Abstract
Mycobacterium avium is a cause of disseminated disease in AIDS patients. A need for a better understanding of possible sources and routes of transmission of this organism has arisen. This study utilized a PCR typing method designed to amplify DNA segments located between the insertion sequences IS1245 and IS1311 to compare levels of relatedness of M. avium isolates found in patients and foods. Twenty-five of 121 food samples yielded 29 mycobacterial isolates, of which 12 were M. avium. Twelve food and 103 clinical M. avium isolates were tested. A clinical isolate was found to be identical to a food isolate, and close relationships were found between two patient isolates and two food isolates. Relatedness between food isolates and patient isolates suggests the possibility that food is a potential source of M. avium infection. This study demonstrates a rapid, inexpensive method for typing M. avium, possibly replacing pulsed-field gel electrophoresis.
Nontuberculous mycobacteria have become a significant cause of infection with the emergence of AIDS. Until recently, 25 to 50% of patients with AIDS in the United States and Europe were infected with this group of bacteria, primarily Mycobacterium avium, the predominant cause of disseminated mycobacteremia in AIDS patients (6). Because of the increase of morbidity and mortality associated with this infection, there is a need for better understanding of the sources and routes of M. avium transmission.
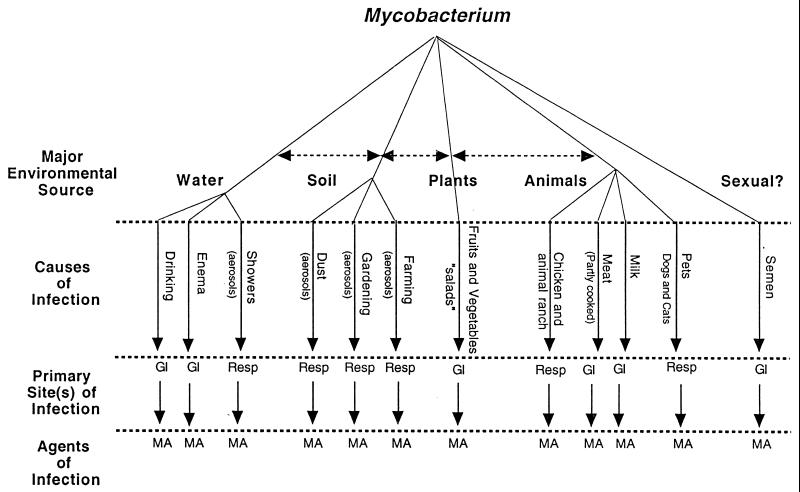
M. avium has been isolated from water, dust, soil, and a variety of animals, including chickens and pigs (6). The environment, not human-to-human transmission, is a primary source of infection, with the portal of entry being either gastrointestinal or pulmonary (2, 6). The spectrum of sources and possible routes of entry and infection are depicted in the flow chart (Fig. 1) (1a). Water has been identified as a potential source of infection by analysis of long restriction fragments by using pulsed-field gel electrophoresis (PFGE) (18).
FIG. 1.
Flow chart of probable routes of M. avium (MA) infection. Resp, respiratory tract; GI, gastrointestinal tract.
PFGE, considered the “gold standard” application in M. avium strain typing (10), has been employed to identify polyclonal infections (16), epidemiologic relationships (18), and community diversity among M. avium strains (4). Lengthy and technically difficult sample preparations make PFGE impractical for simple, rapid typing.
Insertion sequences (ISs) can also be used for strain typing and epidemiologic studies. IS elements have been found on bacterial plasmids and chromosomes (15), and their mobilities and copy numbers as well as the distances between various IS elements can be used to show genetic and evolutionary relationships (3).
Molecular probes for IS1245 have been compared to PFGE in studying diversity among isolates from human and animal sources (7, 14). The IS elements IS1245 and IS1311, reported to be 85% identical (14), have also been used in epidemiologic studies of M. avium infections in AIDS patients (14). Picardeau et al. developed a discriminating tool for epidemiologic studies involving AIDS patients with possible polyclonal infections by employing a combination PCR assay to amplify DNA sequences and then by comparing the lengths between the two ISs (12, 13).
In this study, we utilized the above-described PCR typing method to compare levels of relatedness of clinical and food M. avium isolates.
MATERIALS AND METHODS
Clinical isolates.
One hundred three isolates obtained from AIDS patients and patients without AIDS were identified as M. avium by using AccuProbe (GenProbe, Inc., San Diego, Calif.) or SNAP (Syngene, San Diego, Calif.) DNA probe kits. Isolates were maintained on Middlebrook 7H10 medium supplemented with 500 μg of cycloheximide (7H10c) (Clinical Research Laboratories, Dominguez Hills, Calif.) per ml prior to performance of PCR studies.
Food isolates.
The methods of collection and processing of food samples are described by Argueta et al. (1). In brief, approximately 1 lb of food was suspended in 500 ml of ultrapure water in a polypropylene bag and shaken for 10 min. The resulting supernatant was filtered into a 1-liter polypropylene bottle. The filtrate was centrifuged, and the pellet was resuspended in Middlebrook 7H9 broth supplemented with 500 μg of cycloheximide/ml and 10% glycerol (Difco Laboratories). Samples were frozen until the time of decontamination. Five-milliliter aliquots were decontaminated with 10 ml of a solution consisting of 5% oxalic acid, 2% NaOH, and 2.9% sodium citrate and incubated at room temperature for 10 min. The reaction was neutralized with 20 ml of phosphate buffer (pH 7.0). The suspension was centrifuged, and the pellet was resuspended in 1 ml of phosphate buffer. One hundred microliters of the resuspended pellet was plated onto Middlebrook 7H10c with 0.002% malachite green medium and incubated at 37°C for 6 to 8 weeks. Auramine-rhodamine stains were done to confirm acid fastness of resulting mycobacterial colonies.
Preparation of DNA for PCR.
Mycobacterial colonies were removed from 7H10c agar plates with sterile wood applicator sticks and placed in 50 μl of TE (10 mM Tris, 1 mM EDTA [pH 7.5]) with 1% Triton X-100 (Research Organics, Inc., Cleveland, Ohio) and incubated at 100°C for 30 min. Lysates were centrifuged briefly to pellet debris, and DNA in the supernatant solution was used as the template for PCR.
Species identification PCR.
Species identification of food isolates was performed by using PCR-restriction fragment length polymorphism (RFLP) (17), and 12 M. avium isolates were confirmed by using DT6 PCR (5).
Typing PCR.
Typing of M. avium was performed by the method developed by Picardeau et al. (13) with the following modifications: amplification was performed in 25-μl reaction mixtures in Ready-To-Go PCR beads (Amersham Pharmacia Biotech, Piscataway, N.J.) by using a PT-200 thermal cycler (MJ Research, Inc., Watertown, Mass.). Reaction mixtures contained 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 2.5 mM MgCl2, 160 μg of bovine serum albumin, 200 μM each deoxynucleoside triphosphate, 1.5 μM each primer (PA, 5′-CAGAGCCTCAGGCGA-3′, and PB, 5′-CAGAGCCTCACGCGGA-3′), and approximately 1.5 U of Taq polymerase. The primers were designed by Picardeau et al. to hybridize to an inverted repeat found in both IS1245 and IS1311 (13), and the PCR was designed to amplify the distances between the two IS elements. To each mix, 2.5 μl of template DNA prepared as described above was added. Amplification involved an initial denaturation at 95°C for 2 min followed by 35 cycles consisting of a 15-s denaturation at 95°C, 15-s annealing at 55°C, and 30-s primer extension at 72°C, followed by a final extension of 5 min at 72°C. Products were maintained at 4°C until analyzed by electrophoresis. One clinical M. avium isolate, W187, was incorporated as a positive control and a water blank was incorporated as a negative control for each PCR mixture to assess reproducibility and consistency. PCR products were analyzed by electrophoresis on 2.5% MetaPhor agarose (FMC BioProducts, Rockland, Maine) and stained with ethidium bromide for 15 min. Gels were visualized and documented by using an UltraLum 302-nm UV transilluminator (UltraLum, Paramont, Calif.). PCR band lengths were sized by using GelReader software (National Center for Supercomputing Applications, Urbana-Champaign, Ill.) and categorized into 56 presence and/or absence groups allowing a 5% product length error tolerance between fragment sizes (12). Values were analyzed to determine relatedness by using Phylogenetic Analysis Using Parsimony (PAUP) software, version 3.1.1 (Laboratory of Molecular Systematics, Smithsonian Institution, Washington, D.C.).
RESULTS
Application of PCR to amplify sequences between IS1245 and IS1311 was found to be a practical method of strain typing M. avium. Twenty-five of 121 food samples yielded 29 mycobacterial isolates (Table 1). Twelve of 29 isolates from 25 foods were positively identified as M. avium by using both RFLP (17) and DT6 species-specific (5) PCR methods (Table 1). Six species of nontuberculous mycobacteria were identified in the 29 isolates recovered from food samples, and M. avium was the predominant one. One hundred fifteen clinical and food isolates were tested by using a modified method of Picardeau et al. (13), producing an average of four PCR bands per isolate (range, 0 to 10 bands per isolate). Ten isolates yielded no bands (eight clinical and two food isolates). Two PCR product sizes were found to be present in a majority of isolates (around 180 and 500 bp). The positive and negative controls produced reproducible and consistent results in each PCR test.
TABLE 1.
Food isolates and species of mycobacteria isolated
| Isolate | Food sample | PCR identification
|
|
|---|---|---|---|
| PCR-RFLP | DT6 | ||
| F1 | Broccoli slaw | M. avium | + |
| F2 | Spinach | ?a | NAb |
| F3 | Spinach | M. avium | + |
| F4 | Spinach | M. avium | + |
| F5 | Red leaf | M. avium | + |
| F6 | Red leaf | M. avium | + |
| F7 | Green leaf salad | ? | NA |
| F8 | Packaged salad | M. avium | + |
| F9 | Broccoli slaw | ? | NA |
| F10 | Red leaf | M. gordonae V | NA |
| F11 | Romaine salad | ? | − |
| F12 | Spinach | ? | NA |
| F18 | Parsley | ? | NA |
| F19-A | Leeks | M. avium | + |
| F19-B | Leeks | M. gordonae VI | NA |
| F35 | Apple cider | M. simiae | NA |
| F36 | Apple berry | M. simiae | NA |
| F37 | Apple cherry | M. simiae | − |
| F38 | Apple pomegranate | M. scrofulaceum | NA |
| F40 | Rhubarb swiss chard | ? | NA |
| F42 | Basil | M. genavense | NA |
| F66 | Basil | M. gordonae III | NA |
| F84-B | Monterey mushrooms | M. avium | + |
| F84-C | Monterey mushrooms | M. avium | + |
| F84-D | Monterey mushrooms | M. flavescens II | NA |
| F97 | Daikon sprouts | M. avium | + |
| F104 | Italian brown mushrooms | M. avium | + |
| F107 | Portabella mushrooms | M. flavescens II | NA |
| F108 | Portabella mushrooms | M. avium | + |
?, isolate yielded no RFLP pattern or a pattern not found on the Taylor algorithm (14).
NA, DT6 PCR identification not applicable for isolate.
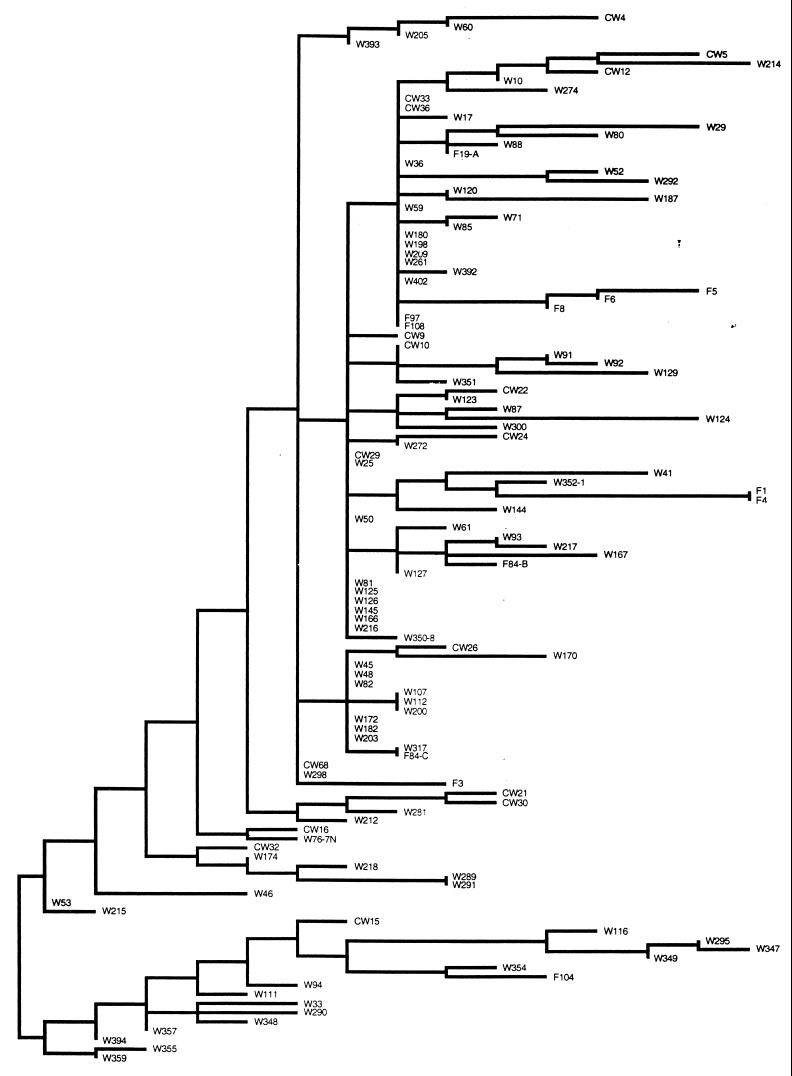
A parsimonious tree was produced during a heuristic search performed by using PAUP (Fig. 2). Similarities between isolates were found by examination of the tree and PCR product sizes (Table 2).
FIG. 2.
Dendrogram of 115 strains typed by using PCR. The isolates that yielded no bands are not shown. F, food isolate; CW and W, patient isolates.
TABLE 2.
Clinical isolates and food isolates exhibiting relatedness
| Isolatea | Mol size (in bp) |
|---|---|
| W167 | 579 442 362 311 258 180 |
| F84-B | 567 525 311 180 |
| W317 | 568 498 374 179 |
| F84-C | 576 500 375 181 |
| W354 | 655 509 441 413 372 168 91 |
| F104 | 650 505 406 367 204 165 122 87 |
| F1 | 437 403 383 347 252 178 161 |
| F4 | 445 410 386 353 255 179 164 |
F, food isolate; W, patient isolate.
Similarities were found in comparisons between clinical samples and food samples and of food samples with other food samples. Clinical isolate W317 was found to be identical to food isolate F84-C. Close relationships were found between two pairs of patient isolates and food isolates (W167 with F84-B and W354 with F104) based on their PCR product size similarities. Two food isolates were found to be closely related to each other as well (F1 with F4). Two isolates from the same food (F84-B and F84-C) yielded different banding patterns (Table 2).
DISCUSSION
This study involves the practical application of a PCR typing method for the comparison of clinical and food isolates of M. avium. This report may be the first to establish a possible relatedness between food isolates and clinical isolates. The demonstration of relatedness between food and patient isolates suggests the possibility that food is a potential source of M. avium.
One disadvantage, however, to extending this method to a variety of local or regional epidemiologic studies may be the instability of ISs. Insertion events may occur over time due to environmental pressures. Different patterns produced over time lead to difficulty in maintaining IS fingerprint libraries. Future research study of IS1245 and IS1311 may define the properties of these ISs and reveal their stability for long-term investigations.
Several epidemiologic studies have suggested that patients with AIDS acquire their disseminated M. avium infections by the oral route rather than through the respiratory system (8, 9, 11). This study strengthens the possibility that the gastrointestinal tract acts as a portal of entry for M. avium infection. The probability of acquiring M. avium infections through the gastrointestinal tract now seems more realistic following the recovery of mycobacteria from a variety of food samples (1), the detection of a close similarity between a clinical isolate and a food isolate, and research suggesting M. avium entry into an intestinal epithelial cell line (2).
In conclusion, this study demonstrates the practical use of a PCR typing method to study relatedness between different ecological strains of M. avium. Picardeau et al. have shown this method to be as effective an M. avium typing method as PFGE (12). This procedure is rapid, can be performed within 8 h, and is inexpensive. This tool for strain typing could replace PFGE for M. avium. Further investigations with this procedure will reveal sources and routes by which these environmental organisms infect patients.
ACKNOWLEDGMENTS
This work was supported by the U.S. Environmental Protection Agency Cooperative Agreement CR-818928.
We thank Jennifer Matos at California State University, Northridge, for her valuable assistance with PAUP and Theresa Sase, Chief Librarian at Northridge Hospital Medical Center, for her generous assistance in literature searches.
REFERENCES
- 1.Argueta, C., S. Yoder, A. Holtzman, T. Aronson, O. G. W. Berlin, P. Tomasek, N. Glover, S. Froman, and G. Stelma, Jr. 1998. Unpublished data.
- 1a.Berlin, O. G. W. Unpublished data.
- 2.Bermudez L E, Petrofsky M, Goodman J. Exposure to low oxygen tension and increased osmolarity enhance the ability of Mycobacterium avium to enter intestinal epithelial (HT-29) cells. Infect Immun. 1997;65:3768–3773. doi: 10.1128/iai.65.9.3768-3773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisercic M, Ochman H. The ancestry of insertion sequences common to Escherichia coli and Salmonella typhimurium. J Bacteriol. 1993;175:7863–7868. doi: 10.1128/jb.175.24.7863-7868.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burki D R, Bernasconi C, Bodmer T, Telenti A. Evaluation of the relatedness of strains of Mycobacterium avium using pulsed-field gel electrophoresis. Eur J Clin Microbiol Infect Dis. 1995;14:212–217. doi: 10.1007/BF02310358. [DOI] [PubMed] [Google Scholar]
- 5.Devallois A, Picardeau M, Goh K S, Sola C, Vincent V, Rastogi N. Comparative evaluation of PCR and commercial DNA probes for detection and identification to species level of Mycobacterium avium and Mycobacterium intracellulare. J Clin Microbiol. 1996;34:2756–2759. doi: 10.1128/jcm.34.11.2756-2759.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falkinham J O., III Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev. 1996;9:177–215. doi: 10.1128/cmr.9.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerrero C, Bernosconi C, Burki D, Bodmer T, Telenti A. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J Clin Microbiol. 1995;33:304–307. doi: 10.1128/jcm.33.2.304-307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hellyer T J, Brown I N, Taylor M B, Allen W, Easmon C S F. Gastrointestinal involvement in Mycobacterium avium-intracellulare infection of patients with HIV. J Infect. 1993;26:55–66. doi: 10.1016/0163-4453(93)96840-m. [DOI] [PubMed] [Google Scholar]
- 9.Horsburgh C R, Metchock B G, McGowan J E, Thompson S E. Clinical implications of recovery of Mycobacterium avium complex from the stool or respiratory tract of HIV-infected individuals. AIDS. 1992;6:512–514. [PubMed] [Google Scholar]
- 10.Maslow J N, Mulligan M E, Albeit R D. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993;17:153–164. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 11.O’Brien R J. The epidemiology of nontuberculous mycobacterial disease. Clin Chest Med. 1989;10:407–418. [PubMed] [Google Scholar]
- 12.Picardeau M, Varnerot A, Lecompte T, Brel F, May T, Vincent V. Use of different molecular typing techniques for bacteriological follow-up in a clinical trial with AIDS patients with Mycobacterium avium bacteremia. J Clin Microbiol. 1997;35:2503–2510. doi: 10.1128/jcm.35.10.2503-2510.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picardeau M, Vincent V. Typing of Mycobacterium avium isolates by PCR. J Clin Microbiol. 1996;34:389–392. doi: 10.1128/jcm.34.2.389-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roiz M P, Palenque E, Guerrero C, Garcia M J. Use of restriction fragment length polymorphism as a genetic marker for typing Mycobacterium avium strains. J Clin Microbiol. 1995;33:1389–1391. doi: 10.1128/jcm.33.5.1389-1391.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shapiro J A. Transposable elements in bacteria. In: Sherratt D J, editor. Mobile genetic elements. New York, N.Y: Oxford University Press; 1995. pp. 6–8. [Google Scholar]
- 16.Slutsky A M, Arbeit R D, Barber T W, Rich J, von Reyn C F, Pieciak W, Barlow M A, Barlow J N. Polyclonal infections due to Mycobacterium avium complex in patients with AIDS detected by pulsed-field gel electrophoresis of sequential clinical isolates. J Clin Microbiol. 1994;32:1773–1778. doi: 10.1128/jcm.32.7.1773-1778.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor T B, Patterson C, Hale Y, Safranek W W. Routine use of PCR-restriction fragment length polymorphism analysis for identification of mycobacteria growing in liquid media. J Clin Microbiol. 1997;35:79–85. doi: 10.1128/jcm.35.1.79-85.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Von Reyn C F, Maslow J N, Barber T W, Falkinham III J O, Arbeit R D. Persistent colonisation of potable water as a source of Mycobacterium avium infection in AIDS. Lancet. 1994;343:1137–1141. doi: 10.1016/s0140-6736(94)90239-9. [DOI] [PubMed] [Google Scholar]