacteria. As shown here, a pure culture of P. putida MB1 that utilizes 232-MB as a sole carbon source was isolated from soil under an Austrian pine (P. nigra), which is not in the group of pines that emit detectable 232-MB (16) but which does contain pools of 232-MB in the needles (16a). Several other bacterial colony types were noted in these 232-MB enrichments; two pure cultures that could utilize 232-MB as a sole carbon source were identified as Arthrobacter sp. and Bacillus sp. (23a). The fact that so many different 232-MB-degrading bacteria have been found in soil associated with pines suggests that 232-MB is a readily available carbon source in these soils.
P. putida MB-1 was found to contain inducible 321-MB and 3-methyl-2-buten-1-al dehydrogenases, indicative of catabolism of 232-MB via 3-methylcrotonic acid. Although strain MB-1 did not grow on 3-methylcrotonic acid as a carbon source, it grew readily on l-leucine, suggesting that 232-MB might lead to the induction of enzymes of the distal part of the pseudomonad leucine degradative pathway where 3-methylcrotonyl-CoA is converted to acetoacetate and acetyl-CoA (21). Notably, pseudomonads that catabolize the acyclic isoprenoids geraniol and farnesol are known to use the methylcrotonyl-CoA pathway (reviewed in reference 21). In this regard P. putida MB-1 was not able to grow on other tested isoprenoid alcohols such as 321-MB, geraniol, linalool, or perillyl alcohols (2), and it seems to be highly adapted to growth on the hemiterpene alcohol 232-MB. In addition, it may be significant that 321-MB was a poor inducer of 321-MB dehydrogenase, and 2-methyl-2-butanol, the saturated structural analog of allylic 232-MB, was not an inducer of the dehydrogenase, suggesting that induction of the 232-MB degradative pathway is relatively specific for 232-MB.
The inducible 321-MB dehydrogenase was purified to near-homogeneity and found to have properties similar to those of known bacterial benzyl alcohol dehydrogenases. This enzyme is soluble, NAD+ dependent, and structurally homologous with other members of the long-chain zinc-containing family of ADHs (17). It was found to be a tetramer of identical 37.7-kDa subunits with a native molecular size of about 151 kDa, resembling other ADHs in subunit mass and quaternary structure. NAD+-dependent, group I (zinc-containing) ADHs have a subunit mass of approximately 40 kDa (24). Quaternary structure is apparently evolutionarily variable, since some group I ADHs are dimeric and others are tetrameric. TOL-BADH, for example, is a homodimer with a subunit mass of 42 kDa (25). AC-BADH is a tetramer of identical subunits with masses of 38.9 kDa (13). Analysis of the N-terminal amino acid sequence of 321-MB dehydrogenase revealed significant similarity with these bacterial benzyl alcohol dehydrogenases.
Like other members of the group I zinc-containing ADHs, 321-MB dehydrogenase was not inhibited by moderate concentrations of metal ion-chelating agents such as EDTA or o-phenanthroline but was sensitive to thiol-blocking agents such as iodoacetamide and N-ethylmaleimide. Similarly, TOL-BADH is known to contain an active-site Zn2+ ion that is either tightly bound or otherwise protected from removal by zinc chelators (26) and is inhibited by thiol-blocking reagents, a common feature in this family of ADHs (19, 25, 29).
The purified 321-MB dehydrogenase exhibited the highest catalytic specificity constants with substrates containing an allylic double bond or with an aromatic ring attached to the carbinol carbon. In this respect the enzyme behaves like AC-BADH. MacKintosh and Fewson (19) noted that the A. calcoaceticus enzyme oxidized not only a range of aromatic alcohols related to benzyl alcohol but also the allylic alcohol moieties in perillyl, cinnamyl, and coniferyl alcohols. They suggested that for the latter two alcohols, the alkenyl group located between the aromatic ring and the carbinol center may help correctly position the alcohol in the active site. Now, with the finding that these benzyl alcohol dehydrogenases, like 321-MB dehydrogenase, are relatively good catalysts for the oxidation of nonaromatic, acyclic, allylic alcohols, a slightly different interpretation can be presented, namely, that the most effective substrates of these ADHs provide π electrons in the allylic double bond or aromatic ring adjacent to the reactive carbinol. The allylic and benzylic resonance of these π electrons with neighboring p orbitals produces similar hybrid orbitals perpendicular to a plane through the carbinol carbon and the adjacent carbon atom (or aromatic ring) (31). The structural consequences of allylic and benzylic resonance may enhance substrate binding to these enzymes, as evidenced, for example, by the fact that the apparent Km values for the nonallylic substrates 3-methyl-1-butanol and 3-methyl-3-buten-1-ol were 7- to 43-fold higher than those for 321-MB when all were assayed under identical conditions (Table 3). Allylic and benzylic substrates might also facilitate catalysis (kcat), since resonance stabilization of the partial positive charge developing on the carbinol carbon may enhance hydride transfer to NAD+. Significant decreases in Vmax (i.e., two- to fivefold) were seen in nonallylic analogs of 321-MB, such as 3-methyl-1-butanol and 3-methyl-3-buten-1-ol (Table 3).
It is pertinent to ask, is 321-MB dehydrogenase actually a benzyl alcohol dehydrogenase that has been recruited to the 232-MB degradative pathway? Its catalytic properties and N-terminal sequence are very similar to those of TOL-BADH. The major difference noted here between 321-MB dehydrogenase and known bacterial benzyl alcohol dehydrogenases is that 321-MB was not induced by the presence of 5 mM benzyl alcohol (Table 1) but was specifically induced by 232-MB, as noted above. Both P. putida PaW1 and A. calcoaceticus contain inducible benzyl alcohol dehydrogenases. PaW1 contains a plasmid-encoded benzyl alcohol dehydrogenase that is induced by toluene, xylenes, and their alcohol derivatives (25). A. calcoaceticus contains a chromosomally encoded benzyl alcohol dehydrogenase that is induced by benzaldehyde or benzyl alcohol (18). Since we failed to detect a plasmid in strain MB-1, it is likely that the 321-MB dehydrogenase is a chromosomally encoded enzyme that is related to the benzyl alcohol dehydrogenase family of genes.
To further test the relationship between 321-MB dehydrogenase and bacterial benzyl alcohol dehydrogenases, we tested whether the latter enzymes would oxidize 321-MB. To this end, we demonstrated that crude extracts from A. calcoaceticus and P. putida PaW1 possess activity for 321-MB dehydrogenase, that this activity is induced by benzyl alcohol in parallel with benzyl alcohol dehydrogenase, and that this activity exactly copurifies with benzyl alcohol dehydrogenase activity. Thus, it appears that these enzymes can also be classified as allylic and benzylic alcohol dehydrogenases. This finding is relevant in that our attempts to produce an enzyme-linked assay for 232-MB could use TOL-BADH or AC-BADH. As the genes for these enzymes have been cloned (13, 15), one could envision their overexpression and production in E. coli and their use in the enzyme-linked assay.
Now that a 321-MB dehydrogenase has been characterized, the critical element in the proposed enzyme-linked assay for 232-MB becomes the putative 232-MB isomerase (Fig. 2). Logic indicates that an enzyme of this type exists, allowing growth on 232-MB to occur by conversion to the primary alcohol 321-MB. In support of this idea, Foss and Harder (12) have shown that anaerobic bacteria that consume linalool, the C10 analog of 232-MB, convert it to the primary alcohol geraniol. However, the putative linalool-geraniol isomerase has not yet been characterized. Since we now have a method for purifying 321-MB dehydrogenase, we are currently attempting coupled assays with excess 321-MB dehydrogenase, using 232-MB as a substrate, to search for the 232-MB isomerase. This approach has revealed the presence of a 232-MB isomerase activity in P. putida MB-1 extracts (8a), so it may soon be possible to test the idea of a coupled enzyme assay for 232-MB.
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation (grant ATM-9633285). V.F.M. was also supported by the Department of the Army.
We thank Tad Koch, Rob Kuchta, and Jens Harder for helpful suggestions, Peter Williams for providing a bacterial strain, Charles Fewson for sharing unpublished data, and Abby Curtis for providing a sample of 321-MB dehydrogenase.
REFERENCES
- 1.Altschul S F, Madden T L, Schäeffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballal N R, Bhattacharyya P K, Rangachari P N. Perillyl alcohol dehydrogenase from a soil pseudomonad. Biochem Biophys Res Commun. 1966;23:473–478. doi: 10.1016/0006-291x(66)90752-2. [DOI] [PubMed] [Google Scholar]
- 3.Banthorpe D V. Terpenoids. In: Mann J, Davidson R S, Hobbs J B, Banthorpe D V, Harborne J B, editors. Natural products: their chemistry and biological significance. Essex, United Kingdom: Longman Scientific & Technical; 1994. pp. 303–359. [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1522. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 6.Brändén C-I, Jörnvall H, Eklund H, Furugren B. Alcohol dehydrogenases. In: Boyer P D, editor. The enzymes. New York, N.Y: Academic Press; 1975. pp. 103–190. [Google Scholar]
- 7.Chalmers R M, Keen J N, Fewson C A. Comparison of benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase from the benzyl alcohol and mandelate pathways in Acinetobacter calcoaceticus and from the TOL-plasmid-encoded toluene pathway in Pseudomonas putida. N-terminal amino acid sequence, amino acid compositions and immunological cross-reactions. Biochem J. 1991;273:99–107. doi: 10.1042/bj2730099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalmers R M, Scott A J, Fewson C A. Purification of the benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase encoded by the TOL plasmid pWW53 of Pseudomonas putida MT53 and their preliminary comparison with benzyl alcohol dehydrogenase and benzaldehyde dehydrogenases I and II from Acinetobacter calcoaceticus. J Gen Microbiol. 1990;136:637–643. [Google Scholar]
- 8a.Curtis, A., and R. Fall. Unpublished data.
- 9.Fall R. Biogenic emissions of volatile organic compounds from higher plants. In: Hewitt C N, editor. Reactive hydrocarbons in the atmosphere. San Diego, Calif: Academic Press; 1999. pp. 41–96. [Google Scholar]
- 10.Fall R R, Brown J L, Schaeffer T S. Enzyme recruitment allows the biodegradation of recalcitrant branched hydrocarbons by Pseudomonas citronellolis. Appl Environ Microbiol. 1979;38:715–722. doi: 10.1128/aem.38.4.715-722.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehsenfeld F, Calvert J, Fall R, Goldan P, Guenther A, Hewitt C, Lamb B, Liu S, Trainer M, Westberg H, Zimmerman P. Emissions of volatile organic compounds from vegetation and the implications for atmospheric chemistry. Global Biogeochem Cycles. 1992;6:389–430. [Google Scholar]
- 12.Foss S, Harder J. Microbial transformation of a tertiary allylalcohol: regioselective isomerization of linalool to geraniol without nerol formation. FEMS Microbiol Lett. 1997;149:71–75. [Google Scholar]
- 13.Gillooly D J, Robertson A G S, Fewson C A. Molecular characterization of benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase II of Acinetobacter calcoaceticus. Biochem J. 1998;330:1375–1381. doi: 10.1042/bj3301375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldan P D, Kuster W C, Fehsenfeld F C, Montzka S A. The observation of a C5 alcohol in a North American pine forest. Geophys Res Lett. 1993;20:1039–1042. [Google Scholar]
- 15.Harayama S, Leppik R A, Rekik M, Mermod N, Lehrbach P R, Reineke W, Timmis K N. Gene order of the TOL catabolic plasmid upper pathway operon and oxidation of both toluene and benzyl alcohol by the xlaA product. J Bacteriol. 1986;167:455–461. doi: 10.1128/jb.167.2.455-461.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harley P, Fridd-Stroud V, Greenberg J, Guenther A, Vasconcellos P. Emission of 2-methyl-3-buten-2-ol by pines: a potentially large natural source of reactive carbon in the atmosphere. J Geophys Res. 1998;103:25. , 479–425, 486. [Google Scholar]
- 16a.Holmes, A., and R. Fall. Unpublished data.
- 17.Jörnvall H, Persson B, Jeffery J. Characteristics of alcohol/polyol dehydrogenases. Eur J Biochem. 1987;176:195–201. doi: 10.1111/j.1432-1033.1987.tb13323.x. [DOI] [PubMed] [Google Scholar]
- 18.MacKintosh R W, Fewson C A. Benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase II from Acinetobacter calcoaceticus. Purification and preliminary characterization. Biochem J. 1988;250:743–751. doi: 10.1042/bj2500743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacKintosh R W, Fewson C A. Benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase II from Acinetobacter calcoaceticus. Substrate specificities and inhibition studies. Biochem J. 1988;255:653–661. [PMC free article] [PubMed] [Google Scholar]
- 20.Malone V F. The purification and characterization of an allylic alcohol dehydrogenase from Pseudomonas putida MB-1. M.S. thesis. Boulder: University of Colorado; 1998. [Google Scholar]
- 21.Massey L K, Sokatch J R, Conrad R S. Branched-chain amino acid catabolism in bacteria. Bacteriol Rev. 1976;40:42–54. doi: 10.1128/br.40.1.42-54.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miozzari G F, Niederberger P, Hütter R. Permeabilization of microorganisms by Triton X-100. Anal Biochem. 1978;90:220–233. doi: 10.1016/0003-2697(78)90026-x. [DOI] [PubMed] [Google Scholar]
- 23.Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: ASM Press; 1996. [Google Scholar]
- 23a.Nemecek-Marshall, M., and R. Fall. Unpublished data.
- 24.Reid M F, Fewson C A. Molecular characterization of microbial alcohol dehydrogenases. Crit Rev Microbiol. 1994;20:13–56. doi: 10.3109/10408419409113545. [DOI] [PubMed] [Google Scholar]
- 25.Shaw J P, Harayama S. Purification and preliminary characterization of TOL plasmid-encoded benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase of Pseudomonas putida. Eur J Biochem. 1990;191:705–714. doi: 10.1111/j.1432-1033.1990.tb19179.x. [DOI] [PubMed] [Google Scholar]
- 26.Shaw J P, Rekik M, Schwager F, Harayama S. Kinetic studies on benzyl alcohol dehydrogenase encoded by TOL plasmid pWW0. A member of the zinc-containing long-chain alcohol dehydrogenase family. J Biol Chem. 1993;268:10842–10850. [PubMed] [Google Scholar]
- 27.Shaw J P, Schwager F, Harayama S. Substrate specificity of benzyl alcohol dehydrogenase and benzyaldehyde dehydrogenase encoded by TOL plasmid pWW0. Metabolic and mechanistic implications. Biochem J. 1992;283:789–794. doi: 10.1042/bj2830789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanier R Y, Palleroni N J, Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 29.Sund H, Theorell H. Alcohol dehydrogenases. In: Boyer P D, Lardy H, Myrback K, editors. The enzymes. New York, N.Y: Academic Press; 1963. pp. 25–83. [Google Scholar]
- 30.Tinoco J I, Sauer K, Wang J C. Physical chemistry. Principles and applications in biological sciences. 3rd ed. Upper Saddle River, N.J: Prentice Hall; 1995. [Google Scholar]
- 31.Vollhardt K P C, Shore N E. Organic chemistry. 2nd ed. New York, N.Y: W. H. Freeman and Company; 1994. [Google Scholar]
- 32.Williams P A, Shaw L M, Pitt C W, Vrecl M. xylUW, two genes at the start of the upper pathway operon of TOL plasmid pWW0, appear to play no essential part in determining its catabolic phenotype. Microbiology. 1997;143:101–107. doi: 10.1099/00221287-143-1-101. [DOI] [PubMed] [Google Scholar]