Abstract
Simple Summary
The prevalence of GJB2-related (MIM: 121011) congenital non-syndromic hearing impairment (NSHI) accounts for close to 50% in populations of Asian and European ancestry. However, in sub-Saharan Africa, except for Ghana, previous data showed that the prevalence of GJB2-associated NSHI is close to zero. To investigate the contribution of GJB2 mutations in autosomal recessive NSHI in Senegal, we screened 129 affected and 143 unaffected individuals from 44 multiplex families, 9 sporadic cases, and 148 hearing controls with no personal or family history of hearing impairment, by targeted gene sequencing. We identified three pathogenic GJB2 variants in 34% (n = 15/44) of multiplex families, of which 80% (n = 12/15) were consanguineous. The most common variant, GJB2: c.94C>T: p.(Arg32Cys), accounted for 27.3% (n = 12/44) of familial cases. We also identified the previously reported “Ghanaian” founder variant, GJB2: c.427C>T: p.(Arg143Trp), in four multiplex Senegalese families. Relatively high allele frequencies of c.94C>T. and c.427C>T variants were observed among the screened hearing controls: 1% (n = 2/148 ∗ 2), and 2% (n = 4/148 ∗ 2), respectively. No GJB6-D13S18 deletion was identified in any of the hearing-impaired participants. The data suggest that GJB2: c.94C>T: p.(Arg32Cys) should be routinely tested in NSHI in Senegal.
Abstract
This study aimed to investigate GJB2 (MIM: 121011) and GJB6 (MIM: 604418) variants associated with familial non-syndromic hearing impairment (HI) in Senegal. We investigated a total of 129 affected and 143 unaffected individuals from 44 multiplex families by segregating autosomal recessive non-syndromic HI, 9 sporadic HI cases of putative genetic origin, and 148 control individuals without personal or family history of HI. The DNA samples were screened for GJB2 coding-region variants and GJB6-D3S1830 deletions. The mean age at the medical diagnosis of the affected individuals was 2.93 ± 2.53 years [range: 1–15 years]. Consanguinity was present in 40 out of 53 families (75.47%). Variants in GJB2 explained HI in 34.1% (n = 15/44) of multiplex families. A bi-allelic pathogenic variant, GJB2: c.94C>T: p.(Arg32Cys) accounted for 25% (n = 11/44 families) of familial cases, of which 80% (n = 12/15) were consanguineous. Interestingly, the previously reported “Ghanaian” founder variant, GJB2: c.427C>T: p.(Arg143Trp), accounted for 4.5% (n = 2/44 families) of the families investigated. Among the normal controls, the allele frequency of GJB2: c.94C>T and GJB2: c.427C>T was estimated at 1% (2/148 ∗ 2) and 2% (4/148 ∗ 2), respectively. No GJB6-D3S1830 deletion was identified in any of the HI patients. This is the first report of a genetic investigation of HI in Senegal, and suggests that GJB2: c.94C>T: p.(Arg32Cys) and GJB2: c.427C>T: p.(Arg143Trp) should be tested in clinical practice for congenital HI in Senegal.
Keywords: hearing impairment, GJB2, GJB2: c.94C>T: p.(Arg32Cys), Senegal, Africa
1. Introduction
Congenital hearing impairment (HI) remains the most disabling condition with the highest rate of age-standardized disability life years [1,2]. Late diagnosis (after 2 years) results in significant sequelae with consequences for language acquisition and cognitive development [3]. The incidence of congenital HI has been estimated at 1 in 1000 live births in developed countries, but a six times higher incidence was observed in sub-Saharan African (SSA) countries [4]. Genetic factors account for 50% of congenital HI cases [5], of which 70% are non-syndromic [6]. Non-syndromic hearing impairment (NSHI) is genetically highly heterogeneous. To date, approximately 170 loci have been mapped and 124 genes have been identified [7]. The DFNB1 locus for autosomal recessive non-syndromic hearing impairment (ARNSHI) was mapped to the 13q11-q12 region [8]. This locus contains the GJB2 and GJB6 genes, which encode connexin 26 (Cx26) and connexin 30 (Cx30), respectively. Pathogenic GJB2 variants are the most common genetic etiology of ARNSHI [9]. The contribution of the GJB2 variants to ARNSHI varies from 0 to 50% in diverse populations [9]. In European and Asian populations, GJB2 variants are the major contributors to ARNSHI [10,11]. However, except for Ghana where the GJB2: c.427C>T: p.(Arg143Trp) founder variant is highly prevalent [12], the prevalence of GJB2-related ARNSHI is close to zero in several SSA populations (Cameroon, South Africa, Nigeria, Sudan and Kenya) [13,14,15,16].
In European populations, up to 50% of individuals with ARNSHI have a pathogenic variant in the GJB2-coding region (exon2) at a heterozygous state [17]. It was suggested that there could be other pathogenic variants in the DFNB1 locus but outside the GJB2 gene. This hypothesis was supported by the finding of a large genomic deletion in the DFNB1 locus outside GJB2, which removes the neighboring GJB6 gene [18], which encodes Cx30, another subunit of the gap-junction channels of the auditory hair cells of the cochlea [19]. Several deletions have been reported [17,20,21]. The largest genomic deletion (342 kb), named del(GJB6-D13S1830), was found in up to 9.7% of affected individuals, either in a homozygous state or a heterozygous state with a GJB2 variant in trans, and constitutes the second most common genetic etiology of ARNSHI. This deletion disrupts GJB2 expression at the transcriptional level by removing putative cis-regulatory elements upstream of GJB6 [22].
The genetic etiology of ARNSHI in Senegal has not been investigated to date. In the present study, we examined the contribution of GJB2 variants and del(GJB6-D13S1830) to ARNSHI in Senegal.
2. Materials and Methods
2.1. Ethical Approvals
The study was performed in accordance with the Declaration of Helsinki regarding medical research on humans. Ethical approval was obtained from the Research Ethics Committee of Cheikh Anta Diop University (CER/UCAD/AD/MSN/034/2020), Dakar, Senegal, and the University of Cape Town, Faculty of Health Sciences’ Human Research Ethics Committee (HREC 104/2018). Written informed consent was obtained from all the adult participants and from the parents or guardians of the minors.
2.2. Study Population
Hearing-impaired patients were recruited from eleven out of the fourteen administrative regions of Senegal, from children’s hospitals, schools for the deaf, as well as from the community by following the procedures previously described in Cameroon and Mali [16,23]. A total of 129 affected and 143 unaffected individuals from 44 multiplex families, segregating ARNSHI, and 9 simplex families with suspected genetic origin of HI were recruited for GJB2 and GJB6 genetic analyses. Pedigrees were drawn for each family through at least three generations.
We performed an otoscopic examination for all the study participants and the cerumen plug was removed before audiological evaluation. The hearing assessments were based on the international standard ISO 8253-1 [24]. The pure tone audiometry (PTA) was performed to evaluate air conduction (250 Hz to 8000 Hz) and bone conduction (250 Hz to 4000 Hz) with a mobile audiometer (KUDUWAVE TM N°0901-04011, Cape Town, South Africa). The hearing threshold was calculated as the average hearing level at 0.5, 1.0, 2.0, and 4.0 kHz. The WHO Global Burden Disease Hearing Loss Expert Group guidelines [25] were used to categorize patients according to the degree of HI. Normal hearing was defined as hearing thresholds up to 25 dB. For children who were too young for a PTA testing, auditory brainstem response (ABR) was performed when applicable.
We also recruited 148 unrelated apparently healthy individuals, who were ethno-linguistically matched, during a blood donation, from four blood banks in four administrative regions of Senegal. A questionnaire was administered to each participant for the exclusion of any personal or familial history of HI.
2.3. Mutation Screening of GJB2 and GJB6
Genomic DNA was extracted from peripheral blood samples, following the manufacturer’s instructions (Puregene Blood Kit®, (Qiagen, Alameda, CA, USA)), at the Division of Human Genetics, Faculty of Medicine, Pharmacy and Odontology of Cheikh Anta Diop University, Dakar, Senegal.
Previously reported primers for GJB2 exon 2 [26] were evaluated with BLAST® software to assess primer specificity. The coding exon of the GJB2 gene (exon 2) was amplified, followed by Sanger sequencing in an ABI 3130XL Genetic Analyzer® (Applied Biosystems, Waltham, MA, USA), at the Division of Human Genetics, University of Cape Town, South Africa. The housekeeping strategy was to sequence the coding region of GJB2 using previously described primers for all recruited probands and affected kindreds. When a pathogenic variant was identified, we sequenced all the other family members to make sure that the identified pathogenic variant was segregated with the HI phenotype.
Subsequently, the detection of del (GJB6-D13S1830) was examined using the previously reported primers GJB6-1R (forward) and BKR-1 (reverse) [18] to amplify a 460 bp fragment corresponding to the sum of 244 bp and 216 bp, flanking the deletion, as well as a second reverse primer, GJB6-2R (5′-TCATCGGGGGTGTCAACAAACA-3′) that is located in the deleted segment, in order to positively detect a 681 bp fragment corresponding to the wild-type product [17].
2.4. Bioinformatic and Statistical Analyses
The AB1 files retrieved from the ABI 3130XL Genetic Analyzer® were manually reviewed using FinchTV v1.4.0, and aligned in UGENE v34.0 [27], to a GJB2 reference sequence [28] (NM_004004.6.; retrieved from NCBI browser). Detected variations were described using Human Genome Variation Society (HGVS) nomenclature [29], and classified using American Society of Medical Genetics’ (ACMG) guidelines [30,31]. The association between allele frequency in affected individuals and controls was assessed using the Chi-square test when applicable, or Fisher’s exact test. A p-value less than 0.05 was considered as significant. Statistical analyses were performed using R software v 4.0.5 (R Core Team, 2020. Vienna, Austria).
3. Results
3.1. Socio-Demographic Data
A total of 129 HI participants belonging to 44 unrelated multiplex families, segregating ARNSHI, and 9 simplex families with a suspected genetic origin of HI were recruited. The average number of participants from whom whole-blood samples were obtained per family was 6 and 3 for multiplex and simplex families, respectively. Consanguinity was present in 40 out of 53 families (75.47%).
The mean age of hearing-impaired participants was 14.80 ± 9.80 years [1–16 years], with a sex ratio of 0.98 (64 males and 65 females). The mean age at medical diagnosis was 2.93 ± 2.53 years [1–15 years].
3.2. Audiological Patterns
Grouping the 124 patients according to the degree of HI showed that the majority (102/124) had a profound HI. Two patients were too young for PTA testing (<2 years), and three patients were not available during the audiological assessment. The age at medical diagnosis was inversely correlated to the degree of HI. Profound HI was associated with an early diagnosis compared to severe and moderate HI (Table 1). The audiometric curve-pattern analysis showed a flat curve in 93 out of 124 patients (75%) and sloping in 31 patients (25%).
Table 1.
Repartition of patients according to the degree of HI and the mean age at medical diagnosis.
| Degree of HI | Number of Patients(n) | Mean Age at Medical Diagnosis |
|---|---|---|
| Moderate (41–60 dB) | 8 (6.42%) | 8.37 ± 3.81 [5–14 years] |
| Severe (61–80 dB) | 14 (11.29%) | 4.25 ± 3.77 [1.5–13 years] |
| Profound (≥81 dB) | 107 (82.26%) | 2.33 ± 1.14 [1–6 years] |
3.3. Molecular Analysis of GJB2 and GJB6
We screened 129 participants from 44 multiplex families and 9 individuals from simplex families, living with congenital sensorineural HI for variants in the coding region of the GJB2 gene and GJB6-D3S1830 deletion. We did not observe any GJB6-D3S1830 deletion in any HI patients (Supplementary Materials Figure S1).
Three variants in GJB2 were identified and classified as pathogenic based on the American College of Medical Genetics (ACMG) guidelines (Supplementary Materials, Table S1). Thirty-four percent (34%, n = 15/44) of multiplex families were positive for a GJB2 pathogenic variant either in a homozygous state or in a compound heterozygous state. The consanguinity rate among GJB2-positive families was estimated at 80% (n = 12/15). The most common variant, GJB2: c.94C>T: p.(Arg32Cys), was in a homozygous state in patients from 11 multiplex families (Table 2).
Table 2.
GJB2 pathogenic variants among 15/44 multiplex families with congenital ARNSHI.
| Genotypes | Multiplex Families | |
|---|---|---|
| n * | % (n/N) | |
| [c.94C>T]; [c.94C>T] | 11 | 25 |
| [c.427C>T]; [c.427C>T] | 2 | 4.54 |
| [c.427C>T]; [c.94C>T] | 1 | 2.27 |
| [c.427C>T]; [c.132G>A] | 1 | 2.27 |
| Total | 15 | 34.09 |
* Number of multiplex families.
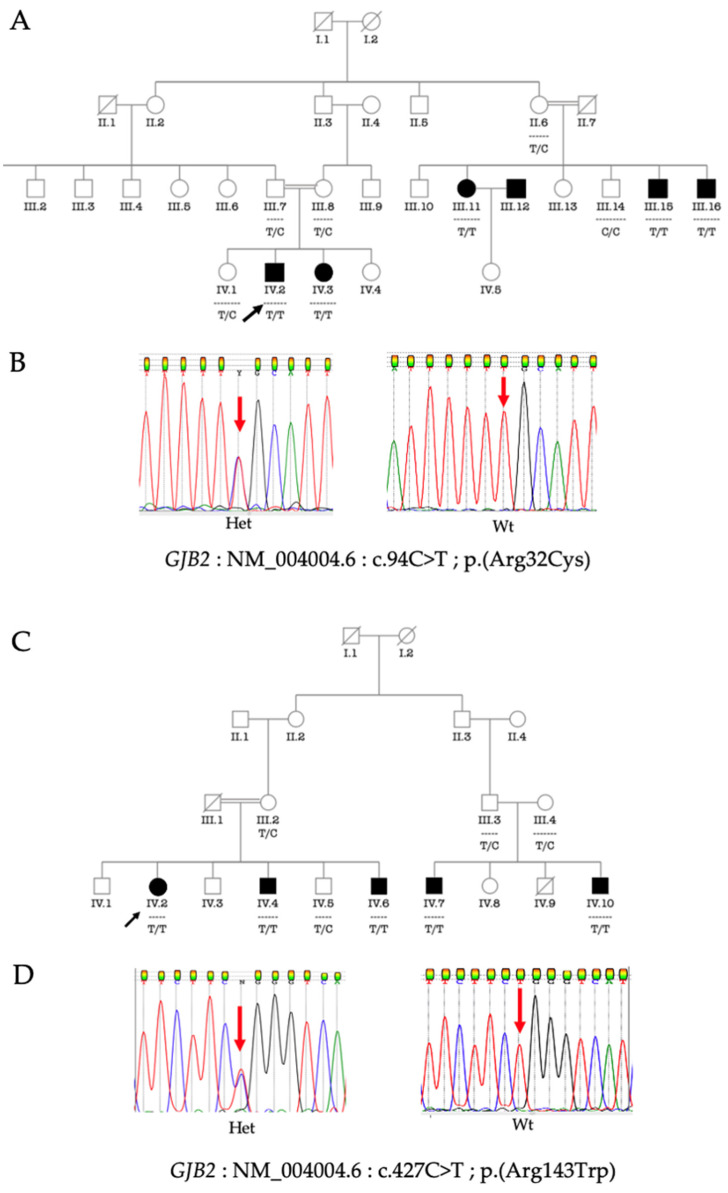
GJB2: c.427C>T: p.(Arg143Trp) and GJB2: c.9C>T: p.(Arg32Cys) segregated with HI either in a homozygous state or a compound heterozygous state. Figure 1 shows the segregation of GJB2: c.427C>T: p.(Arg143Trp) and GJB2: c.9C>T: p.(Arg32Cys) in a homozygous state in two HI multiplex families.
Figure 1.
Pedigree of two multiplex families segregating HI and bi-allelic GJB2: c.94C>T: p.(Arg32Cys) and GJB2: c.427C>T: p.(Arg143Trp), respectively; black arrow indicates the proband (A,C). Electropherograms showing the reference and the pathogenic allele (B,D). The red arrows indicate the nucleotides affected by the variant. Het, heterozygous for the variant allele; Wt, wild type (homozygous for the reference allele) (B,D).
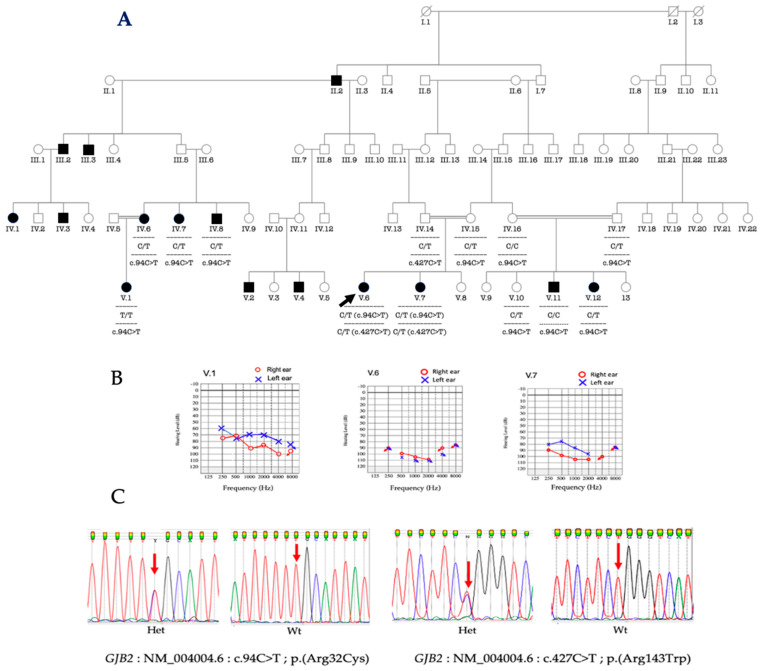
In a particularly large multiplex consanguineous family, some patients were compound heterozygous for GJB2 variants ([c.427C>T: p.(Arg143Trp)]; [c.94C>T: p.(Arg32Cys)], e.g., proband V.6 and her sister V.7 (Figure 2A), while in other branches of the family, different biallelic homozygous variants were found, e.g., cousin (V.1) from the father side and V.11 from the mother side (Figure 2A). The proband as well as her sister presented profound HI (Figure 2B). The proband’s affected uncle (IV.7), aunt (IV.8) and cousin (V.12) were heterozygous for the GJB2 variant (C/T), and his cousin (V.11) was homozygous for the reference allele (Figure 2A), suggesting the implication of another gene in this particular family that can benefit from Whole-Exome-Sequencing (WES) analysis.
Figure 2.
Pedigree of a multiplex family segregating HI with observed genotypes. V.6 is the proband (A); audiological phenotypes of the proband V.6, and her sister V.7, and the cousin from the father’s side, V.1 (B); electropherograms of pathogenic variants in GJB2 (C); Het, heterozygous; Wt, wild type. Black arrow indicates the proband. The red arrows indicate the nucleotides affected by the variant; black arrow indicates the proband.
No pathogenic variant of GJB2 was identified in nine individuals from simplex families with HI.
The three GJB2 pathogenic variants identified in HI patients were also observed in a heterozygote state in the control population. The GJB2: c.427C>T: p.(Arg143Trp) variant is the most frequent (2%; n = 4/148 ∗ 2), followed by the c.94C>T: p.(Arg32Cys) variant (1%; n = 2/148 ∗ 2) (Table 3).
Table 3.
Comparison of GJB2 variants identified in Senegal and other populations from Ensembl database.
| Allele Frequency (n/N) | Allele Frequency from Ensembl | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variants | rs Number | Allele | Cases | Controls | p-Value (Cases vs. Controls) | Global | Africa | America | East Asia | Europe |
| c.94C>T | rs371024165 | C | 0.78 (86/106) | 0.99 (294/296) | <0.0001 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| T | 0.22 (23/106) | 0.01 (2/296) | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | |||
| c.427C>T | rs80338948 | C | 0.94 (100/106) | 0.98 (292/296) | 0.024 | 0.9998 | 1.0000 | 1.0000 | 0.9990 | 1.0000 |
| T | 0.06 (6/106) | 0.02 (4/296) | 0.0002 | 0.0000 | 0.0000 | 0.0010 | 0.0000 | |||
| c.132G>A | rs104894407 | G | 0.99 (105/106) | 0.996 (295/296) | 0.458 | 0.9998 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| A | 0.01 (1/106) | 0.004 (1/296) | 0.0002 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | |||
3.4. Phenotype-Genotype Correlation
It appears that GJB2: c.427C>T: p.(Arg143Trp) in the homozygous or compound heterozygous state was associated with profound HI. Only patients with GJB2: c.94C>T: p.(Arg32Cys) in a homozygous state showed different degrees of HI (Table 4).
Table 4.
Comparison of GJB2 genotypes and the degree of HI.
| Genotypes | Degree of HI | ||
|---|---|---|---|
| Moderate (41–60 dB) |
Severe (61–80 dB) |
Profound (≥81 dB) |
|
| [c.94C>T]; [c.94C>T] | 4 | 3 | 9 |
| [c.427C>T]; [c.427C>T] | 0 | 0 | 7 |
| [c.427C>T]; [c.94C>T] | 0 | 0 | 2 |
| [c.427C>T]; [c.132G>A] | 0 | 0 | 3 |
4. Discussion
To the best of our knowledge, this is the first genetic study of ARNSHI in Senegal, which revealed a surprisingly high proportion (34%, n = 15/44) of pathogenic variants in GJB2 associated with non-syndromic congenital HI. Until recently, Ghana was the exception in SSA, where GJB2 was a major cause of HI. In light of our findings, Senegal is the second SSA country where GJB2 variants significantly contribute to ARNSHI.
The high implication of the GJB2 variants in ARNSHI in Senegal could be supported by the relatively high allele frequency of c.427C>T: p.(Arg143Trp) and c.94C>T: p.(Arg32Cys) in the hearing controls. The carrier frequency of c.427C>T: p.(Arg143Trp) in a control population from Ghana was estimated at 1.4% [12], which is almost half of what we have reported in Senegal (2.7%). This might be due to the control participants being recruited from geographic regions where only 1/3 of affected participants were recruited, therefore not representative of the general population of Senegal. Indeed, the recruitment of cases was based on families segregating HI in at least two affected individuals, and families were recruited nationwide from schools for the deaf, and within the communities, following similar successful methods we previously implemented in both Cameroon and Mali [16,23]. Therefore, we do not expect any significant bias in the sampling of cases. Nevertheless, the recruitment of apparently healthy controls from blood donors did not match the geographical area where families were recruited. Therefore, the ethno-linguistic and geographical origin of controls were likely not representative of the general Senegalese population, and probably biased the carrier-frequency estimates for GJB2-427C>T: p.(Arg143Trp), and c.94C>T: p.(Arg32Cys). This limitation should be alleviated in future studies.
The mean age at medical diagnosis of HI participants was estimated at 2.80 ± 2.53 years. A similar result has been reported in Cameroon by Wonkam et al., with a mean age at medical diagnosis of 3.2 years [16], while a higher mean age of 6 years was reported in Ghana [12]. In Mali, the median age at diagnosis was 12 years [23]. In contrast to SSA, a mean age at diagnosis of less than 6 months has been reported in the United States [32], and between 12 and 36 months in France depending on the degree of HI [33]. This disparity could be explained by limited universal newborn hearing screening (UNHS) in most SSA countries, and none in others, e.g., in Senegal [34]. We also observed an inverse correlation between age at diagnosis and the degree of HI, as previously reported elsewhere [33].
In this study we identified a common variant, GJB2: c.94C>T: p.(Arg32Cys), in 12/44 of multiplex families. Families positive for this variant were recruited across the country, from the western, northern and central geographic regions of Senegal. Ely CMM et al. reported this variant in two consanguineous families in Mauritania, a northern neighboring country of Senegal [35]. GJB2: c.94C>T: p.(Arg32Cys) has also been reported in hearing-impaired individuals in China [36], Japan [37], and South Korea [38]. Owing to the high positivity rate, it might be worth developing an affordable diagnostic method that can be broadly implemented in Senegal, for example, based on RFLP-PCR and following a process that was successfully developed and implemented for the Ghanaian founder variant, GJB2: c.427C>T: p.(Arg143Trp), and included in the public-health-policy decisions in Ghana [39,40].
Contrary to data from Ghana, this study reported a high proportion of consanguinity (75.47%) that favored the enrichment of pathogenic variants, particularly GJB2: c.94C>T: p.(Arg32Cys), which accounted for 25% (n = 11/44 families). Like many west African countries, Senegal has several ethnic groups with a long tradition of consanguineous marriages. In two neighboring countries of Senegal, Mali and Mauritania, consanguinity accounted for 55.5% and 61.33% of familial cases of HI, respectively [23,35]. Consanguinity favors gene identification for numerous recessive conditions [41]. Given the high frequency of the variant among Senegalese consanguineous multiplex families, we postulate that c.94C>T: p.(Arg32Cys) may be a founder variant in Senegal. Future studies should explore this possibility. Indeed, recent data reported that GJB2: c.427C>T: p.(Arg143Trp) evolved in a single individual in Ghana about 10,000 years ago [42].
An unexpected finding was that the “Ghanaian” founder variant, i.e., GJB2: c.427C>T: p.(Arg143Trp), was present in 4.5% (n = 2/44 families) of multiplex families in Senegal. Interestingly, Ghana and Senegal do not share a border, and that variant in GJB2 is absent in populations with HI from Nigeria [13], which is closer to Ghana. It is thus highly speculated that this finding is not due to regional migration, but rather to forced movement of people during the transatlantic slave trade. Indeed, slaves were brought to Gorée [43], an Atlantic island near the Senegalese coastal city of Dakar, before being transported to the Americas. Interestingly, the four families that segregated that variant were all based in Dakar. Future haplotype studies should comparatively investigate haplotypes in GJB2: c.427C>T: p.(Arg143Trp) in families from both Ghana and Senegal to explore this hypothesis. Moreover, the Mayan founder variant, GJB2: c.132G>A: p.(Trp44Ter), reported by Adadey SM et al. [12] in a Ghanaian family was also identified in a Senegalese family, in the compound heterozygous state.
There has been growing evidence of the association between the type of variant and the severity of HI. The degree of GJB2-associated HI depends on the degree of damage to the coding protein Cx26 [44]. Truncating variants, which create a premature stop codon and may result in the absence of any functional Cx26 protein, have been reported to induce a profound HI [45]. In our cohort, patients carrying GJB2: c.132G>A: p.(Trp44Ter), which is a non-sense variant, in a compound heterozygous state, exhibited a profound HI, which is in line with previous reporting in Guatemala [46]. However, the most common variant, GJB2: c.94C>T: p.(Arg32Cys) was associated with variable degrees of HI, ranging from moderate to profound. This variability may reflect a possible effect of modifier genes and/or environmental factors that lead to variable expression [47].
GJB6 is located 50kb upstream of GJB2, and the del(GJB6-D13S1830) variant is the most common deletion of GJB6 and is the second most prevalent ARNSHI variant in western European populations [17]. The deletion occurs in trans in either the homozygous or heterozygous state with pathogenic GJB2 variants, and appears to have an ethnic-specific origin. The del(GJB6-D13S1830) variant was not found in our cohort of HI participants. This is in line with data reported from other African populations [12,16]. However, in a multicentric study, it has been shown that the GJB6-D13S1830 deletion is most frequent in Spain, France, the United Kingdom, Israel, and Brazil (5.9–9.7% of all DFNB1 alleles), is less frequent in the USA, Belgium, and Australia (1.3–4.5% of all DFNB1 alleles), and is very rare in southern Italy [10].
The study also indicates almost 2/3 of multiplex families with HI and all sporadic cases are eligible for next-generation sequencing, due to the highly heterogeneous genetic nature of NSHI. Future research should use high-throughput sequencing platforms that will allow the identification of pathogenic variants in either known genes or novel causative genes.
5. Conclusions
This is the first report of a genetic investigation of HI in Senegal. The study reveals a high consanguinity rate (75.47%) in affected families, and highlights that Senegal is the second country in SSA where GJB2 pathogenic variants significantly contribute to ARNSHI, accounting for 15/44 (34.1%) in multiplex families. The data suggests that GJB2: c.94C>T: p.(Arg32Cys) and GJB2: c.427C>T: p.(Arg143Trp) should be tested in clinical practice for congenital HI in Senegal. Further studies using whole exome or whole genome sequencing approaches are needed to identify the other genes involved in families that are GJB2 negative in Senegal.
Acknowledgments
We thank patients and their families, and controls for their participation in this study. We also thank schools and associations for the deaf in Senegal, the ENT department at Albert Royer Children’s hospital, and specialists on hearing aids for their help during the process of identification and recruitment of families.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology11050795/s1, Table S1: Variant’s interpretation according to ACMG guidelines, Figure S1: PCR-Multiplex products visualized on 2% agarose gel.
Author Contributions
Conceptualization, A.W. and R.N.D.; Resources, Y.D., A.W., O.G.O., R.N.D., J.P.D.D., S.A.B., C.A.T.L., A.R.G.S., P.D.S. and B.K.D.; Investigation, Y.D., S.M.A. and E.T.A. Project administration, A.W., R.N.D., B.K.D. and C.D.K.; Formal Analysis, Y.D. and S.M.A.; Writing Original Draft Preparation, Y.D.; Writing review and editing, S.M.A., E.T.A., O.G.O., J.P.D.D., S.A.B., C.A.T.L., A.R.G.S., P.D.S., C.D.K., B.K.D., R.N.D. and A.W.; Supervision, A.W., R.N.D. and B.K.D.; Funding Acquisition, A.W. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Cheikh Anta Diop University (CER/UCAD/AD/MSN/034/2020), Dakar, Senegal; and the University of Cape Town, Faculty of Health Sciences’ Human Research Ethics Committee (HREC 104/2018).
Informed Consent Statement
Written informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Conflicts of Interest
The authors declare no conflict of interests.
Funding Statement
This research was funded by the NIH, USA, grant number U01-HG-009716 to AW, and the African Academy of Science/Wellcome Trust, grant number H3A/18/001 to AW.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.GBD 2013 DALYs and HALE Collaborators. Murray C.J.L., Barber R.M., Foreman K.J., Ozgoren A.A., Abd-Allah F., Abera S.F., Aboyans V., Abraham J.P., Abubakar I., et al. Global, regional, and national disability-adjusted life years (DALYs) for 306 diseases and injuries and healthy life expectancy (HALE) for 188 countries, 1990–2013: Quantifying the epidemiological transition. Lancet. 2015;386:2145–2191. doi: 10.1016/S0140-6736(15)61340-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO . The Global Burden of Disease: 2004 Update. World Health Organization; Geneva, Switzerland: 2008. [Google Scholar]
- 3.Wolff R., Hommerich J., Riemsma R., Antes G., Lange S., Kleijnen J. Hearing screening in newborns: Systematic review of accuracy, effectiveness, and effects of interventions after screening. Arch. Dis. Child. 2009;95:130–135. doi: 10.1136/adc.2008.151092. [DOI] [PubMed] [Google Scholar]
- 4.Olusanya B.O., Neumann K.J., Saunders J.E. The global burden of disabling hearing impairment: A call to action. Bull. World Health Organ. 2014;92:367–373. doi: 10.2471/BLT.13.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korver A.M.H., Smith R.J.H., Van Camp G., Schleiss M.R., Bitner-Glindzicz M.A.K., Lustig L.R., Usami S., Boudewyns A.N. Congenital hearing loss. Nat. Rev. Dis. Prim. 2017;3:16094. doi: 10.1038/nrdp.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shearer A.E., Hildebrand M.S., Smith R.J. Hereditary Hearing Loss and Deafness Overview. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J., Gripp K.W., editors. GeneReviews® [Internet] University of Washington; Seattle, WA, USA: 1993. [(accessed on 28 March 2022)]. Available online: http://www.ncbi.nlm.nih.gov/books/NBK1434/ [Google Scholar]
- 7.Van Camp G., Smith R.J.H. Hereditary Hearing Loss Homepage. 2018. [(accessed on 17 December 2021)]. Available online: http://hereditaryhearingloss.org.
- 8.Guilford P., Ben Arab S., Blanchard S., Levilliers J., Weissenbach J., Belkahia A., Petit C. A non–syndromic form of neurosensory, recessive deafness maps to the pericentromeric region of chromosome 13q. Nat. Genet. 1994;6:24–28. doi: 10.1038/ng0194-24. [DOI] [PubMed] [Google Scholar]
- 9.Kenneson A., Braun K.V.N., Boyle C. GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: A HuGE review. Genet. Med. 2002;4:258–274. doi: 10.1097/00125817-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Del Castillo F.J., Del Castillo I. DFNB1 Non-syndromic Hearing Impairment: Diversity of Mutations and Associated Phenotypes. Front. Mol. Neurosci. 2017;10:428. doi: 10.3389/fnmol.2017.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adhikary B., Ghosh S., Paul S., Bankura B., Pattanayak A.K., Biswas S., Maity B., Das M. Spectrum and frequency of GJB2, GJB6 and SLC26A4 gene mutations among nonsyndromic hearing loss patients in eastern part of India. Gene. 2015;573:239–245. doi: 10.1016/j.gene.2015.07.050. [DOI] [PubMed] [Google Scholar]
- 12.Adadey S.M., Manyisa N., Mnika K., De Kock C., Nembaware V., Quaye O., Amedofu G.K., Awandare G.A., Wonkam A. GJB2 and GJB6 Mutations in Non-Syndromic Childhood Hearing Impairment in Ghana. Front. Genet. 2019;10:841. doi: 10.3389/fgene.2019.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lasisi A.O., Bademci G., Foster J., Blanton S., Tekin M. Common genes for non-syndromic deafness are uncommon in sub-Saharan Africa: A report from Nigeria. Int. J. Pediatr. Otorhinolaryngol. 2014;78:1870–1873. doi: 10.1016/j.ijporl.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasmelseed N.M.A., Schmidt M., Magzoub M.M.A., Macharia M., Elmustafa O.M., Ototo B., Winkler E., Ruge G., Horstmann R.D., Meyer C.G. Low frequency of deafness-associated GJB2 variants in Kenya and Sudan and novel GJB2 variants. Hum. Mutat. 2004;23:206–207. doi: 10.1002/humu.9216. [DOI] [PubMed] [Google Scholar]
- 15.Kabahuma R.I., Ouyang X., Du L.L., Yan D., Hutchin T., Ramsay M., Penn C., Liu X.-Z. Absence of GJB2 gene mutations, the GJB6 deletion (GJB6-D13S1830) and four common mitochondrial mutations in nonsyndromic genetic hearing loss in a South African population. Int. J. Pediatr. Otorhinolaryngol. 2011;75:611–617. doi: 10.1016/j.ijporl.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tingang Wonkam E., Chimusa E., Noubiap J.J., Adadey S.M.F., Fokouo J.V., Wonkam A. GJB2 and GJB6 Mutations in Hereditary Recessive Non-Syndromic Hearing Impairment in Cameroon. Genes. 2019;10:844. doi: 10.3390/genes10110844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Castillo I., Moreno-Pelayo M.A., Del Castillo F.J., Brownstein Z., Marlin S., Adina Q., Moreno F., Cockburn D.J., Pandya A., Siemering K.R., et al. Prevalence and evolutionary origins of the del(GJB6-D13S1830) mutation in the DFNB1 locus in hearing-impaired subjects: A multicenter study. Am. J. Hum. Genet. 2003;73:1452–1458. doi: 10.1086/380205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.del Castillo I., Villamar M., Moreno-Pelayo M.A., del Castillo F.J., Alvarez A., Tellería D., Sc M., Ibis Menéndez M.D., Felipe Moreno P.D. A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. N. Engl. J. Med. 2002;346:243–249. doi: 10.1056/NEJMoa012052. [DOI] [PubMed] [Google Scholar]
- 19.Zhao H.-B., Kikuchi T., Ngezahayo A., White T.W. Gap Junctions and Cochlear Homeostasis. J. Membr. Biol. 2006;209:177–186. doi: 10.1007/s00232-005-0832-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Castillo F.J., Rodríguez-Ballesteros M., Alvarez A., Hutchin T., Leonardi E., De Oliveira C.A., Azaiez H., Brownstein Z., Avenarius M.R., Marlin S., et al. A novel deletion involving the connexin-30 gene, del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. J. Med. Genet. 2005;42:588–594. doi: 10.1136/jmg.2004.028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A Bliznets E., Makienko O.N., Okuneva E.G., Markova T.G., Poliakov A.V. New recurrent extended deletion, including GJB2 and GJB6 genes, results in isolated sensorineural hearing impairment with autosomal recessive type of inheritance. Genetika. 2014;50:474–480. [PubMed] [Google Scholar]
- 22.Moisan S., Le Nabec A., Quillévéré A., Le Maréchal C., Férec C. Characterization of GJB2 cis-regulatory elements in the DFNB1 locus. Qual. Life Res. 2019;138:1275–1286. doi: 10.1007/s00439-019-02068-8. [DOI] [PubMed] [Google Scholar]
- 23.Yalcouyé A., Traoré O., Taméga A., Maïga A.B., Kané F., Oluwole O.G., Guinto C.O., Kéita M., Timbo S.K., DeKock C., et al. Etiologies of Childhood Hearing Impairment in Schools for the Deaf in Mali. Front. Pediatr. 2021;9:726726. doi: 10.3389/fped.2021.726776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acoustics—Audiometric Test Methods—Part 1: Basic Pure Tone Air and Bone Conduction Audiometry. International Organization for Standardization; Geneva, Switzerland: 2010. [Google Scholar]
- 25.Olusanya B.O., Davis A.C., Hoffman H.J. Hearing loss grades and the International classification of functioning, disability and health. Bull. World Health Organ. 2019;97:725–728. doi: 10.2471/BLT.19.230367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosch J., Lebeko K., Nziale J.J.N., Dandara C., Makubalo N., Wonkam A. In Search of Genetic Markers for Nonsyndromic Deafness in Africa: A Study in Cameroonians and Black South Africans with the GJB6 and GJA1 Candidate Genes. OMICS A J. Integr. Biol. 2014;18:481–485. doi: 10.1089/omi.2013.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okonechnikov K., Golosova O., Fursov M., The UGENE Team Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 28.Homo Sapiens Gap Junction Protein Beta 2 (GJB2), mRNA. 2021 [Cited 29 September 2021] [(accessed on 1 March 2022)]; Available online: http://www.ncbi.nlm.nih.gov/nuccore/NM_004004.6.
- 29.Den Dunnen J.T., Dalgleish R., Maglott D.R., Hart R.K., Greenblatt M.S., McGowan-Jordan J., Roux A.F., Smith T., Antonarakis S.E., Taschner P.E.M., et al. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum Mutat. 2016;37:564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 30.Oza A.M., DiStefano M.T., Hemphill S.E., Cushman B.J., Grant A.R., Siegert R.K., Shen J., Chapin A., Boczek N.J., Schimmenti L.A., et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum. Mutat. 2018;39:1593–1613. doi: 10.1002/humu.23630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grosse S.D., Mason C.A., Gaffney M., Thomson V., White K.R. What Contribution Did Economic Evidence Make to the Adoption of Universal Newborn Hearing Screening Policies in the United States? Int. J. Neonatal Screen. 2018;4:25. doi: 10.3390/ijns4030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masson E. Évolution De L’âge Du Diagnostic Des Surdités Congénitales. EM-Consulte. [Cited 12 February 2022] [(accessed on 30 March 2022)]. Available online: https://www.em-consulte.com/article/287793/evolution-de-lage-du-diagnostic-des-surdites-conge.
- 34.Engelman D. Ph.D. Thesis. City University of New York; New York, NY, USA: 2014. The Status of Neonatal Hearing Screening in Sub-Saharan Africa: A Systematic Review. [Google Scholar]
- 35.Moctar E.C.M., Riahi Z., El Hachmi H., Veten F., Meiloud G., Bonnet C., Abdelhak S., Errami M., Houmeida A. Etiology and associated GJB2 mutations in Mauritanian children with non-syndromic hearing loss. Eur. Arch. Oto-Rhino-Laryngol. 2016;273:3693–3698. doi: 10.1007/s00405-016-4036-z. [DOI] [PubMed] [Google Scholar]
- 36.Zheng J., Ying Z., Cai Z., Sun D., He Z., Gao Y., Zhang T., Zhu Y., Chen Y., Guan M.-X. GJB2 Mutation Spectrum and Genotype-Phenotype Correlation in 1067 Han Chinese Subjects with Non-Syndromic Hearing Loss. PLoS ONE. 2015;10:e0128691. doi: 10.1371/journal.pone.0128691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kasakura-Kimura N., Masuda M., Mutai H., Masuda S., Morimoto N., Ogahara N., Misawa H., Sakamoto H., Saito K., Matsunaga T. WFS1andGJB2mutations in patients with bilateral low-frequency sensorineural hearing loss. Laryngoscope. 2017;127:E324–E329. doi: 10.1002/lary.26528. [DOI] [PubMed] [Google Scholar]
- 38.Kim S.Y., Kim A.R., Kim N.K.D., Lee C., Kim M.Y., Jeon E.H., Park W.Y., Choi B.Y. Unraveling of Enigmatic Hearing-Impaired GJB2 Single Heterozygotes by Massive Parallel Sequencing: DFNB1 or Not? Medicine. 2016;95:e3029. doi: 10.1097/MD.0000000000003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adadey S.M., Quaye O., Amedofu G.K., Awandare G.A., Wonkam A. Screening for GJB2-R143W-Associated Hearing Impairment: Implications for Health Policy and Practice in Ghana. Public Health Genom. 2020;23:184–189. doi: 10.1159/000512121. [DOI] [PubMed] [Google Scholar]
- 40.Adadey S.M., Wonkam E.T., Aboagye E.T., Quansah D., Asante-Poku A., Quaye O., Amedofu G.K., Awandare G.A., Wonkam A. Enhancing Genetic Medicine: Rapid and Cost-Effective Molecular Diagnosis for a GJB2 Founder Mutation for Hearing Impairment in Ghana. Genes. 2020;11:132. doi: 10.3390/genes11020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fareed M., Afzal M. Genetics of consanguinity and inbreeding in health and disease. Ann. Hum. Biol. 2016;44:99–107. doi: 10.1080/03014460.2016.1265148. [DOI] [PubMed] [Google Scholar]
- 42.Aboagye E.T., Adadey S.M., Esoh K., Jonas M., De Kock C., Amenga-Etego L., Awandare G.A., Wonkam A. Age Estimate of GJB2-p.(Arg143Trp) Founder Variant in Hearing Impairment in Ghana, Suggests Multiple Independent Origins across Populations. Biology. 2022;11:476. doi: 10.3390/biology11030476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fofana D.M. Senegal, the African Slave Trade, and the Door of No Return: Giving Witness to Gorée Island. Humanities. 2020;9:57. doi: 10.3390/h9030057. [DOI] [Google Scholar]
- 44.Cryns K., Orzan E., Murgia A., Huygen P.L.M., Moreno F., Del Castillo I., Chamberlin G.P., Azaiez H., Prasad S., Cucci R.A., et al. A genotype-phenotype correlation for GJB2 (connexin 26) deafness. J. Med. Genet. 2004;41:147–154. doi: 10.1136/jmg.2003.013896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X., Huang L., Zhao X., Wang X., Cheng X., Du Y., Liu D. Children with GJB2 gene mutations have various audiological phenotypes. Biosci. Trends. 2018;12:419–425. doi: 10.5582/bst.2018.01159. [DOI] [PubMed] [Google Scholar]
- 46.Carranza C., Menendez I., Herrera M., Castellanos P., Amado C., Maldonado F., Rosales L., Escobar N., Guerra M., Alvarez D., et al. A Mayan founder mutation is a common cause of deafness in Guatemala. Clin. Genet. 2015;89:461–465. doi: 10.1111/cge.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nadeau J.H. Modifier genes in mice and humans. Nat. Rev. Genet. 2001;2:165–174. doi: 10.1038/35056009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.