Abstract
From a pool of 600 temperature-sensitive transposon mutants of Pseudomonas putida P8, 1 strain was isolated that carries a mini-Tn5 insertion within the cytochrome c operon. As a result, genes involved in the attachment of heme to cytochrome c-type proteins are turned off. Accordingly, cytochrome c could not be detected spectrophotometrically. The mutant also exhibited a remarkable reduction of cis-trans isomerization capability for unsaturated fatty acids. Consistent with the genetic and physiological data is the detection of a cytochrome c-type heme-binding motif close to the N terminus of the predicted polypeptide of the cis/trans isomerase (cti) gene (CVACH; conserved amino acids in italics). The functional significance of this motif was proven by site-directed mutagenesis. A possible mechanism of heme-catalyzed cis-trans isomerization of unsaturated fatty acids is discussed.
cis-trans isomerization of unsaturated fatty acids enables bacterial cells to escape from being damaged by various environmental stress factors, e.g., exposure to elevated temperature, high levels of harmful compounds, and excess salinity (21). Conversion of unsaturated fatty acids from cis to trans is brought about by direct isomerization of the double bond without a shift of its position (7, 24). Since the acyl chains of phospholipids are the substrates for the reaction, membrane fluidity is concomitantly reduced due to altered steric conformations of cis and trans unsaturated fatty acid constituents (27). This rapid change of membrane fluidity is independent of de novo fatty acid and lipid biosynthesis (12, 13); it allows bacteria to adapt to, tolerate, and grow on otherwise toxic levels of substances such as ethanol or phenol. The cti gene encoding the cis/trans isomerase (CTI) of unsaturated fatty acids was isolated and sequenced from Pseudomonas putida P8 only quite recently. When introduced into a corresponding mutant, the gene restored the wild-type phenotype in P. putida P8 but also conferred the capability for cis-trans isomerization of unsaturated fatty acids to Escherichia coli transformants which are normally devoid of CTI activity (14). The enzyme is still only poorly characterized. A 9-hexadecenoic acid cis/trans isomerase was purified to homogeneity from Pseudomonas sp. strain E-3, and its features were determined. Enzymatic activity was negatively affected by catecholic antioxidants such as α-tocopherol and nordihydroguaiaretic acid, known inhibitors of lipoxygenases, which are nonheme-iron-containing proteins (37). Reaction mechanisms of both lipoxygenase and CTI were thus suggested to share similarities (26).
In this study, a mutant of P. putida P8 was isolated which is severely impaired in its capability for cis-trans isomerization of unsaturated fatty acids due to the insertion of a mini-Tn5 into the cytochrome c operon. Further striking evidence that the enzyme of P. putida P8 encoded by the cti locus is a cytochrome c-type protein is provided by site-directed mutagenesis and biochemical studies of the cloned cti gene product. Based on these findings, a possible mechanism for heme-catalyzed cis-trans isomerization of unsaturated fatty acids is discussed.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. putida P8 wild type and transposon mutants were grown in minimal medium (21) supplemented with 0.2% succinate as the carbon source at 30°C on a rotary shaker (200 rpm, model G76; New Brunswick Scientific). E. coli JM107 (40) served as a cloning host, and E. coli M15pRep4 (Qiagen, Hilden, Germany) was used for expression of the cis/trans isomerase. E. coli strains were routinely grown in Luria-Bertani (LB) medium (31). Solid (1.5% agar-agar) and liquid media were supplemented, when required, with 50 to 100 μg of ampicillin ml−1 and 25 to 50 μg of kanamycin ml−1, respectively.
TABLE 1.
Bacteria and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli JM107 | E. coli K-12 supE Δ(lac-proAB) hskR17 F′ traΔ36 proAB+ lacIR lacZ ΔM15 | 40 |
| E. coli M15 | E. coli K-12 Strr F− ΔlacZ (pREP4) | 29 |
| E. coli SM10(λpir) | E. coli K-12 Kmrthr leu tonA lacY supE recA::RP4-2-Tc::Mu λpir | 23 |
| P. putida P8 | Wild type | 2 |
| P. putida A9 | Kmr, cti::Tn5 | 14 |
| P. putida S12 | Kmr, ccmC::Tn5 | This study |
| Plasmids | ||
| pS12-1 | pUCBM20, carrying 3.7-kb Sau3A fragment of ccmA ccmB ccmC with inserted Tn5-Km | This study |
| pUCBM20 | Apr | 3 |
| pREP4 | KmrlacI | 29 |
| pQE60 | Apr | 29 |
| pQE60cti | Aprcti | This study |
| pQE60cti-C46S | Aprcti C46S | This study |
| pUT mini-Tn5 Km | Apr Kmr, delivery plasmid for mini-Tn5 Km | 6 |
Tn5 transposon mutagenesis.
Plasmid pUT-mini-Tn5-Km (6) was transferred from donor strain E. coli SM10(λpir) into P. putida P8 by a filter-mating technique (5). Filters with a mixture of donor and recipient strains at a 1:2 ratio were incubated for 12 to 15 h at 30°C on complete medium. Cells were subsequently suspended in 10 mM MgSO4, and appropriate dilutions were plated on minimal medium containing kanamycin.
Colonies to be checked for the presence of trans fatty acids were applied by toothpick onto minimal medium and replica plated; the petri dishes were then incubated at 30, 37, and 40°C. Colonies were also plated onto media containing different phenol concentrations (0.5, 0.8, and 1 g liter−1), with succinate as a carbon source, and that is why it was possible to screen for cells which no longer tolerated phenol rather than those that simply had lost the ability to degrade it. All clones which did not grow or grew poorly, both at temperatures above 37°C and on phenol at concentrations of ≥0.8 g liter−1, were tested for their ability for cis-trans isomerization of unsaturated fatty acids (see also reference 14).
Determination of fatty acids.
Extraction of lipids from cells was done as described previously (14). Fatty acid methylesters were analyzed by gas chromatography as described by Diefenbach et al. (8).
Recombinant DNA techniques.
Molecular cloning procedures were carried out according to the method of Sambrook et al. (31). DNA sequence analysis was done by using IRD41-labelled universal and reverse primers with the Thermo Sequenase cycle sequencing kit with 7-deaza-dGTP (Amersham Buchler, Braunschweig, Germany) and an automatic LI-COR sequencer (LI-COR, Inc., Lincoln, Nebr.).
Plasmid construction.
For the expression and proper isolation of the protein, a plasmid containing the cti coding region with a C-terminal His tag was constructed. For this purpose, a DNA fragment was amplified by PCR that contained an NcoI site at codon 1 of cti and a BamHI site at the end of the coding region to clone it in frame with six additional histidine codons. The fragment was amplified with primer 1 (5′-CAGGACTTTTCGCCCATGGTGCATCGTATC-3′) and primer 2 (5′-AGATCTGGATCCGAGGTTCT CATA-3′) (NcoI and BamHI sites are underlined). The PCR product was cloned as an NcoI-BamHI fragment into pQE60 (Qiagen, Hilden, Germany), thereby adding the His tag fragment present in pQE60 to the C terminus of the cis/trans isomerase.
Site-directed mutagenesis.
Plasmid pQE60cti was used for site-directed mutagenesis to exchange the second cysteine (C46) of the putative heme-binding site within the Cti protein by serine. Oligonucleotides 3 (5′-GAGCGTAAAGAGCCACTTCTAG-3′) and 4 (5′-GCCAGCCATGCCTGCAAC-3′) were used to amplify the whole plasmid pQE60cti. Amplified plasmid DNA was ligated and transferred to E. coli M15(pRep4). The mutation was verified by sequencing. Since C46 was exchanged by a serine, the resulting plasmid was designated pQE60cti-C46S.
Expression of Cti.
E. coli M15(pREP4) cells were transformed with the expression plasmid pQE60cti and grown in 1.6 liters of LB medium containing ampicillin (100 μg ml−1) and kanamycin (50 μg ml−1) at 30°C to an optical density of 1 (at 600 nm). After IPTG (isopropyl-β-d-thiogalactopyranoside) induction (final concentration, 0.005 mM), cells were cultivated for an additional 4 h and were subsequently harvested by centrifugation. After sonification (Branson Sonifier 250; Sonic Power Co.), the protein was purified by nickel affinity chromatography (Ni-NTA agarose; Qiagen) according to the instructions of the manufacturer with the addition of 0.1% Tween 20 to all buffers.
Electrophoresis of proteins.
Proteins were analyzed by polyacrylamide gel electrophoresis (PAGE) as described by Laemmli (19). Heme proteins were detected in sodium dodecyl sulfate (SDS)-polyacrylamide gels by staining for heme peroxidase activity (10) with 3,3′,5,5′-tetramethylbenzidine (TMBZ) and hydrogen peroxide. Protein staining in gels was performed with Coomassie brilliant blue (38).
Western blot.
Proteins separated by SDS-PAGE (11% acrylamide) were transferred to nitrocellulose filters by using a semidry electroblotter (Biometra, Göttingen, Germany); gels were equilibrated with buffer (25 mM Tris–200 mM glycine containing 5% [vol/vol] methanol) prior to transfer of the polypeptides at 5 mA cm−2. Blots on nitrocellulose were blocked with 5% milk powder in TBST (50 mM Tris-HCl [pH 7] containing 500 mM NaCl and 0.2% Tween 20) for 30 min at room temperature. Cti-His tag proteins were detected by immunoblotting with an anti-His tag-svFv-alkaline phosphatase conjugate (20) that was used at a 1:500 dilution in TBST with 0.5% milk powder. Detection of bands was achieved by using the luminescence substrate CSPD (Boehringer, Mannheim, Germany).
Nucleotide sequence accession number. The sequenced part of the cytochrome c operon of P. putida P8 was sent to EMBL Heidelberg and given accession no. AJ131925.
RESULTS
Isolation of a transposon mutant.
In previous studies we isolated a temperature-sensitive mini-Tn5 mutant (A9) of P. putida P8 that could not grow at 37°C and which concomitantly had completely lost its capability for cis-trans isomerization of unsaturated fatty acids. In strain A9 insertion of the mini-Tn5 occurred in the 5′ noncoding region of the cti locus, thereby preventing transcription of the gene (14). In order to find additional genetic loci involved in the cis-trans isomerization of fatty acids, we looked for more mutants whose growth is affected at elevated temperatures (37°C) and, simultaneously, whose ability to isomerize unsaturated fatty acids is also affected. From a pool of 600 temperature-sensitive mutants (14), 1 additional strain (S12) was identified that was severely but not completely impaired in its capability for cis-trans isomerization. The CTI activity of this mutant is compared to those of wild-type P. putida P8 and the previously obtained cti mutant A9 in Table 2.
TABLE 2.
Effect of ethanol on cis-trans isomerization of unsaturated fatty acids in different strainsa
| Strain | Ethanol addition | Fatty acid content (%)b
|
|||||
|---|---|---|---|---|---|---|---|
| 16:0 | 16:1 cis | 16:1 trans | 18:0 | 18:1 cis | 18:1 trans | ||
| P. putida P8 | − | 33.9 ± 1.7 | 33.4 ± 4.6 | 8.1 ± 3.8 | 2.1 ± 0.3 | 20.0 ± 2.2 | 2.3 ± 1.7 |
| + | 33.4 ± 1.0 | 21.2 ± 2.4 | 18.8 ± 4.5 | 2.9 ± 1.7 | 17.9 ± 2.1 | 3.5 ± 2.2 | |
| P. putida A9 | − | 32.5 ± 0.8 | 44.8 ± 1.2 | 0 | 1.7 ± 0.3 | 20.9 ± 0.3 | 0 |
| + | 32.4 ± 1.8 | 43.6 ± 1.7 | 0 | 1.8 ± 0.5 | 21.5 ± 0.7 | 0 | |
| P. putida S12 | − | 34.0 ± 2.0 | 33.3 ± 1.5 | 1.4 ± 1.6 | 2.7 ± 0.4 | 27.4 ± 2.0 | 1.1 ± 0.7 |
| + | 33.9 ± 1.6 | 29.9 ± 2.0 | 5.0 ± 1.7 | 2.5 ± 0.3 | 27.0 ± 1.9 | 1.5 ± 0.5 | |
Strains were grown in minimal medium to an optical density at 600 nm of 0.3 at 30°C before ethanol was added to a final concentration of 10%. Fatty acid determination was done 2 h thereafter in ethanol-treated (+) and control (−) cells.
Fatty acids were assayed as described in Materials and Methods; values for only the relevant fatty acids are given. The values are the means from at least quadruplicate experiments; standard deviations are also indicated.
Table 2 shows that mutant S12 has a remarkably low trans fatty acid content before a membrane active substance, such as ethanol, is added, and after the addition of alcohol the trans 16:1 fatty acids increased to only 5%, whereas P. putida P8 wild type reached approximately 18%. Mutant A9, which served as a control, is completely devoid of trans fatty acids, as was found previously (14). Restriction enzyme and Southern blot analyses with mini-Tn5 as a probe revealed the insertion locus to be different from that of the cti mutant A9 (data not shown). Since this newly identified mini-Tn5 mutant S12 is clearly different from strain A9, both with respect to cis-trans isomerization capability and with respect to the affected gene, it appeared to be a good candidate for the isolation of an additional genetic locus involved in the cis-trans isomerization of unsaturated fatty acids in P. putida P8.
Molecular characterization of transposon mutant S12.
Bulk DNA from the mutant was isolated, partially digested with restriction endonuclease Sau3A, and subsequently ligated into the BamHI site of plasmid pUCBM20. We used E. coli JM107 as a recipient in transformation experiments. Selection of the aph genetic trait of the mini-Tn5 with kanamycin-containing medium resulted in a transformant that carried hybrid plasmid pS12-1, which consists of pUCBM20 with a 3.7-kb insertion. The insert comprises a 1.9-kb Tn5 fragment and a 1.8-kb genomic flank of the P. putida P8 transposon mutant S12. The latter sequence was completely determined by using subclones of plasmid pS12-1 by applying universal and reverse sequencing primers.
Three open reading frames were identified in computer-aided searches. These open reading frames evidently constitute a part of the cytochrome c operon because of striking similarities to the ccmA, ccmB, and ccmC genes of Pseudomonas fluorescens (93% on DNA level) and E. coli (65%). Gene products of the ccmABC genes were shown to be involved in the transport of heme groups across the cytoplasmic membrane (36). In P. fluorescens and E. coli these genes are part of a corporate operon in which they are followed in both bacterial species by the ccmDEFGH genes, all of which are instrumental in the maturation and installation of the heme group into cytochrome c-type proteins (35).
In mutant S12, mini-Tn5 is inserted into the last codon of the ccmC gene. As described previously for E. coli (36) and P. fluorescens (9), insertion of a transposon into this position results in an almost complete loss of cytochrome c-type proteins due to the prevention of transcription of the ccmDEFGH genes.
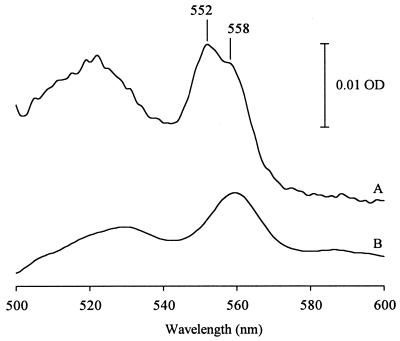
Thus, we checked the lack of cytochrome c-type proteins in mutant S12 by measuring difference spectra of cell extracts in comparison to the wild-type strain (Fig. 1). As depicted in Fig. 1, the characteristic cytochrome c peak at 552 nm, which can be seen for the wild-type strain P. putida P8, is below the detection limit in mutant S12. The peak for cytochrome b-type proteins at 558 nm is, however, present in both strains.
FIG. 1.
Reduced-minus-oxidized difference spectra of cell extracts (soluble proteins obtained after disintegration of cells by sonification) from P. putida P8 and mutant S12. Samples were reduced with a pinch of dithionite and oxidized in the reference cuvette with potassium ferricyanide. (A) P. putida P8 wild type. (B) P. putida S12. The protein concentration was 10 mg ml−1 for P8 and 12 mg ml−1 for S12. OD, optical density.
Heme peroxidase activity of the cis/trans isomerase.
Based on the above findings, we searched for a heme cytochrome c-type binding motif (CXXCH [22]) within the cti locus of P. putida P8 (14). Indeed, at positions 43 to 47 such a motif exists in the predicted polypeptide (CVACH [conserved amino acids in boldface]).
Heme-containing proteins have peroxidase activity which can easily be demonstrated by using chromogenic substrates, such as TMBZ (34). However, demonstration of such an activity requires, as a prerequisite, sufficient amounts of the enzyme. Since it was not possible to produce adequate quantities of the Cti protein from P. putida P8, presumably due to low expression of the gene, a His tag was added to the cloned cti coding region and the protein was expressed in E. coli M15 by using plasmid pQE60cti, which was constructed as described in Materials and Methods (see also Table 1).
The His tag facilitated enrichment and rapid isolation of a functional protein because the isolated enzyme exhibited cis-trans isomerization activity for free unsaturated fatty acids (data not shown). The molecular mass of the protein obtained was ca. 85 kDa as determined by PAGE, a finding which is in good accordance with the calculated molecular mass of the predicted Cti polypeptide (86.7 kDa) (Fig. 2).
FIG. 2.
SDS-PAGE of recombinant Cti-His tag proteins isolated with Ni affinity chromatography under nondenaturating conditions from E. coli M15(pREP4) transformants. (A) Heme peroxidase activity staining with TMBZ. (B) Immunological detection of His-tagged proteins. Lane 1, native Cti; lane 2, Cti with amino acid exchange C46S.
Cytochrome c-type proteins retain their heme groups due to covalent linkages even under denaturating conditions. Thus, the peroxidase activity of the heme constituent can be demonstrated within denaturating SDS gels, as shown in Fig. 2 for the cloned P. putida cti gene product.
The functional significance of the putative heme-binding motif (CXXCH) was confirmed by site-directed mutagenesis (30); replacement of the highly conserved and, thus, presumably the essential C residue at position 46 by S resulted in vector pQE60cti-C46S; for construction of the plasmid, see Materials and Methods. Purified wild-type and mutant His tag proteins were tested for CTI activity by using 16:1 free fatty acids as the substrate. In contrast to the native Cti protein, there was no cis/trans isomerase activity (data not shown) or peroxidase activity (Fig. 2) of the protein encoded by pQE60cti-C46S.
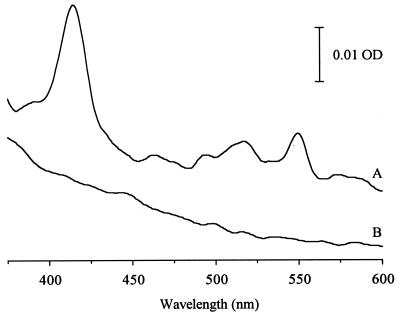
Expression of the Cti protein and its mutated derivative in pQE60cti-C46S was verified by both Western blots with heterologously expressed antibodies against the His tag (Fig. 2) and spectroscopic analysis of purified His tag proteins (Fig. 3).
FIG. 3.
Reduced-minus-oxidized-difference spectra of Cti-His tag proteins isolated from E. coli M15(pREP4) transformants by Ni affinity chromatography under nondenaturing conditions. (A) Wild-type enzyme. (B) C46S mutant protein. Samples were treated as described for Fig. 1. OD, optical density.
Ni affinity chromatography under native conditions of the mutated protein did not give rise to a distinct band in PAGE, but only degraded polypeptides could be detected with antibodies against the His tag.
The typical reduced-minus-oxidized difference spectrum for cytochrome c-type proteins was obtained only for the purified wild-type His tag enzyme, whereas the protein with the mutated heme-binding site (C46S) lacks the typical peaks at ca. 415, 520, and 550 nm (Fig. 3).
DISCUSSION
cis-trans isomerization of unsaturated fatty acids was clearly affected in the obtained mini-Tn5 transposon mutant S12 of P. putida P8; the mini-Tn5 was found to reside within the cytochrome c operon. Striking similarities of the cloned genomic flank to cytochrome c operons from P. fluorescens and E. coli, together with the fact that cytochrome c-type proteins could hardly be detected in mutant S12, suggested a similar structure of the P. putida P8 operon. Accordingly, the synthesis and transport of heme across the cytoplasmic membrane is presumably not affected, but the installation of the heme residue into cytochrome c-type proteins is no longer possible (35).
Attachment of heme groups to cytochrome c proteins is brought about by proteins encoded by ccmEFGH genes on the periplasmic face of cell membranes (35). The expression of the latter genes is prevented in transposon mutant S12. Nevertheless, the mutant is still capable of cis-trans isomerization at a low level (Table 2). Comparable findings were reported for P. fluorescens in which insertion of a Km cassette into the ccmC gene reduced cytochrome c-type proteins to the level of 10% (9). Low-level expression can be explained generally by fortuitous promoter activity within the ccmEFGH genes. It can also not be excluded that an additional, although less efficient, set of cytochrome c biogenesis genes is present, as was shown for E. coli (15), in which the nrf operon, required for formate-dependent nitrite reduction, encodes genes homologous to ccmF (nrfE) and ccmH (nrfF) (36).
Prokaryotic cytochrome c-type proteins have in common two main characteristics: (i) a signal sequence and (ii) the conserved CXXCH motif for covalent heme attachment. Due to the signal sequence, c-type cytochromes of gram-negative bacteria are routinely found in the periplasm or are associated with the periplasmic face of the cytoplasmic membrane. As shown earlier, the predicted Cti polypeptide has a signal sequence followed by a proline at position +1 of the signal peptidase cleavage site (14), which leads to an uncleavable signal peptide (1, 28). Thus, irrespective of the overall hydrophilicity of Cti, the amino-terminal 22 residues of the signal peptide may function as a hydrophobic domain that tethers the protein in the outer leaflet of the cytoplasmic membrane. However, it cannot be excluded at present that an alternative cleavage site is used.
If the heme peroxidase and CTI activity of the cloned cti gene product is also taken into consideration, there is convincing evidence that the enzyme is a cytochrome c-type protein. In addition, the functional significance of the heme-binding site was proven by site-directed mutagenesis. Consistent with these experiments is also the fact that the best match in database searches (46% similarity on the amino acid level for positions 30 to 100 in the Cti polypeptide) is to the cytochrome c1 from Entosphenus tridentatus (the EMBL accession number for the latter is P00028).
Essentially the same amino acid exchange as in this study was done for cytochrome c550 of Paracoccus denitrificans (30), resulting in a level of cytochrome c550 that could hardly be detected. As for the cti gene product, the mutated cytochrome c proteins had lost peroxidase activity and could only be detected immunologically.
Beside the strikingly reduced cis-trans isomerization capability of mutant S12, there is a shift in the length of the acyl chains in phospholipids toward the long-chained fatty acids. In wild-type P. putida P8 and the cti mutant A9 the ratio of 18:1/16:1 unsaturated fatty acids is approximately 0.5, whereas it is 0.8 in mutant S12 (Table 2). In this mutant, the characteristic peak of functional cytochrome c-type proteins at 552 nm could not be detected. The majority of cytochrome c proteins are membrane associated, as are the enzymes encoded by the ccmEFGH genes (35), which are not expressed in mutant S12. Compensation of the lack of this considerable number of different membrane proteins might be the reason for the observed shift in length toward long-chained fatty acids of phospholipids in mutant S12.
For an enzyme preparation from Pseudomonas sp. strain E3, which is presumably homologous to the cti gene product of P. putida P8, it was assumed that iron plays a crucial role in the catalytic reaction (26). Known inhibitors of lipoxygenases, such as α-tocopherol and nordihydroguaiaretic acid (16, 37), were shown to act on that enzyme, and that is why those authors suggested similarities concerning the catalytic mechanism of cis/trans isomerases and lipoxygenases, the latter being non-heme-iron-containing enzymes. For example, lipoxygenases catalyze the oxidation of arachidonic acid, resulting in hydroperoxyeicosatetraenic acid. The reaction involves the removal of hydrogen (H), followed by the addition of molecular oxygen (O2) and the subsequent reattachment of the hydrogen (H). There are, however, reports that heme also has lipoxygenase activity which is subject to inhibition by phenolic antioxidants (17, 18). Oxygenation of unsaturated fatty acids by heme groups was also reported by Chan and Newby (4). Peroxidation of unsaturated fatty acids in membrane phospholipids by iron-protoporphyrin IX (heme) appeared to be sensitive to α-tocopherol (33). Thus, the above-mentioned findings reported by Okuyama et al. (26) for the cis/trans isomerase of Pseudomonas sp. strain E3 are not contradictory to a heme group being the catalytic domain, for which experimental evidence is presented in the present study.
Heme-containing cytochrome P450 enzymes catalyze the hydroxylation of unsaturated fatty acids; they are also instrumental in desaturating fatty acids, reactions which are, however, NADPH dependent (11, 25). Cti activity is independent of additional factors such as ATP, NADPH, or O2 (7, 26); it differs in this respect from all other known heme-containing enzymes acting on fatty acids as substrates. There is, however, no need of a cofactor because no net electron power is consumed.
From a mechanistic point of view, cis-trans isomerization of carbon-carbon double bonds may be a coupled hydration-dehydration reaction, as was demonstrated for β-oxidation enzymes, which show 2-cis-enoyl- and 3-trans-enoyl-coenzyme A isomerase activities (39). Involvement of a heme group, independent of cofactors, however, points to a different mechanism of an enzyme, which does not shift the position of the double bond. cis-trans isomerization of unsaturated fatty acids can also be catalyzed nonenzymatically by iron, which is attached to the double bond during the reaction (32). Though the mechanism is not completely understood, cleavage of the double bond is presumably facilitated by the formation of a substrate-catalyst complex, enabling rotation about the carbon-carbon single bond.
A direct attack on the double bond in unsaturated fatty acids by the electrophilic heme iron (Fe2+ or Fe3+) may result in the removal of an electron from the double bond and might transform the sp2 linkage of the carbon atoms to the sp3 type, which can be rotated. The concomitantly arising positive charge of one of the involved C atoms could be stabilized by a negatively charged amino acid. The double bond must then be reconstituted after rotation has occurred. Future work will focus on the elucidation of the mechanism underlying cis-trans isomerization, e.g., by demonstration of the postulated protein-stabilized fatty acid radical intermediate that possibly occurs transiently during the reaction by electron spin resonance analysis. A different approach would be the isolation and comparison of a number of cti genes and their gene products from a variety of bacteria in order to identify highly conserved regions or residues within the protein, the function of which can subsequently be studied with molecular tools.
ACKNOWLEDGMENTS
Experimental work was supported by the Deutsche Forschungsgemeinschaft, Bonn Bad Godesberg.
We thank H. Pape, Institut für Mikrobiologie, Universität Münster, Münster, Germany, and B. Witholt, Institute of Biotechnology, ETH-Zürich, Zurich, Switzerland, for critically reading the manuscript.
Footnotes
This work is dedicated to Karl Esser on the occasion of his 75th birthday.
REFERENCES
- 1.Barkocy-Gallagher G A, Bassford P J. Synthesis of precursor maltose-binding protein with proline in the +1 position of the cleavage site interferes with the activity of Escherichia coli signal peptidase I in vivo. J Biol Chem. 1992;267:1231–1238. [PubMed] [Google Scholar]
- 2.Bettmann H, Rehm H J. Degradation of phenol by polymer entrapped microorganisms. Appl Microbiol Biotechnol. 1984;22:389–393. [Google Scholar]
- 3.Boehringer Mannheim. Biochemicals for molecular biology. Boehringer Mannheim GmbH. Mannheim, Germany: Biochemica; 1990. [Google Scholar]
- 4.Chan H W, Newby V K. Haemoprotein- and transition metal ion-catalysed oxidation of linoleic acid. Selectivity of the position of oxygenation. Biochim Biophys Acta. 1980;617:353–362. doi: 10.1016/0005-2760(80)90001-6. [DOI] [PubMed] [Google Scholar]
- 5.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived mini-transposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 6.de Lorenzo V, Herrero M, Jacubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diefenbach R, Keweloh H. Synthesis of trans unsaturated fatty acids in Pseudomonas putida P8 by direct isomerization of the double bond of lipids. Arch Microbiol. 1994;162:120–125. doi: 10.1007/s002030050112. [DOI] [PubMed] [Google Scholar]
- 8.Diefenbach R, Heipieper H J, Keweloh H. The conversion of cis into trans unsaturated fatty acids in Pseudomonas putida P8: evidence for a role in the regulation of membrane fluidity. Appl Microbiol Biotechnol. 1992;38:382–387. [Google Scholar]
- 9.Gaballa A, Koedam N, Cornelis P. A cytochrome c biogenesis gene involved in pyoverdine production in Pseudomonas fluorescens ATCC17400. Mol Microbiol. 1996;21:777–785. doi: 10.1046/j.1365-2958.1996.391399.x. [DOI] [PubMed] [Google Scholar]
- 10.Goodhew C F, Brown K R, Pettigrew G W. Haem straining in gels, a useful tool in the study of bacterial c-type cytochromes. Biochim Biophys Acta. 1986;852:288–294. [Google Scholar]
- 11.Guan X, Fisher M B, Lang D H, Zheng Y M, Koop D R, Rettie A E. Cytochrome P450-dependent desaturation of lauric acid: isoform selectivity and mechanism of formation of 11-dodecenoic acid. Chem-Biol Interact. 1998;110:103–121. doi: 10.1016/s0009-2797(97)00145-2. [DOI] [PubMed] [Google Scholar]
- 12.Heipieper H J, Weber F J, Sikkema J, Keweloh H, de Bont J A M. Mechanisms of resistance of whole cells to toxic organic solvents. Trends Biotechnol. 1994;12:409–425. [Google Scholar]
- 13.Heipieper H J, Diefenbach R, Keweloh H. Conversion of cis unsaturated fatty acids to trans, a possible mechanism for the protection of phenol-degrading Pseudomonas putida P8 from substrate toxicity. Appl Environ Microbiol. 1992;58:1847–1852. doi: 10.1128/aem.58.6.1847-1852.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holtwick R, Meinhardt F, Keweloh H. cis-trans isomerization of unsaturated fatty acids: cloning and sequencing of the cti gene from Pseudomonas putida P8. Appl Environ Microbiol. 1997;63:4292–4297. doi: 10.1128/aem.63.11.4292-4297.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussain H, Grove J, Griffith L, Busby S, Cole J. A seven-gene operon essential for formate-dependent nitrite reduction to ammonia by enteric bacteria. Mol Microbiol. 1994;12:153–163. doi: 10.1111/j.1365-2958.1994.tb01004.x. [DOI] [PubMed] [Google Scholar]
- 16.Kemal C, Louis-Flamberg P, Krupinski-Olsen R, Shorter A L. Reductive inactivation of soyabean lipoxygenase 1 by catechols: a possible mechanism for regulation of lipoxygenase activity. Biochemistry. 1987;26:7064–7072. doi: 10.1021/bi00396a031. [DOI] [PubMed] [Google Scholar]
- 17.Kühn H, Götze R, Schewe T, Rapoport S M. Quasi-lipoxygenase activity of haemoglobin. Eur J Biochem. 1981;120:161–168. doi: 10.1111/j.1432-1033.1981.tb05684.x. [DOI] [PubMed] [Google Scholar]
- 18.Kühn H, Hache A, Sklenar H, Schewe H, Rapoport S M. A structural model for the interaction of haem with unsaturated fatty acids explaining its quasi-lipoxygenase activity. Quantum chemical calculations. Biomed Biochim Acta. 1983;42:175–176. [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lindner P, Bauer K, Krebber A, Nieba L, Kremmer E, Krebber C, Honegger A, Klinger B, Mocikat R, Plückthun A. Specific detection of His-tagged proteins with recombinant anti-His tag scFv-phosphatase or scFc-phage fusions. BioTechniques. 1997;22:140–149. doi: 10.2144/97221rr01. [DOI] [PubMed] [Google Scholar]
- 21.Loffeld B, Keweloh H. cis/trans isomerization of unsaturated fatty acids as possible control mechanism of membrane fluidity in Pseudomonas putida P8. Lipids. 1996;31:811–815. doi: 10.1007/BF02522976. [DOI] [PubMed] [Google Scholar]
- 22.Mathews F. The structure, function and evolution of cytochromes. Prog Biophys Mol Biol. 1985;45:1–56. doi: 10.1016/0079-6107(85)90004-5. [DOI] [PubMed] [Google Scholar]
- 23.Miller V, Mekalanos J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morita N, Shibahara A, Yamamoto K, Shinkai K, Kajimoto G, Okuyama H. Evidence for cis-trans isomerization of a double bond in the fatty acids of the psychrophilic bacterium Vibrio sp. strain ABE-1. J Bacteriol. 1993;175:916–918. doi: 10.1128/jb.175.3.916-918.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murataliev M B, Klein M, Fulco A, Feyereisen R. Functional interactions in cytochrome P450BM3: flavin semiquinone intermediates, role of NADP(H), and mechanism of electron transfer by flavoprotein domain. Biochemistry. 1997;36:8401–8412. doi: 10.1021/bi970026b. [DOI] [PubMed] [Google Scholar]
- 26.Okuyama H, Uneno A, Enari D, Morita N, Kusano T. Purification and characterization of 9-hexadecenoic acid cis-trans isomerase from Pseudomonas sp. strain E-3. Arch Microbiol. 1998;169:29–35. doi: 10.1007/s002030050537. [DOI] [PubMed] [Google Scholar]
- 27.Okuyama H, Okajima N, Sasaki S, Higashi S, Murata N. The cis/trans isomerization of the double bond of a fatty acid as a strategy for adaption to changes in ambient temperature in the psychrophilic bacterium Vibrio sp. strain ABE-1. Biochim Biophys Acta. 1991;1084:13–20. doi: 10.1016/0005-2760(91)90049-n. [DOI] [PubMed] [Google Scholar]
- 28.Plückthun A, Knowles J R. The consequences of stepwise deletions from signal-processing site of β-lactamase. J Biol Chem. 1987;262:3951–3957. [PubMed] [Google Scholar]
- 29.Qiagen. The QIAexpressionist. A handbook for high level expression and purification of 6xHis-tagged proteins. Hilden, Germany: Qiagen; 1997. [Google Scholar]
- 30.Sambongi Y, Stoll R, Ferguson S J. Alteration of haem-attachment and signal-cleavage site for Paracoccus denitrificans cytochrome c550 probes pathway of c-type cytochrome biogenesis in Escherichia coli. Mol Microbiol. 1996;19:1193–1204. doi: 10.1111/j.1365-2958.1996.tb02465.x. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Selzer S. cis-trans isomerization. In: Boyer P D, editor. The enzymes. Vol. 6. New York, N.Y: Academic Press, Inc.; 1972. pp. 381–406. [Google Scholar]
- 33.Sugioka Y, Suzuki M, Sugioka K, Nakano M. A ferriprotoporphyrin IX-chloroquine complex promotes membrane phospholipid peroxidation. A possible mechanism for antimalarial action. FEBS Lett. 1987;223:251–254. doi: 10.1016/0014-5793(87)80299-5. [DOI] [PubMed] [Google Scholar]
- 34.Thomas P E, Ryan D, Levin W. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 35.Thöny-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thöny-Meyer L, Fischer F, Künzler P, Ritz D, Hennecke H. Escherichia coli genes required for cytochrome c maturation. J Bacteriol. 1995;177:4321–4326. doi: 10.1128/jb.177.15.4321-4326.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vliegenthart J F G, Veldink G A. Lipoxygenase. In: Pryor W A, editor. Free radicals in biology. Vol. 5. New York, N.Y: Academic Press; 1982. pp. 29–64. [Google Scholar]
- 38.Weber K, Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969;244:4406–4412. [PubMed] [Google Scholar]
- 39.Yang S-Y, Elzinga M. Association of both enoyl coenzyme A hydratase and 3-hydroxyacyl coenzyme A epimerase with an active site in the amino-terminal domain of the multifunctional fatty acid oxidation protein for Escherichia coli. J Biol Chem. 1993;268:6588–6592. [PubMed] [Google Scholar]
- 40.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]